Abstract
Despite significant advances in recent years in culture-independent molecular microbiology methods, the detailed study of individual bacterial species still relies on having pure cultures in the laboratory. Yet, more than a third of the approximately 700 bacterial taxa found in the human oral cavity are as yet uncultivated in vitro. One such taxon, Tannerella sp. HOT-286 (phylotype BU063), is the focus of much interest since it is associated with periodontal health, while Tannerella forsythia, its closest phylogenetic neighbor, is strongly associated with periodontal disease. HOT-286, however, has remained uncultivated despite the efforts of several research groups, spanning over a decade. The aim of this study was to cultivate Tannerella sp. HOT-286. A heavily diluted sample of subgingival plaque was inoculated onto culture plates supplemented with siderophores (pyoverdines-Fe complex or desferricoprogen) or a neat plaque suspension. After 8 d of anaerobic incubation, microcolonies and colonies showing satellitism were passaged onto fresh culture plates cross-streaked with potential helper strains or onto cellulose-acetate membranes placed over lawn cultures of helper strains. Subcultured colonies were identified by 16S rRNA gene sequencing, and purity was confirmed by sequencing 20 clones per library prepared from a single colony. Three colonies of interest (derived from pyoverdines- and plaque-supplemented plates) were identified as Tannerella sp. HOT-286. The isolates were found to be incapable of independent growth, requiring helpers such as Propionibacterium acnes and Prevotella intermedia for stimulation, with best growth on membranes over “helper” lawns. A representative isolate was subjected to phenotypic characterization and found to produce a range of glycosidic and proteolytic enzymes. Further comparison of this novel “periodontal health-associated” taxon with T. forsythia will be valuable in investigating virulence factors of the latter and possible health benefits of the former.
Keywords: culture, microbiome, periodontitis, isolation, dental plaque, bacteriology
Introduction
Although the advent of next-generation sequencing has revealed the true diversity of the human oral microbiome, the need for laboratory culture for the comprehensive physiologic and pathologic characterization of individual bacterial species remains. Approximately 700 bacterial taxa/species have been identified in the human oral cavity, based on 16S rRNA gene sequence data (Human Oral Microbiome Database, release 13; www.homd.org). However, of these, nearly 250 are as yet uncultivated in vitro (Chen et al. 2010; Dewhirst et al. 2010). Examples of uncultivated oral bacterial taxa include all members of the candidate bacterial divisions SR1 and GN02 (Camanocha and Dewhirst 2014). Until recently, there were also no cultivated oral phylotypes from the phyla TM7 and Chloroflexi—a single oral strain from each phylum has now been successfully cultivated (He et al. 2015; Vartoukian et al. 2016).
The recently cultivated TM7 strain TM7x has a reduced genome of 705 kb, which lacks the genes necessary for essential amino acid biosynthesis; consequently, it is incapable of independent growth and leads an obligately symbiotic relationship with another bacterium, Actinomyces odontolyticus (He et al. 2015). Davis et al. (2013) also showed that gene loss in bacteria is associated with auxotrophy for purine, pyrimidine, fatty acid, and amino acid synthetic pathways. Bacteria that are metabolically dependent on others may be impossible to grow in pure culture. Conversely, in vitro cultivation of bacteria in consortia can enable the isolation of previously uncultivated bacteria (Vartoukian et al. 2010; Tanaka and Benno 2015). In particular, species within biofilm communities, such as dental plaque, may depend on one another for metabolic cooperation and intercellular signals (Vartoukian et al. 2010; Stewart 2012; Mihai et al. 2015). Kummerli and coworkers (Kummerli et al. 2009; Kummerli et al. 2014) reported that the sharing of metabolites such as iron-scavenging siderophores is particularly prevalent in structured bacteria-host environments. It has been suggested that “unculturable” bacteria may have lost the ability to produce siderophores (Lewis et al. 2010) and so depend on provision from neighboring bacteria. Indeed, there is evidence that adding siderophores to culture media stimulates the growth of previously uncultivated organisms (Guan and Kamino 2001; D’Onofrio et al. 2010; Vartoukian et al. 2016).
Tannerella forsythia is strongly associated with periodontitis (Socransky et al. 1998), possesses several virulence factors (Sharma 2010), and is the only cultivable taxon from the genus Tannerella. The as-yet-uncultivated oral phylotype Tannerella sp. HOT-286 (clone BU063) is phylogenetically closely related to T. forsythia but is associated with periodontal health rather than disease (Leys et al. 2002; Kumar et al. 2003; de Lillo et al. 2004). Fodor et al. (2012) included Tannerella sp. HOT-286 on their high-priority microorganisms “most wanted” list for genome sequencing; de Lillo et al. (2004) suggested more than a decade ago that work to enable culture of Tannerella sp. HOT-286 should be urgently prioritized.
The aim of this study was to cultivate the previously uncultivated oral phylotype Tannerella sp. HOT-286 through several approaches: growth in consortia, addition of siderophores, cross-streaking with helper strains, and growth on membranes over helper lawns.
Materials and Methods
Ethical approval for the study was granted by the South West London REC 3 Research Ethics Committee (10/H0803/161). A 50-y-old female subject with chronic periodontitis, who had not received periodontal or antimicrobial therapy within the previous 3 mo, was recruited for the study with her informed consent. Subgingival plaque was collected with a sterile curette from 2 deep periodontal pockets (7 to 8 mm), pooled, and suspended in reduced transport medium (Bowden and Hardie 1971).
The sample was transported within 45 min of collection to an anaerobic workstation (Don Whitley Scientific Ltd.) with an atmosphere of 80% nitrogen, 10% hydrogen, and 10% carbon dioxide at 37 °C. It was diluted to 10-6 in reduced transport medium after vortexing for 1 min, and 50 μL of the diluted plaque suspension was used to inoculate multiple prereduced Blood Agar Base No. 2 (Lab M) / 5% horse blood (blood agar [BA]) plates. A well was made in the center of each agar plate to which was added 150 μL of 0.1-mg/mL solutions of pyoverdines-Fe complex (Sigma-Aldrich) or desferricoprogen (EMC Microcollections), 150 μL of neat plaque suspension, or 150 μL of sterile water.
After 8 d of anaerobic incubation, the mixed cultures were inspected under a plate-dissecting microscope for microcolonies and colonies satelliting around/on larger colonies. The colonies of interest were passaged onto BA plates cross-streaked with potential helper strains (Fusobacterium nucleatum subsp. polymorphum NCTC 10562 or Propionibacterium acnes ATCC 6919) and onto 0.45-μm-pore cellulose acetate membranes (Sartorius, 1110650ACN) overlying fresh F. nucleatum or 48-h P. acnes lawn cultures.
Growth on secondary plates was purified where necessary and DNA extracted with the GenElute Bacterial Genomic DNA kit (Sigma-Aldrich) with the protocol for Gram-positive bacteria prior to end point polymerase chain reaction (PCR) with “universal” primers 27FYM and 1492R (Lane 1991) as described previously (Vartoukian et al. 2009). For secondary plates showing minimal growth, direct “touch” PCR of single colonies with “universal” primers (Vartoukian et al. 2009) was performed. PCR products were subjected to partial 16S rRNA gene sequencing with primer 519R (Vartoukian et al. 2009).
For cultures identified as Tannerella sp. HOT-286 (phylotype BU063), purity was confirmed by sequencing 20 cloned inserts from a library prepared (as described by Vartoukian et al. 2009) with the amplification product of 16S rRNA gene “touch” PCR of a single colony with “universal” primers. Subsequently, the full length of the 16S rRNA gene was sequenced with multiple primers for triple coverage (Vartoukian et al. 2009).
Colonial and cellular morphology of Tannerella sp. HOT-286 strains were determined by examination under a dissecting microscope, light microscopy after Gram staining, and transmission electron microscopy (TEM). For TEM, isolated colonies were gently suspended in 10mM Tris-HCL buffer (pH 7.4) at a concentration of about 108 cells/mL. Samples were negatively stained with 1% (wt/vol) phosphotungstic acid (pH 6.5) for 20 to 30 s. The specimens were examined with a JEOL model JEM-1200EX transmission electron microscope operating at 100 kV.
Enzyme profiles were determined for Tannerella sp. HOT-286 isolate SP18_24 and T. forsythia FDC 338T with the API ZYM test (BioMerieux) and the Rapid ID 32 A anaerobe identification kit (BioMerieux) in duplicate.
Susceptibility to penicillin (1 U), amoxicillin (10 μg), ampicillin (2 μg), erythromycin (5 μg), tetracycline (10 μg), metronidazole (5 μg), ceftazidime (30 μg), gentamycin (10 μg), chloramphenicol (10 μg), and ciprofloxacin (1 μg; Oxoid) was determined in duplicate with the disc diffusion method for 1) Tannerella sp. HOT-286 SP18_24 with cultures cross-streaked with P. acnes and 2) T. forsythia FDC 338T.
Growth characteristics of Tannerella sp. HOT-286 SP18_24 were investigated as follows, with duplicate testing in all cases.
The ability of pyoverdines-Fe to stimulate growth of this strain on BA under anaerobic conditions was assessed 1) by adding either 150 μL of 0.1 mg/mL of pyoverdines-Fe or an equivalent volume of sterile water (negative control) to a central well on the plates or 2) by applying a small circular inoculum of live P. acnes (positive control) to the center of the plates.
The effect on growth of SP18_24 of P. acnes culture supernatant (CS) or cell-free extract (CFE) was assessed with a method similar to that described above, with addition of test (CS or CFE) or negative control (Nutrient Broth No. 2 [NB]; Oxoid) or phosphate-buffered saline [PBS]) agents to a central well or inoculation with live P. acnes as positive control. CS was prepared from a 4-d NB culture of P. acnes by centrifuging the culture and passing the supernatant through a 0.2-μm-pore filter. CFE was prepared from the same 25-mL broth culture by resuspending the pellet in 5 mL of PBS, sonicating the suspension for 3 pulses of 2 min, and centrifuging and filtering the supernatant.
The effect of CS/CFE on growth of SP18_24 was also assessed in broth culture. Briefly, SP18_24 was cultured in NB + 1% yeast extract, with or without the following: CS (50%, v/v), CFE (25%, v/v), or equivalent volumes of plain NB or PBS as controls. Growth was assessed over 16 d with spectrophotometric turbidity measurements at 600 nm.
Finally, a panel of 7 oral bacteria was evaluated alongside P. acnes for their stimulatory effect on the growth of SP18_24 as lawn cultures on BA. Small circular inocula of the following bacterial strains were applied to plates: Streptococcus oralis (NCTC 7864), Veillonella dispar (NCTC 11831), Actinomyces oris (ATCC 19246), Parvimonas micra (ACTC 33270), Porphyromonas gingivalis (ATCC 33277), Prevotella intermedia (ATCC 25611), P. acnes (ATCC 6919), and F. nucleatum (NCTC 10562). After 7 d of anaerobic incubation, growth stimulatory effect was graded arbitrarily as 0, +, ++, or +++.
Results
A heavily diluted subgingival plaque sample was inoculated onto culture plates supplemented with either siderophores or a neat suspension of the plaque sample. Forty-six isolates forming microcolonies or exhibiting satellite growth around other colonies were passaged to fresh plates cross-streaked with helper strains and onto membranes overlying lawn cultures of helpers. Three isolates (2 from a pyoverdines-Fe-supplemented plate and 1 from a plaque-supplemented plate) formed several large cream-colored colonies on membranes overlying P. acnes lawns but showed no or limited growth (1 to 2 tiny colonies) on 1) secondary plates cross-streaked with F. nucleatum and P. acnes as helpers or 2) membrane cultures over F. nucleatum lawns. The isolates were identified as Tannerella sp. HOT-286 (phylotype BU063), and the cultures were confirmed pure by sequence analysis of multiple cloned amplicons derived from a single colony of each isolate. The full-length 16S rRNA gene sequences of the 3 isolates were found to be identical and 99.2% similar over 1,450 bases to Tannerella clone BU063, accession number AY008308. The novel sequences were deposited in the GenBank nucleotide sequence database with the following accession numbers: Tannerella sp. HOT-286 isolate SP18_4, KT861600; Tannerella sp. HOT-286 SP18_24, KT861601; and Tannerella sp. HOT-286 SP18_26, KT861602.
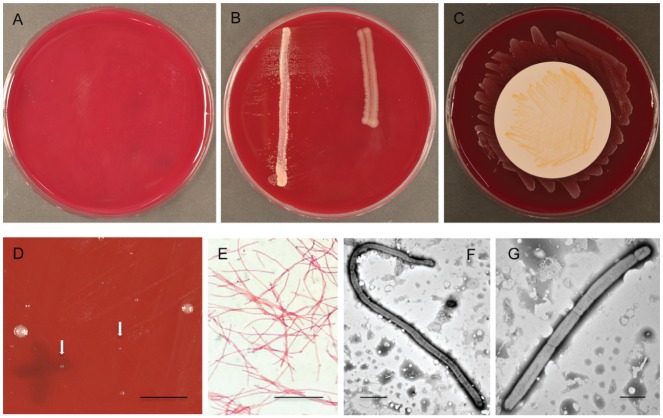
The 3 isolates showed limited independent growth (Fig. 1A) and were significantly stimulated by P. acnes but not F. nucleatum (Fig. 1B). Culture on membranes over P. acnes lawns resulted consistently in stronger growth than that observed after culture directly on media with P. acnes cross-streaks (Fig. 1B, C). The isolates were successfully revived after storage in broth/glycerol at −80 °C, although growth was initially sparse, consisting of tiny colonies approximately 0.2 mm in diameter and larger colonies of the same type (1 to 1.5 mm; Fig. 1D). After 2 passages, colonies of Tannerella sp. HOT-286 on BA measured, on average, approximately 0.5 mm in diameter after 8 d growth and had a circular or slightly irregular shape, undulate edge, convex profile, and convoluted surface. Colonies were gray/off-white in color and speckled with opaque cream internal flecks.
Figure 1.
Growth characteristics, colony, and cellular morphology of Tannerella sp. HOT-286. (A–C) Six-day cultures of Tannerella sp. HOT-286: (A) no visible growth in absence of helper strain; (B) satellitism beside Propionibacterium acnes streak on left, no growth beside Fusobacterium nucleatum streak on right; (C) strong growth on membrane over P. acnes lawn. (D) Nine-day culture of Tannerella sp. HOT-286 after revival from −80 °C storage showing sparse growth of colonies of variable size. Arrows indicate position of tiny colonies. Bar = 5 mm. (E) Cellular morphology of Tannerella sp. HOT-286 by Gram staining. Bar = 10 μm. (F, G) Transmission electron photomicrographs of 2 cells of Tannerella sp. HOT-286 (strain SP18_24). (F) Long cell about 50 μm in length. Bar = 5 μm. (G) Cell showing segments of variable size. Bar = 2 μm.
Gram staining and TEM revealed that cells of Tannerella sp. HOT-286 were Gram negative and filamentous (Fig. 1E), measuring 1.2 μm in width, and composed of segments of varying sizes (Fig. 1F, G). Cells ranged in length from 4 to >50 μm (Fig. 1F). Pili, flagella, or other surface structures were not observed (Fig. 1F, G).
There was insufficient independent growth of Tannerella sp. HOT-286 SP18_24 on BA to perform API ZYM and Rapid ID 32 A tests according to the manufacturer’s instructions. Therefore, tests were repeated with SP18_24 biomass harvested from 7-d cultures cross-streaked with P. acnes and compared against results of equivalent tests for P. acnes. For some tests, the result was positive for SP18_24 but negative for P. acnes and vice versa (Table), lending credence to the validity of the results. SP18_24 exhibited proteolytic and glycolytic activity and was positive for alkaline phosphatase, acid phosphatase, esterase, esterase lipase, and naphthol-AS-BI-phosphohydrolase.
Table.
Results of API ZYM and Rapid ID 32 A Tests.
| Tannerella sp. HOT-286 (SP18_24)a | Tannerella forsythia (FDC 338T) | Propionibacterium acnes (ATCC 6969) | |
|---|---|---|---|
| API ZYM | |||
| Alkaline phosphatase | + | + | – |
| Esterase (C4) | + | + | – |
| Esterase lipase (C8) | + | + | – |
| Lipase (C14) | – | – | – |
| Leucine arylamidase | + | + | – |
| Valine arylamidase | – | – | – |
| Cystine arylamidase | – | – | – |
| Trypsin | + | + | – |
| α-chymotrypsin | – | – | – |
| Acid phosphatase | + | + | + |
| Naphthol-AS-BI-phosphohydrolase | + | + | – |
| α-Galactosidase | – | – | – |
| β-Galactosidase | – | – | + |
| β-Glucuronidase | – | + | – |
| α-Glucosidase | + | – | – |
| β-Glucosidase | – | – | – |
| N-acetyl-β-glucosaminidase | – | + | + |
| α-Mannosidase | – | – | + |
| α-Fucosidase | – | + | – |
| Rapid ID 32 A | |||
| Urease | – | – | – |
| Arginine dihydrolase | – | – | + |
| α-Galactosidase | – | – | – |
| β-Galactosidase | – | + | + |
| β-Galactosidase-6-phosphate | – | + | – |
| α-Glucosidase | + | – | – |
| β-Glucosidase | – | + | – |
| α-Arabinosidase | – | – | – |
| β-Glucuronidase | – | – | – |
| N-acetyl-β-glucosaminidase | – | + | + |
| Mannose fermentation | – | – | + |
| Raffinose fermentation | – | – | – |
| Glutamic acid decarboxylase | – | – | – |
| α-Fucosidase | + | + | – |
| Reduction of nitrates | – | – | + |
| Indole | + | – | + |
| Alkaline phosphatase | + | + | – |
| Arginine arylamidase | + | + | + |
| Proline arylamidase | – | – | + |
| Leucyl glycine arylamidase | + | + | – |
| Phenylalanine arylamidase | + | – | – |
| Leucine arylamidase | + | + | – |
| Pyroglutamic acid arylamidase | + | – | – |
| Tyrosine arylamidase | + | + | – |
| Alanine arylamidase | + | + | + |
| Glycine arylamidase | – | – | + |
| Histidine arylamidase | + | + | – |
| Glutamyl glutamic acid arylamidase | – | – | – |
| Serine arylamidase | – | – | + |
Reactions graded on a scale of 0 to 5, with values of 3 to 5 reported as a positive result (+) as recommended in the manufacturer’s guidelines.
Cross-streaks of Propionibacterium acnes present on source plates.
Tannerella sp. HOT-286 SP18_24 was susceptible to amoxicillin, ampicillin, erythromycin, tetracycline, metronidazole, and ceftazidime (with zones of inhibition measuring ≥30 mm in diameter); weakly susceptible to penicillin and chloramphenicol (zones of inhibition, 14 to 20 mm); and resistant to gentamycin and ciprofloxacin (no zone). The antimicrobial susceptibility profile for T. forsythia FDC 338T was the same as that of SP18_24 except that it was strongly susceptible to penicillin, with a 65-mm zone of inhibition.
Neither pyoverdines-Fe nor the CS or CFE of P. acnes showed any stimulatory effect on the growth of SP18_24 relative to negative controls. Furthermore, SP18_24 did not grow in broth culture, with or without P. acnes CS/CFE.
P. acnes and P. intermedia showed the strongest growth stimulation (+++) of SP18_24, with dense satelliting growth around P. acnes (Fig. 2g) and with the development of large colonies of SP18_24 at a distance of up to 25 mm from P. intermedia, as well as satellite growth (Fig. 2f). A. oris and F. nucleatum showed moderate growth stimulation (++; Fig. 2d, h). V. dispar and P. gingivalis were able to weakly stimulate growth of SP18_24 (+; Fig. 2b, e), whereas S. oralis and P. micra showed no stimulatory capacity (Fig. 2a, c).
Figure 2.
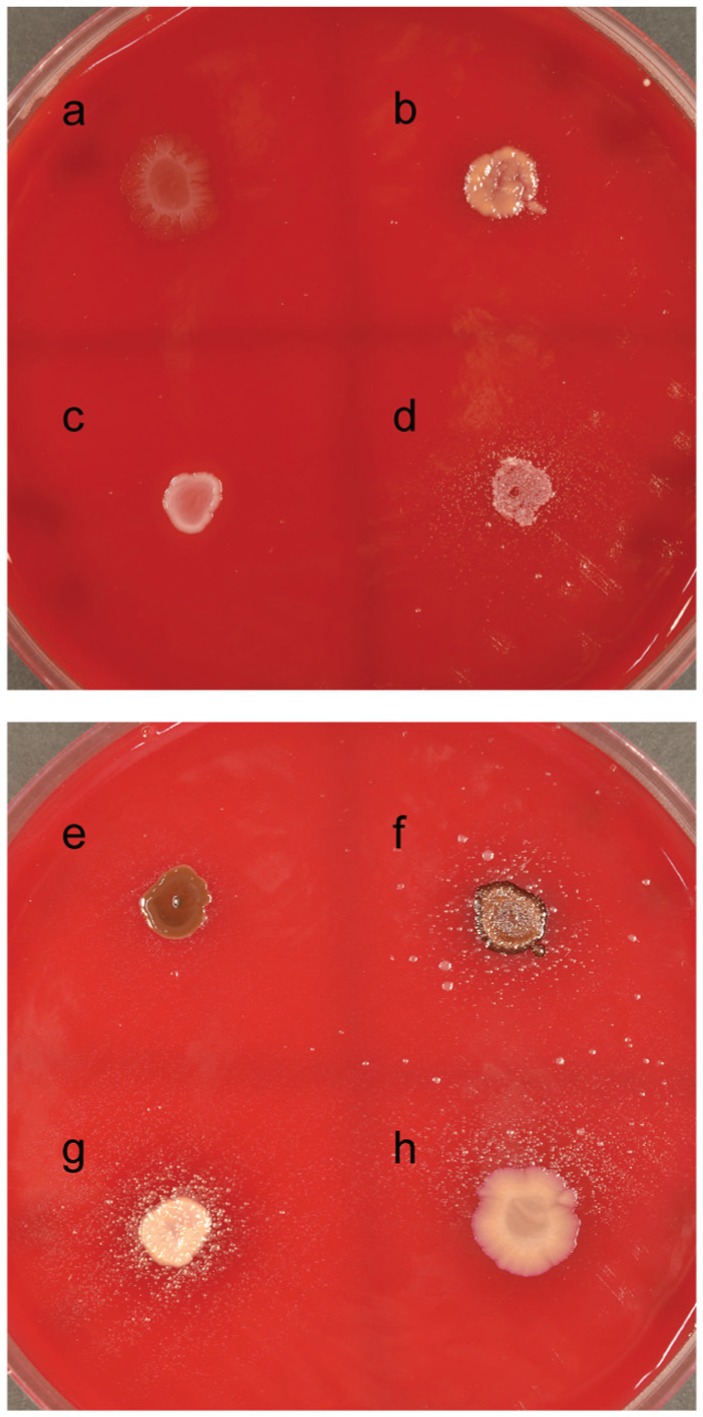
Seven-day cultures of Tannerella sp. HOT-286 (SP18_24) showing growth stimulation by several of the 8 potential helper strains tested: (a) Streptococcus oralis, (b) Veillonella dispar, (c) Parvimonas micra, (d) Actinomyces oris, (e) Porphyromonas gingivalis, (f) Prevotella intermedia, (g) Propionibacterium acnes, and (h) Fusobacterium nucleatum.
The novel Tannerella sp. HOT-286 strains have been deposited into culture collections as follows: Tannerella sp. HOT-286 SP18_4, DSM 102836, JCM 31301; Tannerella sp. HOT-286 SP18_24, DSM 102893, JCM 31302; and Tannerella sp. HOT-286 SP18_26, DSM 102894, JCM 31303.
Discussion
Several authors have highlighted the need to cultivate Tannerella sp. HOT-286 (Leys et al. 2002; de Lillo et al. 2004; Zuger et al. 2007). Although it was reported that Tannerella sp. HOT-286 had been successfully cultivated as part of a consortium (Duran-Pinedo et al. 2011), the consortium was lost before a pure culture could be obtained (Frias-Lopez, personal communication). In this study, we used growth in consortia, growth with a cross-streaked helper organism, growth on a membrane over a helper organism lawn, and supplementation with siderophores to allow culture of Tannerella sp. HOT-286, leading to its successful isolation in purity. Two of the 3 novel Tannerella isolates were cultured on pyoverdines-Fe-supplemented plates, although growth stimulation by pyoverdines-Fe was not confirmed. Pyoverdines-Fe has been shown to be strongly stimulatory to the difficult-to-culture bacterium Prevotella sp. HOT-376 (Vartoukian et al. 2016), demonstrating that growth enhancement by siderophores is a selective phenomenon.
Tannerella sp. HOT-286 was found to be dependent for growth on the proximity of a helper strain, P. acnes. It has been observed over the years within the Wade and other laboratories (Davis et al. 2014) that P. acnes stimulates the growth of a number of previously uncultivated bacteria, although the mechanisms of action are unknown. Interestingly, although coculture with live P. acnes had a strong growth-promoting effect on Tannerella sp. HOT-286, this effect was not observed with P. acnes CS or CFE, suggesting that the stimulating factor is labile. Furthermore, growth was enhanced more strongly by culturing Tannerella sp. HOT-286 on the surface of a membrane over a lawn culture of P. acnes than by culturing the strain directly on agar with P. acnes cross-streaks. This would imply either 1) that a greater amount of “helper” signal, as provided by the larger surface area of a lawn culture than of a narrow cross-streak, is needed for growth or 2) that separation from the agar surface by a membrane helps to protect the recipient from potential growth inhibitors present in the agar medium. It has been shown that hydrogen peroxide, produced during autoclave sterilization of media that includes phosphate and agar, can inhibit bacterial growth (Tanaka et al. 2014).
Six of 8 oral bacterial species representing 4 phyla stimulated the growth of Tannerella sp. HOT-286 SP18_24. In general, growth stimulation was observed as satellitism immediately surrounding the helper strain, although P. intermedia effected the emergence of several large outlier colonies of SP18_24 at a distance from the helper. Although beyond the scope of this study, a future challenge will be to determine by what mechanism these different helpers stimulate growth of SP18_24 and whether there is a universal or specific mode of action.
Cells of Tannerella sp. HOT-286 were found to be segmented filaments of variable length, confirming the observations of Zuger et al. (2007) following fluorescence in situ hybridization (FISH) analysis of BU063 cells. However, their impression that individual segments of cells are of equal length and that, consequently, overall cell length is a reflection of the total number of segments present was not confirmed by our TEM images: whereas the 16-μm cell shown in Figure 1G had 6 segments, the longer 50-μm cell in Figure 1F had only 4.
Tannerella sp. HOT-286 is found in high prevalence but low abundance in periodontal disease–associated plaques (Zuger et al. 2007), and the relative abundance of this phylotype is estimated to be around 0.05% of subgingival bacteria (Human Oral Microbiome Database, release 13; Beall et al. 2014). Evidence from several studies has indicated that, unlike T. forsythia, Tannerella sp. HOT-286 is primarily associated with periodontal health (Leys et al. 2002; Kumar et al. 2003; de Lillo et al. 2004); Leys et al (2002) reported odds ratios for prevalence in periodontitis of Tannerella sp. HOT-286 and T. forsythia of 0.1 and 9.9, respectively. The apparent phenotypic dichotomy between these closely related taxa is clearly of interest, and comparative studies of Tannerella sp. HOT-286 and T. forsythia could provide some insight into factors involved in the latter’s virulence.
To this end, an enzymatic profile of Tannerella sp. HOT-286 was generated and compared with that of T. forsythia. The profiles of the 2 Tannerella taxa were similar, despite their different clinical phenotypes. Both taxa produced a range of proteolytic, hydrolytic, lipolytic, and saccharolytic enzymes. A comparison of the enzyme activity of the 2 taxa did not reveal any obvious differences relevant to the virulence of T. forsythia, although clearly its virulence could be related to factors unconnected to the tests included in the API ZYM and Rapid ID 32 A kits. Antimicrobial susceptibility profiles were also similar for the 2 taxa.
Beall and coworkers (2014) isolated individual cells of Tannerella sp. HOT-286 by flow cytometry and used multiple displacement amplification to generate a collection of single cell–amplified genomes with predicted sizes from 3.44 to 4.07 Mb. Putative virulence genes of T. forsythia were detected by comparative analysis with the HOT-286 genomes and included genes encoding the PrtH, BspA, NanH, and KLIKK proteases (Beall et al. 2014; Ksiazek et al. 2015). Beall et al. (2014) reported a surprisingly high level of strain polymorphism and substantial nucleotide divergence among the various genomes of Tannerella sp. HOT-286. Given that multiple displacement amplification can result in uneven amplification of the genome (Lasken 2012), complete genome sequences are being generated for the 3 Tannerella sp. HOT-286 strains isolated in this study, to enable further comparative genomic analysis with T. forsythia.
Leys and coworkers (2002) showed that subgingival plaque samples were less likely to be dual colonized with Tannerella sp. HOT-286 and T. forsythia than that expected by chance. They suggested a specific exclusionary mechanism, with the possibility that Tannerella sp. HOT-286 may provide protection from acquisition of T. forsythia. Inverse associations among oral bacteria as a result of antagonistic interactions have been reported for Streptococcus mutans and Streptococcus sanguinis (Kreth et al. 2005). If confirmed, this could have far-reaching implications in the management of periodontitis. With health-associated Tannerella sp. HOT-286 having finally been cultivated and available for study, such exciting therapeutic possibilities may now be explored.
Author Contributions
S.R. Vartoukian, W.G. Wade, contributed to conception, design, and data analysis, drafted the manuscript; R.V. Moazzez, contributed to conception and design, critically revised the manuscript; B.J. Paster, F.E. Dewhirst, contributed to conception, design, and data analysis, critically revised the manuscript. All authors gave final approval and agree to be accountable for all aspects of the work.
Acknowledgments
The authors wish to thank Dr. J. Aldridge Taylor for collecting the subgingival plaque sample used in this study.
Footnotes
Research reported in this publication was supported by the National Institute of Dental and Craniofacial Research of the National Institutes of Health under awards R37DE016937 and R01DE 024468. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
References
- Beall CJ, Campbell AG, Dayeh DM, Griffen AL, Podar M, Leys EJ. 2014. Single cell genomics of uncultured, health-associated tannerella bu063 (oral taxon 286) and comparison to the closely related pathogen tannerella forsythia. PLoS One. 9(2):e89398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden G, Hardie J. 1971. Anaerobic organisms from the human mouth. In: Isolation of anaerobes. London (UK): Academic Press. [Google Scholar]
- Camanocha A, Dewhirst FE. 2014. Host-associated bacterial taxa from chlorobi, chloroflexi, gn02, synergistetes, sr1, tm7, and wps-2 phyla/candidate divisions. J Oral Microbiol. 2014:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Yu WH, Izard J, Baranova OV, Lakshmanan A, Dewhirst FE. 2010. The human oral microbiome database: a web accessible resource for investigating oral microbe taxonomic and genomic information. Database (Oxford). 2010:bAQ3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis IJ, Bull C, Horsfall A, Morley I, Harris S. 2014. The unculturables: targeted isolation of bacterial species associated with canine periodontal health or disease from dental plaque. BMC Microbiol. 14:196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis JJ, Xia F, Overbeek RA, Olsen GJ. 2013. Genomes of the class erysipelotrichia clarify the firmicute origin of the class mollicutes. Int J Syst Evol Microbiol. 63(Pt 7):2727–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lillo A, Booth V, Kyriacou L, Weightman AJ, Wade WG. 2004. Culture-independent identification of periodontitis-associated porphyromonas and tannerella populations by targeted molecular analysis. J Clin Microbiol. 42(12):5523–5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Yu WH, Lakshmanan A, Wade WG. 2010. The human oral microbiome. J Bacteriol. 192(19):5002–5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Onofrio A, Crawford JM, Stewart EJ, Witt K, Gavrish E, Epstein S, Clardy J, Lewis K. 2010. Siderophores from neighboring organisms promote the growth of uncultured bacteria. Chem Biol. 17(3):254–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran-Pinedo AE, Paster B, Teles R, Frias-Lopez J. 2011. Correlation network analysis applied to complex biofilm communities. PLoS One. 6(12):e28438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fodor AA, DeSantis TZ, Wylie KM, Badger JH, Ye Y, Hepburn T, Hu P, Sodergren E, Liolios K, Huot-Creasy H, et al. 2012. The “most wanted” taxa from the human microbiome for whole genome sequencing. PLoS One. 7(7):e41294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan LL, Kamino K. 2001. Bacterial response to siderophore and quorum-sensing chemical signals in the seawater microbial community. BMC Microbiol. 1:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, McLean JS, Edlund A, Yooseph S, Hall AP, Liu SY, Dorrestein PC, Esquenazi E, Hunter RC, Cheng G, et al. 2015. Cultivation of a human-associated tm7 phylotype reveals a reduced genome and epibiotic parasitic lifestyle. Proc Natl Acad Sci U S A. 112(1):244–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreth J, Merritt J, Shi W, Qi F. 2005. Competition and coexistence between streptococcus mutans and streptococcus sanguinis in the dental biofilm. J Bacteriol. 187(21):7193–7203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ksiazek M, Mizgalska D, Eick S, Thogersen IB, Enghild JJ, Potempa J. 2015. Klikk proteases of tannerella forsythia: putative virulence factors with a unique domain structure. Front Microbiol. 6:312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar PS, Griffen AL, Barton JA, Paster BJ, Moeschberger ML, Leys EJ. 2003. New bacterial species associated with chronic periodontitis. J Dent Res. 82(5):338–344. [DOI] [PubMed] [Google Scholar]
- Kummerli R, Jiricny N, Clarke LS, West SA, Griffin AS. 2009. Phenotypic plasticity of a cooperative behaviour in bacteria. J Evol Biol. 22(3):589–598. [DOI] [PubMed] [Google Scholar]
- Kummerli R, Schiessl KT, Waldvogel T, McNeill K, Ackermann M. 2014. Habitat structure and the evolution of diffusible siderophores in bacteria. Ecol Lett. 17(12):1536–1544. [DOI] [PubMed] [Google Scholar]
- Lane D. 1991. 16s/23s rrna sequencing. In: Nucleic acid techniques in bacterial systematics. Chichester (UK): John Wiley & Sons. [Google Scholar]
- Lasken RS. 2012. Genomic sequencing of uncultured microorganisms from single cells. Nat Rev Microbiol. 10(9):631–640. [DOI] [PubMed] [Google Scholar]
- Lewis K, Epstein S, D’Onofrio A, Ling LL. 2010. Uncultured microorganisms as a source of secondary metabolites. J Antibiot (Tokyo). 63(8):468–476. [DOI] [PubMed] [Google Scholar]
- Leys EJ, Lyons SR, Moeschberger ML, Rumpf RW, Griffen AL. 2002. Association of bacteroides forsythus and a novel bacteroides phylotype with periodontitis. J Clin Microbiol. 40(3):821–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihai MM, Holban AM, Giurcaneanu C, Popa LG, Oanea RM, Lazar V, Chifiriuc MC, Popa M, Popa MI. 2015. Microbial biofilms: impact on the pathogenesis of periodontitis, cystic fibrosis, chronic wounds and medical device-related infections. Curr Top Med Chem. 15(16):1552–1576. [DOI] [PubMed] [Google Scholar]
- Sharma A. 2010. Virulence mechanisms of tannerella forsythia. Periodontol 2000. 54(1):106–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL., Jr. 1998. Microbial complexes in subgingival plaque. J Clin Periodontol. 25(2):134–144. [DOI] [PubMed] [Google Scholar]
- Stewart EJ. 2012. Growing unculturable bacteria. J Bacteriol. 194(16):4151–4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Benno Y. 2015. Application of a single-colony coculture technique to the isolation of hitherto unculturable gut bacteria. Microbiol Immunol. 59(2):63–70. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Kawasaki K, Daimon S, Kitagawa W, Yamamoto K, Tamaki H, Tanaka M, Nakatsu CH, Kamagata Y. 2014. A hidden pitfall in the preparation of agar media undermines microorganism cultivability. Appl Environ Microbiol. 80(24):7659–7666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartoukian SR, Adamowska A, Lawlor M, Moazzez R, Dewhirst FE, Wade WG. 2016. In vitro cultivation of “unculturable” oral bacteria, facilitated by community culture and media supplementation with siderophores. PLoS One. 11(1):e0146926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartoukian SR, Palmer RM, Wade WG. 2009. Diversity and morphology of members of the phylum “Synergistetes” in periodontal health and disease. Appl Environ Microbiol. 75(11):3777–3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartoukian SR, Palmer RM, Wade WG. 2010. Cultivation of a Synergistetes strain representing a previously uncultivated lineage. Env Microbiol. 12(4):916–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuger J, Luthi-Schaller H, Gmur R. 2007. Uncultivated tannerella bu045 and bu063 are slim segmented filamentous rods of high prevalence but low abundance in inflammatory disease-associated dental plaques. Microbiology. 153(Pt 11):3809–3816. [DOI] [PubMed] [Google Scholar]