Abstract
A common genetic variation in the transmembrane protein 106B (TMEM106B) gene has been suggested to be a risk factor for frontotemporal lobar degeneration (FTLD) with inclusions of transactive response DNA-binding protein-43 (TDP-43) (FTLD-TDP), the most common pathological subtype in FTLD. Furthermore, previous studies have shown that TMEM106B levels are up-regulated in the brains of FTLD-TDP patients, although the significance of this finding remains unknown. In this study, we show that the overexpression of TMEM106B and its N-terminal fragments induces cell death, enhances oxidative stress-induced cytotoxicity, and causes the cleavage of TDP-43, which represents TDP-43 pathology, using cell-based models. TMEM106B-induced death is mediated by the caspase-dependent mitochondrial cell death pathways and possibly by the lysosomal cell death pathway. These findings suggest that the up-regulation of TMEM106B may increase the risk of FTLD by directly causing neurotoxicity and a pathological phenotype linked to FTLD-TDP.
Keywords: amyotrophic lateral sclerosis (ALS) (Lou Gehrig's disease), cell death, lysosome, neurodegenerative disease, TAR DNA-binding protein 43 (TDP-43) (TARDBP), Frontotemporal lobar degeneration, Transmembrane protein 106B
Introduction
Frontotemporal lobar degeneration (FTLD)2 is the second leading cause of presenile dementia, typically with an onset of disease before 65 years of age (1, 2). It is pathologically characterized by degeneration of the frontal and anterior temporal lobes of the cerebral cortices. Clinical characteristics include changes in personality and behavior and difficulties with language. Approximately 25–50% of FTLD cases are genetically inherited, and several genes, such as GRN (the gene encoding progranulin), MAPT, VCP, CHAMP2B, TARDBP, FUS, and C9ORF72, have been identified as FTLD-causative genes (3). FTLD has been recently recognized as a disease that has a common pathogenetic background with amyotrophic lateral sclerosis (ALS) (4). ALS is an incurable motor neuron degenerative disease, characterized by loss of both upper and lower motor neurons (5, 6). Approximately 15% of FTLD patients develop motor neuron disease, and more than 15% of ALS patients have cognitive and behavioral impairments (4, 7). Mutations in several genes, such as TARDBP and C9ORF72, lead to the development of both FTLD and ALS (1, 4, 8). An important pathological hallmark of FTLD and ALS is a cytoplasmic transactive response DNA-binding protein-43 (TDP-43) inclusion. TDP-43 is the major component of inclusions in ∼50% of FTLD patients (subtype FTLD-TDP) and the majority of ALS patients (9, 10). However, currently, the pathogenetic mechanisms underlying these neurodegenerative diseases are not fully understood.
A genome-wide association study and cohort studies have identified three SNPs (rs1990622, rs6966915, and rs1020004) in the transmembrane protein 106B (TMEM106B) gene region as genetic risk modifiers for FTLD-TDP (11–14). The risk association is more prominent in FTLD-TDP cases with GRN and C9ORF72 mutations (11, 13–17). The risk rs1990622 genotype has also been shown to be associated with cognitive impairment in ALS (18).
TMEM106B is a glycosylated type 2 transmembrane protein, located in late endosomes and lysosomes, with uncharacterized function (19–22). Several clinical studies have shown that TMEM106B levels are significantly up-regulated in FTLD-TDP cases, especially in those with a GRN mutation (11, 13, 20, 23). In agreement with this, the expression level of the TMEM106B risk variant tends to be up-regulated, compared with that encoded by the non-risk TMEM106B variant in vitro (22). These observations suggest that the up-regulation of TMEM106B expression is closely linked to the onset of FTLD-TDP. However, how the up-regulation of TMEM106B increases the risk for FTLD-TDP remains to be elucidated.
In this study, we show that the overexpression of TMEM106B induces cell death, enhances oxidative stress-induced cytotoxicity, and causes the cleavage of TDP-43, a representative TDP-43 pathology observed in FTLD-TDP, using cell-based models. TMEM106B-induced cell death is mediated by the caspase-dependent mitochondrial cell death pathways and possibly by the lysosomal cell death pathway. These findings suggest that the up-regulation of TMEM106B increases the risk of FTLD by directly causing neurotoxicity.
Results
A TMEM106B Antibody Recognizes the TMEM106B Protein
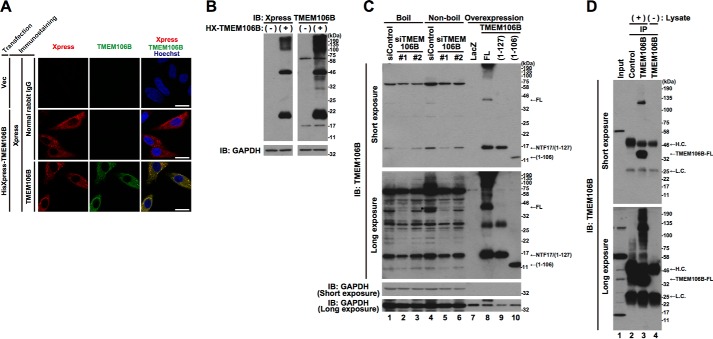
Following the transient overexpression of N-terminally HisXpress (HX)-tagged human TMEM106B-full length (FL) in HeLa cells, we detected its presence by immunofluorescence analysis and immunoblotting analysis using Xpress and TMEM106B antibodies (Fig. 1, A and B). Immunofluorescence analysis indicated that HX-TMEM106B-FL showed a cytoplasmic granular localization pattern (Fig. 1A), which has been shown to be a lysosomal localization pattern in previous reports (19–22). Immunoblotting analysis indicated that multiple proteins were stained with the Xpress and the TMEM106B antibodies. They included smeared high molecular mass proteins, a 46-kDa protein, and a 20-kDa protein (Fig. 1B). Because the predicted molecular mass of TMEM106B is 31 kDa and it is a highly glycosylated protein (20–22), the 46-kDa protein in Fig. 1B was thought to be TMEM106B-FL. Based on the finding that TMEM106B tends to be multimerized (24), the smeared high molecular mass proteins may be TMEM106B multimers. The 20-kDa protein appears to correspond to the N-terminal fragment (NTF) of TMEM106B, as reported in a previous study (25).
FIGURE 1.
A TMEM106B antibody recognizes the TMEM106B protein. A, HeLa cells transiently overexpressing HisXpress-tagged TMEM106B-FL were fixed and immunostained with Xpress (red) and TMEM106B (green) antibodies. Normal rabbit IgG was used as a negative control for the TMEM106B antibody. Nuclei were stained with Hoechst 33258 (blue). White bar, 20 μm. B, the lysates from HeLa cells transiently overexpressing HX-tagged TMEM106B-FL (+) were subjected to immunoblotting analysis (IB) using Xpress and TMEM106B antibodies. The empty vector was used as a negative control (−). C, HeLa cells, seeded on 6-well plates at 4 × 104 cells/well, were transfected with 5 nm control siRNA or TMEM106B-1 or -2 siRNA. At 70 h after the transfection, cell lysates were prepared with or without boiling and subjected to immunoblotting analysis using the TMEM106B antibody (lanes 1–6). For references, the lysates from HeLa cells overexpressing TMEM106B-FL, TMEM106B(1–127), and TMEM106B(1–106) were loaded (lanes 7–10). To avoid film overexposure of overexpressed TMEM106B proteins, the quantities of the lysates loaded were reduced (lanes 7–10). *, endogenous TMEM106B-FL. D, endogenous TMEM106B-FL in HeLa cell lysates was immunoprecipitated (IP) using the TMEM106B antibody that recognized an N-terminal region of TMEM106B, followed by immunoblotting analysis using the same antibody (lane 3). Normal rabbit IgG was used as a negative control for the immunoprecipitation (lane 2). To detect TMEM106B antibody-derived background signals, a lysis buffer not containing the cell lysate was used for a mock immunoprecipitation using the TMEM106B antibody (lane 4). The amount of the HeLa cell lysate loaded for input (lane 1) was 12.5 μg, and the amount used for each immunoprecipitation was 500 μg. H.C., heavy chain; L.C., light chain.
We then examined whether the TMEM106B antibody is capable of detecting endogenous TMEM106B by immunoblotting analysis. To address whether the detected protein is TMEM106B, we also knocked down the endogenous TMEM106B using two types of specific siRNA against human TMEM106B before we conducted immunoblotting analysis. Because the TMEM106B protein has been reported to be very sensitive to heat and become undetectable by immunoblotting analysis after heat denaturation (20), cell lysates were either boiled (Fig. 1C, lanes 1–3) or left unboiled (Fig. 1C, lanes 4–6) in the SDS sample buffer, before the SDS-PAGE. A protein with a molecular mass of ∼40 kDa (indicated as FL and with an asterisk in Fig. 1C), migrating in a fashion nearly identical to exogenous TMEM106B-FL (Fig. 1C, Long exposure, lane 8), was detected in non-boiled cell lysates (Fig. 1C, Long exposure, lane 4). The intensity of the band was markedly reduced by two kinds of siRNA (Fig. 1C, Long exposure, lanes 5 and 6). As expected, the level of the protein with a molecular mass of ∼40 kDa was quite low in the boiled lysates (Fig. 1C, Long exposure, lanes 1–3). To further confirm that the protein is the endogenous TMEM106B-FL, we immunoprecipitated it from the cell lysates using the TMEM106B antibody, followed by immunoblotting analysis of the immunoprecipitates with the same antibody (Fig. 1D). We found that the antibody efficiently immunoprecipitated endogenous TMEM106B (Fig. 1D, lane 3). Thus, these experiments show that the TMEM106B antibody is capable of detecting endogenous TMEM106B-FL. The other proteins that were recognized by immunoblotting analysis by the TMEM106B antibody, such as the proteins with a approximate molecular mass of 70 or 17 kDa, were not greatly affected by boiling (Fig. 1C, Short exposure, cf. lanes 1 and 4) or not immunoprecipitated with the TMEM106B antibody (Fig. 1D, lane 3). These results suggest that they are nonspecific artifacts of the TMEM106B antibody.
TMEM106B Is Proteolytically Cleaved into Unstable N-terminal Fragments
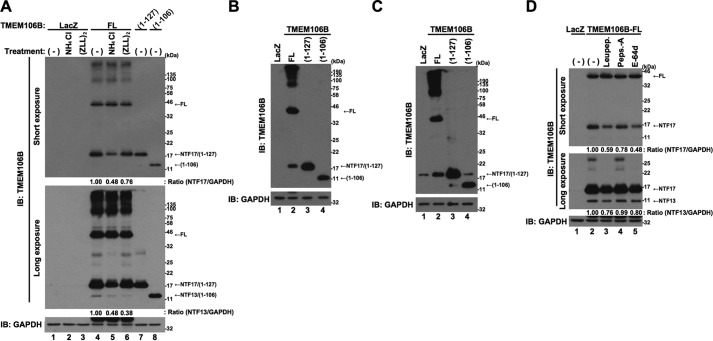
It has been reported that some uncharacterized lysosomal protease(s) and SPPL2a cleave TMEM106B at around the 127th and 106th amino acids from the N terminus, respectively, to generate two NTFs of TMEM106B (25). The precise cleavage sites have not been identified. In agreement with this, a considerable amount of protein with a molecular mass of ∼20 kDa was recognized by the Xpress antibody in the cell lysates from the cells overexpressing HX-TMEM106B-FL (Fig. 1B). To confirm that this protein is a cleaved derivative of TMEM106B, we overexpressed non-tagged TMEM106B-FL in HeLa cells and treated the cells with NH4Cl and (ZLL)2-ketone, inhibitors of the cleavage of TMEM106B (25) (Fig. 2A). Again, the overexpression of non-tagged TMEM106B-FL resulted in the generation of two similar fragments with molecular masses of ∼17 and 13 kDa. Because they were recognized by the TMEM106B antibody, raised against the 14-amino acid peptide of the N-terminal region of TMEM106B (see “Experimental Procedures”), they were believed to be NTFs and were designated NTF17 and NTF13, respectively (Fig. 2A, lane 4). Based on the findings of a previous study (25), we constructed vectors to express the two TMEM106B-NTFs corresponding to TMEM106B(1–127) and TMEM106B(1–106) as references. Immunoblotting analysis showed that the apparent SDS-PAGE migration distances of NTF17 and NTF13 were identical to those of TMEM106B(1–127) and TMEM106B(1–106), respectively (Fig. 2A, cf. lane 4 and lane 7 or 8). Furthermore, specific protease inhibitors, NH4Cl and (ZLL)2-ketone, partially inhibited the generation of NTF17 and NTF13 (Fig. 2A, cf. lanes 4–6). These data together led us to conclude that NTF17 and NTF13 correspond to TMEM106B(1–127) and TMEM106B(1–106), respectively. NTF17 was also generated in neuronal cells, NSC34 cells and Neuro2a cells that overexpress TMEM106B-FL (Fig. 2, B and C, lanes 2), whereas NTF13 was not detected in these cells, probably because of relatively low expression of TMEM106B-FL. We failed to identify the precise cleavage sites because a small deletion of TMEM106B made it quite unstable (data not shown). Treatment of the cells overexpressing TMEM106B-FL with lysosomal protease inhibitors leupeptin, pepstatin A, and E-64d partially inhibited the generation of NTF17 (Fig. 2D, cf. lanes 2 and 3–5). Leupeptin and E-64d, but not pepstatin A, also partially inhibited the generation of NTF13 (Fig. 2D, cf. lanes 2 and 3–5). These results suggest that lysosomal protease(s) are involved in the cleavage of TMEM106B.
FIGURE 2.
TMEM106B is proteolytically cleaved into N-terminal fragments. A, HeLa cells, seeded on 6-well plates at 5 × 104 cells/well, were infected with adenovirus vectors encoding TMEM106B-FL at an MOI of 800 or TMEM106B(1–127) or TMEM106B(1–106) at an MOI of 100. At 24 h after the start of infection, the cells were treated with or without 5 mm NH4Cl or 0.1 μm (ZLL)2-ketone. At 48 h after the start of infection, the cells were harvested for immunoblotting analysis (IB) using the TMEM106B antibody. Calculated intensities of TMEM106B-NTF17 and -NTF13, normalized with that of GAPDH, are shown. B and C, NSC34 cells (B) or Neuro2a cells (C), seeded on 6-well plates at 1 × 105 cells/well, were infected with adenovirus vectors encoding TMEM106B-FL at an MOI of 800 or TMEM106B(1–127) or TMEM106B(1–106) at an MOI of 400. At 24 h after the start of infection, media were replaced with DMEM/N2 supplement. At 24 h after the replacement of media, the cells were harvested for immunoblotting analysis using the TMEM106B antibody. D, HeLa cells, seeded on 6-well plates at 5 × 104 cells/well, were infected with adenovirus vectors encoding TMEM106B-FL at an MOI of 800. At 18 h after the start of infection, cells were co-incubated with or without 50 μg/ml leupeptin (Leupep.), 10 μg/ml pepstatin A (Peps.-A), or 5 μg/ml E-64d. At 24 h after the start of the co-incubation, the cells were harvested for immunoblotting analysis using the TMEM106B antibody. Calculated intensities of TMEM106B-NTF17 and -NTF13, normalized with that of GAPDH, are shown.
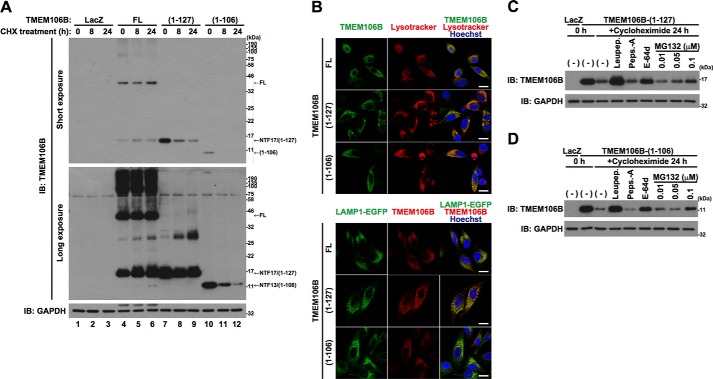
In contrast to the NTFs, generated from the exogenously expressed TMEM106B, endogenous NTFs were not clearly detected (Fig. 1, C and D, lane 3). Because the NTF17-like protein in the input lanes (Fig. 1D, Long exposure, lane 1) was not immunoprecipitated with the TMEM106B antibody (Fig. 1D, lane 3) that does immunoprecipitate exogenous TMEM106B(1–127) (data not shown), this was believed to be a background artifact unrelated to the NTFs of TMEM106B. These results suggest that the endogenous TMEM106B-NTFs are too unstable to be detected. To confirm this, HeLa cells overexpressing TMEM106B-FL, TMEM106B(1–127), or TMEM106B(1–106) were treated with cycloheximide for 8 or 24 h (Fig. 3A). As expected, TMEM106B(1–127) (lanes 7–9) and TMEM106B(1–106) (lanes 10–12) were more rapidly degraded than TMEM106B-FL (lanes 4–6). Immunocytochemistry analysis showed that TMEM106B(1–127) and TMEM106B(1–106) as well as TMEM106B-FL partially colocalized with the lysosomal markers, Lysotracker and LAMP1-EGFP (Fig. 3B), suggesting that lysosomal proteases are involved in the degradation of TMEM106B-NTFs. Lysosomal protease inhibitors leupeptin and E-64d, but not pepstatin A, and ubiquitin-proteasome inhibitor, MG132, inhibited their degradation (Fig. 3, C and D). These results suggest that TMEM106B-NTFs are degraded through the lysosome and ubiquitin-proteasome degradation pathways.
FIGURE 3.
TMEM106B-NTFs are rapidly degraded by lysosomal and proteasomal degradation pathways. A, HeLa cells, seeded on 6-well plates at 5 × 104 cells/well, were infected with adenovirus vectors encoding TMEM106B-FL at an MOI of 800 or TMEM106B-(1–127) or TMEM106B-(1–106) at an MOI of 200. At 18 h after the start of infection, cells were treated with 5 μg/ml cycloheximide (CHX) for 8–24 h. Then the cells were harvested for immunoblotting analysis (IB) using the TMEM106B antibody. B, top, HeLa cells overexpressing non-tagged TMEM106B-FL, TMEM106B(1–127), or TMEM106B(1–106) were incubated with 100 nm Lysotracker (red) at 37 °C for 1 h just before fixation. Then the fixed cells were immunostained with the TMEM106B antibody (green). Bottom, HeLa cells overexpressing LAMP1-EGFP (green) and non-tagged TMEM106B-FL, TMEM106B(1–127), or TMEM106B(1–106) were fixed and immunostained with the TMEM106B antibody (red). Nuclei were stained with Hoechst 33258 (blue). White bar, 20 μm. C and D, HeLa cells, seeded on 6-well plates at 5 × 104 cells/well, were infected with adenovirus vectors encoding TMEM106B(1–127) (C) or TMEM106B(1–106) (D) at an MOI of 200. After the infection, the cells were co-incubated with or without 50 μg/ml leupeptin (Leupep.), 10 μg/ml pepstatin A (Peps.-A), 5 μg/ml E-64d, or 0.01–0.1 μm MG132. At 18 h after the start of infection, the cells were co-incubated with or without 5 μg/ml cycloheximide for 24 h in the presence or absence of 50 μg/ml leupeptin, 10 μg/ml pepstatin A, 5 μg/ml E-64d, or 0.01–0.1 μm MG132. At 24 h after the start of the CHX treatment, the cells were harvested for immunoblotting analysis using the TMEM106B antibody.
Overexpression of TMEM106B Induces Cell Death
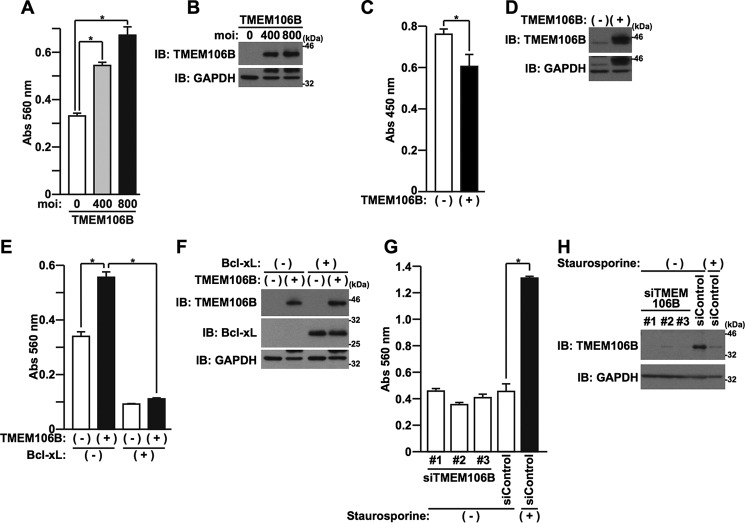
It has been reported that levels of TMEM106B are up-regulated in the brains of FTLD-TDP patients, especially in those with GRN mutations (11, 13, 20, 23). This finding suggests that the overexpression of TMEM106B is linked to pathogenesis in these patients. To investigate this, we first examined the direct effect of overexpression of TMEM106B-FL on the viability of HeLa cells and primary cortical neurons (PCNs). Cytotoxicity was evaluated by a lactate dehydrogenase (LDH) release cell death assay or WST-8 cell viability assay. We found that the overexpression of TMEM106B-FL induced cell death in HeLa cells in an expression level-dependent manner (Fig. 4, A and B) and decreased cell viability in PCNs (Fig. 4, C and D). The cell death, induced by the overexpression of TMEM106B, was inhibited by overexpression of Bcl-xL (Fig. 4, E and F). These data indicated that increased expression of TMEM106B causes cell death that is mediated by the mitochondrial apoptosis pathway. In contrast, the reduction in expression of TMEM106B, caused by specific siRNAs against TMEM106B, did not induce cell death (Fig. 4, G and H).
FIGURE 4.
Overexpression of TMEM106B induces cell death. A and B, HeLa cells, seeded on 6-well plates at 5 × 104 cells/well, were infected with adenovirus vectors encoding TMEM106B-FL at MOIs of 0–800. At 18 h after the start of infection, media were replaced with DMEM/N2 supplement. At 24 h after the replacement of media, LDH release was measured (A), and the cells were harvested for immunoblotting analysis (IB) using the TMEM106B antibody (B). *, p < 0.05. C, PCNs, seeded on 96-well plates at 5 × 104 cells/well, were infected with adenovirus vectors encoding LacZ (−) or TMEM106B-FL (+) at an MOI of 200. At 120 h after the start of the infection, the WST-8 cell viability assay was performed. *, p < 0.05. D, PCNs, seeded on 6-well plates at 1 × 106 cells/well, were infected with adenovirus vectors encoding LacZ (−) or TMEM106B-FL (+) at an MOI of 200. At 120 h after the start of the infection, the cell lysates were subjected to immunoblotting analysis using the TMEM106B antibody. E and F, HeLa cells, seeded on 6-well plates at 5 × 104 cells/well, were co-infected with LacZ (−) or TMEM106B (+) adenovirus vector at an MOI of 600 in association with LacZ (−) or Bcl-xL (+) adenovirus vector at an MOI of 50. At 18 h after the start of the infection, media were replaced with DMEM/N2 supplement. At 24 h after the replacement of media, LDH release was measured (E), and immunoblotting analysis was performed with the antibodies indicated (F). *, p < 0.05. G and H, HeLa cells, seeded on 6-well plates at 5 × 104 cells/well, were transfected with 5 nm control siRNA or TMEM106B-1, -2, or -3 siRNA using Lipofectamine2000 reagent. At 48 h after the start of transfection, media were replaced with DMEM/N2 supplement containing (+) or not containing (−) 0.05 μm staurosporine. At 24 h after the replacement of media, LDH release was measured (G), and immunoblotting analysis was performed with the TMEM106B antibody (H). Staurosporine was used as a positive control for the induction of cell death. The expression of endogenous TMEM106B was reduced by the treatment with staurosporine, probably due to the increase in the cleavage or degradation by some undetermined proteases that are activated by staurosporine. *, p < 0.05. Error bars, S.D.
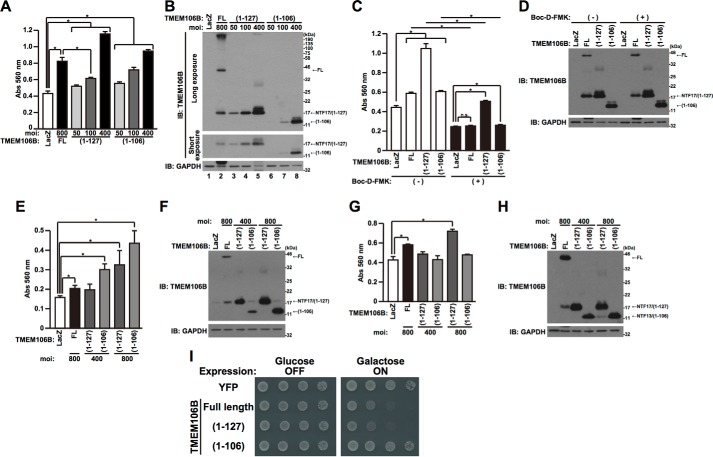
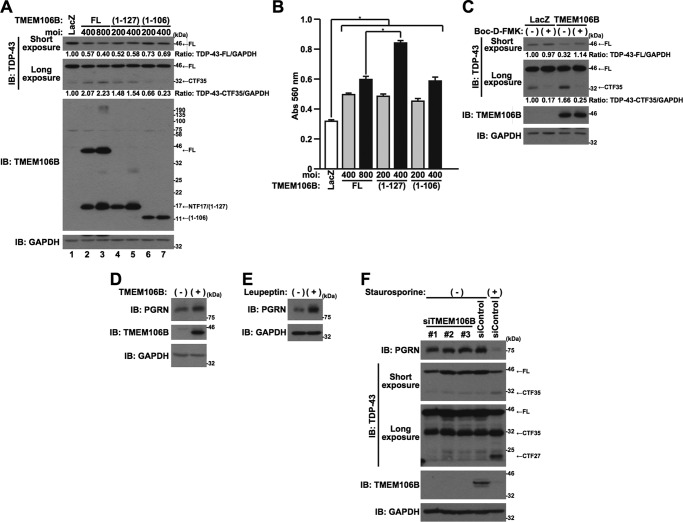
The overexpression of TMEM106B resulted in the appearance of NTF17 and NTF13, in addition to TMEM106B-FL (Fig. 2). Therefore, it was not clear which forms of TMEM106B protein, TMEM106B-FL and/or cleaved fragments, were responsible for inducing cell death. To investigate this, we generated adenovirus vectors that express TMEM106B(1–127) and TMEM106B(1–106). These adenovirus vectors express each encoded protein more efficiently than the TMEM106B-FL-encoding adenovirus vector does (Fig. 5B). The overexpression of TMEM106B(1–127) and TMEM106B(1–106) also induced cell death in an expression level-dependent manner (Fig. 5A). Adjusting the multiplicity of infection (MOI) of each adenovirus vector, we overexpressed comparable levels of TMEM106B-FL, TMEM106B(1–127), and TMEM106B(1–106) and compared their cell death-inducing activity (Fig. 5A). The cell death-inducing activity of TMEM106B (1–127) at an MOI of 100 was weaker than that of TMEM106B-FL at an MOI of 800 (Fig. 5A). However, the level of TMEM106B(1–127) was higher than that of NTF17 derived from TMEM106B-FL (Fig. 5B, Short exposure, cf. lanes 2 and 4). This result indicates that TMEM106B-FL and/or high molecular mass TMEM106B, in addition to NTF17 and NTF13, possess cytotoxic activity. TMEM106B-FL and TMEM106B(1–106)-induced cell death is attenuated by the treatment with a broad range caspase inhibitor, Boc-D-FMK (Fig. 5, C and D). TMEM106B(1–127)-induced cell death is partially inhibited by Boc-D-FMK (Fig. 5, C and D). These results indicate that caspases are involved in the TMEM106B cytotoxicity.
FIGURE 5.
TMEM106B-FL and TMEM106B-NTFs cause cytotoxicity in vitro and in vivo. A and B, HeLa cells, seeded on 6-well plates at 5 × 104 cells/well, were infected with adenovirus vectors encoding TMEM106B-FL at an MOI of 800 or TMEM106B(1–127) or TMEM106B(1–106) at MOIs of 0–400. At 18 h after the start of infection, media were replaced with DMEM/N2 supplement. At 24 h after the replacement of media, LDH release was measured (A), and the cells were harvested for immunoblotting analysis (IB) using the TMEM106B antibody (B). *, p < 0.05. C and D, HeLa cells, seeded on 6-well plates at 5 × 104 cells/well, were infected with adenovirus vectors encoding TMEM106B-FL at an MOI of 800 or TMEM106B(1–127) or TMEM106B(1–106) at an MOI of 300. After the infection, cells were co-incubated with (+) or without (−) 50 μm Boc-D-FMK. At 18 h after the start of infection, media were replaced with DMEM/N2 supplement containing (+) or not containing (−) 50 μm Boc-D-FMK. At 24 h after the replacement of media, LDH release was measured (C), and the cells were harvested for immunoblotting analysis using the TMEM106B antibody (D). *, p < 0.05. E–H, NSC34 cells (E and F) or Neuro2a cells (G and H), seeded on 6-well plates at 1 × 105 cells/well, were infected with adenovirus vectors encoding TMEM106B-FL at an MOI of 800 or TMEM106B(1–127) or TMEM106B(1–106) at an MOI of 400 or 800. At 24 h after the start of infection, media were replaced with DMEM/N2 supplement. At 24 h after the replacement of media, LDH release was measured (E and G), and the cells were harvested for immunoblotting analysis using the TMEM106B antibody (F and H). *, p < 0.05. I, BY4741 yeast strains, transformed with the vectors indicated, were plated onto the culture dishes containing glucose (expression off) or galactose (expression on) and incubated for 48 h for the glucose-containing plate or 72 h for the galactose-containing plate, respectively, at 30 °C. The YFP backbone vector was used as a negative control. Error bars, S.D.
We also examined cytotoxicity of TMEM106B in neuronal cells, NSC34 cells (Fig. 5, E and F), and Neuro2a cells (Fig. 5, G and H). Overexpression of TMEM106B-FL and TMEM106B(1–127) induced cell death in both NSC34 cells and Neuro2a cells (Fig. 5, E–H). On the other hand, overexpression of TMEM106B(1–106) induced cell death in only NSC34 cells and not in Neuro2a cells (Fig. 5, E–H).
To further demonstrate that the overexpression of TMEM106B is toxic in vivo, we performed a yeast growth retardation assay using a galactose-inducible TMEM106B-encoding vector. Overexpression of TMEM106B-FL and TMEM106B(1–127) was extremely toxic, whereas yeast growth was unaffected by the expression of TMEM106B (1–106) (Fig. 5I).
TMEM106B-NTF-induced Cell Death Is Leupeptin-sensitive
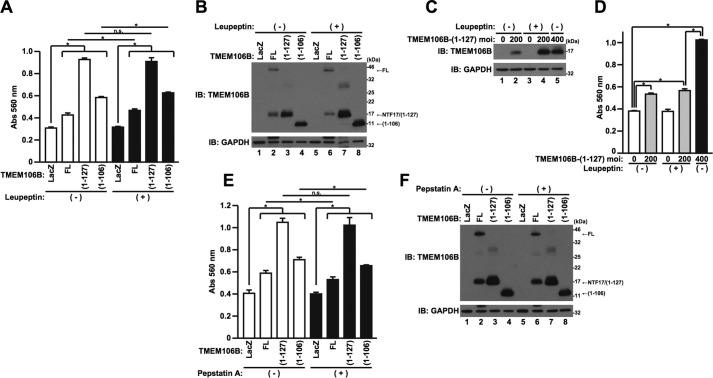
Because TMEM106B-FL and -NTFs localize in the lysosome (Fig. 3B), we next hypothesized that TMEM106B-induced cell death is mediated by lysosomal protease(s). To examine this, HeLa cells overexpressing TMEM106B-FL, TMEM106B(1–127), or TMEM106B(1–106) were treated with lysosomal protease inhibitor, leupeptin or pepstatin A. The treatment with leupeptin did not appear to affect TMEM106B toxicity in the LDH release assay (Fig. 6A). However, the expression levels of TMEM106B(1–127) and TMEM106B(1–106) increased when cells were treated with leupeptin (Fig. 6B, cf. lanes 3 and 7 and cf. lanes 4 and 8), possibly due to the suppression of the degradation, as already shown above (Fig. 3, C and D). These results led us to speculate that TMEM106B(1–127)- and TMEM106B(1–106)-induced toxicity was actually inhibited by leupeptin if the levels of the TMEM106B-NTFs were adjusted to be equal. Therefore, we next increased the TMEM106B(1–127) expression level in the cells not co-incubated with leupeptin, by increasing the MOI of the adenoviral vector for comparison. Indeed, the level of TMEM106B(1–127) expression in the cells infected with the TMEM106B(1–127) adenoviral vector at an MOI of 400, in the absence of leupeptin co-incubation, became largely identical to that of TMEM106B(1–127) expression in the cells infected with the adenoviral vector at an MOI of 200, in the presence of leupeptin co-incubation (Fig. 6C, cf. lanes 4 and 5). Notably, TMEM106B(1–127) at an MOI of 200 in the presence of leupeptin induced cell death more weakly than did TMEM106B(1–127) at an MOI of 400 in the absence of leupeptin (Fig. 6D). These results indicated that TMEM106B (1–127)-induced toxicity and possibly TMEM106B(1–106)-induced toxicity were inhibited by leupeptin and leupeptin-sensitive protease(s) are therefore involved in the cytotoxicity of TMEM106B-NTFs. On the other hand, TMEM106B-FL-induced cell death was not apparently inhibited by leupeptin (Fig. 6, A and B).
FIGURE 6.
TMEM106B-NTF-induced cell death is leupeptin-sensitive. A and B, HeLa cells, seeded on 6-well plates at 5 × 104 cells/well, were infected with adenovirus vectors encoding TMEM106B-FL at an MOI of 800 or TMEM106B(1–127) or TMEM106B(1–106) at an MOI of 300. After the infection, cells were co-incubated with (+) or without (−) 50 μg/ml leupeptin. At 18 h after the start of infection, media were replaced with DMEM/N2 supplement containing (+) or not containing (−) 50 μg/ml leupeptin. At 24 h after the replacement of media, LDH release was measured (A), and the cells were harvested for immunoblotting analysis (IB) using the TMEM106B antibody (B). *, p < 0.05. n.s., not significant. C and D, HeLa cells, seeded on 6-well plates at 5 × 104 cells/well, were infected with adenovirus vectors encoding TMEM106B-(1–127) at an MOI of 200 or 400. After the infection, cells were co-incubated with (+) or without (−) 50 μg/ml leupeptin. At 18 h after the start of infection, media were replaced with DMEM/N2 supplement containing (+) or not containing (−) 50 μg/ml leupeptin. At 24 h after the replacement of media, the cells were harvested for immunoblotting analysis using the TMEM106B antibody (C), and LDH release was measured (D). *, p < 0.05. E and F, HeLa cells, seeded on 6-well plates at 5 × 104 cells/well, were infected with adenovirus vectors encoding TMEM106B-FL at an MOI of 800 or TMEM106B(1–127) or TMEM106B(1–106) at an MOI of 300. After the infection, cells were co-incubated with (+) or without (−) 5 μg/ml pepstatin A. At 18 h after the start of infection, media were replaced with DMEM/N2 supplement containing (+) or not containing (−) 5 μg/ml pepstatin A. At 24 h after the replacement of media, LDH release was measured (E), and the cells were harvested for immunoblotting analysis using the TMEM106B antibody (F). *, p < 0.05. n.s., not significant. Error bars, S.D.
Also, treatment with pepstatin A mildly inhibited TMEM106B-FL- and TMEM106B(1–106)-induced cell death but not TMEM106B(1–127)-induced cell death (Fig. 6E). However, it also slightly reduced their expression levels (Fig. 6F, cf. lanes 2 and 6 and cf. lanes 4 and 8). These results suggest that pepstatin A does not affect TMEM106B cytotoxicity.
Low Grade Overexpression of TMEM106B Enhances Oxidative Stress-induced Cytotoxicity
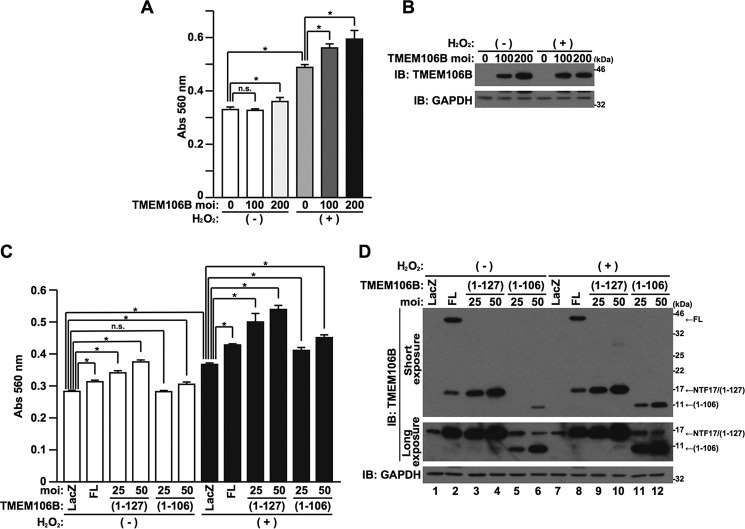
Oxidative stress is implicated in the pathogenesis of neurodegenerative diseases (26). Given that the TMEM106B genetic variation or the overexpression of TMEM106B increases susceptibility to FTLD-TDP as a risk factor, it is hypothesized that the low level overexpression of TMEM106B potentiates oxidative stress-induced cell death. To investigate this, we lowered the expression levels of TMEM106B by decreasing MOIs of adenovirus vector and found that low grade overexpression of TMEM106B alone at MOIs of 100–200 was unable to cause cell death or caused cell death only marginally (Fig. 7, A and B). Importantly, however, it significantly enhanced cell death, induced by hydrogen peroxide (H2O2), a well known oxidative stress inducer (Fig. 7, A and B). TMEM106B(1–127) and TMEM106B(1–106) also potentiated H2O2-induced cell death (Fig. 7, C and D).
FIGURE 7.
Low grade overexpression of TMEM106B-FL and TMEM106B-NTFs enhances oxidative stress-induced cell death. A and B, HeLa cells, seeded on 6-well plates at 5 × 104 cells/well, were infected with adenovirus vectors encoding TMEM106B-FL at MOIs of 0–200. At 18 h after the start of infection, media were replaced with DMEM/N2 supplement with (+) or without (−) 50 μm H2O2. At 24 h after the replacement of media, LDH release was measured (A), and the cells were harvested for immunoblotting analysis (IB) using the TMEM106B antibody (B). *, p < 0.05. n.s., not significant. C and D, HeLa cells, seeded on 6-well plates at 5 × 104 cells/well, were infected with adenovirus vectors encoding TMEM106B-FL at an MOI of 100 or TMEM106B(1–127) or TMEM106B(1–106) at an MOI of 25 or 50. At 18 h after the start of infection, media were replaced with DMEM/N2 supplement with (+) or without (−) 50 μm H2O2. At 24 h after the replacement of media, LDH release was measured (C), and the cells were harvested for immunoblotting analysis using the TMEM106B antibody (D). *, p < 0.05. n.s., not significant. Error bars, S.D.
Lysosomal Localization Is Important for TMEM106B Toxicity
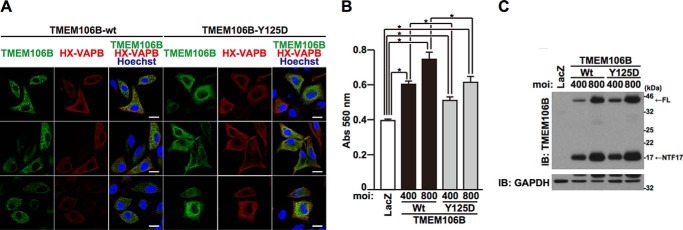
TMEM106B-FL predominantly localizes to lysosomes (Fig. 3B). To examine whether lysosomal localization of TMEM106B is necessary for TMEM106B to induce cell death, we expressed TMEM106B-Y125D, which mislocalizes to the endoplasmic reticulum (ER) (25). ER localization was confirmed by the colocalization with vesicle-associated membrane protein-associated protein B (VAPB), an ER marker (Fig. 8A) (27). TMEM106B-Y125D colocalized with HX-tagged VAPB more efficiently than TMEM106B-WT (weighted colocalization coefficient: Y125D = 0.746 ± 0.090, WT = 0.571 ± 0.052, p < 0.001). An intracytoplasmic granular localization, indicative of lysosomal localization of TMEM106B, was still observed even in cells expressing TMEM106B-Y125D. The putative lysosomal localization of TMEM106B-Y125D was assumed to be largely caused by lysosome-localizing TMEM106B-NTFs (Fig. 3B) that were derived from TMEM106B-Y125D-FL (Fig. 8C). We then compared the extent of cell death induced by TMEM106B-Y125D with that caused by TMEM106B-WT (Fig. 8, B and C). TMEM106B-Y125D had a weaker cell death-inducing activity than TMEM106B-WT (Fig. 8, B and C), although TMEM106B-Y125D is still capable of inducing mild cell death (Fig. 8B). Given that a considerable amount of TMEM106B-NTFs that are capable of inducing cell death (Fig. 5A) was present in TMEM106B-Y125D-expressing cells (Fig. 8C), it could be assumed that TMEM106B-NTFs, derived from TMEM106B-Y125D, may be responsible for the induction of cell death. These data together suggest that the lysosomal localization is important for the induction of cell death caused by TMEM106B.
FIGURE 8.
Lysosomal localization of TMEM106B is important for TMEM106B cytotoxicity. A, HeLa cells transiently overexpressing TMEM106B-FL or -Y125D together with HX-tagged VAPB were fixed and immunostained with TMEM106B (green) and Xpress (red) antibodies. HX-VAPB was used as an ER marker. Nuclei were stained with Hoechst 33258 (blue). White bar, 20 μm. Three representative views per sample are shown. B and C, HeLa cells, seeded on 6-well plates at 5 × 104 cells/well, were infected with adenovirus vectors encoding TMEM106B-WT or -Y125D at MOIs of 0–800. At 18 h after the start of infection, media were replaced with DMEM/N2 supplement. At 24 h after the replacement of media, LDH release was measured (B), and the cells were harvested for immunoblotting analysis (IB) using the TMEM106B antibody (C). *, p < 0.05. Error bars, S.D.
Overexpression of TMEM106B Induces Cleavage of TDP-43 but Does Not Decrease the Progranulin Levels
We next examined whether the up-regulation of TMEM106B is linked to the TDP-43 pathology in FTLD-TDP. Notably, the overexpression of TMEM106B-FL and TMEM106B(1–127) reduced the level of full-length TDP-43 (TDP-43-FL) and increased the level of the C-terminal fragment (CTF) of TDP-43 (TDP-43-CTF35) (Fig. 9A, lanes 1–5). If we consider that the extents of TMEM106B(1–127)-induced cell death were larger than those of TMEM106B-FL-induced cell death (Fig. 9B), we could conclude that the TMEM106B(1–127)-induced potentiation of the TDP-43 cleavage is less prominent than TMEM106B-FL. On the other hand, although the overexpression of TMEM106B(1–106) mildly reduced the level of TDP-43-FL, it did not increase the level of TDP-43-CTF35 (Fig. 9A, lanes 6 and 7). These results suggest that the overexpression of TMEM106B-FL and TMEM106B(1–127) accelerated the cleavage of TDP-43, an essential initiator of TDP-43 pathology (28). Given that the cleavage of TDP-43 is mediated by caspase activation (29) and that caspases are involved in the TMEM106B-induced cell death (Fig. 5, C and D), it could be hypothesized that the TMEM106B-induced potentiation of the cleavage of TDP-43 is mediated by caspases. In accordance with this notion, a caspase inhibitor attenuated the cleavage of TDP-43, induced by TMEM106B-FL (Fig. 9C, cf. lanes 3 and 4).
FIGURE 9.
Overexpression of TMEM106B induces the cleavage of TDP-43 but does not decrease the progranulin level. A and B, HeLa cells, seeded on 6-well plates at 5 × 104 cells/well, were infected with adenovirus vectors encoding TMEM106B-FL at an MOI of 400 or 800 or TMEM106B (1–127) or TMEM106B (1–106) at an MOI of 200 or 400. At 18 h after the start of infection, media were replaced with DMEM/N2 supplement. At 24 h after the replacement of media, the cells were harvested for immunoblotting analysis (IB) using the antibodies indicated (A), and LDH release was measured (B). *, p < 0.05. CTF35, C-terminal fragment of 35 kDa. Calculated intensities of TDP-43-FL and -CTF35, normalized with that of GAPDH, are shown. C, HeLa cells, seeded on 6-well plates at 5 × 104 cells/well, were infected with adenovirus vectors encoding TMEM106B-FL at an MOI of 800. After the infection, cells were co-incubated with (+) or without (−) 50 μm Boc-D-FMK. At 18 h after the start of infection, media were replaced with DMEM/N2 supplement containing (+) or not containing (−) 50 μm Boc-D-FMK. At 24 h after the replacement of media, the cells were harvested for immunoblotting analysis using the antibodies indicated. Calculated intensities of TDP-43-FL and -CTF35, normalized with that of GAPDH, are shown. D, HeLa cells, seeded on 6-well plates at 5 × 104 cells/well, were infected with LacZ (−) or TMEM106B-FL (+) adenovirus vector at an MOI of 800. At 18 h after the start of infection, media were replaced with DMEM/N2 supplement. At 24 h after the replacement of media, the cells were harvested for immunoblotting analysis using the antibodies indicated. E, HeLa cells, seeded on 6-well plates at 5 × 104 cells/well, were treated with (+) or without (−) 50 μg/ml leupeptin. At 18 h after the treatment, cells were replaced with DMEM/N2 supplement and treated with (+) or without (−) 50 μg/ml leupeptin. At 24 h after the second treatment, the cells were harvested for immunoblotting analysis using a progranulin (PGRN) antibody. F, HeLa cells, seeded on 6-well plates at 5 × 104 cells/well, were transfected with 5 nm control siRNA or TMEM106B-1, -2, or -3 siRNA using Lipofectamine2000 reagent. At 48 h after the start of transfection, media were replaced with DMEM/N2 supplement containing (+) or not containing (−) 0.05 μm staurosporine. At 24 h after the replacement of media, the cells were harvested for immunoblotting analysis using the antibodies indicated. Staurosporine was used as a positive control for the induction of cleavage of TDP-43. The expression of endogenous progranulin and TMEM106B was reduced by the treatment with staurosporine, probably due to the increase in the cleavage or degradation of progranulin and TMEM106B by undetermined proteases that are activated by staurosporine. CTF27, C-terminal fragment of 27 kDa. Error bars, S.D.
We also found that the overexpression of TMEM106B-FL slightly increased the intracellular level of progranulin (Fig. 9D). Treatment of the cells with a lysosomal protease inhibitor, leupeptin, markedly increased the level of progranulin (Fig. 9E). These results suggest that the overexpression of TMEM106B may impair lysosomal function and increase the level of progranulin that is degraded by lysosomal enzymes (30). In contrast, reduced expression of TMEM106B by siRNAs did not affect either the cleavage of TDP-43 or the level of progranulin expression (Fig. 9F).
Discussion
Several clinical studies have shown that both mRNA and protein levels of TMEM106B increase in the brains of FTLD-TDP patients, particularly in patients with GRN mutations (11, 13, 20, 23). In agreement with this, the level of TMEM106B, encoded by the risk variant of the TMEM106B gene, tends to be up-regulated, compared with that encoded by the non-risk TMEM106B gene (22). On the other hand, some studies have provided data contrary to this notion (12, 14). Because all of these studies have been conducted using samples derived from a relatively small number of FTLD-TDP patients, this issue needs to be further investigated before a final conclusion can be drawn. In the current study, supported by some clinical data (11, 13, 20, 23) and in vitro findings (22), we hypothesized that the level of TMEM106B is elevated in FTLD-TDP and examined the effect of overexpression of TMEM106B on cell survival. We found that the up-regulation of TMEM106B causes cell death in vitro and in vivo (Figs. 4 and 5), and the low grade up-regulation of TMEM106B enhances oxidative stress-induced cytotoxicity (Fig. 7). In contrast, the loss of TMEM106B does not affect cell viability (Fig. 4, G and H) (31).
The molecular mechanism underlying the TMEM106B-induced cell death has not been fully elucidated. In the current study, we have shown that the caspase-dependent mitochondrial cell death pathways are involved in the TMEM106B-induced cell death (Figs. 4 and 5). We have also demonstrated that the lysosomal localization is important for TMEM106B-mediated toxicity (Fig. 8). In agreement, very recently, Busch et al. (32) found that increased expression of TMEM106B causes cytotoxicity that requires lysosome localization. Furthermore, some earlier studies showed that lysosomal function and morphology are impaired by TMEM106B overexpression (19, 20, 24). Collectively, these data suggest that the TMEM106B-induced cell death is at least partially mediated by lysosomal cell death (33). Given that the lysosomal cell death pathway is mediated by the caspase-dependent mitochondrial cell death pathway (33), it is highly likely that this notion is correct. In support, we also found that TMEM106B-NTFs induced caspase-dependent (Fig. 5, C and D) and leupeptin-sensitive (Fig. 6, A–D) cell death. This result indicates that the TMEM106B-NTF-induced cell death is mediated by the lysosomal cell death pathway.
The low grade overexpression of TMEM106B that is incapable of causing cell death predisposes cells to vulnerability to oxidative stress-induced cell death (Fig. 7). According to a previous finding that oxidative stress causes the lysosomal cell death (34) and the finding in the present study that the lysosomal cell death pathway may be triggered by the overexpression of TMEM106B (Fig. 8), this result could be expected. Most importantly, this finding may reflect the in vivo physiological effect of low grade overexpression of TMEM106B as a risk factor of FTLD-TDP.
In the current study, we have shown that the overexpression, but not the knockdown, of TMEM106B-FL and TMEM106B(1–127) increases the caspase-dependent cleavage of TDP-43 (Fig. 9, A–C). Aggregated TDP-43 in the TDP-43 pathology mainly consists of unfolded TDP-43 CTFs with multiple phosphorylations (9, 10). Based on the finding that the overexpression of TMEM106B increased the fragmentation of TDP-43 (Fig. 9A), we could speculate that the TDP-43 inclusion body is more easily formed when TMEM106B is overexpressed. This mechanism may explain the TDP-43 pathology that occurs in FTLD-TDP patients. In support, previous studies have shown that the levels of TMEM106B are elevated in both GRN and cathepsin-D (CTSD) gene knock-out mice. The CTSD knock-out mice recapitulated neuronal ceroid lipofuscinosis, a lysosomal storage disorder (23). Interestingly, both knock-out mice are also associated with the TDP-43 pathology (23, 35, 36). Therefore, it could be postulated that the increased expression of TMEM106B contributes to the formation of the TDP-43 inclusion bodies, even in mice.
It has generally been recognized that mutations in the GRN gene cause familial FTLD-TDP, possibly by down-regulating the progranulin levels (37, 38). Recent clinical studies have further demonstrated that TMEM106B levels inversely correlate with progranulin levels (13) and the TMEM106B risk genotypes are associated with decreased progranulin levels (12, 13). Thus, it could be hypothesized that the up-regulation of TMEM106B increases the risk of FTLD-TDP, particularly the risk of FTLD-TDP, accompanied by a GRN mutation, by down-regulating progranulin. In the current study, however, TMEM106B overexpression did not down-regulate progranulin in vitro. Conversely, it appeared to increase the progranulin level slightly (Fig. 9D). Similar results were also demonstrated in previous in vitro studies (19, 20, 22). Given that the degradation of progranulin occurs in lysosomes (30), the effect of the TMEM106B overexpression on the progranulin level in the current study may reflect the impairment of lysosomal function, caused by the overexpression of TMEM106B, as reported previously (19, 20, 24). In support of this idea, the treatment with the lysosomal protease inhibitor leupeptin increased progranulin levels (Fig. 9E).
TMEM106B is cleaved to generate NTFs by lysosome-localizing protease(s) (Fig. 2) (25). TMEM106B-NTFs are more rapidly degraded than TMEM106B-FL (Fig. 3A) through the lysosome and ubiquitin-proteasome degradation pathways (Fig. 3, C and D). Consequently, we have failed to detect endogenous NTFs of TMEM106B in normal cultured cells (Fig. 1, C and D) and normal mouse brain tissues (data not shown). Thus, the physiological and pathological relevance of NTFs still remains undetermined. However, the disruption of the gene for the lysosomal protein cathepsin D increases the TMEM106B level and causes the TDP-43 pathology (23). In addition, the lysosomal dysfunction, the up-regulation of TMEM106B expression, and the TDP-43 pathology simultaneously occur in aged GRN knock-out mice (23, 35, 36). Also, oxidative stress, which causes the lysosomal cell death (34) and is implicated in the pathogenesis of neurodegenerative diseases (26), increases the expression levels of TMEM106B-NTFs (Fig. 7D). Finally, multiple genes related to the lysosomal and ubiquitin-proteasome degradation pathway, such as CHAMP2B, SQSTM1, VCP, and UBQLN2, have been identified as familial FTLD-causative genes (3). These data suggest that the dysfunction of the lysosome and/or ubiquitin-proteasome degradation pathways, as observed in FTLD-TDP patients (23, 39), may increase the steady-state levels of TMEM106B-FL and TMEM106B-NTFs, which probably contributes to the pathogenesis of some FTLD cases.
TMEM106B(1–127) and TMEM106B-FL possess a cytotoxic effect in all cells examined, including neuronal cells and yeast cells (Fig. 5). In contrast, TMEM106B(1–106) does not cause cell death in Neuro2a cells and yeast cells, whereas it does in HeLa cells and NSC34 cells (Fig. 5). These results indicate a cell type specificity in the cell death-inducing activity of TMEM106B(1–106), although the mechanism underlying it remains to be investigated.
In summary, TMEM106B-FL or TMEM106B-NTF overexpression causes cytotoxicity via the caspase-dependent mitochondrial cell death pathway and the cleavage of TDP-43. It also increases vulnerability of cells to oxidative stress-induced cell death. These results provide an important insight into the pathogenesis of FTLD-TDP. The in vitro cell death assay established in this study will serve as a new model for the investigation of FTLD-TDP.
Experimental Procedures
Antibodies and Compounds
A rabbit polyclonal TMEM106B antibody was generated by immunizing a synthetic peptide corresponding to amino acid residues 14–27 (KEDAYDGVTSENMR) of human TMEM106B, followed by affinity purification. The following antibodies were purchased from suppliers: GAPDH (catalog no. 2118) (Cell Signaling Technology, Beverly, MA), TDP-43 (catalog no. 12892-1-AP) (ProteinTech Group, Inc., Chicago, IL), Bcl-xS/L (catalog no. sc-634) and normal rabbit IgG (catalog no. sc-2027) (Santa Cruz Biotechnology, Inc.), Xpress (catalog no. R910-25) and progranulin (catalog no. 40-3400) (Thermo Fisher Scientific), HRP-conjugated goat anti-rabbit secondary antibody (catalog no. 170-6515) and HRP-conjugated goat anti-mouse secondary antibody (catalog no. 170-6516) (Bio-Rad), and HRP-conjugated Protein A (catalog no. NA9120) (Amersham Biosciences). (ZLL)2-ketone was purchased from Peptide Institute, Inc. (Osaka, Japan). Cycloheximide, pepstatin A, Boc-D-FMK, staurosporine, and MG132 were purchased from Calbiochem (Darmstadt, Germany). Leupeptin and E-64d were purchased from Sigma.
Plasmid Constructs
Human TMEM106B-T185S-encoding cDNAs were amplified from HeLa cell cDNA by PCR using a forward primer (5′-CGGGATCCACCATGGGAAAGTCTCTTTCTC-3′) and a reverse primer (5′-GCTCTAGATTACTGTTGTGGCTGAAGTAC-3′). WT or FL TMEM106B-S185T and TMEM106B-S185T/Y125D cDNAs were generated using the KOD-Plus-Mutagenesis kit (Toyobo, Osaka, Japan). cDNAs encoding TMEM106B(1–127) and TMEM106B(1–106) were generated by PCR amplification or mutagenesis. These cDNAs were subcloned into the pEF1/Myc-His vector (Invitrogen) and the pEF4/His vector (Invitrogen) for the non-tagged (by using the native stop codon of TMEM106B) and the N-terminal HisXpress-tagged expression vector, respectively. Primer sequences for subcloning and mutagenesis are available on request. The N-terminal HisXpress-tagged VAPB-WT-encoding vector has been described previously (27). LAMP1 cDNA, kindly provided by Dr. Daniela Rotin (Hospital for Sick Children, Toronto, Canada), was subcloned into the pEGFP-N3 vector (Clontech).
Adenoviral Vector-mediated Expression
The adenovirus expression vector systems were purchased from TaKaRa (Shiga, Japan). LacZ, Cre, and Cre-Bcl-xL adenovirus vectors were used as described previously (29). The cDNAs encoding non-tagged TMEM106B-FL, TMEM106B(1–127), TMEM106B(1–106), and TMEM106B-Y125D were inserted into the SwaI site of a cosmid adenoviral vector, pAxCALNLw. In this vector, a stuffer DNA fragment, sandwiched between two loxP sequences, is located just upstream of cDNA and interferes with gene expression. If an adenovirus vector expressing Cre-recombinase is co-introduced into the cells, the stuffer is removed, and the gene is expressed. All viruses were grown in HEK293 cells and purified using CsCl2 gradient ultracentrifugation. Cells were incubated with media containing adenovirus vector at the indicated MOI at 37 °C for 1 h with constant agitation. All samples were co-infected with Cre-recombinase adenovirus vector at an MOI of 40. To keep the total MOIs of viruses constant, appropriate MOIs of LacZ adenovirus vectors were added for each infection.
Cell Culture and Transfection
NSC34 cells, a hybrid cell line established from a mouse neuroblastoma cell line and mouse embryo spinal cord cells, were a kind gift from Dr. Neil Cashman (University of Toronto). HeLa cells (purchased from ATCC), NSC34 cells, mouse neuronal Neuro2a cells (purchased from ATCC), and HEK293 cells were grown in DMEM, supplemented with 10% FBS (Invitrogen). PCNs were prepared as described previously (40). Transfection was performed using Lipofectamine2000 (Invitrogen) according to the manufacturer's protocol.
Cell Death Assay and Cell Viability Assay
Cells, seeded on 6-well plates, were incubated with virus-containing media at the indicated MOI at 37 °C for 1 h with agitation. At 18–24 h after infection, media of cells were replaced by DMEM with N2 supplement (Invitrogen) to conduct an LDH release assay. About 24 h from the replacement of media, LDH releases from cells were measured with an LDH assay kit (Wako, Osaka, Japan). Absorbance of the mixtures at the 560-nm wavelength was measured by a 2030 ARVOTM X5 multilabel reader (PerkinElmer Life Sciences). Cell viability was measured using WST-8 cell viability assays. The WST-8 assay, conducted using Cell Counting Kit-8 (Dojindo, Osaka, Japan), was based on the ability of cells to convert a water-soluble 2-(2-methoxy-4-nitrophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium monosodium salt into a water-soluble formazan. Cells were treated with WST-8 reagent at 37 °C, and 450-nm absorbance was measured by a 2030 ARVOTM X5 multilabel reader.
Western Blotting Analysis
Cells were homogenized and solubilized by sonication in a 4% SDS-containing sample buffer or a cell lysis buffer (10 mm Tris-HCl, pH 7.4, 1% Triton X-100, 1 mm EDTA, protease inhibitors, phosphatase inhibitors) and through a freeze-thaw cycle. The samples were boiled for 5 min at 95 °C in the SDS-containing sample buffer, fractionated by SDS-PAGE, and blotted onto polyvinylidene fluoride membranes. Immunoreactive bands were detected using ECL Western blotting detection reagents (Amersham Biosciences) or Clarity Western ECL substrate (Bio-Rad). GAPDH was visualized as an internal control. Intensities of immunodetected signals were densitometrically measured with ImageJ software. The intensity of a band was normalized with the intensity of GAPDH.
siRNA-mediated Knockdown
siRNAs against TMEM106B and a non-targeting control siRNA were purchased from RNAi Co., Ltd. (Tokyo, Japan). The siRNA sequences for TMEM106B-1, -2, and -3 are 5′-CAGUAUGUCGACUGUGGAAGA-3′, 5′-GUACUAGGAUCUUUUACUUGA-3′, and 5′-GUUCAGAAGCGUACAAUUUAU-3′, respectively. HeLa cells were transfected using Lipofectamine2000 (Invitrogen) according to the manufacturer's reverse transfection protocol. Briefly, 4 × 104 cells/well on 6-well plates were combined with the 5 nm siRNA and Lipofectamine2000 reagent complexes.
Immunoprecipitation
HeLa cells were lysed by sonication in a lysis buffer containing 150 mm NaCl, 20 mm HEPES (pH 7.4), 1 mm EDTA, 1 mm DTT, 0.1% Triton X-100, and protease inhibitors. After centrifugation at 12,000 × g for 15 min, the cell lysates were precleared with Sepharose beads (Amersham Biosciences) for 1.5 h. The cleared supernatants were then incubated for 2–3 h with TMEM106B antibody and precipitated for 1–2 h with protein G-Sepharose (Amersham Biosciences) at 4 °C. After four washes with the lysis buffer, the precipitates were fractionated by SDS-PAGE, followed by immunoblotting analysis.
Immunocytochemistry
HeLa cells were transfected using Lipofectamine2000 (Invitrogen) according to the manufacturer's protocol. Forty-eight hours after transfection, the cells were fixed with 4% paraformaldehyde PBS and immunostained using the TMEM106B antibody and Xpress antibody for the first antibody and fluorescein isothiocyanate or Texas Red-conjugated goat anti-rabbit polyclonal antibody and Texas Red-conjugated goat anti-mouse monoclonal antibody (Jackson ImmunoResearch Laboratories, West Grove, PA) for the secondary antibody. Nuclei were stained with Hoechst 33258 (Sigma). LysoTracker Red DND-9 (100 nm) (Thermo Fisher Scientific) was incubated at 37 °C for 1 h before fixation. The cells were observed with an LSM510 confocal microscope (Carl Zeiss, Oberkochen, Germany). The weighted colocalization coefficient, which is the colocalizing fluorescence intensities relative to the total fluorescence intensities above the background, was analyzed by ZEN software (Carl Zeiss).
Yeast Growth Retardation Assay
Human TMEM106B-FL cDNA was subcloned into a galactose-inducible pRS416-Gal-YFP vector (provided by Dr. James Shorter, University of Pennsylvania) using the native stop codon of TMEM106B. TMEM106B(1–127) and TMEM106B(1–106) were generated using the KOD-Plus-Mutagenesis kit (Toyobo). Primer sequences for subcloning and mutagenesis are available on request. BY4741 yeast strains were transformed with expression vectors indicated using the Direct transformation kit (Wako). The transformants, serially diluted, were spotted onto glucose- or galactose-containing SC agar plates and incubated at 30 °C.
Statistical Analysis
All values are shown as means ± S.D. with n = 3 for each experiment unless indicated otherwise. Statistical significance was determined using Student's t test (*, p < 0.05).
Author Contributions
H. S. conceived, designed, performed, and analyzed the experiments and wrote the paper. M. M. conceived, designed, and analyzed the experiments; wrote the paper; and coordinated the study. Both authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We are especially grateful to Takako Hiraki and Tomoko Yamada for essential assistance throughout the study. We thank Dr. James Shorter and Dr. Daniela Rotin for providing plasmids and Dr. Neil Cashman for providing NSC34 cells.
This work was supported in part by Grant-in-Aid for Scientific Research (B) 15H04689 (to M. M.) and Grant-in-Aid for Scientific Research (C) 25460342 (to H. S.) from the Japan Society for the Promotion of Science, by a grant from the Japan ALS association (JALSA) (to H. S.), by a Naito Foundation Natural Science Scholarship (to M. M.), and by a research grant from the Akaeda Medical Research Foundation (to M. M.). The authors declare that they have no conflicts of interest with the contents of this article.
- FTLD
- frontotemporal lobar degeneration
- ALS
- amyotrophic lateral sclerosis
- LDH
- lactate dehydrogenase
- MOI
- multiplicity of infection
- PCN
- primary cultured cerebral cortical neuron
- Boc-D-FMK
- t-butoxycarbonyl-Asp(O-Me)-fluoromethyl ketone
- HX
- HisXpress
- FL
- full-length
- NTF
- N-terminal fragment
- CTF
- C-terminal fragment
- ER
- endoplasmic reticulum
- VAPB
- vesicle-associated membrane protein-associated protein B.
References
- 1. Rademakers R., Neumann M., and Mackenzie I. R. (2012) Advances in understanding the molecular basis of frontotemporal dementia. Nat. Rev. Neurol. 8, 423–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sieben A., Van Langenhove T., Engelborghs S., Martin J. J., Boon P., Cras P., De Deyn P. P., Santens P., Van Broeckhoven C., and Cruts M. (2012) The genetics and neuropathology of frontotemporal lobar degeneration. Acta Neuropathol. 124, 353–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pottier C., Ravenscroft T. A., Sanchez-Contreras M., and Rademakers R. (2016) Genetics of FTLD: overview and what else we can expect from genetic studies. J. Neurochem. 138, 32–53 [DOI] [PubMed] [Google Scholar]
- 4. Ling S. C., Polymenidou M., and Cleveland D. W. (2013) Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron 79, 416–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ferraiuolo L., Kirby J., Grierson A. J., Sendtner M., and Shaw P. J. (2011) Molecular pathways of motor neuron injury in amyotrophic lateral sclerosis. Nat. Rev. Neurol. 7, 616–630 [DOI] [PubMed] [Google Scholar]
- 6. Peters O. M., Ghasemi M., and Brown R. H. Jr. (2015) Emerging mechanisms of molecular pathology in ALS. J. Clin. Invest. 125, 1767–1779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cruts M., Gijselinck I., Van Langenhove T., van der Zee J., and Van Broeckhoven C. (2013) Current insights into the C9orf72 repeat expansion diseases of the FTLD/ALS spectrum. Trends Neurosci. 36, 450–459 [DOI] [PubMed] [Google Scholar]
- 8. Renton A. E., Chiò A., and Traynor B. J. (2014) State of play in amyotrophic lateral sclerosis genetics. Nat. Neurosci. 17, 17–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Arai T., Hasegawa M., Akiyama H., Ikeda K., Nonaka T., Mori H., Mann D., Tsuchiya K., Yoshida M., Hashizume Y., and Oda T. (2006) TDP-43 is a component of ubiquitin-positive Tau-negative inclusions in frontotemporal lobardegeneration and amyotrophic lateral sclerosis. Biochem. Biophys. Res. Commun. 351, 602–611 [DOI] [PubMed] [Google Scholar]
- 10. Neumann M., Sampathu D. M., Kwong L. K., Truax A. C., Micsenyi M. C., Chou T. T., Bruce J., Schuck T., Grossman M., Clark C. M., McCluskey L. F., Miller B. L., Masliah E., Mackenzie I. R., Feldman H., et al. (2006) Ubiquitinated TDP-43 in frontotemporal lobar degeneration and, amyotrophic lateral sclerosis. Science 314, 130–133 [DOI] [PubMed] [Google Scholar]
- 11. Van Deerlin V. M., Sleiman P. M., Martinez-Lage M., Chen-Plotkin A., Wang L. S., Graff-Radford N. R., Dickson D. W., Rademakers R., Boeve B. F., Grossman M., Arnold S. E., Mann D. M., Pickering-Brown S. M., Seelaar H., Heutink P., et al. (2010) Common variants at 7p21 are associated with frontotemporal lobar degeneration with TDP-43 inclusions. Nat. Genet. 42, 234–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cruchaga C., Graff C., Chiang H. H., Wang J., Hinrichs A. L., Spiegel N., Bertelsen S., Mayo K., Norton J. B., Morris J. C., and Goate A. (2011) Association of TMEM106B gene polymorphism with age at onset in granulin mutation carriers and plasma granulin protein levels. Arch. Neurol. 68, 581–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Finch N., Carrasquillo M. M., Baker M., Rutherford N. J., Coppola G., Dejesus-Hernandez M., Crook R., Hunter T., Ghidoni R., Benussi L., Crook J., Finger E., Hantanpaa K. J., Karydas A. M., Sengdy P., et al. (2011) TMEM106B regulates progranulin levels and the penetrance of FTLD in GRN mutation carriers. Neurology 76, 467–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. van der Zee J., Van Langenhove T., Kleinberger G., Sleegers K., Engelborghs S., Vandenberghe R., Santens P., Van den Broeck M., Joris G., Brys J., Mattheijssens M., Peeters K., Cras P., De Deyn P. P., Cruts M., and Van Broeckhoven C. (2011) TMEM106B is associated with frontotemporal lobar degeneration in a clinically diagnosed patient cohort. Brain 134, 808–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. van Blitterswijk M., Mullen B., Nicholson A. M., Bieniek K. F., Heckman M. G., Baker M. C., DeJesus-Hernandez M., Finch N. A., Brown P. H., Murray M. E., Hsiung G. Y., Stewart H., Karydas A. M., Finger E., Kertesz A., et al. (2014) TMEM106B protects C9ORF72 expansion carriers against frontotemporal dementia. Acta Neuropathol. 127, 397–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gallagher M. D., Suh E., Grossman M., Elman L., McCluskey L., Van Swieten J. C., Al-Sarraj S., Neumann M., Gelpi E., Ghetti B., Rohrer J. D., Halliday G., Van Broeckhoven C., Seilhean D., Shaw P. J., et al. (2014) TMEM106B is a genetic modifier of frontotemporal lobar degeneration with C9orf72 hexanucleotide repeat expansions. Acta Neuropathol. 127, 407–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lattante S., Le Ber I., Galimberti D., Serpente M., Rivaud-Péchoux S., Camuzat A., Clot F., Fenoglio C., French research network on FTD and FTD-ALS, Scarpini E., Brice A., and Kabashi E. (2014) Defining the association of TMEM106B variants among frontotemporal lobar degeneration patients with GRN mutations and C9orf72 repeat expansions. Neurobiol. Aging 35, 2658.e1–2658.e5 [DOI] [PubMed] [Google Scholar]
- 18. Vass R., Ashbridge E., Geser F., Hu W. T., Grossman M., Clay-Falcone D., Elman L., McCluskey L., Lee V. M., Van Deerlin V. M., Trojanowski J. Q., and Chen-Plotkin A. S. (2011) Risk genotypes at TMEM106B are associated with cognitive impairment in amyotrophic lateral sclerosis. Acta Neuropathol. 121, 373–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Brady O. A., Zheng Y., Murphy K., Huang M., and Hu F. (2013) The frontotemporal lobar degeneration risk factor, TMEM106B, regulates lysosomal morphology and function. Hum. Mol. Genet. 22, 685–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen-Plotkin A. S., Unger T. L., Gallagher M. D., Bill E., Kwong L. K., Volpicelli-Daley L., Busch J. I., Akle S., Grossman M., Van Deerlin V., Trojanowski J. Q., and Lee V. M. (2012) TMEM106B, the risk gene for frontotemporal dementia, is regulated by the microRNA-132/212 cluster and affects progranulin pathways. J. Neurosci. 32, 11213–11227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lang C. M., Fellerer K., Schwenk B. M., Kuhn P. H., Kremmer E., Edbauer D., Capell A., and Haass C. (2012) Membrane orientation and subcellular localization of transmembrane protein 106B (TMEM106B), a major risk factor for frontotemporal lobar degeneration. J. Biol. Chem. 287, 19355–19365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nicholson A. M., Finch N. A., Wojtas A., Baker M. C., Perkerson R. B. 3rd, Castanedes-Casey M., Rousseau L., Benussi L., Binetti G., Ghidoni R., Hsiung G. Y., Mackenzie I. R., Finger E., Boeve B. F., Ertekin-Taner N., et al. (2013) TMEM106B p. T185S regulates TMEM106B protein levels: implications for frontotemporal dementia. J. Neurochem. 126, 781–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Götzl J. K., Mori K., Damme M., Fellerer K., Tahirovic S., Kleinberger G., Janssens J., van der Zee J., Lang C. M., Kremmer E., Martin J. J., Engelborghs S., Kretzschmar H. A., Arzberger T., Van Broeckhoven C., Haass C., and Capell A. (2014) Common pathobiochemical hallmarks of progranulin-associated frontotemporal lobar degenerationand neuronal ceroid lipofuscinosis. Acta Neuropathol. 127, 845–860 [DOI] [PubMed] [Google Scholar]
- 24. Stagi M., Klein Z. A., Gould T. J., Bewersdorf J., and Strittmatter S. M. (2014) Lysosome size, motility and stress response regulated by fronto-temporal dementia modifier TMEM106B. Mol. Cell. Neurosci. 61, 226–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Brady O. A., Zhou X., and Hu F. (2014) Regulated intramembrane proteolysis of the frontotemporal lobar degeneration risk factor, TMEM106B, by signal peptide peptidase-like 2a (SPPL2a). J. Biol. Chem. 289, 19670–19680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Uttara B., Singh A. V., Zamboni P., and Mahajan R. T. (2009) Oxidative stress and neurodegenerative diseases: a review of upstream and downstream antioxidant therapeuticoptions. Curr. Neuropharmacol. 7, 65–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Suzuki H., Kanekura K., Levine T. P., Kohno K., Olkkonen V. M., Aiso S., and Matsuoka M. (2009) ALS-linked P56S-VAPB, an aggregated loss-of-function mutant of VAPB, predisposes motor neurons to ER stress-related death by inducing aggregation of co-expressed wild-type VAPB. J. Neurochem. 108, 973–985 [DOI] [PubMed] [Google Scholar]
- 28. Li Q., Yokoshi M., Okada H., and Kawahara Y. (2015) The cleavage pattern of TDP-43 determines its rate of clearance and cytotoxicity. Nat. Commun. 6, 6183. [DOI] [PubMed] [Google Scholar]
- 29. Suzuki H., Lee K., and Matsuoka M. (2011) TDP-43-induced death is associated with altered regulation of BIM and Bcl-xL and attenuated by caspase-mediated TDP-43 cleavage. J. Biol. Chem. 286, 13171–13183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hu F., Padukkavidana T., Vægter C. B., Brady O. A., Zheng Y., Mackenzie I. R., Feldman H. H., Nykjaer A., and Strittmatter S. M. (2010) Sortilin-mediated endocytosis determines levels of the frontotemporal dementia protein, progranulin. Neuron 68, 654–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schwenk B. M., Lang C. M., Hogl S., Tahirovic S., Orozco D., Rentzsch K., Lichtenthaler S. F., Hoogenraad C. C., Capell A., Haass C., and Edbauer D. (2014) The FTLD risk factor TMEM106B and MAP6 control dendritic trafficking of lysosomes. EMBO J. 33, 450–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Busch J. I., Unger T. L., Jain N., Tyler Skrinak R., Charan R. A., and Chen-Plotkin A. S. (2016) Increased expression of the frontotemporal dementia risk factor TMEM106B causes C9orf72-dependent alterations in lysosomes. Hum. Mol. Genet. 10.1093/hmg/ddw127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Aits S., and Jäättelä M. (2013) Lysosomal cell death at a glance. J. Cell Sci. 126, 1905–1912 [DOI] [PubMed] [Google Scholar]
- 34. Boya P., and Kroemer G. (2008) Lysosomal membrane permeabilization in cell death. Oncogene 27, 6434–6451 [DOI] [PubMed] [Google Scholar]
- 35. Wils H., Kleinberger G., Pereson S., Janssens J., Capell A., Van Dam D., Cuijt I., Joris G., De Deyn P. P., Haass C., Van Broeckhoven C., and Kumar-Singh S. (2012) Cellular ageing, increased mortality and FTLD-TDP-associated neuropathology in progranulin knockout mice. J. Pathol. 228, 67–76 [DOI] [PubMed] [Google Scholar]
- 36. Tanaka Y., Chambers J. K., Matsuwaki T., Yamanouchi K., and Nishihara M. (2014) Possible involvement of lysosomal dysfunction in pathological changes of the brain in aged progranulin-deficient mice. Acta Neuropathol. Commun. 2, 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Baker M., Mackenzie I. R., Pickering-Brown S. M., Gass J., Rademakers R., Lindholm C., Snowden J., Adamson J., Sadovnick A. D., Rollinson S., Cannon A., Dwosh E., Neary D., Melquist S., Richardson A., Dickson D., Berger Z., Eriksen J., Robinson T., Zehr C., Dickey C. A., Crook R., McGowan E., Mann D., Boeve B., Feldman H., and Hutton M. (2006) Mutations in progranulin cause Tau-negative frontotemporal dementia linked to chromosome 17. Nature 442, 916–919 [DOI] [PubMed] [Google Scholar]
- 38. Cruts M., Gijselinck I., van der Zee J., Engelborghs S., Wils H., Pirici D., Rademakers R., Vandenberghe R., Dermaut B., Martin J. J., van Duijn C., Peeters K., Sciot R., Santens P., De Pooter T., et al. (2006) Null mutations in progranulin cause ubiquitin-positive frontotemporal dementia linked to chromosome 17q21. Nature 442, 920–924 [DOI] [PubMed] [Google Scholar]
- 39. Ju J. S., and Weihl C. C. (2010) Inclusion body myopathy, Paget's disease of the bone and fronto-temporal dementia: a disorder of autophagy. Hum. Mol. Genet. 19, R38–R45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Suzuki H., Shibagaki Y., Hattori S., and Matsuoka M. (2015) Nuclear TDP-43 causes neuronal toxicity by escaping from the inhibitory regulation by hnRNPs. Hum. Mol. Genet. 24, 1513–1527 [DOI] [PubMed] [Google Scholar]