Abstract
The transcriptional co-activator Yki (Yorkie), a member of the Hippo pathway, regulates cell proliferation or apoptosis, depending on its nuclear or cytoplasmic location. However, the upstream factors regulating the subcellular localization of Yki are unclear. We found that the steroid hormone 20-hydroxyecdysone (20E) induces phosphorylation of Yki, causing it to remain in the cytoplasm, where it promotes apoptosis in the midgut of the lepidopteran insect Helicoverpa armigera. Yki is expressed in various tissues, with an increase in the epidermis and midgut during early metamorphic molting. Yki is localized mainly in the nucleus of feeding larval midgut cells but is mainly localized in the cytoplasm of metamorphic molting larval midgut cells. The knockdown of Yki in the feeding larvae promotes larval-pupal transition, midgut programmed cell death, and repressed IAP1 (inhibitor of apoptosis 1) expression. Knockdown of Yki in the epidermal cell line (HaEpi) induced increased activation of Caspase3/7. Overexpressed Yki in HaEpi cells was mainly localized in the nucleus and induced cell proliferation. 20E promotes the cytoplasmic localization of Yki, reducing the expression of the IAP1, resulting in apoptosis. 20E promotes cytoplasmic retention of Yki by increasing Yki phosphorylation levels and promoting the interaction between Yki and the adaptor protein 14-3-3-ϵ. This regulation of Yki suppresses cell proliferation and induces cell apoptosis.
Keywords: apoptosis, cell proliferation, Hippo pathway, insect, steroid hormone, 20-hydroxyecdysone, Yorkie, cytoplasmic localization
Introduction
In flies, Yki (Yorkie), the homolog of YAP (yes-associated protein) in mammal, is a critical factor that regulates cell fate by promoting either cell proliferation or cell death under different conditions in the Hippo signaling pathway (1). Yki promotes cell proliferation when it is localized in the nucleus, whereas the cytoplasmic localization of Yki inhibits its cell proliferation function and causes apoptosis. Hippo regulates Wts (Warts) phosphorylation (2) and Wts directly phosphorylates Yki to retain Yki in the cytoplasm (3). When the Hippo pathway is repressed, Yki cannot be phosphorylated; Yki, as a co-transcription factor, moves into the nucleus and interacts with the transcription factor scalloped to initiate the expression of the IAP1 (inhibitor of apoptosis 1) to prevent cell death (4). Conversely, phosphorylated Yki is retained in the cytoplasm by interacting with the adaptor protein 14-3-3; thus the function of Yki in promoting cell proliferation and inhibiting cell apoptosis is lost (5). Therefore, the subcellular localization of Yki is critical for cell fate. However, the upstream factors regulating the expression and subcellular localization of Yki remain unknown.
Apoptosis occurs in the larval midgut during the larval-pupal transition (metamorphosis) in Bombyx mori (6) and in the lepidopteran agricultural pest Helicoverpa armigera (7). The apoptotic midgut shrinks inward and becomes separated from the newly formed imaginal midgut; further apoptosis occurs in the midgut during metamorphosis (8). The steroid hormone 20-hydroxyecdysone (20E)3 is produced in insects (9) and plants (10). In insects, 20E promotes apoptosis and metamorphosis (11). At the end of the larval stage, 20E causes apoptosis in the midgut (12). The caspase inhibitor DIAP1 (Drosophila IAP1) inhibits caspase protein activities before the pupal stage (13). The down-regulation of IAP1 is essential for salivary apoptosis in Drosophila (14). The inhibition of IAP1 is also necessary to promote 20E-induced cell death (15). IAP1 expression is up-regulated by Yki (4); therefore, 20E might repress IAP1 expression by inhibiting Yki activity. This hypothesis prompted us to investigate 20E as a new upstream factor that regulates subcellular localization of Yki. Previous work revealed that Hippo is involved in 20E-induced metamorphosis via promoting the phosphorylation and cytoplasmic retention of Yki, causing suppressed expression of the IAP (inhibitor of apoptosis) in H. armigera (16). However, the mechanism of 20E regulation of Yki function is unclear. The insect midgut is a good model that can be used to investigate the function and mechanism of Yki in steroid hormone-induced apoptosis.
We investigated the role and hormonal regulatory mechanism of Yki during midgut apoptosis in H. armigera. We found that 20E increased the phosphorylation of Yki, which caused Yki to be retained in the cytoplasm, thereby inhibiting Yki nuclear function; thus IAP1 expression was repressed, and apoptosis was promoted. This study provides new insights into the steroid hormonal regulation of Yki function and subcellular localization.
Results
Yki Expression Increased in the Early Metamorphic Stage
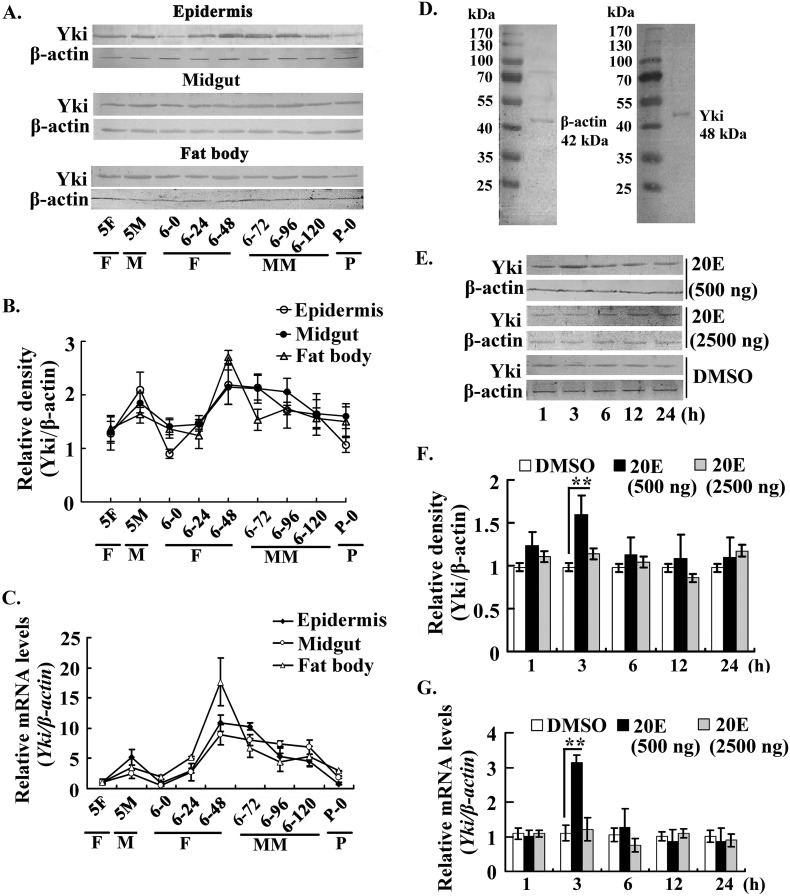
We examined the expression profile of Yki from the fifth instar larval stage to the pupation stage to study the function of Yki in insect larval development. Western blotting analysis revealed that Yki was expressed in the epidermis, midgut, and fat body with varying expression profiles. Yki expression in the epidermis showed a dramatic decrease at the sixth instar 0-h stage, probably because the larvae had just finished ecdysis. Yki levels recovered to the normal level at the sixth instar 24-h stage, obviously increased from the sixth instar 48-h to 96-h stages and then decreased in the prepupation stage from the sixth instar 120-h stage to pupation. Yki expression level in the midgut increased from the sixth instar 48-h to 96-h stages and then decreased from the sixth instar 120-h to pupation stages. Yki was expressed in the fat body at a relatively steady level in the feeding, molting, and metamorphic stages, except an increased expression at sixth instar 48-h stage (Fig. 1A). The relative density of Western blotting from three independent experiments was calculated (Fig. 1B). Quantitative real time PCR (qRT-PCR) showed that the relative mRNA levels of Yki in different tissues were consistent with the Western blotting results (Fig. 1C). The molecular masses of β-actin (GenBankTM accession number KT186238) and Yki (GenBankTM accession number KT153628) were determined with the midgut proteins as 42 and 48 kDa, respectively (Fig. 1D). This expression profile of Yki suggested that Yki expression is up-regulated at the earlier metamorphic stage.
FIGURE 1.
Expression profile and hormonal regulation of Yki in tissues during larval development. A, Yki expression profiles in the epidermis, midgut, and fat body, as shown by Western blotting using a polyclonal antibody against H. armigera Yki. The gel concentration was 12.5%. 5F, fifth instar feeding; 5M, fifth instar molting; 6-0 to 6-120, sixth instar 0 h to sixth instar 120 h; P-0, pupal stage day 0; F, feeding; M, molting; MM, metamorphic molting; P, pupation. β-actin was used as a quantitative control using an antibody against H. armigera β-actin. B, quantitation of the images of Western blotting from three independent experiments using ImageJ software. C, qRT-PCR detected the mRNA levels of Yki in the epidermis, midgut, and fat body. D, protein ladder (Thermo Fisher Scientific, Lithuania) was used to identify the molecular weight of β-actin and Yki, respectively. The proteins in the midgut were used for Western blotting. The gel concentration was 12.5%. E, effect of 20E on Yki expression. Five μl of 20E (100 ng/μl or 500 ng/μl) was injected into the larval hemocoel of the sixth instar 6-h larvae for different times. An equal volume of diluted DMSO was injected as the control. The proteins in the midgut were detected by Western blotting. The gel concentration was 12.5%. F, the statistical analysis of the pictures in E from three independent experiments using ImageJ software. The values are expressed as the means ± S.D. (n = 3). **, p < 0.01 indicates a significant difference by Student's t test. G, qRT-PCR detected the mRNA levels of Yki after 20E induction. The experimental method was same as with E. β-actin was used as the control. Asterisks indicate significant differences (*, p < 0.05; **, p < 0.01), assessed using Student's t test based on three replicates (n = 3).
We examined the induction of Yki expression by 20E, because the 20E titer is higher during metamorphosis in lepidopteran insects (11). Western blotting showed that 20E increased Yki expression at 3 h; however, 20E neither continued to up-regulate Yki expression nor repressed its expression from 6 to 24 h at a low dose (500 ng/larva). When the dose of 20E was increased to 2500 ng/larva, Yki expression levels were neither increased nor decreased significantly (Fig. 1, E and F). In mRNA levels, Yki was also only up-regulated by 20E (500 ng/larva) at 3 h (Fig. 1G). These results suggested that 20E could transiently increase Yki expression at a low dose.
The Subcellular Localization of Yki Varied in the Larval Midgut
We examined the localization of Yki in the midgut to identify the function of Yki. Immunohistochemistry showed that Yki was detected in the nuclei of the midgut cells of the sixth instar 48-h larvae at the feeding stage. However, Yki was detected in the cytoplasm of the larval midgut cells at the sixth instar 96-h point in the metamorphic stage (Fig. 2A). These data suggested that Yki is located in the larval midgut; however, its subcellular localization is different at the feeding and metamorphic molting stages.
FIGURE 2.
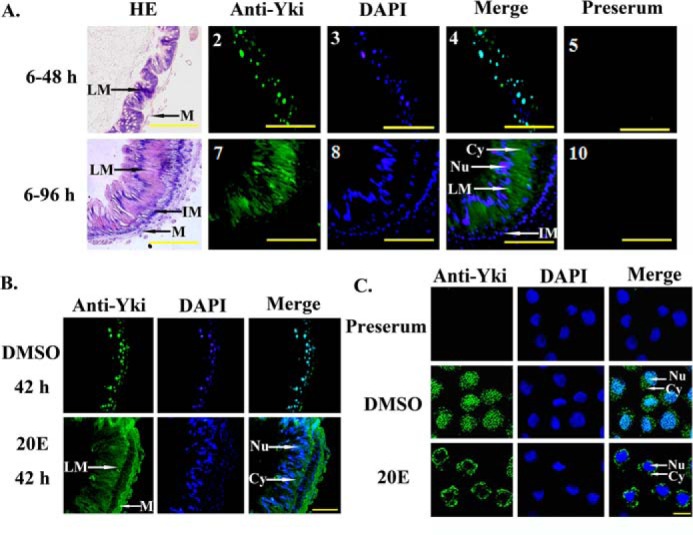
Subcellular localization of Yki in the larval midgut and HaEpi cells. A, Yki is located in the cytoplasm during metamorphosis. Panels 1–5, midgut of the sixth instar 48-h (6–48 h) larvae; panels 6–10, midgut of the sixth instar 96 h larvae (6–96 h). HE, HE staining; Anti-Yki, antibody against H. armigera Yki and Alexa 488-labeled goat anti-rabbit secondary antibodies; DAPI, cell nuclei stained with DAPI; Preserum, preimmune serum. LM, larval midgut; IM, imaginal midgut; M, muscle; Cy, cytoplasm; Nu, nucleus. The scale bars represent 50 μm. B, 20E regulated the cytoplasmic location of Yki in larval midgut cells. Five μl of 20E (500 ng/μl) was injected into the sixth instar 6-h larvae for 42 h. The same volume of DMSO was injected as a control. The scale bar represents 50 μm. C, the location of endogenous Yki in HaEpi cells. The final concentration of 20E was 5 μm. An equal volume of DMSO was used as a control. The endogenous Yki was detected using the polyclonal antibody against H. armigera Yki. Anti-Yki, indicates Yki detected by anti-Ha-Yki and Alexa 488-labeled goat anti-rabbit secondary antibodies; DAPI, cell nuclei stained with DAPI. The scale bar represents 25 μm.
To examine the regulation of 20E on Yki localization in the midgut, we injected 20E into the sixth instar 6-h feeding larvae for 42 h, with an equal volume of DMSO injection as the control. Immunohistochemistry showed that Yki was mainly located in the nucleus in the DMSO treatment control, but treatment with 20E induced Yki to locate to the cytoplasm (Fig. 2B). In HaEpi cells, the endogenous Yki was distributed uniformly in the cells in the DMSO hormone solvent control. However, Yki was mostly localized in the cytoplasm after 5 μm 20E treatment (Fig. 2C). These data suggested that 20E induces the cytoplasmic localization of Yki.
Knockdown of Yki Accelerated the Larval-Pupal Transition and Midgut Remodeling
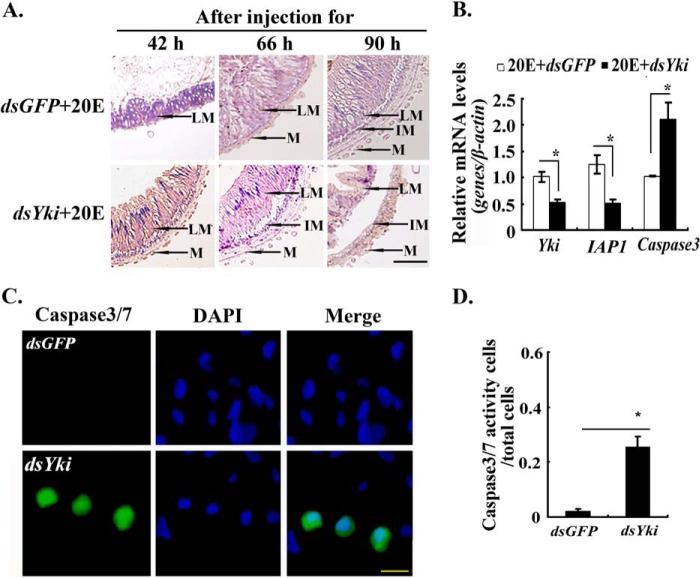
We knocked down Yki in larvae by injecting dsRNA into the hemocoel of the sixth instar 6-h feeding larvae to explore the function of Yki in metamorphosis and midgut remodeling. After knockdown of Yki at the larval feeding stage, 30% of the larvae formed abnormal larva-pupa, 31% died, and 39% formed normal pupae (Fig. 3, A and B). The efficiency of RNA interference was confirmed by Western blotting (Fig. 3C). Yki knockdown accelerated the 20E-promoted pupation by 16 h (Fig. 3D). The larval midgut also showed accelerated remodeling under 20E treatment after Yki knockdown, with the larval midgut separating from the newly formed imaginal midgut, compared with the dsGFP+20E-treated control group (Fig. 4A). The expression level of IAP1 was down-regulated, and the expression level of apoptosis-related gene Caspase3 was up-regulated after Yki knockdown (Fig. 4B). These results suggested that Yki functions to prevent metamorphosis and midgut remodeling by keeping IAP1 expression and repressing Caspase3 expression.
FIGURE 3.
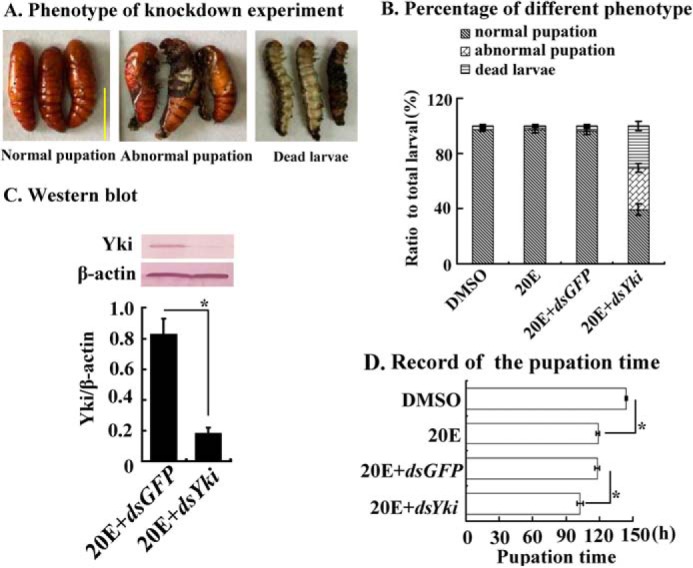
Yki knockdown accelerated metamorphosis. Five μl of dsYki and dsGFP (800 ng/μl) were injected separately into the hemocoel of sixth instar 6-h larvae three times at 24-h intervals. In the last injection of dsRNA, 20E (500 ng) was injected into the hemocoel together. A, different phenotypes of Yki knockdown in larvae. The scale bar indicates 1 cm. B, percentages of the different phenotypes in A. C, Western blotting revealed the efficiency of Yki knockdown. Protein was extracted from midgut at the sixth instar 72 h. The gel concentration was 12.5%. β-actin was used as the control. The image data were processed by ImageJ software. The values are expressed as the means ± S.D. (n = 3). D, pupation time following Yki knockdown. The asterisks denote significant differences (0.01 < p < 0.05, via Student's t test) (3 × 30 larval samples).
FIGURE 4.
Yki knockdown accelerated midgut apoptosis. A, midgut morphology after dsYki or dsGFP injection as detailed in Fig. 3. LM, larval midgut; IM, imaginal midgut; M, muscle. The scale bar represents 50 μm. B, qRT-PCR showing the mRNA levels of IAP1 and Caspase3 after dsYki injection for 66 h in larval midgut in the above treatment. β-actin was used as the control. The asterisks denote significant differences (0.01 < p < 0.05, via Student's t test) based on three replicates. C, Yki knockdown increased Caspase3/7 activity in HaEpi cells. Caspase3/7 activity was detected in dsGFP and dsYki cells (dsRNA 2 μg/ml). Green fluorescence represents the Caspase3/7 activity, as assessed using a Caspase3/7 activity detection kit. Blue fluorescence indicates DAPI-stained nuclei. “Merge” is the superimposed images of the green and blue fluorescence. Fluorescence was observed using an Olympus 257 BX51 fluorescence microscope. The scale bar represents 25 μm. D, statistical analyses of the ratio of Caspase3/7 activity cells in the total cells by ImageJ software. The values are expressed as the means ± S.D. (n = 3). The asterisks indicate significant difference calculated by Student's t test from three independent experiments.
Caspase3/7 activity was thus detected in the Yki knockdown HaEpi cells to confirm Yki function to prevent Caspase3 activity and cell apoptosis. A lower Caspase3/7 signal was detected in the dsGFP-treated cells. However, increased Caspase3/7 activity was detected in the dsYki-treated cells (Fig. 4C). The statistical analysis confirmed the significant difference in the Caspase3/7 activities between dsGFP- and dsYki-treated cells (Fig. 4D). These results suggested that Yki prevents apoptosis.
Yki Promotes Cell Proliferation When Located in the Nucleus
We overexpressed Yki in HaEpi cells by fusing Yki with GFP to determine the mechanism of prevention of apoptosis by Yki. GFP was overexpressed as a control. The 5-ethynyl-2′-deoxyuridine (EdU) signal indicating cell proliferation was lower in the GFP overexpressing cells were treated with DMSO or 20E. In contrast, the EdU signals increased in the Yki-GFP-His-overexpressing cells under DMSO treatment, when Yki-GFP-His was mainly located in the nucleus. However, the EdU signals decreased in the Yki-GFP-His-overexpressing cells under 20E treatment when Yki-GFP-His was partially transferred into the cytoplasm (Fig. 5A). The statistical analysis indicated a significant difference in the EdU signals in the Yki-GFP-His-overexpressing cells under DMSO and 20E treatments (Fig. 5B). Meanwhile, accompanying Yki-GFP-His overexpression, the mRNA level of IAP1 increased in the DMSO control but decreased under 20E treatment (Fig. 5C). These results suggested that Yki promotes cell proliferation and IAP1 expression when localized in the nucleus, and 20E regulates Yki by partially redistributing it into the cytoplasm to suppress both IAP1 expression and cell proliferation.
FIGURE 5.
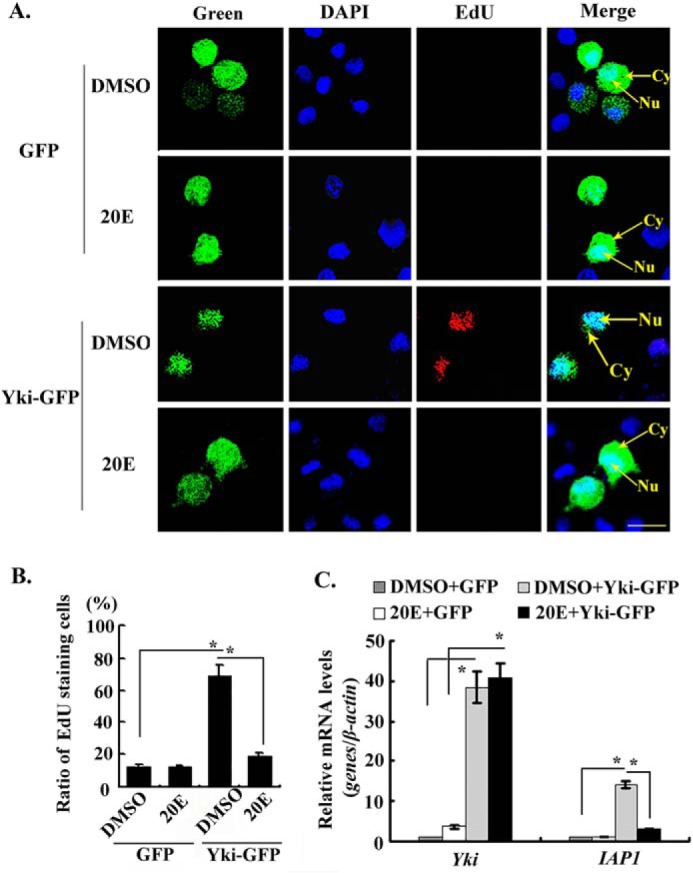
20E suppressed the proliferation function of Yki. A, the overexpression of GFP-His and Yki-GFP-His in HaEpi cells and detection of cell proliferation by EdU. Green, GFP-His and Yki-GFP-His. 20E (5 μm) treated cells for 24 h. The same volume of DMSO was used as the control. Blue, nucleus stained with DAPI; red, EdU; Merge, the overlapped red, green, and blue. Bar, 20 μm. B, statistical analysis of the percentage of EdU staining cells in various treatments using ImageJ software. The values are expressed as the means ± S.D. (n = 3). The asterisks indicate significant differences between the groups (p < 0.05). Student's t test was based on three independent experiments. C, detection of the expression level of IAP1 in various treatments by qRT-PCR. The asterisks indicate significant differences calculated by Student's t test from three independent experiments.
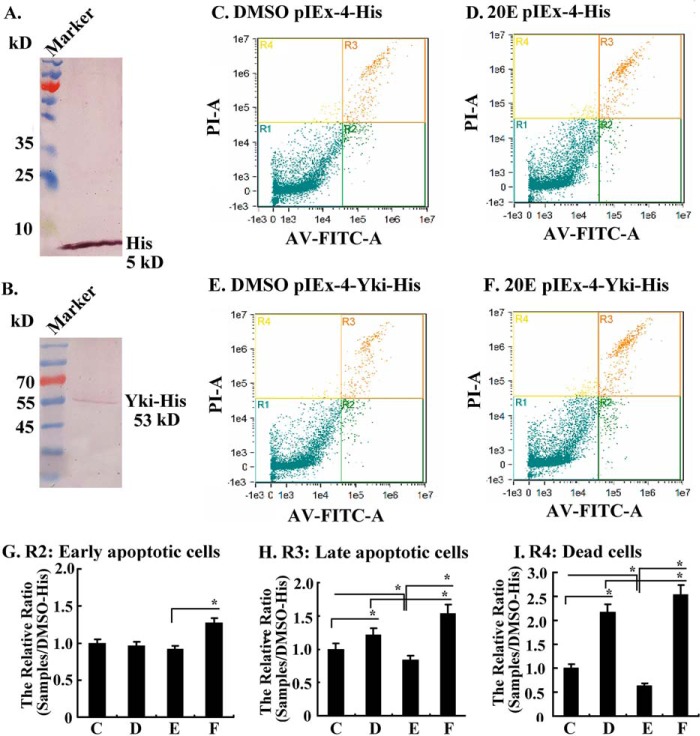
Flow cytometry was performed with HaEpi cells under various treatments to confirm the function of Yki in preventing apoptosis and repression of Yki function by 20E. His tag and Yki-His were overexpressed in HaEpi cells via pIEx-4-His and pIEx-4-Yki-His plasmids (Fig. 6, A and B). Compared with DMSO treatment, 5 μm 20E induced more late apoptosis (R3) and cell death (R4) in the pIEx-4-His-transfected control cells (Fig. 6, C and D). In contrast, in pIEx-4-Yki-His transfected cells, less late apoptosis and cell death were detected in the DMSO control compared with DMSO- or 20E-treated pIEx-4-His-transfected cells, suggesting that overexpression of Yki decreased apoptosis. However, more earlier and late apoptosis, and cell death, were detected under 20E treatment in pIEx-4-Yki-His transfected cells, suggesting that 20E repressed Yki function (Fig. 6. E and F). Statistical analysis of the apoptosis and cell death in Fig. 6 (C–F) confirmed that 20E induced early (R2) and late apoptosis (R3) and cell death (R4). However, the overexpression of Yki decreased the late apoptosis and cell death in the DMSO control cells, but 20E increased the late apoptosis and cell death in Yki-overexpressing cells (Fig. 6, G–I). These results suggested that Yki prevents apoptosis and 20E represses the anti-apoptotic function of Yki.
FIGURE 6.
20E suppressed Yki function to induce cell apoptosis, as detected by Amnis flow cytometry. A and B, Western blotting showing the overexpression of His tag and Yki-His in HaEpi cells. C and D, His tag expressing cells induced with DMSO or 20E (5 μm) for 72 h in Grace's medium, respectively. E and F, Yki-His expressing cells induced with DMSO or 20E (5 μm) for 72 h in Grace's medium, respectively. AV-FITC-V, annexin V-FITC to detect the early apoptotic cells; PI-A, propidium iodide (PI) to detect the late apoptotic cells; R1, living cells (PI-negative, FITC-negative); R2, early apoptotic cells (PI-negative, FITC-positive); R3, late apoptotic cells (PI-positive, FITC-positive); R4, dead cells (PI-positive, FITC-negative). G–I, statistics of cells in R2, R3, and R4 from C–F. The values are expressed as the means ± S.D. (n = 3). The asterisks indicate significant differences (*, p < 0.05; **, p < 0.01) via Student's t test based on three replicates.
20E Regulated Yki Phosphorylation and Cytoplasmic Localization
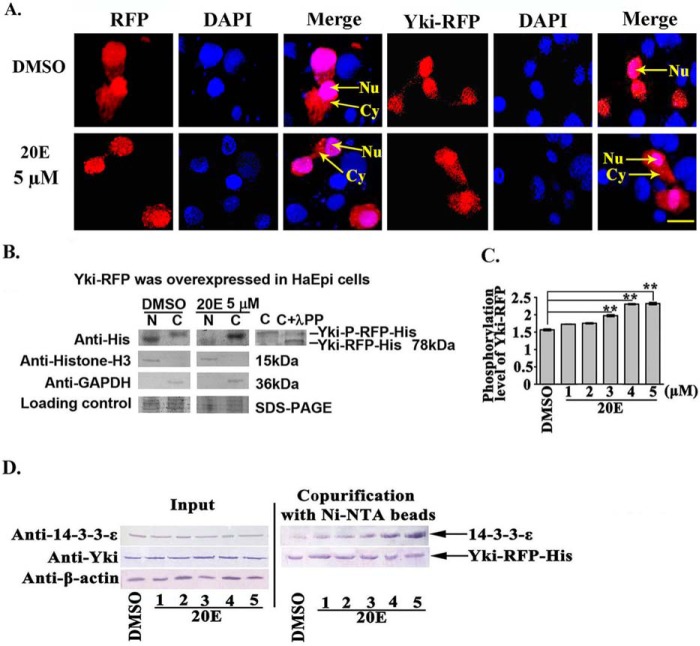
We examined the subcellular localization and mechanism of Yki under 20E induction by overexpressing Yki-RFP in HaEpi cells to confirm 20E regulation of Yki cytoplasmic localization. The subcellular localization of RFP did not change obviously; however, the overexpressed Yki-RFP was partially localized in the cytoplasm after 20E treatment (Fig. 7A), which suggested that 20E induces Yki-RFP cytoplasmic localization.
FIGURE 7.
20E arrested Yki in the cytoplasm by regulating Yki phosphorylation and interaction with 14-3-3-ϵ. A, the location of Yki-RFP-His in HaEpi cells. Red fluorescence represents RFP and the recombinant protein annexin-RFP, whereas blue represents DAPI-stained nuclei. Merge shows the superimposed red and blue fluorescence. Nu, nucleus; Cy, cytoplasm. The pictures were obtained after 6 h of 20E (5 μm) incubation; DMSO was used as the solvent control. Pictures were taken using a Zeiss LSM 700 laser confocal microscope. The scale bar represents 20 μm. B, Western blotting analysis of Yki-RFP-His expression in the cytoplasm and nucleus following pIEx-Yki-RFP-His overexpression. The gel concentration was 7.5%. N, nucleus; C, cytoplasm. λPP, λ-phosphatase. Anti-histone-H3 and anti-GAPDH were used to control the separation of the cytoplasmic and nuclear proteins, respectively. Loading control indicated the quantity of the protein loading. C, Yki phosphorylation levels detected using a phosphoprotein phosphate estimation kit. The cells were treated with 20E (1–5 μm) for 6 h. The error bars represent the standard deviations of three replicates. The asterisks denote significant differences (p < 0.01, via Student's t test). D, interaction between Yki and 14-3-3-ϵ (30 kDa). Input, protein expression levels of 14-3-3-ϵ and Yki-RFP-His in the cell lysates after various treatments. The co-purified 14-3-3-ϵ was detected using a rabbit antiserum against Helicoverpa 14-3-3-ϵ prepared in our laboratory. β-actin was used as the quantitative control.
The cytoplasmic and nuclear proteins were extracted separately and analyzed by Western blotting to confirm the immunocytochemical observations. The results showed that the overexpressed Yki-RFP was mainly localized in the nucleus, with a little localized in the cytoplasm in the DMSO control. In contrast, Yki-RFP was mainly localized in the cytoplasm, with a little localized in the nucleus after 20E induction for 12 h. In addition, the Yki localized in the cytoplasm had a slightly higher molecular mass than that of the nuclear-Yki: the molecular weight of the cytoplasmic-located Yki could be decreased by lambda phosphatase (Fig. 7B), suggesting that the cytoplasmic-located Yki was phosphorylated.
Yki phosphorylation levels were examined using a phosphoprotein phosphate estimation kit to confirm the induction of Yki phosphorylation by 20E. Yki phosphorylation levels increased with increasing 20E concentration (Fig. 7C). This result confirmed that 20E regulates Yki phosphorylation.
Previously, cytoplasmic-located Yki was demonstrated to interact with 14-3-3 (17); therefore, we examined the interaction between Yki and 14-3-3-ϵ. Using affinity purification of Yki-RFP-His using nickel-nitrilotriacetic acid beads, increased levels of 14-3-3-ϵ were detected as the concentration of 20E increased (Fig. 7D). These results suggested that 20E induced the interaction between Yki and 14-3-3-ϵ, thereby retaining Yki in the cytoplasm.
Discussion
Yki promotes cell proliferation when localized in the nucleus and promotes cell apoptosis when localized in the cytoplasm (2). However, the upstream regulators, especially the steroid hormones regulating the subcellular localization of Yki, are poorly understood. This work reported that the steroid hormone 20E promotes Yki phosphorylation, causing Yki to be retained in the cytoplasm, which consequently suppresses IAP1 expression and induces apoptosis. This study presents a new upstream factor, steroid hormone 20E, which regulates the cytoplasmic location of Yki, using H. armigera as a model.
Nuclear-localized Yki Promotes Cell Proliferation
Yki and YAP were identified previously to promote cell proliferation and inhibit apoptosis through their functions as transcription co-factors in the nucleus (5). Yki promotes cell proliferation and inhibits apoptosis in the nucleus by initiating IAP1 expression (4). Our study also showed that nuclear-located Yki prevents apoptosis and upregulates IAP1 expression. A previous study indicated that the H. armigera larval midgut cells are proliferating at the sixth instar 48-h stage but undergo apoptosis at the sixth instar 96-h stage (18). Therefore, the knockdown of Yki in sixth instar 6-h larvae would block cell proliferation, accelerating the 20E-mediated midgut apoptosis and larval-pupal transition.
In addition, we observed the increased expression of Caspase3 after Yki knockdown at the sixth instar 6-h larvae, and the activity of Caspase3/7 was increased in HaEpi cells after Yki knockdown. These data indicated that Yki prevents apoptosis not only by upregulating IAP1 expression but also by repressing Caspase3 expression and activation.
20E Promotes Yki Phosphorylation and Cytoplasmic Localization
Yki activity is promoted by multiple upstream signaling pathways, such as the insulin signaling pathway, which activates Yki to promote cell proliferation (19). The JNK signaling pathway increases Yki activity for wing regeneration (20). Recently, lysophosphatidic acid (21), sphingosine 1-phosphophate (22), and stimulation of protease-activated receptors (23) were found to stimulate the nuclear localization of YAP and increase cell proliferation. By contrast, epinephrine and glucagon increase the phosphorylation and cytoplasmic localization of YAP via a GPCR (24). In HEK293 cells, YAP1 that is c-Abl-phosphorylated on tyrosine can interact with p73 to co-activate proapoptotic genes in response to DNA damage (25). Phosphorylation of YAP by the Lats tumor suppressor kinase leads to cytoplasmic translocation and inactivation of the YAP oncoprotein (26).
Hippo regulates Wts phosphorylation, after which Wts regulates Yki phosphorylation, thus retaining Yki in the cytoplasm (3). Our previous work showed that Hippo is highly expressed during metamorphosis to regulate Yki phosphorylation and its cytoplasmic localization by 20E induction (16). This work further revealed that 20E regulates Yki phosphorylation and its interaction with 14-3-3-ϵ in a concentration-dependent manner, thereby retaining Yki in the cytoplasm. This result suggested that a higher concentration of steroid hormone 20E could promote the phosphorylation-dependent cytoplasmic location of Yki.
The Cytoplasmic Localization of Yki Results in Apoptosis
Increased amounts of cytoplasmic YAP induced apoptosis strongly in HEK293T cells (27). In this study, we also found that 20E promoted Yki cytoplasmic localization in midgut and in HaEpi cells to induce apoptosis. Overexpressed Yki was mainly localized in the nucleus. However, 20E treatment caused the endogenous or overexpressed Yki to be retained in the cytoplasm. YAP is a pro-oncogenic factor. Recently, its activity was found to be regulated by metabolic pathways, such as aerobic glycolysis and mevalonate synthesis pathways (28). Inhibitors of YAP1 are thought to be beneficial in cancer therapy (29). 20E is produced in insects (30) and plants (31), representing a target for repression of Yki activity.
Conclusion
Using H. armigera as a model, we observed that Yki was located in the nuclei of feeding larval midgut cells and HaEpi cells, where it increased IAP1 expression and promoted cell proliferation. The steroid hormone 20E regulates Yki phosphorylation and its interaction with 14-3-3-ϵ to retain Yki in the cytoplasm, which blocks Yki function in the nucleus, thus inhibiting cell proliferation and promoting cell apoptosis. These results provide a new insight into the mechanism whereby steroid hormone 20E promotes the cytoplasmic location of Yki to promote apoptosis.
Experimental Procedures
Experimental Animals
The experimental cotton bollworms were reared under a 14-h:10-h light:dark cycle. The cotton bollworms were fed with an artificial diet composed of wheat and soybean (32). The cotton bollworms were obtained from Henan Agricultural University (Zhengzhou, China).
Cloning of the ORF of Yki and Sequencing Analysis
The ORF of Yki was obtained from the transcriptome sequence of the epidermal cell line (HaEpi) of H. armigera, established in our laboratory (33). A pair of specific primers Yki-F and Yki-R (Table 1) was designed to amplify the open reading frame of Yki. Using the Basic Local Alignment Search Tool (BLAST), the Yki cDNA was shown to have 89% identity with B. mori Yki.
TABLE 1.
Oligonucleotide sequences of PCR primers
| Primer name | Oligonucleotide sequence (5′ → 3′) |
|---|---|
| qRT-PCR | |
| Yki-RT-F | ggttccaagtcgccatctgt |
| Yki-RT-R | cagcggcgtaagtttgttgc |
| IAP1-RT-F | gcttccgaagctgcgcgtttg |
| IAP1-RT-R | cagctgcacgtaggcgcagcg |
| Caspase3-RT-F | ggagacaagggtggagaa |
| Caspase3-RT-R | ggaaggcgtgtatgtggt |
| β-actinF | cctggtattgctgaccgtatgc |
| β-actinR | ctgttggatggtggagagggaa |
| RNA interference | |
| GFPRNAi-F | gcgtaatacgactcactataggtggtcccaattctcgtggaac |
| GFPRNAi-R | gcgtaatacgactcactataggcttgaagttgaccttgatgcc |
| YkiRNAi-F | gcgtaatacgactcactatagggcctcgccgcagaacatacc |
| YkiRNAi-R | gcgtaatacgactcactatagggcgttcaacaggtcgtcagtagagt |
| Prokaryotic expression | |
| Yki-exp-F | tactcagcggccgcaaacgaccgctacaggtg |
| Yki-exp-R | tactcagaattcctgcgccgccaacgtctt |
| Overexpression | |
| YkiOEF-F | tactcagagctcatggctctcaattcggacgct |
| YkiOEF-R | tactcagtcgaccagccacgtcagcacgttgtc |
| Full-length clone | |
| Yki-F | atggctctcaattcggacgctg |
| Yki-R | tacagccacgtcagcacgttg |
Expression of Recombinant Yki Protein in Escherichia coli and Antiserum Preparation
A 450-bp fragment of Yki was amplified from the double-strand full-length cDNA using primers Yki-exp-F, Yki-exp-R (Table 1). The Yki fragment was inserted into the expression vector pGEX-4T-1. The recombinant vector was subsequently transformed into competent E. coli BL21 host cells. Isopropyl-β-d-thiogalactopyranoside (0.5 mm) was used to induce the host cells to produce the target protein in LB medium with ampicillin (100 μg/ml). The recombinant protein was expressed as a soluble protein. The soluble protein samples were purified using an affinity column with a glutathione S-transferase tag. The purified samples were injected into a rabbit to produce rabbit polyclonal antibodies against Yki (anti-Yki). The immunization method was the same as a previously described procedure (34).
Protein Extraction and Western Blotting
The tissues used for protein extraction were ground in 40 mm Tris-HCl, pH 7.5, with 1 mm phenylmethanesulfonyl fluoride. The suspension was centrifuged at 10,000 × g at 4 °C. The supernatant containing the proteins was collected, and the protein concentration was detected using the Bradford method (35). Proteins (50 μg) of each sample were subjected to SDS-PAGE. Afterward, the proteins were transferred onto a 0.22-μm nitrocellulose membrane; the membrane was blocked for 1 h in blocking buffer (2% nonfat dry milk in TBS: 10 mm Tris-Cl, 150 mm NaCl, pH 7.5) at room temperature. The membrane was then incubated with the antiserum against H. armigera Yki (1:100 in blocking buffer) for 2 h. The membrane was then washed three times with TBS containing Tween (TBST, 0.1% Tween 20 in TBS) for 10 min and three times with TBS for 5 min each time and then probed with HRP-conjugated goat anti-rabbit IgG (Sungene, Tianjin, China) (1:10,000 dilution in blocking buffer) for 2 h at room temperature. The target protein signal was visualized using 4-Chloro-1-naphthol, which was used as an HRP substrate to visualize the peroxidase activity.
In Vivo Hormone Induction
The sixth instar 6-h larvae were selected and subjected to in vivo hormone induction. Five μl of 20E (500 ng) was injected into the hemocoel of larvae; the same volume of diluted DMSO was injected into the control larvae. The larvae were sacrificed at 1, 3, 6, 12, and 24 h after the hormone was added, and the total protein was extracted for Western blotting.
Immunohistochemistry
The midgut of the sixth instar 48- and 96-h larvae were taken out and fixed overnight in 4% paraformaldehyde at 4 °C. The tissue was gradient-dehydrated and then embedded in paraffin for 16 h. The paraffin-embedded tissues were sliced into 7-μm serial sections by a paraffin slicing machine. The serial histological sections were mounted on gelatin-coated glass slides, which were dried overnight at 42 °C. The slides were dewaxed with dimethylbenzene and gradient-rehydrated. The slides were then digested with proteinase K (20 μm) for 10 min at 37 °C, blocked in 2% BSA at 37 °C for 30 min, incubated with polyclonal antibody against Yki (1:500 ratio, diluted in blocking buffer) at 4 °C overnight, washed with PBS (10 mm Na2HPO4, 1.8 mm KH2PO4, 140 mm NaCl, 2.7 mm KCl, pH 7.4), and incubated with goat anti-rabbit IgG-Alexa Fluor 488 (Eugene, OR) (1:1000 dilution in blocking buffer) at 37 °C for 1 h. The nuclei were stained using DAPI (AnaSpec Inc., San Jose, CA; 1 μg/ml in PBS) for 10 min at room temperature. Preimmune rabbit serum was used as the negative control. Fluorescence was observed using an Olympus BX51 fluorescence microscope (Shinjuku-ku, Tokyo, Japan).
RNA Interference of Yki in Larvae
Double-strand RNAs of GFP and Yki were synthesized using primers GFPRNAi-F, GFPRNAi-R, and primers YkiRNAi-F and YkiRNAi-R (Table 1). A total of 120 sixth instar 6-h larvae were selected and divided equally into four groups. The four groups were individually injected with DMSO, 20E, double-stranded RNA of GFP (dsGFP) with 20E, and double-stranded RNA of Yki (dsYki) with 20E. The details were as follows: dsGFP or dsYki (800 ng) were injected into the larval hemocoel three times at the sixth instar 6-h, 30-h, and 42-h stages, respectively, and mixed with DMSO or 20E (500 ng) at the third time injection. The survival, development, and pupation conditions of the larvae were recorded. Total proteins were extracted to determine efficiency. Moreover, the phenotypes were analyzed statistically. The histological sections of the midgut after dsYki injection for 42, 66, and 90 h were stained with hematoxylin and eosin (HE). Three replications were performed independently for statistical analysis.
HE Staining
The histological sections were prepared, dewaxed, and rehydrated as described in the previous sections. The histological sections were stained with hematoxylin for 5 min to 10 min at room temperature and washed with water. The sections were treated with Scott liquid (0.35 g of NaHCO3, 2 g of MgSO4, and 100 ml of H2O) for 1 min, differentiation liquid (0.5 ml of HCl and 49.5 ml of 70% ethanol) for 20 s, Scott liquid for 1 min, and 0.5% water-soluble eosin dye solution for 30 s. The sections were immediately washed with running water. The slices were sealed and observed using an Olympus BX51 fluorescence microscope (Shinjuku-ku, Tokyo, Japan).
qRT-PCR
The total RNA of the treated cells was extracted using the Unizol reagent in accordance with the manufacturer's instructions (Biostar, Shanghai, China). The cDNA was reverse transcribed using a reverse transcription kit (Tiangen, Beijing, China) in accordance with the manufacturer's instructions. Four pairs of primers, β-actinF and β-actinR, Yki-RT-F and Yki-RT-R, IAP1-RT-F and IAP1-RT-R, and Caspase3-RT-F and Caspase3-RT-R, were designed using software Primer 5.0. A final volume of 10 μl was used in the qRT-PCR analysis (CFX96TM real time system; Bio-Rad). The reaction was performed as follows: 95 °C for 10 min for the initial denaturation and then 40 cycles of 95 °C for 10 s, 60 °C for 1 min, and 75 °C 2 s for plate reading; and the melt curve was analyzed from 65 to 95 °C at 0.5 °C increments. β-actin was used as the quantity and quality control. The data were calculated using r = 2−ΔΔCT (7, 36).
Caspase 3/7 Activity Detection
The activity of Caspases3/7 in HaEpi cells was detected with the Cell EventTM Caspase3/7 Green detection reagent (Invitrogen), following the manufacturer's instructions. Briefly, as the HaEpi cell density reached 70% in the 6-well cell culture plates in Grace's medium, the cells were transfected with 2 μg dsGFP or dsYki for 48 h. The same time and concentration of dsGFP treatment were used as the control. The cells were then treated with 2 μm Cell EventTM Caspase3/7 Green detection reagent in fresh Grace's medium with 10% FBS at 37 °C for 1 h. The nuclei were stained with DAPI (1 μg/ml in DPBS) for 10 min at room temperature in the dark. Finally, the cells were observed using an Olympus BX51 fluorescence microscope. Images were captured to count the number of cells that were stained with Caspase3/7 Green using ImageJ software. Three replicates were performed for statistical analysis by Student's t test.
Overexpression of Yki in HaEpi Cells
The ORF of Yki was amplified using the overexpression primers YkiOEF-F and YkiOEF-R (Table 1). The fragment was then separately inserted into the pIEx-4-RFP-His, pIEx-4-GFP-His, and pIEx-4-His plasmids, respectively, using a previously described method (37). The constructed plasmids were subsequently transfected into the HaEpi cells using Cellfection II reagent (Invitrogen), in accordance with the manufacturer's instructions. After 48 h of incubation, 5 μm 20E and DMSO were added into Grace's medium with 10% FBS for 12 h. The changes in the subcellular localizations of Yki-RFP-His were observed using a Zeiss LSM 700 laser confocal microscope (Zeiss).
EdU Detection
EdU is a novel thymidine analog that is incorporated into the DNA of dividing cells to indicate DNA synthesis (38). An EdU kit (Ribobio, Guangzhou, China) was used to detect cell proliferation levels according to the manufacturer's protocol. Yki-GFP and GFP tags were overexpressed from the pIEx-4-Yki-GFP His and pIEx-4-GFP-His plasmids, respectively, in HaEpi cells for 24 h, respectively. Then the cells were treated with 5 μm 20E for 24 h. The same volume of DMSO was used as the control. For EdU detection, first the cells were incubated with 100 μl of EdU substrate (50 μm, Grace's: buffer A = 1000: 1) for 2 h. After being washed with DPBS for 5 min, the cells were fixed in the dark using 100 μl of 4% paraformaldehyde for 30 min at room temperature. The cells were then incubated with 100 μl of 2 mg/ml glycine for 5 min with shaking and washed by DPBS for 5 min twice. Second, the cells were incubated with 1× Apollo staining reaction liquid for 30 min at shaking device in the dark and washed by DPBS for 10 min twice. Lastly, the cells were incubated in the dark with 1 × Hoechst 33342 staining reaction (deionized water: Hoechst 33342 = 100: 1) for 30 min with shaking and then washed by DPBS for 5 min twice. The cells were observed using a Zeiss LSM 700 laser Confocal microscope (Zeiss).
Flow Cytometry Detection of Cell Apoptosis
Yki-His and His tag were overexpressed from the pIEx-4-His and pIEx-4-Yki-His plasmids, respectively, in HaEpi cells for 24 h. The cells were then treated with 20E (final concentration, 5 μm) for 72 h. An equal volume of DMSO was added to the control cells. An annexin V-FITC/PI apoptosis detection kit (Wanleibio, Tianjin, China) was used to detect the early apoptosis and late apoptosis levels in accordance with the manufacturer's instructions. Flow cytometric detection (Amnis) of the fluorescence signal was used to detect the levels of apoptosis. Data analysis software The IDEAS software (Amnis) was used to perform the data analysis.
λ-Phosphatase Treatment
Yki-RFP-His protein was overexpressed using pIEx-Yki-RFP-His plasmid in HaEpi cells for 24 h. The cells were then treated with 5 μm 20E for 6 h. The protein was purified using His-Bind resin. The nuclear and cytoplasmic proteins were extracted using a nuclear and cytoplasmic protein extraction kit (Beyotime, Haimen, China). 40 μl of protein (2 mg/μl) was incubated with 0.5 μl of λ-phosphatase, 5 μl of buffer, and 5 μl of MnCl2 at 30 °C for 30 min, according to the manufacturer's specifications (Millipore, Temecula, CA). The proteins were subjected to SDS-PAGE with 7.5% low concentration gels and Western blotting analysis. Protein phosphorylation was examined by molecular mass variation.
Detection of Yorkie Phosphorylation Levels
Yki-RFP-His was overexpressed in HaEpi cells by transfecting with the pIEx-Yki-RFP-His plasmid. The protein was purified by His-Bind resin (50 μl of resin) after different treatments. The phosphorylation levels of purified Yki-RFP were analyzed in a 96-well microplate using a phosphoprotein phosphate estimation assay kit (Sangon Biotechnology, Shanghai, China), based on the alkaline hydrolysis of phosphate from Ser1 and Thr1 residues in phosphoproteins. The released phosphates were then quantified using malachite green and ammonium molybdate, in accordance with the manufacturer's instructions.
Co-purification
HaEpi cells were seeded into a 6-well plate. When the cells covered ∼80% of each plate, Yki overexpression was induced; after 24 h, the hormones were added. After 6 h of incubation with the hormones, the culture medium was discarded, and 800 μl of the cell lysis solution radioimmune precipitation assay buffer (0.1 m Tris-HCl buffer, pH 8.0, containing 0.15 m NaCl, 1% Nonidet P-40) was added to each well, and the cells were dissociated at 4 °C for at least 30 min. The lysates were incubated with nickel-nitrilotriacetic acid beads for 1 h (Novagen, Darmstadt, Germany). Subsequently, the supernatant was discarded after centrifugation at 1,000 rpm for 3 min. The collected nonspecific proteins were washed with 400 μl of binding buffer (0.5 m NaCl, 20 mm Tris-Cl, and 5 mm imidazole, pH 7.9) three times. The supernatant was discarded after centrifugation at 1,000 rpm for 3 min. The specific Yki-RFP-His protein was stripped using 150 μl of stripping solution (0.5 m NaCl, 20 mm Tris-Cl, and 100 mm EDTA). The specific proteins in the supernatant were separated after centrifugation at 12,000 rpm for 3 min. Thereafter, 70 μl of protein-processing liquid was added to the final supernatant, and the mixture was boiled for 10 min. The samples were loaded onto SDS-PAGE for Western blotting.
Author Contributions
The main work was contributed by D. W. and X.-R. L. D.-J. D. did experiments related to Hippo. The foundation work was performed by H. H. This work was conceived and supervised by J.-X. W. and X.-F. Z.
This work was supported by the National Natural Science Foundation of China Grants 31230067 and 31372261 and Ph.D. Programs Foundation of the Ministry of Education of China Grant 20120131110025. The authors declare that they have no conflicts of interest with the contents of this article.
- 20E
- 20-hydroxyecdysone
- EdU
- 5-ethynyl-2′-deoxyuridine
- qRT-PCR
- quantitative real time PCR
- HE
- hematoxylin and eosin
- DPBS
- Dulbecco's PBS
- PI
- propidium iodide.
References
- 1. Moroishi T., Hansen C. G., and Guan K. L. (2015) The emerging roles of YAP and TAZ in cancer. Nat. Rev. Cancer 15, 73–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Huang J., Wu S., Barrera J., Matthews K., and Pan D. (2005) The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila homolog of YAP. Cell 122, 421–434 [DOI] [PubMed] [Google Scholar]
- 3. Dong J., Feldmann G., Huang J., Wu S., Zhang N., Comerford S. A., Gayyed M. F., Anders R. A., Maitra A., and Pan D. (2007) Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell 130, 1120–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhang L., Ren F., Zhang Q., Chen Y., Wang B., and Jiang J. (2008) The TEAD/TEF family of transcription factor Scalloped mediates Hippo signaling in organ size control. Dev. Cell 14, 377–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Oh H., and Irvine K. D. (2008) In vivo regulation of Yorkie phosphorylation and localization. Development 135, 1081–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tettamanti G., Grimaldi A., Casartelli M., Ambrosetti E., Ponti B., Congiu T., Ferrarese R., Rivas-Pena M. L., Pennacchio F., and Eguileor M. (2007) Programmed cell death and stem cell differentiation are responsible for midgut replacement in Heliothis virescens during prepupal instar. Cell Tissue Res. 330, 345–359 [DOI] [PubMed] [Google Scholar]
- 7. Liu C. Y., Liu W., Zhao W. L., Wang J. X., and Zhao X. F. (2013) Upregulation of the expression of prodeath serine/threonine protein kinase for programmed cell death by steroid hormone 20-hydroxyecdysone. Apoptosis 18, 171–187 [DOI] [PubMed] [Google Scholar]
- 8. Cai M. J., Liu W., He H. J., Wang J. X., and Zhao X. F. (2012) Mod(mdg4) participates in hormonally regulated midgut programmed cell death during metamorphosis. Apoptosis 17, 1327–1339 [DOI] [PubMed] [Google Scholar]
- 9. Fraenkel G. S. (1935) A hormone causing pupation in the blowfly Calliphora erythrocephala. Proc. R. Soc. Lond. B Biol. Sci. 118, 1–12 [Google Scholar]
- 10. Ogawa S., and Nishimoto N. (1967) Isolation of the moulting hormones of insects from Achyranthis radix. Yakugaku Zasshi 87, 325–327 [DOI] [PubMed] [Google Scholar]
- 11. Riddiford L. M., Hiruma K., Zhou X., and Nelson C. A. (2003) Insights into the molecular basis of the hormonal control of molting and metamorphosis from Manduca sexta and Drosophila melanogaster. Insect Biochem. Mol. Biol. 33, 1327–1338 [DOI] [PubMed] [Google Scholar]
- 12. Jiang C., Baehrecke E. H., and Thummel C. S. (1997) Steroid regulated programmed cell death during Drosophila metamorphosis. Development 124, 4673–4683 [DOI] [PubMed] [Google Scholar]
- 13. Orme M., and Meier P. (2009) Inhibitor of apoptosis proteins in Drosophila: gatekeepers of death. Apoptosis 14, 950–960 [DOI] [PubMed] [Google Scholar]
- 14. Yin V. P., Thummel C. S., and Bashirullah A. (2007) Down-regulation of inhibitor of apoptosis levels provides competence for steroid-triggered cell death. J. Cell Biol. 178, 85–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Martin S. J. (2002) Destabilizing influences in apoptosis: sowing the seeds of IAP destruction. Cell 109, 793–796 [DOI] [PubMed] [Google Scholar]
- 16. Dong D. J., Jing Y. P., Liu W., Wang J. X., and Zhao X. F. (2015) The steroid hormone 20-hydroxyecdysone upregulates Ste-20 family serine/threonine kinase Hippo to induce programmed cell death. J. Biol. Chem. 290, 24738–24746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Muslin A. J., and Xing H. (2000) 14-3-3 proteins: regulation of subcellular localization by molecular interference. Cell Signal. 12, 703–709 [DOI] [PubMed] [Google Scholar]
- 18. Liu C. Y., Zhao W. L., Wang J. X., and Zhao X. F. (2015) Cyclin-dependent kinase regulatory subunit 1 promotes cell proliferation by insulin regulation. Cell Cycle 14, 3045–3057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Strassburger K., Tiebe M., Pinna F., Breuhahn K., and Teleman A. A. (2012) Insulin/IGF signaling drives cell proliferation in part via Yorkie/YAP. Dev. Biol. 367, 187–196 [DOI] [PubMed] [Google Scholar]
- 20. Sun G., and Irvine K. D. (2013) Ajuba family proteins link JNK to Hippo signaling. Sci. Signal. 6, ra81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hsueh Y. J., Chen H. C., Wu S. E., Wang T. K., Chen J. K., and Ma D. H. (2015) Lysophosphatidic acid induces YAP-promoted proliferation of human corneal endothelial cells via PI3K and ROCK pathways. Mol. Ther. Methods Clin. Dev. 2, 15014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Miller E., Yang J., DeRan M., Wu C., Su A. I., Bonamy G. M., Liu J., Peters E. C., and Wu X. (2012) Identification of serum-derived sphingosine-1-phosphate as a small molecule regulator of YAP. Chem. Biol. 19, 955–962 [DOI] [PubMed] [Google Scholar]
- 23. Mo J. S., Yu F. X., Gong R., Brown J. H., and Guan K. L. (2012) Regulation of the Hippo-YAP pathway by protease-activated receptors (PARs). Genes Dev. 26, 2138–2143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yu F. X., Zhao B., Panupinthu N., Jewell J. L., Lian I., Wang L. H., Zhao J., Yuan H., Tumaneng K., Li H., Fu X. D., Mills G. B., and Guan K. L. (2012) Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling. Cell 150, 780–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Levy D., Adamovich Y., Reuven N., and Shaul Y. (2008) Yap1 phosphorylation by c-Abl is a critical step in selective activation of proapoptotic genes in response to DNA damage. Mol. Cell 29, 350–361 [DOI] [PubMed] [Google Scholar]
- 26. Zhao B., Wei X., Li W., Udan R. S., Yang Q., Kim J., Xie J., Ikenoue T., Yu J., Li L., Zheng P., Ye K., Chinnaiyan A., Halder G., Lai Z. C., et al. (2007) Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Gene Dev. 21, 2747–2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wennmann D. O., Vollenbröker B., Eckart A. K., Bonse J., Erdmann F., Wolters D. A., Schenk L. K., Schulze U., Kremerskothen J., Weide T., and Pavenstädt H. (2014) The Hippo pathway is controlled by angiotensin II signaling and its reactivation induces apoptosis in podocytes. Cell Death Dis. 5, e1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Santinon G., Pocaterra A., and Dupont S. (2016) Control of YAP/TAZ activity by metabolic and nutrient-sensing pathways. Trends Cell Biol. 26, 289–299 [DOI] [PubMed] [Google Scholar]
- 29. Nagashima S., Bao Y., and Hata Y. (2016) The Hippo pathway as drug targets in cancer therapy and regenerative medicine. Curr. Drug. Targets, in press [DOI] [PubMed] [Google Scholar]
- 30. Gilbert L. I. (2004) Halloween genes encode P450 enzymes that mediate steroid hormone biosynthesis in Drosophila melanogaster. Mol. Cell Endocrinol. 215, 1–10 [DOI] [PubMed] [Google Scholar]
- 31. Boo K. H., Lee D., Jeon G. L., Ko S. H., Cho S. K., Kim J. H., Park S. P., Hong Q., Lee S. H., Lee D. S., and Riu K. Z. (2010) Distribution and biosynthesis of 20-hydroxyecdysone in plants of Achyranthes japonica Nakai. Biosci. Biotechnol. Biochem. 74, 2226–2231 [DOI] [PubMed] [Google Scholar]
- 32. Zhao X. F., An X. M., Wang J. X., Dong D. J., Du X. J., Sueda S., and Kondo H. (2005) Expression of the Helicoverpa cathepsin B-like proteinase during embryonic development. Arch. Insect Biochem. Physiol. 58, 39–46 [DOI] [PubMed] [Google Scholar]
- 33. Shao H. L., Zheng W. W., Liu P. C., Wang Q., Wang J. X., and Zhao X. F. (2008) Establishment of a new cell line from lepidopteran epidermis and hormonal regulation on the genes. PLoS One 3, e3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sui Y. P., Wang J. X., and Zhao X. F. (2009) The impacts of classical insect hormones on the expression profiles of a new digestive trypsin-like protease (TLP) from the cotton bollworm, Helicoverpa armigera. Insect Mol. Biol. 18, 443–452 [DOI] [PubMed] [Google Scholar]
- 35. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 36. Schmittgen T. D., and Livak K. J. (2008) Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3, 1101–1108 [DOI] [PubMed] [Google Scholar]
- 37. Liu W., Cai M. J., Zheng C. C., Wang J. X., and Zhao X. F. (2014) Phospholipase Cγ1 connects the cell membrane pathway to the nuclear receptor pathway in insect steroid hormone signaling. J. Biol. Chem. 289, 13026–13041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chehrehasa F., Meedeniya A. C., Dwyer P., Abrahamsen G., and Mackay-Sim A. (2009) EdU, a new thymidine analog for labelling proliferating cells in the nervous system. J. Neurosci. Methods 177, 122–130 [DOI] [PubMed] [Google Scholar]