Abstract
In this study bacterial isolates from Ventricina del Vastese sausage, previously identified as Lactobacillus (L.) sakei, were characterised genotypically, physiologically and on the basis of some technologically relevant traits. A total of 70 L. sakei isolates from sausages manufactured with spontaneous fermentation in the same producing plant were taken into account. Six genotypic groups were distinguished on the basis of Rep-polymerase chain reaction with the GTG5 primer, some of which were found only in the sausages ripened at temperatures lower than 10°C for the first two months and lower than 16°C for the remaining three months, according to the traditional ripening process. Six strains were selected as representative of the genotypic profiles and further characterised. A high diversity in their fermentation profiles was observed, and different groups were separated on the basis of growth and acidifying capacity in meat extract. None of the strains produced histamine or tyramine in vitro. One strain was able to slightly inhibit Listeria (L.) monocytogenes and L. innocua and all six strains were able to slightly inhibit Enterobacteriaceae isolated from Ventricina del Vastese sausages in vitro. Results showed that most L. sakei strains can have a role in improving the safety of low acidity fermented sausages, even though a limited acidifying capacity was observed in a meat-like substrate, and that L. sakei strains able to produce biogenic amines are unlikely to occur in spontaneously fermented meat products.
Key words: Lactobacillus sakei, Sausages, Ventricina del Vastese
Introduction
Although initially isolated from rice wine (Kandler and Weiss, 1986), Lactobacillus (L.) sakei is the predominant Lactobacillus species in fermented meat and fish products and also in Southern European sausages (Montel, 1999). In fermented sausages, L. sakei is more competitive than other lactobacilli, showing a shorter lag phase, higher growth rate and higher cell numbers (Dossmann et al., 1996). L. sakei is psychrotrophic and can tolerate high salt concentrations; the capacity of some strains to grow at 4°C and in the presence of 6.5% NaCl, was reported (Ammor et al., 2005). The salt tolerance has been attributed to the ability of L. sakei to efficiently accumulate compatible solutes such as betaine and carnitine, and the cold tolerance was explained with the presence in the genome of L. sakei of more cold-stress genes than any other Lactobacillus spp. (Chaillou et al., 2005). L. sakei possesses a heme-dependent catalase (Ammor et al., 2005), which can prevent the green colour defect caused by H2O2 production by some lactic acid bacteria in meat products, and actively contributes to the hydrolysis of the sarcoplasmic proteins and to the subsequent decomposition of peptides into amino acids (Fadda et al., 1999). It has been demonstrated that L. sakei, for its rapid growth and acid production, greatly reduces the accumulation of biogenic amines (BAs) in fermented sausages by preventing the development of amine-producing bacteria (Bover-Cid et al., 2001). The occurrence of bacteriocinogenic strains of L. sakei inhibitory to Listeria (L.) monocytogenes has been known for long time (Shillinger et al., 1991), though the effect of non-bacteriocinogenic strains on this respect was little investigated. One study compared the antagonistic features of bacteriocinogenic and non-bacteriocinogenic L. sakei isolates toward bacterial contaminants when inoculated in fresh vacuum packaged meat (Jones et al., 2009) but there are no data available on the antagonistic activity on non-bacteriocinogenic L. sakei strains in fermented meat products.
L. sakei has been isolated from human feces and it is therefore able to survive the passage through the human gastrointestinal tract (GIT) (Walter et al., 2001), the studies on its probiotic properties are underway (Park et al., 2008; Garriga et al., 2015).
In a previous study on the identification of the pro-technological bacterial species isolated from Ventricina del Vastese, a traditional sausage of the Abruzzi and Molise regions made from pork meat cut into cubes of 2-5 cm sides, mixed with 20-25% lard, stuffed in natural casings to obtain sausages with 11-12 mm diameter and ripened for at least 150 days, it was found that all except one among 71 Lactobacillus spp. isolates belonged to the species Lactobacillus sakei, as ascertained by species-specific polymerase chain reaction (PCR) (Amadoro et al., 2013). The L. sakei isolates, obtained at different ripening times, came from sausages manufactured by a single producer and ripened in natural environmental conditions or in controlled conditions in plug-in showcase cabinets. The sausages ripened in the natural way, manufactured in December, had been exposed to temperatures lower than 10°C for the first 60 days and lower than 16°C for the remaining 90 days of ripening, while the sausages ripened in controlled conditions had been kept at 18°C in the first week of ripening and at 12°C later on. The predominance of the species L. sakei highlighted its importance for the traditional product considered and suggested to study the diversity of the isolates and the activities relevant for ripening and safety. Therefore, they were characterised genotypically by Rep-PCR and strains representing distinct genotypes were selected for the examination of physiological features. Moreover, safety aspects, such as the ability to inhibit Listeria spp. and Enterobacteriaceae isolated from Ventricina del Vastese and the absence of BAs production, were assessed in vitro.
Materials and Methods
Bacterial strains and culture methods
In this study 70 L. sakei isolates identified in a previous investigation were used. For routine sub-culturing the strains were propagated anaerobically in De Man Rogosa Sharp (MRS) broth (Biolife Italiana Srl., Milan, Italy) at 30°C. Listeria monocytogenes ATCC 7644, L. monocytogenes 19111, L. innocua ATCC 33090 and 12 still unidentified Enterobacteriaceae isolates from Ventricina del Vastese sausage were propagated aerobically in Tryptone Soy Broth [TSB; Oxoid Spa., Rodano (MI), Italy] at 30 and 37°C, respectively. Fermentation profiles were obtained using the API 50 CHL system [BioMerieux Italia Spa., Bagno a Ripoli (FI), Italy], after incubation for 48 h at 30°C in anaerobiosis.
The strains of L. sakei were inoculated in a culture broth consisting of 10 g/L of Lab Lemco powder (Oxoid) to assess the growth capacity in different conditions in a substrate similar to meat. The cultures were then incubated at 28°C for 48 h.
The acidifying ability was determined by measuring the pH of 10 g/L Lab Lemco suspensions inoculated with 1% (v/v) of fresh culture at 0, 3, 4, 6, 12, 24, 48 and 72 h. Growth was followed by measuring the OD600 with a UV/Vis Spectrometer Jasco V-530 [Jasco Europe Srl., Cremella (LC), Italy].
The capacity to grow at 5 and 15°C and in presence of salt at concentrations 0, 5, 7.5 and 10% w/v was tested during an incubation period of 14 days. Growth was considered to be positive when an OD600 of at least 0.150 was reached.
The type of fermentative metabolism was assayed in homofermentative-heterofermentative differential (HHD) broth (Biolife Italiana Srl.).
The ability to produce histamine and tyramine was assayed as described by Joosten and Northolt (1989). The antagonistic activity was tested by the agar spot deferred method on MRS modified according to Schillinger and Lücke (1989), i.e. with ten fold less glucose (final concentration 2 g/L), as follows: 5 µL of L. sakei fresh culture were placed on the surface of modified MRS in a Petri plate and let to develop overnight aerobically at 30°C. Then 5 mL of tryptic soy agar (TSA), containing 5 g/L agar, inoculated with 50 µL fresh culture of indicator strain, Listeria spp. or Enterobacteriaceae isolates, were poured on the modified MRS with the L. sakei spots. Incubation was carried out for 48 h at 30°C and at 37°C for Listeria spp. and Enterobacteriaceae, respectively.
Molecular techniques
Genomic DNA was extracted from 1 ml of fresh bacterial culture by the Genomic DNA Extraction kit, according to manufacturer’s protocol (RBC Bioscience, New Taipei City, Taiwan). For genotyping of the 70 L. sakei isolates, the Rep-PCR with the primer GTG5 and agarose gel separation were applied as described by Versalovic et al. (1994). Identification to species level was confirmed by the PCR test reported by Berthier and Ehrlich (1999) and by sequencing of the 16S rRNA gene. Amplification of a region of the 16S rRNA gene of c.a. 1500 bp was carried out according to Bringel et al. (2005) and sequencing was done on the PCR fragments purified with the HiYield Gel/PCR Fragment Extraction Kit (RBC Bioscience) by Eurofins Genomics (Ebersberg, Germany) with the same oligonucleotides used for the amplification. The species identification was obtained by BLAST alignment with the public database.
Results
Seventy L. sakei isolates (Amadoro et al., 2013) were typed by Rep-PCR and six genotypes could be distinguished, so that six strains, each representative of one genotype, were retained for further characterisation (Figure 1). Noticeably, most of the strains with similar profiles, namely L. sakei LS1, LS2, and LS3 were isolated from sausages ripened in controlled conditions, while L. sakei LS4 and the most genetically dissimilar, L. sakei LS5 and LS6, that represented a minority of the isolates, 5 and 3, respectively, were isolated from sausages ripened in the traditional way (Amadoro et al., 2013).
Figure 1.
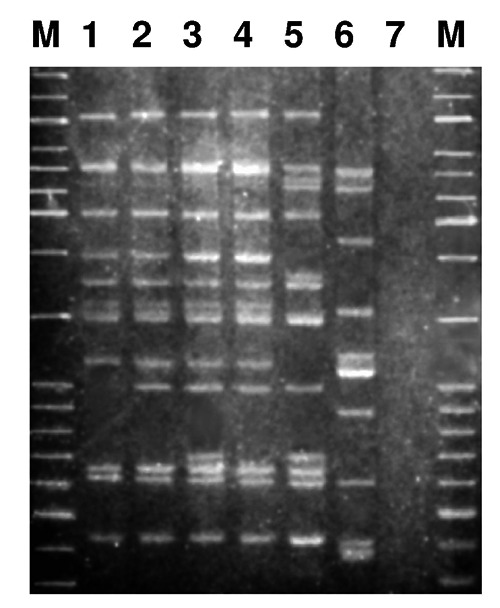
Electrophoretic profiles corresponding to the six strains representative of the six Lactobacillus sakei genotypes (lane 1, strain LS1; lane 2, strain LS2; lane 3, strain LS3; lane 4, strain LS4; lane 5, strain LS5; lane 6, strain LS6) identified in Ventricina del Vastese sausages manufactured in the same producing plant. Lane 7, negative control; M, mixture of GeneRuler 1 kb Plus and 100 bp DNA Ladders [Carlo Erba Reagents, Cornaredo (MI), Italy].
The strains exhibited also distinct fermentation profiles showing different carbohydrates utilisation profiles (Table 1). All fermented D-Ribose, D-Galactose, D-Glucose, D-fructose, D-Mannose, N-Acetylglucosamine, Amygdalin, D-Melibiose, Sucrose, D-Trehalose and hydrolysed Esculin ferric citrate. L. sakei LS6 exhibited the widest substrate utilisation capacity. All strains gave a homofermentative reaction when grown in HHD medium except L. sakei LS5. The ability to grow at temperature as low as 5°C in meat extract suspension was common to all strains, however L. sakei LS4 and LS6 were inhibited by salt in this medium, even at the lowest concentration assayed of 5% (w/v). L. sakei LS4 was inhibited by 5% (w/v) NaCl also at 15°C, while strain LS6 was not (data not shown). The six strains showed different growth behaviors in meat extract, and their growth capacity was limited in this medium. All strains except LS6 acidified the medium with a similar trend, comprising a rapid drop of about 1 pH unit in 3 h and a new increase to the initial levels starting after 6 h. Only for strain L. sakei LS6, pH reached lower values and remained rather stable until 72 h (Figure 2). None of the strains produced histamine or tyramine in vitro.
Table 1.
Physiological characteristics of Lactobacillus sakei strains isolated from Ventricina del Vastese sausages.
| Physiological characteristics | Strain | |||||
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | |
| Sorbitol | - | - | - | - | - | + |
| Methyl-αD-mannopyranoside | - | - | - | - | - | + |
| Methyl-αD-glucopyranoside | - | - | + | - | - | - |
| Amylose | - | - | - | - | - | + |
| Arbutin | - | - | - | - | - | + |
| Salicin | - | - | - | - | + | + |
| D-Cellobiose | + | + | + | + | - | + |
| D-Maltose | - | - | - | - | - | + |
| D-Lactose | - | - | - | - | + | + |
| D-Melezitose | - | - | - | - | - | + |
| D-Raffinose | - | - | - | - | - | + |
| Gentiobiose | ± | ± | + | ± | - | + |
| Potassium gluconate | ± | ± | ± | ± | ± | + |
| D-Turanose | - | - | - | - | - | + |
| D-Arabitol | - | - | - | - | - | ± |
| NaCl 10%, 5°C | + | + | + | - | + | - |
| NaCl 5%, 15°C | + | + | + | - | + | + |
| NaCl 10%, 15°C | + | + | + | - | + | - |
±, uncertain reaction. Only traits found to be variable are reported.
Figure 2.
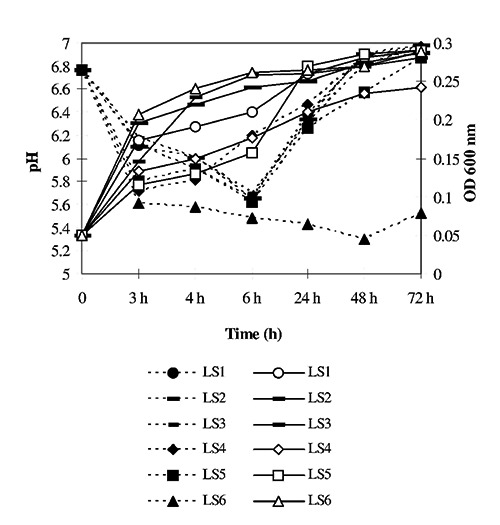
Growth and acidification of Lactobacillus sakei strains in 1% (w/v) meat extract suspension. Open symbols correspond to growth levels and solid symbols to pH values. Data are the average of two measurements.
The L. sakei strains were tested for inhibiting capacity against 12 Enterobacteriaceae isolates previously obtained from Ventricina del Vastese sausage at 10 days of ripening (unpublished), and against L. monocytogenes and L. innocua strains. None of the strains showed a strong inhibiting capacity. Nevertheless, a small inhibition zone was visible around all the L. sakei spots in the plates inoculated with all the Enterobacteriaceae isolates tested as in the example shown in Figure 3A. Very slight inhibition towards L. monocytogenes and L. innocua was noticed only for L. sakei LS5 (Figure 3B).
Figure 3.
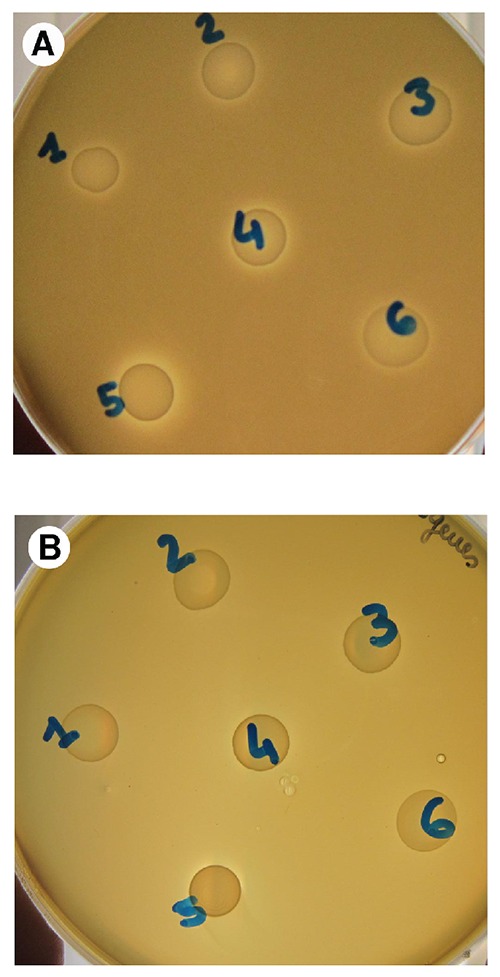
Antagonistic effect of the six Lactobacillus sakei strains in vitro against an Enterobacteriaceae isolate (A) and Listeria monocytogenes ATCC 19111 (B).
Discussion
The genetic heterogeneity of L. sakei has been reported for strains isolated from different sources, i.e., meat, fish and plant material (Chaillou et al., 2009). However, in this study, the occurrence of different biotypes was demonstrated even in the same product from a single producing plant. In addition, it was shown that different environmental conditions can drive the selection of particular strains. The results are in agreement with the low levels of genetic relatedness among L. sakei strains found in other studies that proposed two subspecies, L. sakei subsp. sakei and L. sakei subsp. carnosus, and, more recently, three main evolutionarily divergent lineages (Klein et al., 1996; Torriani et al., 1996; Chaillou et al., 2009, 2013). However, the distinction at the subspecies level was not investigated in this study, since the molecular techniques applied were different from those proposed to attribute the L. sakei strains to the two subspecies and biochemical tests cannot be used to this purpose (Klein et al., 1996).
The strains examined exhibited also physiological variability, in accordance with that observed in other studies reporting phenotypic differences that rendered difficult the classification of L. sakei strains on the basis of fermentation profiles (Klein et al., 1996; Torriani et al., 1996).
Strain L. sakei LS6 had the widest fermentation spectrum and was able to ferment substrates not previously reported for L. sakei strains (Nyquist et al., 2011). Moreover, the ability of L. sakei LS5 and LS6 to ferment lactose, that was previously observed for strains isolated from Spanish sausages (Nyquist et al., 2011), is interesting for application in sausages where this sugar is added. A particular finding is the heterofermentative-like behaviour of L. sakei LS5 in HHD medium, considered that such phenotype was not observed before (Ammor et al., 2005).
Strain L. sakei LS6 is the most versatile among the biotypes characterised in this study, and also the one that permitted a better and long lasting acidification in meat extract (Figure 2). However it exhibited a low tolerance to NaCl (Table 1). This may be the reason why it was retrieved only from sausages ripened at low temperature according to the traditional process, where slower drying may have favoured its persistence.
The strains studied exhibited some capacity to inhibit the growth of Enterobacteriaceae and strain LS5 also slightly inhibited Listeria spp. Therefore a general capacity of L. sakei to contrast the growth of Enterobacteriaceae in fermented meat products can be hypothesised and requires confirmation by testing different species and by examining their inhibition during sausage ripening. On the other hand, evidence that L. sakei strains, even if non bacteriocinogenic, are capable to retard spoilage and to reduce the number of bacterial pathogens by developing dominant populations is presently available for vacuum packaged fresh meat (Jones et al., 2009). An explanation for the inhibitory effects exerted by non bacteriocinogenic strains might be the competition for nutrients and better colonising ability of meat, given the specialisation of L. sakei for this ecological niche (Nyquist et al., 2011). However, a low level of bacteriocin production cannot be ruled out for the strains examined in this study and should be investigated by PCR based assays for the presence of bacteriocin encoding genes and, eventually, for their expression during sausage ripening. Indeed, on the basis of what reported by Todorov et al. (2013), the low inhibition effect observed in this study might be due to a low bacteriocin production determined by the low concentration of glucose in the test medium.
In particular, the ability to produce bacteriocins should be investigated for L. sakei LS5, which exhibited a slight anti-listerial effect in vitro. Indeed, recent findings confirmed that the anti-listerial potential of bacteriocinogenic L. sakei represents an efficient tool to contrast the development of L. monocytogenes in food even at low temperatures (Martinez et al., 2015).
Evidences obtained in this study indicated that for an appropriate evaluation of the role of L. sakei on Ventricina del Vastese microbiological safety, challenge tests with different bacterial contaminants, co-inoculated with single strain or combined L. sakei cultures, should be carried out.
Finally, the inability to form BAs is in line with the fact that this physiological trait was never reported to occur in L. sakei strains. The capacity of autochthonous L. sakei strains to become dominant in the microbial population associated to Ventricina del Vastese sausage suggested that they can reduce the risk of accumulation of these hazardous compounds also in this product by outcompeting BA forming bacterial species.
Conclusions
This study represents a contribution for knowledge on the genotypic, phenotypic and technological characteristics of L. sakei biotypes occurring in Italian sausages. It emerged that the L. sakei population of a traditional sausage production, for the same manufacturer and for a single production cycle, is genotypically and phenotypically heterogeneous and that some strains could be isolated only in the product which was subjected to the traditional ripening process at low temperature. The limited growth and pH lowering capacity shown by most strains in pure cultures and in meat extract alone suggests that these bacterial species need nutritional sources that are made available by other components of the sausage microbiota for optimal development. However, some strains can be selected, such as L. sakei LS6 that are endowed with a good acidification capacity.
All strains were able to slightly inhibit Enterobacteriaceae isolated from the same product by mechanisms different from acidification. This suggested the possibility that authochtonous L. sakei strains play a role in the decline of such microbial groups and prevent the formation of undesired metabolites. However, most strains did not inhibit L. monocytogenes, indicating that only some L. sakei strains can have an effect in limiting this pathogen in the product.
Acknowledgements
This study was funded by the project FILVEA, under the Programme of Rural Development 2007/2013 of the Abruzzi region, Italy.
References
- Amadoro C, Rossi F, Piccirilli M, Berardino L, Colavita G, 2013. Studio della flora microbica pro-tecnologica nella ventricina. Ing Alim 50:51-4. [Google Scholar]
- Ammor S, Dufour E, Zagorec M, Chaillou S, Chevallier I, 2005. Characterization and selection of Lactobacillus sakei strains isolated from traditional dry sausage for their potential use as starter cultures. Food Microbiol 22:529-38. [Google Scholar]
- Berthier F, Ehrlich SD, 1999. Genetic diversity within Lactobacillus sakei and Lactobacillus curvatus and design of PCR primers for its detection using randomly amplified polymorphic DNA. Int J Syst Bacteriol 49:997-1007. [DOI] [PubMed] [Google Scholar]
- Bover-Cid S, Izquierdo-Pulido M, Vidal-Carou MC, 2001. Effectiveness of a Lactobacillus sakei starter culture in the reduction of biogenic amine accumulation as a function of the raw material quality. J Food Protect 64:367-73. [DOI] [PubMed] [Google Scholar]
- Bringel F, Castioni A, Olukoya DK, Felis GE, Torriani S, Dellaglio F, 2005. Lactobacillus plantarum subsp. argentoratensis subsp. nov., isolated from vegetable matrices. Int J Syst Evol Micr 55:1629-34. [DOI] [PubMed] [Google Scholar]
- Chaillou S, Champomier-Vergès MC, Cornet M, Crutz-Le Coq AM, Dudez AM, Martin V, Beaufils S, Darbon-Rongère E, Bossy R, Loux V, Zagorec M, 2005. The complete genome sequence of the meat-borne lactic acid bacterium Lactobacillus sakei 23K. Nat Biotechnol 23:1527-33. [DOI] [PubMed] [Google Scholar]
- Chaillou S, Daty M, Baraige F, Dudez AM, Anglade P, Jones R, Alpert CA, Champomier-Vergès MC, Zagorec M, 2009. Intraspecies genomic diversity and natural population structure of the meat-borne lactic acid bacterium Lactobacillus sakei. Appl Environ Microb 75:970-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaillou S, Lucquin I, Najjari A, Zagorec M, Champomier-Vergès MC, 2013. Population genetics of Lactobacillus sakei reveals three lineages with distinct evolutionary histories. PLoS One 8:e73253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dossmann MU, Vogel RF, Hammes WP, 1996. Mathematical description of the growth of Lactobacillus sake and Lactobacillus pentosus under conditions prevailing in fermented sausages. Appl Microbiol Biot 46:334-9. [DOI] [PubMed] [Google Scholar]
- Fadda S, Sanz Y, Vignolo G, Aristoy M, Oliver G, Toldrá F, 1999. Hydrolysis of pork muscle sarcoplasmic proteins by Lactobacillus curvatus and Lactobacillus sake. Appl Environ Microb 65:578-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garriga M, Rubio R, Aymerich T, Ruas-Madiedo P, 2015. Potentially probiotic and bioprotective lactic acid bacteria starter cultures antagonise the Listeria monocytogenes adhesion to HT29 colonocyte-like cells. Benef Microb 6:337-43. [DOI] [PubMed] [Google Scholar]
- Jones RJ, Zagorec M, Brightwell G, Tagg JR, 2009. Inhibition by Lactobacillus sakei of other species in the flora of vacuum packaged raw meats during prolonged storage. Food Microbiol 26:876-81. [DOI] [PubMed] [Google Scholar]
- Joosten HMLJ, Northolt MD, 1989. Detection, growth, and amine-producing capacity of lactobacilli in cheese. Appl Environ Microb 55:2356-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandler O, Weiss N, 1986. Genus lactobacillus Beijerinck. Sneath PHA, Mair NS, Sharpe ME, Holt JG, eds. Bergey’s manual of systematic bacteriology. Williams and Wilkins, Baltimore, MA, USA, pp 1209-34. [Google Scholar]
- Klein G, Dicks LMT, Pack A, Hack B, Zimmerman K, Dellaglio F, Reuter G, 1996. Emended descriptions of Lactobacillus sake (Katagiri, Kitahara, and Fukami) and Lactobacillus curvatus (Abo-Elnaga and Kandler): numerical classification revealed by protein fingerprinting and identification based on biochemical patterns and DNA-DNA hybridizations. Int J Syst Bacteriol 46:367-76. [Google Scholar]
- Martinez RC, Staliano CD, Vieira AD, Villarreal ML, Todorov SD, Saad SM, Franco BD, 2015. Bacteriocin production and inhibition of Listeria monocytogenes by Lactobacillus sakei subsp. sakei 2a in a potentially synbiotic cheese spread. Food Microbiol 48:143-52. [DOI] [PubMed] [Google Scholar]
- Montel MC, 1999. Fermented meat products. Batt CA, Patel PD, eds. Fermented foods, Encyclopedia of food microbiology. Academic Press, London, UK, pp 744-53. [Google Scholar]
- Nyquist OL, McLeod A, Brede DA, Snipen L, Aakra Å, Nes IF, 2011. Comparative genomics of Lactobacillus sakei with emphasis on strains from meat. Mol Genet Genomics 285:297-311. [DOI] [PubMed] [Google Scholar]
- Park CW, Youn M, Jung YM, Kim H, Jeong Y, Lee HK, Kim HO, Lee I, Lee SW, Kang KH, Park YH, 2008. New functional probiotic Lactobacillus sakei probio 65 alleviates atopic symptoms in the mouse. J Med Food 11:405-12. [DOI] [PubMed] [Google Scholar]
- Schillinger U, Lücke FK, 1989. Antibacterial activity of Lactobacillus sake isolated from meat. Appl Environ Microb 55:1901-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todorov SD, Vaz-Velho M, de Melo Franco BDG, Holzapfel WH, 2013. Partial characterization of bacteriocins produced by three strains of Lactobacillus sakei, isolated from salpicao, a fermented meat product from North-West of Portugal. Food Control 30:111-21. [Google Scholar]
- Torriani S, Van Reenen CA, Klein G, Reuter G, Dellaglio F, Dicks LMT, 1996. Lactobacillus curvatus subsp. curvatus nov. and Lactobacillus curvatus subsp. melibiosus nov. and Lactobacillus sake subsp. sake subsp. nov. and Lactobacillus sake subsp. carnosus subsp. nov., new subspecies of Lactobacillus curvatus Abo-Elnaga and Kandler 1965 and Lactobacillus sake Katagiri, Kitahara, and Fukami 1934 (Klein etal. 1996, emended descriptions), respectively. Int J Syst Bacteriol 46:1158-63. [DOI] [PubMed] [Google Scholar]
- Versalovic J, Schneider M, de Bruijn FJ, Lupski JR, 1994. Genomic fingerprinting of bacteria using repetitive sequence-based polymerase chain reaction. Method Mol Cell Biol 5:25-40. [Google Scholar]
- Walter J, Hertel C, Tannock GW, Lis CM, Munro K, Hammes WP, 2001. Detection of Lactobacillus, Pediococcus, Leuconostoc, and Weissella species in human feces by using group-specific PCR primers and denaturing gradient gel electrophoresis. Appl Environ Microb 67:2578-85. [DOI] [PMC free article] [PubMed] [Google Scholar]


