Abstract
Sponges have generated significant interest as a source of bioactive and elaborate secondary metabolites that hold promise for the development of novel therapeutics for the control of an array of human diseases. However, research and development of marine natural products can often be hampered by the difficulty associated with obtaining a stable and sustainable production source. Herein we report the first successful characterization and utilization of the microbiome of a marine invertebrate to identify a sustainable production source for an important natural product scaffold. Through molecular-microbial community analysis, optimization of fermentation conditions and MALDI-MS imaging, we provide the first report of a sponge-associated bacterium (Micromonospora sp.) that produces the manzamine class of antimalarials from the Indo-Pacific sponge Acanthostrongylophora ingens (Thiele, 1899) (Class Demospongiae, Order Haplosclerida, Family Petrosiidae). These findings suggest that a general strategy of analysis of the macroorganism’s microbiome could significantly transform the field of natural products drug discovery by gaining access to not only novel drug leads, but the potential for sustainable production sources and biosynthetic genes at the same time.
Keywords: marine natural product, microbiome, manzamine, natural product drug discovery, MALDI MS imaging
INTRODUCTION
Over 60% of approved pharmaceuticals are either natural products, derived from natural products via semi-synthesis and synthesis, or modeled after natural product prototypes (Cragg et al., 1997; Cragg and Newman, 2013). Marine invertebrates have introduced a wealth of complex secondary metabolites as potential drug leads; however, the sustainable supply of these marine invertebrate metabolites for development and clinical use remains a challenge. With the development of metagenomics techniques it is possible to assess large amounts of genetic information directly from environmental samples, and natural products drug discovery has moved toward a new frontier in discovery and development. Mining the genetic information associated with the microbiome of macroorganisms has provided unique opportunities to interrogate the role of the host’s microbial community in the production of secondary metabolites (Owen et al., 2013).
Although the oceans represent the largest biosphere on Earth and contain extraordinary biological and chemical diversity for the discovery of drug leads, the majority of the potential drug-producing groups of marine organisms come from extremely delicate ecosystems that are vulnerable to over-harvesting, pollution and climate change (Hughes et al., 2005; Ibrahim et al., 2013). Presented here is a minimally invasive technique for sustainable natural products exploration involving a phylogenetic analysis of the entire microbiome followed by culture approaches to interrogate the most likely drug-producing organisms. The approach presented here illustrates the use of exploration of the phylogeny of the microbiome leading to the rational selection of culture approaches including a focus on biomimetic approaches to yield the biosynthesis and production of the active metabolites from a host associated microbe. This approach offers opportunities to address both the discovery of unique metabolites from silent genes as well as the possibility of a sustainable supply from rare and fragile natural product producing groups and in both cases is minimally invasive.
Marine invertebrates, in particular, are composed of diverse microbiomes that are challenging to characterize and culture because of their unique relationships developed between the host and the environment (Taylor, 2007; Waters et al., 2010). Most of these invertebrates have co-evolved with microbial assemblages that have become highly specialized and, in many cases, obligate hence the need for community analysis and biomimetics to achieve successful microbial cultivation and metabolite production (Ruby et al., 2004). The isolation of the same groups of natural products from both macroorganisms and microorganisms has led to the hypothesis that the true producers are the microorganisms associated with the invertebrates; however, it has been difficult to identify a producing microorganism that is truly responsible for the biosynthesis of these complex secondary metabolites found in the invertebrate (Piel, 2004). Sponges have been equated to natural microbial fermenters of the ocean and represent a unique opportunity for mining sustainable microbial sources for marine natural products (Hentschel et al., 2006). Exploring these unique environmental conditions gives us yet another piece of the puzzle to develop efficient biomimetic conditions for the cultivation of previously unculturable microbes or the activation of silent genes in established cultures. The manza-mine class of alkaloids is a prime example regarding the useful drug potential of unique marine structural classes as well as the challenges that arise in supply, optimization and preclinical development of complex marine molecules.
The manzamine class of alkaloids offer some remarkable opportunities based on both the chemistry and therapeutic potential for this group of metabolites. Manzamine A was originally described as a possible cancer lead, but has shown significant activity against the malaria parasite Plasmodium falciparum including a 2–7 fold greater activity compared to artemisinin in vitro and a single dose clearance of the parasite from the host (Sakai et al., 1986; Peng, 2010). Mycobacterium tuberculosis and serine/threonine kinases, such as GSK3-β and CDK5, are also inhibited by manzamine A or derivatives (Kasanah and Hamann, 2004; Peng, 2010). Treatment with manzamine A impedes metastasis in cancer cell lines and sensitizes them to TNF-related apoptosis-inducing ligand (TRAIL)-induced apoptosis (Guzmán, 2011). Wright et al. filed a patent for manzamine A showing that when added to a cancer treatment regimen cancer cells became susceptible to other treatments, but more importantly cell invasion and tumor metastasis were also inhibited by the inclusion of manzamine A (Guzmán et al., 2014). Manzamine A has also shown promise as a suppressor of hyper-lipidemia and atherosclerosis in vivo (Eguchi et al., 2013). The manzamine family now consists of more than 80 β-carboline-containing manzamine and manzamine-related alkaloids isolated across nine genera of marine sponges from distinct geographical areas (Radwan et al., 2012). This broad distribution across distantly related sponges suggested the putative role of a microorganism for actual biosynthesis of the manzamine alkaloids (Faulkner, 2000; Hu et al., 2003) (Supplementary Figure 1 and Tables 1, 2).
Although the manzamine alkaloids possess potent bioactivity, their medicinal potential is limited in part by a lack of constant supply of manzamine as well as key precursors for medicinal chemistry, optimization and preclinical development (Rao et al., 2004). While there are several common sponges that produce large yields of manzamine alkaloids, harvesting of wild populations of marine invertebrates is neither practical, nor environmentally sustainable. The structural complexity of manzamine and lack of a cost-effective synthesis is currently challenging for both medicinal chemistry studies and drug production of a potential antimalarial agent (Jakubec et al., 2012). Community analysis and biomimetic fermentation approaches hold significant promise for a solution to the supply challenges and the availability of a sustainable source for the chemically complex precursors that are critical to future medicinal chemistry studies.
MATERIALS AND METHODS
SAMPLE COLLECTION
Several different specimens of A. ingens were collected by SCUBA diving in Manado Bay, Indonesia at depths between 6 and 33 m. Sponges were transferred directly to plastic bags containing seawater. Sponge tissue was stored frozen at −80°C until used. The sponge tissue was lyophilized prior to molecular manipulation. Water samples were collected in sterile 20-liter containers adjacent to the sponges sampled. Water samples (10 L) were filtered through 0.22 μm-pore-size Sterivex filters (Millipore). Nucleic acids were extracted from bacteria retained in the filters as described by Somerville et al. (1989).
TOTAL DNA EXTRACTION FROM A. INGENS
DNA was extracted from lyophilized sponge tissue using a bead-beater method adapted from Pitcher et al. (1989) Dried tissue (4 g) was ground with a mortar and pestle and resuspended in 16 mL of TE buffer and 4 mL of isoamyl alcohol. The solution was transferred to a 50 mL bead-beater chamber (Biospec Products, Inc.). Zirconia/silica beads (0.1 and 1 mm) were added to 1/4 volume of the chamber and homogenized 4 times in 1 min via the bead-beater. The solution was transferred to a 50 mL tube, 10 mL of guanidium thiocyanate buffer was added, mixed gently, and transferred to ice. Ammonium acetate (10 M) was added to a 2.5 M final concentration. Standard phenol/chloroform extraction and chloroform/isoamyl alcohol extraction were followed. DNA was precipitated with cold isopropanol, cleaned with 70% (vol/vol) ethanol, and resuspended in TE buffer (pH 8). DNA was quantified using a UV/Vis spectrophotometer.
BACTERIAL CLONE LIBRARY OF A. INGENS
PCR was performed using 100 ng of DNA with universal 16S rRNA gene primers 8-27f (Weisburg et al., 1991) and 1492r (Reysenbach et al., 1992) using Hi-Fi Platinum® Taq (Invitrogen). Cycling conditions were as follow: initial denaturation at 94°C for 5 min, 20 cycles of 94°C for 30 s, 48°C for 2 min, 72°C for 1.5 min, and a final extension of 5 min at 72°C in a PTC-200 MJ-research thermal cycler (Bio-Rad). PCR products were purified by electrophoresis in a 1% (wt/vol) agarose gel and bands of approximately 1500 bp were excised and recovered using a gel extraction kit (Qiagen, Inc.). Purified PCR products were cloned with a TOPO®-XL cloning kit (Invitrogen) according to the manufacturer’s instructions. Sequence of clones (>500 bp) were manually aligned with Phydit software (Chun, 1995). The tree was generated by the neighbor-joining algorithm (Saitou and Nei, 1987) implemented in Phydit. The robustness of inferred tree topologies was evaluated after 1000 bootstrap resamplings of the neighbor-joining data, and only values >50% were shown. A similar approach was used to more deeply investigate the bacterial community closely related to Micromonospora strain M42. In this case, the PCR reaction was performed with a primer M42-16S (5′-GACCGTGAAAACCTGGGGC-3′) derived from the 16S rRNA gene sequence of Micromonospora sp. strain M42 together with reverse primer 1492r.
DENATURING GRADIENT GEL ELECTROPHORESIS (DGGE) AND FLUORESCENT IN SITU HYBRIDIZATION ANALYSIS (FISH)
DGGE analysis was performed to compare the bacterial communities associated with A. ingens to that in the surrounding seawater. The procedure followed was described previously (Enticknap et al., 2006). The numbers of bacteria associated with A. ingens that are closely related to Actinobacteria was determined by FISH analysis following the method described by Enticknap et al. (2006) using the probe HGC69a (Roller et al., 1994).
ISOLATION AND IDENTIFICATION OF SPONGE-ASSOCIATED BACTERIA
A sponge microbial extract was obtained by grinding 1 cm3 of sponge tissue in sterile artificial seawater using a mortar and pestle. The extract was plated in a serial dilution (100 to 10−4) on three different media types and incubated at 30°C for up to 3 weeks. Marine Agar 2216 (Difco), a non-selective medium, ISP2 (Difco) and starch casein agar, both improve the growth of Actinobacteria and were supplemented with a final concentration of nalidixic acid (10 μg/ml), cycloheximide (10 μg/ml), nystatin (25 μg/ml) and 2% (wt/vol) NaCl. One morphotype of each bacterial colony was selected and subcultured until pure. Isolates were identified by 16S rRNA gene sequence analysis. Bacterial isolates were grown at 30°C in 50 mL broth of the medium from which they were originally isolated. After 5 days of growth, DNA was extracted from cell pellets using the Ultra-Clean® Microbial DNA Isolation kit (Mo Bio Laboratories Inc.). The 16S rRNA gene fragments were amplified by PCR using Platinum® Taq DNA polymerase High Fidelity, primers 8-27f (Weisburg et al., 1991), 1492r (Reysenbach et al., 1992) and sequenced. Phylogenetic analysis of isolates sequence was carried out as described above.
MANZAMINE A IDENTIFICATION AND EXTRACTION FROM MICROMONOSPORA SP. M42 AND PRODUCTION OPTIMIZATION
All sponge microbes were grown in 500 mL of ISP2 media supplemented with 2% NaCl and grown at 28°C for 14 days with aeration. At the end of the fermentation time period, the cultures were extracted with ethyl acetate three times (v/v). Extracts were then screened using silica gel TLC (mobile phase CHCl3: MeOH 9:1) and detected with the alkaloid specific Draggendorf stain using the sponge-isolated manzamine A and 8 hydroxymanzamine A as standards. The Micromonospora sp. strain M42 was the only strain to show production and was consequently cultured again in shake flasks and a 20-liter fermenter in ISP2 liquid medium with vigorous aeration at 30°C. The media was partitioned three times with ethyl acetate, dried and subjected to silica gel vacuum-liquid chromatography and eluted beginning with hexane (100%), hexane–acetone (9:1, 3:1, 1:1), acetone (100%), chloroform–methanol (1:1) and finally with methanol (100%). Fraction 3, eluted with hexane–acetone (3:1), was subjected to HPLC chromatography (Phenomenex Luna 5 μM 250 × 10.0 mm column, flow rate 3 ml/min, λ 254, 360 nm) using a gradient solvent system of acetonitrile and water both with 0.1% TFA to obtain manzamine A. To optimize production several different media and growth conditions were explored including ISP2 media (4 g yeast extract, 10 g malt extract, 4 g dextrose/L) with 2% NaCl, ISP2 media with 2% artificial ocean, ISP2 with no salts added, YM media (3 g yeast extract, 3 g malt extract, 3 g dextrose, 5 g peptone/L) with 2% artificial ocean, M4 media (10 g glucose, 2 g casamino acid, 2 g yeast extract, 2 g beef extract/L) with 2% artificial ocean, ½ dilution of YM media, marine broth 2216 (Difco), and marine broth 2216 (Difco) supplemented with 2 g glucose/L.
PRECURSOR DIRECTED BIOSYNTHETIC EXPERIMENTS
Commercially available tryptamine and sponge isolated ircinal A were fed to M42. A 1 L culture of M42 in ISP2 media supplemented with 2% (wt/vol) NaCl was shaken at 28°C and 150 rpm for two days followed by addition of tryptamine and ircinal A (each 25 mg) and then continued incubation for an additional three days. Cells and supernatant were separated by centrifugation. The supernatant was extracted with chloroform and evaporated under vacuum. The crude extract was passed though SPE C-8 column before analysis using HPLC, LC-TOF and NMR. These experiments were repeated four times yielding an average 5-fold increase in manzamine A production. The same experiments were repeated to obtain 15N-labeled manzamine A by substituting 15N-labeled tryptamine.
LC-TOF- MS AND NMR ANALYSIS
LC-MS analysis was carried out on a Bruker microTOF with electrospray ionization. Solutions of standard manzamine A from the sponge and manzamine A and B obtained from a microbial source were prepared in methanol and subjected to LCMS using a reverse-phase C8 column (Phenomenex, 5μ, 4.6 × 150 mm) eluting at 0.4 mL/min flow rate with 20 min linear gradient from 20 to 100% phase B. Phase A was water and phase B was acetonitrile. The NMR of both the sponge and bacteria associated manzamine A were obtained on a Bruker 400 MHz NMR in CDCl3.
EA-IRMS FOR 15N-LABELED MANZAMINE A
Samples and references were weighed into tin capsules, sealed, and loaded into an auto-sampler on a Europa Scientific elemental analyzer. They are dropped in sequence into a furnace held at 1000°C and combusted in the presence of oxygen. The tin capsules flash combust, raising the temperature in the region of the sample to ∼1700°C. The combusted gases are swept in a helium stream over combustion catalyst (Cr2O3), copper oxide wires (to oxidize hydrocarbons), and silver wool to remove sulfur and halides. The resultant gases, N2, NOx, H2O, O2, and CO2 are swept through a reduction stage of pure copper wires held at 600°C. This removes any oxygen and converts NOx species to N2. A magnesium perchlorate chemical trap is used to remove water. Nitrogen and carbon dioxide are separated using a packed column gas chromatograph held at a constant temperature of 65°C. The resultant nitrogen peak enters the ion source of the Europa Scientific 20–20 IRMS first, where it is ionized and accelerated. Nitrogen gas species of different mass are separated in a magnetic field then simultaneously measured using a Faraday cup collector array to measure the isotopomers of N2 at m/z 28, 29, and 30. After a delay the carbon dioxide peak enters the ion source and is ionized and accelerated. Carbon dioxide gas species of different mass are separated in a magnetic field then simultaneously measured using a Faraday cup collector array to measure the isotopomers of N2 at m/z 44, 45, and 46. Both references and samples are converted to N2 and analyzed using this method. The analysis proceeds in a batch process by which a reference is analyzed followed by a number of samples and then another reference.
MALDI-MS IMAGING EXPERIMENTS AND HRMS
To prepare colonies for MALDI-MS imaging, 9–15 mL thick ISP2 plates with 2% artificial ocean were poured and allowed to set overnight. 5 μL of Micromonospora sp. M42 was plated in 6 distinct places on the plate. The bacterium was allowed to grow at 30°C in an incubator for 0–10 days. The plates were removed from the incubator, individual colonies and appropriate controls were cut from the agar and placed on a Bruker™ MTP 384 ground steel plate using a spatula. This was done slow enough to avoid the trapping of air bubbles underneath the media. Then sinapic acid (Sigma Aldrich) was sprinkled on top of the culture using a 20 μm sieve (Hogentogler & Co.) Then the colonies were dried in a vacuum oven (VWR 1410) for 8 h at 38°C. Then the sample plate was inserted into Bruker™ Autoflex II mass spectrometer equipped with Compass 1.1 software suite (consists of FlexImaging 3.0, FlexControl 2.4, and FlexAnalysis 2.4). The samples were analyzed in linear positive mode, with 200 μm intervals. The spectra were collected in 360–1200 m/z mass range at 200 Hz laser frequency. After data acquisition, the data were analyzed using the FlexImaging software (Yang et al., 2009; Roland et al., 2011; Xu et al., 2012).
The MALDI-MS imaging used for this analysis only provides a low resolution mass image thus six colonies were grown for each day tested above. One colony was used for MALDI-MS imaging, and the remaining five colonies for each day through the entire time course. The colonies were combined and extracted with CHCl3 three times with 30 min of sonication with each extraction. The extract was then eluted through a silica gel flash column with 100% hexane, 90% hexane:10% acetone, 75% hexane:25% acetone and 100% acetone. The 75% hexane fraction which is the correct polarity for manzamine A based on previous sponge extractions was then subjected to high-resolution MS using a Bruker microTOF ESI.
RESULTS
ISOLATION AND IDENTIFICATION OF SPONGE-ASSOCIATED BACTERIA PRODUCING MANZAMINE A
Herein we report the isolation of manzamine A from a Micromonospora sp. M42 isolated from the Indonesian sponge A. ingens (Figures 1A,B) using molecular-microbial community analysis, targeted microbiology, and Matrix Assisted Laser Desorption Ionization-Mass Spectrometry (MALDI-MS) imaging experiments to identify and validate it as a producer of the manzamine alkaloids. We began by investigating the entire sponge-associated bacterial community using a 16S rRNA gene sequence analysis of the sponge-associated bacterial community on a library of 117 clones. This yielded three dominating groups, deltaproteobacteria (30%, n = 36), chloroflexi (19%, n = 23) and actinobacteria (16%, n = 19), the remainder of the community includes acidobacteria, gammaproteobacteria, alphaproteobacteria, spirochaetes, and cytophaga-flexibacter-bacteroides (CFB) related bacteria (Figure 2). As is commonly found in bacterial community analysis of sponge-associated bacteria, in many cases the closest relatives of clones derived from A. ingens were uncultured bacteria and bacteria found in community analysis of other sponge samples.
FIGURE 1. Confirmation of manzamine A production by Micromonospora sp. M42.
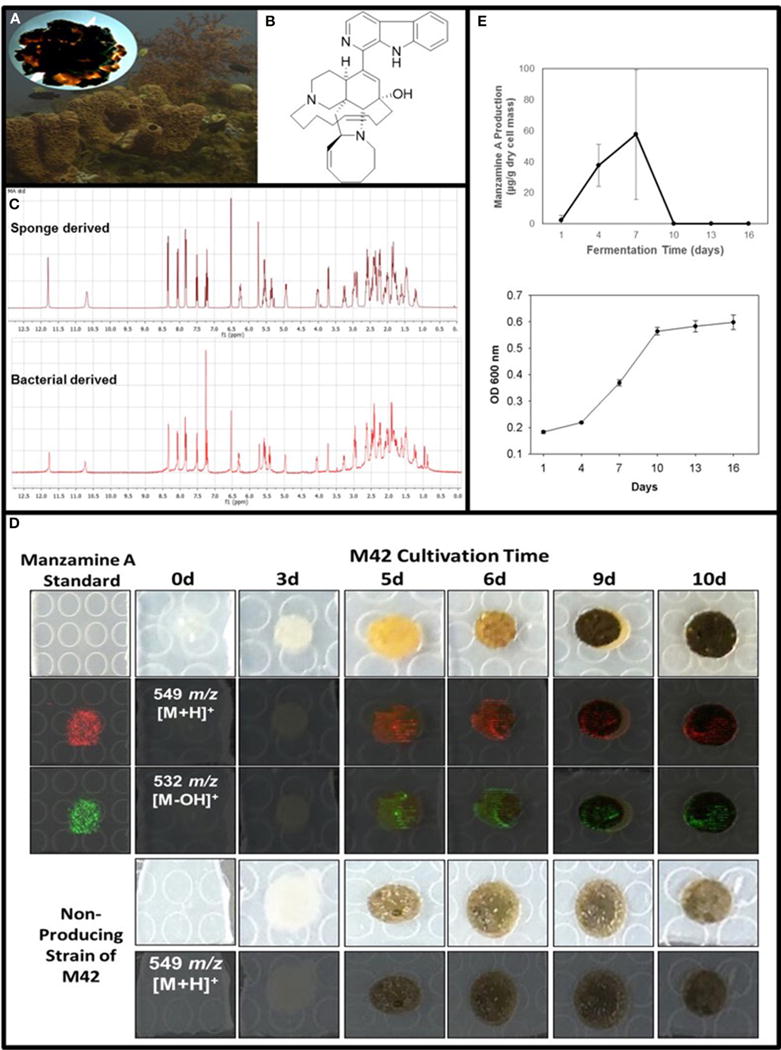
(A) Acanthostrongylophora ingens, a common Indo-Pacific manzamine producing sponge with the image of a colony of Micromonospora sp. strain M42 with orange mycelial growth and black spores. (B) Structure of manzamine A. (C) 1H NMR analysis comparison of sponge derived and bacterial derived manzamine A. (D) MALDI-MS imaging of manzamine A production on ISP2 over 10 days. 1st column: manzamine A standard control (10 μg on ISP2 agar); 1st row: Producing M42 colonies; 2nd row: MALDI-MS imaging for [M+H]+ (549 m/z); 3rd row: MALDI-MS imaging for the [M−OH]+ fragment (532 m/z); 4th row: non-producing M42 which sporulates faster; 5th row: MALDI-MS imaging for [M+H]+ (549 m/z) showing no detectable production of manzamine A. The extraction of replicate colonies yielded a HRESIMS signal of [M+H]+ = 549.3607 m/z (mean error: −2.86 ppm for manzamine A). (E) Production curve of manzamine A from liquid ISP2 culture based on LC-TOF-MS and growth curve of the bacteria based on OD600 measurements.
FIGURE 2.
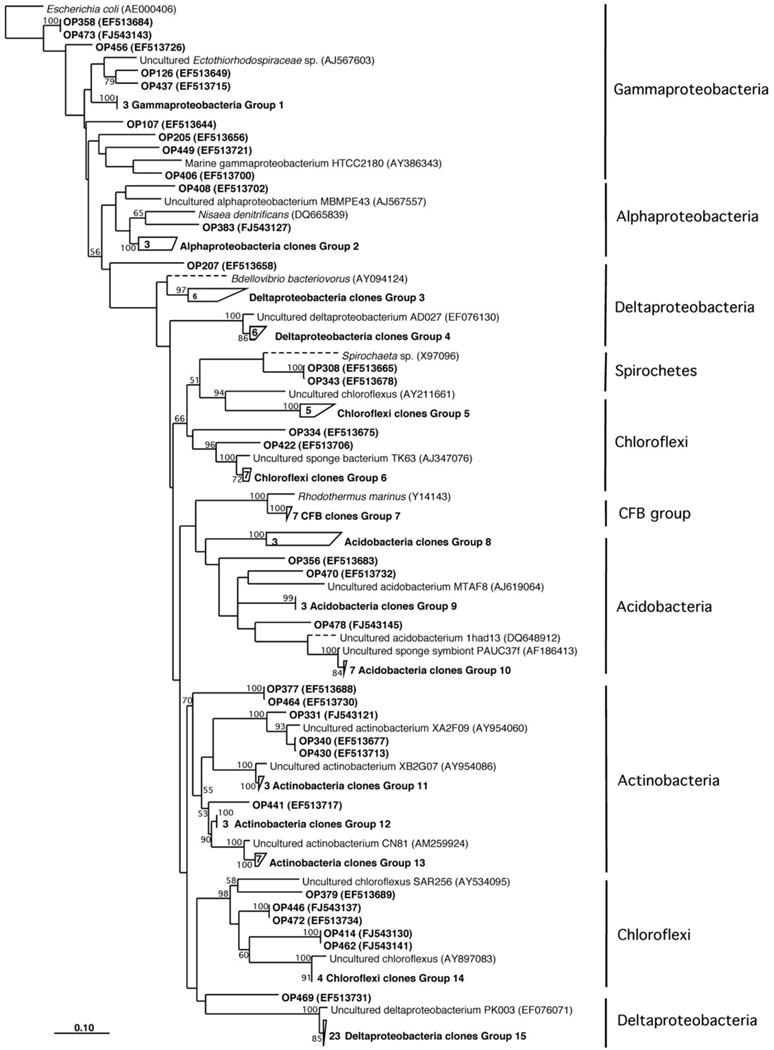
Neighbor-joining phylogenetic tree based on 16S rRNA gene sequence analysis showing the bacterial diversity of Acanthostrongylophora sp.
DGGE comparison of the sponge bacterial community to the surrounding water column (Figure 3) showed it to be both distinct and more diverse. Isolation of the heterotrophic bacteria yielded eleven pure cultures: one gammaproteobacteria (M37), two alphaproteobacteria (M31, M36), five firmicutes (M28, M29, M39, M34, M40) and three actinomycetes (M41, M42, M62). Marine agar 2216 yielded eight isolates (M28, M29, M31, M34, M36, M37, M39, M40), starch casein one isolate (M41) and ISP2 two isolates (M42, M62). Each A. ingens isolate was tested for the presence of manzamines by TLC (Figure 4). LC-TOF-MS analysis quickly identified a single bacterial strain (M42) producing a compound with identical retention time and mass as manzamine A. This strain was identified as a Micromonospora sp., closely related to Micromonospora chalcea (X92594) through phylogenetic analysis, and thus named Micromonospora sp. M42 (Figure 5). In order to further investigate the presence of bacteria closely related to Micromonospora sp. M42 in the sponge, a nested PCR approach was used. Hundreds of colonies of which 96 were picked and 50 were sequenced. The phylogenetic analysis revealed that this clone library comprised a majority of Micromonosporaceae (60%) and also contained gamma-, delta-proteobacteria and Acidobacteria.
FIGURE 3. Confirmation of unique bacterial presence in the sponge tissue.
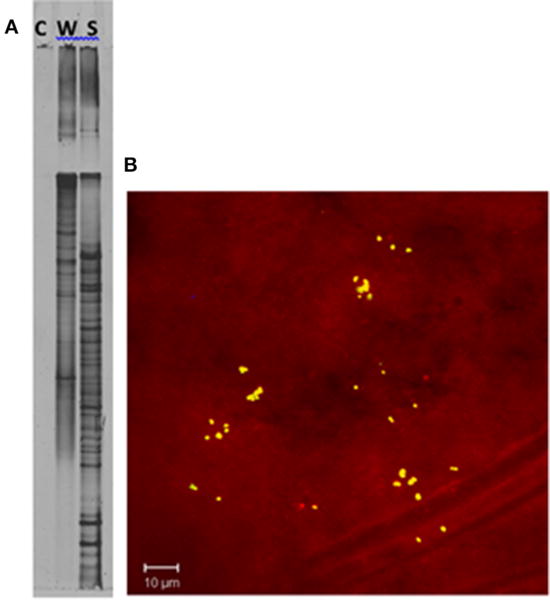
(A) DGGE analysis of a PCR negative control (C), seawater (W), and Acanthostrongylophora sp. (S). The increase in bands in the sponge sample indicates a higher diversity in the sponge than in the surrounding water column. (B) Epifluorescence micrograph section of Acanthostrongylophora sp. mesohyl tissue visualized by FISH. The mesohyl region was hybridized with Cy-3 labeled HGC69a probe. Numerous high GC bacteria (possibly actinobacteria) were undetectable in the water column, but clearly visible in the sponge. Visualization of actinobacteria within A. ingens tissue by FISH using a high-GC probe specific for actinobacteria (Montalvo, 2005) revealed distribution throughout the sponge. Clusters of 20–50 high-GC bacteria were randomly distributed throughout the mesohyl.
FIGURE 4.
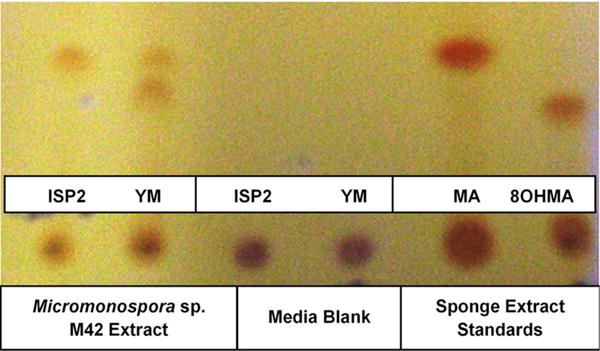
TLC of alkaloids produced by M42 in either ISP2 or YM compared to media and manzamine A and 8 hydroxymanzamine A standards obtained from the sponge.
FIGURE 5. Phylogenetic analysis based on 16S rRNA gene sequence of cultured bacteria isolated from sponge Acanthostrongylophora sp. noted in bold.
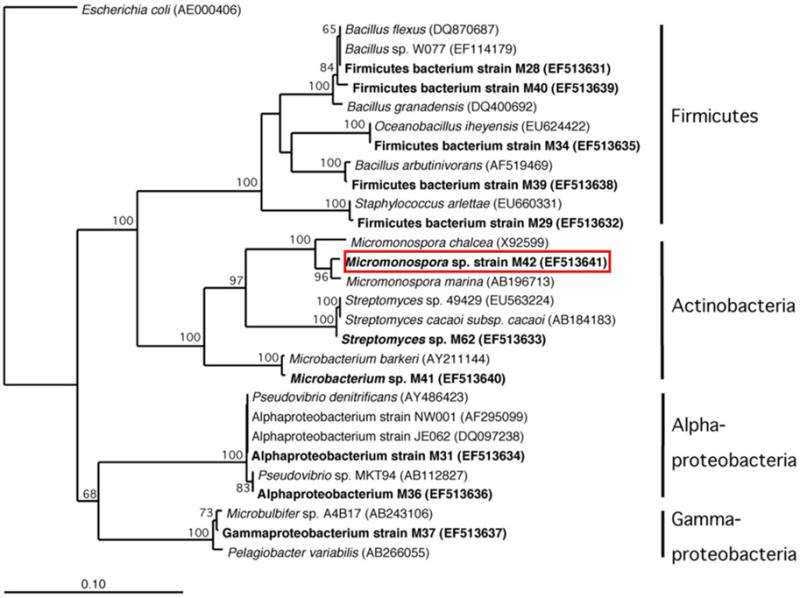
The red box indicates the location of the manzamine A producing strain.
OPTIMIZATION OF CULTURE METHODS FOR MICROMONOSPORA SP. M42 TO PRODUCE MANZAMINE A
After exhaustive fermentation and isolation studies of M42, it yielded a pure compound with the correlating mass and 1H NMR of the sponge derived manzamine A (Figure 1C). To optimize production several different media and growth conditions were explored including ISP2 media with NaCl, ISP2 media with artificial ocean, ISP2 with no salts added, YM media with artificial ocean, M4 media with artificial ocean, ½ dilution of YM media, marine broth 2216, and marine broth 2216 supplemented with glucose. Other growing conditions such as submerged fermentation, solid state fermentation, fermentation with various resins (XAD2, XAD7, XAD 1180 and HP20) and fermentation with extracted sponge fiber were also explored to optimize yield. The hypothesis behind the use of the different resins and extracted sponge fiber was that they would simulate the action of the sponge by sequestering manzamine A from the culture thus eliminating any feedback mechanism to limit production. The highest yield achieved was several mg/L in ISP2 liquid media grown for 7 days at 28°C and 150 rpm and the whole culture extracted twice with chloroform via liquid-liquid partition. However, the most reproducible production yield of manzamine A was closer to and frequently less than 1 mg/L utilizing the same conditions.
CONFIRMATION OF MANZAMINE A PRODUCTION
To further verify the production of manzamine A, MALDI-MS imaging was performed (Figure 1D). After many experiments, sinapic acid was the matrix that gave the best ionization with the least interference (Figure 6). Manzamine A production over 16 days in liquid culture revealed that manzamine A is initially produced in a linear fashion and then metabolized by the organisms resulting in undetectable levels of manzamine A after day 10 (Figure 1E). The MALDI-MS imaging results qualitatively show the same production pattern over a 10 day time course. Production of manzamine A is not detectable until day 4, peaks around day 6 and then is excreted and eventually metabolized by day 10. The two main signals seen in the manzamine A standard are 549 and 532 m/z. These same masses are observed being produced in the colonies in overlapping areas starting at day 5. The MALDI-MS imaging used for this analysis is only low resolution mass, thus additional colonies were cultured, extracted, purified and subjected to HRMS analysis. This yielded a HRMS for the microbial extract with a signal at 549.3607 m/z which corresponds to the [M+H]+ of manzamine A (mean error: −2.86 ppm). The standard sponge isolated manzamine A was measured after this microbial product and showed a signal at 549.3638 m/z which corresponds to the [M+H]+ of manzamine A (mean error: −8.06 ppm) (Figure 7). Thus, providing further evidence that the low resolution signal seen in the MALDI-MS imaging is manzamine A.
FIGURE 6. Mass spectra of manzamine A standard vs. sinapic acid matrix on MALDI.
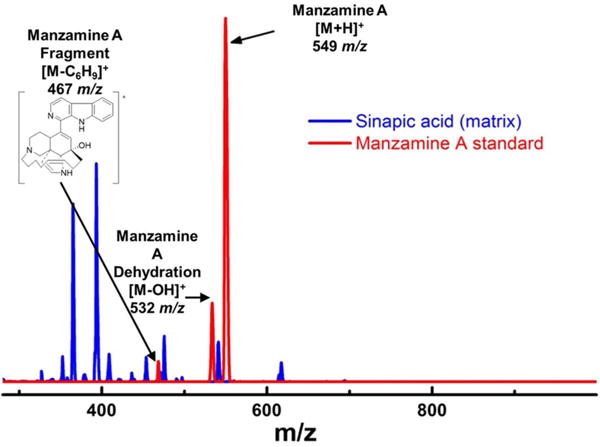
There were challenges in choosing the appropriate matrix for MALDI MS imaging. After attempting several different MALDI matrixes, sinapic acid was chosen due to its ability to ionize manzamine A well, but without any interfering peaks. The fragment at 467 m/z was not used for imaging due to its low intensity and overlap with the matrix peaks.
FIGURE 7.
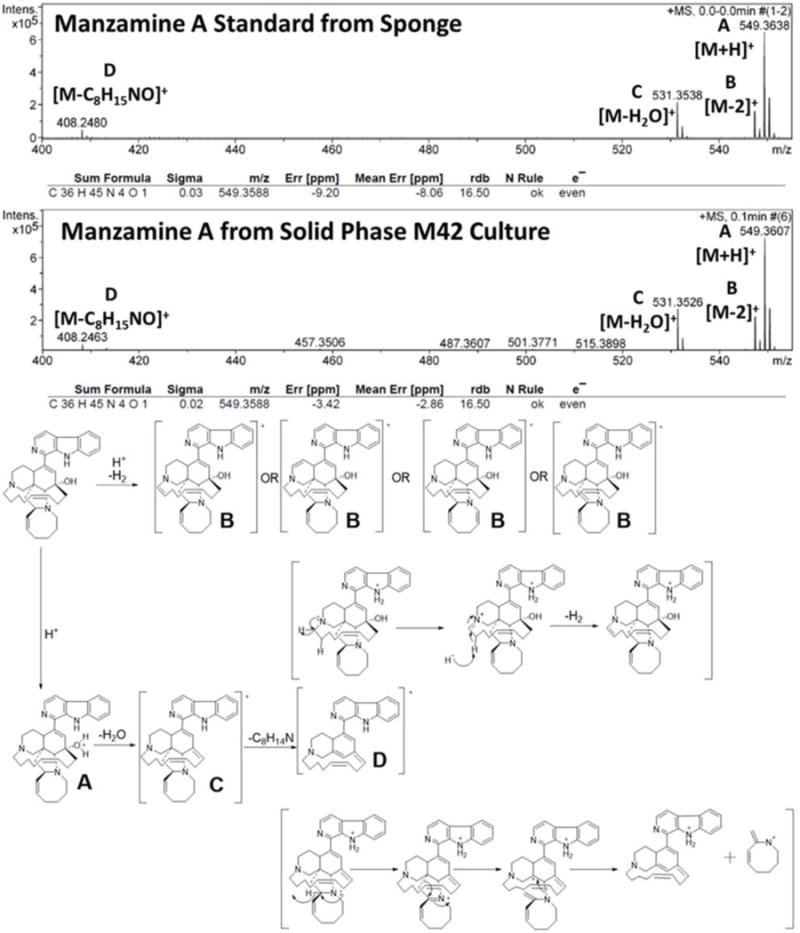
HRESIMS from extracts of solid phase growth of M42 on ISP2.
Further support of the biogenesis of manzamine A by M42 was obtained through preliminary biosynthetic studies. Tryptamine and ircinal A were added to M42 cultures in ISP2 media and this resulted in an approximate 5-fold increase in manzamine A production demonstrating the potential ability of M42 to convert precursors to manzamine A (Figure 8). This same increase could not be seen when tryptophan replaced the tryptamine in culture. As a control experiment, perillaldehyde and tryptamine were added to the culture to see if the Pictet-Spengler reaction would occur with a similar aldehyde. This experiment yielded none of the expected products.
FIGURE 8. Feeding ircinal and tryptamine results in an increase in manzamine A production.
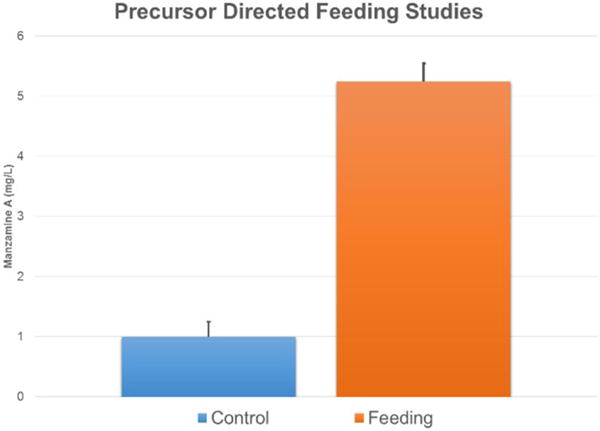
The error bars represent the range of the positive and negative standard deviation.
Several attempts have been made to insert a labeled atom into the production media of M42 in order to obtain a labeled manzamine A molecule. All attempts at producing manzamine A from the bacteria in minimal media failed. Manzamine A could only be obtained under rich media conditions and any dilution of these media results in drastic reduction or elimination of manzamine A production. Therefore, attempts were made to insert a label into the rich media utilizing [U-13C] glucose. While studies showed that the bacteria produced manzamine A in the presence of [U-13C] glucose, no incorporation could be detected by MS or NMR due to the low incorporation of labeled isotope. In addition to the labeled glucose several forms of labeled acetate were also tested (CD3COONa, CH3COO15NH4, and ) and gave similar production of manzamine A, but not detectable incorporation by MS or NMR. These results are not unexpected due to the richness of the media and other starting materials being available for the biosynthesis of manzamine A. Finally, 15N labeled tryptamine was added to the culture in a similar fashion to the precursor directed feeding experiments outlined above. This yielded successful incorporation of 15N based on Elemental Analysis Isotope Ratio Mass Spectrometry (EA-IRMS). Table 1 outlines the part per thousand and standard deviations for M42 produced manzamine A, sponge isolated manzamine A, and 15N enriched manzamine A obtained from the above conditions. The data shows an increase in the presence of 15N compared to the other unenriched samples. The 2.5 times incorporation of 15N was enough to be seen via EA-IRMS, but not less sensitive techniques such as MS or NMR.
Table 1.
Analysis of 15N Incorporation by EA-IRMS of 15N-labeled tryptamine by M42 to produce 15N labeled manzamine A (MA).
| Sample | Mean d13CV-PDB (‰) | SD (‰) | Mean d15NAir (‰) | SD (‰) | Mean 15N/14N | SD |
|---|---|---|---|---|---|---|
| M42 MA | −22.20 | 0.10 | 4.21 | 0.22 | 0.003692 | 0.000001 |
| 15N M42 MA | −21.99 | 0.02 | 1424 | 125 | 0.0089 | 0.0005 |
| Sponge MA | −22.54 | 0.05 | 4.26 | 0.16 | 0.003692 | 0.000001 |
DISCUSSION
Through molecular-microbial community analysis, optimization of fermentation conditions and MALDI-MS imaging, the first report of a sponge-associated bacterium (Micromonospora sp. M42) that produces the manzamine class of antimalarials from the Indo-Pacific sponge A. ingens is presented. The 16S rRNA gene sequence analysis of the sponge-associated bacterial community yielded deltaproteobacteria, chloroflexi, actinobacteria, acidobacteria, gammaproteobacteria, alphaproteobacteria, spirochaetes and CFB related bacteria (Figure 2). As seen in previously reported sponge community analysis (Webster et al., 2001), actinobacteria was one of the major components of the bacterial community. Bacterial isolation efforts yielded ten pure cultures only one of which, Micromonospora sp. M42, was shown to produce manzamine A.
MALDI-MS imaging was chosen to confirm manzamine A production because it gives the widest available mass range, as well as having the best established techniques and protocols for experimental development and design. The main benefit of this analytical method is the ability to perform a time course experiment and see the production of the metabolite in question being produced and consumed in its native environment or under culture conditions. Since the first report of MALDI-MS imaging on microorganisms was published in 2008, the technique has grown in popularity and shown its diverse utility in discovery of new natural products; characterization of the ecological role via spatial localization experiments within colonies; exploration of biosyn-thetic origins of natural products and metabolic profiles related to phenotypic studies (Esquenazi et al., 2008, 2011; Simmons et al., 2008; Taniguchi et al., 2009; Yang et al., 2009; Kersten et al., 2011; Liu et al., 2011; Xu et al., 2012; Jaeger et al., 2013; Shih et al., 2014). While MALDI-MS imaging has numerous benefits, it is important to be aware of the challenges. The first of which is the application of matrix to the sample in order for the ionization process to occur. This was a particularly challenging aspect for confirming manzamine A production. A matrix has to be identified that would allow manzamine A to ionize effectively, but not produce interfering signals. For this reason both ISP2 and YM media from above were analyzed with 4 different matrixes (2,5-dihydoxybenzoic acid, sinapic acid, 2,5-dihydroxyacetophenone, and α-cyano-4-hydroxycinnamic acid). As different experiments show, the production of manzamine A has a finite window for detection. Stability studies revealed that degradation of the metabolite is not caused by solvent; however, we cannot rule out external enzymatic degradation. Another limitation of MALDI-MS imaging is that is can only produce qualitative results. This is due to the inherently imprecise application of matrix leading to appreciable pixel-to-pixel variability (Watrous and Dorrestein, 2011; Yang et al., 2012). To minimize these effects on the overall qualitative analysis, all colonies (day 0–10) were run at the same time using the same MALDI plate leading to only one uniform application of matrix.
The microbial production appears to typically produce only manzamine A and under specific conditions small amounts of the 8-hydroxy manzamine A. Despite the fact that over 80 manzamine alkaloids are reported in the literature from sponges, it is apparent that all could be formed by plausible biotransformation mechanisms involving oxidations and rearrangement. Figure 9 provides an overview of the known and plausible biotransformation reactions leading to the manzamine alkaloids isolated from sponges beginning with just manzamine A (Kasanah et al., 2003).
FIGURE 9. Potential routes for A. ingens derived metabolites.
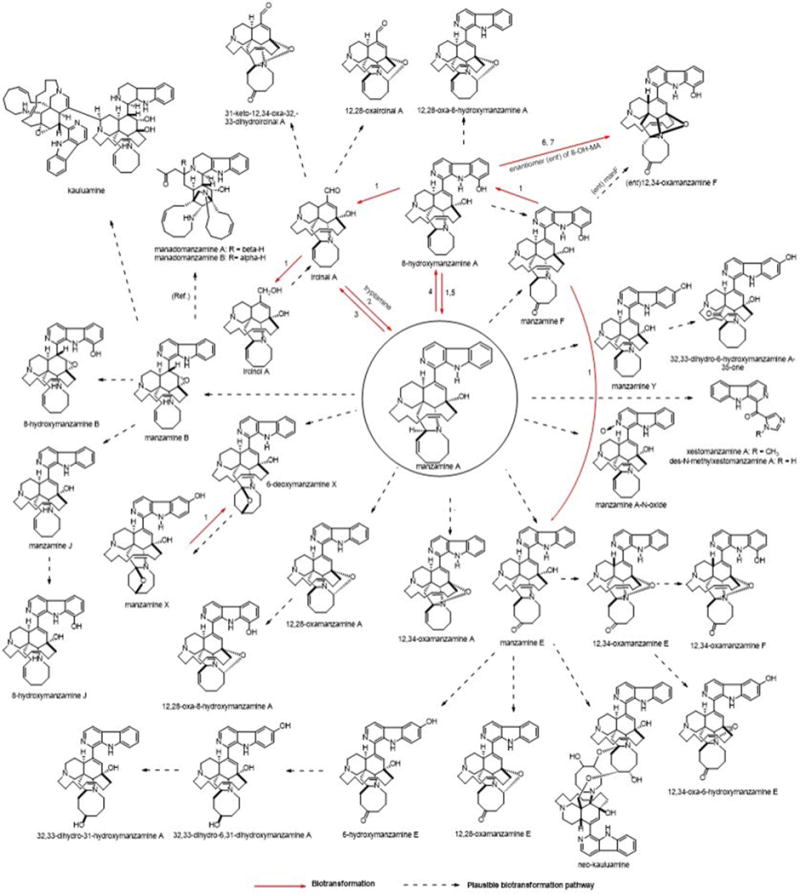
Biotransformation results using manzamine standards are displayed as well as plausible explanation for related metabolites. Microorganisms: (1) Fusarium solani F0007, (2) Micromonospora M42, (3) Fusarium oxysporium f.sp. gladioli ATCC 1113713, (4) Bacillus sp. VAN35 (M28), (5) Streptomyces seokies, (6) Fusarium oxysporium ATCC 760114, (7) Nocardia sp. ATCC 2114514 were capable of transforming MA into a related metabolite (Ref. = Peng, 2003, Plausible biosynthetic route for manadomanzamines in this reference).
As with all natural products produced from microbial sources, identifying and having the ability to manipulate the biosynthetic pathway would help to solve the production and stability issues plaguing the manzamine story. In the first biosynthetic scheme proposed for manzamine A (Baldwin et al., 1996), a pyridinium dimer undergoes a Diels-Alder reaction followed by hydrolysis to generate an aldehyde similar to ircinal A. This aldehyde can react with tryptamine through a Pictet-Spengler reaction, producing manzamine D in a similar biochemical mechanism as plant strictosidine synthase (Maresh et al., 2008). Further oxidation would yield the aromatic β-carboline ring system. The preliminary biosynthetic confirmation experiments presented here show that M42 is capable of increasing the production of manzamine A when proposed precursors (tryptamine and iricinal A) are added to the culture. Using substrates with similar active functional groups such as perillaldehyde instead of iricinal A did not yield any of the expected Pictet-Spengler products. This further supports that the production is specific to the substrates and will not work on just any aldehyde tryptamine combination.
Consistent and stable production of manzamine A by M42 has proven to be challenging. In subsequent fermentations over the decade since M42 was originally isolated, a strain of M42 was identified that does not produce detectable levels of manzamine. However, when returning to the original 2002 M42 strain for MALDI-MS imaging, production could again be detected. Figure 1D shows the non-detectable production of manzamine A in the later strains in contrast to the original glycerol stocks of M42. It is clear from this image that the non-producing strain sporolates at least 24 h before the original strain and never obtains the deep orange color characteristic of M42. Partial 16S rRNA gene sequences from both strains were amplified and sequenced using the primers 27F 5′-AGAGTTTGATCMTGGCTCAG-3′and 1492R 5′-TACGGYTACCTTGTTACGACTT-3′ (Lane, 1991). The sequence was deposited in Genbank under accession number EF513641. The 16S rRNA analysis of these strains showed both to be identical. Further genetic analysis in the differences of these strains is ongoing and may shed light on the biosynthesis of the manzamines. It is possible that the biosynthetic machinery is plasmid derived and can be lost or acquired through horizontal gene transfer. The wide distribution of manzamine alkaloids in different sponge species suggests that the machinery is able to be acquired by multiple microorganisms, most suitably in plasmid form. Other research suggests that secondary metabolite production can decrease and even become non-existent after multiple generations of subcultures as is the case with the camptothecins (Kusari et al., 2009).
The symbiotic relationship between the invertebrate and the bacteria has successfully evolved to yield g/L scale production in the sponge vs. the less than mg/L scale by the microbe. We have seen that manzamine A inhibits the growth of M42 at 12 μg/mL; much larger concentrations are found in the sponge, thus supporting the proposed sequestration of large amounts of manza-mine alkaloids in a manner that is not toxic to the bacterium (Figure 10). These data indicate that M42 could provide chemical defense for A. ingens in exchange for habitat or acting as a natural continuous-flow bioreactor. A reasonable explanation for the data presented is that manzamine A is produced at low levels by M42 within the sponge but accumulates over the lifetime of the organism to the concentrations seen in the field. A second possibility is that M42 is present at a high population within the sponge; however, this seems unlikely due to the lack of representation in the 16S rRNA gene clone library from the sponge and detailed FISH analysis. A third possibility is that M42 is not the only producer but rather the only bacterium that has been cultivated. To examine this hypothesis, a primer based on M42 16S rRNA gene was designed and coupled to a eubacterial universal primer 1492r. Using a nested PCR approach, a clone library of 16S rRNA genes enriched for those related to M42 were obtained. One clone, OPM11 was closely related to Micromonospora spp. No sequences matching exactly that of Micromonospora sp. strain M42 was recovered. The analysis revealed several sequences closely related to M42 that were not previously seen (Figure 11). The bacterial community associated with A. ingens could be elucidated in more detail by using next generation sequencing approaches and bacterial species of interest could be better quantified by additional FISH analyses in future work. Yet another possibility is the transfer of genes horizontally between unrelated bacteria, which would produce multiple producers that would avoid detection by homology based approaches. All of these are possible explanations for the discrepancy in total yields between the laboratory and the field.
FIGURE 10. Inhibition of M42 growth by manzamine A.
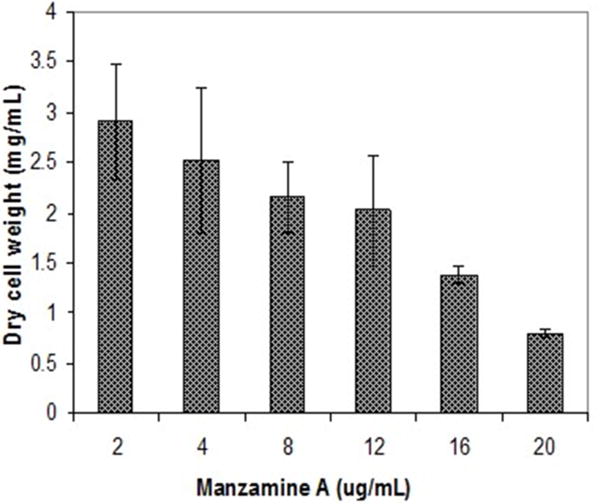
Increasing manzamine A concentration inhibits the growth of M42 in a dose dependent fashion. The error bars represent the range of the positive and negative standard deviation.
FIGURE 11. Increased amplification of actinobacteria.
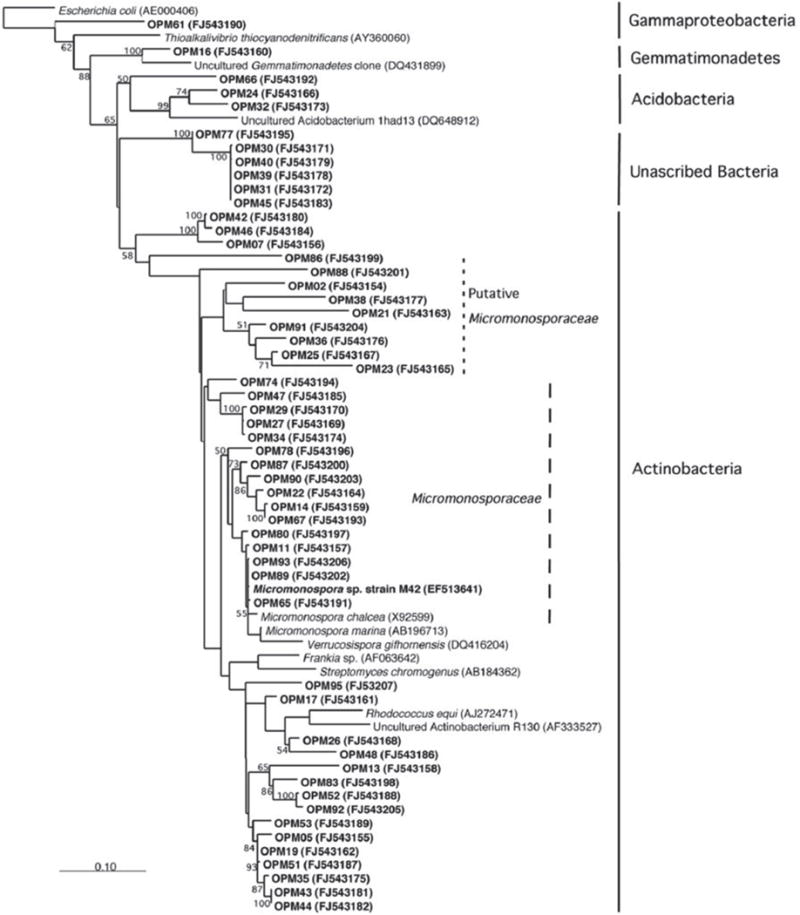
Using a universal primer as well as an actinobacteria primer resulted in a higher amplification of actinobacteria 16s RNA. This result is consistent with those published by Farris and Olson (2007).
With the use of community analysis studies, Micromonospora sp. M42 is the first sponge associated actinomycete bacteria that has been shown to produce a sponge derived metabolite (Hill et al., 2005). A culturable producing strain provides opportunities for overcoming future supply challenges that hinder marine natural products development into usable drugs; however, significant improvements in culture and production methods will need to be developed. Despite the successful production from a single pure culture, it has become quite evident that significant barriers in drug production and successful biomimetics for culture conditions still remain. The cumulative data presented confirms that M42 is capable of producing manzamine A, and although the yields are not yet sufficient to provide a practical solution for drug development, it provides an important starting point for chemical biology, precursor directed feeding experiments and improvements in gene expression to generate higher producing strains. This study also illustrates the unique potential for applying molecular phylogeny for the exploration of invertebrate microbiomes in the discovery and development of natural products.
Supplementary Material
Acknowledgments
For Amanda L. Waters, this investigation was supported in part by a National Science Foundation Graduate Research Fellowship under Grant No. 1144250. This investigation supported in part by NCCAM RO1AT007318, NIAID RO1AI36596 and the Medicines for Malaria Venture. Research was conducted in a facility constructed with support from Research Facilities Improvement Program Grant Number C06 RR-14503-01 from the National Center for Research Resources, National Institutes of Health. For James W. Sims, this material is based upon work supported by Grant Number 1F32AI083157-01A1 from the National Institute of Allergy and Infectious Diseases, National Institutes of Health. MK thanks Lori Bell, Patrick Colin, Rob van Soest, Jane Fromont, Francis Schmitz, and Brad Carté for discussions regarding the identity of several questionable specimens in the published literature, NIWA for on-going research support in marine biodiversity, and the Coral Reef Research Foundation, Palau, Micronesia, for permission to use their images and field data. Michelle Kelly also thanks Blayne Herr of TCMedia, Auckland, for production of the color figure. Systematics research was funded by NIWA under Coasts and Oceans Research Programme 2 Marine Biological Resources: Discovery and definition of the marine biota of New Zealand (2012/13 SCI). Appreciation to Iso-Analytical for running the EA-IRMS. We would also like to thank Profs William Fenical and William Gerwick for their thoughtful suggestions and reading this manuscript.
Footnotes
Edited by:
Donatella De Pascale, National Research Council- CNR, Italy
Reviewed by:
Aldo Nicosia, National Research Council- CNR, Italy Angelina Lo Giudice, University of Messina, Italy
AUTHOR CONTRIBUTIONS
Amanda L. Waters, Olivier Peraud, Noer Kasanah, James W. Sims, Samuel H. Abbas, Karumanchi V. Rao, Michelle Kelly, Amala Dass, Russell T. Hill and Mark T. Hamann designed experiments and wrote the manuscript. Olivier Peraud analyzed the microbial community. Noer Kasanah isolated and validated the presence of manzamine A in liquid culture. Amanda L. Waters prepared M42 for MALDI-MS imaging and Nuwan Kothalawala obtained the MALDI-MS imaging. James W. Sims and Samuel H. Abbas conducted the biosynthetic gene cluster analysis. Matthew A. Anderson conducted the feeding studies. Michelle Kelly provided taxonomic information on the sponge material.
SUPPLEMENTARY MATERIAL
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fmars.2014.00054/abstract
Conflict of Interest Statement: The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Baldwin JE, Claridge TDW, Culshaw AJ, Heupel FA, Smrcková S, Whitehead RC. A biomimetic approach to the manzamine alkaloids. Tetrahedron Lett. 1996;37:6919–6922. doi: 10.1016/0040-4039(96)01516-X. [DOI] [Google Scholar]
- Chun J. PhD thesis. Newcastle University; 1995. Computer-assisted Classification and Identification of Actinomycetes. Available online at: http://hdl.handle.net/10443/410. [Google Scholar]
- Cragg GM, Newman DJ. Natural products: a continuing source of novel drug leads. Biochim Biophys Acta. 2013;1830:3670–3695. doi: 10.1016/j.bbagen.2013.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cragg GM, Newman DJ, Snader KM. Natural products in drug discovery and development. J Nat Prod. 1997;60:52–60. doi: 10.1021/np9604893. [DOI] [PubMed] [Google Scholar]
- Eguchi K, Fujiwara Y, Hayashida A, Horlad H, Kato H, Rotinsulu H, et al. Manzamine A, a marine-derived alkaloid, inhibits accumulation of cholesterol ester in macrophages and suppresses hyperlipidemia and atherosclerosis in vivo. Bioorg Med Chem. 2013;21:3831–3838. doi: 10.1016/j.bmc.2013.04.025. [DOI] [PubMed] [Google Scholar]
- Enticknap JJ, Shanks MK, Peraud O, Hill RT. Characterization of a culturable alphaproteobacterial symbiont common to many marine sponges and evidence for vertical transmission through the germline. Appl Environ Microbiol. 2006;72:3724–3732. doi: 10.1128/AEM.72.5.3724-3732.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esquenazi E, Coates C, Simmons L, Gonzalez D, Gerwick WH, Dorrestein PC. Visualizing the spatial distribution of secondary metabolites produced by marine cyanobacteria and sponges via MALDI-TOF imaging. Mol Biosyst. 2008;4:562–570. doi: 10.1039/b720018h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esquenazi E, Jones AC, Byrum T, Dorrestein PC, Gerwick WH. Temporal dynamics of natural product biosynthesis in marine cyanobacteria. Proc Natl Acad Sci USA. 2011;108:5226–5231. doi: 10.1073/pnas.1012813108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farris MH, Olson JB. Detection of actinobacteria cultivated from environmental samples reveals bias in universal primers. Lett Appl Microbiol. 2007;45:376–381. doi: 10.1111/j.1472-765X.2007.02198.x. [DOI] [PubMed] [Google Scholar]
- Faulkner DJ. Marine pharmacology. Antonie Van Leeuwenhoek. 2000;77:135–145. doi: 10.1023/A:1002405815493. [DOI] [PubMed] [Google Scholar]
- Guzmán EA. A novel activity from an old compound: Manzamine A reduces the metastatic potential of AsPC-1 pancreatic cancer cells and sensitizes them to TRAIL-induced apoptosis. Invest New Drugs. 2011;29:777–785. doi: 10.1007/s10637-010-9422-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzmán E, Johnson J, Wright AE. Use of Manzamine Compounds in Anti-Cancer Therapeutic Regimens. Boca Raton, FL: Florida Atlantic University Board of Trustees; US8691801 B2 US Patent. 2014
- Hentschel U, Usher KM, Taylor MW. Marine sponges as microbial fermenters. FEMS Microbiol Ecol. 2006;55:167–177. doi: 10.1111/j.1574-6941.2005.00046.x. [DOI] [PubMed] [Google Scholar]
- Hill RT, Hamann MT, Peraud O, Kasanah N. Manzamine Producing Actinomycetes. College Park, MD: University, MS: University of Maryland Biotechnology Institute and University of Mississippi; US20050244938 A1 US Patent. 2005
- Hu JF, Hamann MT, Hill R, Kelly M. The manzamine alkaloids. Alkaloids chem Biol. 2003;60:207–285. doi: 10.1016/S0099-9598(03)60004-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes TP, Bellwood DR, Folke C, Steneck RS, Wilson J. New paradigms for supporting the resilience of marine ecosystems. Trends Ecol Evol. 2005;20:380–386. doi: 10.1016/j.tree.2005.03.022. [DOI] [PubMed] [Google Scholar]
- Ibrahim MA, Na M, Oh J, Schinazi RF, McBrayer TR, Whitaker T, et al. Significance of endangered and threatened plant natural products in the control of human disease. Proc Natl Acad Sci USA. 2013;110:16832–16837. doi: 10.1073/pnas.1311528110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger RJ, Lamshöft M, Gottfried S, Spiteller M, Spiteller P. HR-MALDI-MS imaging assisted screening of β-carboline alkaloids discovered from Mycena metata. J Nat Prod. 2013;76:127–134. doi: 10.1021/np300455a. [DOI] [PubMed] [Google Scholar]
- Jakubec P, Hawkins A, Felzmann W, Dixon DJ. Total synthesis of manzamine A and related alkaloids. J Am Chem Soc. 2012;134:17482–17485. doi: 10.1021/ja308826x. [DOI] [PubMed] [Google Scholar]
- Kasanah N, Hamann MT. Development of antibiotics and the future of marine microorganisms to stem the tide of antibiotic resistance. Curr Opin Investig Drugs. 2004;5:827–837. [PMC free article] [PubMed] [Google Scholar]
- Kasanah N, Rao KV, Yousaf M, Wedge DE, Hamann MT. The biocatalytic conversion of 8-hydroxymanzamine A to manzamine A. Tetrahedron Lett. 2003;44:1291–1293. doi: 10.1016/S0040-4039(02)02816-2. [DOI] [Google Scholar]
- Kersten RD, Yang YL, Xu Y, Cimermancic P, Nam SJ, Fenical W, et al. A mass spectrometry–guided genome mining approach for natural product peptidogenomics. Nat Chem Biol. 2011;7:794–802. doi: 10.1038/nchem-bio.684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusari S, Zühlke S, Spiteller M. An endophytic fungus from camptotheca acuminata that produces camptothecin and analogues. J Nat Prod. 2009;72:2–7. doi: 10.1021/np800455b. [DOI] [PubMed] [Google Scholar]
- Lane DJ. 16S/23S rRNA sequencing. In: Stachebrandt E, Goodfellow M, editors. Nucleic Acid Techniques in Bacterial Systematics. New York, NY: John Wiley and Sons; 1991. pp. 115–175. [Google Scholar]
- Liu WT, Kersten RD, Yang YL, Moore BS, Dorrestein PC. Imaging mass spectrometry and genome mining via short sequence tagging identified the anti-infective agent arylomycin in Streptomyces roseosporus. J Am Chem Soc. 2011;133:18010–18013. doi: 10.1021/ja2040877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maresh JJ, Giddings LA, Friedrich A, Loris EA, Panjikar S, Trout BL, et al. Strictosidine synthase: mechanism of a Pictet-Spengler catalyzing enzyme. J Am Chem Soc. 2008;130:710–723. doi: 10.1021/ja077190z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montalvo NF. Novel actinobacteria from marine sponges. Antonie Van Leeuwenhoek. 2005;87:29–36. doi: 10.1007/s10482-004-6536-x. [DOI] [PubMed] [Google Scholar]
- Owen JG, Reddy BV, Ternei MA, Charlop-Powers Z, Calle PY, Kim JH, et al. Mapping gene clusters within arrayed metagenomic libraries to expand the structural diversity of biomedically relevant natural products. Proc Natl Acad Sci USA. 2013;110:11797–11802. doi: 10.1073/pnas.1222159110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J. Manadomanzamines A and B: a novel alkaloid ring system with potent activity against mycobacteria and HIV-1. J Am Chem Soc. 2003;125:13382–13386. doi: 10.1021/ja030087z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J. Structure-activity relationship and mechanism of action studies of manzamine analogues for the control of neuroinflammation and cerebral infections. J Med Chem. 2010;53:61–76. doi: 10.1021/jm900672t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piel J. Metabolites from symbiotic bacteria. Nat Prod Rep. 2004;21:519–538. doi: 10.1039/b310175b. [DOI] [PubMed] [Google Scholar]
- Pitcher DG, Saunders NA, Owen RJ. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett Appl Microbiol. 1989;8:151–156. doi: 10.1111/j.1472-765X.1989.tb00262.x. [DOI] [Google Scholar]
- Radwan M, Hanora A, Khalifa S, Abou-El-Ela SH. Manzamines: a potential for novel cures. Cell Cycle. 2012;11:1765–1772. doi: 10.4161/cc.20135. [DOI] [PubMed] [Google Scholar]
- Rao KV, Kasanah N, Wahyuono S, Tekwani BL, Schinazi RF, Hamann MT. Three new manzamine alkaloids from a common Indonesian sponge and their activity against infectious and tropical parasitic diseases. J Nat Prod. 2004;67:1314–1318. doi: 10.1021/np0400095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reysenbach AL, Giver LJ, Wickham GS, Pace NR. Differential amplification of rRNA genes by polymerase chain reaction. Appl Environ Microbiol. 1992;58:3417–3418. doi: 10.1128/aem.58.10.3417-3418.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roland DK, Yu-Liang Y, Yuquan X, Peter C, Sang-Jip N, William F, et al. A mass spectrometry–guided genome mining approach for natural product peptidogenomics. Nat Chem Biol. 2011;7:794–802. doi: 10.1038/nchem-bio.684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roller C, Wagner M, Amann R, Ludwig W, Schleifer KH. In situ probing of Gram-positive bacteria with high DNA G+C content using 23S rRNA-targeted oligonucleotides. Microbiology. 1994;140:2849–2858. doi: 10.1099/00221287-140-10-2849. [DOI] [PubMed] [Google Scholar]
- Ruby E, Henderson B, Mcfall-Ngai M. We get by with a little help from our (little) friends. Science. 2004;303:1305–1307. doi: 10.1126/science.1094662. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sakai R, Higa T, Jefford CW, Bernardinelli G. Manzamine A, a novel antitumor alkaloid from a sponge. J Am Chem Soc. 1986;108:6404–6405. doi: 10.1021/ja00280a055. [DOI] [Google Scholar]
- Shih CJ, Chen PY, Liaw CC, Lai YM, Yang YL. Bringing microbial interactions to light using imaging mass spectrometry. Nat Prod Rep. 2014;31:739–755. doi: 10.1039/c3np70091g. [DOI] [PubMed] [Google Scholar]
- Simmons TL, Coates RC, Clark BR, Engene N, Gonzalez D, Esquenazi E, et al. Biosynthetic origin of natural products isolated from marine microorganism–invertebrate assemblages. Proc Natl Acad Sci USA. 2008;105:4587–4594. doi: 10.1073/pnas.0709851105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville CC, Knight IT, Straube WL, Colwell RR. Simple, rapid method for direct isolation of nucleic acids from aquatic environments. Appl Environ Microbiol. 1989;55:548–554. doi: 10.1128/aem.55.3.548-554.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi M, Nunnery JK, Engene N, Esquenazi E, Byrum T, Dorrestein PC, et al. Palmyramide A, a cyclic depsipeptide from a palmyra atoll collection of the marine cyanobacterium Lyngbya majuscula. J Nat Prod. 2009;73:393–398. doi: 10.1021/np900428h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MW. Sponge-associated microorganisms: evolution, ecology, and biotechnological potential. Microbiol Mol Biol Rev. 2007;71:295–347. doi: 10.1128/MMBR.00040-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiele J. Studien über pazifische Spongien. II. Ueber einige Spongien von Celebes. Zoologica. Original-Abhandlungen aus dem Gesamtgebiete der Zoologie. Stuttgart. 1899;24:1–33. pls I–V. [Google Scholar]
- Waters AL, Hill RT, Place AR, Hamann MT. The expanding role of marine microbes in pharmaceutical development. Curr Opin Biotechnol. 2010;21:780–786. doi: 10.1016/j.copbio.2010.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watrous JD, Dorrestein PC. Imaging mass spectrometry in microbiology. Nat Rev Microbiol. 2011;9:683–694. doi: 10.1038/nrmicro2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster NS, Wilson KJ, Blackall LL, Hill RT. Phylogenetic diversity of bacteria associated with the marine sponge Rhopaloeides odor-abile. Appl Environ Microbiol. 2001;67:434–444. doi: 10.1128/AEM.67.1.434-444.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Kersten RD, Nam SJ, Lu L, Al-Suwailem AM, Zheng H, et al. Bacterial biosynthesis and maturation of the didemnin anti-cancer agents. J Am Chem Soc. 2012;134:8625–8632. doi: 10.1021/ja301735a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JY, Phelan VV, Simkovsky R, Watrous JD, Trial RM, Fleming TC, et al. Primer on agar-based microbial imaging mass spectrometry. J Bacteriol. 2012;194:6023–6028. doi: 10.1128/JB.00823-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YL, Xu Y, Straight P, Dorrestein PC. Translating metabolic exchange with imaging mass spectrometry. Nat Chem Biol. 2009;5:885–887. doi: 10.1038/nchembio.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


