Abstract
The yeast Set2 histone methyltransferase is a critical enzyme that plays a number of key roles in gene transcription and DNA repair. Recently, the human homologue, SETD2, was found to be recurrently mutated in a significant percentage of renal cell carcinomas, raising the possibility that the activity of SETD2 is tumor-suppressive. Using budding yeast and human cell line model systems, we examined the functional significance of two evolutionarily conserved residues in SETD2 that are recurrently mutated in human cancers. Whereas one of these mutations (R2510H), located in the Set2 Rpb1 interaction domain, did not result in an observable defect in SETD2 enzymatic function, a second mutation in the catalytic domain of this enzyme (R1625C) resulted in a complete loss of histone H3 Lys-36 trimethylation (H3K36me3). This mutant showed unchanged thermal stability as compared with the wild type protein but diminished binding to the histone H3 tail. Surprisingly, mutation of the conserved residue in Set2 (R195C) similarly resulted in a complete loss of H3K36me3 but did not affect dimethylated histone H3 Lys-36 (H3K36me2) or functions associated with H3K36me2 in yeast. Collectively, these data imply a critical role for Arg-1625 in maintaining the protein interaction with H3 and specific H3K36me3 function of this enzyme, which is conserved from yeast to humans. They also may provide a refined biochemical explanation for how H3K36me3 loss leads to genomic instability and cancer.
Keywords: cancer, chromatin, enzyme, histone methylation, mutant, H3K36, SETD2, Set2
Introduction
Cancer is increasingly characterized by alterations in chromatin-modifying enzymes (1). SETD2, a non-redundant histone H3 lysine 36 (H3K36)4 methyltransferase (2), has been found to be mutated in a growing list of tumor types, most notably in clear cell renal cell carcinoma (ccRCC) (1, 3, 4), but also in high grade gliomas (5), breast cancer (6), bladder cancer (7), and acute lymphoblastic leukemia (8–10). Recent studies exploring intratumor heterogeneity in ccRCC identified distinct mutations in SETD2 from spatially distinct subsections of an individual tumor, suggesting that mutation of SETD2 is a critical and selected event in ccRCC cancer progression (11). Mutations in SETD2 are predominantly inactivating, such as early nonsense or frameshift mutations, which lead to non-functional protein and global loss of H3K36 trimethylation (H3K36me3) (4, 11, 12). Missense mutations tend to cluster in two domains (1, 4, 12, 13): the SET domain, which catalyzes H3K36me3 (14), and the Set2 Rpb1 interaction (SRI) domain, which mediates the interaction between SETD2 and the hyperphosphorylated form of RNA polymerase II (RNAPII) (13).
SETD2 and its yeast counterpart, Set2, both associate with RNAPII in a co-transcriptional manner (13, 15, 16). In yeast, Set2 mediates all H3K36 methylation states (H3K36me1/me2/me3) (17) and regulates the recruitment of chromatin-remodeling enzymes (Isw1b) and a histone deacetylase (Rpd3) (18) that functions to keep gene bodies deacetylated, thereby maintaining a more compact chromatin structure (19, 20) that is more resistant to inappropriate and bidirectional transcription (18, 21). The Set2/SETD2 pathway is also important for DNA repair (22–27) in both yeast and humans, as well as for proper mRNA splicing (12, 28, 29). Although yeast Set2 can mediate all forms of H3K36 methylation, SETD2 only trimethylates H3K36 (13). Other methyltransferases (e.g. NSD2 and ASH1L) mediate mono- and dimethylation (14), indicating an increased complexity of H3K36 regulation in higher eukaryotes. Consistent with a more diverse role, H3K36me3 recruits a variety of effector proteins in addition to those that are recruited in yeast, including DNMT3b, which regulates gene body methylation (30); LEDGF, which functions in DNA repair (31); and ZMYND11, which regulates co-transcriptional splicing and transcription elongation (32, 33).
The structural and functional similarities between SETD2 and Set2 provide an exceptional opportunity in which existing assays in Saccharomyces cerevisiae can be applied to investigate the functional consequences of SETD2 mutations reported in human cancer. In this work, we characterized cancer-associated SETD2 mutations that occur at evolutionarily conserved residues in functionally important domains (i.e. the SET and SRI domains). We discovered that a missense mutation in the SET domain of SETD2 (R1625C) altered the capacity of this mutant to engage H3, leading to reduced protein stability, and a complete loss of H3K36me3. Strikingly, the same mutation in yeast Set2 (R195C) resulted in an identical effect on H3K36me3 but not H3K36me1 or H3K36me2 levels (or biological outcomes associated with these lower methylation states). Further biological studies in human cells revealed that loss of H3K36me3 in the R1625C mutant leads to DNA repair defects, thereby revealing a greater understanding of how this recurrent mutation probably leads to a loss of SETD2 tumor-suppressive activity.
Results
SETD2 and Set2 Share a High Degree of Structural and Sequence Homology at Their SET and SRI Domains
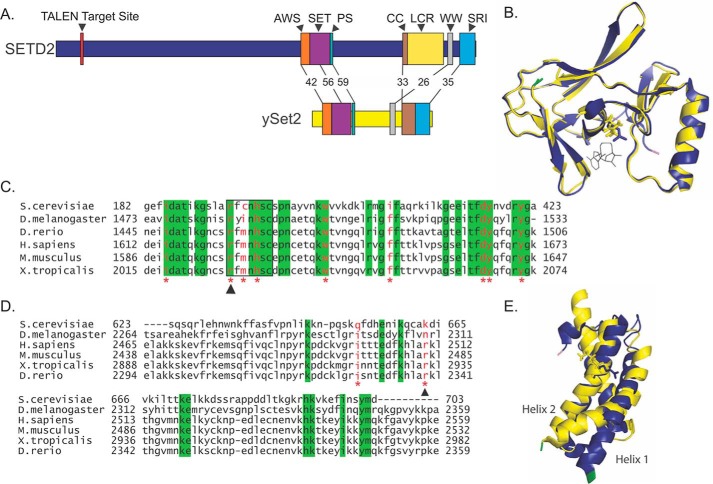
SETD2 and Set2 share significant structural and functional homology. SETD2 demonstrates strong sequence conservation at all of the annotated functional domains present in yeast Set2: AWS (associated with SET), 42%; SET (Su(var)3–9, Enhancer-of-zeste, Trithorax), 56%; PS (post-SET), 59%; coiled-coil, 33%; WW, 26%; and SRI (Set2 Rpb1-interacting), 35% (Fig. 1A). Given this similarity, we compared the structure of the SETD2 and Set2 SET domains to identify highly conserved residues for further study. The structure of the SET domain in SETD2 has been solved by crystallography (34), whereas the SET domain of Set2 was predicted here using I-TASSER (35–38). When the predicted structure of the Set2 SET domain was aligned with the crystal structure of the SETD2 SET domain, the structures were strikingly similar (Fig. 1B). We then examined the conservation of amino acids previously reported to be mutated in human ccRCC (1, 4, 11, 12) across six organisms (Homo sapiens, Drosophila melanogaster, S. cerevisiae, Mus musculus, Xenopus tropicalis, and Danio rerio). Seven of the nine ccRCC mutations occur at residues that are conserved across all model organisms (Fig. 1C). Additionally, three of these seven mutations occur in a region previously identified to act as the catalytic site for lysine methylation (13). One of these mutations, R1625C, is found in a location that is adjacent to the S-adenosyl-l-methionine (SAM) binding site in the structure and thus would be predicted to impact catalytic activity (Fig. 1B). This residue is the most common site of missense mutation reported in both CBioPortal (39, 40) and COSMIC (41). The specific arginine to cysteine mutation is found in both glioma (5) and ccRCC (1). Significantly, mutation of the corresponding residue in S. cerevisiae is known to affect Set2 catalytic activity (42). Given its location and mutation frequency, we chose this mutation for further analysis.
FIGURE 1.
SETD2 and yeast Set2 show high sequence and structural conservation. A, comparison of SETD2 and yeast Set2 (ySet2) annotated protein structure. The percentage of conserved residues within the BLAST-aligned domain sequence is indicated. Annotated domains include AWS, SET, PS, CC, LCR, WW, and SRI. Numbers represent percent conservation. B, alignment of human SET domain crystal structure (blue) with I-TASSER protein structure prediction for yeast SET domain (yellow). The N terminus is marked in green, the C terminus is marked in pink, and residues mutated are shown as sticks. C, partial SET domain sequence alignment across multiple species. Amino acids 1612–1673 of the human SET domain (amino acids 1550–1667) are shown. Residues mutated in ccRCC are shown in red and marked with an asterisk. The arrow indicates Arg-1625, the residue mutated for study. The black box indicates residues previously shown to be an important catalytic site. Residues that are conserved across species are indicated in green. D, SRI domain sequence alignment across multiple species. Residues mutated in ccRCC are in red and marked with an asterisk. The arrow indicates Arg-2510, the residue mutated for study. Residues that are conserved across species are indicated in green. E, alignment of the human SRI domain crystal structure (blue) with the yeast SRI domain crystal structure (yellow). The N terminus is marked in green, the C terminus is marked in pink, and residues mutated are shown as sticks.
We then examined the sequence and structural conservation of the SRI domain and the location of ccRCC-associated missense mutations. In contrast to the SET domain, primary sequence of the SRI domain is less conserved across model organisms (Fig. 1D). However, the aligned crystal structures of yeast (43) and human (44) SRI domains display structural conservation (Fig. 1E). In particular, the predicted site of SETD2 and RNAPII interaction was previously suggested to be the concave surface between α helix 1 and α helix 2 (44). The physical relationship of these helices appears conserved between Set2 and SETD2. We therefore selected the Arg-2510 residue for further study because this amino acid is recurrently mutated (R2510H and R2510L) in ccRCC (1, 12) and is predicted to be important for SETD2-RNAPII interaction by in vitro peptide interaction assays (44).
SET Domain Mutation Destabilizes SETD2 in Cells
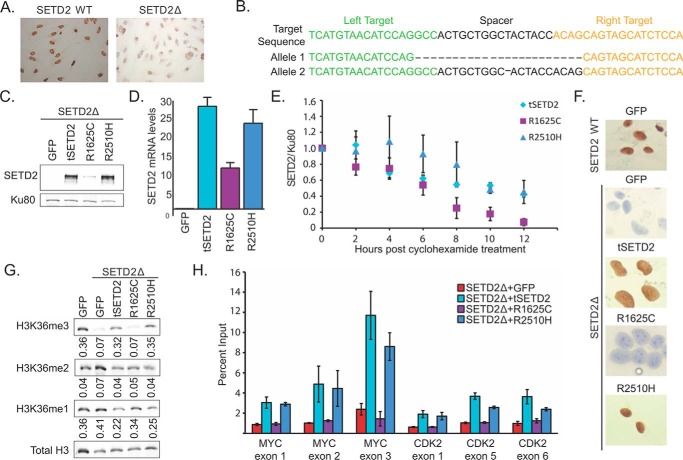
To establish a human cell system in which to study the function of SETD2 mutants, we generated SETD2-deficient cells (SETD2Δ). TAL effector nucleases (TALENs) (45, 46) targeting exon 3 of SETD2 were introduced into two immortalized kidney cell lines (human SV40 immortalized proximal tubule kidney cells (HKC) (47) and 293T). Individual clones of TALEN-treated cells were isolated, and loss of H3K36me3 was demonstrated by immunocytochemistry (Fig. 2A). We verified inactivation of both alleles of SETD2 via Sanger sequencing. Representative allelic sequencing is shown (Fig. 2B).
FIGURE 2.
ccRCC-specific mutations in SETD2 have separate effects on H3K36me3. A, immunocytochemistry of HKC SETD2 wild-type (top) and SETD2Δ cells for H3K36me3. B, Sanger sequencing results of TALEN target sequence in exon 3 of SETD2. Two allelic variants in one HKC SETD2Δ clone are represented. C, immunoblot displaying protein expression level 72 h after transfection in 293T cells. Ku80 acts as a loading control. D, average quantification of SETD2/Ku80 over 12 h following cycloheximide treatment in three independent Western blots (left). Right, average half-life of mutant SETD2 proteins. E, average RNA levels of tSETD2, R1625C, or R2510H, as determined by qPCR for tSETD2 levels. F, anti-H3K36me3 immunocytochemistry on HKC at 72 h post-transfection following reintroduction of GFP, wild-type tSETD2, R1625C, or R2510H. G, anti-H3K36 methylation immunoblot displaying levels of methylation at 72 h post-transfection following reintroduction of GFP, wild-type tSETD2, R1625C, or R2510H. Quantification of H3K36me3/H3 levels is shown below the blot. H, ChIP-qPCR displaying H3K36me3 levels at exonic locations in CDK2 (left) and MYC (right), displayed as ChIP signal/input. Error bars, S.E. Significance comparisons were made with SETD2-inactive + GFP.
We then exogenously expressed a truncated wild-type FLAG-tagged form of SETD2 (amino acids 1323–2564; tSETD2), which includes all known functional domains. The R1625C or R2510H mutants were generated in tSETD2. Relative to tSETD2 and R2510H, R1625C protein levels were reduced (Fig. 2C). R1625C mutant mRNA levels were also less abundant (Fig. 2D). We examined protein stability after treatment with the protein synthesis inhibitor cycloheximide. The R1625C protein demonstrated a significantly shorter half-life compared with that of wild type (Fig. 2E). In contrast, the half-life of the R2510H mutant was unchanged (Fig. 2E). These data suggest that the decreased protein level of the R1625C SET domain mutant results from both decreased RNA and a shortened protein half-life.
Histone H3 Lysine 36 Trimethylation Is Linked to SETD2 Mutational Status
We interrogated H3K36 methylation status in cells transiently transfected with either tSETD2 or the mutants R1625C and R2510H. Using immunocytochemistry, we found that transfection of tSETD2 resulted in global restoration of H3K36me3 (Fig. 2F), demonstrating that the N terminus is not required for catalytic activity of SETD2. Transfection of the R1625C (SET domain) mutant construct failed to restore H3K36me3. In contrast, expression of the R2510H SRI mutant globally restored H3K36me3.
We next examined the H3K36 methylation status by Western blotting analysis. Consistent with findings from immunocytochemistry, SETD2Δ cells show complete loss of H3K36me3. Trimethylation was restored to wild-type levels by expression of either the tSETD2 or the SRI mutant. In contrast, the SET domain mutant failed to trimethylate H3K36 (Fig. 2G). Monomethylation (H3K36me1) and dimethylation (H3K36me2) were unaffected by SETD2 loss or expression of SETD2 variants (Fig. 2G). These results are in agreement with the findings that SETD2 is the exclusive H3K36 trimethyltransferase in mammalian cells.
We then asked whether expression of either tSETD2 or R2510H restored H3K36me3 to levels similar to those of wild-type cells at specific loci. H3K36me3 levels have been shown to increase along the gene body with preference for exons (48). Using ChIP-qPCR, we examined the H3K36me3 levels at multiple exons of two genes, CDK2 and MYC, which had previously been described (49). As expected, SETD2Δ cells displayed low H3K36me3 levels at all sites (Fig. 2H). Expression of tSETD2 recapitulated the previously described pattern for H3K36me3 in wild-type cells (49) at both CDK2 and MYC, with higher signal at exons 5 and 6 relative to exon 1 in CDK2, and in exons 2 and 3 relative to exon 1 in MYC. Cells expressing the R1625C SET domain mutant displayed loss of H3K36me3 at levels similar to that of SETD2Δ cells. Finally, expression of the R2510H mutant also showed greater signal at later exons, indicating that this point mutation restores not only the levels of methylation but also the spatial placement of these methyl marks on actively transcribed genes.
The SETD2 R1625C Variant Is Enzymatically Inactive in Vitro and Has Diminished Substrate Binding
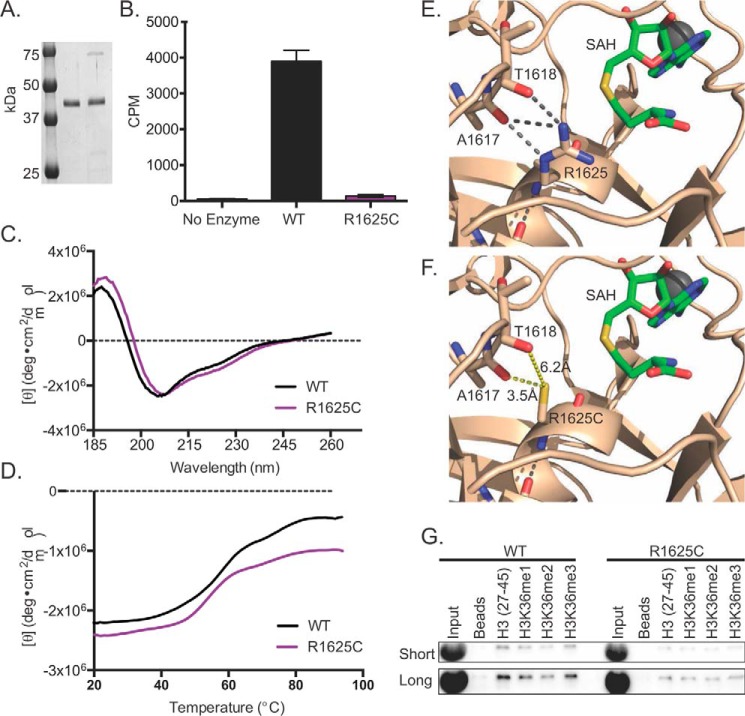
Given that the R1625C SETD2 variant is associated with loss of H3K36me3 in cells, we asked whether the R1625C mutation disrupts the methyltransferase activity of SETD2 in vitro. To do this, we expressed and purified from bacteria a wild-type or R1625C mutated fragment of SETD2 (residues 1345–1711) containing the SET domain. Both the wild-type and the R1625C SET domain constructs yielded soluble proteins that were >90% pure as assessed by SDS-PAGE (Fig. 3A). Methyltransferase activity was then assessed using a radiometric assay with chicken oligonucleosomes as the substrate. Whereas wild-type SETD2 displayed robust activity, the R1625C variant displayed little enzymatic activity over the no enzyme control (Fig. 3B).
FIGURE 3.
The SETD2 R1625C variant is catalytically inactive and has reduced substrate-binding capacity. A, Coomassie Blue-stained SDS-polyacrylamide gel of 1 μg of purified wild type or R1625C variant SETD2 construct containing amino acids 1345–1711 (42 kDa). Precision Plus protein standards (Bio-Rad) are annotated. B, radiometric histone methyltransferase assays comparing the catalytic activity of the wild type and R1625C variant when chicken oligonucleosomes were the substrate. The amount of 3H-methyl incorporated is quantified as cpm, and error bars represent S.E. (n = 3). A reaction without enzyme served as a negative control. C, CD absorbance spectra (plotted as the molar ellipticity ([Θ]) as a function of wavelength) comparing the secondary structure of wild type SETD2 (black) and the R1625C variant (purple). D, thermal melt curves showing the change in CD absorbance at 207 nm over the temperature range from 20 to 95 °C for wild type SETD2 (black) and the R1625C variant (purple). E, structural analysis of Arg-1625. The crystal structure of the SETD2 SET domain (show in tan) bound to SAH (shown in green) near the active site. Hydrogen bonds are shown as gray dashed lines (Protein Data Bank code 4H12). F, in silico mutagenesis analysis (performed in PyMOL, Schrödinger Inc.)). The distances between the R1625C thiol and the carbonyl oxygens of Ala-1617 and Thr-1618 were measured in PyMOL (yellow dashed lines). G, peptide pull-down assays comparing the binding of the wild type and the R1625C variant with the indicated histone H3 peptides. All peptides were biotinylated at the C terminus and were unmodified or were modified as indicated. Streptavidin-coated magnetic beads without peptide served as the negative control. Short (top) and Long (bottom) refer to exposure length.
We next sought to determine why the R1625C variant is catalytically inactive. We first considered whether this mutation results in a misfolded protein, thereby inactivating the SET domain. We compared the CD spectra of the wild-type SETD2 with the R1625C variant. The CD spectra in the low UV range (185–260 nm) of the wild type and the R1625C variant were nearly indistinguishable, suggesting that the R1625C substitution does not alter the secondary structure of the SET domain (Fig. 3C). To determine whether the R1625C variant alters the thermal stability of the SET domain, we monitored the CD signal at the 207-nm peak over a temperature range from 20 to 95 °C. Both the wild type and the R1625C variant showed highly similar thermal melt curves with a melting temperature (Tm) of ∼55 °C (Fig. 3D). Together, these results suggest that the loss of catalytic activity observed for the R1625C variant is not due to protein misfolding or reduced thermal stability.
Structural analysis of the SETD2 SET domain shows that Arg-1625 is positioned within the active site, opposite the SAM binding pocket, and is located about 7 Å away from the sulfur group of S-adenosyl-l-homocysteine (SAH) (Fig. 3E). Although substitution of Arg-1625 with cysteine would not be expected to directly disrupt SAM binding, the Arg-1625 side chain engages in three hydrogen bonds with the backbone carbonyl oxygens of Ala-1617 and Thr-1618 (Fig. 3E). Substituting cysteine for Arg-1625 using in silico mutagenesis showed that every possibly cysteine rotamer would cause steric clashes. The cysteine side chain would not recapitulate the hydrogen bonding network of Arg-1625 when oriented in the same direction as the Arg-1625 side chain observed in the crystal structure (Fig. 3F). Although no structure of the SETD2 SET domain ternary complex containing histone H3 is available, the location of Arg-1625 in close proximity to, but opposite, the SAM binding pocket suggests that Arg-1625 may directly or indirectly engage H3 or may maintain local structural integrity that aids substrate binding.
To determine whether the R1625C variant has altered substrate binding, we performed peptide pull-down experiments using histone H3 peptides that were unmodified or were methylated at Lys-36. The pull-down experiments showed that the R1625C variant associated with all of the histone peptides to a lesser degree compared with the wild-type SETD2 SET domain, suggesting that the R1625C substitution weakens substrate binding (Fig. 3G). Taken together, our results suggest that the R1625C substitution impairs enzymatic activity by reducing substrate binding, which is probably a consequence of fine structural disturbances induced by loss of the Arg-1625 hydrogen bonding network within the active site.
Domain-specific Mutations in Yeast Set2 Separate Roles of H3K36 Methylation States
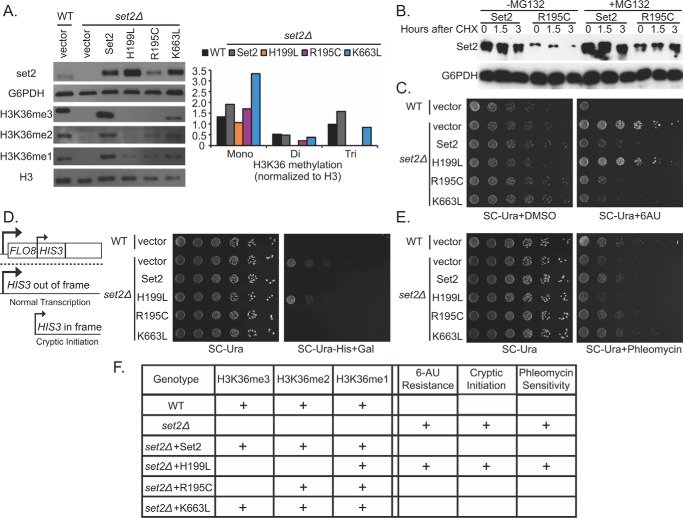
To further explore the functional significance of ccRCC-associated mutations, we took advantage of several well characterized phenotypic assays in S. cerevisiae. Using set2Δ cells, which are devoid of all H3K36 methylation (42), we created strains that either contained vector alone or exogenously expressed either wild-type or mutated forms of Set2. Mutant forms of Set2 included the homologous SETD2 SET domain mutant (R195C), the homologous SETD2 SRI mutant (K663L), or a control SET domain mutant (H199L) previously characterized to disrupt both tri- and dimethylation while retaining monomethylation activity (42). As expected, Set2 loss resulted in the complete absence of mono-, di-, and trimethylation of H3K36, which was rescued upon the addition of wild-type SET2 (Fig. 4A). As shown previously, the H199L mutant only restored monomethylation. In contrast, whereas the K663L mutant restored all H3K36 methylation states, the R195C only restored H3K36 mono- and dimethylation. Intriguingly, the restoration of H3K36 mono- and dimethylation by the R195C mutant mimics the status of SETD2-deficient human cells (i.e. both have a selective loss of H3K36me3). Because the SETD2 R1625C mutant demonstrated decreased protein stability in human cells, we examined protein levels of the R195C mutant in the yeast cells. Following cycloheximide treatment, we observed decreased protein levels of the R195C mutant relative to wild-type Set2, particularly at 3 h post-treatment. This effect was rescued by treatment with the proteosome inhibitor MG132 (Fig. 4B). This suggests that, as in humans, the R195C variant is less stable than wild type in yeast cells.
FIGURE 4.
Modeling of ccRCC specific mutations in Set2 results in separate effects based on H3K36 methylation status. A, anti-H3K36 methylation immunoblots displaying levels of methylation in set2Δ yeast cells as well as yeast with the indicated Set2 mutation. Quantification of H3K36me3/H3 is shown as a bar graph. B, Western results for Set2 and R195C protein levels after treatment with cyclohexamide (100 μg/ml) and MG132 (75 μm). C, 6-AU treatment of wild-type or set2Δ yeast cells expressing the indicated Set2 mutations. D, phleomycin treatment of wild-type or set2Δ yeast cells expressing the indicated Set2 mutations. E, cryptic initiation assay of wild-type or set2Δ yeast cells expressing the indicated Set2 mutations. F, table summary of yeast phenotypes for each of the Set2 mutants
Loss of Set2 has been implicated in various phenotypes in S. cerevisiae, including transcription elongation defects, cryptic initiation, and sensitivity to DNA-damaging agents (42). We asked whether the R195C Set2 mutant would be associated with any of these phenotypes. To examine transcriptional elongation, we performed a spotting assay in the presence of the transcription elongation inhibitor 6-azauracil (6-AU). This assay has been used previously to assay for the presence of transcriptional elongation defects in yeast (16). As expected, wild-type yeast were sensitive to 6-AU (200 μg/ml), whereas set2Δ cells were resistant to this drug (Fig. 4C) (16). Although expression of wild-type SET2 restored sensitivity to 6-AU, the H199L mutant did not. The R195C and K663L mutants behaved similar to wild-type Set2. These data suggest that H3K36me2 is primarily responsible for the sensitivity to inhibitors of transcriptional elongation.
Cryptic initiation has been previously associated with Set2 loss (18). We therefore assessed the effects of our Set2 mutations in a cryptic transcription reporter assay. This assay monitors the growth of yeast cells that contain the HIS3 gene with a cryptic start site that exists in the FLO8 gene. Importantly, the cryptic start site is out of frame when the 5′ promoter is used, and a functional transcript is only produced if the 3′ cryptic start site is utilized. In this setting, cryptic transcription results in expression of HIS3, which can restore growth in medium lacking histidine. Consistent with previous results (18), loss of Set2 permits growth in the absence of histidine (Fig. 4D). No growth was observed in the cells expressing the R195C or K663L mutants. However, cell growth occurred in the presence of the H199L mutant (Fig. 4D). These data indicate that trimethylation is dispensable for preventing cryptic initiation, whereas dimethylation is required to suppress this phenotype.
Recent studies have demonstrated that yeast lacking Set2 cannot properly activate the DNA damage checkpoint (42, 50). To determine whether the RCC-associated SET domain mutation impacts this phenotype, we assessed the impact of the Set2 point mutants on growth in the presence of phleomycin, a double strand break-inducing agent. As expected, set2Δ cells displayed increased sensitivity to phleomycin relative to Set2 wild-type yeast (Fig. 4E). set2Δ cells expressing either wild-type Set2 or the R195C or K663L mutants showed sensitivity similar to that of the wild-type rescue. However, yeast expressing the H199L mutant showed a level of sensitivity similar to that of the set2Δ cells (Fig. 4E). Taken together, these data indicate that the cellular phenotypes associated with Set2 loss in yeast are associated with H3K36me2 and that H3K36me3 is dispensable for these activities (summarized in Fig. 4F).
Human Kidney Cells Display an H3K36 Trimethylation-dependent DNA Damage Response
Because of the exclusivity of SETD2 in mediating trimethlyation in human cells, we studied phenotypes similar to those examined in yeast in human cells that express ccRCC-relevant mutants. We first examined the effect of the transcriptional elongation inhibitor 5,6-dichlorobenzimidazole 1-β-d-ribofuranoside (DRB) on cell survival. DRB inhibits CDK9, which results in premature termination of transcription (51). Assessing viability at 12 h time points for 3 days, we observed that DRB-associated toxicity did not differ between SETD2 wild-type and SETD2Δ cells (Fig. 5A).
FIGURE 5.
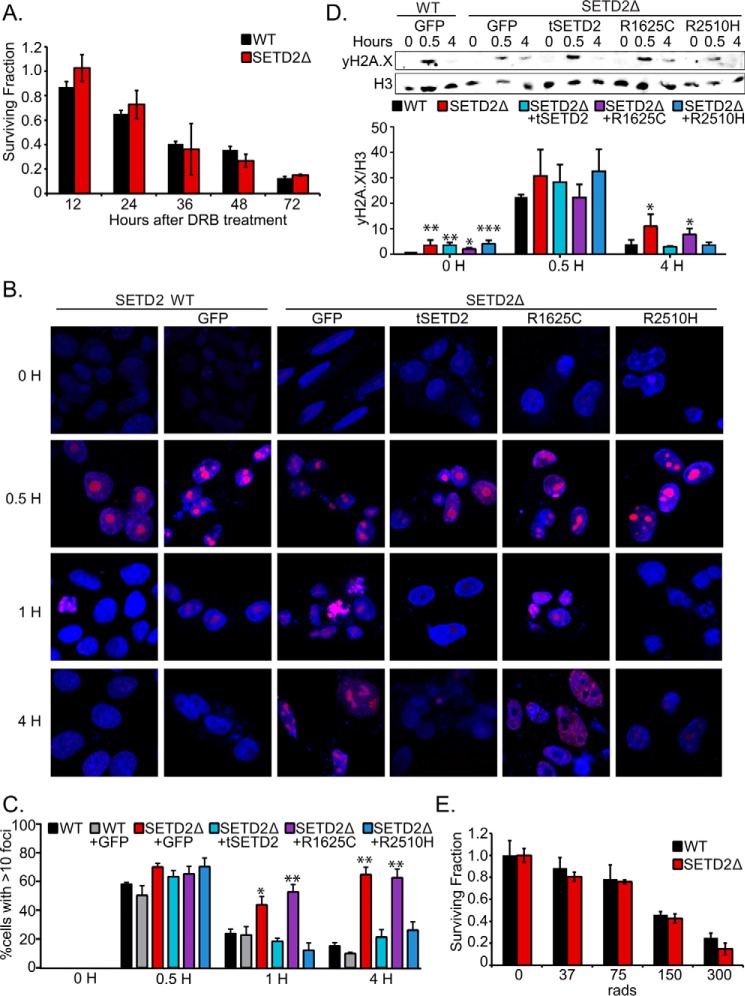
H3K36me3 loss delays γH2A.X foci resolution after DNA damage but does not alter viability. A, surviving fraction of cells at 12, 24, 36, 48, and 72 h post-treatment with 100 μm DRB. Fraction was determined compared with an untreated control. Error bars, S.D. of triplicate treatments. B, γH2A.X foci formation at 0, 0.5, 1, and 4 h after irradiation (2 Gy) in HKC wild-type or SETD2-inactivated cells transfected with GFP, tSETD2, R1625C mutant, or R2510H mutant. The nuclei were visualized by DAPI staining. Representative immunofluorescence images are shown. Scale bar, 10 μm. At least five fields were taken from each condition, and four independent experiments were performed. C, percentage of HKC with >10 γH2A.X foci/cell at 0, 0.5, 1, and 4 h after irradiation (2 Gy). Error bars, S.E. *, p < 0.05; **, p < 0.01 (two-sided t test, comparison with HKC wild type). D, immunoblotting analysis for the expression of γH2A.X and H3 (loading control) from the 293 wild-type or SETD2-inactivated cells transfected with GFP, tSETD2, SET domain R1625C mutant, or SRI domain R2510H mutant. The cells were irradiated by 2 Gy, and histones were acid-extracted at various time points. Shown is average quantification of γH2A.X/H3 after irradiation in three independent Western blots. Error bars, S.E. *, p < 0.05; **, p < 0.01; ***, p < 0.001 (two-sided t test, comparison with 293 wild type + GFP). E, radiation foci formation assay. The surviving fraction represents ratio of treatment (37, 75, 150, and 300 rads) to 0 rad comparison. Error bars, S.D. of triplicate results.
Several recent studies have examined the effect of SETD2 loss in human cells on the response to DNA damage (22–26). To further explore the role of H3K36me3 in the DNA damage response, we irradiated HKC to 2 Gy and then performed immunofluorescence for γH2A.X, a marker of DNA damage. At 30 min postirradiation, γH2A.X foci were seen in all cell types at similar levels (Fig. 5B). In untransfected and in control transfected wild-type cells, the number of foci greatly decreased by 1 h and largely resolved by 4 h. However, in SETD2Δ cells, the number of γH2A.X foci remained elevated at both 1 and 4 h. Expression of tSETD2 in SETD2Δ cells led to resolution of foci at time points similar to wild-type cells. Cells expressing the SRI mutant, R2510H, also showed rapid foci resolution. However, foci persisted in cells expressing the R1625C mutant (Fig. 5B). Quantification of these results demonstrated that both SETD2Δ cells and R1625C-expressing cells had a significantly higher percentage of cells with greater than 10 foci compared with the other conditions (Fig. 5C).
We quantified γH2A.X by immunoblotting, enabling us to account for changes in total protein and histone levels. These studies were performed in 293T cells, as additional validation of results in HKC. As observed with HKC, regardless of SETD2 status, 293T cells showed increased total γH2A.X at 30 min postirradiation (Fig. 5D). By 4 h, γH2A.X levels returned to baseline in cells with H3K36 trimethylation (WT, tSETD2, and R2510H). However, elevated levels of γH2A.X were observed in cells lacking H3K36me3 associated with SETD2 loss or R1625C expression. Finally, we examined the effect of irradiation on viability using a colony formation assay. The fraction of surviving colonies did not differ between SETD2 wild-type and SETD2Δ cells (Fig. 5E). Overall, these findings demonstrate that SETD2-mediated H3K36me3 is coupled to the efficient resolution of double strand breaks. Corresponding to results in yeast, loss of trimethylation is not associated with enhanced sensitivity due to inhibition of transcriptional elongation or from DNA damage.
Discussion
In an effort to explore the function of missense mutations identified in human cancers, we examined several recurrent mutations that occur at evolutionarily conserved residues in yeast and human cell lines. The striking homology between SETD2 and Set2 creates an opportunity to compare the effects of mutations while taking advantage of the strengths of each model system. Indeed, a recent study (52) also modeled cancer mutations in yeast, highlighting the utility and power of yeast to be a robust model system to aid in human protein analyses. In this paper, we investigated how two highly conserved SETD2 residues that are commonly mutated in cancer affect the functions of this enzyme. Limited studies have explored the potential roles that SETD2 loss may play in cancer development. We found that mutation of the SET domain, but not the SRI domain, resulted in effects in the human and yeast assays. Specifically, we identified R1625C as a critical mutation that impacted SETD2 enzymatic activity and protein stability in cells, an effect also noted when this mutation was modeled in yeast Set2. Loss of H3K36me3 in human cells also led to defects in DNA repair, indicating a potential mechanism by which SETD2 functions as a tumor suppressor. To our knowledge, this study is the first to dissect the impact of cancer-associated mutations in SETD2 and further validate using yeast as a model to complement human cell analyses.
A key discovery emerged from the study of the R1625C mutant. In contrast to the human R1625C variant, which was catalytically inactive in vitro, the homologous substitution in yeast Set2 led to an uncoupling of di- and trimethylating activities. This suggests that this residue may be important for the specific trimethylating activity of the enzyme. Moreover, many substrate binding interactions govern stability. Thus, the reduced protein stability (in the absence of other thermal instability) may reflect a structural role that differentiates mono-, di-, and trimethylating activity. Because of this unique mode of regulation, the R195C mutation allowed us to examine the functions specifically associated with the trimethylated state of H3K36 in cells (i.e. impaired transcriptional elongation, cryptic initiation, and impaired survival in the face of DNA damage). Consistent with other reports that examined cryptic initiation (19, 53), we found that H3K36me3 is dispensable, whereas H3K36me1/me2 is required to suppress cryptic initiation as well as for transcription elongation and DNA damage survival phenotypes. In contrast, the SET domain mutation in SETD2 had a similar impact on H3K36me3 levels but resulted in a clear DNA damage response phenotype.
These studies offer a rationale for differences in observed phenotypes in SETD2 deficient human cells associated with Set2 loss in yeast, including cryptic initiation and impaired transcriptional elongation. However, impaired response to DNA damage, as we observed, has been reported for both mammalian systems and Set2 in yeast, linking this feature with H3K36 trimethylation. The γH2A.X results suggest that resolution of DNA strand breaks in human cells requires H3K36me3. (Due to the presence of multiple H3K36 dimethylating enzymes in mammalian cells, absence of H3K36me2 is rarely encountered in human models.) H3K36me2 is induced by ionizing radiation and improves association of early DNA repair components with an induced break and improved repair by non-homologous end joining in human cells (27). Although our data show that the resolution of strand breaks, as measured by clearance of γH2A.X foci, was delayed in the absence of H3K36me3, our data also shows that H3K36me3 loss does not affect viability after radiation in mammalian cells. Thus, loss of dimethylation may convey a sensitivity to DNA damage that is not present in the absence of SETD2 trimethylating activity. It is important to consider that multiple factors may influence cell death in transformed cells. However, these distinct findings in yeast and mammalian systems indicate a complex level of regulation of DNA repair mediated by the histone code at H3 lysine 36. Multiple studies have concluded that the loss of SETD2 confers a variety of types of genomic instability, ranging from microsatellite instability to impairment of non-homologous end joining (50, 54, 55). Our data agree with these results.
Through modeling disease-relevant SETD2 mutations, we were able to gain insight into H3K36me3 function and dissect the roles of H3K36 dimethylation and trimethylation. Future studies will further explore the roles of the SETD2 SRI domain and examine the effects of additional mutations and will further define the role of SETD2 loss in the development of kidney cancer and other tumor types.
Experimental Procedures
Modeling SETD2 and Set2
The primary protein sequences of Set2 from S. cerevisiae and SETD2 from H. sapiens were compared via BLAST alignment analysis, and the percentage of homology between annotated domains was determined using the percentage overlap of the BLAST-aligned regions. The primary sequences of the SET and SRI domains of the enzyme responsible for H3K36 methylation from H. sapiens, S. cerevisiae, X. tropicalis, D. melanogaster, D. rerio, and M. musculus were aligned using ClustalOmega (56) and annotated with reported SETD2 mutations in ccRCC (1, 4, 11, 12). The structures of the SETD2 SET domain (34), SETD2 SRI domain (44), and Set2 SRI domain (43) have been reported previously. To predict the structure of the yeast SET domain, the amino acid sequence (UniProtKB, P46995) was submitted to I-TASSER using the default parameters (35–38). The ribbon structures were aligned using the align command in the PyMOL Molecular Graphics System (57).
Mammalian Cell Lines Transfections and Phenotypic Assays
293T human embryonic kidney cells were generously provided by Dr. Jenny Ting (University of North Carolina, Chapel Hill, NC). The SV40 transformed human renal tubule epithelial cell line (referred to as HKC) was obtained from Dr. Lorraine Racusen (Johns Hopkins University, Baltimore, MD) (47). A pair of vectors containing TALENs targeting exon 3 of SETD2 was generated using the REAL (restriction enzyme and ligation) assembly method. Component plasmids were obtained from Addgene. Briefly, target sites were selected, and TALENs were designed using Zifit, followed by assembly (46). The TALEN target sequences are 5′-TCATGTAACATCCAGGCC-3′ and 5′-ACAGCAGTAGCATCTCCA-3′.
An expression construct containing an N-terminal truncated form of SETD2 (amino acids 1323–2564; tSETD2) was sequence-optimized for expression in human cells, tagged with the FLAG sequence on the C terminus, and synthesized by Life Technologies, Inc. tSETD2 was specifically used because it models the yeast protein in domain structure, and expression of full-length SETD2 was technically unfeasible. tSETD2 was subcloned into the pINDUCER20 vector (58). Disease-relevant SETD2 mutations were introduced into the tSETD2 pINDUCER20 construct using the QuikChange II site-directed mutagenesis kit according to the manufacturer's instructions (Agilent Technologies). Mutations were verified through direct DNA sequence analysis.
293T and HKC human renal cells were transfected with the TALEN constructs, tSETD2 construct, and mutation constructs using the Amaxa® cell line Nucleofector® kit V (Lonza). For the protein stability assay, cycloheximide (100 ng/ml) was applied to cells 72 h post-transfection. For the DRB transcription inhibition assay, 1000 cells/well were plated in triplicate on a 96-well plate and treated with 100 μm DRB for 72 h, with viability being measured every 12 h by Cell Titer Glo (Promega).
Sequencing and Allelic Analysis
DNA was extracted, and the SETD2 TALEN target site was PCR-amplified (primers 5′-ACAGGGACGACAGAAGGTGTCATT-3′ and 5′-ACTGGTGCTGGTGATGAGAGTGTT-3′), sequenced (Applied Biosystems 3730xl Genetic Analyzers, Life Technologies) and analyzed (Sequencher DNA analysis software version 5.0, Gene Codes Corp.). Allelic analysis was performed by subcloning individual PCR products (TOPO TA Cloning® kit, 45-0641, Life Technologies). DNA from individual clones was PCR-amplified, sequenced, and analyzed as described above.
Immunoblotting Analysis
To isolate mammalian cellular proteins, cells were lysed in mammalian protein extraction reagent (Pierce) supplemented with Complete Mini Protease Inhibitor Mixture (Roche Applied Science). Histones were extracted using an overnight acid extraction protocol (Abcam). For yeast immunoblots, asynchronously grown mid-log (0.6–0.8 OD) phase cultures were lysed in SUMEB buffer using glass bead methods described on the Gottschling laboratory website.
Antibodies
Antibodies used include SETD2 (HPA042451, Sigma-Aldrich), Ku80 (ab3107, Abcam), H3K36me3 (ab9050, Abcam), total H3 (Abcam, ab10799; Epicypher, 13-0001), H3K36me2 (39255, Active Motif), H3K36me1 (ab9048, Abcam), γH2A.X (ab2893, Abcam), GST (EpiCypher, catalog no. 13-0022), and Set2 (raised in the Strahl laboratory). Secondary antibodies used in human studies were anti-mouse and anti-rabbit IRDye secondary antibodies from LI-COR Biosciences (Lincoln, NE). HRP-conjugated donkey anti-rabbit secondary antibody was used (Amersham Biosciences) for yeast studies. Human antibodies were detected using the Odyssey IR imager (LI-COR Biosciences), and densitometry analysis was performed using ImageStudio version 2.0. The yeast immunoblots were developed using ECL-Prime (Amersham Biosciences), and densitometry analysis was done using ImageJ (National Institutes of Health).
Immunocytochemistry
Cells were fixed with 4% paraformaldehyde for 15 min and permeabilized using 0.25% Triton X-100 in PBS. Endogenous peroxidase activity was blocked by incubation in 1% H2O2. Cells were then blocked in 5% bovine serum albumin, followed by incubation in primary antibody. The Vectastain ABC kit (PK6101, Vector Laboratories) was used for secondary antibody and HRP conjugation, followed by the DAB peroxidase substrate kit (SK-3100, Vector Laboratories) and hematoxylin staining.
Chromatin Immunoprecipitation
Cells were fixed in 1% formaldehyde for 10 min, quenched with 125 mm glycine treatment, and homogenized in hypotonic solution (10 mm Tris, pH 7.4, 15 mm NaCl, 60 mm KCl, 1 mm EDTA, 0.1% Nonidet P-40, 5% sucrose, 1× protease inhibitors). Nuclei were separated by centrifugation through a sucrose pad (10 mm Tris, pH 7.4, 15 mm NaCl, 60 mm KCl, 10% sucrose, 1× protease inhibitors) and then resuspended in ChIP buffer (10 mm Tris, pH 7.4, 100 mm NaCl, 60 mm KCl, 1 mm EDTA, 0.1% Nonidet P-40, 1× protease inhibitors, 0.05% SDS) and sonicated to obtain DNA between 200 bp and 1 kb. DNA was immunoprecipitated with H3K36me3 antibody prebound to protein A/G beads. Immunoprecipitated complexes were washed and RNase- and Proteinase K-treated, and protein-DNA cross-links were reversed by overnight incubation at 65 °C.
Quantitative RT-PCR
Total RNA was extracted using the Qiagen RNeasy minikit. cDNA was made from total RNA using random primers and Superscript II reverse transcriptase reagents (Invitrogen). Primers used for RT-PCR are listed in Table 1.
TABLE 1.
Primers
| Target | Forward primer (5′–3′) | Reverse primer (5′–3′) |
|---|---|---|
| tSETD2 | CACCATGACACAGGGCCA | GGGTGTCCTTGATGCTGTT |
| c-Myc exon 1 | GCCGCATCCACGAAACTTT | TCCTTGCTCGGGTGTTGTAAG |
| c-Myc exon 2 | TGCCCCTCAACGTTAGCTTC | GGCTGCACCGAGTCGTAGTC |
| c-Myc exon 3 | CCTGAGCAATCACCTATGAACTTG | CAAGGTTGTGAGGTTGCATTTG |
| CDK2 exon 1 | GTCGGGAACTCGGTGGGAG | AGAAGGCGGACCCTGGCTC |
| CDK2 exon 5 | CATCTGGAGCCTGGGCTGCA | TGGGGAGGAGAGGGAGGGGG |
| CDK2 exon 6 | CCCTATTCCCTGGAGATTCTG | CTCCGTCCATCTTCATCCAG |
Expression and Purification of Human SETD2
An E. coli codon-optimized synthetic gene corresponding to human SETD2 (UniProtKB ID Q9BYW2) residues 1345–1711 followed by a stop codon was cloned into the pGEX-6P-2 expression vector (GE Healthcare) using standard procedures. The protein was expressed in soluBL21 (DE3) (Amsbio) cells by growing cells in Terrific Broth II medium (MP Biomedicals) at 37 °C until an A600 of ∼0.6 and then chilling the cells for 30 min at 4 °C before inducing them with 1 mm isopropyl 1-thio-β-d-galactopyranoside in the presence of 25 μm ZnCl2 for 20 h at 16 °C. Cells were harvested by centrifugation, and pellets were flash-frozen in liquid nitrogen. For purification, thawed cell pellets were resuspended in binding buffer (50 mm Tris, pH 7.3, 300 mm NaCl, 4 mm DTT, 10% glycerol, and 1 μm ZnCl2) supplemented with 1 Complete mini-EDTA-free protease inhibitor tablet (Roche Applied Science), 0.1 mm PMSF, 0.5 mg/ml chicken egg lysozyme, and 0.2% (v/v) Triton X-100 and incubated on ice for 45 min and then lysed with sonication and clarified by centrifugation. Clarified lysates were diluted 1:2 with binding buffer and applied to a 5-ml glutathione-agarose gravity flow column (pre-equilibrated with 10 column volumes of binding buffer) at a flow rate of ∼0.5 ml/min at 4 °C. The bound protein was washed with 10 column volumes of binding buffer and then eluted from the column with 35 ml of elution buffer (50 mm Tris, pH 8.0, 300 mm NaCl, 4 mm DTT, 10% glycerol, and 10 mm reduced l-glutathione). The eluted protein was mixed with Precision Protease (GE Healthcare) and exhaustively dialyzed against binding buffer, without ZnCl2, over the course of 20 h at 4 °C. The cleaved protein sample was applied to a pre-equilibrated 5-ml glutathione-agarose gravity flow column at a flow rate of ∼0.5 ml/min at 4 °C, and the flow-through was collected and concentrated using an Amicon-Ultra 15 concentrator (Millipore). The Bradford assay and SDS-PAGE analysis were used to determine the quantity and purity of the protein samples, respectively. The SETD2 R1625C mutant was generated by site-directed mutagenesis using the QuikChange kit (Agilent) and expressed and purified as described above. Note that a small amount of GST-SETD2 WT and R1625C was not treated with Precision Protease but was extensively dialyzed against binding buffer and then used for peptide pull-down experiments (see below).
Histone Methyltransferase Assays
Histone methyltransferase assays were performed by incubating wild type SETD2 or the R1625C variant at a final concentration of 500 nm with 1 μg of chicken oligonucleosomes (EpiCypher) and 1 μCi of [3H]AdoMet (PerkinElmer Life Sciences) in a buffer containing 50 mm HEPES, pH 8.0, 150 mm NaCl, 2.5 mm MgCl2, 1 μm ZnCl2, and 2.5% glycerol for 16 h at room temperature (total reaction volume was 20 μl). The reactions were quenched with 0.5% TFA and then spotted onto Whatman filter paper, air-dried, and washed four times with ∼200 ml of a sodium bicarbonate (pH 9.0) solution. Then they were air-dried again and added to liquid scintillation vials containing 5 ml of Ultima Gold F (PerkinElmer Life Sciences). Samples were counted for 1 min each using an all-purpose Beckman Coulter liquid scintillation counter in 3H mode. A reaction without enzyme was used as the negative control and to determine background counts.
CD Spectroscopy
For CD experiments, proteins were exhaustively dialyzed into a buffer containing 20 mm sodium phosphate (pH 7.0), 150 mm sodium fluoride (NaF), and 0.2 mm tris(2-carboxyethyl)phosphine at 4 °C. CD spectra were collected using a 0.1-cm quartz cuvette and a Chirascan Plus instrument (Applied Photophysics Inc.) at 20 °C over the wavelength range 185–260 nm with a step size of 0.5 nm. A sample of the buffer was collected over the same wavelength scan, and absorbance values were subtracted from the final data sets. Each sample was scanned three times, and the final plots represent the average scan with the CD signal (in millidegrees) converted to molar ellipticity. For thermal melt curves, the CD absorbance at 207 nm was collected over the temperature range from 20 to 95 °C with 1 °C temperature ramping and a temperature tolerance range set to 0.2 °C. Proteins were diluted to 0.25 mg/ml for all CD data collection (protein stock concentrations determined by A280).
Peptide Pull-downs
A total of 50 pmol of GST-tagged wild type SETD2 or the R1625C variant was incubated with 500 pmol of each biotinylated histone peptide for 1 h at 4 °C in peptide binding buffer (50 mm Tris, pH 8.0, 300 mm NaCl, 0.1% Nonidet P-40) supplemented with 2 mm DTT and 1 μm ZnCl2. Following incubation, the protein/peptide mixture was incubated with streptavidin-coated magnetic beads (Pierce), pre-equilibrated with peptide binding buffer, for an additional 1 h at 4 °C. The beads were washed three times with peptide binding buffer, and bound complexes were eluted with 1× SDS loading buffer, resolved via SDS-PAGE, and transferred to a PVDF membrane. The membrane was probed with anti-GST antibody diluted to 1:4000 in 5% BSA in PBS-T. The peptides contained the budding yeast histone H3 residues 27–45 and were mono-, di-, or trimethylated at Lys-36. In this region, the human and budding yeast H3 sequences differ by an Ala to Ser substitution at residue 31 and by an Arg to Lys substitution at residue 42.
Yeast Growth Assays
Parental yeast strains were transformed with the indicated plasmids and were grown to saturation in appropriate selection medium. Saturated cultures were diluted to an A600 of 0.5 and 5-fold serially diluted and plated with or without 6-AU or plated with or without phleomycin; pictures were taken 2–3 days after spotting. Similarly, strains with an integrated cryptic initiation cassette (as shown in Fig. 4) were serially diluted and plated on −URA-HIS plates with or without galactose for 3 days to detect growth. Growth on −URA acted as a control for equivalent growth for all of the strains.
Immunofluorescence Staining for γH2A.X
HKC were cultured for 16–18 h followed by 2-Gy radiation (RS 2000 Biological Research Irradiator), fixed for 15 min in 4% paraformaldehyde, washed with cold PBS, and permeabilized (0.25% Triton X-100 in PBS) for 10 min. After blocking with 1% hydrogen peroxide and then 5% BSA, cells were incubated with anti-γH2A.X (1:500) at 4 °C overnight. Cells were washed using PBST and incubated with goat anti-rabbit IgG (1:500) Cy5 for 1 h at room temperature. After washing, cells were counterstained using DAPI. Fluorescence signals were visualized using confocal microscopy (LSM 700, Zeiss), and the number of foci per cell was analyzed using Zen (LSM 700, Zeiss). 5 images/coverslip (total of 15 images) were collected in three independent experiments. For the radiation colony formation assay, cells were diluted to a single cell suspension, and 300 cells were plated on a 10-cm plate. Plates were irradiated at 0, 37, 75, 150, and 300 rads; allowed to grow for 10 days; and then stained with crystal violet. Colonies were counted manually.
Author Contributions
K. E. H., C. C. F., S. A. S., Y. C. C., D. K. J., I. J. D., B. D. S., and W. K. R. conceived and designed experiments. A. H. V. and J. A. S. generated TALEN constructs. K. E. H. and C. C. F. generated data comprising Figs. 1 and 2. S. A. S. generated data comprising Fig. 3. J. V. D. completed the experiments of Fig. 4. Y.-C. J. C. and C. C. F. completed the experiments of Fig. 5. K. E. H., C. C. F., and S. A. S. wrote the manuscript, and C. C. F. generated final figures and coordinated paper completion. All authors reviewed the results, provided editing for the manuscript, and approved the final version of the manuscript.
Acknowledgments
We thank the extended membership of the Davis, Strahl, Rathmell, and William Kim laboratories for many fruitful conversations. We thank Dr. Ashutosh Tripathy (University of North Carolina Macromolecular Interactions Facility) for help collecting the CD data. The confocal microscopy service was supported by the University of North Carolina, Chapel Hill, core facility for the Microscopy Services Laboratory.
This work was supported by the V Foundation Translational Research Grant (to B. D. S., I. J. D., and W. K. R.), National Institutes of Health (NIH) Grants R01 CA198482 (to B. D. S., I. J. D., and W. K. R.) and K24 CA172355 (to W. K. R.), UNC Medical Scientist Training Program Grant T32 GM008719-12 (to K. E. H. and C. C. F.), National Research Service Award (NRSA) Training Grant TR000085-05 (to C. C. F.), NRSA Award F30 CA192643-02 (to C. C. F.), a UNC Lineberger Cancer Center Postdoctoral Fellowship Award (to S. A. S.), NIGMS, NIH, award 5T32 GM007092 (to J. V. D.), and NIH Grant R01 HL124232 (to J. A. S.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- H3K36
- histone H3 Lys-36
- me1
- monomethylation
- me2
- dimethylation
- me3
- trimethylation
- ccRCC
- clear cell renal cell carcinoma
- SET
- Su(var)3–9, Enhancer-of-zeste, Trithorax
- AWS
- associated with SET
- PS
- post-SET
- CC
- coiled-coil
- LCR
- low charge region
- SRI
- Set2 Rpb1 interaction
- RNAPII
- RNA polymerase II
- SAH
- S-adenosyl-l-homocysteine
- SAM
- S-adenosyl-l-methionine
- TALEN
- TAL effector nuclease
- 6-AU
- 6-azauracil
- DRB
- 5,6-dichlorobenzimidazole 1-β-d-ribofuranoside
- HKC
- human SV40 immortalized proximal tubule kidney cell(s)
- tSETD2
- truncated SETD2
- qPCR
- quantitative PCR
- Gy
- gray(s).
References
- 1. Cancer Genome Atlas Research Network (2013) Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 499, 43–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Edmunds J. W., Mahadevan L. C., and Clayton A. L. (2008) Dynamic histone H3 methylation during gene induction: HYPB/Setd2 mediates all H3K36 trimethylation. EMBO J. 27, 406–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Duns G., van den Berg E., van Duivenbode I., Osinga J., Hollema H., Hofstra R. M., and Kok K. (2010) Histone methyltransferase gene SETD2 is a novel tumor suppressor gene in clear cell renal cell carcinoma. Cancer Res. 70, 4287–4291 [DOI] [PubMed] [Google Scholar]
- 4. Dalgliesh G. L., Furge K., Greenman C., Chen L., Bignell G., Butler A., Davies H., Edkins S., Hardy C., Latimer C., Teague J., Andrews J., Barthorpe S., Beare D., Buck G., Campbell P. J., et al. (2010) Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature 463, 360–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fontebasso A. M., Schwartzentruber J., Khuong-Quang D.-A., Liu X.-Y., Sturm D., Korshunov A., Jones D. T. W., Witt H., Kool M., Albrecht S., Fleming A., Hadjadj D., Busche S., Lepage P., Montpetit A., et al. (2013) Mutations in SETD2 and genes affecting histone H3K36 methylation target hemispheric high-grade gliomas. Acta Neuropathol. 125, 659–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Al Sarakbi W., Sasi W., Jiang W. G., Roberts T., Newbold R. F., and Mokbel K. (2009) The mRNA expression of SETD2 in human breast cancer: correlation with clinico-pathological parameters. BMC Cancer 9, 290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cancer Genome Atlas Research Network (2014) Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 507, 315–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhang J., Ding L., Holmfeldt L., Wu G., Heatley S. L., Payne-Turner D., Easton J., Chen X., Wang J., Rusch M., Lu C., Chen S.-C., Wei L., Collins-Underwood J. R., Ma J., et al. (2012) The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature 481, 157–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mar B. G., Bullinger L. B., McLean K. M., Grauman P. V., Harris M. H., Stevenson K., Neuberg D. S., Sinha A. U., Sallan S. E., Silverman L. B., Kung A. L., Lo Nigro L., Ebert B. L., and Armstrong S. A. (2014) Mutations in epigenetic regulators including SETD2 are gained during relapse in paediatric acute lymphoblastic leukaemia. Nat. Commun. 5, 3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhu X., He F., Zeng H., Ling S., Chen A., Wang Y., Yan X., Wei W., Pang Y., Cheng H., Hua C., Zhang Y., Yang X., Lu X., Cao L., et al. (2014) Identification of functional cooperative mutations of SETD2 in human acute leukemia. Nat. Genet. 46, 287–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gerlinger M., Rowan A. J., Horswell S., Larkin J., Endesfelder D., Gronroos E., Martinez P., Matthews N., Stewart A., Tarpey P., Varela I., Phillimore B., Begum S., McDonald N. Q., Butler A., et al. (2012) Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 366, 883–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Simon J. M., Hacker K. E., Singh D., Brannon A. R., Parker J. S., Weiser M., Ho T. H., Kuan P.-F., Jonasch E., Furey T. S., Prins J. F., Lieb J. D., Rathmell W. K., and Davis I. J. (2014) Variation in chromatin accessibility in human kidney cancer links H3K36 methyltransferase loss with widespread RNA processing defects. Genome Res. 24, 241–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sun X.-J., Wei J., Wu X.-Y., Hu M., Wang L., Wang H.-H., Zhang Q.-H., Chen S.-J., Huang Q.-H., and Chen Z. (2005) Identification and characterization of a novel human histone H3 lysine 36-specific methyltransferase. J. Biol. Chem. 280, 35261–35271 [DOI] [PubMed] [Google Scholar]
- 14. Wagner E. J., and Carpenter P. B. (2012) Understanding the language of Lys36 methylation at histone H3. Nat. Rev. Mol. Cell Biol. 13, 115–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yoh S. M., Lucas J. S., and Jones K. A. (2008) The Iws1:Spt6:CTD complex controls cotranscriptional mRNA biosynthesis and HYPB/Setd2-mediated histone H3K36 methylation. Genes Dev. 22, 3422–3434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kizer K. O., Phatnani H. P., Shibata Y., Hall H., Greenleaf A. L., and Strahl B. D. (2005) A novel domain in Set2 mediates RNA polymerase II interaction and couples histone H3 K36 methylation with transcript elongation. Mol. Cell. Biol. 25, 3305–3316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Venkatesh S., Smolle M., Li H., Gogol M. M., Saint M., Kumar S., Natarajan K., and Workman J. L. (2012) Set2 methylation of histone H3 lysine 36 suppresses histone exchange on transcribed genes. Nature 489, 452–455 [DOI] [PubMed] [Google Scholar]
- 18. Lickwar C. R., Rao B., Shabalin A. A., Nobel A. B., Strahl B. D., and Lieb J. D. (2009) The Set2/Rpd3S pathway suppresses cryptic transcription without regard to gene length or transcription frequency. PLoS One 4, e4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li B., Jackson J., Simon M. D., Fleharty B., Gogol M., Seidel C., Workman J. L., and Shilatifard A. (2009) Histone H3 lysine 36 dimethylation (H3K36me2) is sufficient to recruit the Rpd3s histone deacetylase complex and to repress spurious transcription. J. Biol. Chem. 284, 7970–7976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Quan T. K., and Hartzog G. A. (2010) Histone H3K4 and K36 methylation, Chd1 and Rpd3S oppose the functions of Saccharomyces cerevisiae Spt4-Spt5 in transcription. Genetics 184, 321–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Carrozza M. J., Li B., Florens L., Suganuma T., Swanson S. K., Lee K. K., Shia W.-J., Anderson S., Yates J., Washburn M. P., and Workman J. L. (2005) Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 123, 581–592 [DOI] [PubMed] [Google Scholar]
- 22. Carvalho S., Vítor A. A. C. A., Sridhara S. C. S., Martins F. B., Raposo A. C., Desterro J. M. P., Ferreira J., de Almeida S. F., Filipa B. M., Ana C. R., Desterro J. M. P., Ferreira J., de Almeida S. F., Martins F. B., Raposo A. C., Desterro J. M. P., Ferreira J., and de Almeida S. F. (2014) SETD2 is required for DNA double-strand break repair and activation of the p53-mediated checkpoint. Elife 3, e02482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Daugaard M., Baude A., Fugger K., Povlsen L. K., Beck H., Sørensen C. S., Petersen N. H. T., Sorensen P. H. B., Lukas C., Bartek J., Lukas J., Rohde M., and Jäättelä M. (2012) LEDGF (p75) promotes DNA-end resection and homologous recombination. Nat. Struct. Mol. Biol. 19, 803–810 [DOI] [PubMed] [Google Scholar]
- 24. Pfister S. X., Ahrabi S., Zalmas L. P., Sarkar S., Aymard F., Bachrati C. Z., Helleday T., Legube G., La Thangue N. B., Porter A. C. G., and Humphrey T. C. (2014) SETD2-dependent histone H3K36 trimethylation is required for homologous recombination repair and genome stability. Cell Rep. 7, 2006–2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kanu N., Grönroos E., Martinez P., Burrell R. A., Yi Goh X., Bartkova J., Maya-Mendoza A., Mistrík M., Rowan A. J., Patel H., Rabinowitz A., East P., Wilson G., Santos C. R., McGranahan N., et al. (2015) SETD2 loss-of-function promotes renal cancer branched evolution through replication stress and impaired DNA repair. Oncogene 34, 5699–5708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Aymard F., Bugler B., Schmidt C. K., Guillou E., Caron P., Briois S., Iacovoni J. S., Daburon V., Miller K. M., Jackson S. P., and Legube G. (2014) Transcriptionally active chromatin recruits homologous recombination at DNA double-strand breaks. Nat. Struct. Mol. Biol. 21, 366–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fnu S., Williamson E. A., De Haro L. P., Brenneman M., Wray J., Shaheen M., Radhakrishnan K., Lee S.-H., and Nickoloff J. A., and Hromas R. (2011) Methylation of histone H3 lysine 36 enhances DNA repair by nonhomologous end-joining. Proc. Natl. Acad. Sci. U.S.A. 108, 540–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Luco R. F., Pan Q., Tominaga K., Blencowe B. J., Pereira-Smith O. M., and Misteli T. (2010) Regulation of alternative splicing by histone modifications. Science 327, 996–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sorenson M. R., Jha D. K., Ucles S. A., Flood D. M., Strahl B. D., Stevens S. W., and Kress T. L. (2016) Histone H3K36 methylation regulates pre-mRNA splicing in Saccharomyces cerevisiae. RNA Biol. 13, 412–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dhayalan A., Rajavelu A., Rathert P., Tamas R., Jurkowska R. Z., Ragozin S., and Jeltsch A. (2010) The Dnmt3a PWWP domain reads histone 3 lysine 36 trimethylation and guides DNA methylation. J. Biol. Chem. 285, 26114–26120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pradeepa M. M., Sutherland H. G., Ule J., Grimes G. R., and Bickmore W. A. (2012) Psip1/Ledgf p52 binds methylated histone H3K36 and splicing factors and contributes to the regulation of alternative splicing. PLoS Genet. 8, e1002717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wen H., Li Y., Xi Y., Jiang S., Stratton S., Peng D., Tanaka K., Ren Y., Xia Z., Wu J., Li B., Barton M. C., Li W., Li H., and Shi X. (2014) ZMYND11 links histone H3.3K36me3 to transcription elongation and tumour suppression. Nature 508, 263–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Guo R., Zheng L., Park J. W., Lv R., Chen H., Jiao F., Xu W., Mu S., Wen H., Qiu J., Wang Z., Yang P., Wu F., Hui J., Fu X., et al. (2014) BS69/ZMYND11 reads and connects histone H3.3 lysine 36 trimethylation-decorated chromatin to regulated pre-mRNA processing. Mol. Cell. 56, 298–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zheng W., Ibáñez G., Wu H., Blum G., Zeng H., Dong A., Li F., Hajian T., Allali-Hassani A., Amaya M. F. M., Siarheyeva A., Yu W., Brown P. J., Schapira M., Vedadi M., et al. (2012) Sinefungin derivatives as inhibitors and structure probes of protein lysine methyltransferase SETD2. J. Am. Chem. Soc. 134, 18004–18014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yang J., Yan R., Roy A., Xu D., Poisson J., and Zhang Y. (2015) The I-TASSER Suite: protein structure and function prediction. Nat. Methods 12, 7–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Roy A., Kucukural A., and Zhang Y. (2010) I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 5, 725–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang Y. (2008) I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yang J., and Zhang Y. (2015) I-TASSER server: new development for protein structure and function predictions. Nucleic Acids Res. 43(W1), W174–W181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gao J., Aksoy B. A., Dogrusoz U., Dresdner G., Gross B., Sumer S. O., Sun Y., Jacobsen A., Sinha R., Larsson E., Cerami E., Sander C., and Schultz N. (2013) Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6, pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cerami E., Gao J., Dogrusoz U., Gross B. E., Sumer S. O., Aksoy B. A., Jacobsen A., Byrne C. J., Heuer M. L., Larsson E., Antipin Y., Reva B., Goldberg A. P., Sander C., and Schultz N. (2012) The cBio Cancer Genomics Portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Forbes S. A., Bhamra G., Bamford S., Dawson E., Kok C., Clements J., Menzies A., Teague J. W., Futreal P. A., and Stratton M. R. (2008) The catalogue of somatic mutations in cancer (COSMIC). Curr. Protoc. Hum. Genet. Genet. 10.1002/0471142905.hg1011s57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jha D. K., and Strahl B. D. (2014) An RNA polymerase II-coupled function for histone H3K36 methylation in checkpoint activation and DSB repair. Nat. Commun. 5, 3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Vojnic E., Simon B., Strahl B. D., Sattler M., and Cramer P. (2006) Structure and carboxyl-terminal domain (CTD) binding of the Set2 SRI domain that couples histone H3 Lys36 methylation to transcription. J. Biol. Chem. 281, 13–15 [DOI] [PubMed] [Google Scholar]
- 44. Li M., Phatnani H. P., Guan Z., Sage H., Greenleaf A. L., and Zhou P. (2005) Solution structure of the Set2-Rpb1 interacting domain of human Set2 and its interaction with the hyperphosphorylated C-terminal domain of Rpb1. Proc. Natl. Acad. Sci. U.S.A. 102, 17636–17641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cermak T., Doyle E. L., Christian M., Wang L., Zhang Y., Schmidt C., Baller J. A., Somia N. V., Bogdanove A. J., and Voytas D. F. (2011) Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 39, e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sander J. D., Cade L., Khayter C., Reyon D., Peterson R. T., Joung J. K., and Yeh J.-R. J. (2011) Targeted gene disruption in somatic zebrafish cells using engineered TALENs. Nat. Biotechnol. 29, 697–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Racusen L. C., Monteil C., Sgrignoli A., Lucskay M., Marouillat S., Rhim J. G. S., and Morin J.-P. P. (1997) Cell lines with extended in vitro growth potential from human renal proximal tubule: characterization, response to inducers, and comparison with established cell lines. J. Lab. Clin. Med. 129, 318–329 [DOI] [PubMed] [Google Scholar]
- 48. Kolasinska-Zwierz P., Down T., Latorre I., Liu T., Liu X. S., and Ahringer J. (2009) Differential chromatin marking of introns and expressed exons by H3K36me3. Nat. Genet. 41, 376–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Carvalho S., Raposo A. C., Martins F. B., Grosso A. R., Sridhara S. C., Rino J., Carmo-Fonseca M., and de Almeida S. F. (2013) Histone methyltransferase SETD2 coordinates FACT recruitment with nucleosome dynamics during transcription. Nucleic Acids Res. 41, 2881–2893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pai C.-C., Deegan R. S., Subramanian L., Gal C., Sarkar S., Blaikley E. J., Walker C., Hulme L., Bernhard E., Codlin S., Bähler J., Allshire R., Whitehall S., and Humphrey T. C. (2014) A histone H3K36 chromatin switch coordinates DNA double-strand break repair pathway choice. Nat. Commun. 5, 4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhu Y., Pe'ery T., Peng J., Ramanathan Y., Marshall N., Marshall T., Amendt B., Mathews M. B., and Price D. H. (1997) Transcription elongation factor p tefb is required for hiv 1 tat transactivation in vitro. Genes Dev. 11, 2622–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sun S., Yang F., Tan G., Costanzo M., Oughtred R., Hirschman J., Theesfeld C. L., Bansal P., Sahni N., Yi S., Yu A., Tyagi T., Tie C., Hill D. E., Vidal M., et al. (2016) An extended set of yeast-based functional assays accurately identifies human disease mutations. Genome Res. 26, 670–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Youdell M. L., Kizer K. O., Kisseleva-Romanova E., Fuchs S. M., Duro E., Strahl B. D., and Mellor J. (2008) Roles for Ctk1 and Spt6 in regulating the different methylation states of histone H3 lysine 36. Mol. Cell. Biol. 28, 4915–4926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Awwad S. W., and Ayoub N. (2015) Overexpression of KDM4 lysine demethylases disrupts the integrity of the DNA mismatch repair pathway. Biol. Open 4, 498–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Li F., Mao G., Tong D., Huang J., Gu L., Yang W., Li G.-M. (2013) The histone mark H3K36me3 regulates human DNA mismatch repair through its interaction with MutSα. Cell. 153, 590–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., Thompson J. D., and Higgins D. G. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. DeLano W. L. (2016) The PyMOL Molecular Graphics System, version 1.8, Schrödinger, LLC, New York [Google Scholar]
- 58. Meerbrey K. L., Hu G., Kessler J. D., Roarty K., Li M. Z., Fang J. E., Herschkowitz J. I., Burrows A. E., Ciccia A., Sun T., Schmitt E. M., Bernardi R. J., Fu X., Bland C. S., Cooper T. A., et al. (2011) The pINDUCER lentiviral toolkit for inducible RNA interference in vitro and in vivo. Proc. Natl. Acad. Sci. U.S.A. 108, 3665–3670 [DOI] [PMC free article] [PubMed] [Google Scholar]