Summary
The small molecules produced by environmental bacteria have been mainstays of both chemical and biological research for decades, and some have lead to important therapeutic interventions. These small molecules have been shaped by natural selection as they evolved to fulfill changing functional roles in their native environments. This minireview describes some recent systematic studies providing illustrative examples that involve the acquisition and alteration of genetic information for molecular innovation by bacteria in well-defined environments. Two different bacterial genera are featured, Pseudonocardia and Salinispora, and while the small molecule repertoires of both have benefited from horizontal gene transfer, Pseudonocardia spp. have relied on plasmid-based tactics while Salinispora spp. have relied on chromosomally integrated genomic islands.
Introduction
Bacteria produce an extraordinary array of small molecules that formed the historical basis for much of chemistry and biology, and later became the major discovery paradigm for the pharmaceutical industry. Defining their complex molecular structures prompted major advances in both analytical and synthetic chemistry and understanding their biological functions led to the identification of their macromolecular targets and biological mechanisms. Together, these advances combined to create a greatly increased understanding of biological mechanisms and supply of therapeutic agents. The resulting aura surrounding naturally occurring molecules, which seem so different from their vastly more numerous relatives created in the laboratory, obscures the recognition that their most significant feature is their evolutionary history. They represent the current survivors of numerous and continuing rounds of modification, selection, and amplification on a limited set of starting materials with a modest set of chemical reactions. Ironically, while we know and continue to learn a great deal about the chemistry and biology of these molecules, we know relatively little about their evolutionary histories. What were the genetic mechanisms that led to the structures seen today? Several recent reports have begun to provide preliminary answers to these questions.
The antibiotic erythromycin, the anticancer agent bleomycin, the immunomodulator rapamycin, along with many other important molecules are produced by the sequential operation of enzymes, and the ordered sequence of conversions leading from a starting material to a final product is called a biosynthetic pathway. The enzymes that make up a pathway are, of course, genetically encoded, and in bacteria a pathway’s genes are typically clustered on a contiguous stretch of DNA. This collection of genes is referred to as a biosynthetic gene cluster or BGC. How do BGCs evolve? Gradually accumulating changes in a lineage, a process familiar from animal and plant evolution, would be one possibility. The spatial organization of the genes that make up a typical BGC, however, show hallmarks of recombination. A single BGC can often be divided into distinct subclusters, and each subcluster is defined by its own independent evolutionary history [1, 2]. A typical bacterial BGC is best described as an assembly of smaller parts that have both independent and co-evolutionary histories, and this summation model in which biosynthetic parts are mixed to create new molecules appears to be the path most often followed to create molecular diversity. High quality genome sequences have increasingly confirmed that horizontal gene transfer (HGT) is the major contributor to the genetic diversity seen in bacteria [3-5], and this extensive genetic exchange argues that modifications to BGCs are strongly dependent on the environment. In this minireview, we first compare two general evolutionary trajectories for the way in which small molecules originate and spread, and then we review recent observations that illustrate how HGT contributes to the catalogue of small molecules within specialized bacterial niches.
General considerations for biosynthetic pathway evolution
BGCs evolve in many different ways. Changes to the numbers or functions of their genes, the nucleotide sequences, genetic organization and context in which they appear all can have profound effects on the small molecule products and the levels of their production. The major distinctions between the genes that encode for a small molecule are reflected in the way it evolves. The most well-studied classes of small molecules are: the ribosomally-synthesized and post-translational modified peptides (RiPPs), non-ribosomal peptides, and polyketides. As the name suggests, the precursors of RiPPs are genetically-encoded by a single gene that is heavily modified by additional enzymes after translation[6]. In this case, evolution can occur rapidly at the single gene level to diversify or finely tune a chemical structure. One prominent example is the patellamide family of small molecules, which are produced by cyanobacterial symbionts of ascidians. A metagenomic survey of the patellamide family BGCs from sponge samples revealed that the large number of distinct small molecules that have been isolated result from a large number of naturally-occurring genetic variants; however, the variation is confined to two regions within the single gene that encodes the precursor[7]. In contrast, the non-ribosomal peptides and polyketides are typically encoded by large clusters with a number of multimoldular enzymes that have evolved by gene duplication and recombination events[2]. Notably, while each of these mechanisms contribute to the molecular innovations of as single organism, the whole bacterial ecosystem and the gene flow that exists between bacteria potentiate these evolutive mechanisms.
Genesis and Dissemination of Bacterially-Produced Small Molecules
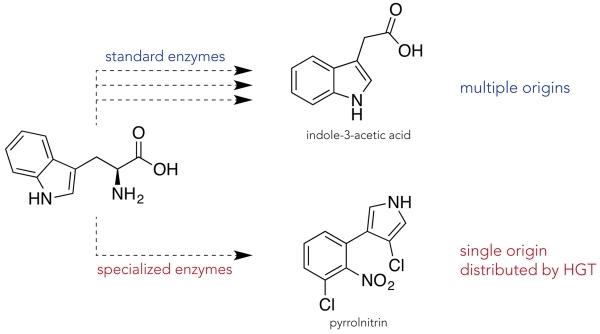
Naturally-occurring small molecules vary markedly in their distribution. Some, like the amino acid tryptophan, can be found in all living organisms – either biosynthesized or acquired – while others have limited distribution. These limits also vary widely. Indole-3-acetic acid (IAA), best known as a plant auxin, is widespread in plants, fungi, and bacteria, while and pyrrolnitrin, an antifungal agent, has been reported in Proteobacteria (Figure 1)[8-10]. Both IAA and pyrrolonitrin are generated from tryptophan. IAA is the product of at least five different bacterial biosynthetic pathways, indicating that the widespread distribution of this important molecule is due, in part, to convergent evolution[11]. Pyrrolnitrin production, in contrast, results from a single known four-gene operon[12] that is highly conserved[13]. An analysis of the distribution of the pyrrolnitrin (prn) operon argues for a single molecular origin that has spread in the environment through HGT[14]. This differential distribution undoubtedly reflects their differential biological functions. IAA is well known as a plant hormone, but it also plays roles in bacterial physiology[11, 15]. Pyrrolonitrin, by contrast is only known as an antifungal antibiotic.
Figure 1.
Two paths to the widespread distribution of a small molecule: multiple origins or HGT. At the top, the arrows indicate that there are several biosynthetic pathways for IAA that evolved independently by repurposing enzymes that catalyze standard chemical reactions. Along the bottom, a single arrow represents the single evolutionary origin of the biosynthetic pathway for pyrrolnitrin, which requires more specialized enzymes.
There are additional features that differentiate IAA and pyrrolonitrin. IAA’s structure is remarkably similar to its tryptophan starting material, and the enzymes that produce it are similar to enzymes involved in primary metabolism. They catalyze the same reactions on different substrates. As a result, the multiple independent pathways leading to IAA can evolve relatively easily by repurposing existing enzymes. In contrast, pyrrolnitrin’s structure is a highly modified product of tryptophan, and some of the enzymes used in its creation are quite unusual as highlighted by the incorporation of relatively rare chlorine atoms, a nitro functionality, and the destruction and creation of heterocyclic rings. Evolving these new enzymatic functions, in particular the heterocyclic ring chemistry, creates a high barrier for chemical innovation. This barrier leads to limited distribution, and this limited distribution simplifies documenting the spread of a highly specialized pathway like pyrrolnitrin’s relative to those of the multiple convergent pathways like IAA. This is also evident from the high level of conservation of the BGC, which suggests that it is more likely that the genes that encode for pyrrolnitrin have been and will be shared by HGT rather than be re-invented.
Both IAA and pyrrolnitrin have relatively small structures and correspondingly small BGCs for their production. In the growing catalogue of bacterially-produced small molecules, most examples have significantly more complex structures and larger modular BGCs. Studying how these BGCs evolved describes past events, but the tactics could be useful for future endeavors. Early recognition of the modular nature of BGCs inspired the idea of a ‘biosynthetic code’ that could be harnessed to the create new molecules through combinatorial biosynthesis[16]. Recently, synthetic biologists, operating with an expanded set of technologies and data, are revisiting the idea of deciphering and re-engineering or refactoring BGCs[17]. These efforts face many challenges, among which will be the ability to find and predict compatible parts with which to increase diversity and expand biological function. It’s likely that studying how Nature recombines modules will reveal important lessons for the application of synthetic biology to pathway engineering.
Lessons from chemical ecology
A focus on bacterially produced small molecules that are likely to be distributed by HGT simplifies studying the genetic changes involved in molecular innovation, and a focus on specialized bacterial populations in a defined environment provides a route to understanding the motivation for molecular innovation. A true understanding of an evolutionary history requires knowledge of both the method of distribution and the selection principles. Two recently discovered groups of Actinobacteria exemplify this analytical approach. One is the bacterial symbionts (genus Pseudonocardia) of fungus-growing ants and the second is bacteria from marine sediment (genus Salinispora). Both became prominent ~15 years ago[18, 19] and are now among the best characterized genera of Actinobacteria. Studies of these two bacterial genera have begun to address the challenge posed years ago by Carl R. Woese[20]: “Our task now is to resynthesize biology; put the organism back into its environment; connect it again to its evolutionary past; and let us feel that complex flow that is organism, evolution and environment united.”
Pseudonocardia: acquiring new molecular traits through plasmid-encoded BGCs
Bacteria adapt to their environments, and an individual’s genetic makeup reflects this adaptation. These niche-adapted genes contribute to what is often referred to as the “flexible genome” of an individual – the genes it has that are not generally found in a larger group of related bacteria. For this minireview, plasmids encoding niche-adaptive small molecules are particularly important example of the flexible genome.
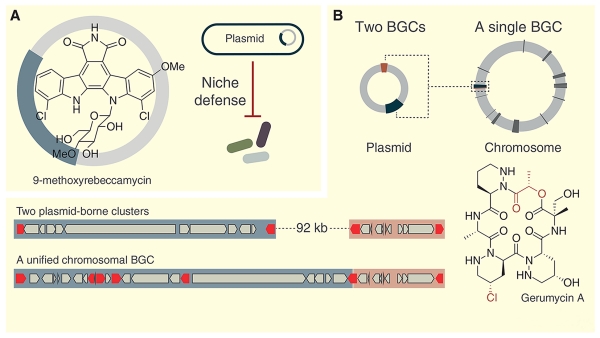
Genome sequencing has repeatedly revealed that molecular diversity, not conservation, is the rule at the genus and even species level. This diversity appears even within a highly specialized ecological niche. In the case of the ant-associated Pseudonocardia, the side-by-side growth of closely related bacterial isolates revealed antagonistic interactions[21], and inoculation of laboratory-reared ants with distinct Pseudonocardia provided variable protection from a common fungal pathogen[22]. These phenomenological studies implicate both bacterially-produced antibacterial and antifungal molecules and demonstrate that significant molecular diversity exists within this group of symbionts. The molecular and genetic basis of bacterial antagonism, perhaps more appropriately described as niche defense by one Pseudonocardia isolate against another, has recently been established[23]. A Pseudonocardia isolated from Apterostigma dentigerum living on the Barro Colorado Island in Panama was able to inhibit the growth of other closely-related isolates from the same region. When compared to it’s closest relative, this Pseudonocardia differed only in plasmid content, and a single 119 kb plasmid contained the BGC for the antibacterial molecule, 9-methoxyrebeccamycin (Figure 2A).
Figure 2.
Plasmids are a major reservoir of mobile BGCs. (A) The plasmid-encoded rebeccamycin analogue was discovered through the investigation of intraspecies interactions. (B) The two BGCs reported for the gerumycins include a split biosynthetic pathway encoded by two plasmid-borne clusters and a unified version that was observed within a chromosomal genomic island. The blue and red boxes highlight the distinct plasmid-borne clusters that are unified on the chromosome, and the molecule is colored to match the genes responsible for the product.
Additional plasmid-encoded traits appear to be widespread in the ant-associated Pseudonocardia. The BGC for the pseudonocardones[24], a family of quinone-containing molecules, is encoded by an ~850 kb megaplasmid, and the genes responsible for the production of the cyclic depsipeptides gerumycin A and B are encoded by a 297 kb plasmid[25-28]. To date, all but one of the sequenced ant-associates carry plasmids that encode for BGCs. Although this is a small sample size, the frequency is not uncommon for Actinobacteria, which have a relatively high occurrence of plasmid-borne BGCs. To provide a comparison, inspection of sequences available in the NCBI plasmid database using antiSMASH[29] shows that only ~11% of the entries encode a BGC.
The previous examples illustrate that bacteria acquire plasmids from the environment to amass a larger pool of molecular diversity. Beyond the initial acquisition, the fates of these plasmids are largely unknown. The piperazic acid-containing cyclic depsipeptides gerumycin A-C provide an interesting case study that addresses the mobility and enrichment of BGCs within an ecological niche (Figure 2B). The BGCs were encoded on either a plasmid or within a chromosomal genomic island in two different Pseudonocardia isolates. The difference in genetic context was not reflected in the shared gene content and nucleotide sequences which were conserved at ~99% identity. The arrangement of the biosynthetic genes on the plasmid was particularly unusual in that they were not assembled at a single locus. Instead, two gene clusters, separated by ~92 kb, encode for the plasmid-borne gerumycins, and remarkably, these two clusters were found adjacent to each other (as a single cluster) on the chromosomal genomic island. Overall, the example of the cyclic depsipeptides demonstrates not only the recruitment of a common molecular trait to organisms within an ecological niche it also highlights the importance of plasmids, and the observation of two very distinct gene cluster organizations shows the importance of genetic exchange and recombination in small molecule evolution.
Split clusters and BGC evolution
The two-piece, or split, biosynthetic pathway for gerumycin both identifies a shortcoming in the conventional “cluster” framework for the production of small molecules and provides further evidence for the mix-and-match logic of biosynthetic pathways. Gerumycin’s split pathway is part of a small but growing catalogue of examples in which spatially separated and co-dependent genes or gene clusters have been reported for a single molecule. A biosynthetic strategy that is dependent on more than a single genetic locus is often referred to as BGC ‘crosstalk,’ and appears to be relatively common for some molecular scaffolds, most notably siderophores (iron-binding small molecules). The hydroxamate siderophore erythrochelin produced by Saccharopolyspora erythraea, for example, is encoded by two loci, one for the core scaffold and one for a critical acetyltransferase[30]. Similarly, the genes required for the production and incorporation of 2,3-dihydroxybenzoate are often encoded just once within a bacterial genome despite possible incorporation into multiple natural products within a single organism. The production of the serratiochelins by Serratia plymuthica V4, for example, relies on two BGCs, only one of which encodes for the catechol[31]. And, the siderophore rhodochelin from Rhodococcus jostii RHA1 uses three BGCs to generate a non-ribosomal peptide scaffold that includes a 2,3-dihydroxybenzoic acid and two hydroxamates[32]. The range of molecules whose biosynthesis requires more than one pathway is unknown, but it is likely that the reports on this genetic strategy are underrepresented in the literature. Genome-mining efforts compensate for this deficiency. For example, the production of the glycosylated terpene sioxanthin from Salinispora tropica CNB-400 is dependent on four genetic loci[33], and a single promiscuous enzyme is required for glycosylation during the biosynthesis of a plecomacrolide and methyl-rhamnosylated phenazines in Kitasatospora sp. MBT166[34]. Finally, and perhaps the most unusual example of this phenomenon is the reciprocally-dependent BGCs in Streptomyces netropsis DSM40846 that encode for a suite of at least three pyrrolamides[35, 36]. This all-for-one strategy, described as a natural example of combinatorial biosynthesis[36], is a stark contrast to the one-for-all or diversity-generating biosynthetic pathways that have been dissected for decades, and it provides another confirmation of the summation model for assembling BGCs.
Salinospora: Small molecules encoded on genomic islands
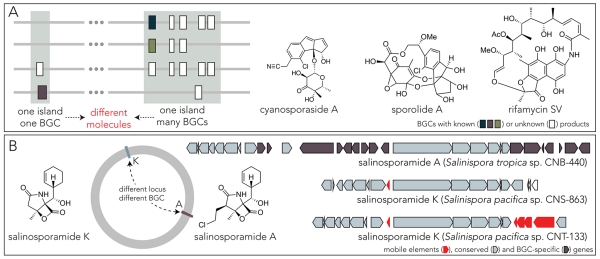
While the ant-associated Pseudonocardia exploit a plasmid-based strategy for molecular acquisition, the small molecule repertoire of Salinispora is primarily encoded within genomic islands[4, 5] – large stretches of DNA containing multiple genes and evidence of HGT. The documentation of these genomic islands reinforce some of the earliest observations made studying the longtime model organism, Streptomyces[3]. In Salinispora, fewer than 10% of the predicted BGCs are conserved or thought to be shared by the common ancestor of the genus[5]. The majority of the BGCs are encoded within conserved recombination hotspots (genomic islands) that are known to turn over gene content. In this regard, the Salinispora genomes are particularly enlightening, showing that the same genomic island can encode from one to five different BGCs depending on the individual, and that the identity of a BGC within a specific site can differ dramatically (Figure 3A) [5].
Figure 3.
(A) The plasticity of genomic islands allows for the integration of variable and distinct BGCs. Three examples from Salinispora are drawn. The cyanosporasides and sporolide BGCs are found at the same genetic locus in distinct bacterial isolates (blue and green clusters) whereas the rifamycin BGC (purple) resides in a separate island that is also characterized by variable BGC content. (B) The mobility of BGCs can result in chemical diversity. A comparison of salinosporamide A and K BGCs shows the presence of mobile elements and the loss of biosynthetic genes in the latter.
The genetic exchange that populates and alters the content of these genomics islands in Salinispora also provides an example of how the process can result in the molecular diversification of a single class of small molecule (Figure 3B). The salinosporamides, a family of β-lactam-γ-lactone-containing molecules produced by Salinispora pacifica and Salinispora tropica, are a particularly interesting example that correlates BGC mobility and molecular diversity. A direct comparison of the BGCs for salinosporamide A and K reveal a presence or absence of the biosynthetic genes for the chloroethylmalonyl-CoA required to produce the former[37, 38]. In the minimal salinosporamide K clusters, a number of transposases surround key biosynthetic genes. Moreover, the alignment of high quality sequences from a number of Salinispora revealed that the genetic loci that encode for the salinosporamides is not conserved between the pacifica and tropica groups[5]. Interestingly, the observed genetic change correlates with the pattern of molecular diversity in this family of small molecules, which is most often manifested as a difference in the substituents appended on the γ-lactam ring[39]. Altogether, the observed plasticity and turnover of BGCs in the Salinispora genomic islands further implicate HGT and recombination as major contributors to the natural history of small molecules, and the correlation between substituent and genetic diversity mirrors the bulk observation from sequencing data that subcluters are at least partially discrete evolutionary units.
Conclusion
This minireview reifies the longstanding microbial apothegm from Baas Becking: “Everything is everywhere, but the environment selects.” Plasmids, genomic islands, and HGT have long been recognized to play dominant roles in bacterial speciation, pathogenesis, and symbiosis [40, 41]. For example, a plasmid found in the bacterial symbionts of legume roots confers the ability to nodulate and fix-nitrogen[42], and encodes for a bacteriocin that likely contributes to the host organism’s competitive fitness[43]. The latter is analogous to observations with the Pseudonocardia strains, which seem to rely largely on plasmids for chemical diversity and defense. The marine sediment dwelling Salinispora seem to rely more heavily on genomic islands for molecular diversification in a manner reminiscent of the virulence factors that characterize pathogenic bacteria. For example, pathogenic Escherichia coli and other Enterobacteriaceae can produce two small molecules associated with virulence and disease: the colibactins, DNA-damaging agents[44], and yersiniabactin, a siderophore, both of which are encoded for on a single genomic island[45]. Both plasmids and the contents of genomic islands are spread by HGT. The molecular repertoires of bacterial populations are in constant flux, changing with the influence of the local environment and microbial neighbors. As a collective, the microbial genomes in each environment provide raw materials, the genomic building blocks, that individuals can mix and match to create the BGCs that provide selective advantages. Adding ecological considerations to genetic and molecular studies allows the selection principles to be identified and fills out a molecule’s natural history, providing mechanistic insight into the manner in which BGCs are acquired and evolve.
In Brief.
The small molecules produced by environmental bacteria have been mainstays of both chemical and biological research for decades, and have been shaped by natural selection as they evolved to fulfill changing functional roles in their native environments. This minireview describes some recent systematic studies providing illustrative examples that involve the acquisition and alteration of genetic information for molecular innovation by bacteria in well-defined environments.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Fischbach MA, Walsh CT, Clardy J. The evolution of gene collectives: How natural selection drives chemical innovation. Proc Natl Acad Sci U S A. 2008;105:4601–4608. doi: 10.1073/pnas.0709132105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Medema MH, Cimermancic P, Sali A, Takano E, Fischbach MA. A systematic computational analysis of biosynthetic gene cluster evolution: lessons for engineering biosynthesis. PLoS Comput Biol. 2014;10:e1004016. doi: 10.1371/journal.pcbi.1004016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Choulet F, Aigle B, Gallois A, Mangenot S, Gerbaud C, Truong C, Francou FX, Fourrier C, Guerineau M, Decaris B, et al. Evolution of the terminal regions of the Streptomyces linear chromosome. Mol Biol Evol. 2006;23:2361–2369. doi: 10.1093/molbev/msl108. [DOI] [PubMed] [Google Scholar]
- 4.Penn K, Jenkins C, Nett M, Udwary DW, Gontang EA, McGlinchey RP, Foster B, Lapidus A, Podell S, Allen EE, et al. Genomic islands link secondary metabolism to functional adaptation in marine Actinobacteria. ISME J. 2009;3:1193–1203. doi: 10.1038/ismej.2009.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ziemert N, Lechner A, Wietz M, Millan-Aguinaga N, Chavarria KL, Jensen PR. Diversity and evolution of secondary metabolism in the marine actinomycete genus Salinispora. Proc Natl Acad Sci U S A. 2014;111:E1130–1139. doi: 10.1073/pnas.1324161111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arnison PG, Bibb MJ, Bierbaum G, Bowers AA, Bugni TS, Bulaj G, Camarero JA, Campopiano DJ, Challis GL, Clardy J, et al. Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat Prod Rep. 2013;30:108–160. doi: 10.1039/c2np20085f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Donia MS, Hathaway BJ, Sudek S, Haygood MG, Rosovitz MJ, Ravel J, Schmidt EW. Natural combinatorial peptide libraries in cyanobacterial symbionts of marine ascidians. Nat Chem Biol. 2006;2:729–735. doi: 10.1038/nchembio829. [DOI] [PubMed] [Google Scholar]
- 8.Arima K, Imanaka H, Kousaka M, Fukuda A, Tamura G. Studies on pyrrolnitrin, a new antibiotic. I. Isolation and properties of pyrrolnitrin. J Antibiot (Tokyo) 1965;18:201–204. [PubMed] [Google Scholar]
- 9.Gerth K, Trowitzsch W, Wray V, Hofle G, Irschik H, Reichenbach H. Pyrrolnitrin from Myxococcus fulvus (Myxobacterales) J Antibiot (Tokyo) 1982;35:1101–1103. doi: 10.7164/antibiotics.35.1101. [DOI] [PubMed] [Google Scholar]
- 10.el-Banna N, Winkelmann G. Pyrrolnitrin from Burkholderia cepacia: antibiotic activity against fungi and novel activities against streptomycetes. J Appl Microbiol. 1998;85:69–78. doi: 10.1046/j.1365-2672.1998.00473.x. [DOI] [PubMed] [Google Scholar]
- 11.Spaepen S, Vanderleyden J. Auxin and plant-microbe interactions. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a001438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hammer PE, Hill DS, Lam ST, Van Pee KH, Ligon JM. Four genes from Pseudomonas fluorescens that encode the biosynthesis of pyrrolnitrin. Appl Environ Microbiol. 1997;63:2147–2154. doi: 10.1128/aem.63.6.2147-2154.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hammer PE, Burd W, Hill DS, Ligon JM, van Pee K. Conservation of the pyrrolnitrin biosynthetic gene cluster among six pyrrolnitrin-producing strains. FEMS Microbiol Lett. 1999;180:39–44. doi: 10.1111/j.1574-6968.1999.tb08775.x. [DOI] [PubMed] [Google Scholar]
- 14.Costa R, van Aarle IM, Mendes R, van Elsas JD. Genomics of pyrrolnitrin biosynthetic loci: evidence for conservation and whole-operon mobility within gram-negative bacteria. Environ Microbiol. 2009;11:159–175. doi: 10.1111/j.1462-2920.2008.01750.x. [DOI] [PubMed] [Google Scholar]
- 15.Patten CL, Blakney AJ, Coulson TJ. Activity, distribution and function of indole-3-acetic acid biosynthetic pathways in bacteria. Crit Rev Microbiol. 2013;39:395–415. doi: 10.3109/1040841X.2012.716819. [DOI] [PubMed] [Google Scholar]
- 16.Cane DE, Walsh CT, Khosla C. Harnessing the biosynthetic code: combinations, permutations, and mutations. Science. 1998;282:63–68. doi: 10.1126/science.282.5386.63. [DOI] [PubMed] [Google Scholar]
- 17.Smanski MJ, Zhou H, Claesen J, Shen B, Fischbach MA, Voigt CA. Synthetic biology to access and expand nature’s chemical diversity. Nat Rev Microbiol. 2016;14:135–149. doi: 10.1038/nrmicro.2015.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Currie CR, Scott JA, Summerbell RC, Malloch D. Fungus-growing ants use antibiotic-producing bacteria to control garden parasites. Nature. 1999;398:701–704. [Google Scholar]
- 19.Mincer TJ, Jensen PR, Kauffman CA, Fenical W. Widespread and persistent populations of a major new marine actinomycete taxon in ocean sediments. Appl Environ Microbiol. 2002;68:5005–5011. doi: 10.1128/AEM.68.10.5005-5011.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Woese CR. A new biology for a new century. Microbiol Mol Biol Rev. 2004;68:173–186. doi: 10.1128/MMBR.68.2.173-186.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poulsen M, Erhardt DP, Molinaro DJ, Lin TL, Currie CR. Antagonistic bacterial interactions help shape host-symbiont dynamics within the fungus-growing ant-microbe mutualism. PLoS One. 2007;2:e960. doi: 10.1371/journal.pone.0000960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poulsen M, Cafaro MJ, Erhardt DP, Little AE, Gerardo NM, Tebbets B, Klein BS, Currie CR. Variation in Pseudonocardia antibiotic defence helps govern parasite-induced morbidity in Acromyrmex leaf-cutting ants. Environ Microbiol Rep. 2010;2:534–540. doi: 10.1111/j.1758-2229.2009.00098.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Arnam EB, Ruzzini AC, Sit CS, Currie CR, Clardy J. A Rebeccamycin Analog Provides Plasmid-Encoded Niche Defense. J Am Chem Soc. 2015;137:14272–14274. doi: 10.1021/jacs.5b09794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carr G, Derbyshire ER, Caldera E, Currie CR, Clardy J. Antibiotic and antimalarial quinones from fungus-growing ant-associated Pseudonocardia sp. J Nat Prod. 2012;75:1806–1809. doi: 10.1021/np300380t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haeder S, Wirth R, Herz H, Spiteller D. Candicidin-producing Streptomyces support leaf-cutting ants to protect their fungus garden against the pathogenic fungus Escovopsis. Proc Natl Acad Sci U S A. 2009;106:4742–4746. doi: 10.1073/pnas.0812082106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oh DC, Poulsen M, Currie CR, Clardy J. Dentigerumycin: a bacterial mediator of an ant-fungus symbiosis. Nat Chem Biol. 2009;5:391–393. doi: 10.1038/nchembio.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barke J, Seipke RF, Gruschow S, Heavens D, Drou N, Bibb MJ, Goss RJ, Yu DW, Hutchings MI. A mixed community of actinomycetes produce multiple antibiotics for the fungus farming ant Acromyrmex octospinosus. BMC Biol. 2010;8:109. doi: 10.1186/1741-7007-8-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sit CS, Ruzzini AC, Van Arnam EB, Ramadhar TR, Currie CR, Clardy J. Variable genetic architectures produce virtually identical molecules in bacterial symbionts of fungus-growing ants. Proc Natl Acad Sci U S A. 2015;112:13150–13154. doi: 10.1073/pnas.1515348112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blin K, Medema MH, Kazempour D, Fischbach MA, Breitling R, Takano E, Weber T. antiSMASH 2.0--a versatile platform for genome mining of secondary metabolite producers. Nucleic Acids Res. 2013;41:W204–212. doi: 10.1093/nar/gkt449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lazos O, Tosin M, Slusarczyk AL, Boakes S, Cortes J, Sidebottom PJ, Leadlay PF. Biosynthesis of the putative siderophore erythrochelin requires unprecedented crosstalk between separate nonribosomal peptide gene clusters. Chem Biol. 2010;17:160–173. doi: 10.1016/j.chembiol.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 31.Seyedsayamdost MR, Cleto S, Carr G, Vlamakis H, Joao Vieira M, Kolter R, Clardy J. Mixing and matching siderophore clusters: structure and biosynthesis of serratiochelins from Serratia sp. V4. J Am Chem Soc. 2012;134:13550–13553. doi: 10.1021/ja304941d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bosello M, Robbel L, Linne U, Xie X, Marahiel MA. Biosynthesis of the siderophore rhodochelin requires the coordinated expression of three independent gene clusters in Rhodococcus jostii RHA1. J Am Chem Soc. 2011;133:4587–4595. doi: 10.1021/ja1109453. [DOI] [PubMed] [Google Scholar]
- 33.Richter TK, Hughes CC, Moore BS. Sioxanthin, a novel glycosylated carotenoid, reveals an unusual subclustered biosynthetic pathway. Environ Microbiol. 2015;17:2158–2171. doi: 10.1111/1462-2920.12669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu C, Medema MH, Lakamp RM, Zhang L, Dorrestein PC, Choi YH, van Wezel GP. Leucanicidin and Endophenasides Result from Methyl-Rhamnosylation by the Same Tailoring Enzymes in Kitasatospora sp. MBT66. ACS Chem Biol. 2016;11:478–490. doi: 10.1021/acschembio.5b00801. [DOI] [PubMed] [Google Scholar]
- 35.Hao C, Huang S, Deng Z, Zhao C, Yu Y. Mining of the pyrrolamide antibiotics analogs in Streptomyces netropsis reveals the amidohydrolase-dependent “iterative strategy” underlying the pyrrole polymerization. PLoS One. 2014;9:e99077. doi: 10.1371/journal.pone.0099077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vingadassalon A, Lorieux F, Juguet M, Le Goff G, Gerbaud C, Pernodet JL, Lautru S. Natural combinatorial biosynthesis involving two clusters for the synthesis of three pyrrolamides in Streptomyces netropsis. ACS Chem Biol. 2015;10:601–610. doi: 10.1021/cb500652n. [DOI] [PubMed] [Google Scholar]
- 37.Eustaquio AS, McGlinchey RP, Liu Y, Hazzard C, Beer LL, Florova G, Alhamadsheh MM, Lechner A, Kale AJ, Kobayashi Y, et al. Biosynthesis of the salinosporamide A polyketide synthase substrate chloroethylmalonyl-coenzyme A from S-adenosyl-L-methionine. Proc Natl Acad Sci U S A. 2009;106:12295–12300. doi: 10.1073/pnas.0901237106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eustaquio AS, Nam SJ, Penn K, Lechner A, Wilson MC, Fenical W, Jensen PR, Moore BS. The discovery of salinosporamide K from the marine bacterium “Salinispora pacifica” by genome mining gives insight into pathway evolution. Chembiochem. 2011;12:61–64. doi: 10.1002/cbic.201000564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jensen PR, Moore BS, Fenical W. The marine actinomycete genus Salinispora: a model organism for secondary metabolite discovery. Nat Prod Rep. 2015;32:738–751. doi: 10.1039/c4np00167b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dobrindt U, Hochhut B, Hentschel U, Hacker J. Genomic islands in pathogenic and environmental microorganisms. Nat Rev Microbiol. 2004;2:414–424. doi: 10.1038/nrmicro884. [DOI] [PubMed] [Google Scholar]
- 41.Polz MF, Alm EJ, Hanage WP. Horizontal gene transfer and the evolution of bacterial and archaeal population structure. Trends Genet. 2013;29:170–175. doi: 10.1016/j.tig.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Johnston AWB, Beynon JL, Buchanan-Wollaston AV, Setchell SM, Hirsch PR, Beringer JE. High frequency transfer of nodulating ability between strains and species of Rhizobium. Nature. 1978;276:634–636. [Google Scholar]
- 43.Hirsch PR. Plasmid-determined Bacteriocin Production by Rhizobium leguminosarum. Journal of General Microbiology. 1979;113:219–228. [Google Scholar]
- 44.Nougayrede JP, Homburg S, Taieb F, Boury M, Brzuszkiewicz E, Gottschalk G, Buchrieser C, Hacker J, Dobrindt U, Oswald E. Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science. 2006;313:848–851. doi: 10.1126/science.1127059. [DOI] [PubMed] [Google Scholar]
- 45.Putze J, Hennequin C, Nougayrede JP, Zhang W, Homburg S, Karch H, Bringer MA, Fayolle C, Carniel E, Rabsch W, et al. Genetic structure and distribution of the colibactin genomic island among members of the family Enterobacteriaceae. Infect Immun. 2009;77:4696–4703. doi: 10.1128/IAI.00522-09. [DOI] [PMC free article] [PubMed] [Google Scholar]