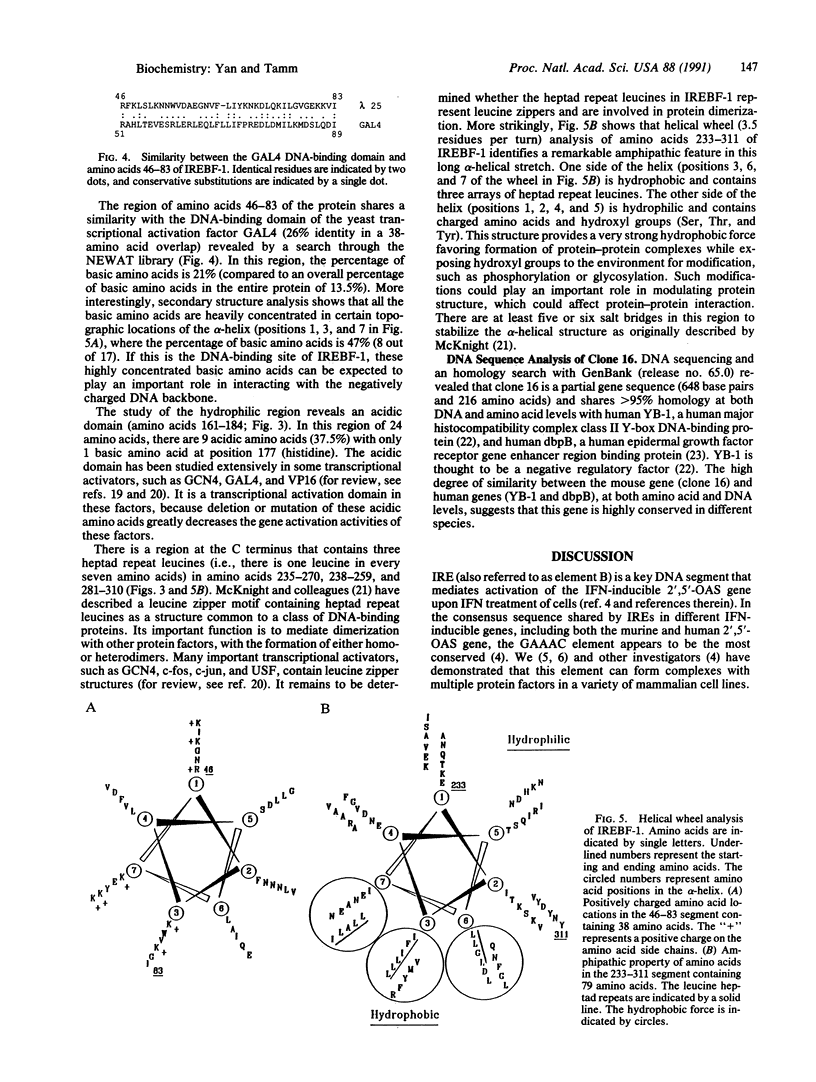
Abstract
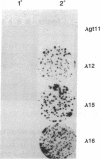
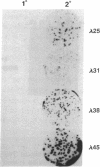
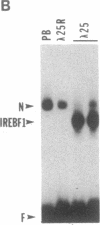
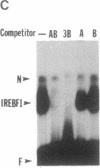
Seven clones encoding interferon response element binding factors have been isolated from a mouse fibroblast lambda gt11 cDNA library by using a 32P end-labeled tandem trimer of the mouse (2'-5')oligoadenylate synthetase gene interferon response element as a probe. Clone 16 shares strong similarity (95%) at both DNA and amino acid level with YB-1, a human major histocompatibility complex class II Y-box DNA-binding protein, and with dbpB, a human epidermal growth factor receptor gene enhancer region binding protein. The product of the gene represented by clone 16 may represent a factor that regulates multiple genes by binding to a variety of 5' regulatory elements. Clone 25 is a 2407-base-pair-long cDNA and contains a putative 311-amino acid open reading frame corresponding to an estimated mass of 35.5 kDa. This putative protein, designated as interferon response element binding factor 1 (IREBF-1), contains an acidic domain, three heptad repeat leucine arrays, and a region that shares similarity with the yeast transcriptional factor GAL4 DNA-binding domain. Furthermore, the C terminus of IREBF-1 shows an unusual amphipathic property: within a 79-amino acid range, one side of the alpha-helical region contains a preponderance of hydrophobic amino acids and the other side contains hydrophilic amino acids. This type of structure provides a strong hydrophobic force for protein-protein interaction.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cohen B., Peretz D., Vaiman D., Benech P., Chebath J. Enhancer-like interferon responsive sequences of the human and murine (2'-5') oligoadenylate synthetase gene promoters. EMBO J. 1988 May;7(5):1411–1419. doi: 10.1002/j.1460-2075.1988.tb02958.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didier D. K., Schiffenbauer J., Woulfe S. L., Zacheis M., Schwartz B. D. Characterization of the cDNA encoding a protein binding to the major histocompatibility complex class II Y box. Proc Natl Acad Sci U S A. 1988 Oct;85(19):7322–7326. doi: 10.1073/pnas.85.19.7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driggers P. H., Ennist D. L., Gleason S. L., Mak W. H., Marks M. S., Levi B. Z., Flanagan J. R., Appella E., Ozato K. An interferon gamma-regulated protein that binds the interferon-inducible enhancer element of major histocompatibility complex class I genes. Proc Natl Acad Sci U S A. 1990 May;87(10):3743–3747. doi: 10.1073/pnas.87.10.3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada H., Fujita T., Miyamoto M., Kimura Y., Maruyama M., Furia A., Miyata T., Taniguchi T. Structurally similar but functionally distinct factors, IRF-1 and IRF-2, bind to the same regulatory elements of IFN and IFN-inducible genes. Cell. 1989 Aug 25;58(4):729–739. doi: 10.1016/0092-8674(89)90107-4. [DOI] [PubMed] [Google Scholar]
- Hope I. A., Mahadevan S., Struhl K. Structural and functional characterization of the short acidic transcriptional activation region of yeast GCN4 protein. Nature. 1988 Jun 16;333(6174):635–640. doi: 10.1038/333635a0. [DOI] [PubMed] [Google Scholar]
- Johnson P. F., McKnight S. L. Eukaryotic transcriptional regulatory proteins. Annu Rev Biochem. 1989;58:799–839. doi: 10.1146/annurev.bi.58.070189.004055. [DOI] [PubMed] [Google Scholar]
- Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986 Jan 31;44(2):283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- Landschulz W. H., Johnson P. F., McKnight S. L. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science. 1988 Jun 24;240(4860):1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- Levy D. E., Kessler D. S., Pine R., Darnell J. E., Jr Cytoplasmic activation of ISGF3, the positive regulator of interferon-alpha-stimulated transcription, reconstituted in vitro. Genes Dev. 1989 Sep;3(9):1362–1371. doi: 10.1101/gad.3.9.1362. [DOI] [PubMed] [Google Scholar]
- Levy D. E., Kessler D. S., Pine R., Reich N., Darnell J. E., Jr Interferon-induced nuclear factors that bind a shared promoter element correlate with positive and negative transcriptional control. Genes Dev. 1988 Apr;2(4):383–393. doi: 10.1101/gad.2.4.383. [DOI] [PubMed] [Google Scholar]
- Ma J., Ptashne M. Deletion analysis of GAL4 defines two transcriptional activating segments. Cell. 1987 Mar 13;48(5):847–853. doi: 10.1016/0092-8674(87)90081-x. [DOI] [PubMed] [Google Scholar]
- Miller P. F., Hinnebusch A. G. Sequences that surround the stop codons of upstream open reading frames in GCN4 mRNA determine their distinct functions in translational control. Genes Dev. 1989 Aug;3(8):1217–1225. doi: 10.1101/gad.3.8.1217. [DOI] [PubMed] [Google Scholar]
- Mitchell P. J., Tjian R. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science. 1989 Jul 28;245(4916):371–378. doi: 10.1126/science.2667136. [DOI] [PubMed] [Google Scholar]
- Miyamoto M., Fujita T., Kimura Y., Maruyama M., Harada H., Sudo Y., Miyata T., Taniguchi T. Regulated expression of a gene encoding a nuclear factor, IRF-1, that specifically binds to IFN-beta gene regulatory elements. Cell. 1988 Sep 9;54(6):903–913. doi: 10.1016/s0092-8674(88)91307-4. [DOI] [PubMed] [Google Scholar]
- Pestka S., Langer J. A., Zoon K. C., Samuel C. E. Interferons and their actions. Annu Rev Biochem. 1987;56:727–777. doi: 10.1146/annurev.bi.56.070187.003455. [DOI] [PubMed] [Google Scholar]
- Pine R., Decker T., Kessler D. S., Levy D. E., Darnell J. E., Jr Purification and cloning of interferon-stimulated gene factor 2 (ISGF2): ISGF2 (IRF-1) can bind to the promoters of both beta interferon- and interferon-stimulated genes but is not a primary transcriptional activator of either. Mol Cell Biol. 1990 Jun;10(6):2448–2457. doi: 10.1128/mcb.10.6.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakura H., Maekawa T., Imamoto F., Yasuda K., Ishii S. Two human genes isolated by a novel method encode DNA-binding proteins containing a common region of homology. Gene. 1988 Dec 20;73(2):499–507. doi: 10.1016/0378-1119(88)90514-8. [DOI] [PubMed] [Google Scholar]
- Tamm I., Lin S. L., Pfeffer L. M., Sehgal P. B. Interferons alpha and beta as cellular regulatory molecules. Interferon. 1987;9:13–74. [PubMed] [Google Scholar]
- Vinson C. R., LaMarco K. L., Johnson P. F., Landschulz W. H., McKnight S. L. In situ detection of sequence-specific DNA binding activity specified by a recombinant bacteriophage. Genes Dev. 1988 Jul;2(7):801–806. doi: 10.1101/gad.2.7.801. [DOI] [PubMed] [Google Scholar]
- Yan C., Sehgal P. B., Tamm I. Signal transduction pathways in the induction of 2',5'-oligoadenylate synthetase gene expression by interferon alpha/beta. Proc Natl Acad Sci U S A. 1989 Apr;86(7):2243–2247. doi: 10.1073/pnas.86.7.2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu-Lee L. Y., Hrachovy J. A., Stevens A. M., Schwarz L. A. Interferon-regulatory factor 1 is an immediate-early gene under transcriptional regulation by prolactin in Nb2 T cells. Mol Cell Biol. 1990 Jun;10(6):3087–3094. doi: 10.1128/mcb.10.6.3087. [DOI] [PMC free article] [PubMed] [Google Scholar]