Summary
Spinal and bulbar muscular atrophy (SBMA), a progressive degenerative disorder, is caused by a CAG/glutamine expansion in the androgen receptor (polyQ AR). Recent studies demonstrate that skeletal muscle is an important site of toxicity that contributes to the SBMA phenotype. Here, we sought to identify critical pathways altered in muscle that underlie disease manifestations in AR113Q mice. This led to the unanticipated identification of gene expression changes affecting regulators of carbohydrate metabolism, similar to those triggered by denervation. AR113Q muscle exhibits diminished glycolysis, altered mitochondria, and an impaired response to exercise. Strikingly, the expression of genes regulating muscle energy metabolism is rescued following peripheral polyQ AR gene silencing by antisense oligonucleotides (ASO), a therapeutic strategy that alleviates disease. Our data establish the occurrence of a metabolic imbalance in SBMA muscle triggered by peripheral expression of the polyQ AR, and indicate that alterations in energy utilization contribute to non-neuronal disease manifestations.
Graphical abstract
Introduction
The progressive degeneration of the neuromuscular system that occurs in spinal and bulbar muscular atrophy (SBMA) results in skeletal muscle weakness and atrophy and in the loss of lower motor neurons in the spinal cord and brainstem (Chua and Lieberman, 2013; Lieberman and Fischbeck, 2000). Clinical onset typically occurs in young to midlife adults and is slowly progressive. The disease is X-linked and caused by expansion of a CAG/glutamine tract in the coding region of the first exon of the gene encoding the androgen receptor (AR). Ligand binding to the polyglutamine AR (polyQ AR) promotes nuclear translocation and protein unfolding. Both of these steps are required for pathogenesis and underlie the occurrence of disease only in men (Katsuno et al., 2002; Takeyama et al., 2002). While the polyglutamine tract expansion leads to a partial loss of the AR’s transactivation function (Chamberlain et al., 1994; Kazemi-Esfarjani et al., 1995; Lieberman et al., 2002; Mhatre et al., 1993), and this may contribute to features of androgen insensitivity, neuromuscular degeneration is mediated by a toxic gain-of-function conferred by the misfolded protein. As with all of the polyglutamine disorders, the mechanisms that lead to progressive functional impairment are poorly understood and disease-modifying therapies are currently unavailable.
Studies of SBMA patients and genetic mouse models have established that skeletal muscle is an important site of toxicity and an attractive therapeutic target. These studies have demonstrated both denervation and myofiber degeneration in muscle biopsies from SBMA subjects (Soraru et al., 2008). This mixed pathology reflects the occurrence of myopathic symptoms in SBMA patients, including early onset muscle cramps and elevated levels of serum creatine kinase that exceed those detected in pure denervating diseases (Atsuta et al., 2006; Rhodes et al., 2009). Similarly, analysis of a mouse model of SBMA generated through gene targeting (AR113Q mice) demonstrated the presence of both myopathic and denervating changes in skeletal muscle (Yu et al., 2006a). Highlighting the contribution of skeletal muscle to neuromuscular degeneration are transgenic mice that over-express the wild type AR only in muscle and exhibit an SBMA-like phenotype (Johansen et al., 2009; Monks et al., 2007). Similar results have been reported recently in mice over-expressing the polyQ AR only in muscle (Ramzan et al., 2015). Moreover, genetic over-expression of IGF-1 in skeletal muscle alleviates disease in a transgenic mouse model of SBMA (Palazzolo et al., 2009), emphasizing the potential utility of targeting peripheral tissues in this disease. This notion is supported by treatment of AR113Q or polyQ AR BAC transgenic mice with subcutaneously administered antisense oligonucleotides (ASO) targeted against AR (Lieberman et al., 2014). This treatment diminishes polyQ AR expression in the periphery but not CNS and rescues disease. These findings are corroborated by genetic studies demonstrating that deletion of a floxed polyQ AR allele only in skeletal muscle prevents disease in BAC transgenic mice (Cortes et al., 2014). Taken together, this body of work has established the importance of polyQ AR expression in skeletal muscle to the SBMA phenotype.
Here, we sought to define critical pathways altered in AR113Q muscle that contribute to the disease phenotype. To accomplish this, we mined gene expression data from skeletal muscle of wild type and AR113Q males. This approach led to the unanticipated identification of a cohort of genes that regulate carbohydrate metabolism in muscle that are expressed at diminished levels in AR113Q mice. We provide evidence of decreased glycolysis and altered mitochondria in AR113Q muscle, and demonstrate that these changes are functionally significant by impacting the response to exercise. Moreover, we establish that subcutaneous administration of ASO to AR113Q males rescues the expression of diverse genes controlling energy metabolism in muscle. Our data provide evidence of metabolic alterations in SBMA muscle and demonstrate that these changes are triggered by peripheral expression of the polyQ AR.
Results
Decreased expression of carbohydrate metabolism-related genes in AR113Q muscle
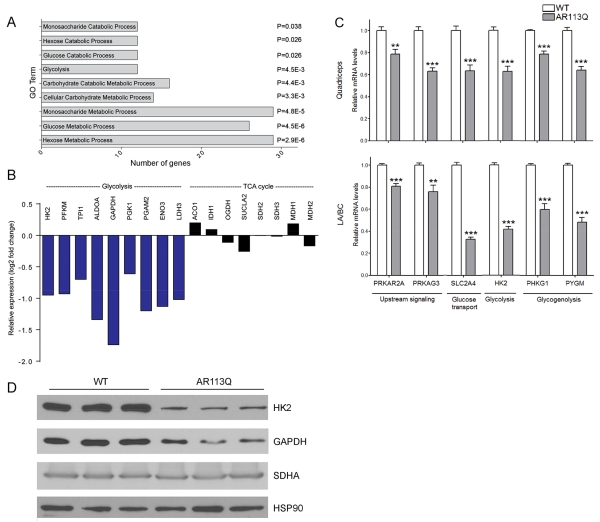
We sought to understand the mechanisms by which expression of the polyQ AR in skeletal muscle contributes to the phenotype of AR113Q knock-in mice. To uncover pathways that are altered in disease, we harvested quadriceps muscle from wild type (WT) and AR113Q males at 14 weeks of age. RNA-seq was used to characterize gene expression, and pairwise analysis uncovered 909 genes differentially expressed by at least 1.5 fold in quadriceps muscle of AR113Q compared to WT mice (Tables S1 and S2). Pathway analysis by gene ontology terms (GO terms) revealed altered expression in AR113Q muscle of regulators of carbohydrate metabolism (Fig. 1A). Detailed examination uncovered significantly decreased expression of nine genes encoding enzymes in the glycolytic pathway (Fig. 1B). In contrast, none of the genes encoding enzymes in the tricarboxylic acid cycle (TCA cycle) were expressed at significantly different levels in AR113Q muscle. Broad effects on the expression of genes regulating energy metabolism were confirmed in independent cohorts of WT and AR113Q mice by analyzing both a hindlimb muscle (quadriceps) and a pelvic floor muscle that expresses high levels of the polyQ AR (levator ani/bulbocavernosus; LA/BC). We confirmed by quantitative real time RT-PCR (qPCR) significantly decreased expression of genes encoding upstream signaling regulators of carbohydrate metabolism (Prkar2a, Prkag3), and mediators of glucose uptake (Slc2a4), glycolysis (Hk2), and glycogenolysis (Phkg1, Pygm) (Fig. 1C). Similarly, western blot demonstrated decreased expression of glycolytic enzymes HK2 and GAPDH, but not the mitochondrial respiratory chain component SDHA in AR113Q muscle (Fig. 1D). These data demonstrate an unexpected decrease in the expression of carbohydrate metabolism-related genes in AR113Q skeletal muscle.
Figure 1. Decreased expression of carbohydrate metabolism genes in AR113Q skeletal muscle.
(A, B) Quadriceps muscles from wild type or AR11Q males (n = 3/group) were harvested at 14 weeks of age and gene expression was analyzed by RNA-seq. (A) Go terms of differentially expressed genes (≥1.5-fold) related to carbohydrate metabolism. P values by false discovery rate are indicated. (B) Relative expression of genes encoding glycolytic or TCA cycle enzymes in AR113Q vs WT muscle. Shown in blue are genes whose expression is changed by ≥1.5-fold and in black genes whose expression is changed by <1.5 fold. (C) Expression of carbohydrate metabolism-related genes in quadriceps and LA/BC muscles of WT and AR113Q mice (mean ± SEM), n = 5/group. **P < 0.01; ***P < 0.001 by Student’s t-test. (D) Western blots of HK2, GAPDH and SDHA in lysates from LA/BC muscles of WT and AR113Q mice.
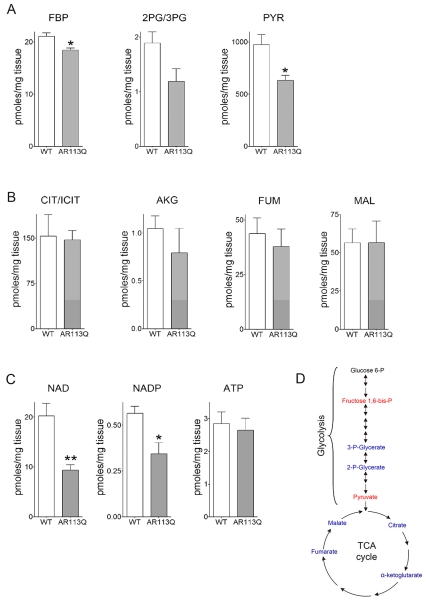
To glean insight into the functional effect of these gene expression changes, WT and AR113Q mice were run on an inclined treadmill for 15 minutes at increasing speed, and quadriceps muscle was harvested for metabolomics analysis. This intense exercise protocol was designed to favor anaerobic metabolism (Koch and Britton, 2001). Tissues were collected under continuous anesthesia and flash frozen in liquid nitrogen using a procedure that maintains unaltered levels of metabolites in muscle (Overmyer et al., 2015). Samples were analyzed by liquid chromatography–mass spectrometry (LC-MS) and gas chromatography–mass spectrometry (GC-MS). In accordance with gene expression analysis, AR113Q muscle harvested during exercise contained significantly lower levels of glycolytic intermediates fructose 1,6-bisphosphate (FBP), the product of the rate limiting and irreversible step in glycolysis, and pyruvate (PYR) (Fig. 2A, D). A similar trend was detected in levels of 2- and 3-phosphyoglycerate (2PG/3PG), but these did not reach statistical significance because of variability between samples. In contrast, TCA cycle intermediates citrate/isocitrate (CIT/ICIT), α-ketoglutarate (AKG), fumarate (FUM) and malate (MAL) were unchanged between cohorts (Fig. 2B, D). These data are indicative of a more severe functional impairment of glycolysis than the TCA cycle, and are consistent with the broad decrease in expression of carbohydrate metabolism-related genes in AR113Q muscle (Fig. 1). Metabolomics additionally identified significantly diminished levels of NAD and NADP in AR113Q muscle, findings that are suggestive of lower metabolism through the glycolytic pathway (Fig. 2C). In contrast, no difference in levels of ATP was observed (Fig. 2C), consistent with the notion that the TCA cycle was relatively intact in mutant muscle.
Figure 2. Energy metabolites in skeletal muscle during exercise.
WT and AR113Q male mice at 14 weeks were run at increasing speeds on an inclined treadmill for 15 min and then quadriceps muscles was harvested for analysis. (A) Glycolytic metabolites fructose 1,6-bis-P (FBP), 2- and 3-phosphyglycerate (2PG/3PG) and pyruvate (PYR) (B) TCA cycle metabolites citrate/isocitrate (CIT/ICIT), α-ketoglutarate (AKG), fumarate (FUM), and malate (MAL); (C) NAD, NADP and ATP levels. (mean ± SEM; n = 4/group). *P < 0.05, **P < 0.01 by Student’s t-test. (D) Schematic highlighting measured glycolytic and TCA cycle metabolites. Metabolites significantly different between WT and AR113Q are in red, unchanged in blue, and not measured in black.
Mitochondrial defects in AR113Q skeletal muscle
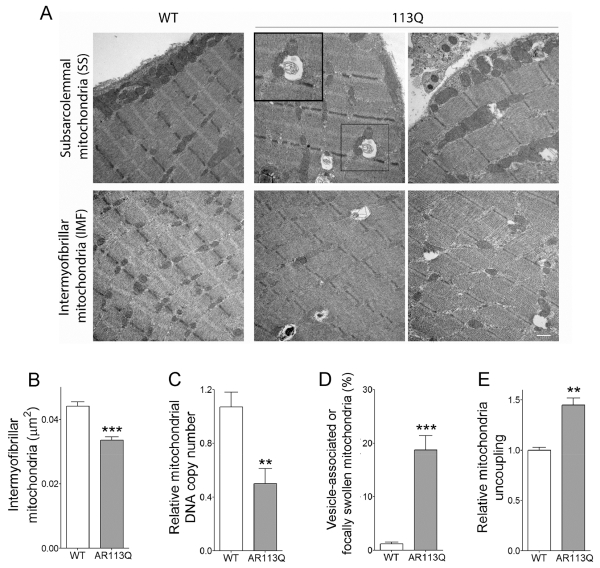
We next sought to establish the extent to which mitochondrial abnormalities accompanied alterations in the expression of carbohydrate metabolism-related genes in AR113 muscle. Prior studies identified mitochondrial defects in cellular models of SBMA and showed diminished expression of several mitochondrial genes in AR113Q muscle (Ranganathan et al., 2009). We performed transmission electron microscopy (TEM) on WT and AR113Q soleus muscles (Fig. 3A). The soleus is rich in slow-twitch oxidative fibers that contain a large number of mitochondria. While sarcomere architecture was intact in mutants, we observed a significant decrease in the mean area occupied by intermyfibrillar mitochondria (Fig. 3A, B). Similarly, mitochondrial DNA copy number was significantly decreased in the LA/BC (Fig. 3C). Moreover, ~20% of mitochondria in AR113Q muscle were either focally swollen or tightly associated with vesicles, many of which resembled autophagosomes (Fig. 3A, D; Fig. S1). This finding is consistent with the robust activation of macroautophagy previously demonstrated in AR113Q skeletal muscle (Chua et al., 2014; Yu et al., 2011) and suggests the occurrence of increased mitophagy. To test functional status, mitochondria were isolated from WT or AR113Q muscle for Seahorse analysis. Mitochondria from quadriceps muscles of AR113Q mice were significantly more uncoupled than those from WT mice (Fig. 3E). These findings suggest that a fraction of mitochondria in AR113Q muscle are dysfunctional.
Figure 3. Mitochondrial defects in the skeletal muscle of AR113Q mice.
(A) Representative TEMs of soleus muscle from WT and AR113Q males at 14 weeks. Inset shows vesicle-associated mitochondria. Scale bar = 500nm. (B) Mean area ± SEM of intermyofibrillar mitochondria (n = 3 mice/group; ≥ 90 mitochondria/mouse). ***P < 0.001 by Student’s t-test. (C) Mitochondrial DNA copy number in the LA/BC muscle of WT and AR113Q mice measured as a ratio between mitochondrial DNA (D-loop) and genomic DNA (β2m) (mean ± SEM; n = 8/group). **P < 0.01 by Student’s t-test. (D) Percentage of mitochondrial with focal swellings or associated with vesicles (mean ± SEM; n = 3/group; ≥90 mitochondria per mouse). ***P < 0.001 by Student’s t-test. (E) Relative uncoupling of mitochondria isolated from quadriceps muscles of WT and AR113Q mice (mean ± SEM; n = 3/group). **P < 0.01 by Student’s t-test.
Altered response of AR113Q mice to exercise
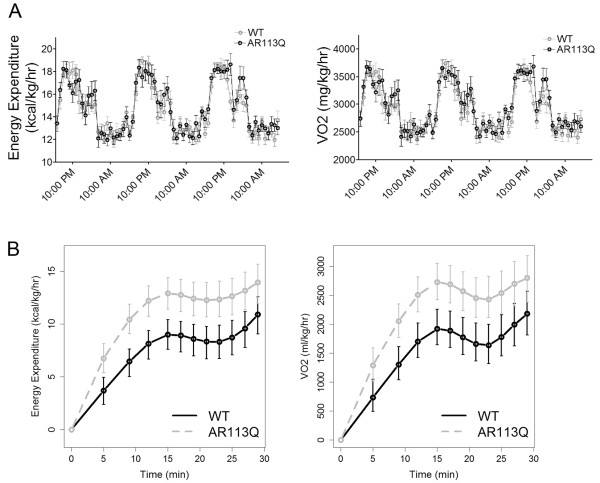
Our observations of altered carbohydrate metabolism-related gene expression and mitochondrial abnormalities led us to compare metabolic rate and substrate utilization in WT and AR113Q mice. Oxygen consumption (VO2, mg/kg/hr) and energy expenditure (kcal/kg/hr) were quantified by CLAMS analysis for three consecutive days. No significant differences in energy metabolism during home cage activity were detected between the two groups (Fig. 4A). Similarly, serum glucose and insulin levels as measured during a glucose tolerance test were unchanged in AR113Q males (Fig. S2). However, when mice were challenged to perform acute exercise, significant differences between these cohorts emerged (Fig. 4B). Mice were tasked to run on an inclined treadmill at increasing speeds for 30 minutes or until exhaustion. While both groups of mice ran the same time and distance, AR113Q mice expended more energy and consumed more oxygen to complete this task. These differences were consistent with partial mitochondrial uncoupling in AR113Q muscle (Fig. 3E), and may reflect enhanced utilization of fats or amino acids as alternative energy sources to carbohydrates.
Figure 4. Enhanced energy expenditure and oxygen consumption in AR113Q mice during exercise.
(A) Home cage energy expenditure (kcal/kg/hr) and O2 consumption (mg/kg/hr) in WT and AR113Q mice at 14 weeks, as measured during three consecutive days (mean ± SEM; n = 9/group). (B) Energy expenditure (kcal/kg/hr) and O2 consumption (mg/kg/hr) in WT and AR113Q mice during acute running at increasing speeds on an inclined treadmill (mean ± SEM; n = 9/group). *P< 0.05 for all time points from 5 to 25 minutes.
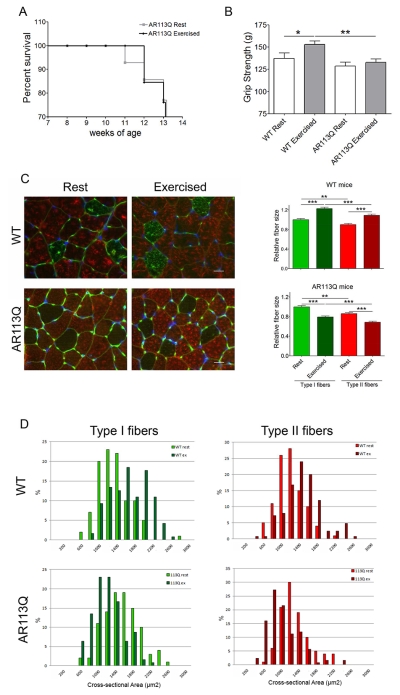
The metabolic alterations in AR113Q muscle after intense, acute exercise prompted us to determine whether mutant males would respond aberrantly to modest, chronic exercise. To test this possibility, WT and AR113Q males were randomly assigned to exercise or rest cohorts, with exercise cohorts completing a mild running regimen from 8 to 14 weeks of age. Mice were run 5 days/week on a horizontal treadmill for 30 minutes at 10 meters/minute, while control mice were placed on the immobile apparatus for the same duration to control for confounding effects from environmental enrichment. This low-intensity endurance exercise protocol was designed to favor oxidative metabolism. All WT mice completed the six weeks of training while only 7/10 AR113Q mice in the exercise cohort (70%) and 6/8 AR113Q mice in the rest cohort (75%) survived until the end of the study (Fig. 5A). This is consistent with previous analysis of lifespan in AR113Q mice (Yu et al., 2006a) and indicates that exercise did not impact survival. While chronic exercise significantly improved forelimb grip strength of WT males, it had no effect on AR113Q mice (Fig. 5B). Moreover, analysis of soleus and gastrocnemius muscles showed that both type I and type II fibers were significantly reduced in size in exercised AR113Q mice, whereas WT muscle showed the opposite effect (Fig. 5C, D). Notably, exercise did not alter polyQ AR protein levels or aggregation in mutant muscle (Fig. S3). These data demonstrate an aberrant response to chronic, low intensity exercise in AR113Q males.
Figure 5. Effects of chronic exercise in AR113Q mice.
WT and AR113Q mice were run on a treadmill 5 times/week for 30 min at 10 meters/minute for 6 weeks, from 8 – 14 weeks of age. Control mice (“rest”) were placed on an immobile treadmill apparatus for an equivalent time. (A) Survival of AR113Q males was not altered by exercise. P > 0.05 by log-rank analysis. (B) Forelimb grip strength (g) of WT and AR113Q mice assessed at the end of the six weeks training (mean ± SEM; n ≥ 4 WT mice/group; n ≥ 6 AR113Q mice/group. *P < 0.05; **P < 0.01 by one-way ANOVA. (C) Left, Immunofluorescence staining for type I slow-twitch heavy-chain myosin (green) and type II fast-twitch heavy-chain myosin (red) in soleus/gastrocnemius muscles. Basement membrane was stained with wheat germ agglutinin (green); nuclei were stained by DAPI (blue). Scale bar: 20 μm. Right, relative mean fiber size ± SEM; n ≥ 4 mice/group, ≥ 25 fibers/mouse. **P < 0.01; ***P < 0.001 by one-way ANOVA. (D) Fiber size distribution of type I and type II fibers in WT and AR113Q mice (n ≥ 4 mice/group, ≥ 25 fibers/mouse).
Peripheral polyQ AR gene suppression rescues expression of muscle metabolism genes
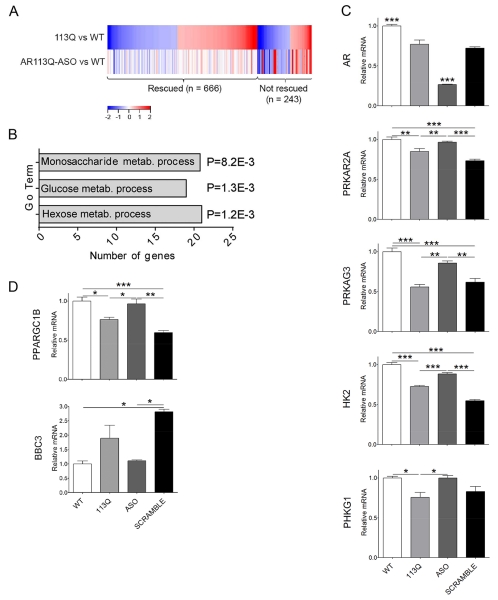
We hypothesized that peripheral expression of the polyQ AR contributed to altered expression of carbohydrate metabolism-related genes in AR113Q muscle. This notion was based on recent studies demonstrating that subcutaneous administration of Ar-targeted ASO decreases expression of the mutant protein in skeletal muscle, but not in spinal cord, and ameliorates disease (Lieberman et al., 2014). To test this hypothesis, AR113Q males received a therapeutic dose of Ar-targeted ASO (50mg/kg/week, s.c.) from 6 to 14 weeks of age. Quadriceps muscles were harvested and gene expression determined by RNA-seq. This analysis demonstrated that nearly three-quarters of the gene expression differences detected in the comparison of AR113Q versus WT muscle were rescued by peripheral treatment with ASO (Fig. 6A, Tables S1 and S3). As this intervention alleviates the disease phenotype, we focused on the 666 genes whose expression was responsive to ASO administration. Analysis of GO terms demonstrated that many of the carbohydrate metabolism-related genes whose expression was altered in AR113Q muscle were rescued by ASO treatment (Fig. 6B). Notably, of the 21 ASO-responsive genes identified in the GO term “hexose metabolic process”, 19 were expressed at lower levels in AR113Q muscle. Moreover, one of the genes expressed at higher levels in mutant muscle was Cpt1a, encoding a protein that is essential for uptake of long-chain fatty acids into mitochondria. In addition, of the nine genes encoding glycolytic enzymes that were expressed at lower levels in AR113Q muscle (Fig. 1B), all except Gapdh and Pgk1 were rescued by treatment with ASO (Tables S1, S2). We confirmed that treatment with Ar-targeted ASO, but not a non-targeted control, diminished expression of AR mRNA and rescued the expression of carbohydrate metabolism-related genes in skeletal muscle (Fig.6C). In addition, we identified ASO-responsive expression of a limited set of mitochondria-related genes including Ppargc1b (Peroxisome Proliferator-Activated Receptor Gamma, Coactivator 1 Beta, PGC-1β), a transcriptional co-activator of genes regulating mitochondrial function (Villena, 2015), and Bbc3 (BCL2 Binding Component 3), a BCL-2 family member that induces mitochondrial outer membrane permeabilization (Huang and Strasser, 2000) (Fig. 6D).
Figure 6. Peripheral ASO treatment rescues expression of metabolic genes in AR113Q muscle.
Male mice (n = 3/group) were treated with ASO (50mg/kg/week, s.c.) or vehicle from 6-14 weeks of age, and gene expression in skeletal muscle was analyzed by RNA-seq. (A) Heat map shows genes differentially expressed (≥1.5-fold) in quadriceps muscle of AR113Q versus wild type (WT) mice (top row). Differential expression in ASO-treated AR113Q versus WT muscle (bottom row) highlights genes rescued by ASO (expression difference <1.5-fold). Activated genes are shown in red and repressed genes in blue. (B) Go terms of differentially expressed genes (≥1.5-fold) related to carbohydrate metabolism that are rescued by treatment with ASO. P values by false discovery rate are indicated. (C, D) Relative mRNA levels in quadriceps muscle of WT, AR113Q and AR113Q mice treated with AR-targeted ASO or control ASO (SCRAMBLE) from 6-14 weeks of age (mean ± SEM), n = 3/group. *P < 0.05; **P < 0.01; ***P < 0.001 by one-way ANOVA. Shown in (C) is expression of AR and carbohydrate metabolism-related genes, and in (D) mitochondrial-related genes.
Altered expression of upstream regulators of energy homeostasis
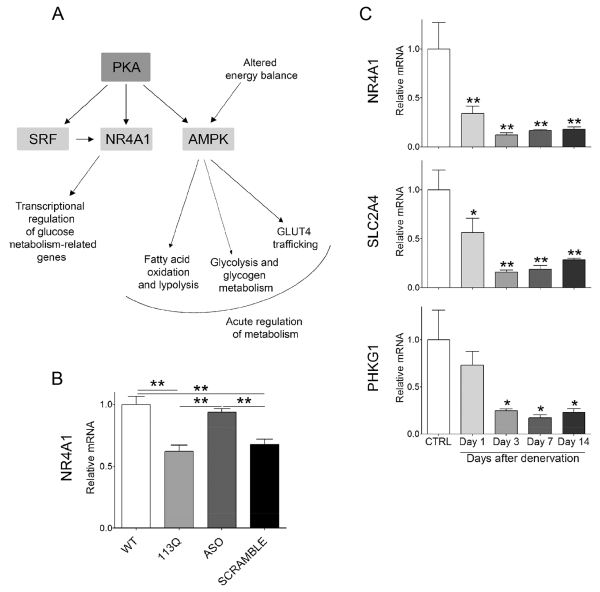
Given the large number of genes related to carbohydrate metabolism whose expression was diminished in AR113Q muscle, we reasoned that activity of upstream regulators of this pathway might be altered. To explore this possibility, we analyzed the set of ASO-responsive genes identified through RNA-seq by Ingenuity Pathway Analysis (IPA). This analysis identified several interrelated nodes of canonical pathways and upstream regulators that converge to influence energy homeostasis (Fig. 7A). Among these pathways were genes associated with protein kinase A (PKA, Prkar2a; P=4.51×10−6) (Table S3) and AMPK (5’ AMP-activated protein kinase) signaling, including the γ3 regulatory subunit of AMPK (Prkag3; P=7.72×10−14). In skeletal muscle, AMPK regulates glucose transport as well as mitochondrial function and biogenesis (reviewed in (Lantier et al., 2014)). IPA also identified transcription factors SRF (serum response factor; P=7.55×10−12) and NR4A1 (nuclear receptor subfamily 4, group A, member 1; P=9.18×10−13). NR4A1 is an orphan receptor in the steroid-thyroid hormone-retinoid receptor superfamily that is induced by diverse stimuli in skeletal muscle including PKA signaling (Kanzleiter et al., 2009; Maxwell et al., 2005) as well as the transcription factor SRF (Iacono et al., 2013; Jin et al., 2011). As NR4A1 regulates skeletal muscle expression of genes influencing glucose transport, insulin signaling, glycolysis and glycogenolysis (Chao et al., 2009; Chao et al., 2007; Kanzleiter et al., 2010; Maxwell et al., 2005), we sought to confirm its altered expression in AR113Q muscle. Significantly decreased expression of NR4A1 mRNA was found in AR113Q versus WT quadriceps, and this defect was rescued by treatment with Ar-targeted, but not control ASO (Fig. 7B). In contrast, no changes in the expression of genes encoding NR4A1 or HK2 were detected in spinal cord of AR113Q mice (Fig. S4), indicating that altered expression of carbohydrate metabolism-related genes was a phenotype more robustly manifest by skeletal muscle. Analysis of RNA-seq data indicated that 17 genes reported to respond to alterations in NR4A1 expression (Pearen and Muscat, 2010) were differentially expressed in AR113Q versus WT quadriceps (Tables S1, S4), suggesting that the activity of this transcription factor was decreased in mutant muscle. Notably, the majority of these genes encoded proteins involved in carbohydrate metabolism, and the expression of 13 of them was restored by ASO treatment.
Figure 7. Regulators of energy metabolism are sensitive to peripheral polyQ AR expression.
(A) Gene clusters identified through Ingenuity Pathway Analysis. Shown in dark grey are canonical pathways and in light grey are upstream regulators. (B) NR4A1 mRNA levels in quadriceps muscle of WT, AR113Q and AR113Q mice treated with AR-targeted ASO or control ASO (SCRAMBLE) from 6-14 weeks of age (mean ± SEM; n = 3/group). **P < 0.01 by one-way ANOVA. (C) Expression of NR4A1 and target genes that regulate carbohydrate metabolism in gastrocnemius muscles of control WT male mice following sciatic nerve transection (mean ± SEM; n = 3/group). *P < 0.05; **P < 0.01 by one-way ANOVA.
Prior studies have demonstrated that expression of NR4A1 is significantly reduced in skeletal muscle after denervation (Chao et al., 2007). We confirmed this observation in gastrocnemius muscle of WT mice following sciatic nerve transection and additionally demonstrated decreased expression of NR4A1-responsive genes Scl2a4 and Phkg1 (Fig. 7C). As these genes were also down-regulated in AR113Q muscle and restored by ASO, we conclude that peripheral polyQ AR induces changes in muscle that mimic those triggered by denervation to influence the expression of metabolic genes. This likely reflects an underlying impairment of communication between skeletal muscle and innervating motor neurons that influences the expression of regulators of skeletal muscle energy metabolism and the response to exercise in AR113Q mice.
Discussion
We demonstrate previously unrecognized alterations in skeletal muscle energy metabolism that are a consequence of peripheral polyQ AR expression (Fig. S5). RNA-seq data led to the discovery of decreased expression of numerous genes regulating carbohydrate metabolism in skeletal muscle of mutant males. These changes are accompanied by evidence of diminished activity of regulators of glucose utilization, including upstream signaling pathways controlled by PKA and AMPK and downstream transcription factors such as SRF and NR4A1. We suggest that diminished energy utilization through glycolysis leads to a shift toward alternative energy sources such as lipids, and that this contributes to mitochondrial damage and dysfunction. The consequences of these metabolic alterations are evident during intense, acute exercise that favors anaerobic metabolism and following chronic, low intensity endurance exercise that favors oxidative metabolism.
Interestingly, we note that similar alterations in gene expression occur following sciatic nerve transection in wild type mice. Our observations are consistent with prior reports indicating that loss of β-adrenergic signaling following denervation contributes to diminished PKA activity and reduced expression of NR4A1 and its target genes (Chao et al., 2007). Metabolic alterations characterized by a glycolytic to oxidative shift also occur in skeletal muscle of transgenic mice expressing mutant G86R SOD1 (Palamiuc et al., 2015), and clinical studies have demonstrated that approximately two thirds of patients with amyotrophic lateral sclerosis are hypermetabolic (Desport et al., 2005). These findings support the concept that muscle energy metabolism is altered in degenerative motor neuron diseases as a consequence of denervation. Our data indicate that similar changes occur in AR113Q mice due to peripheral expression of the polyQ AR. The identification of ASO-sensitive changes in the expression of carbohydrate metabolism genes complements previously characterized changes in markers of denervation, including acetylcholine receptor-α subunit and myogenin mRNAs, that are also abnormal in AR113Q muscle and rescued by peripheral gene knockdown (Lieberman et al., 2014). Taken together, these findings indicate that peripherally expressed polyQ AR disrupts communication between motor neurons and skeletal muscle, and recapitulates gene expression and metabolic alterations in skeletal muscle that have been previously characterized to occur following injury to motor neurons or their axons. Prior studies have demonstrated impaired retrograde labeling of spinal motor neurons in AR113Q mice despite the absence of morphological abnormalities at the neuromuscular junction that are evident by light microscopy (Kemp et al., 2011), suggesting that disruption of nerve-muscle communication may initially reflect functional, rather than structural changes.
The model of pathogenesis that we present (Fig. S5) places alterations in carbohydrate metabolism upstream of the mitochondrial abnormalities that were identified in AR113Q muscle, including diminished mitochondrial number, increased mitophagy and partial mitochondrial uncoupling. This latter effect may be a consequence of damage by reactive oxygen species (Mailloux and Harper, 2012), perhaps arising due to the use of fats as an important, alternative energy source. This model is based on gene expression analysis that showed a large number of changes involving regulators of glucose metabolism compared to components of the TCA cycle, and metabolomics analysis that showed significantly lower levels of glycolytic intermediates without alterations in TCA cycle metabolites. We note that it is also possible that the polyQ AR may directly damage mitochondria, as previously suggested (Ranganathan et al., 2009), and that in this setting alterations in glycolysis may occur secondary to mitochondrial damage. Indeed, it is possible that both pathways are directly targeted by the polyQ AR. Further analysis to clarify the relationship between changes in glycolysis and mitochondria in SBMA muscle will be an important focus of future studies.
The occurrence of metabolic abnormalities in AR113Q muscle underlying an impaired response to exercise suggests that these changes are important aspects of the neuromuscular phenotype of SBMA patients. Indeed, a recent report showed decreased levels of HK2, PFKFB3 and GAPDH mRNA in skeletal muscle from SBMA patients, accompanied by a glycolytic to oxidative shift in muscle fibers (Rocchi et al., 2016). These alterations may have contributed to the modest effects of exercise observed in a randomized study of SBMA patients (Shrader et al., 2015), and in the poor tolerance of SBMA patients to aerobic exercise in an observational study (Dahlqvist and Vissing, 2015). Moreover, skeletal muscle plays a critical role in energy homeostasis for the organism, accounting for approximately 25% of energy expenditure in the resting state. As such, altered energy utilization by skeletal muscle may play a part in the systemic manifestations of disease. Recent studies have demonstrated elevated fasting glucose as well as increased total cholesterol, low-density lipoprotein cholesterol and triglyceride levels in a cohort of SBMA patients (Querin et al., 2015). These findings add to a growing appreciation of non-neuronal phenotypes in this disorder, and raise the possibility that targeting metabolic alterations may alleviate disease manifestations. In support of this hypothesis is recent work demonstrating that feeding mice a high fat diet diminishes aspects of the SBMA phenotype (Rocchi et al., 2016). Additional evidence suggests that enhancing anabolic support of muscle is also beneficial in SBMA mice. Abrogation of SUMO modification of the polyQ AR enhances its activity as a ligand-dependent transcription factor, increases the expression of genes related to mitochondria function in skeletal muscle, and rescues both exercise endurance and type I muscle fiber atrophy in mutant mice (Chua et al., 2015).
Derangements of energy utilization and homeostasis have emerged as important components of the pathophysiology of many age-dependent neurodegenerative disorders and of aging itself. The changes documented here in AR113Q skeletal muscle indicate that similar processes occur in SBMA. Moreover, our analysis suggests that these changes arise due to expression of the polyQ AR in muscle and demonstrates that they are mitigated by peripherally targeted therapies. The extent to which similar metabolic alterations also impair function of lower motor neurons in SBMA patients remains to be more fully explored, although our initial analysis of AR113Q mice suggests less severe gene expression changes in spinal cord than in skeletal muscle. As such, we speculate that targeting the metabolic phenotype trigged by expression of the polyQ AR may be particularly effective in rescuing non-neuronal manifestations of the disease, including important aspects of skeletal muscle dysfunction.
Experimental Procedures
Mice
Gene targeted mice with an Ar allele containing 113 CAG repeats (AR113Q mice) were previously described (Yu et al., 2006a; Yu et al., 2006b). Mice were group housed in a specific pathogen–free (SPF) facility, provided with chow and water ad libitum and maintained on a constant 12 hr light/12 hr dark cycle. Genotypes were verified by PCR of DNA harvested from tail biopsies obtained shortly after weaning, using the forward primer 5’-CCAGAATCTGTTCCAGAGCGTG-3’ and the reverse primer 5’-TGTTCCCCTGGACTCAGATG-3’.
ASO treatment
AR113Q male mice and wild type littermates were treated for 8 weeks with antisense oligonucleotides (ASO). The 16-mer, 2’,4’-constrained ethyl gapmer ASO complementary to human and mouse AR transcripts and the 20-mer control has been described previously (Lieberman et al., 2014). Subcutaneous administration of 50 mg/kg body weight of AR targeted ASO (ASO1 from ref) or control ASO (SCRAMBLE; Control ASO from ref) was performed once a week from 6 to 14 weeks of age (n = 3 per group). Quadriceps muscle was harvested at 14 weeks for analysis of mRNA expression.
Sciatic nerve transection
C57BL/6J male mice at 7 weeks of age were used for studies of denervated muscle as detailed in the Supplemental Experimental Procedures.
All procedures involving mice were approved by the University of Michigan Committee on Use and Care of Animals, in accord with the NIH Guidelines for the Care and Use of Experimental Animals.
Gene expression
Total RNA was isolated from quadriceps and LA/BC muscles with TRIzol Reagent (Life Technologies) according to the manufacturer’s instructions. RNA (1 μg) was reverse transcribed to cDNA using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Gene-specific primers with FAM-labeled probes were from Applied Biosystems: Nr4a1, Mm01300401_m1; Prkar2a, Mm00435916_m1; Prkag3, Mm00463997_m1; Hk2, Mm00443385_m1; Slc2a4, Mm01245502_m1; Phkg1, Mm02580948_m1; Pygm, Mm00478582_m1; Ppargc1a, Mm01208835_m1; Ppargc1b, Mm00504720_m1; Bbc3, Mm00519268_m1. AR-specific forward primer 5’-CCAGTCCCAATTGTGTCAAA-3’ and reverse primer 5’-TCCCTGGTACTGTCCAAACG-3’ (Life Technologies) were used with Roche Universal Probe Library FAM-labeled probe #58 (cat.no. 04688554001). Cpsf2-specific primers with VIC-labeled probes (Applied Biosystems; Mm00489754_m1) were used as internal control. Quantitative PCR (qPCR) was carried out using FastStart TaqMan Probe Master Mix (Roche) on software supplied with a 7500 Real-Time PCR SDS System (Applied Biosystems). PCR cycling conditions were as follows: 50°C for 2 min, 95°C for 10 min, 40 cycles at 95°C for 15 s and 60°C for 1min.
RNA-seq data
For RNA-Seq, cDNA was prepared into sequencer-ready fragment libraries using the Illumina TruSeq mRNA-Seq Sample Prep Kit according to manufacturer’s protocols, and sequenced on the Illumina HiSeq 2000 using manufacturer’s protocols for paired-end, 100 nt sequencing. Samples were multiplexed into pools of six samples each, and each pool was sequenced on one lane of the sequencer. The software package Tuxedo Suite was used for alignment, differential expression analysis, and postanalysis diagnostics (Langmead et al., 2009; Trapnell et al., 2013; Trapnell et al., 2009). The reference transcriptome (UCSC mm10) (http://genome.ucsc.edu/) was used for alignment. Expression quantitation and differential expression analysis were performed using CuffDiff (Cufflinks version 2.1.1). Genes and transcripts were identified as differentially expressed based on 3 criteria: test status = OK, FDR <0.05, and fold change ≥ 1.5. Analysis of differentially expressed genes to identify significantly enriched functional categories was performed using DAVID and Ingenuity Pathway Analysis (Qiagen). The heat map was plotted in R using the gplots and RcolorBrewer packages.
Transmission electron microscopy
Mice were perfused with 2.5% glutaraldehyde in 0.1M phosphate buffer, pH 7.4, and soleus muscle was collected. Ultra-thin tissue sections were obtained with a diamond knife, placed on copper grids and stained with uranyl acetate and lead citrate. Image acquisition was performed with a Philips CM-100 transmission electron microscope. Total mitochondria number and vesicle-associated mitochondria were manually counted (n = 3 mice/group, ≥ 500 mitochondria/group). Mitochondria area (μm2) was measured by ImageJ software (n = 3 mice/group, ≥ 250 mitochondria/group).
Mitochondrial DNA copy number
Genomic and mitochondrial DNA was isolated from LA/BC using the QIAamp® DNA Micro Kit (Qiagen). D-loop forward primer 5’- GACCAACATAACTGTGGTGTCA-3’ and reverse primer 5’- ATTCTTCTCCGTAGGTGCGTCTAG-3’ were designed to quantify mitochondrial DNA; Beta-2 microglobulin (B2m) forward primer 5’- TGTCAGATATGTCCTTCAGCAAGG-3’ and reverse primer 5’- TGCTTAACTCTGCAGGCGTATG-3’ were designed to quantify genomic DNA as an internal control. qPCR was carried out using SYBR Select Master mix (Life Technologies) on software supplied with a 7500 Real-Time PCR SDS System (Applied Biosystems). PCR cycling conditions were as follows: 50°C for 2 min, 95°C fo r 10 min, 40 cycles at 95°C for 15 s and 60°C for 1min. Data were expressed as Ct values and used for the relative quantification of targets with the ΔΔCt calculation to give N-fold differences. Data were transformed through the equation 2−ΔΔCt.
Mitochondria uncoupling
Mitochondria were isolated from quadriceps muscles of WT and AR113Q mice, and uncoupling was assessed as detailed in the Supplemental Experimental Procedures.
CLAMS and glucose tolerance test
Oxygen consumption (VO2), carbon dioxide production (VCO2), spontaneous motor activity and food intake were measured using the Comprehensive Laboratory Monitoring System (CLAMS, Columbus Instruments), an integrated open-circuit calorimeter equipped with an optical beam activity monitor. This analysis and glucose tolerance testing was performed by the University of Michigan Metabolomics Core and is described in detail in the Supplemental Experimental Procedures.
Metabolomics
WT and AR113Q male mice ran on an Exer3/6 treadmill (Columbus Instruments) with 25°C slope for 15 minutes at increasing speeds (as described for CLAMS analysis). Animals were induced into anesthesia at a dose of 5% isoflurane, and then maintained by continuous inhalation of 2% isoflurane. Quadriceps muscles were collected and flash frozen in liquid nitrogen for analysis of metabolites using LC-MS and GC-MS, by the University of Michigan Metabolomics Core Facility, as detailed in the Supplemental Experimental Procedures.
Chronic exercise
WT and AR113Q mice were run on an Exer3/6 treadmill (Columbus Instruments) from 8 – 14 weeks of age, 30 min/day, 5 days/week, at 10 m/min fixed-speed. Control mice were placed on an immobile treadmill apparatus for the same amount of time. To analyze exercise capacity, a protocol for treadmill running was used as previously described (Chua et al., 2015). Briefly, mice underwent 2 days of training on a treadmill, with day 1 consisting of 5 minutes of running at 8 m/min and day 2 consisting of 5 minutes of running at 8 m/min followed by 5 minutes at 10 m/min. On day 3, mice ran using a graded protocol consisting of 10 m/min for 40 minutes, then increasing speed by increments of 1 m/min every 10 minutes for a total of 30 minutes. This was followed by increasing speed at increments of 1 m/min every 5 min until mice were exhausted. Exhaustion was defined as mice making no attempt to exercise for 5 seconds. Distance run and time to exhaustion were recorded. At the end of the six weeks training, body weight (g) and forelimb grip strength (g) were also measured.
Immunoprecipitation and western blotting
Quadriceps muscles were homogenized in RIPA buffer containing 1 tablet/10 ml of Complete Mini-Protease Inhibitor Mix (Roche), rotated for 2 hours at 4°C and centrifuged at 13,000g for 10 minutes at 4°C. The supernatant fraction (5 00 μg) was incubated on a rotator overnight at 4°C with 10 μl of AR antibody (Millipore, PG-21) or nonimmune rabbit IgG (Santa Cruz Biotechnology). Prewashed protein A–agarose beads (20 μl, Santa Cruz Biotechnology) were added, and samples were incubated for 1 additional hour at 4°C. Protein-antibody-bead complexes were washed 6 times in RIPA buffer in filtered spin columns (Thermo Scientific) and proteins were eluted by boiling in SDS-loading buffer for 5 minutes at 100°C. Samples were then resolved on a 7.5% SDS-PAGE gel.
Immunofluorescence staining
Quadriceps and soleus/gastrocnemius muscles were placed on cork and embedded in O.C.T. compound (Optimal Cutting Temperature, Tissue-Tek) immediately after collection. Cryosections were prepared using a Cryocut 1800 cryostat (Leica) at 10 μm and placed on Superfrost® Plus Microscope Slides (Fischer Scientific). Briefly, slides were incubated in acetone for 10 min, then non-specific binding was blocked for 30 min at 37°C with 5% donkey serum in PBS-T (Tween 0.1% in PBS) and samples were incubated overnight at 4°C with the indicated antibodies in 1.5% donkey serum in PBS-T. Fluorescent secondary antibody was then applied for 1 hour at 37°C in 1.5% donkey seru m in PBS-T. Basement membrane was stained with wheat germ agglutinin (WGA, green) at 5 μg/mL in PBS for 10 min at RT. The slides were mounted using Vectashield with DAPI (Vector Laboratories) and sealed with nail polish. Muscle fiber size was measured by ImageJ (n = 4 mice/group, ≥ 25 fibers/mouse).
Antibodies
The following antibodies were used: anti-AR (PG-21, Merck Millipore; N20, Santa Cruz), anti-GAPDH (6C5, Novus Biologicals), type I/slow-twitch heavy-chain myosin (A4.840, DSHB, University of Iowa, Iowa City, Iowa, USA), and type II/fast-twitch heavy-chain myosin (Ab-2, Thermo Scientific). Horseradish peroxidase–conjugated secondary antibodies used for Western blot analysis were from Bio-Rad. Goat anti-mouse IgG Alexa Fluor 555, goat anti-rabbit Alexa Fluor 488, and wheat germ agglutinin Alexa Fluor 488 conjugate antibodies used for immunofluorescence were from Life Technologies. FITC-conjugated donkey anti-mouse IgM was from Jackson ImmunoResearch.
Statistics
Data were analyzed using unpaired 2-tailed Student’s t test or one-way ANOVA with Newman-Keuls multiple comparison test in Prism 4 (GraphPad). Statistical significance was considered at P < 0.05. Low-rank thin-plate splines were used to estimate the curves of energy expenditure and VO2 over time (Ruppert et al., 2003). The model includes a random effect for the intercept to account for correlations between measurements on the same subject. At a time point, comparisons between groups (WT vs. AR113Q mice) were made using posterior distributions of the estimated outcome (such as VO2) for each group. Statistical significance was determined by a 95% credible set for the difference. If the credible set does not include zero, the difference is statistically significant. WinBUGS (Version 1.4) was used for the analysis.
Supplementary Material
Acknowledgements
We thank Dr. Mark Schultz for help with figure preparation. Work in this project utilized services from the University of Michigan Metabolomics Core and the University of Michigan Microscopy & Image Analysis Laboratory.
Funding
This work was supported by the United States National Institutes of Health (R21 NS089516, R01 NS055746 to A.P.L.; U24 DK097153 to the University of Michigan Metabolomics Core) and Telethon-Italy and Provincia Autonoma di Trento (TCP12013 to M.P).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
EG, ZY, JPC, RS: performed experiments
LZ, FZ, YG: performed statistical analysis
SV, MP, GH, APL: planned experiments, interpreted data
GH: supplied essential reagents
EG, APL: wrote paper
Accession Numbers
The GEO accession number for the RNA-Seq data is GSE60691.
References
- Atsuta N, Watanabe H, Ito M, Banno H, Suzuki K, Katsuno M, Tanaka F, Tamakoshi A, Sobue G. Natural history of spinal and bulbar muscular atrophy (SBMA): a study of 223 Japanese patients. Brain. 2006;129:1446–1455. doi: 10.1093/brain/awl096. [DOI] [PubMed] [Google Scholar]
- Chamberlain NL, Driver ED, Miesfeld RL. The length and location of CAG trinucleotide repeats in the androgen receptor N-terminal domain affect transactivation function. Nucleic Acids Res. 1994;22:3181–3186. doi: 10.1093/nar/22.15.3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao LC, Wroblewski K, Zhang Z, Pei L, Vergnes L, Ilkayeva OR, Ding SY, Reue K, Watt MJ, Newgard CB, et al. Insulin resistance and altered systemic glucose metabolism in mice lacking Nur77. Diabetes. 2009;58:2788–2796. doi: 10.2337/db09-0763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao LC, Zhang Z, Pei L, Saito T, Tontonoz P, Pilch PF. Nur77 coordinately regulates expression of genes linked to glucose metabolism in skeletal muscle. Mol Endocrinol. 2007;21:2152–2163. doi: 10.1210/me.2007-0169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua JP, Lieberman AP. Pathogenic mechanisms and therapeutic strategies in spinobulbar muscular atrophy. CNS Neurol Disord Drug Targets. 2013;12:1146–1156. [PMC free article] [PubMed] [Google Scholar]
- Chua JP, Reddy SL, Merry DE, Adachi H, Katsuno M, Sobue G, Robins DM, Lieberman AP. Transcriptional activation of TFEB/ZKSCAN3 target genes underlies enhanced autophagy in spinobulbar muscular atrophy. Hum Mol Genet. 2014;23:1376–1386. doi: 10.1093/hmg/ddt527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua JP, Reddy SL, Yu Z, Giorgetti E, Montie HL, Mukherjee S, Higgins J, McEachin RC, Robins DM, Merry DE, et al. Disrupting SUMOylation enhances transcriptional function and ameliorates polyglutamine androgen receptor-mediated disease. J Clin Invest. 2015;125:831–845. doi: 10.1172/JCI73214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes CJ, Ling SC, Guo LT, Hung G, Tsunemi T, Ly L, Tokunaga S, Lopez E, Sopher BL, Bennett CF, et al. Muscle expression of mutant androgen receptor accounts for systemic and motor neuron disease phenotypes in spinal and bulbar muscular atrophy. Neuron. 2014;82:295–307. doi: 10.1016/j.neuron.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlqvist JR, Vissing J. Exercise Therapy in Spinobulbar Muscular Atrophy and Other Neuromuscular Disorders. J Mol Neurosci. 2016;58:388–393. doi: 10.1007/s12031-015-0686-3. [DOI] [PubMed] [Google Scholar]
- Desport JC, Torny F, Lacoste M, Preux PM, Couratier P. Hypermetabolism in ALS: correlations with clinical and paraclinical parameters. Neurodegener Dis. 2005;2:202–207. doi: 10.1159/000089626. [DOI] [PubMed] [Google Scholar]
- Huang DC, Strasser A. BH3-Only proteins-essential initiators of apoptotic cell death. Cell. 2000;103:839–842. doi: 10.1016/s0092-8674(00)00187-2. [DOI] [PubMed] [Google Scholar]
- Iacono G, Altafini C, Torre V. Early phase of plasticity-related gene regulation and SRF dependent transcription in the hippocampus. PLoS One. 2013;8:e68078. doi: 10.1371/journal.pone.0068078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin W, Goldfine AB, Boes T, Henry RR, Ciaraldi TP, Kim EY, Emecan M, Fitzpatrick C, Sen A, Shah A, et al. Increased SRF transcriptional activity in human and mouse skeletal muscle is a signature of insulin resistance. J Clin Invest. 2011;121:918–929. doi: 10.1172/JCI41940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen JA, Yu Z, Mo K, Monks DA, Lieberman AP, Breedlove SM, Jordan CL. Recovery of function in a myogenic mouse model of spinal bulbar muscular atrophy. Neurobiol Dis. 2009;34:113–120. doi: 10.1016/j.nbd.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanzleiter T, Preston E, Wilks D, Ho B, Benrick A, Reznick J, Heilbronn LK, Turner N, Cooney GJ. Overexpression of the orphan receptor Nur77 alters glucose metabolism in rat muscle cells and rat muscle in vivo. Diabetologia. 2010;53:1174–1183. doi: 10.1007/s00125-010-1703-2. [DOI] [PubMed] [Google Scholar]
- Kanzleiter T, Wilks D, Preston E, Ye J, Frangioudakis G, Cooney GJ. Regulation of the nuclear hormone receptor nur77 in muscle: influence of exercise-activated pathways in vitro and obesity in vivo. Biochim Biophys Acta. 2009;1792:777–782. doi: 10.1016/j.bbadis.2009.05.002. [DOI] [PubMed] [Google Scholar]
- Katsuno M, Adachi H, Kume A, Li M, Nakagomi Y, Niwa H, Sang C, Kobayashi Y, Doyu M, Sobue G. Testosterone reduction prevents phenotypic expression in a transgenic mouse model of spinal and bulbar muscular atrophy. Neuron. 2002;35:843–854. doi: 10.1016/s0896-6273(02)00834-6. [DOI] [PubMed] [Google Scholar]
- Kazemi-Esfarjani P, Trifiro MA, Pinsky L. Evidence for a repressive function of the long polyglutamine tract in the human androgen receptor: possible pathogenetic relevance for the (CAG)n-expanded neuronopathies. Hum Mol Genet. 1995;4:523–527. doi: 10.1093/hmg/4.4.523. [DOI] [PubMed] [Google Scholar]
- Kemp MQ, Poort JL, Baqri RM, Lieberman AP, Breedlove SM, Miller KE, Jordan CL. Impaired motoneuronal retrograde transport in two models of SBMA implicates two sites of androgen action. Hum Mol Genet. 2011;20:4475–4490. doi: 10.1093/hmg/ddr380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch LG, Britton SL. Artificial selection for intrinsic aerobic endurance running capacity in rats. Physiol Genomics. 2001;5:45–52. doi: 10.1152/physiolgenomics.2001.5.1.45. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lantier L, Fentz J, Mounier R, Leclerc J, Treebak JT, Pehmoller C, Sanz N, Sakakibara I, Saint-Amand E, Rimbaud S, et al. AMPK controls exercise endurance, mitochondrial oxidative capacity, and skeletal muscle integrity. Faseb J. 2014;28:3211–3224. doi: 10.1096/fj.14-250449. [DOI] [PubMed] [Google Scholar]
- Lieberman AP, Fischbeck KH. Triplet repeat expansion in neuromuscular disease. Muscle Nerve. 2000;23:843–850. doi: 10.1002/(sici)1097-4598(200006)23:6<843::aid-mus2>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- Lieberman AP, Harmison G, Strand AD, Olson JM, Fischbeck KH. Altered transcriptional regulation in cells expressing the expanded polyglutamine androgen receptor. Hum Mol Genet. 2002;11:1967–1976. doi: 10.1093/hmg/11.17.1967. [DOI] [PubMed] [Google Scholar]
- Lieberman AP, Yu Z, Murray S, Peralta R, Low A, Guo S, Yu XX, Cortes CJ, Bennett CF, Monia BP, et al. Peripheral androgen receptor gene suppression rescues disease in mouse models of spinal and bulbar muscular atrophy. Cell Rep. 2014;7:774–784. doi: 10.1016/j.celrep.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailloux RJ, Harper ME. Mitochondrial proticity and ROS signaling: lessons from the uncoupling proteins. Trends Endocrinol Metab. 2012;23:451–458. doi: 10.1016/j.tem.2012.04.004. [DOI] [PubMed] [Google Scholar]
- Maxwell MA, Cleasby ME, Harding A, Stark A, Cooney GJ, Muscat GE. Nur77 regulates lipolysis in skeletal muscle cells. Evidence for cross-talk between the beta-adrenergic and an orphan nuclear hormone receptor pathway. J Biol Chem. 2005;280:12573–12584. doi: 10.1074/jbc.M409580200. [DOI] [PubMed] [Google Scholar]
- Mhatre AN, Trifiro MA, Kaufman M, Kazemi-Esfarjani P, Figlewicz D, Rouleau G, Pinsky L. Reduced transcriptional regulatory competence of the androgen receptor in X-linked spinal and bulbar muscular atrophy. Nat Genet. 1993;5:184–188. doi: 10.1038/ng1093-184. [DOI] [PubMed] [Google Scholar]
- Monks DA, Johansen JA, Mo K, Rao P, Eagleson B, Yu Z, Lieberman AP, Breedlove SM, Jordan CL. Overexpressioin of wild-type androgen receptor in muscle recapitulates polyglutamine disease. Proc Natl Acad Sci U S A. 2007;104:18259–18264. doi: 10.1073/pnas.0705501104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overmyer KA, Thonusin C, Qi NR, Burant CF, Evans CR. Impact of anesthesia and euthanasia on metabolomics of mammalian tissues: studies in a C57BL/6J mouse model. PLoS One. 2015;10:e0117232. doi: 10.1371/journal.pone.0117232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palamiuc L, Schlagowski A, Ngo ST, Vernay A, Dirrig-Grosch S, Henriques A, Boutillier AL, Zoll J, Echaniz-Laguna A, Loeffler JP, et al. A metabolic switch toward lipid use in glycolytic muscle is an early pathologic event in a mouse model of amyotrophic lateral sclerosis. EMBO Mol Med. 2015;7:526–546. doi: 10.15252/emmm.201404433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palazzolo I, Stack C, Kong L, Musaro A, Adachi H, Katsuno M, Sobue G, Taylor JP, Sumner CJ, Fischbeck KH, et al. Overexpression of IGF-1 in muscle attenuates disease in a mouse model of spinal and bulbar muscular atrophy. Neuron. 2009;63:316–328. doi: 10.1016/j.neuron.2009.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearen MA, Muscat GE. Minireview: Nuclear hormone receptor 4A signaling: implications for metabolic disease. Mol Endocrinol. 2010;24:1891–1903. doi: 10.1210/me.2010-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Querin G, Bertolin C, Da Re E, Volpe M, Zara G, Pegoraro E, Caretta N, Foresta C, Silvano M, Corrado D, et al. Non-neural phenotype of spinal and bulbar muscular atrophy: results from a large cohort of Italian patients. J Neurol Neurosurg Psychiatry. 2015;87:810–816. doi: 10.1136/jnnp-2015-311305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramzan F, McPhail M, Rao P, Mo K, Halievski K, Swift-Gallant A, Mendoza-Viveros L, Cheng HY, Monks DA. Distinct Etiological Roles for Myocytes and Motor Neurons in a Mouse Model of Kennedy’s Disease/Spinobulbar Muscular Atrophy. J Neurosci. 2015;35:6444–6451. doi: 10.1523/JNEUROSCI.3599-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranganathan S, Harmison GG, Meyertholen K, Pennuto M, Burnett BG, Fischbeck KH. Mitochondrial abnormalities in spinal and bulbar muscular atrophy. Hum Mol Genet. 2009;18:27–42. doi: 10.1093/hmg/ddn310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes LE, Freeman BK, Auh S, Kokkinis AD, La Pean A, Chen C, Lehky TJ, Shrader JA, Levy EW, Harris-Love M, et al. Clinical features of spinal and bulbar muscular atrophy. Brain. 2009;132:3242–3251. doi: 10.1093/brain/awp258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocchi A, Milioto C, Parodi S, Armirotti A, Borgia D, Pellegrini M, Urciuolo A, Molon S, Morbidoni V, Marabita M, et al. Glycolytic-to-oxidative fiber-type switch and mTOR signaling activation are early-onset features of SBMA muscle modfied by high-fat diet. Acta Neuropathol. 2016;132:127–144. doi: 10.1007/s00401-016-1550-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruppert D, Wand M, Carroll . Semiparametric regression. Cambridge University Press; Cambridge: 2003. [Google Scholar]
- Shrader JA, Kats I, Kokkinis A, Zampieri C, Levy E, Joe GO, Woolstenhulme JG, Drinkard BE, Smith MR, Ching W, et al. A randomized controlled trial of exercise in spinal and bulbar muscular atrophy. Ann Clin Transl Neurol. 2015;2:739–747. doi: 10.1002/acn3.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soraru G, D’Ascenzo C, Polo A, Palmieri A, Baggio L, Vergani L, Gellera C, Moretto G, Pegoraro E, Angelini C. Spinal and bulbar muscular atrophy: skeletal muscle pathology in male patients and heterozygous females. J Neurol Sci. 2008;264:100–105. doi: 10.1016/j.jns.2007.08.012. [DOI] [PubMed] [Google Scholar]
- Takeyama K, Ito S, Yamamoto A, Tanimoto H, Furutani T, Kanuka H, Miura M, Tabata T, Kato S. Androgen-dependent neurodegeneration by polyglutamine-expanded human androgen receptor in Drosophila. Neuron. 2002;35:855–864. doi: 10.1016/s0896-6273(02)00875-9. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. 2013;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villena JA. New insights into PGC-1 coactivators: redefining their role in the regulation of mitochondrial function and beyond. FEBS J. 2015;282:647–672. doi: 10.1111/febs.13175. [DOI] [PubMed] [Google Scholar]
- Yu Z, Dadgar N, Albertelli M, Gruis K, Jordan C, Robins DM, Lieberman AP. Androgen-dependent pathology demonstrates myopathic contribution to the Kennedy disease phenotype in a mouse knock-in model. J Clin Invest. 2006a;116:2663–2672. doi: 10.1172/JCI28773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Dadgar N, Albertelli M, Scheller A, Albin RL, Robins DM, Lieberman AP. Abnormalities of Germ Cell Maturation and Sertoli Cell Cytoskeleton in Androgen Receptor 113 CAG Knock-In Mice Reveal Toxic Effects of the Mutant Protein. Am J Pathol. 2006b;168:195–204. doi: 10.2353/ajpath.2006.050619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Wang AM, Adachi H, Katsuno M, Sobue G, Yue Z, Robins DM, Lieberman AP. Macroautophagy Is Regulated by the UPR-Mediator CHOP and Accentuates the Phenotype of SBMA Mice. PLoS Genet. 2011;7:e1002321. doi: 10.1371/journal.pgen.1002321. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.