Abstract
Mitochondrial complex II or succinate dehydrogenase (SDH) is at the crossroads of oxidative phosphorylation and the tricarboxylic acid cycle. It has been shown that Sdh5 (SDHAF2/SDH5 in mammals) is required for flavination of the subunit Sdh1 (SDHA in human cells) in yeast. Here we demonstrate that in human breast cancer cells, SDHAF2/SDH5 is dispensable for SDHA flavination. In contrast to yeast, CRISPR-Cas9 nickase-mediated SDHAF2 KO breast cancer cells feature flavinated SDHA and retain fully assembled and functional complex II, as well as normal mitochondrial respiration. Our data show that SDHA flavination is independent of SDHAF2 in breast cancer cells, employing an alternative mechanism.
Keywords: cancer biology, cell biology, mammal, mitochondria, mitochondrial respiratory chain complex, SDH assembly factor, SDHA, complex II assembly, flavinylation, mitochondrial complex II, succinate dehydrogenase, flavin adenine dinucleotide, flavination, assembly factor, SDHAF2, cancer cells
Introduction
A major role of mitochondria, generation of ATP, is carried out by oxidative phosphorylation (OXPHOS),3 comprising four respiratory complexes of the electron transfer chain, complex I (CI), CII, CIII, and CIV, plus CV with ATP synthase activity. CII, also known as succinate dehydrogenase (SDH), contains four subunits: the hydrophilic SDHA (a flavoprotein) and SDHB (an iron-sulfur protein), plus the hydrophobic SDHC and SDHD (1, 2). CII is an important source of reactive oxygen species in mitochondria (3–6) and serves as a critical step in both the tricarboxylic acid (TCA) cycle and OXPHOS, linking these two essential cellular pathways (1, 2, 7–9). In the TCA cycle, SDH oxidizes succinate to fumarate (SDH activity) in a reaction that depends on the presence of FAD in the active site of SDHA (1, 2, 8, 10). Within OXPHOS, CII transfers electrons from succinate via [Fe-S] clusters of SDHB to ubiquinone in SDHC/SDHD, reducing it to ubiquinol; this is the succinate-quinone reductase (SQR) activity of CII (2, 8, 10, 11).
As SDH is a mitochondrial enzyme involved in both the TCA cycle and OXPHOS, mutations in its subunits have a profound effect on clinical presentation. Diseases associated with CII dysfunction include Leigh syndrome, renal cell carcinoma, gastrointestinal tumors, and the neuro-endocrine tumors paraganglioma (PGL) and pheochromocytoma (12–16). In recent years, the “list” of germ-line mutations in SDH genes and associated pathological condition has been growing rapidly due to the introduction of next generation sequencing (17, 18). However, many important aspects, including the molecular mechanism that links many of the mutations with their associated phenotypes, remain largely unknown. Furthermore, emerging data have shown that post-translational modifications could also modulate the activity of SDH (19, 20).
CII is formed by “maturation” of individual subunits and their stepwise assembly into a holo-complex. SDH subunits are all encoded by the nuclear genome, synthesized in the cytosol, and post-translationally imported into mitochondria as unfolded proteins. They fold inside mitochondria and bind FAD, [Fe-S] clusters, and heme prosthetic groups that are mandatory for the maturation of the fully functional complex (21). Assembly of large protein complexes often requires the assistance of proteins called assembly factors, i.e. proteins that assist with the formation of the complex, although not forming a part of the final functional unit. In the last decade, several assembly factors have been identified and shown to be required for the correct formation of CII, viz. Sdh5-Sdh8 in yeast and SDHAF1-SDHAF4 in humans (21–23). In yeast mitochondria, additional proteins with less defined function are also required for assembly of SDH, including the flavin transporter Flx1 and the “chaperone-like” protein Tcm62 (24).
The flavoprotein SDHA is the catalytic subunit of CII, converting succinate to fumarate. Sdh5 has been identified in yeast as the assembly factor responsible for flavination of Sdh1, which involves covalent attachment of the redox cofactor FAD (25). The G78R mutation in human SDH5/SDHAF2 gene was shown to be associated with PGL2 (25), and we and others have reported that the mutation affects interaction of SDHAF2/SDH5 with SDHA (25–28). SDHAF2/SDH5, which is not in complex with SDHA, is degraded by the Lon protease as part of the mitochondrial protein homeostasis (26, 28). Initially, it was hypothesized that Sdh5 may be necessary and sufficient for flavination of Sdh1, although an NMR study showed that Sdh5 cannot bind FAD (27). A biochemical study revealed that bacterial SdhE (ortholog of human SDH5) is capable of binding FAD, whereas a genetic study documented the presence of flavinated SdhA in SdhE-deleted bacteria (29–31). This indicates that data from yeast and bacteria are not fully compatible. In addition, thermophilic bacteria that lack SdhE feature flavinated SdhA. In this group of bacteria, flavination of SdhA does not require an assembly factor; rather, thermal energy and dicarboxylic acids drive this process (32). Also, it is not currently clear whether the SDH5 protein that carries out SDHA flavination in yeast and some bacteria is generally necessary and/or sufficient for flavination of SDHA in higher organisms.
In this study, we explored the role of the succinate dehydrogenase assembly factor-2 (SDHAF2) in flavination of SDHA in breast cancer cells. We report that SDHA is flavinated and CII is functional in SDHAF2 KO cells, suggesting that SDHAF2 is dispensable for flavination of SDHA in breast cancer cells. This points to an alternative mechanism of SDHA flavination in breast cancer cell mitochondria.
Results
Generation of Human SDHAF2 Knock-out Cells
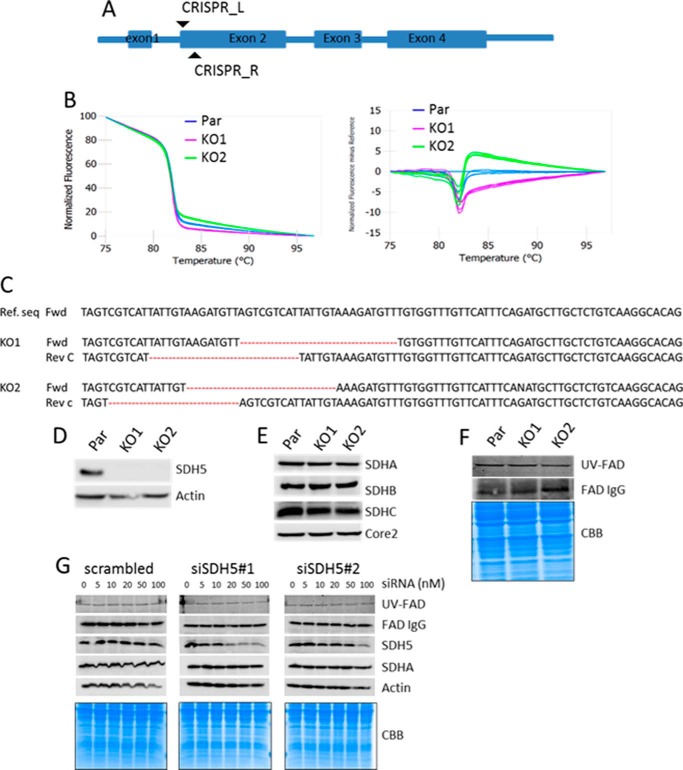
CRISPR-Cas9 has greatly facilitated “gene editing” by means of nucleases that are sequence-specific for the target locus of interest. We used CRISPR-Cas9 nickase with low off-target effects in MDA-MB-231 cells to generate SDHAF2 KO cells (Fig. 1A). Initially, colonies were screened for deletion of the SDHAF2 gene using high-resolution melting (HRM), which was further confirmed by Western blotting (WB) and Sanger sequencing. HRM analysis showed that the SDHAF2 alleles in two clones (termed KO1 and KO2) were found to show a difference in melting curve (Fig. 1B). Both positive and negative strands were then sequenced using the Sanger method. Fig. 1C shows that these cell lines have a 17–24-nucleotide deletion in intron 1 and exon 2 junction of SDHAF2. Using an SDS-PAGE/WB analysis approach, we did not detect the SDHAF2 protein in either the SDHAF2 KO1 or the KO2 sub-lines (Fig. 1D).
FIGURE 1.
Generation of SDHAF2 knock-out cell lines and SDHA flavination analysis. A, scheme showing SDHAF2 gene structure and sgRNA target. B, HRM curve analysis of parental cells (Par) and two selected clones. C, analysis of the nucleotide sequence spanning the SDHAF2-targeted region in the SDHAF2 gene present in the first intron and second coding exon. Reverse complement of sequence (Rev C) was used for antisense strand analyses. Ref. seq, reference sequence; Fwd, forward; Rev, reverse. D, WB demonstrating absence of SDHAF2 in KO1 and KO2 cell lines. Actin was used as a loading control. E, WB showing the steady state levels of protein subunits of CII with anti-Core2 as a loading control. F, SDHA flavination analysis using anti-FAD in WB (upper band) and fluorescent signal of FAD (middle band) from SDS-PAGE gel exposed to UV light. Coomassie Brilliant Blue R staining was used as a loading control (bottom bands). G, MDA-MB-231 cells treated with increasing concentrations of two SDHAF2 siRNAs were probed for flavination of SDHA using the UV method and probing anti-FAD IgG, as well as for the level of SDHAF2 and SDHA using WB. Coomassie Brilliant Blue R (CBB) staining and actin were used as loading control.
Knock-out of SDHAF2 Does Not Affect Flavination and Steady State of SDHA
We next assessed the effect of SDHAF2 gene disruption on the steady state levels of several subunits of respiratory complexes. Fig. 1E shows the steady state levels of selected electron transfer chain proteins, as assessed by SDS-PAGE/WB, revealing that they were similar to each other irrespective of the SDHAF2 status. Interestingly, and rather surprisingly, we found that SDHA is flavinated in the absence of SDHAF2, as documented by a UV fluorescence assay and by WB using an antibody raised against FAD in both SDHAF2 KO1 and KO2 cells (Fig. 1F).
RNA Interference Approach Recapitulates the SDHAF2 KO Results
To independently confirm that SDHAF2 is redundant for SDHA flavination, we knocked down SDHAF2 using two different siRNAs, and as a control, universal negative siRNA was used. Fig. 1G shows that knocking down the SDHAF2 protein by two different siRNAs did not affect either the level of SDHA, or its flavination status.
SDHAF2-deficient Cells Maintain SDH Activity
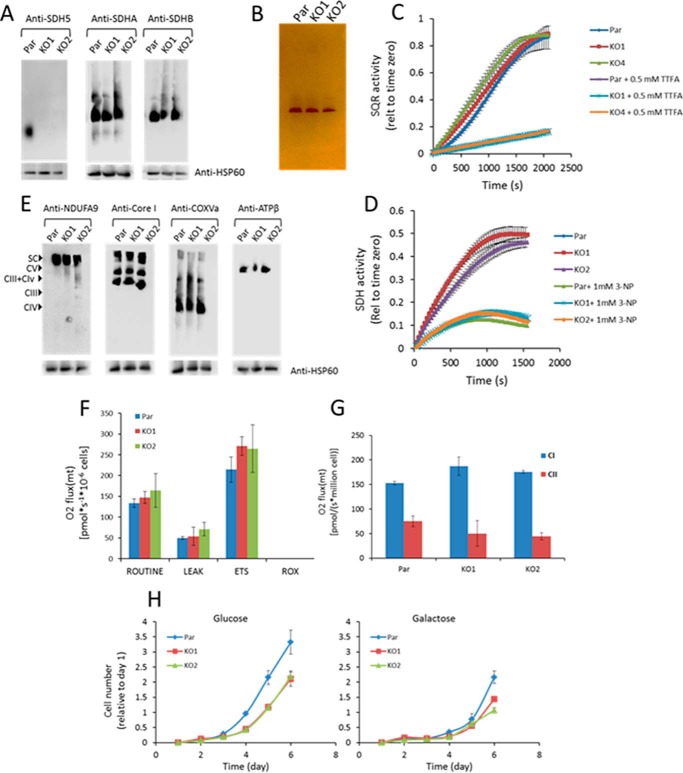
We examined the assembly of the SDH complex in SDHAF2 KO cells. As shown in Fig. 2A, BN-PAGE followed by WB using anti-SDHA and anti-SDHB revealed fully assembled SDH complex in SDHAF2 KO cells, comparable with parental cells. Using anti-SDHAF2 IgG and native gels, we confirmed the absence of the assembly factor in the KO lines (Fig. 2A). We next tested whether the assembled SDH complex in SDHAF2 KO cells is functional using an in-gel activity assay. As indicated in Fig. 2B, the in-gel assay revealed that assembled CII in both SDHAF2 KO cell lines is active. CII features two enzymatic activities, i.e. the SDH activity, whereby the catalytic SDHA subunit in mitochondrial matrix oxidizes succinate to fumarate, and the SQR activity, by means of which electrons generated by succinate oxidation are transferred via the SDHB [Fe-S] clusters to ubiquinone, reducing it to ubiquinol (10, 11). We next tested whether SDH and SQR activities are compromised in SDHAF2 KO cells. We performed analysis of SDH and SQR activities in the absence and presence of their specific inhibitors 3-nitropropionic acid (3-NP) and thenoyl trifluoroacetic acid (TTFA), respectively, and found no difference in either of the two activities between parental and SDHAF2 KO cells (Fig. 2, C and D). Taken together, SDHAF2 KO breast cancer cells maintain functionally active CII.
FIGURE 2.
Respiratory complex assembly and activity in the absence of SDHAF2. A and E, mitochondria isolated from control and SDHAF2 knock-out cell lines were solubilized in digitonin and analyzed by BN-PAGE for the assembly of respiratory complexes detected by the indicated antibodies. Par, parental cells. B, in-gel activity of complex II following hrCNE-3 was analyzed using 10 μg digitonin-solubilized mitochondria. C and D, SQR (C) and SDH (D) activity was assessed by the change in absorbance of DCPIP at 600 nm. F and G, analysis of routine respiration (F) and complex I- and II-driven respiration (G) was performed in permeabilized cell using the Oxygraph-2k. ETS, electron transfer system; ROX, residual oxygen consumption. H, parental and SDHAF2 KO cells were cultured in glucose- or galactose-containing media and analyzed for cell proliferation using the crystal violet method. Error bars indicate means ± S.D.
Oxygen Consumption Rate and Respiratory Complex Assembly Are Not Compromised in SDHAF2 KO Cells
To find whether the absence of SDHAF2 affects other mitochondrial respiratory complexes, we assessed the assembly of CI, CIII, CIV, and CV using BN-PAGE. We found that all complexes as well as supercomplexes assembled irrespective of the SDHAF2 status (Fig. 2E). Oxygen consumption was then assessed using the Oxygraph-2k apparatus. Once again, we observed differences in neither routine nor CI-/CII-dependent respiration between parental and SDHAF2 KO cells (Fig. 2, F and G).
SDHAF2-deficient Cells Proliferate in Both Glycolytic and OXPHOS Media
Yeast cells with deleted Sdh5 feature proliferation defects and cannot grow in non-oxidative media; they also lack SDH activity (25, 27). We observed a decrease in glucose-dependent proliferation in SDHAF2 KO cells as compared with their parental counterparts (Fig. 2H, left panel). We then tested whether SDHAF2 KO cells proliferate in galactose-containing media as a read-out of OXPHOS activity. We found only a slight difference between the growth of SDHAF2 KO and parental cell lines in galactose-containing media (Fig. 2H, right panel). These results indicate that knock-out of SDHAF2/SDH5 has a mild effect on the proliferation rate, but this effect appears to be independent of OXPHOS.
Discussion
SDH integrates two essential mitochondrial pathways, OXPHOS and the TCA cycle. The flavoprotein SDHA catalyzes oxidation of succinate to fumarate. For this, a covalent insertion of FAD (flavination) to SDHA subunit is essential. The yeast Sdh5 was identified as a protein mediating flavination of Sdh1 (SDHA in mammals) (25). However, studies from certain bacterial strains and plants indicate that the function of the SDH5 family proteins may be species-dependent. In this study, we generated an SDHAF2 KO human breast cancer cell line, and unexpectedly, identified SDHAF2 to be dispensable for flavination of SDHA and assembly of the complex. We examined whether SDHA is flavinated using FAD autofluorescence and an antibody raised against FAD (Fig. 1F). Both approaches resulted in identification of flavinated SDHA in SDHAF2 KO cells, and this was independently confirmed in SDHAF2 knockdown cells. We also detected both SQR and SDH activity in SDHAF2 KO cells, indicating functionally active CII (Fig. 2, B–D). In agreement with our data, mouse SDH5 KO (33) and plant cells (34) are viable. Albeit reduced, functional CII was detected on the basis of enzymatic activities of SDH and SQR and in-gel activity following BN-PAGE in SDHAF2 KO Arabidopsis (34).
To date, studies on the role of SDHAF2 in mammalian systems have been limited. Fully assembled CII is critical for both TCA cycle and OXPHOS functions. For example, SDHD KO mice were found embryonically lethal (35). SDHB KO mammalian cell lines failed to exert CII activity and featured a reduced proliferation rate (36, 37). A seminal study linking mutations in SDHAF2 and PGL2 ascribed the potential loss of SDHAF2 function to the lack of SDHA flavination. When human SDHAF2 was expressed in Sdh5 KO yeast, Sdh1 was again flavinated and CII assembled (25). SDHAF2/SDH5 was also shown to flavinate SDHA in a cell-free system (38). On the contrary, our data using breast cancer cells clearly document that flavination of SDHA and assembly of CII occur in the absence of SDHAF2, indicating that SDHAF2 is dispensable for flavination of succinate dehydrogenase.
We have recently shown that the G78R mutation in the SDHAF2 gene, previously identified in a PGL2 patient (25), affects association of the assembly factor with SDHA (26). SDHAF2 is now classified as a tumor suppressor. The following question arises: What is the role of SDHAF2 in the context of CII and how is SDHA flavinated in breast cancer cells? It cannot be excluded that the species-dependent differences in the role of SDHAF2 in flavination of SDHA reflect evolutional aspects, and there might be functional redundancy at least in certain mammalian cells epitomized here by breast cancer cells. Notwithstanding the above unresolved questions, our finding clearly points to the notion that we are far from understanding all the details of genetics, biology, and function of mitochondrial respiratory complexes, as exemplified here by the relatively simple heterotetrameric protein complex CII.
Experimental Procedures
Cell Lines and Media
MDA-MB-231 cells were cultured in DMEM (Gibco) supplemented with 10% FBS (Sigma) and antibiotic-anti-mycotic mixture. Cells were grown at 37 °C under atmosphere of 5% CO2.
CRISPR-Cas9-mediated Knock-out
Cells were transfected with a CRISPR-Cas9 nickase (Cas9n D10A) construct targeting exon 2 of the human SDHAF2 gene. For this, the Cas9 expression plasmid CP-C9NI-02 and the sgRNA plasmid (pCRISPR-SG01) with guide sequences AAT GAT GTC ACA CTG AGC AA and CAG CCC AAC AGA TTC CCA AA were used (GeneCopoeia). To facilitate co-selection of cells expressing the two vectors, each vector contained a distinct selection marker for neomycin and hygromycin to select cells expressing Cas9 and sgRNA, respectively. MDA-MB-231 cells were transfected with a 1:1:1 mixture of the sgRNA construct (L and R) and vectors encoding Cas9 using Lipofectamine® 3000 reagent (Invitrogen) as recommended by the manufacturer. Two days after transfection, cells were selected in 500 μg/ml hygromycin B and 700 μg/ml neomycin for 72 h, followed by plating at ∼1 cell/well in 96-well plates for clonal expansion. Knock-out clones were identified by high-resolution melting curve, which was confirmed by WB, using SDHAF2-specific antibodies and Sanger sequencing.
RNA Interference
Two sets of mission siRNA (Sigma) for human SDHAF2 (SASI_Hs01_00053252 and SASI_Hs01_00053255) and universal scrambled RNA (SIC002) were used. siRNAs were transfected into 50% confluent MB-MDA-231 cells in 6-well plates using Lipofectamine® 3000 reagent (Invitrogen) according to the manufacturer's protocol. Transfected cells were harvested after 72 h for analysis.
Western Blotting
Protein samples were denatured, separated by SDS-PAGE, and transferred onto PVDF membranes (Bio-Rad). The membranes were blocked with 5% skim milk and incubated with anti-SDH5 (1:1000, Cell Signaling), anti-FAD (1:500, MyBioSource), anti-SDHA, anti-SDHB anti-actin, anti-HSP60 (1:1000, Abcam), anti-NDUFA9, anti-Core I, or anti-COX5a IgG (1:1000, Thermo Scientific). Secondary HRP antibodies were then applied, and the membranes were visualized using the ECL WB substrate (Thermo Scientific).
Isolation of Mitochondria and Blue Native Gel Electrophoresis
Mitochondria were isolated from cultured cell lines as described previously (32, 39, 40). Digitonin-solubilized mitochondrial proteins (5–10 μg) were applied and run on 4–16% gradient BN gels. After electrophoresis, the complexes were blotted onto PVDF membranes and sequentially probed with specific antibodies against CI (anti-NDUFA9), CIII (anti-Core I), CIV (anti-COX5a), and CII (anti-SDHA and anti-SDHB).
In-gel CII Assay
Digitonin-solubilized mitochondrial proteins were separated using high-resolution clear native electrophoresis 3 (hrCNE-3) as described previously (41). The gel was then stained in 5 mm Tris-HCl, pH 7.4, 2.5 mg/ml nitro blue tetrazolium, 10 mm succinate, and 0.2 mm phenazine methosulfate, and the activity was stopped with 50% methanol and 10% acetic acid.
SQR and SDH Activity
SQR activity was assessed as described previously (11, 42, 43). Briefly, whole cell lysate (10 μg) was preincubated with 10 mm succinate, 0.2 mm ATP, and 4 μm rotenone with/without 1 mm TTFA for 10 min before adding master mix (20 mm KPO4, pH 7.5, 1 mm EDTA, 80 μm 2,6-dichlorophenolindophenol (DCIP), and 50 μm decylubiquinone). DCIP reduction was monitored at 600 nm. SDH activity was assessed using isolated mitochondria (11, 44). Digitonin-permeabilized mitochondria were incubated with 2 mm KCN, 4 μg/ml antimycin A, and 10 mm succinate with/without 1 mm 3-NP in phosphate buffer at 30 °C. SDH activity was monitored at 600 nm following the addition of 1.6 mm phenazine methosulfate and 80 μm DCPIP.
FAD Analysis
Flavination of SDHA was analyzed as described previously (45). In brief, proteins (30 μg) were separated by SDS-PAGE. Following electrophoresis, the gel was briefly rinsed with Milli-Q water and incubated for 20 min in 10% acetic acid. Flavination of SDHA was monitored on a UV transilluminator using the ChemiDoc system (Bio-Rad). The gel was stained with Coomassie Blue as a loading control.
Oxygen Consumption Measurements
Cellular oxygen consumption was assessed using the Oxygraph-2k apparatus (Oroboros Instruments) according to a standard procedure (46, 47) with the following agents: 1 μm oligomycin, 1.2 μm carbonyl cyanide 4-(trifluoromethoxy) phenyl-hydrazone (FCCP), and 0.5 μm rotenone. For CI- and CII-dependent respiration, digitonin-permeabilized cells were used.
Cell Proliferation
For the cell growth assay, 10,000 cells were seeded in a 12-well culture plate. On the next day, cells were rinsed twice in PBS and either 5 mm galactose or 24 mm glucose DMEM was added to each well, and then the plates were incubated at 37 °C. Cell density was determined by staining with crystal violet.
Statistics
All experiments were repeated at least three times, and results were expressed as mean ± S.D. Representative images of gels or traces are shown. Quantitated data are the average of ≥3 experiments.
Author Contributions
A. B.-G. designed and performed experiments, and analyzed data, A. B.-G., L. D., J. R., and J. N. conceived the research, interpreted the data, and wrote the manuscript.
This work was supported by a grant from the Australian Research Council, a Griffith University New Researcher grant, Czech Research Foundation Grants 15-02203S and 16-22823S, and funding from BIOCEV (Grant CZ.1.05/1.1.00/02.0109). The authors declare that they have no conflicts of interest with the contents of this article.
- OXPHOS
- oxidative phosphorylation
- CI
- complex I
- DCPIP
- 2,6-dichlorophenolindophenol
- HRM
- high-resolution melting
- PGL
- paraganglioma
- SDH
- succinate dehydrogenase
- SDHA
- succinate dehydrogenase subunit A
- SQR
- succinate-quinone reductase
- TCA
- tricarboxylic acid
- TTFA
- thenoyl trifluoroacetic acid
- UbQ
- ubiquinone
- WB
- western blotting
- 3-NP
- 3-nitropropionic acid
- BN-PAGE
- blue native-PAGE
- sgRNA
- sub-genomic RNA
- hrCNE-3
- high-resolution clear native electrophoresis 3.
References
- 1. Cecchini G. (2003) Function and structure of complex II of the respiratory chain. Annu. Rev. Biochem. 72, 77–109 [DOI] [PubMed] [Google Scholar]
- 2. Sun F., Huo X., Zhai Y., Wang A., Xu J., Su D., Bartlam M., and Rao Z. (2005) Crystal structure of mitochondrial respiratory membrane protein complex II. Cell 121, 1043–1057 [DOI] [PubMed] [Google Scholar]
- 3. Kluckova K., Sticha M., Cerny J., Mracek T., Dong L., Drahota Z., Gottlieb E., Neuzil J., and Rohlena J. (2015) Ubiquinone-binding site mutagenesis reveals the role of mitochondrial complex II in cell death initiation. Cell Death Dis. 6, e1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dong L. F., Jameson V. J., Tilly D., Cerny J., Mahdavian E., Marín-Hernández A., Hernández-Esquivel L., Rodríguez-Enríquez S., Stursa J., Witting P. K., Stantic B., Rohlena J., Truksa J., Kluckova K., Dyason J. C., et al. (2011) Mitochondrial targeting of vitamin E succinate enhances its pro-apoptotic and anti-cancer activity via mitochondrial complex II. J. Biol. Chem. 286, 3717–3728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Siebels I., and Dröse S. (2013) Q-site inhibitor induced ROS production of mitochondrial complex II is attenuated by TCA cycle dicarboxylates. Biochim. Biophys. Acta 1827, 1156–1164 [DOI] [PubMed] [Google Scholar]
- 6. Quinlan C. L., Orr A. L., Perevoshchikova I. V., Treberg J. R., Ackrell B. A., and Brand M. D. (2012) Mitochondrial complex II can generate reactive oxygen species at high rates in both the forward and reverse reactions. J. Biol. Chem. 287, 27255–27264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ackrell B. A. (2000) Progress in understanding structure-function relationships in respiratory chain complex II. FEBS Lett. 466, 1–5 [DOI] [PubMed] [Google Scholar]
- 8. Grimm S. (2013) Respiratory chain complex II as general sensor for apoptosis. Biochim. Biophys. Acta 1827, 565–572 [DOI] [PubMed] [Google Scholar]
- 9. Kluckova K., Bezawork-Geleta A., Rohlena J., Dong L., and Neuzil J. (2013) Mitochondrial complex II, a novel target for anti-cancer agents. Biochim. Biophys. Acta 1827, 552–564 [DOI] [PubMed] [Google Scholar]
- 10. Miyadera H., Shiomi K., Ui H., Yamaguchi Y., Masuma R., Tomoda H., Miyoshi H., Osanai A., Kita K., and Omura S. (2003) Atpenins, potent and specific inhibitors of mitochondrial complex II (succinate-ubiquinone oxidoreductase). Proc. Natl. Acad. Sci. U.S.A. 100, 473–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Guzy R. D., Sharma B., Bell E., Chandel N. S., and Schumacker P. T. (2008) Loss of the SdhB, but not the SdhA, subunit of complex II triggers reactive oxygen species-dependent hypoxia-inducible factor activation and tumorigenesis. Mol. Cell. Biol. 28, 718–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bardella C., Pollard P. J., and Tomlinson I. (2011) SDH mutations in cancer. Biochim. Biophys. Acta 1807, 1432–1443 [DOI] [PubMed] [Google Scholar]
- 13. Birch-Machin M. A., Taylor R. W., Cochran B., Ackrell B. A., and Turnbull D. M. (2000) Late-onset optic atrophy, ataxia, and myopathy associated with a mutation of a complex II gene. Ann. Neurol. 48, 330–335 [PubMed] [Google Scholar]
- 14. Bourgeron T., Rustin P., Chretien D., Birch-Machin M., Bourgeois M., Viegas-Péquignot E., Munnich A., and Rötig A. (1995) Mutation of a nuclear succinate dehydrogenase gene results in mitochondrial respiratory chain deficiency. Nat. Genet. 11, 144–149 [DOI] [PubMed] [Google Scholar]
- 15. Favier J., Amar L., and Gimenez-Roqueplo A. P. (2015) Paraganglioma and phaeochromocytoma: from genetics to personalized medicine. Nat. Rev. Endocrinol. 11, 101–111 [DOI] [PubMed] [Google Scholar]
- 16. Parfait B., Chretien D., Rötig A., Marsac C., Munnich A., and Rustin P. (2000) Compound heterozygous mutations in the flavoprotein gene of the respiratory chain complex II in a patient with Leigh syndrome. Hum. Genet. 106, 236–243 [DOI] [PubMed] [Google Scholar]
- 17. Hechtman J. F., Zehir A., Mitchell T., Borsu L., Singer S., Tap W., Oultache A., Ladanyi M., and Nafa K. (2015) Novel oncogene and tumor suppressor mutations in KIT and PDGFRA wild type gastrointestinal stromal tumors revealed by next generation sequencing. Genes Chromosomes Cancer 54, 177–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Italiano A., Chen C. L., Sung Y. S., Singer S., DeMatteo R. P., LaQuaglia M. P., Besmer P., Socci N., and Antonescu C. R. (2012) SDHA loss of function mutations in a subset of young adult wild-type gastrointestinal stromal tumors. BMC Cancer 12, 408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Acín-Pérez R., Carrascoso I., Baixauli F., Roche-Molina M., Latorre-Pellicer A., Fernández-Silva P., Mittelbrunn M., Sanchez-Madrid F., Pérez-Martos A., Lowell C. A., Manfredi G., and Enríquez J. A. (2014) ROS-triggered phosphorylation of complex II by Fgr kinase regulates cellular adaptation to fuel use. Cell Metab. 19, 1020–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nath A. K., Ryu J. H., Jin Y. N., Roberts L. D., Dejam A., Gerszten R. E., and Peterson R. T. (2015) PTPMT1 inhibition lowers glucose through succinate dehydrogenase phosphorylation. Cell Rep. 10, 694–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Van Vranken J. G., Na U., Winge D. R., and Rutter J. (2015) Protein-mediated assembly of succinate dehydrogenase and its cofactors. Crit. Rev. Biochem. Mol. Biol. 50, 168–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ghezzi D., Goffrini P., Uziel G., Horvath R., Klopstock T., Lochmüller H., D'Adamo P., Gasparini P., Strom T. M., Prokisch H., Invernizzi F., Ferrero I., and Zeviani M. (2009) SDHAF1, encoding a LYR complex-II specific assembly factor, is mutated in SDH-defective infantile leukoencephalopathy. Nat. Genet. 41, 654–656 [DOI] [PubMed] [Google Scholar]
- 23. Maio N., Ghezzi D., Verrigni D., Rizza T., Bertini E., Martinelli D., Zeviani M., Singh A., Carrozzo R., and Rouault T. A. (2016) Disease-causing SDHAF1 mutations impair transfer of Fe-S clusters to SDHB. Cell Metab. 23, 292–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rutter J., Winge D. R., and Schiffman J. D. (2010) Succinate dehydrogenase: assembly, regulation and role in human disease. Mitochondrion 10, 393–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hao H. X., Khalimonchuk O., Schraders M., Dephoure N., Bayley J. P., Kunst H., Devilee P., Cremers C. W., Schiffman J. D., Bentz B. G., Gygi S. P., Winge D. R., Kremer H., and Rutter J. (2009) SDH5, a gene required for flavination of succinate dehydrogenase, is mutated in paraganglioma. Science 325, 1139–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bezawork-Geleta A., Brodie E. J., Dougan D. A., and Truscott K. N. (2015) LON is the master protease that protects against protein aggregation in human mitochondria through direct degradation of misfolded proteins. Sci. Rep. 5, 17397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Eletsky A., Jeong M. Y., Kim H., Lee H. W., Xiao R., Pagliarini D. J., Prestegard J. H., Winge D. R., Montelione G. T., and Szyperski T. (2012) Solution NMR structure of yeast succinate dehydrogenase flavinylation factor Sdh5 reveals a putative Sdh1 binding site. Biochemistry 51, 8475–8477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bezawork-Geleta A., Saiyed T., Dougan D. A., and Truscott K. N. (2014) Mitochondrial matrix proteostasis is linked to hereditary paraganglioma: LON-mediated turnover of the human flavinylation factor SDH5 is regulated by its interaction with SDHA. FASEB J. 28, 1794–1804 [DOI] [PubMed] [Google Scholar]
- 29. McNeil M. B., Clulow J. S., Wilf N. M., Salmond G. P., and Fineran P. C. (2012) SdhE is a conserved protein required for flavinylation of succinate dehydrogenase in bacteria. J. Biol. Chem. 287, 18418–18428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. McNeil M. B., and Fineran P. C. (2013) The conserved RGxxE motif of the bacterial FAD assembly factor SdhE is required for succinate dehydrogenase flavinylation and activity. Biochemistry 52, 7628–7640 [DOI] [PubMed] [Google Scholar]
- 31. Maklashina E., Rajagukguk S., Starbird C. A., McDonald W. H., Koganitsky A., Eisenbach M., Iverson T. M., and Cecchini G. (2016) Binding of the covalent flavin assembly factor to the flavoprotein subunit of complex II. J. Biol. Chem. 291, 2904–2916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kounosu A. (2014) Analysis of covalent flavinylation using thermostable succinate dehydrogenase from Thermus thermophilus and Sulfolobus tokodaii lacking SdhE homologs. FEBS Lett. 588, 1058–1063 [DOI] [PubMed] [Google Scholar]
- 33. Liu J., Gao L., Zhang H., Wang D., Wang M., Zhu J., Pang C., and Wang C. (2013) Succinate dehydrogenase 5 (SDH5) regulates glycogen synthase kinase 3β-β-catenin-mediated lung cancer metastasis. J. Biol. Chem. 288, 29965–29973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Huang S., Taylor N. L., Ströher E., Fenske R., and Millar A. H. (2013) Succinate dehydrogenase assembly factor 2 is needed for assembly and activity of mitochondrial complex II and for normal root elongation in Arabidopsis. Plant J. 73, 429–441 [DOI] [PubMed] [Google Scholar]
- 35. Piruat J. I., Pintado C. O., Ortega-Sáenz P., Roche M., and López-Barneo J. (2004) The mitochondrial SDHD gene is required for early embryogenesis, and its partial deficiency results in persistent carotid body glomus cell activation with full responsiveness to hypoxia. Mol. Cell. Biol. 24, 10933–10940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cardaci S., Zheng L., MacKay G., van den Broek N. J., MacKenzie E. D., Nixon C., Stevenson D., Tumanov S., Bulusu V., Kamphorst J. J., Vazquez A., Fleming S., Schiavi F., Kalna G., Blyth K., et al. (2015) Pyruvate carboxylation enables growth of SDH-deficient cells by supporting aspartate biosynthesis. Nat. Cell. Biol. 17, 1317–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lussey-Lepoutre C., Hollinshead K. E., Ludwig C., Menara M., Morin A., Castro-Vega L. J., Parker S. J., Janin M., Martinelli C., Ottolenghi C., Metallo C., Gimenez-Roqueplo A. P., Favier J., and Tennant D. A. (2015) Loss of succinate dehydrogenase activity results in dependency on pyruvate carboxylation for cellular anabolism. Nat. Commun. 6, 8784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zafreen L., Walker-Kopp N., Huang L. S., and Berry E. (2016) In-vitro, SDH5-dependent flavinylation of immobilized human respiratory complex II flavoprotein. Arch. Biochem. Biophys. 604, 47–56 [DOI] [PubMed] [Google Scholar]
- 39. Tan A. S., Baty J. W., Dong L. F., Bezawork-Geleta A., Endaya B., Goodwin J., Bajzikova M., Kovarova J., Peterka M., Yan B., Pesdar E. A., Sobol M., Filimonenko A., Stuart S., Vondrusova M., et al. (2015) Mitochondrial genome acquisition restores respiratory function and tumorigenic potential of cancer cells without mitochondrial DNA. Cell Metab. 21, 81–94 [DOI] [PubMed] [Google Scholar]
- 40. Vondrusova M., Bezawork-Geleta A., Sachaphibulkij K., Truksa J., and Neuzil J. (2015) The effect of mitochondrially targeted anticancer agents on mitochondrial (super)complexes. Methods Mol. Biol. 1265, 195–208 [DOI] [PubMed] [Google Scholar]
- 41. Wittig I., Karas M., and Schägger H. (2007) High resolution clear native electrophoresis for in-gel functional assays and fluorescence studies of membrane protein complexes. Mol. Cell. Proteomics 6, 1215–1225 [DOI] [PubMed] [Google Scholar]
- 42. Dong L. F., Low P., Dyason J. C., Wang X. F., Prochazka L., Witting P. K., Freeman R., Swettenham E., Valis K., Liu J., Zobalova R., Turanek J., Spitz D. R., Domann F. E., Scheffler I. E., et al. (2008) α-Tocopheryl succinate induces apoptosis by targeting ubiquinone-binding sites in mitochondrial respiratory complex II. Oncogene 27, 4324–4335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yan B., Stantic M., Zobalova R., Bezawork-Geleta A., Stapelberg M., Stursa J., Prokopova K., Dong L., and Neuzil J. (2015) Mitochondrially targeted vitamin E succinate efficiently kills breast tumour-initiating cells in a complex II-dependent manner. BMC Cancer 15, 401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lemarie A., Huc L., Pazarentzos E., Mahul-Mellier A. L., and Grimm S. (2011) Specific disintegration of complex II succinate:ubiquinone oxidoreductase links pH changes to oxidative stress for apoptosis induction. Cell Death Differ. 18, 338–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bafunno V., Giancaspero T. A., Brizio C., Bufano D., Passarella S., Boles E., and Barile M. (2004) Riboflavin uptake and FAD synthesis in Saccharomyces cerevisiae mitochondria: involvement of the Flx1p carrier in FAD export. J. Biol. Chem. 279, 95–102 [DOI] [PubMed] [Google Scholar]
- 46. Kluckova K., Dong L. F., Bajzikova M., Rohlena J., and Neuzil J. (2015) Evaluation of respiration of mitochondria in cancer cells exposed to mitochondria-targeted agents. Methods Mol. Biol. 1265, 181–194 [DOI] [PubMed] [Google Scholar]
- 47. Pesta D., and Gnaiger E. (2012) High-resolution respirometry: OXPHOS protocols for human cells and permeabilized fibers from small biopsies of human muscle. Methods Mol. Biol. 810, 25–58 [DOI] [PubMed] [Google Scholar]