Abstract
In the present study, the effectiveness of two loop-mediated isothermal amplification (LAMP) assays was evaluated. Samples of romaine lettuce, strawberries, cherry tomatoes, green onions and sour berries were inoculated with known dilutions (100-108 CFU/g of produce) of S. Enteritidis and L. monocytogenes. With LAMP, assay pathogens can be detected in less than 60 min. The limits of detection of S. Enteritidis and L. monocytogenes depended on the food sample tested and on the presence of enrichment step. After enrichment steps, all food samples were found positive even at low initial pathogen levels. The developed LAMP, assays, are expected to become a valuable, robust, innovative, powerful, cheap and fast monitoring tool, which can be extensively used for routine analysis, and screening of contaminated foods by the food industry and the Public Food Health Authorities.
Key words: Loop-mediated isothermal amplification (LAMP), Ready-to-eat fresh produce, Salmonella, Listeria
Introduction
The consumption of ready-to-eat (RTE) produce, such as salads, fresh fruits and raw vegetables has been increased significantly. RTE fresh produce can become a vehicle for the transmission of pathogens capable of causing human illness (Abadias et al., 2008). Numerous outbreaks in RTE fresh produce have been reported due to extensive human handling during their preparation or by cross-contamination (Escalona et al., 2010). L. monocytogenes and Salmonella spp. are important pathogens detected in vegetables and fruits (Beuchat, 1999; Bracket, 1999). Salmonella has been shown to survive or grow in a wide variety of products (Hanning et al., 2009). An increasing number of product-associated foodborne outbreaks in the United States associated with bacterial contamination are primarily from Salmonella (Tauxe et al., 1997; Harris et al., 2003). L. monocytogenes has been isolated from 15% of environmental samples in New York, emphasizing the prevalence of this pathogen on product fields (Strawn et al., 2013). Prevalence estimates reported in the literature for products grown in the United States range from 0% to more than 35% (Hoelzer et al., 2014). Salmonella remains the most frequently detected causative agent in the food-borne outbreaks reported (22.5 % of total outbreaks). For Listeria foodborne outbreaks a total of 13 outbreaks have been reported in 2013, which was slightly higher than in the previous years (EFSA and ECDC, 2015).
Their control and prevention is essential throughout the food chain, as they can be related with many foodborne outbreaks registered in the European Union (European Food Safety Authority, 2012). Moreover, in case of foodborne outbreaks they cause high mortality rates within high-risk population groups (Kotzekidou, 2013). The fatality rate of L. monocytogenes is relatively rare but can reach 30% (Ponniah et al., 2010).
Salmonellosis constitutes a major foodborne infectious disease worldwide and is caused by 2500 serovars. It is most often attributed to the consumption of contaminated foods such as poultry, beef, pork, eggs, milk, seafood, nut products, and fresh products (Techathuvanan et al., 2010; Wang et al., 2008). S. enterica serovar Enteritidis is often associated with salmonellosis outbreaks (Techathuvanan and D’Souza, 2012). Traditionally, Salmonella detection includes pre-enrichment, then selective enrichment, culture plating, biochemical and serological tests, which in total require approximately 5 days for completion (Kokkinos et al., 2014). Therefore, it is time consuming, cost and labor intensive to meet the criteria for food safety control in routine analysis in food companies (Techathuvanan et al., 2010).
Listeriosis is another major foodborne infectious disease that has the potential to cause serious life-threatening diseases, like septicemia, meningitis, meningoencephalitis and abortion (Dogany, 2003). Listeriosis can lead to a severe illness for individuals with compromised immune systems as the elderly, pregnant women, and young children (Ahmed et al., 2015). In USA approximately 1600 illnesses and 260 deaths have been caused each year due to listeriosis (Scallan et al., 2011). It is well known that a zero tolerance policy for L. monocytogenes in RTE fresh product exists (Gombas et al., 2003). However, for RTE foods which are able to support the growth of L. monocytogenes, EU Regulation 2073 specifies that the 100 CFU/g criterion applies if the manufacturer is able to demonstrate, to the satisfaction of the competent authority, that the product will not exceed the limit of 100 CFU/g throughout the shelf-life and the absence in 25 g criterion applies only when the manufacturer is not able to demonstrate, to the satisfaction of the competent authority, that the product will not exceed the limit of 100 CFU/mL throughout the shelf-life (Koutsoumanis and Angelidis, 2007; EFSA, 2007). Traditionally, food companies test the existence of L. monocytogenes with routine culture methods, which demand at least 5 days for completion. The lack of any heating step prior to consumption of the RTE produce emphasizes the need for taking appropriate sanitary measures for the prevention and reduction of the microbial load, but simultaneously without altering the fresh status and the organoleptic and quality properties of the products, intended to be consumed raw (Baert et al., 2009). Increased public awareness related to health and economic impacts of foodborne diseases has resulted in greater efforts in order to develop sensitive, rapid and inexpensive screening methods of pathogens detection (Wang et al., 2008). To satisfy the expected rapidity, molecular methods have been developed. However, they still require time and expensive, sophisticated equipment, as well as expertise (Techathuvanan et al., 2011). Loop-mediated isothermal amplification (LAMP) is a novel nucleic acid amplification assay that is rapid, specific, and simple, and can be selected as a new alternative for molecular diagnosis (Notomi et al., 2000). The advantages of LAMP are high specificity and sensitivity, simple operation, and low cost, which constitutes a potentially valuable tool for rapid diagnosis of food-borne pathogens (Shao et al., 2011). LAMP works under isothermal conditions between 60 and 65°C resulting in 109 copies of target DNA within an hour (Parida et al., 2008). The aim of the present study was the evaluation of two specific LAMP assays which are simple, cost-effective, one-step, single-tube, for the detection of S. Enteritidis and L. monocytogenes seperately, on a series of artificially inoculated fresh ready-to-eat (RTE) produce samples. The main objective of our study was the application and further evaluation of two diagnostic LAMP-based assays, and the simultaneous-complementary testing with classic ISO methods for the detection of different concentrations of S. Enteritidis and L. monocytogenes in RTE produce, and their potential use in routine food analysis by the food industry and the Public Food Authorities.
Materials and Methods
Bacterial strains
Bacterial strains used were S. Enteritidis NCTC 6676 and L. monocytogenes NCTC 11994 [Health Protection Agency (HPA), Colingdale, UK]. Lenticules with the microorganisms were rehydrated in 9 mL of peptone saline (0.1%) (Oxoid, Basingstoke, UK), and after 20 min, working cultures were streaked onto Tryptic Soy Agar (TSA; Oxoid), incubated at 37°C for 24 h, and stored at 4°C.
Culture preparation
Each bacterial type was cultured in 20 mL Tryptone Soya Broth (TSB; Merck KGaA, Darmstadt, Germany) at 37°C for 17 h, harvested by centrifugation at 4000 x g for 20 min at 4°C and washed three times with buffered peptone water (BPW; Oxoid). The final pellets were resuspended in BPW, corresponding to approximately 108-109 CFU/mL.
Food samples collection and processing
Five different RTE fresh produce, were purchased from a local supermarket (Patras, Greece) the day of the experiment, and stored under refrigerated conditions (4°C) until the time of the experiment. Fresh RTE produce, such as romaine lettuce (Lactuca sativa L. var. longifolia), strawberries (Fragaria x ananassa), cherry tomatoes (Solanum lycopersicum var. cerasiforme), green onions (Allium spp), and sour berries (Prunus cerasus), were used for the specificity and evaluation tests of the developed LAMP assay.
Sample inoculation
All RTE fresh produce samples were rinsed with sterile water to remove some of the natural microbiota or any other matter (e.g. soil) before treatment (Bermúdez-Aguirre and Barbosa-Cánovas, 2013). For the inoculation of the samples, a spot-inoculation method was applied to inoculate the bacteria on their surface. Briefly, 100 µL of S. Enteritidis and L. monocytogenes corresponding to concentrations of 108 CFU/g to 100 CFU/g (which were determined by culture method) were spotted separately with a micropipette on 10 different areas of the surface of each RTE fresh produce sample weighing 10g, in order to simulate real conditions. After spiking, the samples were dried, for 1 hour at 22±2°C, to allow bacterial attachment. All processes were performed in a class II biosafety cabinet (Cytair 155; FluFrance, Wissous, France).
Culture-based confirmation of Salmonella spp. detection
The method used was based on protocol ISO 6579:2002 (ISO, 2002), with slight modifications. Artificially inoculated RTE fresh produce (10 g) were added to 90 mL Buffered Peptone Water (BPW; Oxoid). Secondary enrichment was performed using only Rappaport-Vassiliadis medium (RVS broth; Oxoid). Finally, plating out was performed using xylose lysine desoxycholate agar (XLD; Oxoid). At the same time a LAMP-friendly secondary enrichment with YPCE broth (Knuttson et al., 2002) was assessed, with incubation for 24h at 37°C (Figure 1).
Figure 1.
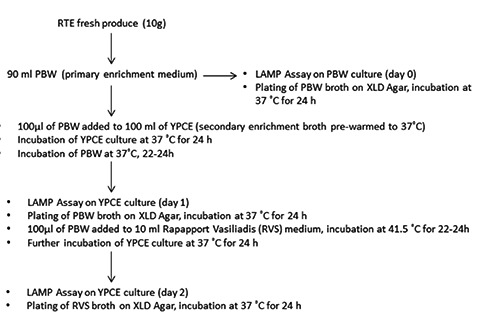
Flowchart of loop-mediated isothermal amplification/International Organization for Standardization methods for Salmonella Enteritidis detection.
Culture-based confirmation of Listeria spp. detection
The method used was based on protocol ISO 11290-1:1996 (ISO, 1996). Artificially inoculated RTE fresh produce (10 g) were added to 90 mL Half Fraser Broth (HFB) (Oxoid) and enrichment for 22-24h at 30°C was followed. Secondary enrichment was performed with Fraser Broth (FB) (Oxoid) for 24h at 37°C. Finally, plating out was performed using Oxford Listeria selective agar (Oxford Listeria; Oxoid). Samples from different stages (samples without enrichment, samples from first enrichment, and samples from secondary enrichment) were taken for culture and LAMP assays (Figure 2). L. monocytogenes nucleic acids were extracted using the Genomic DNA from tissue (Nucleospin tissue; Macherey-Nagel, Düren, Germany), according to the manufacturer’s instructions.
Figure 2.
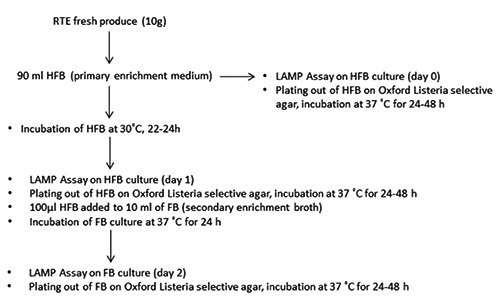
Flowchart of loop-mediated isothermal amplification/International Organization for Standardization methods for Listeria monocytogenes detection.
Sample preparation for loop-mediated isothermal amplification methods
Buffered Peptone Water (90 mL) and Half Fraser Broth (90 mL) were added to 10 g of each RTE fresh produce sample prior inoculation with S. Enteritidis and L. monocytogenes, respectively. The samples were homogenized for Salmonella and Listeria testing, respectively. Sample was taken at day 0 (fresh produce sample with PBW or HFB). Then, 100µL were added to 100ml of selective enrichment broth YPCE and enrichment at 37°C for 24-48h (day 1 and day 2 respectively) followed for Salmonella detection. The LAMP friendly enrichment broth was prepared according to D’Agostino et al. (2015). For L. monocytogenes detection, enrichment of samples with HFB at 37°C for 24h (day 1) followed and finally 0.1 µL of HFB was added to 10ml of FB and the samples were enriched at 37°C for 24h (day 2).
Loop-mediated isothermal amplification assay for Salmonella Enteritidis detection
In this study, six primers (two inner primers, two outer primers and two loop primers), targeting Salmonella enterica invasion protein (invA) gene were used for the LAMP reactions (Hara-Kudo et al., 2005; Ziros et al., 2012). The reaction was carried out in a total of 25µL and contained 16µL of Tin Isothermal Mastermix, 25 µM invASalm FIP, 25 µM invASalm BIP, 5 µM invASalmF3, 5 µM invASalmB3, 12,5 µM invASalmF-Loop, 12,5 µM invASalmB-Loop and 3µL of template DNA (D’Agostino et al., 2015). The primers were high-performance liquid chromatography-purified. The thermal profile of the reaction was the following: 95°C for 2 min (cell lysis), followed by 65°C for 50 min (nucleic acid amplification). Amplicon annealing profiling was performed by heating to 98°C then cooling to 80°C at a rate of 0.05°C sec-1. The samples were analyzed using a LightCycler Nano Instrument (Roche, Basel, Switzerland). Positive and negative controls were included in each run. Aliquots of 10 µL of LAMP products were electrophoresed on 2% agarose gels and were visualized by ethidium bromide (Sigma, St. Louis, MO, USA) staining. The amplified products were also detected by adding 1 µL of 1000 X SYBR green dye to each reaction tube. After incubation for 15 min in the dark at room temperature, a yellowish green colour indicated a positive reaction, while a reddish orange (the colour of the unbound dye) indicated a negative reaction.
Loop-mediated isothermal amplification assay for Listeria monocytogenes detection
This study used six primers (two inner primers, two outer primers and two loop primers) targeting hlyA gene of L. monocytogenes. The primers were high-performance liquid chromatography-purified. Positive and negative controls were included in each run. The LAMP reaction was carried out in a total volume of 25 µL. The optimal conditions as well as the thermal profile were based on the assay of Wang et al. (2012). The samples were analyzed using a LightCycler Nano Instrument (Roche). Aliquots of 10 µL of LAMP products were electrophoresed on 2% agarose gels and were visualized by ethidium bromide (Sigma) staining with UV light transillumination. The amplified products were also detected by adding 1 µL of 1000 X SYBR green dye to each reaction tube. After incubation for 15 min in the dark at room temperature, a yellowish green colour indicated a positive reaction, while a reddish orange (the colour of the unbound dye) indicated a negative reaction.
Prevention of polymerase chain reaction carryover contamination
To avoid any LAMP carryover contamination strict laboratory practices were followed throughout the experimental procedure. The manipulations before the LAMP assay (DNA isolation and LAMP set-up) were performed in a clean UV room that was separated from the LAMP PCR machine and the post-LAMP processing area. Negative controls were run with all assays, and no indications of contamination were detected. Attention was paid when the caps of the used reaction tubes were opened for the addition of SYBR green dye or subsequent electrophoresis.
Specificity and sensitivity of the loop-mediated isothermal amplification assays
The specificity of the developed LAMP assays in real-world situations were evaluated in the context of the present study by analyzing different fresh produce matrices, after inoculation of non-Salmonella and non-Listeria DNA. Strains other than Salmonella and Listeria (human adenovirus-35, hAdV35; human adenovirus 40/41, hAdV40/41; E. coli, NCTC 9001) were selected in order to check if cross-reaction was observed in LAMP assays. The negative controls which have been used in the study were the following: i) for every batch of analyzed food samples, a control sample (non-inoculated with the target microorganism) was tested to investigate any potential initial contamination or contamination during the enrichment procedures; ii) for every LAMP assay, two negative controls with the aforementioned non-target microorganisms, and one additional negative control consisting of water (ddH2O) (instead of the target microorganism), were used. In addition, ten-fold dilutions of Salmonella and Listeria, which were previously quantified by culture methods, were inoculated in RTE fresh produce samples to determine the sensitivity of the assay for food analysis. Finally, for measuring the limit of detection (LOD) of the LAMP assay, RTE fresh produce samples were tested in triplicate and the lowest concentration of colony forming units (CFUs) was taken as the limit when all of the triplicate samples (for each dilution) were positive.
Statistical analysis
All experiments were carried out in triplicates. During each experiment two samples were run in agarose gels (2%) or tested after the addition of SYBR Green. The data for the detection of two pathogens with LAMP assays were analyzed for statistical significance using SPSS 21.0 (SPSS Inc., Chicago, IL, USA). Results were compared by an analysis of variance (ANOVA) followed by Tukey’s method with a significance level of P<0.05.
Results and Discussion
In the present study, different RTE fresh produce (romaine lettuce, strawberries, cherry tomatoes, green onions, sour berries) were tested for detection of 2 pathogens (S. Enteritidis and L. monocytogenes) that were prior inoculated on the food samples.
The specific isothermal amplification of the DNA of Salmonella and Listeria strains on food samples generated ladder-like pattern bands on agarose gel. No amplification was observed in LAMP reactions without template DNA (negative control) and in the control reactions with non-Salmonella/Listeria DNA. LAMP assay successfully detected the aforementioned bacteria in food samples within 60 min, when no enrichment step was followed. However, when enrichment steps were used, the detection limit was higher for both methods (Tables 1 and 2) and the total assay time was 25 h and 49 h for primary and secondary enrichment steps, respectively. Moreover, there was no visual detection of LAMP products after SYBR Green addition and observation under UV light (Figure 3) or difference between the LAMP results detected by agarose gel electrophoresis of LAMP products (Figure 4).
Table 1.
Loop-mediated isothermal amplification results of different ready to eat fresh produce samples prior inoculated with different dilutions of S. Enteritidis during a 3-day period.
| RTE fresh produce | Corresponding initial inoculum (CFU/g) | LAMP-SALM (day 0) | LAMP-SALM (day 1) | LAMP-SALM (day 2) |
|---|---|---|---|---|
| Romaine lettuce | 3*10^0 | - | + | + |
| 3*10^1 | - | + | + | |
| 3*10^2 | - | + | + | |
| 3*10^3 | - | + | + | |
| 3*10^4 | + | + | + | |
| 3*10^5 | + | + | + | |
| 3*10^6 | + | + | + | |
| 3*10^7 | + | + | + | |
| 3*10^8 | + | + | + | |
| Strawberries | ||||
| 1*10^0 | - | + | + | |
| 1*10^1 | - | + | + | |
| 1*10^2 | - | + | + | |
| 1*10^3 | - | + | + | |
| 1*10^4 | - | + | + | |
| 1*10^5 | - | + | + | |
| 1*10^6 | - | + | + | |
| 1*10^7 | + | + | + | |
| 1*10^8 | + | + | + | |
| Cherry tomatoes | ||||
| 2*10^0 | - | + | + | |
| 2*10^1 | - | + | + | |
| 2*10^2 | - | + | + | |
| 2*10^3 | - | + | + | |
| 2*10^4 | - | + | + | |
| 2*10^5 | + | + | + | |
| 2*10^6 | + | + | + | |
| 2*10^7 | + | + | + | |
| 2*10^8 | + | + | + | |
| Green onions | ||||
| 2*10^0 | - | + | + | |
| 2*10^1 | - | + | + | |
| 2*10^2 | - | + | + | |
| 2 *10^3 | - | + | + | |
| 2*10^4 | + | + | + | |
| 2*10^5 | + | + | + | |
| 2*10^6 | + | + | + | |
| 2*10^7 | + | + | + | |
| 2*10^8 | + | + | + | |
| Sour berries | 3*10^0 | - | + | + |
| 3*10^1 | - | + | + | |
| 3*10^2 | - | + | + | |
| 3*10^3 | - | + | + | |
| 3*10^4 | - | + | + | |
| 3*10^5 | + | + | + | |
| 3*10^6 | + | + | + | |
| 3*10^7 | + | + | + | |
| 3*10^8 | + | + | + |
RTE, ready to eat; CFU, colony-forming units; LAMP, loop-mediated isothermal amplifications; SALM, Salmonella. Day 0, fresh produce sample with PBW without enrichment; day 1, fresh produce sample in YPCE enriched at 37°C for 24h; day 2, fresh produce sample in YPCE enriched at 37°C for 48h.
Table 2.
Loop-mediated isothermal amplification results of different ready to eat fresh produce samples prior inoculated with different dilutions of L. monocytogenes during a 3-day period.
| RTE fresh produce | Corresponding initial inoculum (CFU/g) | LAMP-LIS (day 0) |
LAMP-LIS (day 1) |
LAMP-LIS (day 2) |
|---|---|---|---|---|
| Romaine lettuce | 1*10^0 | - | - | + |
| 1*10^1 | - | + | + | |
| 1*10^2 | - | + | + | |
| 1*10^3 | - | + | + | |
| 1*10^4 | - | + | + | |
| 1*10^5 | - | + | + | |
| 1*10^6 | - | + | + | |
| 1*10^7 | + | + | + | |
| 1*10^8 | + | + | + | |
| Strawberries | ||||
| 2*10^0 | - | - | + | |
| 2*10^1 | - | - | + | |
| 2*10^2 | - | - | + | |
| 2*10^3 | - | - | + | |
| 2*10^4 | - | + | + | |
| 2*10^5 | - | + | + | |
| 2*10^6 | - | + | + | |
| 2*10^7 | + | + | + | |
| 2*10^8 | + | + | + | |
| Cherry tomatoes | ||||
| 1*10^0 | - | - | + | |
| 1*10^1 | - | - | + | |
| 1*10^2 | - | + | + | |
| 1*10^3 | - | + | + | |
| 1*10^4 | - | + | + | |
| 1*10^5 | - | + | + | |
| 1*10^6 | - | + | + | |
| 1*10^7 | - | + | + | |
| 1*10^8 | + | + | + | |
| Green onions | ||||
| 2*10^0 | - | - | + | |
| 2*10^1 | - | - | + | |
| 2*10^2 | - | - | + | |
| 2*10^3 | - | + | + | |
| 2*10^4 | - | + | + | |
| 2*10^5 | - | + | + | |
| 2*10^6 | - | + | + | |
| 2*10^7 | + | + | + | |
| 2*10^8 | + | + | + | |
| Sour berries | 1*10^0 | - | - | + |
| 1*10^1 | - | - | + | |
| 1*10^2 | - | - | + | |
| 1*10^3 | - | - | + | |
| 1*10^4 | - | + | + | |
| 1*10^5 | - | + | + | |
| 1*10^6 | + | + | + | |
| 1*10^7 | + | + | + |
RTE, ready to eat; CFU, colony-forming units; LAMP, loop-mediated isothermal amplifications; LIS, Listeria. Day 0, fresh produce sample with PBW without enrichment; day 1, fresh produce sample in YPCE enriched at 37°C for 24h; day 2, fresh produce sample in YPCE enriched at 37°C for 48h.
Figure 3.
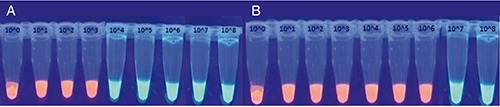
Loop-mediated isothermal amplification products after SYBR green addition and observation under ultra-violet light: ready to eat romaine lettuce at day 0 for Salmonella Enteritidis A) and Listeria monocytogenes (B).
Figure 4.
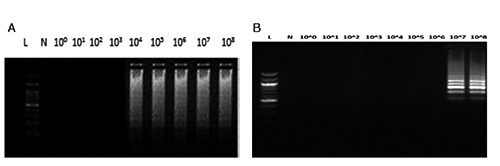
Loop-mediated isothermal amplification results detected by agarose gel electrophoresis: ready to eat romaine lettuce at day 0 for Salmonella Enteritidis A) and Listeria monocytogenes (B). L, 100bp Ladder; N, negative control; 100-108, decimal dilutions of Salmonella Enteritidis loop-mediated isothermal amplification amplicons.
The LAMP-based methods are important for analysis of fresh RTE produce, through screening of uncontaminated or contaminated samples in approximately 24-48 hours less than the standard culture-based method alone (data not shown). It is known that it is necessary for the food companies to certify the fresh RTE products as Salmonella-free or Listeria-free before the products are released to the market. Until now, the time till the final confirmation of the result using the standard cultivation methods, is not in favor of the companies as it is linked with high costs and time consumption.
DNA amplification and reading of results require minimum equipment, thus the technique has great potential for use in diagnosis of foodborne diseases. Simple heating methods, such as chemical heaters or thermal bottles, have been recently designed for removing the dependence upon stable electricity and allowing for LAMP to be conducted at any time in any setting (Curtis et al., 2012; Hatano et al., 2010). As a consequence, LAMP assay has all the characteristics required of real-time assays along with simple operation for easy adaptability on-site.
LAMP has the advantages of simplicity, rapidity, specificity, cost-effectiveness and higher amplification efficiency. Furthermore, since DNA amplification and reading of results require minimum equipment, the technique has great potential for use in low-income countries (Njiru, 2012; Mori et al., 2013).
The specificity and sensitivity of the LAMP assays for Salmonella and L. monocytogenes detection have been previously evaluated (Hara-Kudo et al., 2005; Ziros et al., 2012; Wang et al., 2012). The analytical sensitivity of the LAMP assays for both bacteria depended on the enrichment steps that were followed. Two-step enrichment procedures were used for each pathogen as this can indicate the viability of a presumptive positive (D’ Agostino et al., 2015). After an enrichment step is performed, only a detection result is possible. Any reference to concentrations of target microorganisms throughout this study refers to initial microbial load.
The specific isothermal amplification of the DNA of Salmonella and Listeria on food samples generated ladder-like pattern bands on agarose gel. No amplification was observed in LAMP reactions without template DNA (negative control) and in the control reactions with non-Salmonella and non-Listeria DNA. There was no difference between the LAMP results detected by agarose gel electrophoresis of LAMP products or visual detection of LAMP products after SYBR Green addition and observation under UV light (Figures 3 and 4).
Ten-fold dilutions of 100 to 108 CFU/g of S. Enteritidis and L. monocytogenes in the selected food matrices were used for the sensitivity assays. The analytical sensitivity of the LAMP assay for each food pathogen and each food sample, with and without enrichment was estimated and is illustrated in Tables 1 and 2.
The two enrichment steps that were used for each pathogen increase the possibility of detecting viable cells originally existing in the sample to detectable amounts and increased the sensitivity of the assay (D’Agostino et al., 2015). The likelihood that a LAMP signal will be derived from viable Salmonella can be demonstrated arithmetically (D’Agostino et al., 2015). The LAMP assay for Salmonella can detect ~400 cells per reaction (D’ Agostino et al., 2015), which is in accordance with the results of the present study for romaine lettuce, and onions. Whereas a higher detection limit (approximately 1 log higher) was found for cherry tomatoes, sour berries and strawberries (Table 1). The nature of different food types (smoother or porous surfaces, pH, etc.) could be the reason for the recorded differences in the detection limit. Further studies are needed in order to reveal the mechanisms under these observations. All the enrichment steps exhibited statistically significant results when tested with Paired Samples T-test (P<0.05).
The LAMP assay for Listeria exhibited a higher detection limit (3-4 log higher) compared to Salmonella in all RTE fresh produce, which imposed the necessity of the two enrichment steps. The higher detection level for L. monocytogenes when a longer period of enrichment was used can be explained by the longer generation time of this organism and indicates that selective enrichment with FB is necessary for detection of low initial numbers of cells of this species (Burtscher et al., 1999). Moreover, since L. monocytogenes is a Gram positive bacterium, L. monocytogenes cells are more difficult to extract, which makes DNA recovery less efficient (Burtscher et al., 1999). Obviously, the detection limit of low numbers of cells was increased when the second enrichment broth (FB) was used and incubation for 2 days was followed (Table 2). However, considerably less time than the classic procedures is required, which usually takes at least 4 days (data not shown). In Salmonella LAMP assays, the use of the thermostable heat stable enzyme Thermodesulfotator indicus (Tin) DNA polymerase enzyme allowed the direct addition of the whole Salmonella cells to the LAMP reactions. This is a very important aspect for decreasing time in food companies (D’ Agostino et al., 2015). However, in Listeria LAMP assays the nucleic acid extraction was necessary as no amplification was observed in any sample without prior nucleic acid extraction, compared to Salmonella. Moreover, the boiling extraction method (Wu et al., 2014) exhibited faint signals in gel electrophoresis, which is in accordance with Amagliani et al. (2007), thus a Nucleospin tissue KIT was selected for L. monocytogenes detection, inoculated on RTE fresh produce (data not shown).
In the present study, the robustness of the LAMP assays for the analysis of food samples was shown by the detection of the target Gram negative and Gram positive bacteria in RTE food samples with smoother surface, like cherry tomatoes, green onions, and lettuce, without enrichment step. The detection of Salmonella and Listeria was found to be higher (approximately 2-4 log), compared to that of the samples tested after first and second enrichment step. Moreover, the results for both pathogens were found to be significant (P=0.046), when tested with one-way Anova test, for all the RTE fresh produce tested.
For instance, the detection without enrichment procedure was proved to be slightly lower (approximately 4-7 log) compared to that of the samples tested after an enrichment of 1st and 2nd day. Especially, in more complex food samples (strawberries, sour cherries), which can be categorized as soft fruits with porous surfaces and with rich sugar content, the target DNA was detected only when high levels of initial contamination existed for both pathogens. On the other hand, the detection was achieved in lower levels for vegetables with more firm surfaces, such as lettuce, cherry tomatoes (Dubois et al., 2007; do Nascimento, 2015). However, the results for RTE fresh produce were not significant (P>0.05) for both pathogens tested and for all enrichment steps. This imposes the implementation of enrichment steps, which for Salmonella, one day enrichment was found to be sufficient, whereas for Listeria, two enrichment days were important for the detection of lower levels of pathogen (Tables 1 and 2).
Different researchers have studied LAMP assays for Salmonella detection in RTE fresh produce like lettuce, tomatoes and milk (Zhang et al., 2011; Yang et al., 2015; Wu et al., 2015). More precisely Zhang et al. (2011) have found the detection limit of L. monocytogenes in lettuce (2 CFU/25g) and tomato (19 CFU/25g). Whereas, the detection limit of L. monocytogenes using the ISO 11290-1:1996 method (Amd. 2004) was found to be 5-100 CFU/25g (Zunabovic et al., 2011). Other researchers have studied the detection limit of L. monocytogenes in food samples like chicken and milk (Wu et al., 2014; Wang et al., 2011; Tang et al., 2011).
The present study did not aim at proposing LAMP as an alternative to ISO methods, but aimed to highlight the potential of LAMP assays to complement ISO methods in order to enhance their potential use in routine food analysis by the food industry and the Public Food Authorities. LAMP-based assays may be used as sensitive, specific and cheap tools for a fast initial screening of food samples for the target microorganisms of interest. Negative samples may be rapidly released to the market, while positive samples may be further analyzed and confirmed by ISO methods.
This is to the best of our knowledge the first report on the evaluation of -one Gram negative (Salmonella) and one Gram positive (Listeria) bacterium-specific LAMP assays on different food samples. This is the first time that LAMP assays are used for pathogen detection in five RTE fresh produce.
Conclusions
The results showed that Salmonella spp. and L. monocytogenes were detected successfully on the five fresh RTE fresh produce samples tested (romaine lettuce, strawberries, cherry tomatoes, green onions, sour berries) by both LAMP assays and conventional culture assays. The greatest advantage of the proposed LAMP assays is the reduction of the required analysis time depending on the initial contamination of the products, compared to the classical culture methods. The developed LAMP assays are expected to become a valuable tool and be used extensively for routine analysis and screening of contaminated foods by the food industry and the Public Health Authorities.
Funding Statement
Funding: the work was financially supported by The Greek State Scholarships Foundation-IKY Iky Fellowships of Excellence for Postgraduate Studies in Greece-Siemens Program.
References
- Abadias M, Usall J, Anguera M, Solsona C, Viñas I, 2008. Microbiological quality of fresh, minimally-processed fruit and vegetables, and sprouts from retail establishments. Int J Food Microbiol 123:121-9. [DOI] [PubMed] [Google Scholar]
- Ahmed O, Pangloli P, Hwang C-A, Zivanovic S, Wu T, D'Souza D, Draughon FA, 2015. The occurrence of Listeria monocytogenes in retail ready-to-eat meat and poultry products related to the levels of acetate and lactate in the products. Food Control 52:43-8. [Google Scholar]
- Amagliani G, Giammarini C, Omiccioli E, Brandi G, Magnan M, 2007. Detection of Listeria monocytogenes using a commercial PCR kit and different DNA extraction methods. Food Control 18:1137-42. [Google Scholar]
- European Food Safety Authority, 2012. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2010. EFSA J 10:1-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baert L, Debevere J, Uyttendaele M, 2009. The efficacy of preservation methods to inactivate foodborne viruses. Int J Food Microbiol 131:83-94. [DOI] [PubMed] [Google Scholar]
- Bermúdez-Aguirre D, Barbosa-Cánovas GV, 2013. Disinfection of selected vegetables under nonthermal treatments: chlorine, acid citric, ultraviolet light and ozone. Food Control 29:82-90. [Google Scholar]
- Beuchat LR, 1999. Pathogenic microorganisms associated with fresh produce. J Food Protect 59:204-16. [DOI] [PubMed] [Google Scholar]
- Bracket RE, 1999. Incidence, contributing factors, and control of bacterial pathogens in produce. Postharvest Biol Tech 15:305-11. [Google Scholar]
- Burtscher C, Fall P, Wilderer P, Wuertz S, 1999. Detection of Salmonella spp. and Listeria monocytogenes in Suspended Organic waste by nucleic acid extraction and PCR. Appl Environ Microb 65:2235-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis KA, Rudolph DL, Nejad I, Singleton J, Beddoe A, Weigl B, LaBarre P, Owen SM, 2012. Isothermal amplification using a chemical heating device for point-of-care detection of HIV. PLoS One 7:31432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Agostino M, Diez-Valcarce M, Robles S, Losilla-Garcia B, Cook N, 2015. A Loop-mediated amplification-based method for analyzing animal feed for the presence of Salmonella. Food Anal Method (in press). [Google Scholar]
- Dogany M, 2003. Listeriosis: clinical presentation. FEMS Immunol Med Mic 35:173-5. [DOI] [PubMed] [Google Scholar]
- do Nascimento Nunes MC, 2015. Correlations between subjective quality and physicochemical attributes of fresh fruits and vegetables. Postharvest Biol Tech 107:43-54. [Google Scholar]
- Dubois E, Hennechart C, Merle G, Burger C, Hmila N, Ruelle S, Perelle S, Ferré V, 2007. Detection and quantification by real-time RT-PCR of hepatitis A virus from inoculated tap waters, salad vegetables, and soft fruits: characterization of the method performances. Int J Food Microbiol 117:141-9. [DOI] [PubMed] [Google Scholar]
- EFSA, 2007. Scientific opinion of the panel on biological hazards on a request from the European Commission on Request for updating the former SCVPH opinion on Listeria monocytogenes risk related to ready-to-eat foods and scientific advice on different levels of Listeria monocytogenes in ready-to-eat foods and the related risk for human illness. EFSA J 599:1-42. [Google Scholar]
- EFSA and ECDC, 2015. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2013. EFSA J 13:3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escalona V, Aguayo E, Martínez-Hernández G, Artés F, 2010. UV-C doses to reduce pathogen and spoilage bacterial growth in vitro and in baby spinach. Postharvest Biol Tech 56:223-31. [Google Scholar]
- European Commission, 2005. Commission Regulation of 15 November 2005 on microbiological criteria for foodstuffs, 2073/2005/EC. In: Official Journal, L 338, pp 1-26. [Google Scholar]
- Gombas DE, Chen Y, Clavero RS, Scott VN, 2003. Survey of Listeria monocytogenes in ready-to-eat foods. J Food Protect 66:559-69. [DOI] [PubMed] [Google Scholar]
- Hanning IB, Nutt JD, Ricke SC, 2009. Salmonellosis outbreaks in the United States due to fresh produce: sources and potential intervention measures. Foodborne Pathog Dis 6:6. [DOI] [PubMed] [Google Scholar]
- Hara-Kudo Y, Yoshino M, Kojima T, Ikedo M, 2005. Loop-mediated isothermal amplification for the rapid detection of Salmonella. FEMS Microbiol Lett 253:155-61. [DOI] [PubMed] [Google Scholar]
- Harris LJ, Farber JN, Beuchat LR, Parish ME, Suslow TV, Garrett EH, Busta FF, 2003. Outbreaks associated with fresh produce: incidence, growth, and survival of pathogens in fresh and fresh-cut produce. Compr Rev Food Sci F 2:78-141. [Google Scholar]
- Hatano B, Maki T, Obara T, Fukumoto H, Hagisawa K, Matsushita Y, Okutani A, Bazartseren B, Inoue S, Sata T, Katano H, 2010. LAMP using a disposable pocket warmer for anthrax detection, a highly mobile and reliable method for antibioterrorism. Jpn J Infect Dis 63:36-40. [PubMed] [Google Scholar]
- Hoelzer K, Pouillot R, Van Doren JM, Dennis S, 2014. Reduction of Listeria monocytogenes contamination on produce. A quantitative analysis of common liquid fresh produce wash compounds. Food Control 46:430-40. [Google Scholar]
- ISO, 1996. Microbiology of food and animal feeding stuffs. Horizontal method for the detection and enumeration of Listeria monocytogenes. Part 1: Detection method. ISO Norm 11290-1:1996. International Standardization Organization ed., Geneva, Switzerland. [Google Scholar]
- ISO, 2002. Microbiology of food and animal feeding stuffs. Horizontal method for the detection of Salmonella spp. ISO Norm 6579:2002. International Standardization Organization ed., Geneva, Switzerland. [Google Scholar]
- Knutsson R, Fontanesi M, Grage H, Radstrom P, 2002. Development of a PCR compatible enrichment medium for Yersinia enterocolitica: amplification precision and dynamic detection range during cultivation. Int J Food Microbiol 72:185-201. [DOI] [PubMed] [Google Scholar]
- Kokkinos PA, Ziros PG, Bellou M, Vantarakis A, 2014. Loop-mediated isothermal amplification (LAMP) for the detection of Salmonella in food. Food Anal Method 7:512-26. [Google Scholar]
- Kotzekidou P, 2013. Survey of Listeria monocytogenes, Salmonella spp. and Escherichia coli O157:H7 in raw ingredients and ready-to-eat products by commercial real-time PCR kits. Food Microbiol 35:86-91. [DOI] [PubMed] [Google Scholar]
- Koutsoumanis K, Angelidis A, 2007. Probabilistic modeling approach for evaluating the compliance of ready-to-eat foods with new European Union safety criteria for Listeria monocytogenes. Appl Environ Microb 73:4996-5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori Y, Kanda H, Notomi T, 2013. Loop-mediated isothermal amplification (LAMP): recent progress in research and development. J Infect Chemother 19:404-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T, 2000. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res 28:e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Njiru ZK, 2012. Loop-mediated isothermal amplification technology: towards point of care diagnostics. PLoS Negl Trop Dis 6:e1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parida M, Sannarangaiah S, Dash PK, Rao PV, Morita K, 2008. Loop mediated isothermal amplification (LAMP): a new generation of innovative gene amplification technique; perspectives in clinical diagnosis of infectious diseases. Rev Med Virol 18:407-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponniah J, Robin T, Paie MS, Radu S, Ghazali FM, Kqueen CY, Nishibuchi M, Nakaguchi Y, Kumar Malakar P, 2010. Listeria monocytogenes in raw salad vegetables sold at retail level in Malaysia. Food Control 21:774-8. [Google Scholar]
- Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, Jones JL, Griffin PM, 2011. Foodborne illness acquired in the United States. Major pathogens. Emerg Infect Dis 17:7-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y, Zhu S, Jin C, Chen F, 2011. Development of multiplex loopmediated isothermal amplification-RFLP (mLAMP-RFLP) to detect Salmonella spp. and Shigella spp. in milk. Int J Food Microbiol 148:75-9. [DOI] [PubMed] [Google Scholar]
- Strawn LK, Fortes ED, Bihn EA, Nightingale KK, Grohn YT, Worobo RW, Wiedmann M, Bergholz PW, 2013. Landscape and meteorological factors affecting prevalence of three food-borne pathogens in fruit and vegetable farms. Appl Environ Microb 79:588-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng-Jun T, Sheng Z, Xiao-Yan Z, Jun-Hua P, Qing-Lian G, Xiu-Jun T, 2011. Rapid and sensitive detection of Listeria monocytogenes by loop-mediated isothermal amplification. Curr Microbiol 63:511-6. [DOI] [PubMed] [Google Scholar]
- Tauxe R, Kruse H, Hedberg C, Potter M, Madden J, Wachsmuth K, 1997. Microbial hazards and emerging issues associated with produce, a preliminary report to the national advisory committee on microbiologic criteria for foods. J Food Protect 60:1400-8. [DOI] [PubMed] [Google Scholar]
- Techathuvanan C, Draughon FA, D'Souza DH, 2010. Loop-mediated isothermal amplification (LAMP) for the rapid and sensitive detection of Salmonella typhimurium from pork. J Food Sci 75:165-72. [DOI] [PubMed] [Google Scholar]
- Techathuvanan C, Draughon FA, D'Souza DH, 2011. Comparison of reverse transcriptase PCR, reverse transcriptase loop-mediated isothermal amplification, and culture-based assays for Salmonella detection from pork processing environments. J Food Protect 74:294-301. [DOI] [PubMed] [Google Scholar]
- Techathuvanan C, D'Souza DH, 2012. Reverse-transcriptase loopmediated isothermal amplification as a rapid screening/monitoring tool for Salmonella enterica detection in liquid whole eggs. J Food Sci 77:200-5. [DOI] [PubMed] [Google Scholar]
- Wang L, Shi L, Alam MJ, Geng Y, Li L, 2008. Specific and rapid detection of foodborne Salmonella by loop-mediated isothermal amplification method. Food Res Int 41:69-74. [Google Scholar]
- Wang D, Zhang G, Lu C, Deng R, Zhi A, Guo JG, Dong Z, Zhihong X, 2011. Rapid detection of Listeria monocytogenes in raw milk with loop-mediated isothermal amplification and chemosensor. J Food Sci 76:9. [DOI] [PubMed] [Google Scholar]
- Wang L, Li Y, Chu J, Xu Z, Zhong Q, 2012. Development and application of a simple loop-mediated isothermal amplification method on rapid detection of Listeria monocytogenes strains. Mol Biol Rep 39:445-9. [DOI] [PubMed] [Google Scholar]
- Wu R, Liu X, Guo B, Chen F, Wang X, 2014. Development of double loop-mediated isothermal amplification to detect listeria monocytogenes in food. Curr Microbiol 69:839-45. [DOI] [PubMed] [Google Scholar]
- Wu GP, Chen SH, Levin RE, 2015. Rapid real-time loop-mediated isothermal amplification combined with coated activated carbon for detection of low numbers of Salmonella enterica from lettuce without enrichment. Food Control 56:47-52. [Google Scholar]
- Yang Q, Wang F, Jones KL, Meng J, Prinyawiwatkul W, Ge B, 2015. Evaluation of loop-mediated isothermal amplification for the rapid, reliable, and robust detection of Salmonella in produce. Food Microbiol 46:485-93. [DOI] [PubMed] [Google Scholar]
- Zhang G, Brown EW, González-Escalona N, 2011. Comparison of real-time PCR, reverse transcriptase real-time PCR, loop-mediated isothermal amplification, and the FDA conventional microbiological method for the detection of Salmonella spp. in produce. Appl Environ Microb 77:6495-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziros P, Kokkinos P, Papanotas K, Vantarakis A, 2012. Loop-mediated isothermal amplification (LAMP) for the detection of Salmonella spp. isolated from different food types. J Microbiol Biotechnol Food Sci 2:152-61. [Google Scholar]
- Zunabovic M, Domig KJ, Kneifel W, 2011. Practical relevance of methodologies for detecting and tracing of Listeria monocytogenes in ready-to-eat foods and manufacture environments. A review. LWT - Food Sci Technol 44:351-362. [Google Scholar]


