Abstract
SAMHD1 is a dNTP hydrolase, whose activity is required for maintaining low dNTP concentrations in non-cycling T cells, dendritic cells, and macrophages. SAMHD1-dependent dNTP depletion is thought to impair retroviral replication in these cells, but the relationship between the dNTPase activity and retroviral restriction is not fully understood. In this study, we investigate allosteric activation of SAMHD1 by deoxynucleotide-dependent tetramerization and measure how the lifetime of the enzymatically active tetramer is affected by different dNTP ligands bound in the allosteric site. The EC50dNTP values for SAMHD1 activation by dNTPs are in the 2–20 μm range, and the half-life of the assembled tetramer after deoxynucleotide depletion varies from minutes to hours depending on what dNTP is bound in the A2 allosteric site. Comparison of the wild-type SAMHD1 and the T592D mutant reveals that the phosphomimetic mutation affects the rates of tetramer dissociation, but has no effect on the equilibrium of allosteric activation by deoxynucleotides. Collectively, our data suggest that deoxynucleotide-dependent tetramerization contributes to regulation of deoxynucleotide levels in cycling cells, whereas in non-cycling cells restrictive to retroviral replication, SAMHD1 activation is likely to be achieved through a distinct mechanism.
Keywords: autoimmune disease, human immunodeficiency virus (HIV), innate immunity, nucleoside/nucleotide metabolism, retrovirus
Introduction
SAMHD1 restricts replication of HIV-1, HIV-2, and simian immunodeficiency virus (SIV) in macrophages, dendritic, and resting CD4+ T cells (1–5). SAMHD1 and other members of the HD domain family of enzymes display nucleotide-dependent deoxynucleotide triphosphate hydrolysis (dNTPase) activity (6–10). This conserved enzymatic activity is thought to be central to the immune function of SAMHD1, but the exact relationship between dNTP hydrolysis and SAMHD1-mediated retroviral restriction is not fully understood. The preferred model is that SAMHD1-mediated depletion of dNTPs in non-cycling immune cells such as monocytes, macrophages, and non-cycling T cells inhibits reverse transcription of the viral genome in these cells (9, 11).
Structural and biochemical studies of SAMHD1 revealed that the enzyme can form a tetramer, which requires binding of nucleotides at two distinct allosteric sites A1 and A2, for the total of eight allosteric ligands per SAMHD1 tetramer (12–16). The A1 site is specific for the guanine base and can accommodate either dGTP or GTP, whereas the A2 site requires a deoxynucleotide but can accommodate any of the four dNTPs. The nucleotide-dependent tetramer is the dNTPase active form of SAMHD1, so the specificity of the A1 site explains the dGTP/GTP dependence of the dNTPase activity, whereas deoxynucleotide binding at the A2 site may act as a substrate activation mechanism.
Studies of the regulatory mechanisms controlling SAMHD1 activity yielded several puzzling observations that cannot be easily explained within the framework of the existing model. First, the ability of SAMHD1 to restrict retroviral replication correlates with the phosphorylation state of threonine 592. Phosphomimetic mutations of Thr-592 abolish restriction, but surprisingly, appear to be intact in their ability to deplete dNTPs (17–20). Second, the dNTPase activity of SAMHD1 is regulated via nucleotide-dependent tetramerization (12–16, 21). However, several tetramerization-defective SAMHD1 mutants that display a striking dNTPase defect in vitro can nevertheless deplete cellular dNTPs and are restriction-competent (22, 23). These observations suggest that our understanding of SAMHD1-mediated retroviral restriction is incomplete.
Nucleotide-dependent tetramerization was suggested as a substrate activation mechanism that acts as one of the determinants of cellular dNTP concentrations (12). This putative regulatory mechanism has been difficult to study, because the allosteric activation of the enzyme by the substrate is masked by the high value of the Michaelis-Menten constant (Km >100 μm) (24, 25). To contribute to regulation of cellular dNTPs, the affinity of the allosteric site for the substrate should match the physiological dNTP concentrations, which can range between micromolar in permissive cycling T cells to mid-nanomolar in macrophages and other restrictive cell types (26). Measuring allosteric activation of SAMHD1 as a function of substrate concentration was one of the objectives of this study.
Another unresolved issue is the relationship between SAMHD1 phosphorylation and its ability to hydrolyze and deplete cellular dNTPs. Phosphomimetic mutation T592D does not perturb the tetramerization equilibrium of SAMHD1 in vitro and does not affect the kcat and Km of the enzyme (22, 24, 25). However, it does affect the rate of tetramer disassembly once dNTPs have been depleted (24). The effect of Thr-592 phosphorylation on the lifetime of the catalytically active tetramer has been suggested as the explanation for the inability of the phosphorylated SAMHD1 to restrict retroviral replication (24). In this study, we sought to investigate the dependence of the SAMHD1 tetramer lifetime on the identity of the nucleotide bound in the allosteric site of the enzyme.
We have developed fluorescence-based and NMR-based experimental tools that can be used to investigate the allosteric activation of SAMHD1 by deoxynucleotides (22). In this study, we show that nucleotide-dependent tetramerization requires micromolar concentrations of dNTPs, which is comparable with dNTP concentrations observed in cycling cells. Phosphomimetic mutation T592D does not affect the equilibrium of allosteric activation of SAMHD1 by deoxynucleotides, but has a kinetic effect that is dependent on the identity of the dNTP bound at the allosteric site. Collectively, our data suggest that in restrictive cells, where dNTP concentrations drop below 50 nm, the activation of SAMHD1 is likely to be achieved through a mechanism that is distinct from the nucleotide-dependent tetramerization observed in vitro.
Results
SAMHD1 Tetramer Lifetime and the T592D Effect on Lifetime Depend on What dNTP Is Bound in the Allosteric Site A2
It has been suggested that SAMHD1 is capable of forming a long-lived catalytically active tetramer, which can persist long after cellular dNTPs are depleted (25). The effect of Thr-592 phosphorylation on the lifetime of the active tetramer may thus explain the inability of SAMHD1 to restrict retroviral replication (24). We first set out to determine the lifetime of SAMHD1 tetramer for different dNTP bound in the allosteric site A2. Fluorescence polarization (FP)3 measurements of GTP labeled with a fluorescent ATTO-495 group on its γ-phosphate (GTP-F) can be used to assay formation and dissociation of SAMHD1 tetramers (22). When SAMHD1 is incubated with an equimolar amount of GTP spiked with a small amount of GTP-F, then upon the addition of deoxynucleotides, one can observe an increase in fluorescence polarization readings, indicative of SAMHD1 tetramerization, followed by a slower decrease of polarization back to baseline values as the enzymatically active tetramer gradually disassociates into monomers once deoxynucleotides have been depleted (Fig. 1A). These measurements can be performed with different deoxynucleotides with the exception of dGTP, which readily displaces GTP-F from the allosteric site A1.
FIGURE 1.
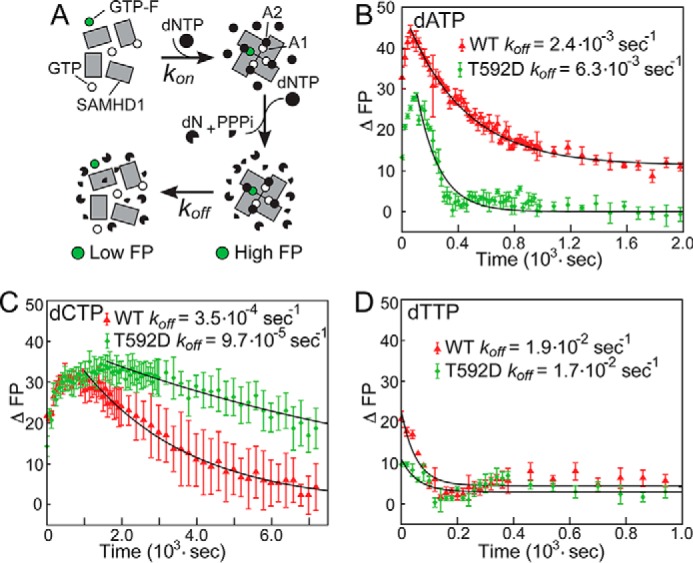
FP studies of SAMHD1 tetramer lifetime. A, SAMHD1 tetramerization requires nucleotide binding at the allosteric sites A1 and A2. When GTP-F is incorporated into SAMHD1 tetramers formed in the presence of unlabeled GTP and dNTPs, FP readings increase. Deoxynucleotides are then rapidly depleted by the enzyme, and the gradual decay of FP values reflects the rate of tetramer dissociation. Dissociation rates were obtained by fitting baseline-corrected FP readings to a simple exponential decay equation (black lines) as explained under “Experimental Procedures.” B, the tetramer dissociation rate of the dATP-bound tetramer is significantly faster for the T592D phosphomimetic mutant than for the WT SAMHD1 construct. C, in contrast, in the dCTP-bound SAMHD1 tetramer, the dissociation rate is slower for the T592D mutant. D, in comparison with the dATP- and dCTP-bound tetramers, the dTTP-mediated SAMHD1 tetramerization displays lower signal amplitude and faster apparent dissociation rates. Error bars indicate means ± S.E.
The results of these measurements reveal that tetramer disassembly rates and the effect of the T592D mutation depend on the identity of the nucleotide occupying the A2 allosteric site (Fig. 1, B–D). For example, the T592D mutation results in a significant increase in the tetramer disassembly rate for the dATP-loaded tetramer (Fig. 1B) in agreement with previous studies (22, 24). In contrast, the phosphomimetic mutation appears to slow down tetramer dissociation for the dCTP-loaded tetramer (Fig. 1C). The amplitude of the FP increase is smaller for dTTP-mediated tetramerization, and the data suggest a faster tetramer dissociation rate. However, the precision of rate determination for the dTTP-bound tetramer is too low to draw any conclusions about the effect of the T592D mutation.
Although the fluorescence polarization experiments offer a convenient experimental approach for studies of SAMHD1 tetramer stability, an important concern in these studies is whether the covalent modification of GTP with a fluorescent label alters SAMHD1 binding and/or affects protein tetramerization in a significant way. To independently verify the FP observations, we have developed an alternative approach, which requires no covalent modifications of the allosteric ligands or substrates. The relatively slow disassembly rates of SAMHD1 tetramers make it possible to separate the enzymatically active protein from free nucleotides by passing the protein through a small-size desalting spin column. A non-equilibrium state of the enzyme, in which SAMHD1 is predominantly tetrameric whereas the free nucleotides are removed, can thus be created in less than 1 min by using a desalting column and a microcentrifuge. The ability to create such a non-equilibrium state of the enzyme permits measurement of the kinetic and equilibrium properties of SAMHD1 that are difficult to determine by other means.
Once free nucleotides are removed, the SAMHD1 tetramer gradually dissociates into monomers until the new equilibrium is reached (Fig. 2A). When dTTP substrate is added to the protein at any time point following nucleotide removal, the dTTP is hydrolyzed by the enzymatically active SAMHD1 tetramer remaining in the sample. However, the dTTP cannot reverse tetramer dissociation because SAMHD1 tetramerization requires GTP or dGTP binding at the A1 site. As a result, the observed dTTP hydrolysis rate is determined by the concentration of the remaining SAMHD1 tetramer and can be used to measure tetramer dissociation rates. For example, when the hydrolysis rate by T592D SAMHD1 is measured as a function of the incubation time between nucleotide removal and dTTP addition, one observes a reduction of the hydrolysis rate from 3.5 s−1 to less than 0.2 s−1 (Fig. 2B). The more than 10-fold decrease of the observed rate upon incubation indicates that nucleotide removal by spin column is remarkably efficient. The reduction of the dNTPase activity with the increasing incubation time was fitted to a simple exponential decay function. Using this approach, we measured rates of SAMHD1 inactivation following nucleotide depletion (designated koff) for all four deoxynucleotides and determined how these rates are affected by the T592D mutation. We interpret these koff rates as rates of dissociation of the enzymatically active SAMHD1 tetramer. Although the individual koff rates measured by this method differ from the koff rates determined by fitting the fluorescence polarization data (Fig. 1), the latter may be affected by the covalent modification of GTP with the fluorescent dye. However, the general trends and key conclusions are in agreement with the fluorescence polarization studies: tetramer dissociation of WT SAMHD1 depends on the identity of the nucleotide bound in the A2 site and is considerably slower for dATP and dCTP than for dTTP and dGTP. Furthermore, we observe that the T592D mutation accelerates dissociation for all nucleotides except dCTP. The dCTP-bound T592D SAMHD1 tetramer is remarkably stable with the dissociation rate less than 10−4 s−1.
FIGURE 2.
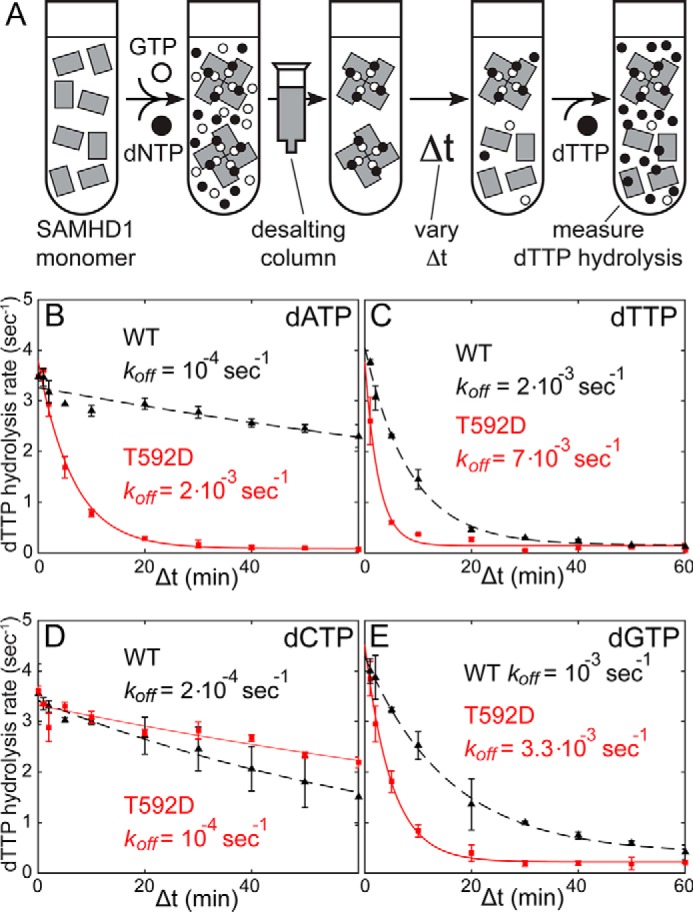
Lifetime measurements of the enzymatically active SAMHD1 tetramers using a non-equilibrium dNTPase assay. A, the enzymatically active SAMHD1 tetramer is formed after incubation with GTP and dNTPs. A non-equilibrium state of the enzyme can subsequently be created using a desalting spin column. Tetramer dissociation is observed as a gradual decrease of the dTTPase hydrolysis rate as a function of the incubation time as described under “Results”. B–E, decay rates (koff) of the SAMHD1 enzymatic activity vary between 10−4 and 10−2 s−1 depending on the dNTP bound at the A2 site. The effect of the T592D mutation is also dNTP-dependent. The solid and dashed lines show the best fits of the data to the simple exponential decay equation. Error bars indicate means ± S.E.
EC50dNTP Values for Allosteric Activation of SAMHD1 by Deoxynucleotides Are in the Low Micromolar Range and Not Affected by the T592D Mutation
The non-equilibrium method described above can also be used to measure the EC50dNTP values for the allosteric activation of SAMHD1 by deoxynucleotides. In this modification of the experiment, the dNTP concentrations during the initial incubation of SAMHD1 with nucleotides are varied, resulting in varying amounts of SAMHD1 tetramer formed in the sample prior to nucleotide removal (Fig. 3A). Once the GTP and dNTP are removed by passing the protein through the desalting column, the dTTP substrate is added and the dTTP hydrolysis rate is measured by NMR. Once again, the dTTP hydrolysis rate is determined by the concentration of the enzymatically active tetramer present in the sample. The EC50dNTP values for the allosteric activation of SAMHD1 can thus be determined from the dependence of the dTTP hydrolysis rate on the dNTP concentration used in the initial incubation of SAMHD1 with nucleotides (Fig. 3, B–E). As in the measurement of the tetramer dissociation rates, the experiment displays good dynamic range with the dTTP hydrolysis rate increasing close to 10-fold with increasing dNTP concentrations. This range demonstrates that GTP is efficiently removed from the sample by the desalting column. The data display a good fit to the simple binding equation and yield EC50dNTP values that range between 2.4 and 23 μm depending on the deoxynucleotide. Notably, these EC50dNTP values are almost 2 orders of magnitude lower than the Km values reported for SAMHD1. In agreement with our recent analytical ultracentrifugation studies (22), we observe that the phosphomimetic mutation T592D has only a minor effect on the allosteric activation of the enzyme by deoxynucleotides.
FIGURE 3.
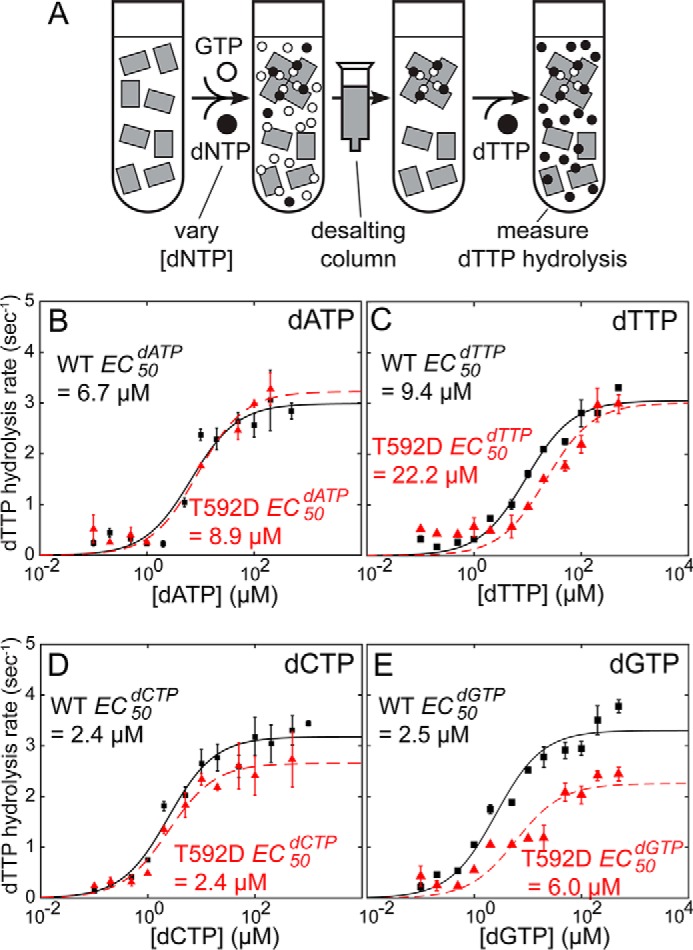
Measurement of the EC50dNTP constants for allosteric activation of SAMHD1 by deoxynucleotides. A, modification of the non-equilibrium dNTPase assay that allows measurement of the EC50dNTP values. dNTP concentrations in the initial incubation are varied, and the fraction of enzymatically active SAMHD1 is determined by measuring the dNTP hydrolysis rate. B–E, EC50dNTP values for all nucleotides are in the low micromolar range, and the effect of the T592D mutation is 2-fold or less. The solid and dashed lines show the best fits of the data to the simple EC50 equation. Error bars indicate means ± S.E.
The Identity of the Allosteric Deoxynucleotide Ligand Does Not Have a Strong Effect on Enzyme Specificity
Finally, we used our experimental scheme to investigate whether the identity of the allosteric ligand bound at the A2 site affects the enzymatic properties of SAMHD1. Substrate specificity of ribonucleotide reductase is known to be altered by different allosteric activators of the enzyme, so we set out to check whether SAMHD1 activation by nucleotide-dependent tetramerization displays similar enzymatic properties. Once again, the relatively long lifetime of the assembled, enzymatically active SAMHD1 tetramer allowed us to first assemble the tetramer using different dNTPs, remove nucleotides using a desalting column, and then immediately add dNTP substrate and measure the rate of its hydrolysis by NMR. In this set of experiments, the hydrolysis rate was measured not only for dTTP, but for each of the four dNTPs individually. The results of these measurements are listed in Table 1. With the exception of dCTP, none of the nucleotides displayed significant differences in the hydrolysis rates when different dNTPs were used as allosteric activators. This was also the case for the phosphomimetic mutant T592D. The hydrolysis rate of dCTP was found to be higher when dATP was used as the allosteric activator for both WT and T592D proteins, but the rate enhancement was less than 2-fold. We conclude that, in contrast to ribonucleotide reductase, the enzymatic specificity of SAMHD1 is not significantly affected by the identity of the allosteric ligand.
TABLE 1.
Allosteric deoxynucleotide activators of SAMHD1 have only a minor effect on substrate specificity
Catalytic rates were measured for different combination of allosteric and substrate deoxynucleotides. Only dCTP hydrolysis displays sensitivity to the allosteric ligand (indicated by bold values), but the effect is only about 2-fold.
| dNTP in A2 site | Substrate hydrolysis rates for WT |
Substrate hydrolysis rates for T592D |
||||||
|---|---|---|---|---|---|---|---|---|
| dGTP | dCTP | dATP | dTTP | dGTP | dCTP | dATP | dTTP | |
| s−1 | s−1 | |||||||
| dGTP | 2.23 | 1.77 | 3.34 | 3.02 | 2.57 | 0.77 | 2.53 | 2.45 |
| dCTP | 1.99 | 1.89 | 2.78 | 3.15 | 2.40 | 0.66 | 3.03 | 3.08 |
| dATP | 1.96 | 2.31 | 2.93 | 2.74 | 2.56 | 1.78 | 2.33 | 2.91 |
| dTTP | 2.11 | 1.79 | 3.07 | 3.19 | 2.33 | 0.39 | 1.29 | 1.73 |
Discussion
SAMHD1 was reported to adopt a long-lived enzymatically active state, which can persist for hours after depletion of deoxynucleotides by the enzyme. The effect of Thr-592 phosphorylation on the ability of SAMHD1 to form such a long-lived active state has been suggested as the explanation for the inability of Thr-592 phosphomimetic mutants of SAMHD1 to restrict retroviral replication. In this study, we used two distinct assays to independently measure the dissociation rate of the nucleotide-loaded SAMHD1 tetramer and the lifetime of the enzymatically active state of SAMHD1 following nucleotide depletion. The results of the two assays are in general agreement, suggesting that the lifetime of the enzymatically active SAMHD1 is determined by the dissociation rate of the SAMHD1 tetramer. We show that these rates can vary between 10−4 and 10−2 s−1 depending on the identity of the deoxynucleotide bound in the A2 site. Although the exact mechanism of nucleotide-dependent SAMHD1 tetramerization and the typical cellular concentrations of SAMHD1 are not known, the observed tetramer dissociation rates are in the range expected for a protein of this size. For example, if one assumes that the association rate of a dimer consisting of 60-kDa monomers is diffusion-controlled (kon ∼ 105 s−1 m−1 is a reasonable ballpark estimate (27)), then for the dimer dissociation constant of 1 nm, the expected dissociation rate is 10−4 s−1. Dissociation rates can be significantly slower for multimers whose association is not diffusion-controlled and involves major structural rearrangements.
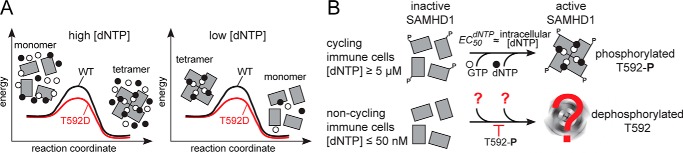
The effect of the phosphomimetic mutation T592D on the lifetime of the enzymatically active SAMHD1 also depends on the identity of the deoxynucleotide bound at the A2 site. For example, the lifetime of the dATP-bound tetramer is significantly shortened by the T592D mutation in agreement with a previous study (Figs. 1B and 2B) (24). In contrast, the lifetime of the dCTP-bound tetramer is not decreased by the T592D mutation, which raises questions about the role of tetramer lifetime in the inability of T592D to restrict retroviral replication (Figs. 1C and 2D). Furthermore, the effect of T592D on SAMHD1 tetramerization is predominantly kinetic, because the equilibrium of nucleotide-dependent tetramerization is changed only about 2-fold or less by the T592D mutation. The most likely explanation for these observations is that the T592D mutation lowers the activation energy of the rate-limiting step in SAMHD1 tetramerization without affecting the free energy difference between the monomeric and the tetrameric states of the enzyme (Fig. 4A). The dependence of the tetramer dissociation rate on the dNTP suggests that this rate-limiting step involves interactions mediated by the base of the allosteric deoxynucleotide.
FIGURE 4.
Implications for SAMHD1 function. A, a predominantly kinetic effect of the T592D mutation is most likely explained by a change in the activation energy for the rate-limiting step in the SAMHD1 tetramerization process. The mutation has only a minor effect on the tetramerization equilibrium. B, EC50dNTP values for SAMHD1 activation by deoxynucleotides match dNTP concentrations in cycling immune cells. The nucleotide-dependent tetramerization of SAMHD1 may thus contribute to dNTP regulation in cycling cells. SAMHD1 activation in non-cycling cells is likely to be mediated by a distinct mechanism.
In this study, we show that the EC50dNTP values of allosteric activation of SAMHD1 by deoxynucleotides are in the low micromolar range, close to the physiological dNTP concentrations observed in cycling T cells. This observation suggests that the affinity of the allosteric site A2 of SAMHD1 for deoxynucleotides may contribute to the establishment and control of stable dNTP concentrations in cycling immune cells. However, such a substrate activation mechanism is not likely to be relevant in the non-cycling immune cells restrictive to retroviral replication, where dNTP concentrations are known to fall below 50 nm. Vpx-mediated depletion of SAMHD1 in macrophages causes a spike in dNTP concentrations in these cells, which strongly suggests that SAMHD1 is enzymatically active even at the low dNTP concentrations present in the long-lived tissue-resident macrophages (11, 28). We suggest that the mechanism of allosteric activation of SAMHD1 in restrictive cells differs in some significant way from the nucleotide-dependent tetramerization observed in vitro (Fig. 4B). For example, the affinity of the A2 site for deoxynucleotides in these cells may be lowered by another posttranslational modification of SAMHD1 or by interaction with another cellular factor. It is also possible that SAMHD1 activation may be achieved by an alternative route that does not involve allosteric deoxynucleotides or protein tetramerization. The existence of such an alternative activation mechanism may explain SAMHD1 mutations that disrupt one of the major tetramerization interfaces in SAMHD1 and impair nucleotide-dependent activation of SAMHD1 in vitro, but do not affect the ability of the enzyme to deplete dNTPs in macrophages and to restrict retroviral replication (22, 23). Furthermore, the effect of the T592D mutation on the EC50dNTP values of SAMHD1 activation by deoxynucleotides in vitro is too small to explain the restriction defect. One can hypothesize that phosphomimetic mutations have a much stronger detrimental effect on the alternative activation mechanism relevant at low dNTP concentrations, which would explain why phosphomimetic mutations abolish retroviral restriction.
In summary, our data support the role of nucleotide-mediated SAMHD1 tetramerization in controlling dNTP levels in cycling cells, but raise questions about the relevance of this mechanism for dNTP depletion needed for restriction of retroviral replication. The properties of phosphomimetic and tetramerization-defective mutants of SAMHD1 suggest the possibility of an alternative activation mechanism responsible for dNTP depletion in macrophages, monocytes, dendritic, and non-cycling T cells.
Experimental Procedures
Protein Production and Purification
All experiments described here were performed on the HD domain constructs of SAMHD1 (residues 115–626). Proteins were expressed using a bacterial expression system and purified as described previously (22).
NMR-based dNTPase Assay
dNTP hydrolysis by SAMHD1 was measured by acquiring a series of 1D NMR spectra at 1-min time intervals as described previously (22). Hydrolysis rates were determined by linear fitting of dNTP and dN NMR peak intensities as a function of time. Curve fitting and data plotting were performed using MATLAB (MathWorks) software.
Fluorescence Polarization
The fluorescent reporter molecule used was γ-(6-aminohexyl)-GTP-ATTO-495 (Jena Biosciences catalog number NU-834-495), referred to as GTP-F in this article. SAMHD1 tetramerization was induced by the addition of dNTP solution (or buffer as a control) to the mixture of SAMHD1, GTP, and GTP-F at time 0, and FP measurements were taken at regular time intervals (22). After the addition of dNTP (or buffer), the samples contained the following final concentrations of reagents: 3 μm SAMHD1, 3 μm GTP, 20 nm GTP-F, and either 500 μm or 0 μm dNTP. All samples were prepared in the buffer containing 50 mm Tris, pH 8.0, 50 mm NaCl, 5 mm MgCl2, and 2 mm DTT. Baseline-corrected FP readings shown in Fig. 1 were obtained by subtracting FP traces of buffer controls from the FP traces of samples to which dNTPs were added. The baseline-corrected data were fitted to a simple exponential decay formula f(x) = A × exp(−time × koff) + B by non-linear least squares regression analysis using GNUplot. The three parameters, A, B, and koff, were fitted except for the dCTP data where B was set to zero. The fits are shown as solid black lines in Fig. 1. The fit lines span the range of data points included in the regression analysis. Data points within the initial rise of the FP readings were excluded from the fit.
Activity Decay Measurements
1 μm SAMHD1 was incubated for 5 min with 1 mm dNTP and 50 μm GTP in the buffer containing 5 mm DTT, 5 mm Mg2+, 150 mm NaCl, 50 mm Tris, pH 8.0. The 50-μl sample(s) were then passed through the 0.5-ml, 7000 molecular weight cut-off Zeba Spin desalting column (Thermo Fisher Scientific, catalog number 89882). After nucleotide removal, the samples were incubated for increasing periods of time, and then SAMHD1 was further diluted into the substrate buffer to yield the final concentrations of 0.2 μm SAMHD1 and 1 mm dTTP in the same buffer. The dTTP hydrolysis rate was then measured using NMR-based detection. The data were fitted to the simple exponential decay equation y(t) = A × exp(−t ×koff) using non-linear least squares regression procedure in MATLAB.
EC50dNTP Measurements
1 μm SAMHD1 was incubated for 5 min with 50 μm GTP and increasing concentrations of dNTPs in the same buffer as above. Nucleotides were then removed using the Zeba column, and the dTTP hydrolysis rates were measured as described in the previous section. The data were fitted to the simple EC50 equation y(x) = A × (x/(x + EC50)), where x is dNTP concentration, using non-linear least squares regression procedure in MATLAB.
Effects of Allosteric dNTPs on Enzyme Specificity
Experiments were performed the same way as for the activity decay measurements, except that there was no incubation after nucleotide removal with the Zeba column. Instead, different deoxynucleotide substrates were immediately added to the samples as described above and the hydrolysis rates were measured for each dNTP substrate individually.
Author Contributions
Z. W. contributed to the experimental design and acquisition and analysis of the data and wrote the manuscript. A. B. contributed to the experimental design and acquisition and analysis of the data and wrote the manuscript. J. V. contributed to the acquisition of data. F. D.-G. contributed to the experimental design and drafting and revising the manuscript. D. N. I. contributed to the experimental design and drafting and revising the manuscript.
Acknowledgments
The NMR Core Facility at the UT Health Science Center at San Antonio is supported in part by the National Institutes of Health P30 CA054174 to the Cancer Therapy and Research Center.
This work was supported in part by National Institutes of Health Grants R01 AI104476 (to D. N. I.) and R01 AI087390 to (F. D.-G.) and a Voelcker Fund Young Investigator Award (to D. N. I.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- FP
- fluorescence polarization
- GTP-F
- γ-(6-aminohexyl)-GTP-ATTO-495.
References
- 1. Baldauf H. M., Pan X., Erikson E., Schmidt S., Daddacha W., Burggraf M., Schenkova K., Ambiel I., Wabnitz G., Gramberg T., Panitz S., Flory E., Landau N. R., Sertel S., Rutsch F., et al. (2012) SAMHD1 restricts HIV-1 infection in resting CD4+ T cells. Nat. Med. 18, 1682–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Berger A., Sommer A. F., Zwarg J., Hamdorf M., Welzel K., Esly N., Panitz S., Reuter A., Ramos I., Jatiani A., Mulder L. C., Fernandez-Sesma A., Rutsch F., Simon V., König R., and Flory E. (2011) SAMHD1-deficient CD14+ cells from individuals with Aicardi-Goutieres syndrome are highly susceptible to HIV-1 infection. PLoS Pathog. 7, e1002425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Descours B., Cribier A., Chable-Bessia C., Ayinde D., Rice G., Crow Y., Yatim A., Schwartz O., Laguette N., and Benkirane M. (2012) SAMHD1 restricts HIV-1 reverse transcription in quiescent CD4+ T-cells. Retrovirology 9, 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hrecka K., Hao C., Gierszewska M., Swanson S. K., Kesik-Brodacka M., Srivastava S., Florens L., Washburn M. P., and Skowronski J. (2011) Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature 474, 658–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Laguette N., Sobhian B., Casartelli N., Ringeard M., Chable-Bessia C., Ségéral E., Yatim A., Emiliani S., Schwartz O., and Benkirane M. (2011) SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature 474, 654–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Beauchamp B. B., and Richardson C. C. (1988) A unique deoxyguanosine triphosphatase is responsible for the optA1 phenotype of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 85, 2563–2567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Goldstone D. C., Ennis-Adeniran V., Hedden J. J., Groom H. C., Rice G. I., Christodoulou E., Walker P. A., Kelly G., Haire L. F., Yap M. W., de Carvalho L. P., Stoye J. P., Crow Y. J., Taylor I. A., and Webb M. (2011) HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature 480, 379–382 [DOI] [PubMed] [Google Scholar]
- 8. Kornberg S. R., Lehman I. R., Bessman M. J., Simms E. S., and Kornberg A. (1958) Enzymatic cleavage of deoxyguanosine triphosphate to deoxyguanosine and tripolyphosphate. J. Biol. Chem. 233, 159–162 [PubMed] [Google Scholar]
- 9. Lahouassa H., Daddacha W., Hofmann H., Ayinde D., Logue E. C., Dragin L., Bloch N., Maudet C., Bertrand M., Gramberg T., Pancino G., Priet S., Canard B., Laguette N., Benkirane M., et al. (2012) SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates. Nat. Immunol. 13, 223–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Seto D., Bhatnagar S. K., and Bessman M. J. (1988) The purification and properties of deoxyguanosine triphosphate triphosphohydrolase from Escherichia coli. J. Biol. Chem. 263, 1494–1499 [PubMed] [Google Scholar]
- 11. Kim B., Nguyen L. A., Daddacha W., and Hollenbaugh J. A. (2012) Tight interplay among SAMHD1 protein level, cellular dNTP levels, and HIV-1 proviral DNA synthesis kinetics in human primary monocyte-derived macrophages. J. Biol. Chem. 287, 21570–21574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ji X., Tang C., Zhao Q., Wang W., and Xiong Y. (2014) Structural basis of cellular dNTP regulation by SAMHD1. Proc. Natl. Acad. Sci. U.S.A. 111, E4305–E4314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ji X., Wu Y., Yan J., Mehrens J., Yang H., DeLucia M., Hao C., Gronenborn A. M., Skowronski J., Ahn J., and Xiong Y. (2013) Mechanism of allosteric activation of SAMHD1 by dGTP. Nat. Struct. Mol. Biol. 20, 1304–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Koharudin L. M., Wu Y., DeLucia M., Mehrens J., Gronenborn A. M., and Ahn J. (2014) Structural basis of allosteric activation of sterile α motif and histidine-aspartate domain-containing protein 1 (SAMHD1) by nucleoside triphosphates. J. Biol. Chem. 289, 32617–32627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yan J., Kaur S., DeLucia M., Hao C., Mehrens J., Wang C., Golczak M., Palczewski K., Gronenborn A. M., Ahn J., and Skowronski J. (2013) Tetramerization of SAMHD1 is required for biological activity and inhibition of HIV infection. J. Biol. Chem. 288, 10406–10417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhu C., Gao W., Zhao K., Qin X., Zhang Y., Peng X., Zhang L., Dong Y., Zhang W., Li P., Wei W., Gong Y., and Yu X. F. (2013) Structural insight into dGTP-dependent activation of tetrameric SAMHD1 deoxynucleoside triphosphate triphosphohydrolase. Nat. Commun. 4, 2722. [DOI] [PubMed] [Google Scholar]
- 17. Cribier A., Descours B., Valadão A. L., Laguette N., and Benkirane M. (2013) Phosphorylation of SAMHD1 by cyclin A2/CDK1 regulates its restriction activity toward HIV-1. Cell Rep. 3, 1036–1043 [DOI] [PubMed] [Google Scholar]
- 18. Welbourn S., Dutta S. M., Semmes O. J., and Strebel K. (2013) Restriction of virus infection but not catalytic dNTPase activity is regulated by phosphorylation of SAMHD1. J. Virol. 87, 11516–11524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Welbourn S., and Strebel K. (2016) Low dNTP levels are necessary but may not be sufficient for lentiviral restriction by SAMHD1. Virology 488, 271–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. White T. E., Brandariz-Nuñez A., Valle-Casuso J. C., Amie S., Nguyen L. A., Kim B., Tuzova M., and Diaz-Griffero F. (2013) The retroviral restriction ability of SAMHD1, but not its deoxynucleotide triphosphohydrolase activity, is regulated by phosphorylation. Cell Host Microbe 13, 441–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhu C. F., Wei W., Peng X., Dong Y. H., Gong Y., and Yu X. F. (2015) The mechanism of substrate-controlled allosteric regulation of SAMHD1 activated by GTP. Acta Crystallogr. D Biol. Crystallogr. 71, 516–524 [DOI] [PubMed] [Google Scholar]
- 22. Bhattacharya A., Wang Z., White T., Buffone C., Nguyen L. A., Shepard C. N., Kim B., Demeler B., Diaz-Griffero F., and Ivanov D. N. (2016) Effects of T592 phosphomimetic mutations on tetramer stability and dNTPase activity of SAMHD1 can not explain the retroviral restriction defect. Sci. Rep. 6, 31353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Brandariz-Nuñez A., Valle-Casuso J. C., White T. E., Nguyen L., Bhattacharya A., Wang Z., Demeler B., Amie S., Knowlton C., Kim B., Ivanov D. N., and Diaz-Griffero F. (2013) Contribution of oligomerization to the anti-HIV-1 properties of SAMHD1. Retrovirology 10, 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Arnold L. H., Groom H. C., Kunzelmann S., Schwefel D., Caswell S. J., Ordonez P., Mann M. C., Rueschenbaum S., Goldstone D. C., Pennell S., Howell S. A., Stoye J. P., Webb M., Taylor I. A., and Bishop K. N. (2015) Phospho-dependent regulation of SAMHD1 oligomerisation couples catalysis and restriction. PLoS Pathog. 11, e1005194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hansen E. C., Seamon K. J., Cravens S. L., and Stivers J. T. (2014) GTP activator and dNTP substrates of HIV-1 restriction factor SAMHD1 generate a long-lived activated state. Proc. Natl. Acad. Sci. U.S.A. 111, E1843–E1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kennedy E. M., Gavegnano C., Nguyen L., Slater R., Lucas A., Fromentin E., Schinazi R. F., and Kim B. (2010) Ribonucleoside triphosphates as substrate of human immunodeficiency virus type 1 reverse transcriptase in human macrophages. J. Biol. Chem. 285, 39380–39391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schlosshauer M., and Baker D. (2004) Realistic protein-protein association rates from a simple diffusional model neglecting long-range interactions, free energy barriers, and landscape ruggedness. Protein Sci. 13, 1660–1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hollenbaugh J. A., Tao S., Lenzi G. M., Ryu S., Kim D. H., Diaz-Griffero F., Schinazi R. F., and Kim B. (2014) dNTP pool modulation dynamics by SAMHD1 protein in monocyte-derived macrophages. Retrovirology 11, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]