Abstract
FAM20C is a newly identified kinase on the secretory pathway responsible for the phosphorylation of serine residues in the Ser-x-Glu/pSer motifs in several enamel matrix proteins. Fam20C-knockout mice showed severe enamel defects very similar to those in the ameloblastin (Ambn)–knockout mice, implying that phosphoserines may have a critical role in AMBN function. To test this hypothesis, we generated amelogenin (Amel) promoter-driven Ambn-transgenic mice, in which Ser48, Ser226, and Ser227 were replaced by aspartic acid (designated as D-Tg) or alanines (designated as A-Tg). The negative charge of aspartic acid is believed to be able to mimic the phosphorylation state of serine, while alanine is a commonly used residue to substitute serine due to their similar structure. Using Western immunoblotting and quantitative polymerase chain reaction, the authors identified transgenic lines expressing transgenes somewhat higher (Tg+) or much higher (Tg++) than endogenous Ambn. The lower incisors collected from 7-d-old and 7-wk-old mice were analyzed by histology, scanning electron microscopy, immunohistochemistry, and Western immunoblotting to examine the morphology and microstructure changes in enamel, as well as the expression pattern of enamel matrix proteins. The A-Tg+ and A-Tg++ mice displayed severe enamel defects in spite of the expression level of transgenes, while the D-Tg+ and D-Tg++ mice showed minor to mild enamel defects, indicating that the D-Tg transgenes disturbed enamel formation less than the A-Tg transgenes did. Our results suggest that the phosphorylation state of serines is likely an essential component for the integrity of AMBN function.
Keywords: phosphoserine, FAM20C, amelogenesis, transgene, extracellular matrix, kinase
Introduction
Dental enamel is the most highly mineralized hard tissue and is unique in its composition and formation. Enamel formation is a strictly controlled stepwise process in which ameloblasts continuously secrete organic extracellular matrix while slowly moving away from the dentinal surface until the desired thickness of the matrix is achieved. In mammals, the enamel formation is artificially divided into presecretion, secretion, and maturation (Hu et al. 2007). During the secretion stage, ameloblasts secrete enamel matrix proteins (EMPs). These include amelogenin (AMEL), ameloblastin (AMBN), enamelin (ENAM), and matrix metalloproteinase 20, a calcium-dependent peptidase that cleaves newly secreted EMPs into various derivative fragments (Bartlett et al. 2004; Simmer et al. 2012). AMEL represents 90% of the matrix deposited, while AMBN and ENAM form the majority of the remaining 10% of the organic matrix.
In humans, the hypomineralization or hypoplastic classes of amelogenesis imperfecta have been related to mutations in the following genes: AMELX (Hart et al. 2002; Kim et al. 2004; Stephanopoulos et al. 2005; Urzúa et al. 2011), ENAM (Hart et al. 2003; Hu and Yamakoshi 2003; Kim et al. 2005; Stephanopoulos et al. 2005), and recently AMBN (Poulter et al. 2014). The mouse model in which exons 5 and 6 are deleted from Ambn showed severe hypoplastic amelogenesis imperfecta that had no enamel formation on the tooth surface (Fukumoto et al. 2004). Ambn overexpression in transgenic mice resulted in abnormal enamel crystallite formation and enamel rod morphology (Paine et al. 2003; Chun et al. 2010) or a generally normal enamel structure when the transgene expression was only somewhat higher than normal (Chun et al. 2010).
Evolutionary analyses have classified AMEL, AMBN, and ENAM into a family named the secretory calcium-binding phosphoproteins, which have ≥1 Golgi casein kinase phosphorylation sites that are recognized by their distinctive Ser-x-Glu/pSer motifs (Brunati et al. 2000; Kawasaki and Weiss 2003). Family with sequence similarity member 20-C (FAM20C) is a newly discovered kinase localized to the Golgi that is believed to be the genuine casein kinase phosphorylating secretory calcium-binding phosphoproteins (Ishikawa et al. 2012; Tagliabracci et al. 2012). Fam20C-knockout mice exhibit severe enamel defects very similar to those in the Ambn- or Enam-knockout mice (Wang et al. 2013; Wang, Wang, Lu, et al. 2012), suggesting that the phosphorylation of serines in EMPs may be an essential posttranslational modification required for their proper function.
The consensus sequences of Ser-x-Glu/pSer motifs are strictly conserved in EMPs (Hu et al. 2005; Al-Hashimi et al. 2010). Three putative phosphoserine (pSer) residues have been identified in the AMBN of pig (Ser17, Ser209, and Ser210), mouse (Ser48, Ser226, and Ser227), and human (Ser17, Ser235, and Ser236). A recent bioinformatic study (Delsuc et al. 2015) indicated that these pSers are highly conserved in AMBN across the species during evolution and thus may have important roles for AMBN function. To examine the functional significance of the pSers in AMBN, we generated Amel promoter-driven Ambn transgenic mice, in which pSer48, pSer226, and pSer227 of the exogenous AMBN were replaced by alanine (Ala) or aspartic acid (Asp) to eliminate or mimic the phosphorylation state of serines.
Materials and Methods
Animals
All animal procedures were approved by the Institutional Animal Care and Use Committee of the Texas A&M Baylor College of Dentistry and performed in accordance with the National Institutes of Health’s Guide for the Care and Use of Laboratory Animals.
The plasmid used for generating Ambn transgenic constructs was a gift from Dr. Jan Hu (Department of Biologic and Materials Sciences, University of Michigan School of Dentistry; Chun et al. 2010). We mutated the cDNA sequences coding Ser48, Ser226, and Ser227 in the Ambn transgene into those encoding Ala (designated as A-Tg) or Asp (designated as D-Tg). The 7.5-kb Ambn transgenes were released from the mutated constructs by restriction digestion with NotI-SrfI, microinjected into fertilized C57BL/6 oocytes, and transferred to recipients at the Transgenic Core Facility of the University of Texas Southwestern Medical Center at Dallas. In total, 6 lines of A-Tg mice and 5 lines of D-Tg mice were generated and mated with C57BL/6 mice. Germline transmission was determined by polymerase chain reaction (PCR) analyses of genomic DNA obtained from tail biopsies with transgene-specific primers, as described previously (Chun et al. 2010). The mice used for enamel morphometric analyses were fed with gel food after weaning.
Assessment of Ambn Transgenic Expression by Western Blotting and Quantitative PCR
To determine the protein expression levels of Ambn transgenes, the molars dissected from 5-d-old mice (3 for each group) were ground into powder in liquid nitrogen and extracted with RIPA buffer (Thermo Fisher Scientific) containing a proteinase inhibitor cocktail (Roche). After bicinchoninic acid protein assay (Thermo Fisher Scientific), equal amounts of lysates from wild-type (WT) and transgenic mice were analyzed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis, followed by Western immunoblotting with anti-AMBN antibody (1/1,000, SC-50534; Santa Cruz Biotechnology) and anti-β-ACTIN antibody (1/3,000, SC-2004; Santa Cruz Biotechnology). The membranes were incubated with horseradish peroxidase–conjugated secondary antibody (Santa Cruz Biotechnology). The blots were visualized with an enhanced chemiluminescence kit (Amersham Biosciences), according to the manufacturer’s instructions.
We also examined the expression levels of ENAM and AMEL in the molar lysates by Western blot using anti-ENAM (1/1,600; Brookes et al. 2011) and anti-AMEL (1/400, SC-32892; Santa Cruz Biotechnology).
To determine the transcriptional levels of Ambn transgenes, total RNAs were isolated from the molars of 5-d-old mice (3 for each group) with an RNeasy Mini Kit (Qiagen) and converted into cDNAs through a Reverse Transcription Kit (Qiagen), according to the manufacturer’s instructions. Quantitative real-time PCR was performed with a Bio-Rad CFX96 system and SYBR Green Master Mix (Stratagene), as previously described (Wang, Wang, Li, et al. 2012). The Ct values were normalized to the reference gene 18s rRNA and expressed as fold changes over the WT controls. The primers for quantitative PCR analysis of AMBN and 18s RNA were purchased from SABiosciences.
Plain X-ray and Backscatter Scanning Electron Microscopy
The lower jaws dissected from 7-wk-old mice (3 for each group) were fixed in 4% paraformaldehyde in PBS overnight and analyzed with plain x-ray radiography (Faxitron Bioptics). Then, the mandibles were dehydrated through gradient concentrations of ethanol (70% to 100%) and embedded in methylmethacrylate. The samples were cut at the position of the first lower molar to cut the incisor transversely, then coated with carbon and examined by field emission scanning electron microscopy (Philips XL30; FEI Company), as previously described (Wang et al. 2015).
Histology and Immunohistochemistry Analyses
The mandibles dissected from 6-d-old mice (3 for each group) were fixed in 4% paraformaldehyde in PBS for 24 h at 4 °C, then decalcified in 8% EDTA/PBS (pH 7.4) at 4 °C for 4 d and embedded in paraffin. Serial sections (5 µm) were prepared for hematoxylin and eosin staining and immunohistochemistry staining, as described previously (Wang et al. 2013). The primary antibodies used for immunohistochemistry staining of AMBN, ENAM, and AMEL were the same as those used for Western blot analyses.
Results
Establishment of A-Tg and D-Tg Ambn Transgenic Lines
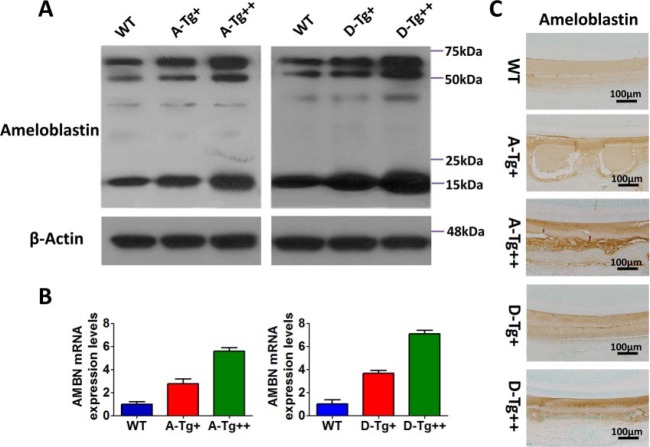
In total, germline transmission was confirmed for 6 lines of A-Tg mice and 5 lines of D-Tg mice. We examined the expression levels of A-Tg and D-Tg in the transgenic lines using quantitative PCR, followed by Western immunoblotting. Four A-Tg lines and 3 D-Tg lines were identified as overexpressing the Ambn transgenes (Appendix Table). The lines overexpressing the transgenes ~2 to 4 folds over WT mice were designated as A-Tg+ and D-Tg+, while those overexpressing the transgenes ~6 to 8 folds over WT mice were designated as A-Tg++ and D-Tg++ (Fig. 1, Appendix Table).
Figure 1.
Evaluation of Ambn A-Tg and D-Tg expression levels in the lower molars from 5-d-old mice. (A) Western immunoblotting identified A-Tg and D-Tg lines in which the total AMBN expression levels were somewhat higher (A/D-Tg+) or much higher (A/D-Tg++) than those of WT mice. (B) Quantitative polymerase chain reaction analysis confirmed that the transcriptional levels of Ambn transgenes were ~2- to 3-fold or ~5- to 7-fold higher than those of WT in each line. (C) Immunohistochemistry staining demonstrated the location of AMBN protein in ameloblasts and enamel matrix in the lower incisor from 7-d-old mice. Scale bars: 100 μm. AMBN, ameloblastin; A-Tg, Ambn transgene bearing serine-alanine mutation; D-Tg, Ambn transgene bearing serine-aspartic acid mutation; WT, wild type.
Gross Enamel Defects in the A-Tg and D-Tg Ambn Transgenic Mice
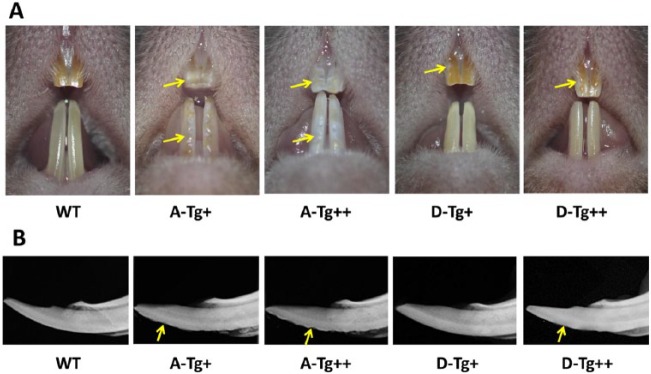
The 7-wk-old A-Tg+ mice showed extensive enamel defects, with discolored pits and dents on the surface of incisors (Fig. 2) and molars (data not shown). X-ray analysis revealed that the incisal edge of A-Tg+ mice appeared to lose the characteristic chisel-like incisal tip exhibited by WT mice—a phenomenon that appeared even more prominent in the A-Tg++ mice (Fig. 2, Appendix Table). The enamel of D-Tg+ mice did not show apparent gross defects except for sporadic pits on the maxillary incisors (Fig. 2), while the enamel of D-Tg++ mice displayed a broader range of defects, which however appeared to be much milder than those in the A-Tg mice (Fig. 2, Appendix Table).
Figure 2.
Gross enamel defects in the lower incisors of 7-wk-old Ambn A-Tg and D-Tg transgenic mice. (A) The A-Tg+ mice showed extensive discolored dents and pits on the enamel surfaces (arrows). The A-Tg++ mice displayed chalky white enamel with patchy lesions on the enamel surfaces (arrows). The enamel of D-Tg+ mice was generally normal except for sporadic discolored lesions on the surfaces (arrow). The enamel surfaces (arrow) of the D-Tg++ mice were more affected than the D-Tg+ mice but not to the same extent as in A-Tg+ and A-Tg++ mice. (B) Lateral x-rays of WT and D-Tg+ incisors displayed similar smooth enamel surface: the enamel of the A-Tg+, A-Tg++, and D-Tg++ mice appeared ridged with evidence of hypoplasia (arrows), with the A-Tg++ mice being the worst affected. A-Tg, Ambn transgene bearing serine-alanine mutation; D-Tg, Ambn transgene bearing serine-aspartic acid mutation; WT, wild type.
Enamel Microstructure of A-Tg and D-Tg Ambn Transgenic Mice
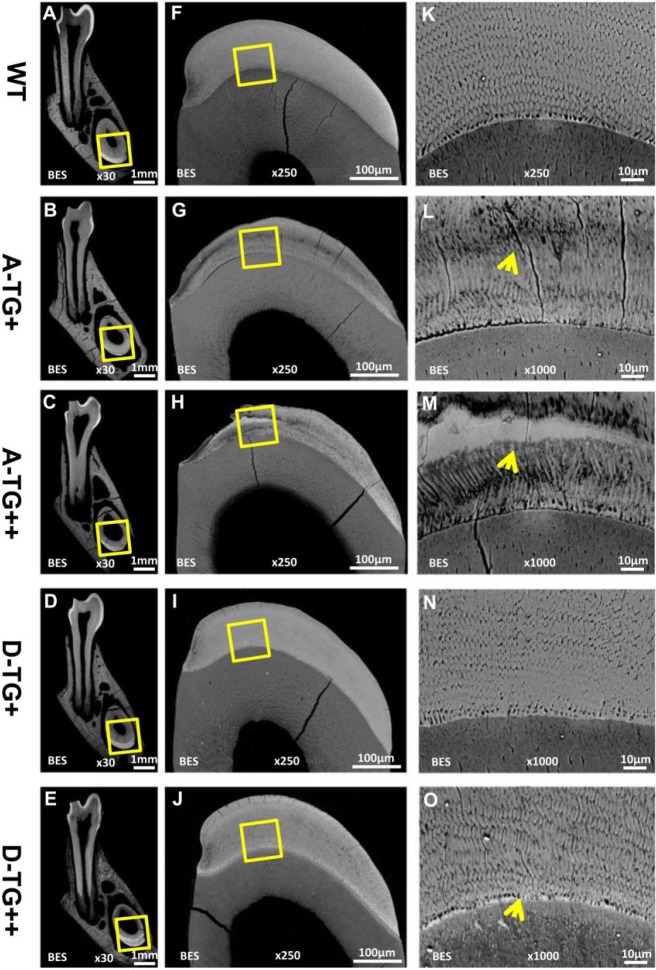
Backscattered scanning electron microscopy analyses revealed a reduced-thickness and disorganized enamel rod and interrod structure mixed with an amorphous middle layer in the enamel of A-Tg+ and A-Tg++ mice, compared with the well-organized enamel structure in WT mice (Fig. 3). The enamel microstructure of D-Tg+ mice appeared more ordered than in A-Tg mice but was less well organized than WT. The enamel microstructure of D-Tg++ mice showed more disturbance than D-Tg+ mice (Fig. 3) but overall was better organized compared with that of the A-Tg mice.
Figure 3.
The enamel microstructure of 7-wk-old A-Tg and D-Tg Ambn transgenic mice. (A–E) Backscatter scanning electron microscopy analyses of the cross sections obtained from a cut through the mesial roots of the first lower molar. (F–J) Higher magnification of the boxed areas in panels A–E. The enamel surfaces of A-Tg+ and A-Tg++ mice were hypoplastic as compared with WT and D-Tg mice. (K–O) Higher magnification of the boxed areas in panels F–J. Compared with the well-organized enamel rods in WT mice, the enamel rods and interrod structures in A-Tg+ mice and A-Tg++ mice were severely disturbed; an area of reduced structural detail (arrows) was located in the middle layer of enamel in these mice. In contrast, the enamel of D-Tg+ and D-Tg++ mice showed an overall better microstructure than that in A-Tg mice, except that the D-Tg mice showed a less defined interrod structure and the enamel rods of D-Tg++ mice were slightly disturbed. The D-Tg mice appeared to have a higher/accelerated mineralization in the enamel matrix at the stage shown in this section. The enamel-dentin junction in the D-Tg++ mice appeared to have a higher mineralization (arrow). Scale bars: 1 mm in panels A–E, 250 μm in panels F–J, 10 μm in panels K–O. A-Tg, Ambn transgene bearing serine-alanine mutation; D-Tg, Ambn transgene bearing serine-aspartic acid mutation; WT, wild type.
Histologic Defects of Enamel in A-Tg and D-Tg Ambn Transgenic Mice
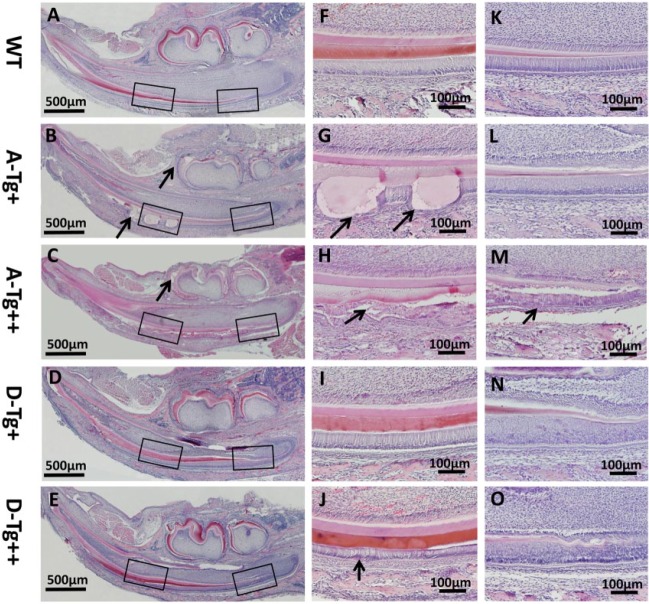
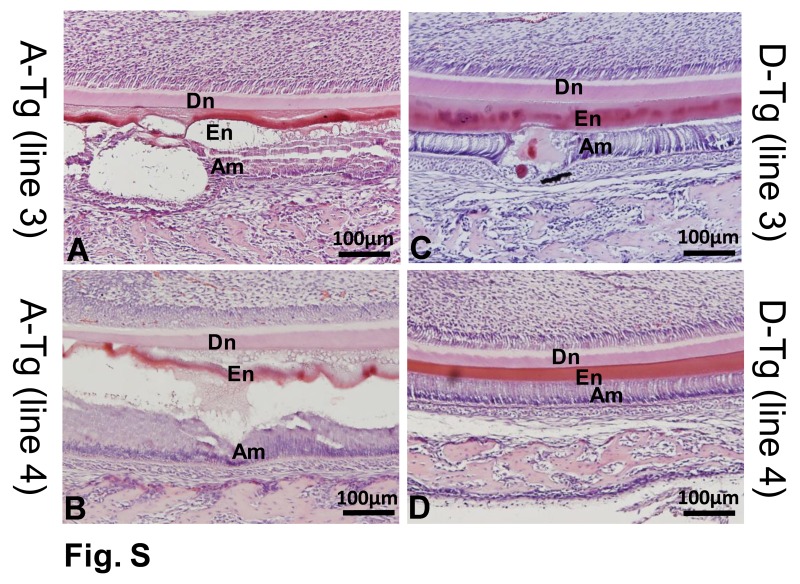
The ameloblasts of 7-d-old A-Tg++ and D-Tg++ mice started to exhibit an abnormal morphology at the presecretory stage (Fig. 4, Appendix Fig., Appendix Table). At the secretory stage, the A-Tg+ mice showed malformed enamel matrix protruding into the ameloblast layer; the A-Tg++ mice displayed disturbed structures in enamel matrix and ameloblasts. In contrast, the D-Tg+ and D-Tg++ mice exhibited generally normal ameloblast morphology and enamel matrix microstructure, except for uneven staining and inconsistent thickness at some locations along the enamel matrix (Fig. 4, Appendix Fig., Appendix Table).
Figure 4.
Histology of the enamel defects in A-Tg and D-Tg Ambn transgenic mice. (A–E) Hematoxylin and eosin staining on the sagittal sections of lower jaws from 7-d-old WT, A-Tg, and D-Tg mice. In the A-Tg mice, the ameloblasts were separated from the enamel matrix at indicated locations in both molars and incisors (arrows). (F–J) Higher magnification of the black boxed areas (~late secretory stage) in panels A–E. The A-Tg+ mice had malformed enamel matrix invading the ameloblast layer (arrows). The A-Tg++ mice showed disorganized enamel matrices and malformed ameloblasts (arrow). The D-Tg+ and D-Tg++ mice showed generally normal ameloblast structure and enamel matrix, except for uneven staining and inconsistent thickness of enamel matrix at indicated locations (arrows). (K–O) Higher magnification of the black boxed areas (~presecretory or early secretory stage) in panels A–E. The ameloblasts of A-Tg++ mice showed disorganized morphology (arrow) as compared with the generally normal ameloblasts in the other lines of Ambn transgenic mice. Scale bars, 500 μm in A–E, 100 μm in panels F–O. A-Tg, Ambn transgene bearing serine-alanine mutation; D-Tg, Ambn transgene bearing serine-aspartic acid mutation; WT, wild type.
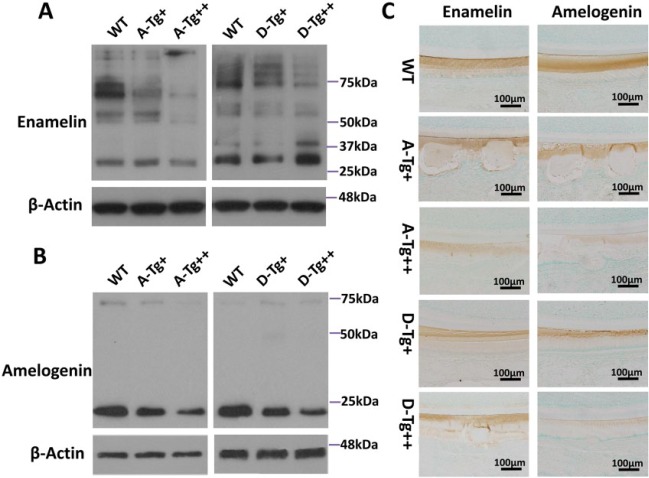
ENAM and AMEL Expression in A-Tg and D-Tg Ambn Transgenic Mouse Molars
The A-Tg and D-Tg mice both showed reduced amounts of ENAM intermediate cleavage fragments migrating at about 50 to 80 kDa as compared with those in the WT mice. The A-Tg mice had a similar amount of 32-kDa ENAM fragments as the WT mice, while the D-Tg++ mice displayed an increased amount of these fragments (Fig. 5). The A-Tg and D-Tg mice both showed less-than-normal amounts of AMEL cleavage fragments (Fig. 5).
Figure 5.
The expression patterns of ENAM and AMEL in 5-d-old A-Tg and D-Tg Ambn transgenic mice. (A) Western immunoblotting of ENAM showed reduced amounts of 50- to 80-kDa intermediate fragments in the lower molars of A-Tg and D-Tg mice versus the WT mice. The D-Tg++ mice appeared to have more 32-kDa fragments. (B) Western immunoblotting of AMEL showed fewer 20-kDa fragments in the lower molars of A-Tg and D-Tg mice than WT mice, and the reduction appeared reversely related to the expression levels of Ambn transgene. (C) Immunohistochemical staining of ENAM (left column) and AMEL (right column) at the secretory stage of the lower incisors from 7-d-old mice. Scale bars: 100 μm. AMEL, amelogenin; A-Tg, Ambn transgene bearing serine-alanine mutation; D-Tg, Ambn transgene bearing serine-aspartic acid mutation; ENAM, enamelin; WT, wild type.
Discussion
In previous studies, we demonstrated that inactivation of FAM20C—a kinase localized to the Golgi apparatus that phosphorylates the serine residues in EMPs—led to severe enamel defects that are similar to those in the Ambn-knockout mice (Wang, Wang, Lu, et al. 2012; Wang et al. 2013), suggesting that the phosphorylation state of serines may be essential to AMBN function. In this study, we generated Ambn transgenic mice in which the pSers in the Ambn transgene were eliminated by replacing the pSers with Ala or mimicked by substituting the pSers with Asp. Ala is the most commonly used amino acid to substitute serine residues due to their similar structures (Dayhoff et al. 1978), while Asp, with its negatively charged side chain, is frequently used for mimicking pSer (Thorsness and Koshland 1987; Pearlman et al. 2011).
A previous study (Chun et al. 2010) showed that overexpression of WT Ambn led to minor or mild enamel defects when the transgene dosage was slightly higher than normal (~3 folds based on the Western blot) or much higher (~6 to 9 folds). As the transgene is randomly inserted into the genomic DNA, no 2 transgenic lines are genetically identical, whereas their phenotypes could be very similar. In this regard, the mice overexpressing WT Ambn (Chun et al. 2010) can serve as controls in comparison with those overexpressing the mutant versions. In our study, D-Tg+ and D-Tg++ mice showed minor and mild enamel defects when the transgene dosage was slightly higher than normal (~3 folds) and much higher (~7 folds). The correlation between the gene dosage and enamel phenotype in D-Tg mice was very similar to that in the controls. In contrast, overexpression of A-mutant Ambn consistently led to severe enamel defects in spite of the expression levels, although the severity of defects increased with gene dosage (Appendix Table). These results suggest that the enamel defects in D-Tg mice are mainly associated with the transgene dosage while those in A-Tg mice are mainly due to the mutation. Theoretically, it might be assumed that when compared with a serine-to-Asp substitution, a serine-to-Ala substitution would have a smaller effect on protein conformation or function due to the similarity in structure between serine and Ala but only if the posttranslational modifications (e.g., phosphorylation) associated with the serine residue are not of significant importance. Since aspartic acids can (partially) mimic the phosphorylation state of serines, our results outlined above (i.e., D-Tg++ mice were relatively unaffected) suggest that the phosphorylation of serines may be an important functional posttranslational modification.
To date, genetically engineered amelogenesis imperfecta mouse models have faithfully recapitulated the phenotypes and inheritance manners of human amelogenesis imperfecta. The Ambn- and Enam-mutant subjects develop autosomal-dominant amelogenesis imperfecta in a dose-dependent manner, while those bearing Amel mutations show X-linked traits. Studies of Ambn knockout (Fukumoto et al. 2004; Wazen et al. 2009) and rescue (Chun et al. 2010) suggest that the amelogenesis imperfecta caused by AMBN truncation was most likely due to haploinsufficiency. However, it remains unclear whether point mutations of AMBN can interfere with WT protein in a dominant negative manner, as point mutation of Ambn has not been reported in humans and animals. In our study, the overdose of A-mutant AMBN caused severe enamel defects, while overexpression of D-mutant protein resulted in only mild defects at the similar dosages, suggesting that the A-mutant AMBN had interfered with WT protein in an unknown manner (e.g., dominant negative) whereas the D-mutant AMBN acted more like the WT version.
The enamel defects of Fam20C-null mice are very similar to those in the Ambn-null mice: both showed severe defects in ameloblasts and enamel matrix and thus impeded enamel formation on the dentin surface. In contrast, the A-Tg mice did not show severe dentoenamel junction defects, and the enamel defects were milder than those in Ambn- and Fam20C-null mice, suggesting that either the endogenous AMBN may alleviate the enamel defects or the mutant protein did not eliminate AMBN function. The role of pSers remains elusive in proper mineralization, cell-amblastin interaction, and AMBN-AMEL interactions; stepwise and tremendous works are needed in future studies using knock-in, cell biology, biochemistry, and biophysics approaches.
A limitation of this study is that the transgenic mice also express endogenous Ambn as well as the A-Tg and D-Tg transgenes, which added complexity to the interpretation of the results. It remains unclear how the mutant AMBN proteins interacted with the endogenous AMBN and how the latter’s function may have been affected in the transgenic mice. A further limitation is that we cannot rule out the possibility that AMBN undergoes a toxic gain of function when serine is replaced by Ala. If this were the case, one other possible explanation for the results obtained may be the toxic effects of A-Tg on amelogenesis (acting in the extracellular matrix or intracellularly). D-Tg may simply be a harmless bystander, while amelogenesis proceeds relatively normal due to the presence of WT AMBN.
These limitations can be overcome in future studies where the effect of endogenous AMBN has been eliminated by generating knock-in mice or crossbreeding the A-Tg and D-Tg mice with Ambn-knockout mice, thus potentially providing a more clear answer to the role of pSers in AMBN function.
The A-Tg and D-Tg mice also displayed fewer 20-kDa AMEL fragments and 50- to 80-kDa ENAM fragments in the molars as compared with those in the WT mice. However, it remains unclear whether the reduction of these intermediate fragments contributed to the enamel defects, because we did not identify a consistent correlation between the severity of enamel defects and the degree by which expression of these proteins was reduced. The ENAM data obtained in this study contrasted with previous data obtained through this specific ENAM antibody. Brookes et al. (2011) showed that the antibody failed to recognize ENAM at 32 kDa, but we observed clear cross reactivity. This interesting difference may be due to the fact that we analyzed developing mouse molars, whereas Brookes et al. analyzed rat lower mandibular incisors. This raises a possibility that ENAM is processed differently in rats and mice; alternatively, it may be processed according to tooth type.
In summary, our results suggest that the pSers in S-x-E motifs are likely essential components for normal AMBN function.
Author Contributions
P. Ma, contributed to data acquisition, analysis, and interpretation, drafted the manuscript; W. Yan, Y. Tian, J. He, and S.J. Brookes, contributed to data acquisition, analysis, and interpretation, critically revised the manuscript; X. Wang, contributed to conception and design, critically revised the manuscript. All authors gave final approval and agree to be accountable for all aspects of the work.
Supplementary Material
Supplementary Material
Acknowledgments
We thank Jingya Wang and Dr. Jian Q. Feng for their supports with the scanning electron microscopy analysis and Jeanne Santa Cruz for her assistance with the editing of this article.
Footnotes
A supplemental appendix to this article is published electronically only at http://jdr.sagepub.com/supplemental.
This study was supported by a grant from the National Institutes of Health (DE23873-01; X.W.) and grants from the Office for Research and Graduate Studies, Texas A&M University Health Science Center, as well as from Department of Biomedical Sciences, Baylor College of Dentistry, Texas A&M University Health Science Center.
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
References
- Al-Hashimi N, Lafont AG, Delgado S, Kawasaki K, Sire JY. 2010. The enamelin genes in lizard, crocodile, and frog and the pseudogene in the chicken provide new insights on enamelin evolution in tetrapods. Mol Biol Evol. 27(9):2078–2094. [DOI] [PubMed] [Google Scholar]
- Bartlett JD, Beniash E, Lee DH, Smith CE. 2004. Decreased mineral content in MMP-20 null mouse enamel is prominent during the maturation stage. J Dent Res. 83(12):909–913. [DOI] [PubMed] [Google Scholar]
- Brookes SJ, Kingswell NJ, Barron MJ, Dixon MJ, Kirkham J. 2011. Is the 32-kDa fragment the functional enamelin unit in all species? Eur J Oral Sci. 119 Suppl 1:345–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunati AM, Marin O, Bisinella A, Salviati A, Pinna LA. 2000. Novel consensus sequence for the Golgi apparatus casein kinase, revealed using proline-rich protein-1 (PRP1)–derived peptide substrates. Biochem J. 351(Pt 3):765–768. [PMC free article] [PubMed] [Google Scholar]
- Chun YH, Lu Y, Hu Y, Krebsbach PH, Yamada Y, Hu JC, Simmer JP. 2010. Transgenic rescue of enamel phenotype in Ambn null mice. J Dent Res. 89(12):1414–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayhoff MO, Schwartz RM, Orcutt B. 1978. A model of evolutionary change in proteins. Atlas of Protein Sequence and Structure. 5(3):345–352; [accessed 2016 Jul 7]. http://ibi.zju.edu.cn/bioinplant/courses/dayhoffetal1978.pdf. [Google Scholar]
- Delsuc F, Gasse B, Sire JY. 2015. Evolutionary analysis of selective constraints identifies ameloblastin (AMBN) as a potential candidate for amelogenesis imperfecta. BMC Evol Biol. 15:148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukumoto S, Kiba T, Hall B, Iehara N, Nakamura T, Longenecker G, Krebsbach PH, Nanci A, Kulkarni AB, Yamada Y. 2004. Ameloblastin is a cell adhesion molecule required for maintaining the differentiation state of ameloblasts. J Cell Biol. 167(5):973–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart PS, Aldred MJ, Crawford PJ, Wright NJ, Hart TC, Wright JT. 2002. Amelogenesis imperfecta phenotype-genotype correlations with two amelogenin gene mutations. Arch Oral Biol. 47(4):261–265. [DOI] [PubMed] [Google Scholar]
- Hart PS, Michalec MD, Seow WK, Hart TC, Wright JT. 2003. Identification of the enamelin (g.8344delG) mutation in a new kindred and presentation of a standardized ENAM nomenclature. Arch Oral Biol. 48(8):589–596. [DOI] [PubMed] [Google Scholar]
- Hu JC, Chun YH, Al Hazzazzi T, Simmer JP. 2007. Enamel formation and amelogenesis imperfecta. Cells Tissues Organs. 186(1):78–85. [DOI] [PubMed] [Google Scholar]
- Hu JC, Yamakoshi Y. 2003. Enamelin and autosomal-dominant amelogenesis imperfecta. Crit Rev Oral Biol Med. 14(6):387–398. [DOI] [PubMed] [Google Scholar]
- Hu JC, Yamakoshi Y, Yamakoshi F, Krebsbach PH, Simmer JP. 2005. Proteomics and genetics of dental enamel. Cells Tissues Organs. 181(3–4):219–231. [DOI] [PubMed] [Google Scholar]
- Ishikawa HO, Xu A, Ogura E, Manning G, Irvine KD. 2012. The Raine syndrome protein FAM20C is a Golgi kinase that phosphorylates bio-mineralization proteins. PLoS One. 7(8):e42988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki K, Weiss KM. 2003. Mineralized tissue and vertebrate evolution: the secretory calcium-binding phosphoprotein gene cluster. Proc Natl Acad Sci U S A. 100(7):4060–4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JW, Seymen F, Lin BP, Kiziltan B, Gencay K, Simmer JP, Hu JC. 2005. ENAM mutations in autosomal-dominant amelogenesis imperfecta. J Dent Res. 84(3):278–282. [DOI] [PubMed] [Google Scholar]
- Kim JW, Simmer JP, Hu YY, Lin BP, Boyd C, Wright JT, Yamada CJ, Rayes SK, Feigal RJ, Hu JC. 2004. Amelogenin p.M1T and p.W4S mutations underlying hypoplastic X-linked amelogenesis imperfecta. J Dent Res. 83(5):378–383. [DOI] [PubMed] [Google Scholar]
- Paine ML, Wang HJ, Luo W, Krebsbach PH, Snead ML. 2003. A transgenic animal model resembling amelogenesis imperfecta related to ameloblastin overexpression. J Biol Chem. 278(21):19447–19452. [DOI] [PubMed] [Google Scholar]
- Pearlman SM, Serber Z, Ferrell JE. 2011. A mechanism for the evolution of phosphorylation sites. Cell. 147(4): 934–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulter JA, Murillo G, Brookes SJ, Smith CE, Parry DA, Silva S, Kirkham J, Inglehearn CF, Mighell AJ. 2014. Deletion of ameloblastin exon 6 is associated with amelogenesis imperfecta. Hum Mol Genet. 23(20):5317–5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmer JP, Richardson AS, Hu YY, Smith CE, Ching-Chun Hu J. 2012. A post-classical theory of enamel biomineralization . . . and why we need one. Int J Oral Sci. 4(3):129–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephanopoulos G, Garefalaki ME, Lyroudia K. 2005. Genes and related proteins involved in amelogenesis imperfect. J Dent Res. 84(12):1117–1126. [DOI] [PubMed] [Google Scholar]
- Tagliabracci VS, Engel JL, Wen J, Wiley SE, Worby CA, Kinch LN, Xiao J, Grishin NV, Dixon JE. 2012. Secreted kinase phosphorylates extracellular proteins that regulate biomineralization. Science. 336(6085):1150–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorsness PE, Koshland DE., Jr. 1987. Inactivation of isocitrate dehydrogenase by phosphorylation is mediated by the negative charge of the phosphate. J Biol Chem. 262(22):10422–10425. [PubMed] [Google Scholar]
- Urzúa B, Ortega-Pinto A, Morales-Bozo I, Rojas-Alcayaga G, Cifuentes V. 2011. Defining a new candidate gene for amelogenesis imperfecta: from molecular genetics to biochemistry. Biochem Genet. 49(1–2):104–121. [DOI] [PubMed] [Google Scholar]
- Wang X, Jung J, Liu Y, Yuan B, Lu Y, Feng JQ, Qin C. 2013. The specific role of FAM20C in amelogenesis. J Dent Res. 92(11):995–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wang J, Liu Y, Yuan B, Ruest LB, Feng JQ, Qin C. 2015. The specific role of FAM20C in dentinogenesis. J Dent Res. 94(2):330–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wang S, Li C, Gao T, Liu Y, Rangiani A, Sun Y, Hao J, George A, Lu Y, et al. 2012. Inactivation of a novel FGF23 regulator, FAM20C, leads to hypophosphatemic rickets in mice. PLoS Genet. 8(5):e1002708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wang S, Lu Y, Gibson MP, Liu Y, Yuan B, Feng JQ, Qin C. 2012. FAM20C plays an essential role in the formation of murine teeth. J Biol Chem. 287(43):35934–35942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wazen RM, Moffatt P, Zalzal SF, Yamada Y, Nanci A. 2009. A mouse model expressing a truncated form of ameloblastin exhibits dental and junctional epithelium defects. Matrix Biol. 28(5):292–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.