Abstract
Reproductive function is controlled by the pulsatile release of hypothalamic gonadotropin-releasing hormone (GnRH), which regulates the expression of the gonadotropins luteinizing hormone and FSH in pituitary gonadotropes. Paradoxically, Fshb gene expression is maximally induced at lower frequency GnRH pulses, which provide a very low average concentration of GnRH stimulation. We studied the role of secreted factors in modulating gonadotropin gene expression. Inhibition of secretion specifically disrupted gonadotropin subunit gene regulation but left early gene induction intact. We characterized the gonadotrope secretoproteome and global mRNA expression at baseline and after Gαs knockdown, which has been found to increase Fshb gene expression (1). We identified 1077 secreted proteins or peptides, 19 of which showed mRNA regulation by GnRH or/and Gαs knockdown. Among several novel secreted factors implicated in Fshb gene regulation, we focused on the neurosecretory protein VGF. Vgf mRNA, whose gene has been implicated in fertility (2), exhibited high induction by GnRH and depended on Gαs. In contrast with Fshb induction, Vgf induction occurred preferentially at high GnRH pulse frequency. We hypothesized that a VGF-derived peptide might regulate Fshb gene induction. siRNA knockdown or extracellular immunoneutralization of VGF augmented Fshb mRNA induction by GnRH. GnRH stimulated the secretion of the VGF-derived peptide NERP1. NERP1 caused a concentration-dependent decrease in Fshb gene induction. These findings implicate a VGF-derived peptide in selective regulation of the Fshb gene. Our results support the concept that signaling specificity from the cell membrane GnRH receptor to the nuclear Fshb gene involves integration of intracellular signaling and exosignaling regulatory motifs.
Keywords: follicle-stimulating hormone (FSH), G protein, G protein-coupled receptor (GPCR), gene regulation, secretion, GnRH, LβT2 cells, VGF, gonadotrope, secretoproteome
Introduction
GnRH3 is a hypothalamic neuropeptide that plays a central role in the regulation of mammalian reproductive functions. Secreted in a pulsatile manner, GnRH stimulates the synthesis and release of the gonadotropins luteinizing hormone (LH) and FSH, which in turn regulate gonadal function and growth. The frequency of GnRH pulses differs at various stages of reproductive development and of the reproductive cycle. During puberty, there is an increase in GnRH pulse frequency, causing a dramatic increase in gonadotropin secretion, resulting in sexual maturation (3). In the mid- to late follicular phase of the female menstrual cycle, an increase in GnRH pulse frequency leads to greater LH synthesis and secretion. Conversely, in the late luteal phase, low GnRH pulse frequency favors FSH production. FSH promotes the development of the ovarian follicle, whereas LH stimulates the secretion of gonadal steroids (4). The sequential and differential release of gonadotropins is important for reproductive physiology.
GnRH analogs are utilized to treat reproductive disorders and hormone-sensitive cancers. GnRH agonists are used to trigger oocyte maturation for in vitro fertilization (5). Idiopathic hypogonadotropic hypogonadism and Kallmann syndrome are reproductive disorders characterized by absent or delayed puberty with low gonadotropin and sex steroid levels. Associated with GnRH deficiency, these syndromes are treated with pulses of GnRH agonist to restore gonadotropin response (for a review, see Ref. 6). Conversely, continuous administration of GnRH agonists and antagonists represents an effective therapy for hormone-dependent cancers like prostate cancer as it causes a decrease in gonadotropin levels and subsequently in sex steroid levels (Ref. 7; for a review, see Ref. 8). GnRH agonists are also used in the treatment of advanced breast cancer in premenopausal women (Ref. 9; for a review, see Ref. 10).
LH and FSH contain a common α-glycoprotein subunit (CGA) and a specific β subunit, namely LHβ and FSHβ (11). Understanding Fshb gene control is particularly important because the major mechanism controlling FSH levels, unlike LH, is biosynthetic and not secretory (12). Furthermore, Fshb gene control shows a paradoxical low GnRH pulse frequency dependence. Lower average GnRH stimulation, which occurs with lower frequency GnRH exposure, causes higher levels of gene induction and FSH secretion (Ref. 13; for a review, see Ref. 12). A widely used experimental model for studying gonadotropin gene regulation is the LβT2 gonadotrope cell line. Mellon and co-workers (14, 15) developed this mature murine gonadotrope cell line using targeted oncogenesis in transgenic mice. The LβT2 cell line, which was isolated from pituitary tumors that developed in these mice, has characteristics of late developmental stage gonadotropes, expressing mRNA for the GnRH receptor, the CGA, and the specific β subunits of LH and FSH (16, 17). In a 10-h experiment using LβT2 cells, for example, exposure to only four brief low concentration GnRH pulses (one every 2 h) leads to about twice the expression of Fshb achieved by a total of 19 pulses (one every 30 min) (1). The difficulty in explaining the preferential induction of Fshb by low frequency GnRH pulses suggests that important regulatory control mechanisms remain to be discovered.
The GnRH receptor belongs to the rhodopsin-like subfamily of G protein-coupled receptors, which activate heterotrimeric GTP-binding proteins (G proteins) to transduce extracellular stimuli into cell signaling responses (18). G proteins consist of G α, β, and γ subunits. Agonist binding to a G protein-coupled receptor activates the Gα subunit, which subsequently exchanges GDP for GTP; Gα-GTP then stimulates downstream effectors (19, 20). The GnRH receptor was shown to activate Gαq/11 (21–23) and Gαs (24, 25) in response to GnRH stimulation.
Although GnRH is the main regulator of gonadotropin subunit gene expression, intrinsic pituitary factors may tune gonadotrope responsiveness to GnRH and thus contribute to the differential regulation of gonadotropin subunits. These include activin, pituitary adenylate cyclase-activating polypeptide, BMPs, inhibin, and growth differentiation factor 9 (GDF9). Activin, for instance, selectively stimulates Fshb gene expression via phosphorylation and nuclear translocation of SMAD2 and SMAD3 in LβT2 cells (26, 27). Pituitary adenylate cyclase-activating polypeptide is thought to be involved in the rat estrus cycle by increasing follistatin transcription, leading to activin sequestration, which results in selective reduction of Fshb mRNA levels (Ref. 28; for a review, see Ref. 29). Several reports have implicated bone morphogenetic proteins (BMPs) in the paracrine/autocrine regulation of FSH expression (Refs. 30–36; for a review, see Ref. 12). As an example, BMP2 was previously shown to synergize with activin A to stimulate Fshb transcription and Fshb mRNA levels in LβT2 cells (33). We previously demonstrated that GnRH activation of Gαs in LβT2 cells promotes Lhb but suppresses Fshb gene expression via secretion of inhibin α (1). Conversely, secreted GDF9 is an autocrine inducer of Fshb and is preferentially suppressed by high frequency GnRH pulses in LβT2 cells (37).
We sought to systematically characterize the role of secretion in gonadotropin regulation and the autocrine regulatory factors involved using a combined proteomic and transcriptomic approach. We identified several novel secreted candidate regulatory factors and identified a VGF nerve growth factor-inducible (VGF)-derived peptide as a GnRH-stimulated autocrine factor that inhibits Fshb gene expression.
Results
Pharmacological Inhibition of Secretion Selectively Suppresses Gonadotropin Gene Regulation
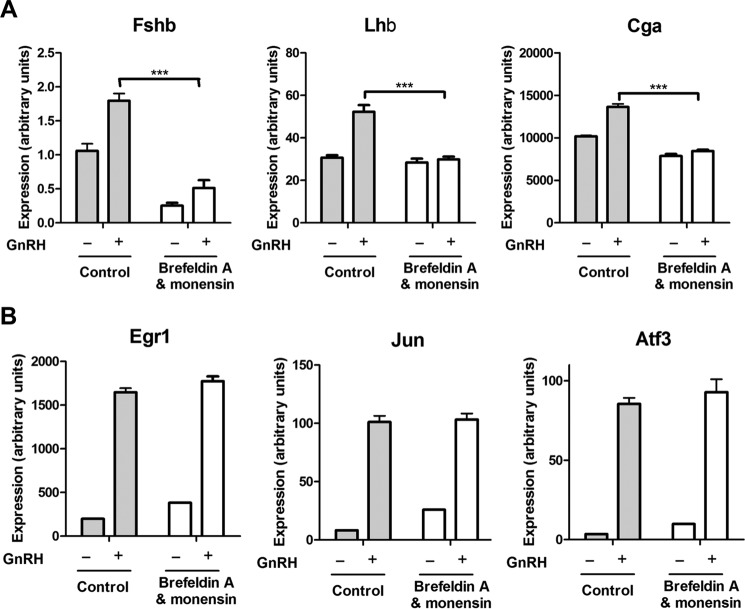
Our recent identification of inhibin α and GDF9 as autocrine regulators of FSHβ gene expression (1, 37) prompted us to study the effects of inhibition of secretion on gonadotropin subunit gene expression. Two known inhibitors of protein secretion, brefeldin A and monensin, were used to treat GnRH-stimulated and unstimulated gonadotrope cells. Inhibition of secretion led to a dramatic decline in both basal and GnRH-induced Fshb mRNA expression (Fig. 1A) without affecting induction of immediate early genes Egr1, Jun, and Atf3 (also known as LRG-21) by GnRH (Fig. 1B). Brefeldin A and monensin treatment also resulted in a significant decrease in GnRH-induced Lhb and Cga expression. These data strongly suggest that locally secreted factors, including but possibly not limited to inhibin α and GDF9, play a key role in the regulation of gonadotropin gene expression by GnRH.
FIGURE 1.
Effect of exocytosis inhibitors on GnRH-induced expression of gonadotropin subunit genes and immediate early genes in LβT2 cells. A, gonadotropin subunit genes. Cells were serum-starved overnight, pretreated with the exocytosis inhibitors brefeldin A and monensin for 30 min, and stimulated with 1 nm GnRH for 2 h; following culture medium replacement, cells were incubated for another 4 h. B, immediate early genes. Cells were serum-starved overnight, pretreated with brefeldin A and monensin for 30 min, and stimulated with 1 nm GnRH for 45 min. mRNA expression levels of the indicated genes were determined by qPCR. Results of three independent experiments performed in triplicate were combined for analysis. Error bars represent S.E. ***, p < 0.001, two-way ANOVA with Bonferroni post-test corrections.
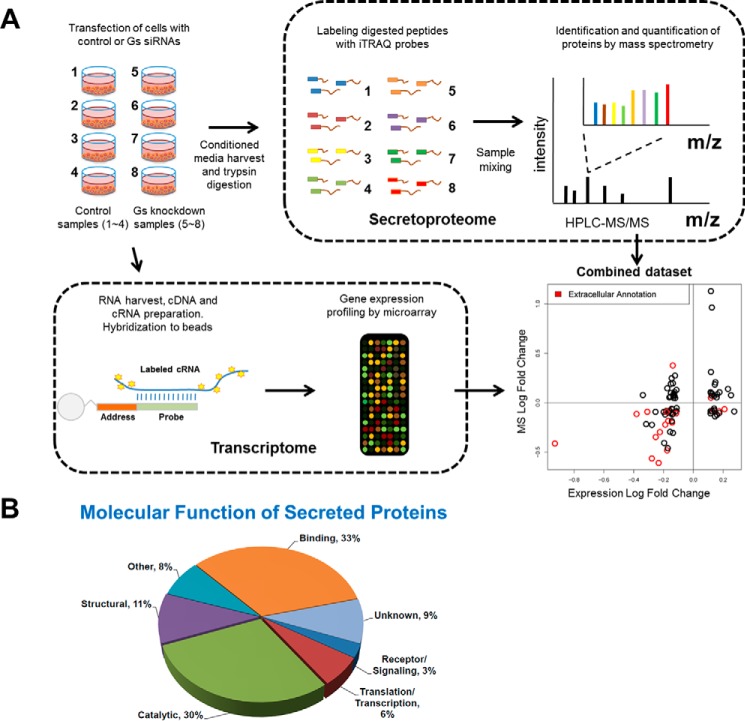
To determine the role of such autocrine factors, we characterized the global secretoproteome and gene expression profile in Gαs siRNA knockdown and control gonadotrope cells (Fig. 2A). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (85) partner repository (www.ebi.ac.uk/pride) with the data set identifier PXD000063. A total of 1077 secreted proteins were detected by mass spectrometry, including structural proteins, catalytic enzymes, receptors and signaling components, and proteins involved in transcription and translation (Fig. 2B). Among those proteins, 34 were identified in this screening study as differentially secreted (up- or down-regulated by over 30%, p < 0.1; supplemental Table S1). Among the 16,000 mRNAs measured in the transcriptome, 98 were potentially differentially expressed (up- or down-regulated by over 30%, p < 0.1). Changes in protein secretion and gene expression showed good concordance (supplemental Table S2), especially for genes annotated as extracellular (Fig. 2A, graph). We next carried out an NCBI analysis of gene ontology terms for each of these candidates and searched the literature for relevance to reproductive endocrinology. Through this process, we selected 19 differentially expressed candidates that either function as ligands, are present in the extracellular space, or are implicated in gonadotrope physiology/reproduction.
FIGURE 2.
LβT2 gonadotrope secretoproteome analysis. A, schematic representation of the experimental design used to identify secreted factors in LβT2 gonadotrope cells. Details are provided under “Experimental Procedures.” The integration of the secretoproteome and transcriptome data is also depicted in the bottom right corner. Briefly, log -fold changes of protein abundance (x axis) were plotted against log -fold changes of mRNA expression (y axis). Genes denoted with gene ontology cellular compartment of “extracellular region” or “secretory granule” were considered as potentially secreted and are denoted in red. Analysis details are provided under “Experimental Procedures.” B, gene ontology analysis of secreted proteins based on molecular function. Panther was used for the analysis.
Secretoproteome Regulation in LβT2 Cells
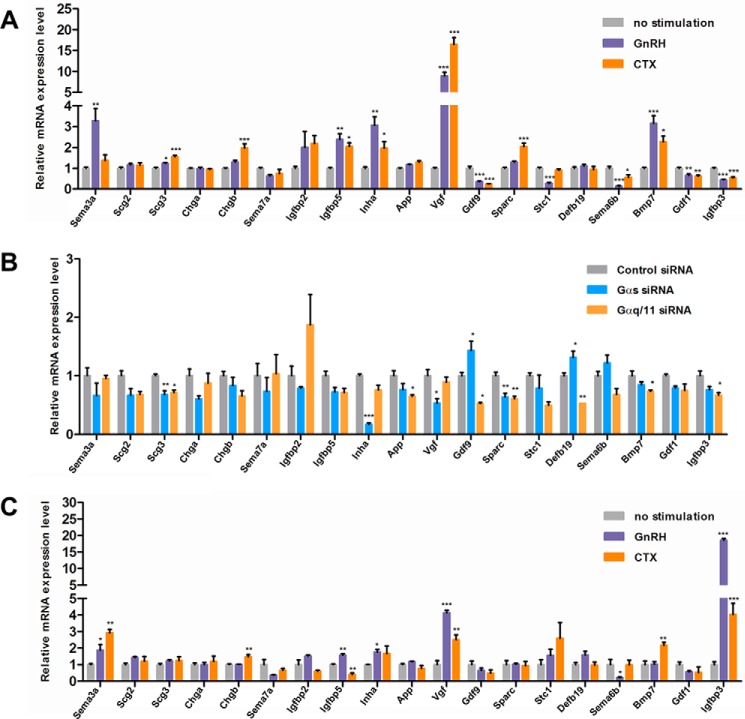
We examined the responsiveness of the 19 candidates to GnRH as well as their dependence on Gαs and Gαq/11 proteins. We previously showed that inhibin α is preferentially induced by high GnRH pulse frequency and is Gαs-dependent (1), whereas GDF9 is considerably suppressed by high frequency GnRH (37). Among the 19 candidates, Vgf showed the largest induction by GnRH (8.9 ± 0.91-fold; Fig. 3A) or by cholera toxin (CTX), a Gαs activator (16.5 ± 1.64-fold). Gαs knockdown caused a significant 47% reduction in Vgf mRNA expression (Fig. 3B). Consonant with previous studies (1, 37), inhibin α (Inha) mRNA expression was stimulated by GnRH and CTX and reduced by Gαs knockdown, whereas Gdf9 was suppressed by GnRH and CTX.
FIGURE 3.
Candidate gene screening for GnRH and CTX responsiveness and Gαs and Gαq/11 protein dependence. A, effect of GnRH or CTX stimulation on the expression of 19 candidate genes in LβT2 cells. Cells were serum-starved overnight and stimulated with either 1 nm GnRH, 5 μg/ml CTX, or vehicle for 10 h. B, effect of Gαs or Gαq/11 silencing on the expression of 19 candidate genes in LβT2 cells. Cells were serum-starved overnight and transfected with either scrambled (control), Gαs, or Gαq/11 siRNA. Transfections were carried out in the Nucleofector 96-well Shuttle as described under” Experimental Procedures.” Cells were harvested 3 days post-transfection. C, effect of GnRH or CTX stimulation on the expression of 19 candidate genes in purified primary gonadotropes. Cells were stimulated with either 1 nm GnRH, 5 μg/ml CTX, or vehicle for 10 h. mRNA expression levels of the indicated genes were determined by qPCR. Data shown are the mean from one experiment performed in triplicate and are representative of three independent experiments. Error bars represent S.E. ***, p < 0.001; **, p < 0.01; *, p < 0.05, one-way ANOVA.
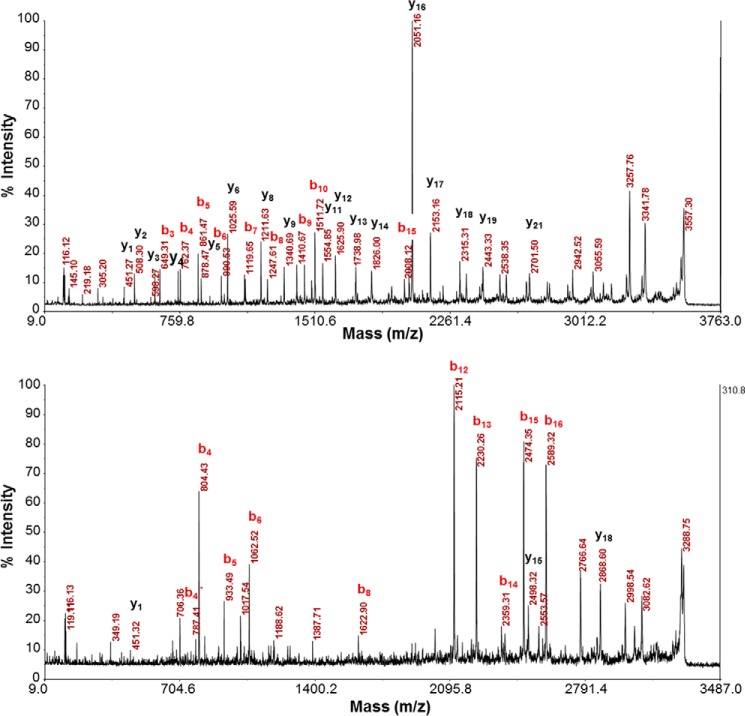
Other regulated secreted factors included secretogranin III (SCG3), insulin-like growth factor-binding protein 2 (IGFBP2), stanniocalcin 1 (STC1), semaphorin 3A (SEMA3A), semaphorin 6B (SEMA6B), BMP7, and GDF1. Similar to results obtained with LβT2 cells, in purified primary mouse gonadotropes we found that mRNAs for Igfbp5 and Sema3a were significantly increased by GnRH. In contrast, Igfbp3 mRNA was decreased by GnRH and CTX in LβT2 cells but was strongly induced by both treatments in purified primary gonadotropes (Fig. 3C). Interestingly, we found that siRNA-mediated knockdown of either SEMA3A, SCG2, SCG3, or STC1 resulted in a significant decrease in GnRH-induced Fshb expression (Table 1), whereas knockdown of either IGFBP3 or IGFBP5 led to a significant decrease in GnRH-induced Lhb expression. Furthermore, fragments of SCG2-derived peptides em66 and secretoneurin were detected by mass spectrometry (Fig. 4). Exogenous em66 and secretoneurin both had a stimulatory effect on GnRH-induced Fshb expression (Table 1). Overall, our results strengthen the hypothesis that secreted elements may play an important role in the gonadotropin response to GnRH stimulus. Because of its dramatic induction by GnRH in both the gonadotrope cell line (Fig. 3A) and primary gonadotrope cells (Fig. 3C), we selected VGF for more extensive study.
TABLE 1.
Effects of candidate siRNA knockdown or ligand addition on Fshb and Lhb mRNA expression
| Name | Regulation by GnRH pulse frequency | Effect of siRNA-mediated knockdown | Effect of ligand additiona |
|---|---|---|---|
| SCG2 | Induced by high pulse frequency | 73% reduction in GnRH-induced FSHβ expression | Increased GnRH-induced FSHβ mRNA expression by SCG2-derived peptides secretoneurin and em66 |
| SCG3 | Induced by high pulse frequency | 18% reduction in GnRH-induced FSHβ expression | Not tested |
| SEMA3A | Not regulated | 33% reduction in GnRH-induced FSHβ expression | No effect (SEMA3A was tested) |
| STC1 | Suppressed by low pulse frequency | 37% reduction in GnRH-induced FSHβ expression | Not tested |
| IGFBP3 | Induced by high pulse frequency | 18% reduction in GnRH-induced LHβ expression | No effect (IGFBP3 was tested) |
| IGFBP5 | Suppressed by high pulse frequency | 35% reduction in GnRH-induced LHβ expression | No effect (IGFBP5 was tested) |
a For each candidate, 1 mg/ml peptide or peptide mixture was tested.
FIGURE 4.
Detection of SCG2-derived peptides em66 and secretoneurin by mass spectrometry. Top panel, fragment ion spectrum of peptide TNEIVEEQYTPQSLATLESVFQELGK derived from endogenously expressed em66 in LβT2 cells. Bottom panel, fragment ion spectrum of peptide ERVDEEQKLYTDDEDDVYK derived from endogenously expressed secretoneurin in LβT2 cells. Both spectra were collected and analyzed using a MALDI-TOF/TOF mass spectrometer after trypsin digestion, iTRAQ labeling, and off-line peptide fractionation as described under “Experimental Procedures.” Major peptide backbone fragmentation ions, b and y, are annotated.
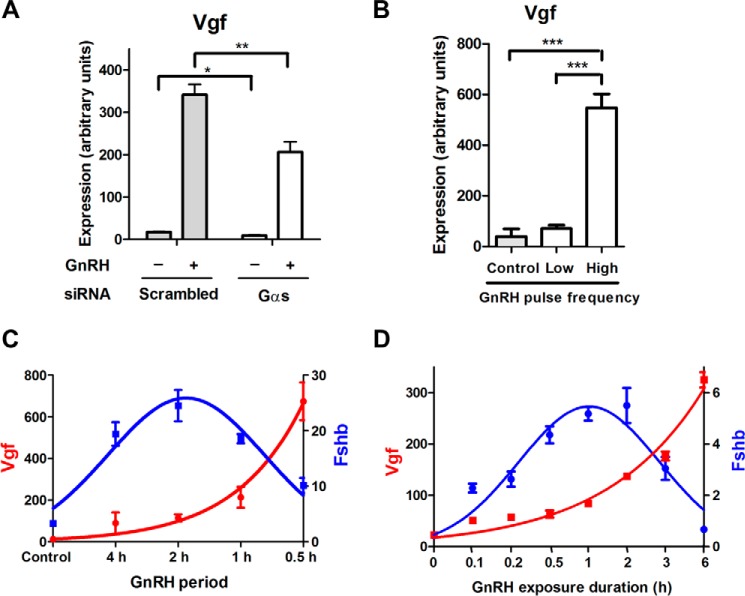
GnRH Induction of Vgf is Gαs-dependent and Pulse Frequency Sensitive
We sought to further characterize the induction of Vgf by GnRH. Gαs knockdown led to a 40% reduction in Vgf gene induction (Fig. 5A), demonstrating that Vgf gene induction is partly Gαs-dependent. We investigated the sensitivity of Vgf induction to GnRH pulse frequency. We used a custom designed and fabricated microprocessor-controlled parallel perifusion system (Ref. 1; see “Experimental Procedures”). Vgf mRNA induction was highly sensitive to pulse frequency. Low frequency GnRH pulses (2-h interpulse period), which are optimal for Fshb gene induction (37), did not significantly regulate Vgf. In contrast, high pulse frequency GnRH pulses (30-min interpulse period), which poorly induce Fshb, increased Vgf gene expression more than 50-fold (Fig. 5, B and C). Notably, with constant GnRH stimulation, Vgf mRNA expression continually increased with the duration of GnRH exposure, reaching a maximum at 6 h (Fig. 5D). Thus, Vgf expression correlated closely with the average levels of GnRH stimulation over time whether due to increased pulse frequency (Fig. 5, B and C) or increased duration (Fig. 5D). Conversely, Fshb was uncorrelated with average GnRH exposure, decreasing at both higher frequency (Fig. 5C) and increased duration exposure (Fig. 5D). Overall, the regulation of Vgf was highly discordant with that of Fshb with respect to the pattern of GnRH exposure.
FIGURE 5.
Effects of Gαs knockdown and varying GnRH pulse frequency on Vgf gene expression in LβT2 cells. A, effect of Gαs silencing on Vgf gene expression in GnRH-stimulated cells. Cells were transfected with either scrambled or Gαs siRNA on day 1. On day 2, cells were serum-starved overnight. On day 3, cells were stimulated with 1 nm GnRH for 2 h followed by 4 h without GnRH or with vehicle. **, p < 0.01; *, p < 0.05, two-way ANOVA with Bonferroni post-test corrections. B, effect of two different GnRH pulse frequencies on Vgf gene expression. Using a perfusion system, cells were stimulated with 1 nm GnRH for 10 h at either low pulse frequency (a 2-h interpulse period) or high pulse frequency (a 30-min interpulse period). ***, p < 0.001, one-way ANOVA with Bonferroni post-test corrections. C, effect of varying GnRH pulse frequency on VGF and Fshb gene expression. Using a perfusion system, cells were stimulated with 1 nm GnRH for 10 h at different interpulse periods (every 30 min, 1 h, 2 h, or 4 h). D, effect of duration of GnRH exposure on Vgf and Fshb gene expression. Cells were serum-starved overnight and stimulated with 1 nm GnRH or vehicle for the indicated times. Cells were harvested 6 h after the first exposure to GnRH. Vgf and Fshb mRNA expression levels were determined by qPCR. Data shown are the mean from one experiment performed in triplicate and are representative of three independent experiments. Error bars represent S.E. Both Vgf and Fshb mRNA levels are expressed in arbitrary units.
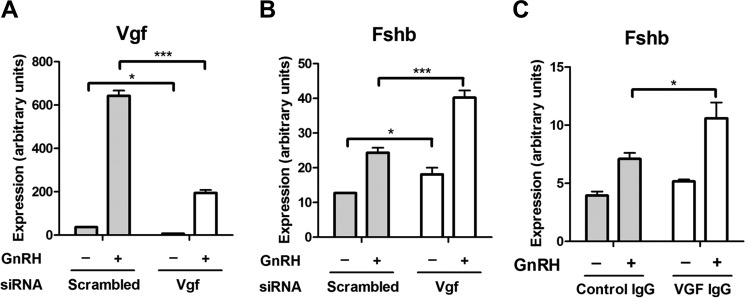
VGF siRNA Knockdown Increases Both Basal and GnRH-induced Fshb Expression
To test whether VGF could serve as an inhibitory regulator of Fshb expression, we carried out VGF siRNA knockdown experiments. Knockdown of Vgf mRNA was highly efficient both at the mRNA (Fig. 6A) and protein levels (data not shown). VGF knockdown led to a significant increase in both basal and GnRH-induced Fshb expression (34 and 65%, respectively; Fig. 6B). Similar to the effect of VGF siRNA knockdown, anti-VGF antibody treatment resulted in a 49% increase in GnRH-induced Fshb expression (Fig. 6C), suggesting that an extracellular VGF-derived peptide may inhibit Fshb expression.
FIGURE 6.
Effect of VGF inactivation on Fshb gene expression in LβT2 cells. A, knockdown efficiency of VGF siRNA in GnRH-stimulated cells. Cells were transfected with either scrambled or VGF siRNA on day 1. On day 2, cells were serum-starved overnight. On day 3, cells were stimulated with 1 nm GnRH for 2 h followed by 4 h without GnRH or with vehicle. B, effect of VGF gene knockdown on Fshb gene expression in GnRH-stimulated cells. C, effect of VGF immunoneutralization on Fshb gene expression in GnRH-stimulated cells. Cells were serum-starved overnight, treated with either control antibodies (Control IgG) or polyclonal antibodies raised against VGF (VGF IgG) for 6 h, and stimulated with 1 nm GnRH for 2 h followed by 4 h without GnRH or with vehicle. Vgf, Fshb, and Lhb mRNA expression levels were determined by qPCR. Data shown are the mean from one experiment performed in triplicate and are representative of three independent experiments. Error bars represent S.E. ***, p < 0.001; *, p < 0.05, two-way ANOVA with Bonferroni post-test corrections.
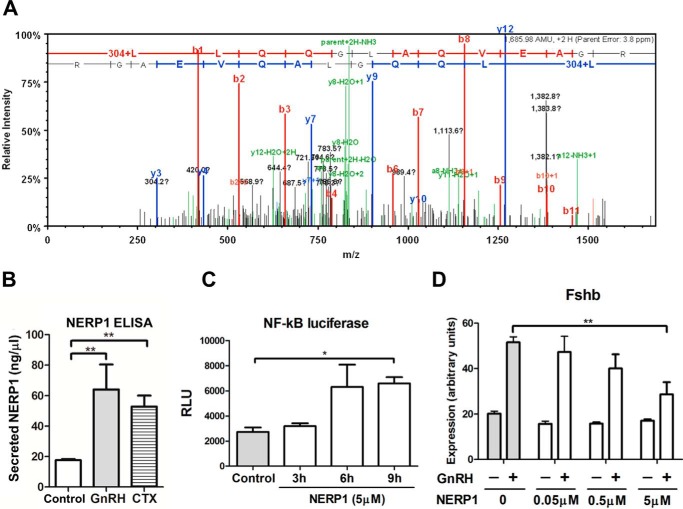
VGF-derived Peptide Neuroendocrine Regulatory Peptide-1 (NERP1) Suppresses GnRH-induced Fshb Expression
More than 10 distinct peptides are processed from the VGF precursor protein (38–40). We sought to determine which VGF-derived peptide might be involved in the regulation of Fshb expression in the gonadotropes. Further analysis of the LβT2 cell secretoproteome by mass spectrometry identified the expression of a 13-amino acid fragment of NERP1, which is located in the central region of VGF (Fig. 7A). Using an anti-VGF antibody raised against the NERP1 sequence for ELISA assays, we found that NERP1 secretion was significantly increased by GnRH stimulation (3.6 ± 0.93-fold) as well as CTX (3.0 ± 0.40-fold; Fig. 7B).
FIGURE 7.
NERP1 negatively regulates GnRH-induced Fshb expression in LβT2 gonadotropes. A, endogenous expression of a 13-amino acid fragment of NERP1 in LβT2 cells. Shown is the fragment ion spectrum of VGF peptide LLQQGLAQVEAGR. Doubly charged peptide ions (parent+2H; 843.99) were fragmented, and the fragment ion spectrum was collected by an LTQ-Orbitrap mass spectrometer in data-dependent acquisition mode. A protein database search of this spectrum matched peptide LLQQGLAQVEAGR with a Mascot ion score of 44.2. Peaks corresponding to peptide backbone fragmentations were annotated as bn and yn for b and y ions (in red and blue, respectively). 304 indicates the N-terminal iTRAQ modification. B, up-regulation of NERP1 secretion in LβT2 cells stimulated with GnRΗ or CTX. Cells were serum-starved overnight and stimulated with either 1 nm GnRH, 5 μg/ml CTX, or vehicle for 6 h. Secreted NERP1 was quantitated by ELISA using a NERP1-specific antibody. **, p < 0.01, one-tailed t test. C, effect of NERP1 on the promoter activity of an NF-κB luciferase reporter vector. LβT2 cells were co-transfected with an NF-κB-firefly luciferase reporter construct and a control TK-Renilla luciferase reporter construct (internal standard for transfection efficiency) on day 1. On day 2, cells were serum-starved overnight. On day 3, cells were treated with 5 μm NERP1 for the indicated times. Firefly luciferase activity was measured and normalized to Renilla luciferase activity. *, p < 0.05, two-way ANOVA; n = 3. RLU, relative light units. D, effect of increasing concentrations of NERP1 on Fshb gene expression in VGF knockdown cells stimulated with GnRH. Cells were transfected with VGF siRNA on day 1. On day 2, cells were serum-starved overnight. On day 3, cells were stimulated with 1 nm GnRH for 2 h followed by 4 h without GnRH or with vehicle. Cells were co-treated with the indicated concentrations of NERP1 for 6 h. Fshb mRNA expression levels were determined by qPCR. ***, p < 0.01, two-way ANOVA with Bonferroni post-test corrections. Data shown are the mean from one experiment performed in triplicate and are representative of three independent experiments. Error bars represent S.E.
NERP1 has been reported to stimulate calcium mobilization in the pituitary (40). We found that NERP1 exposure activated a calcium-sensitive reporter in LβT2 gonadotropes (Fig. 7C). To assess the effects of exogenous VGF-derived peptides on GnRH regulation of Fshb expression, we used VGF siRNA knockdown to reduce the background effects of endogenous NERP1. Exposure of these siRNA-transfected cells to peptides from the N-terminal to central regions of VGF, namely NERP2, NERP3, and NERP4 had no significant effect on Fshb gene regulation by GnRH (data not shown). In contrast, exogenous NERP1 reduced GnRH-induced Fshb mRNA expression in VGF siRNA-transfected cells, causing a concentration-dependent decrease in GnRH-induced Fshb expression (Fig. 7D). These results suggest that central VGF peptide NERP1 is biologically active in the gonadotrope and may act as a negative regulator of GnRH-induced Fshb expression.
Discussion
This study supports the importance of autocrine factors in the regulation of Fshb, identifies several novel putative Fshb autocrine regulatory factors, and implicates a VGF-derived peptide as a new autocrine regulator of Fshb gene expression in the gonadotrope. Vgf is highly induced by high frequency GnRH stimulation. The discordant induction of Fshb and Vgf by different patterns of GnRH stimulation is consistent with the Fshb-suppressing effects of VGF. The VGF-derived peptide NERP1 is synthesized and secreted in response to GnRH. NERP1 suppresses GnRH-induced Fshb expression. Our findings suggest that an autocrine VGF-derived peptide contributes to the differential regulation of gonadotropin genes and provides a potential mechanism for the preferential induction of Fshb expression by low GnRH pulse frequency.
VGF-derived peptides have been implicated in various neuroendocrine functions, including homeostasis and reproduction (for a review, see Ref. 41). VGF pituitary levels are regulated throughout the estrous cycle and increase in response to ovariectomy (42). VGF knock-out mice are infertile (2) and have several deficits in their hypothalamic-pituitary-gonadal axis and severe abnormalities of fat metabolism that may contribute to infertility (2, 43). The same group generated humanized VGF knock-in mouse models expressing either full-length human VGF or a truncated form that included amino acids 1–524. Namely, the truncated human VGF(1–524) was devoid of a series of peptides starting with threonine 554, comprising peptides TLQP-21 and AQEE-30, but retained peptides NERP1 and NERP2. All of these peptides were previously demonstrated to regulate male reproductive behavior and/or female fertility (44–48). Notably, male and female mice from both human VGF lines showed fertility and gonadotropin expression that were indistinguishable from wild-type mice (49), thereby excluding several bioactive C-terminal VGF peptides as being necessary for reproductive function while supporting a biological role for N-terminal to central VGF peptides. These in vivo data are consistent with our in vitro data presenting NERP1 as a putative regulator of gonadotropin gene expression. Additionally, NERP1 and NERP2 evoke a calcium response in the pituitary and hypothalamus (40). Our findings provide a molecular mechanism for the differential regulation of gonadotropins and further substantiate the involvement of VGF in the control of reproduction and the hypothalamic-pituitary-gonadal axis.
Although our data support an inhibitory effect of NERP1 on GnRH-induced Fshb mRNA expression, we note that only in vivo studies can establish a physiological role for NERP1 in regulating Fshb expression. The present investigation uses mouse primary cultured gonadotropes or the murine gonadotrope-derived LβT2 cell line and relies on the measurement of Fshb mRNA expression as a surrogate for regulation of FSH protein. In view of the excellent correlation of pituitary Fshb mRNA expression and blood levels of FSH observed in mouse and rat in vivo studies (50–55), Fshb mRNA is considered a satisfactory assay for changes in FSH biosynthesis in cultured gonadotrope or cell line studies (13, 16, 33, 37, 56–71). This high correlation between gonadotrope Fshb mRNA and secreted FSH exists because FSH regulation occurs at the transcription level and the mature protein is constitutively secreted (72). Nonetheless, in vitro studies may not adequately model in vivo physiology. Establishing the role of NERP1 that is suggested by our in vitro studies would greatly benefit from studying conditional genetic VGF in vivo models in conjunction with measurement of gonadotrope Fshb mRNA and secreted FSH.
In our experiments on LβT2 cells, we see a trend toward suppression of Fshb mRNA induction at extracellular NERP1 concentrations of 50 pmol/ml with higher concentrations required for the suppression to reach significance (See Fig. 7D). In male rat pituitary, NERP1 has been reported to have a level of 50–75 pmol/g (73, 74). This total concentration most likely greatly underestimates the local autocrine gonadotrope concentration of NERP1, especially following up-regulation of Vgf. Thus, we believe that the concentrations of NERP1 tested in our study are physiologically plausible. Notably, although we propose a role for NERP1 in Fshb suppression, our results do not exclude the role of other autocrine VGF peptides in contributing to the regulation of Fshb.
The low GnRH pulse frequency preference of the Fshb gene is important for reproductive physiology; however, a full explanation for this characteristic of the system has eluded decades of experimental and theoretical work. Our data suggest that a VGF-derived peptide may be one factor to help explain this paradox. We propose that under high GnRH pulse frequency both Gαq/11 and Gαs are activated; although Gαs activation promotes Lhb expression, it simultaneously induces an elevated expression and secretion of VGF, which strongly suppresses Gαq/11-dependent Fshb expression. Under low GnRH pulse frequency, activation of Gαq/11 stimulates Fshb expression, which is no longer suppressed by VGF, as its basal level of expression is not significantly altered by this pulse frequency regime. Other components may also contribute to the frequency-dependent gonadotropin gene response patterns. Previous work by Tsutsumi et al. (25) showed that the Gαq/11 pathway, but not Gαs, was desensitized by multiple pulses of GnRH, indicating distinct differences in the responses of the two signaling cascades to pulsatile GnRH stimulation. Using a mouse FSHβ promoter construct, the same group proposed that differential expression of AP1 factors and corepressors ski-oncogene-like protein and TG interacting factor mediated GnRH pulse sensitivity of Fshb expression (75). Another study revealed that inducible cAMP early repressor was preferentially induced by high frequency GnRH stimulation and that it abrogated the GnRH pulse frequency-dependent effects on Fshb gene transcription (58). Secreted prostaglandins have been reported to affect GnRH regulation of Lhb promoter constructs (76, 77). Of interest, VGF may regulate prostaglandin secretion as VGF-derived peptide TLQP-21 also modulates pancreatic exocrine secretion and gastric contractility via stimulation of prostaglandin release (78, 79). Our results also implicate other factors, including inhibin α (1) and GDF9 (see Fig. 2), as potential autocrine modulators of gonadotropin gene regulation by GnRH. Further study and mathematical modeling will be required to understand the relative roles of various regulators and how they integrate to tune the responses of the gonadotrope to GnRH.
The identification of VGF as a regulator of Fshb expression may open new avenues of research for treating reproductive disorders such as polycystic ovary syndrome, endometriosis, and syndromes associated with impaired gonadotropin levels. Modulation of VGF input to the gonadotropes could potentially increase the effectiveness of GnRH analogs in the treatment of gonadal hormone-sensitive diseases. Additionally, endocrine tumors, including pituitary adenomas, were found to have higher levels of VGF precursors than normal tissue (80). Thus, VGF may also serve as a diagnostic marker and a possible therapeutic target for hormone-dependent cancers.
Regulation of a nuclear gene following activation of a cell surface receptor has conventionally been considered to be the function of intracellular signaling pathways. Our work highlights a regulatory mechanism whereby the intracellular signaling cascade triggered by GnRH goes outside the cell via a secreted VGF-derived peptide feedback loop to influence the regulation of the Fshb gene. The large number of secreted and regulated proteins that we have identified in gonadotropes intimates the presence of a robust network of autocrine and paracrine regulatory factors that might rival the intracellular network in its complexity. What is classically considered intracellular signaling does not appear to respect the boundary of the cell membrane. Elucidating how cells decode specific extracellular stimuli, such as GnRH pulses, to direct gene expression patterns may require a broader view of cell signaling that encompasses a complex network involving both intracellular mediators and exosignaling circuits (Fig. 8) (81).
FIGURE 8.
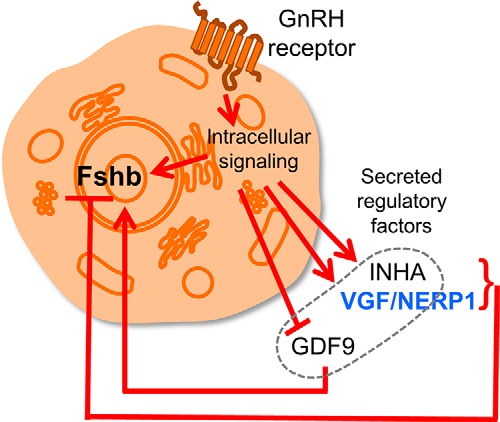
Exosignaling network contributes to Fshb gene regulation. The schematic illustrates the proposed integration of intracellular and extracellular regulatory circuits that coordinate the control of Fshb gene expression by GnRH. INHA, inhibin α.
Experimental Procedures
Materials and Methods
GnRH was purchased from Bachem (Torrance, CA). CTX was ordered from Calbiochem. VGF-derived peptides (NERP1, NERP2, NERP3, and NERP4) were synthesized as reported previously (40, 82); TLQP-30 was obtained from Phoenix Pharmaceuticals, Inc. (Burlingame, CA). VGF and inhibin α siRNAs (On-Target plus siRNA SMARTpool) were ordered from Dharmacon (Denver, CO). The anti-VGF antibody (kindly provided by Dr. S. R. J. Salton) used in immunoneutralization experiments was polyclonal and targeted residues 78–340 of the murine VGF precursor. Brefeldin A and monensin were ordered from BioLegend (San Diego, CA).
Cell Culture
LβT2 cells were obtained from Professor Pamela Mellon (University of California, San Diego, CA). Cells were cultured at 37 °C in DMEM (Mediatech, Herndon, VA) supplemented with 10% fetal bovine serum (FBS; Gemini, Calabasas, CA) in a humidified air atmosphere of 5% CO2. Purified gonadotrope cells were prepared from H2Kk mice as described in our previous study (1).
Perfusion Experiments
The perfusion system was designed and custom-built in the laboratory. The perfusion system maintains a constant internal temperature of 37 °C by means of equipped heating blocks. The system has four cassettes; each cassette holds four chambers where cultured gonadotropes are inserted on a coverslip. Each coverslip in the chamber was perfused with either vehicle (no GnRH control; serum-free DMEM) or 2 nm GnRH via two electronically controlled valves supplying laminar fluid at a constant flow rate (0.6 ml/min/chamber). One million cells were seeded on each coverslip and grown for 2 days in DMEM + 10% FBS. Cells were serum-starved overnight before the perfusion experiment. Cells were subjected to either low (5-min GnRH pulses at intervals/periods of 2 h) or high (5-min GnRH pulses at intervals/periods of 0.5 h) GnRH pulse frequency of GnRH stimulation (2 nm) for 8 h and then harvested for RNA extraction.
siRNA Studies
One million LβT2 cells were transfected with 0.5 μg of siRNA in electroporation cuvettes using Amaxa Cell Line Nucleofector Kit L using SG Buffer and Nucleofector program DS-137 following the manufacturer's instructions (Lonza Inc., Walkersville, MD). Immediately after transfection, cells were seeded in a cell culture plate containing DMEM + 10% FBS and incubated at 37 °C for 24 h. Culture medium was then replaced with serum free-DMEM, and cells were incubated overnight at 37 °C for serum starvation. Cells were next stimulated with vehicle, GnRH, or CTX. Lysis buffer was added to the cells, which were then harvested for quantitative real time PCR.
Quantitative Real Time PCR
For quantitative real time PCR (qPCR) experiments, cells were seeded in 24-well plates at ∼0.5–1 million cells/well. For cell harvest, medium was replaced with 300 μl of RNA lysis buffer (Agilent, Santa Clara, CA). Total RNA was isolated with the Absolutely RNA 96 Microprep kit (Stratagene, La Jolla, CA) according to the manufacturer's protocol. RNA concentrations were determined using a NanoDrop spectrophotometer (Thermo Scientific, Wilmington, DE). After reverse transcription of 1 μg of RNA with Affinity Script reverse transcriptase (Agilent), samples were diluted 1:20 in molecular biology grade H2O (Cellgro, Manassas, VA). Later, SYBR green qPCR assays were performed (40 cycles) using 5 μl of cDNA template and 5 μl of Master Mix containing the specific primers for the targeted gene, Platinum® Taq DNA polymerase, and the required qPCR buffers according to manufacturer's recommendations. Three biological replicates were done. Results were exported as cycle threshold (Ct) values, and cycle threshold values of target genes were normalized to that of Rps11 in subsequent analysis. Data were expressed as arbitrary units by using the formula E = 2500 × 1.93(Rps11 Ct value − gene of interest Ct value) where E is the expression level in arbitrary units. Primer sequences are provided in supplemental Table S3.
Illumina Array
Total RNA was extracted from two million cells transfected with either control or Gαs siRNA. RNA samples were snap frozen in dry ice and sent to the Keck Genomic Facility at Yale University for whole-genome expression profiling analysis using the MouseWG-6 v2.0 Expression BeadChip (Illumina, San Diego, CA). A total of six samples were independently prepared: three samples from control siRNA-treated cells and three samples from Gαs siRNA-treated cells. The control microarray data were reported previously (37). All microarray data were deposited in the Gene Expression Omnibus (GEO) under accession number GSE52631.
Preparation of Conditioned Media and HPLC-Isobaric Tags for Relative and Absolute Quantitation Mass Spectrometry (iTRAQ MS)
Twenty million cells were transfected with either control or Gαs siRNA and then seeded in 100-mm cell culture plates in DMEM + 10% FBS. Two days later, cells were washed twice with prewarmed PBS. Conditioned media were harvested another 24 h later and centrifuged at 20,000 × g for 10 min at 4 °C to remove cell debris. To enrich for secreted proteins in the conditioned media, conditioned media samples were centrifuged using Amicon centrifugal filters with a 3-kDa cutoff (Millipore, Billerica, MA). A total of eight concentrated conditioned media samples were independently prepared: four samples from control siRNA-treated cells and four samples from Gαs siRNA-treated cells. Samples were stored at −70 °C until they were analyzed by iTRAQ labeling and LC-MS/MS.
iTRAQ Labeling
The total protein concentration of each sample was measured using the Bradford assay. A quantity of 100 μg was taken from each sample, denatured in 0.1% SDS, reduced in 50 mm tris(2-carboxyethyl)phosphine, alkylated in 200 mm methyl methanethiosulfonate, and digested overnight with trypsin. The peptides in each sample were labeled with different iTRAQ reagents (AB Sciex, Framingham, MA) following the manufacturer's protocol, combined, dried down to a 50-μl volume, acidified with 1% trifluoroacetic acid (TFA), and desalted with a 100-mg C18 Sep-Pak cartridge (Waters, Milford, MA).
Peptide Fractionation
Combined iTRAQ-labeled peptides were first separated into 12 fractions using a 3100 OFFGEL Fractionator (Agilent) with a 12-well setup and a 12-cm pH 3–10 immobilized pH gradient strip according to the manufacturer's recommendations. Peptides were collected after the focusing voltage reached a maximum plateau (>50 kV-h). The recovered fractions were lyophilized.
LC-MALDI-Time-of-flight/Time-of-flight (TOF/TOF) Analysis
All solvents used in this section were aqueous unless specified and obtained from ThermoFisher Scientific (San Jose, CA). Dried samples were reconstituted in 30 μl of solvent A (2% (v/v) acetonitrile, 0.1% (v/v) TFA), and 10 μl of the resulting solution was injected into an UltimateTM HPLC system with UV detection (LC Packings, Sunnyvale, CA). Solvent B contained 98% (v/v) acetonitrile, 0.1% (v/v) TFA. A C18 trap (5.0 mm × 300-μm inner diameter; LC Packings) and a 15-cm × 100-μm-inner diameter column packed in house with Magic C18 5-μm beads (Michrom Bioresources, Auburn, CA) were used for chromatography. Solvent flow rate was set to 0.700 μl/min. After injection, each sample was loaded and washed with solvent A for 5 min at a flow rate of 0.030 ml/min followed by a 10–50% gradient over 55 min and then ramped to 90% by 65 min. Each sample run was maintained in 90% solvent B until 75 min and returned to 10% at 75.1 min followed by a 10-min equilibration in 10% solvent B. The HPLC system is connected to a modified ProbotTM automated MALDI spotter (LC Packings) to spot eluates on an LC MALDI plate with on-line mixing of α-cyano-4-hydroxycinnamic acid matrix (7.5 mg/ml in 70% acetonitrile). Considering the dead time for the HPLC and spotting system, spot collection time was started 10 min from injection time with a spotting time of 0.33 min per spot. A total of 176 spots was collected.
The sample plate with the resulting spots was loaded onto a 5800 MALDI-TOF/TOF mass spectrometer (AB Sciex) for MS and MS/MS analysis. Batch mode mass calibration with plate alignment was done before data acquisition; thus a separate plate calibration file was generated for each individual plate containing sample spots. Full scan MS spectra were obtained with a total of 1000 laser shots per spectrum. From each spot, the top 20 most intense peaks, weakest precursor first, that were above a signal-to-noise threshold of 10 were selected for successful MS/MS analysis. Each MS/MS spectrum was an average of 1600 laser shots. MS/MS spectra were acquired in the presence of collision gas. The sequence of the VGF NERP1 was confirmed using an LTQ-Orbitrap mass spectrometer (ThermoFisher Scientific).
Data Analysis
ProteinPilot 3.0 (AB Sciex) was used to process the MS/MS spectra for protein identification and quantitation with its searching algorithm Paragon 3.0.0.0 (83). The protein database used for searching was built using UniProt mouse fasta file (release 2010_11). “Thorough” mode was used in the data processing using parameters of “Quantitate” for iTRAQ 8-plex (peptide-labeled), “MMTS” for cysteine alkylation, “Trypsin” for digestion, “Biological modifications” for ID focus, and “Mus musculus” for species. The detected protein threshold was set to 1.3 (95% confidence).
Analysis of the Concordance between Transcript and Protein Levels
Microarray expression data for Gαs siRNA knockdown cells were compared with scrambled siRNA-transfected cells using the limma package (84). Mass spectrometry data were summarized by averaging over log -fold changes computed using each of the eight tags as background. Genes were matched with proteins by their Mouse Genome Informatics Database (MGI) symbol, and genes with an mRNA expression change significant with an uncorrected p value of 0.05 and a mass spectrometry log -fold change of at least 0.1 were selected.
NERP1 ELISA
A NERP1 ELISA kit was purchased from Bachem. One million LβT2 cells were stimulated with vehicle, GnRH, or CTX for 6 h to induce VGF-derived peptide synthesis and secretion. The culture medium was then harvested and subjected to ELISA following the manufacturer's recommendations. The antibody was raised against rat NERP1 and was polyclonal.
Statistical Analysis
Statistical calculations were performed using the GraphPad Prism statistical software package version 5 (GraphPad Inc., San Diego, CA). Data were analyzed for normality followed by calculation of ANOVA. Statistical significance was set as indicated in each figure legend with at least a p value <0.05.
Author Contributions
S. G. C., S. R. J. S., and S. C. S. designed research. S. G. C., Q. W., J. J., and G. D. performed research. K. S., R. W., S. R. J. S., and N. M. contributed new reagents or analytic tools. S. G. C., Q. W., M. C., G. D., R. W., H. P., and S. C. S. analyzed data. S. G. C., H. P., and S. C. S. wrote the paper.
Supplementary Material
This work was supported by National Institutes of Health Grants DK46943, R21/R33 MH083496, RO1 DE021996, RO1 MH086499, P30 NS061777, and S10 RR022415; the Diabetes Action and Education Foundation; and the Hope for Depression Research Foundation. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.

This article contains supplemental Tables S1–S3.
The mass spectrometric raw data and spectral libraries associated with this manuscript are available from ProteomeXchange with the accession number PXD000063.
- GnRH
- gonadotropin-releasing hormone
- CGA
- common α-glycoprotein subunit
- CTX
- cholera toxin
- GDF
- growth differentiation factor
- iTRAQ
- isobaric tags for relative and absolute quantitation
- LH
- luteinizing hormone
- NERP
- neuroendocrine regulatory peptide
- qPCR
- quantitative real time PCR
- VGF
- VGF nerve growth factor-inducible
- SCG
- secretogranin
- IGFBP
- insulin-like growth factor-binding protein
- SEMA
- semaphorin
- STC1
- stanniocalcin 1
- BMP
- bone morphogenetic protein
- ANOVA
- analysis of variance.
References
- 1. Choi S. G., Jia J., Pfeffer R. L., and Sealfon S. C. (2012) G proteins and autocrine signaling differentially regulate gonadotropin subunit expression in pituitary gonadotrope. J. Biol. Chem. 287, 21550–21560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hahm S., Mizuno T. M., Wu T. J., Wisor J. P., Priest C. A., Kozak C. A., Boozer C. N., Peng B., McEvoy R. C., Good P., Kelley K. A., Takahashi J. S., Pintar J. E., Roberts J. L., Mobbs C. V., et al. (1999) Targeted deletion of the Vgf gene indicates that the encoded secretory peptide precursor plays a novel role in the regulation of energy balance. Neuron 23, 537–548 [DOI] [PubMed] [Google Scholar]
- 3. Marshall J. C., and Kelch R. P. (1986) Gonadotropin-releasing hormone: role of pulsatile secretion in the regulation of reproduction. New Engl. J. Med. 315, 1459–1468 [DOI] [PubMed] [Google Scholar]
- 4. Marshall J. C., Eagleson C. A., and McCartney C. R. (2001) Hypothalamic dysfunction. Mol. Cell. Endocrinol. 183, 29–32 [DOI] [PubMed] [Google Scholar]
- 5. Humaidan P., Papanikolaou E. G., Kyrou D., Alsbjerg B., Polyzos N. P., Devroey P., and Fatemi H. M. (2012) The luteal phase after GnRH-agonist triggering of ovulation: present and future perspectives. Reprod. Biomed. Online 24, 134–141 [DOI] [PubMed] [Google Scholar]
- 6. Noel S. D., and Kaiser U. B. (2011) G protein-coupled receptors involved in GnRH regulation: molecular insights from human disease. Mol. Cell. Endocrinol. 346, 91–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kim D. K., Yang J. S., Maiti K., Hwang J. I., Kim K., Seen D., Ahn Y., Lee C., Kang B. C., Kwon H. B., Cheon J., and Seong J. Y. (2009) A gonadotropin-releasing hormone-II antagonist induces autophagy of prostate cancer cells. Cancer Res. 69, 923–931 [DOI] [PubMed] [Google Scholar]
- 8. Van Poppel H., and Klotz L. (2012) Gonadotropin-releasing hormone: an update review of the antagonists versus agonists. Int. J. Urol. 19, 594–601 [DOI] [PubMed] [Google Scholar]
- 9. Hackshaw A., Baum M., Fornander T., Nordenskjold B., Nicolucci A., Monson K., Forsyth S., Reczko K., Johansson U., Fohlin H., Valentini M., and Sainsbury R. (2009) Long-term effectiveness of adjuvant goserelin in premenopausal women with early breast cancer. J. Natl. Cancer Inst. 101, 341–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Del Mastro L., Levaggi A., Giraudi S., and Pronzato P. (2011) Luteinising hormone releasing hormone agonists (LH-RHa) in premenopausal early breast cancer patients: current role and future perspectives. Cancer Treat. Rev. 37, 208–211 [DOI] [PubMed] [Google Scholar]
- 11. Pierce J. G., and Parsons T. F. (1981) Glycoprotein hormones: structure and function. Annu. Rev. Biochem. 50, 465–495 [DOI] [PubMed] [Google Scholar]
- 12. Bernard D. J., Fortin J., Wang Y., and Lamba P. (2010) Mechanisms of FSH synthesis: what we know, what we don't, and why you should care. Fertil. Steril. 93, 2465–2485 [DOI] [PubMed] [Google Scholar]
- 13. Bédécarrats G. Y., and Kaiser U. B. (2003) Differential regulation of gonadotropin subunit gene promoter activity by pulsatile gonadotropin-releasing hormone (GnRH) in perifused LβT2 cells: role of GnRH receptor concentration. Endocrinology 144, 1802–1811 [DOI] [PubMed] [Google Scholar]
- 14. Alarid E. T., Windle J. J., Whyte D. B., and Mellon P. L. (1996) Immortalization of pituitary cells at discrete stages of development by directed oncogenesis in transgenic mice. Development 122, 3319–3329 [DOI] [PubMed] [Google Scholar]
- 15. Thomas P., Mellon P. L., Turgeon J., and Waring D. W. (1996) The LβT2 clonal gonadotrope: a model for single cell studies of endocrine cell secretion. Endocrinology 137, 2979–2989 [DOI] [PubMed] [Google Scholar]
- 16. An B. S., Poon S. L., So W. K., Hammond G. L., and Leung P. C. (2009) Rapid effect of GNRH1 on follicle-stimulating hormone β gene expression in LβT2 mouse pituitary cells requires the progesterone receptor. Biol. Reprod. 81, 243–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Turgeon J. L., Kimura Y., Waring D. W., and Mellon P. L. (1996) Steroid and pulsatile gonadotropin-releasing hormone (GnRH) regulation of luteinizing hormone and GnRH receptor in a novel gonadotrope cell line. Mol. Endocrinol. 10, 439–450 [DOI] [PubMed] [Google Scholar]
- 18. Sealfon S. C., Weinstein H., and Millar R. P. (1997) Molecular mechanisms of ligand interaction with the gonadotropin-releasing hormone receptor. Endocr. Rev. 18, 180–205 [DOI] [PubMed] [Google Scholar]
- 19. Lambert N. A. (2008) Dissociation of heterotrimeric g proteins in cells. Sci. Signal. 1, re5. [DOI] [PubMed] [Google Scholar]
- 20. Oldham W. M., and Hamm H. E. (2008) Heterotrimeric G protein activation by G-protein-coupled receptors. Nat. Rev. Mol. Cell Biol. 9, 60–71 [DOI] [PubMed] [Google Scholar]
- 21. Grosse R., Schmid A., Schöneberg T., Herrlich A., Muhn P., Schultz G., and Gudermann T. (2000) Gonadotropin-releasing hormone receptor initiates multiple signaling pathways by exclusively coupling to Gq/11 proteins. J. Biol. Chem. 275, 9193–9200 [DOI] [PubMed] [Google Scholar]
- 22. Hsieh K. P., and Martin T. F. (1992) Thyrotropin-releasing hormone and gonadotropin-releasing hormone receptors activate phospholipase C by coupling to the guanosine triphosphate-binding proteins Gq and G11. Mol. Endocrinol. 6, 1673–1681 [DOI] [PubMed] [Google Scholar]
- 23. Naor Z., Azrad A., Limor R., Zakut H., and Lotan M. (1986) Gonadotropin-releasing hormone activates a rapid Ca2+-independent phosphodiester hydrolysis of polyphosphoinositides in pituitary gonadotrophs. J. Biol. Chem. 261, 12506–12512 [PubMed] [Google Scholar]
- 24. Liu F., Usui I., Evans L. G., Austin D. A., Mellon P. L., Olefsky J. M., and Webster N. J. (2002) Involvement of both Gq/11 and Gs proteins in gonadotropin-releasing hormone receptor-mediated signaling in LβT2 cells. J. Biol. Chem. 277, 32099–32108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tsutsumi R., Mistry D., and Webster N. J. (2010) Signaling responses to pulsatile gonadotropin-releasing hormone in LβT2 gonadotrope cells. J. Biol. Chem. 285, 20262–20272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bernard D. J. (2004) Both SMAD2 and SMAD3 mediate activin-stimulated expression of the follicle-stimulating hormone β subunit in mouse gonadotrope cells. Mol. Endocrinol. 18, 606–623 [DOI] [PubMed] [Google Scholar]
- 27. Suszko M. I., Balkin D. M., Chen Y., and Woodruff T. K. (2005) Smad3 mediates activin-induced transcription of follicle-stimulating hormone β-subunit gene. Mol. Endocrinol. 19, 1849–1858 [DOI] [PubMed] [Google Scholar]
- 28. Tsujii T., Attardi B., and Winters S. J. (1995) Regulation of α-subunit mRNA transcripts by pituitary adenylate cyclase-activating polypeptide (PACAP) in pituitary cell cultures and αT3–1 cells. Mol. Cell. Endocrinol. 113, 123–130 [DOI] [PubMed] [Google Scholar]
- 29. Winters S. J., and Moore J. P. Jr. (2011) PACAP, an autocrine/paracrine regulator of gonadotrophs. Biol. Reprod. 84, 844–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Faure M. O., Nicol L., Fabre S., Fontaine J., Mohoric N., McNeilly A., and Taragnat C. (2005) BMP-4 inhibits follicle-stimulating hormone secretion in ewe pituitary. J. Endocrinol. 186, 109–121 [DOI] [PubMed] [Google Scholar]
- 31. Ho C. C., and Bernard D. J. (2009) Bone morphogenetic protein 2 signals via BMPR1A to regulate murine follicle-stimulating hormone β subunit transcription. Biol. Reprod. 81, 133–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Huang H. J., Wu J. C., Su P., Zhirnov O., and Miller W. L. (2001) A novel role for bone morphogenetic proteins in the synthesis of follicle-stimulating hormone. Endocrinology 142, 2275–2283 [DOI] [PubMed] [Google Scholar]
- 33. Lee K. B., Khivansara V., Santos M. M., Lamba P., Yuen T., Sealfon S. C., and Bernard D. J. (2007) Bone morphogenetic protein 2 and activin A synergistically stimulate follicle-stimulating hormone β subunit transcription. J. Mol. Endocrinol. 38, 315–330 [DOI] [PubMed] [Google Scholar]
- 34. Nicol L., Faure M. O., McNeilly J. R., Fontaine J., Taragnat C., and McNeilly A. S. (2008) Bone morphogenetic protein-4 interacts with activin and GnRH to modulate gonadotrophin secretion in LβT2 gonadotrophs. J. Endocrinol. 196, 497–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Otsuka F., and Shimasaki S. (2002) A novel function of bone morphogenetic protein-15 in the pituitary: selective synthesis and secretion of FSH by gonadotropes. Endocrinology 143, 4938–4941 [DOI] [PubMed] [Google Scholar]
- 36. Young J. M., Juengel J. L., Dodds K. G., Laird M., Dearden P. K., McNeilly A. S., McNatty K. P., and Wilson T. (2008) The activin receptor-like kinase 6 Booroola mutation enhances suppressive effects of bone morphogenetic protein 2 (BMP2), BMP4, BMP6 and growth and differentiation factor-9 on FSH release from ovine primary pituitary cell cultures. J. Endocrinol. 196, 251–261 [DOI] [PubMed] [Google Scholar]
- 37. Choi S. G., Wang Q., Jia J., Pincas H., Turgeon J. L., and Sealfon S. C. (2014) Growth differentiation factor 9 (GDF9) forms an incoherent feed-forward loop modulating follicle-stimulating hormone β-subunit (FSHβ) gene expression. J. Biol. Chem. 289, 16164–16175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Levi A., Ferri G. L., Watson E., Possenti R., and Salton S. R. (2004) Processing, distribution, and function of VGF, a neuronal and endocrine peptide precursor. Cell. Mol. Neurobiol. 24, 517–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sasaki K., Osaki T., and Minamino N. (2013) Large-scale identification of endogenous secretory peptides using electron transfer dissociation mass spectrometry. Mol. Cell. Proteomics 12, 700–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sasaki K., Takahashi N., Satoh M., Yamasaki M., and Minamino N. (2010) A peptidomics strategy for discovering endogenous bioactive peptides. J. Proteome Res. 9, 5047–5052 [DOI] [PubMed] [Google Scholar]
- 41. Ferri G. L., Noli B., Brancia C., D'Amato F., and Cocco C. (2011) VGF: an inducible gene product, precursor of a diverse array of neuro-endocrine peptides and tissue-specific disease biomarkers. J. Chem. Neuroanat. 42, 249–261 [DOI] [PubMed] [Google Scholar]
- 42. Ferri G. L., Gaudio R. M., Cossu M., Rinaldi A. M., Polak J. M., Berger P., and Possenti R. (1995) The “VGF” protein in rat adenohypophysis: sex differences and changes during the estrous cycle and after gonadectomy. Endocrinology 136, 2244–2251 [DOI] [PubMed] [Google Scholar]
- 43. Fargali S., Scherer T., Shin A. C., Sadahiro M., Buettner C., and Salton S. R. (2012) Germline ablation of VGF increases lipolysis in white adipose tissue. J. Endocrinol. 215, 313–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Aguilar E., Pineda R., Gaytán F., Sánchez-Garrido M. A., Romero M., Romero-Ruiz A., Ruiz-Pino F., Tena-Sempere M., and Pinilla L. (2013) Characterization of the reproductive effects of the Vgf-derived peptide TLQP-21 in female rats: in vivo and in vitro studies. Neuroendocrinology 98, 38–50 [DOI] [PubMed] [Google Scholar]
- 45. Melis M. R., Sanna F., Succu S., Ferri G. L., and Argiolas A. (2012) Neuroendocrine regulatory peptide-1 and neuroendocrine regulatory peptide-2 influence differentially feeding and penile erection in male rats: sites of action in the brain. Regul. Pept. 177, 46–52 [DOI] [PubMed] [Google Scholar]
- 46. Noli B., Brancia C., D'Amato F., Ferri G. L., and Cocco C. (2014) VGF changes during the estrous cycle: a novel endocrine role for TLQP peptides? PLoS One 9, e108456, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pinilla L., Pineda R., Gaytán F., Romero M., García-Galiano D., Sánchez-Garrido M. A., Ruiz-Pino F., Tena-Sempere M., and Aguilar E. (2011) Characterization of the reproductive effects of the anorexigenic VGF-derived peptide TLQP-21: in vivo and in vitro studies in male rats. Am. J. Physiol. Endocrinol. Metab. 300, E837–E847 [DOI] [PubMed] [Google Scholar]
- 48. Succu S., Cocco C., Mascia M. S., Melis T., Melis M. R., Possenti R., Levi A., Ferri G. L., and Argiolas A. (2004) Pro-VGF-derived peptides induce penile erection in male rats: possible involvement of oxytocin. Eur. J. Neurosci. 20, 3035–3040 [DOI] [PubMed] [Google Scholar]
- 49. Sadahiro M., Erickson C., Lin W. J., Shin A. C., Razzoli M., Jiang C., Fargali S., Gurney A., Kelley K. A., Buettner C., Bartolomucci A., and Salton S. R. (2015) Role of VGF-derived carboxy-terminal peptides in energy balance and reproduction: analysis of “humanized” knockin mice expressing full-length or truncated VGF. Endocrinology 156, 1724–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Boerboom D., Kumar V., Boyer A., Wang Y., Lambrot R., Zhou X., Rico C., Boehm U., Paquet M., Céleste C., Kimmins S., and Bernard D. J. (2015) β-Catenin stabilization in gonadotropes impairs FSH synthesis in male mice in vivo. Endocrinology 156, 323–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dalkin A. C., Haisenleder D. J., Ortolano G. A., Ellis T. R., and Marshall J. C. (1989) The frequency of gonadotropin-releasing-hormone stimulation differentially regulates gonadotropin subunit messenger ribonucleic acid expression. Endocrinology 125, 917–924 [DOI] [PubMed] [Google Scholar]
- 52. Fortin J., Boehm U., Deng C. X., Treier M., and Bernard D. J. (2014) Follicle-stimulating hormone synthesis and fertility depend on SMAD4 and FOXL2. FASEB J. 28, 3396–3410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Fortin J., Boehm U., Weinstein M. B., Graff J. M., and Bernard D. J. (2014) Follicle-stimulating hormone synthesis and fertility are intact in mice lacking SMAD3 DNA binding activity and SMAD2 in gonadotrope cells. FASEB J. 28, 1474–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Fortin J., Kumar V., Zhou X., Wang Y., Auwerx J., Schoonjans K., Boehm U., Boerboom D., and Bernard D. J. (2013) NR5A2 regulates Lhb and Fshb transcription in gonadotrope-like cells in vitro, but is dispensable for gonadotropin synthesis and fertility in vivo. PLoS One 8, e59058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Xie H., Hoffmann H. M., Meadows J. D., Mayo S. L., Trang C., Leming S. S., Maruggi C., Davis S. W., Larder R., and Mellon P. L. (2015) Homeodomain Proteins SIX3 and SIX6 regulate gonadotrope-specific genes during pituitary development. Mol. Endocrinol. 29, 842–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bar-Lev T. H., Harris D., Tomić M., Stojilkovic S., Blumenfeld Z., Brown P., Seger R., and Naor Z. (2015) Role of PI4K and PI3K-AKT in ERK1/2 activation by GnRH in the pituitary gonadotropes. Mol. Cell. Endocrinol. 415, 12–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Choi S. G., Ruf-Zamojski F., Pincas H., Roysam B., and Sealfon S. C. (2011) Characterization of a MAPK scaffolding protein logic gate in gonadotropes. Mol. Endocrinol. 25, 1027–1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ciccone N. A., Xu S., Lacza C. T., Carroll R. S., and Kaiser U. B. (2010) Frequency-dependent regulation of follicle-stimulating hormone β by pulsatile gonadotropin-releasing hormone is mediated by functional antagonism of bZIP transcription factors. Mol. Cell. Biol. 30, 1028–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Coss D., Hand C. M., Yaphockun K. K., Ely H. A., and Mellon P. L. (2007) p38 mitogen-activated protein kinase is critical for synergistic induction of the FSHβ gene by gonadotropin-releasing hormone and activin through augmentation of c-Fos induction and Smad phosphorylation. Mol. Endocrinol. 21, 3071–3086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ely H. A., Mellon P. L., and Coss D. (2011) GnRH induces the c-Fos gene via phosphorylation of SRF by the calcium/calmodulin kinase II pathway. Mol. Endocrinol. 25, 669–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Fujii Y., Okada Y., Moore J. P. Jr, Dalkin A. C., and Winters S. J. (2002) Evidence that PACAP and GnRH down-regulate follicle-stimulating hormone-β mRNA levels by stimulating follistatin gene expression: effects on folliculostellate cells, gonadotrophs and LβT2 gonadotroph cells. Mol. Cell. Endocrinol. 192, 55–64 [DOI] [PubMed] [Google Scholar]
- 62. Haisenleder D. J., Burger L. L., Walsh H. E., Stevens J., Aylor K. W., Shupnik M. A., and Marshall J. C. (2008) Pulsatile gonadotropin-releasing hormone stimulation of gonadotropin subunit transcription in rat pituitaries: evidence for the involvement of Jun N-terminal kinase but not p38. Endocrinology 149, 139–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Haisenleder D. J., Dalkin A. C., Ortolano G. A., Marshall J. C., and Shupnik M. A. (1991) A pulsatile gonadotropin-releasing hormone stimulus is required to increase transcription of the gonadotropin subunit genes: evidence for differential regulation of transcription by pulse frequency in vivo. Endocrinology 128, 509–517 [DOI] [PubMed] [Google Scholar]
- 64. Kaiser U. B., Jakubowiak A., Steinberger A., and Chin W. W. (1997) Differential effects of gonadotropin-releasing hormone (GnRH) pulse frequency on gonadotropin subunit and GnRH receptor messenger ribonucleic acid levels in vitro. Endocrinology 138, 1224–1231 [DOI] [PubMed] [Google Scholar]
- 65. Kanasaki H., Mutiara S., Oride A., Purwana I. N., and Miyazaki K. (2009) Pulse frequency-dependent gonadotropin gene expression by adenylate cyclase-activating polypeptide 1 in perifused mouse pituitary gonadotroph LβT2 cells. Biol. Reprod. 81, 465–472 [DOI] [PubMed] [Google Scholar]
- 66. Kanasaki H., Purwana I. N., and Miyazaki K. (2013) Possible role of PACAP and its PAC1 receptor in the differential regulation of pituitary LHβ- and FSHβ-subunit gene expression by pulsatile GnRH stimulation. Biol. Reprod. 88, 35. [DOI] [PubMed] [Google Scholar]
- 67. Lim S., Pnueli L., Tan J. H., Naor Z., Rajagopal G., and Melamed P. (2009) Negative feedback governs gonadotrope frequency-decoding of gonadotropin releasing hormone pulse-frequency. PLoS One 4, e7244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Liu F., Austin D. A., Mellon P. L., Olefsky J. M., and Webster N. J. (2002) GnRH activates ERK1/2 leading to the induction of c-fos and LHβ protein expression in LβT2 cells. Mol. Endocrinol. 16, 419–434 [DOI] [PubMed] [Google Scholar]
- 69. Thompson I. R., Ciccone N. A., Xu S., Zaytseva S., Carroll R. S., and Kaiser U. B. (2013) GnRH pulse frequency-dependent stimulation of FSHβ transcription is mediated via activation of PKA and CREB. Mol. Endocrinol. 27, 606–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Thompson I. R., and Kaiser U. B. (2014) GnRH pulse frequency-dependent differential regulation of LH and FSH gene expression. Mol. Cell. Endocrinol. 385, 28–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wang Y., Ho C. C., Bang E., Rejon C. A., Libasci V., Pertchenko P., Hébert T. E., and Bernard D. J. (2014) Bone morphogenetic protein 2 stimulates noncanonical SMAD2/3 signaling via the BMP type 1A receptor in gonadotrope-like cells: implications for FSH synthesis. Endocrinology 155, 1970–1981 [DOI] [PubMed] [Google Scholar]
- 72. McNeilly A. S., Crawford J. L., Taragnat C., Nicol L., and McNeilly J. R. (2003) The differential secretion of FSH and LH: regulation through genes, feedback and packaging. Reprod. Suppl. 61, 463–476 [PubMed] [Google Scholar]
- 73. Mishiro-Sato E., Sasaki K., Matsuo T., Kageyama H., Yamaguchi H., Date Y., Matsubara M., Ishizu T., Yoshizawa-Kumagaye K., Satomi Y., Takao T., Shioda S., Nakazato M., and Minamino N. (2010) Distribution of neuroendocrine regulatory peptide-1 and -2, and proteolytic processing of their precursor VGF protein in the rat. J. Neurochem. 114, 1097–1106 [DOI] [PubMed] [Google Scholar]
- 74. D'Amato F., Cocco C., Noli B., Cabras T., Messana I., and Ferri G. L. (2012) VGF peptides upon osmotic stimuli: changes in neuroendocrine regulatory peptides 1 and 2 in the hypothalamic-pituitary-axis and plasma. J. Chem. Neuroanat. 44, 57–65 [DOI] [PubMed] [Google Scholar]
- 75. Mistry D. S., Tsutsumi R., Fernandez M., Sharma S., Cardenas S. A., Lawson M. A., and Webster N. J. (2011) Gonadotropin-releasing hormone pulse sensitivity of follicle-stimulating hormone-β gene is mediated by differential expression of positive regulatory activator protein 1 factors and corepressors SKIL and TGIF1. Mol. Endocrinol. 25, 1387–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Naidich M., Shterntal B., Furman R., Pawson A. J., Jabbour H. N., Morgan K., Millar R. P., Jia J., Tomic M., Stojilkovic S., Stern N., and Naor Z. (2010) Elucidation of mechanisms of the reciprocal cross talk between gonadotropin-releasing hormone and prostaglandin receptors. Endocrinology 151, 2700–2712 [DOI] [PubMed] [Google Scholar]
- 77. Naor Z., Jabbour H. N., Naidich M., Pawson A. J., Morgan K., Battersby S., Millar M. R., Brown P., and Millar R. P. (2007) Reciprocal cross talk between gonadotropin-releasing hormone (GnRH) and prostaglandin receptors regulates GnRH receptor expression and differential gonadotropin secretion. Mol. Endocrinol. 21, 524–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Petrella C., Broccardo M., Possenti R., Severini C., and Improta G. (2012) TLQP-21, a VGF-derived peptide, stimulates exocrine pancreatic secretion in the rat. Peptides 36, 133–136 [DOI] [PubMed] [Google Scholar]
- 79. Severini C., La Corte G., Improta G., Broccardo M., Agostini S., Petrella C., Sibilia V., Pagani F., Guidobono F., Bulgarelli I., Ferri G. L., Brancia C., Rinaldi A. M., Levi A., and Possenti R. (2009) In vitro and in vivo pharmacological role of TLQP-21, a VGF-derived peptide, in the regulation of rat gastric motor functions. Br. J. Pharmacol. 157, 984–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Rindi G., Licini L., Necchi V., Bottarelli L., Campanini N., Azzoni C., Favret M., Giordano G., D'Amato F., Brancia C., Solcia E., and Ferri G. L. (2007) Peptide products of the neurotrophin-inducible gene vgf are produced in human neuroendocrine cells from early development and increase in hyperplasia and neoplasia. J. Clin. Endocrinol. Metab. 92, 2811–2815 [DOI] [PubMed] [Google Scholar]
- 81. Pincas H., Choi S. G., Wang Q., Jia J., Turgeon J. L., and Sealfon S. C. (2014) Outside the box signaling: secreted factors modulate GnRH receptor-mediated gonadotropin regulation. Mol. Cell. Endocrinol. 385, 56–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Yamaguchi H., Sasaki K., Satomi Y., Shimbara T., Kageyama H., Mondal M. S., Toshinai K., Date Y., González L. J., Shioda S., Takao T., Nakazato M., and Minamino N. (2007) Peptidomic identification and biological validation of neuroendocrine regulatory peptide-1 and -2. J. Biol. Chem. 282, 26354–26360 [DOI] [PubMed] [Google Scholar]
- 83. Shilov I. V., Seymour S. L., Patel A. A., Loboda A., Tang W. H., Keating S. P., Hunter C. L., Nuwaysir L. M., and Schaeffer D. A. (2007) The Paragon Algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol. Cell. Proteomics 6, 1638–1655 [DOI] [PubMed] [Google Scholar]
- 84. Smyth G. K. (2004) Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3, Article3. [DOI] [PubMed] [Google Scholar]
- 85. Vizcaíno J. A., Csordas A., del-Toro N., Dianes J. A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T., Xu Q. W., Wang R., and Hermjakob H. (2016) 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 44, D447–D456 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.