Abstract
Glucocorticoids (GCs) used as inflammation suppressors have harmful side effects, including induction of hepatic steatosis. The underlying mechanisms of GC-promoted dysregulation of lipid metabolism, however, are not fully understood. GCs could facilitate the accumulation of myeloid-derived suppressor cells (MDSC) in the liver of animals, and the potential role of MDSCs in GC-induced hepatic steatosis was therefore investigated in this study. We demonstrated that granulocytic (G)-MDSC accumulation mediated the effects of GCs on the fatty liver, in which activating transcription factor 3 (ATF3)/S100A9 signaling plays an important role. ATF3-deficient mice developed hepatic steatosis and displayed expansion of G-MDSCs in the liver and multiple immune organs, which shared high similarity with the phenotype observed in GC-treated wild-type littermates. Adoptive transfer of GC-induced or ATF3-deficient G-MDSCs promoted lipid accumulation in the liver, whereas depletion of G-MDSCs alleviated these effects. Mechanistic studies showed that in MDSCs, ATF3 was transrepressed by the GC receptor GR through direct binding to the negative GR-response element. S100A9 is the major transcriptional target of ATF3 in G-MDSCs. Silencing S100A9 clearly alleviated G-MDSCs expansion and hepatic steatosis caused by ATF3 deficiency or GC treatment. Our study uncovers an important role of G-MDSCs in GC-induced hepatic steatosis, in which ATF3 may have potential therapeutic implications.
Keywords: cAMP response element-binding protein, glucocorticoid, immunosuppression, liver metabolism, S100 proteins
Introduction
Hepatic steatosis is characterized by abnormal lipid accumulation in hepatocytes, which was caused by the imbalance between lipogenesis, lipid catabolism, and free fatty acid uptake (1). Clinical administration of exogenous glucocorticoids (GCs)2 to patients, subjected to organ transplantation or suffering from severe inflammation-related diseases, is known to cause side effects, including deregulation of lipid metabolism and hepatic steatosis (2, 3). The mechanisms underlying GC-induced hepatic steatosis are still not fully understood. Recent studies have shown that administration of a synthetic GC dexamethasone (Dex) could cause the expansion of distinctive proinflammatory monocytes; they were CD11b+Gr-1+ and expressed signature molecules of tumor-induced myeloid-derived suppressor cells (MDSCs), both in humans and mice (4). Accumulation of immature myeloid (CD11b+Ly6C+Ly6G−) cells was also observed in mice fed a high fat diet (5). These observations indicate a possible relationship between GC-induced lipid metabolism and myeloid cell differentiation.
MDSCs represent a heterogeneous population consisting of immature myeloid cells and different stages of myeloid progenitors in which their full differentiation into mature myeloid cells has been prevented (6, 7). MDSC expansion has been observed under various pathological conditions, including but not limited to cancer inflammation, infections, and trauma (8–10). Functioning through suppression of other immune cells, MDSCs are believed to be one of the key brakes in the immune system (7). The clinical beneficial effect of GCs on the prevention of graft-versus-host reaction or excessive inflammation is attributed, at least in part, to immune suppressive function of MDSCs (11, 12).
To determine a potential association between MDSCs and GC-induced hepatic steatosis, we have previously performed gene expression analysis of MDSCs purified from liver of Dex-treated and vehicle control mice. We found that the expression of activating transcription factor 3 (ATF3) was dramatically down-regulated in MDSCs upon Dex treatment. ATF3, a basic leucine zipper transcription factor belonging to the ATF/cyclic AMP-response element-binding family is encoded by an adaptive-response gene, i.e. its expression is induced by extracellular or intracellular stress signaling (13, 14). Importantly, ATF3 has been demonstrated as an integrative factor in bridging metabolism, inflammation, and immunity (15).
In this study, we investigated the role of ATF3 in linking MDSCs and GC-induced hepatic steatosis. We demonstrate that MDSCs contribute to GC-induced hepatic steatosis through the ATF3/S100A9 axis. Targeting ATF3 may be promising in therapeutic application of lipid metabolic disorders.
Results
GC-induced Hepatic Steatosis Is Dependent on ATF3 Deficiency
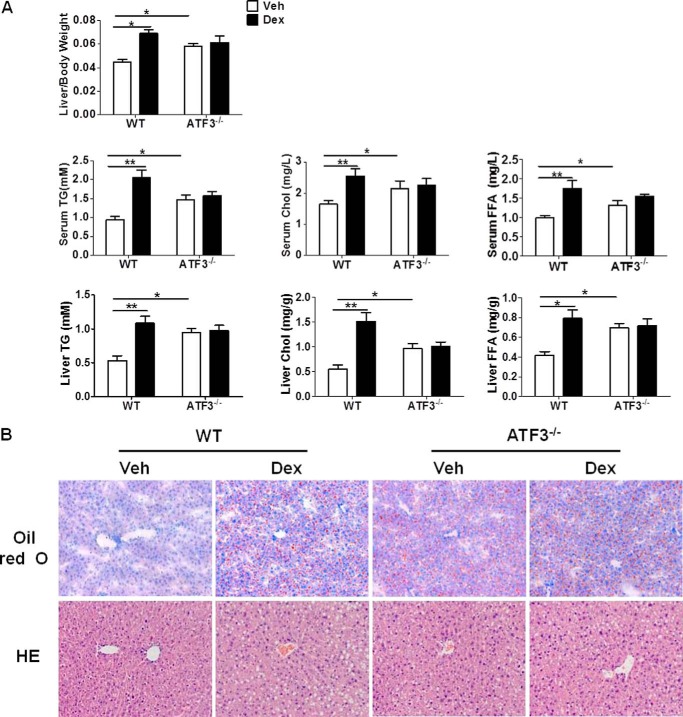
To gain insights into the potential role of ATF3 in GC-induced hepatic steatosis, female wild-type (WT) and ATF3-deficient (ATF3−/−) mice were intraperitoneally injected with synthetic GC, dexamethasone (Dex), or DMSO (Veh). Mice were analyzed for liver weight, as well as lipid contents in the liver and peripheral blood. As expected, in WT mice, Dex induced hepatomegaly and clear elevation of triglyceride, cholesterol, and free fatty acid contents in both liver and circulation. Interestingly, in the absence of Dex, ATF3−/− mice displayed significantly higher levels of liver weight and lipids than those of WT littermates, reaching comparable levels of Dex-treated WT controls. No further increases were observed in ATF3−/− mice after Dex administration (Fig. 1A). The development of liver steatosis was further confirmed by staining with lipid-specific Oil-Red O and hematoxylin/eosin, which revealed small fat droplets in liver sections from ATF3−/− and Dex-treated WT animals (Fig. 1B). The activity of aspartate aminotransferase and alanine aminotransferase in serum, indicative of liver damage, displayed similar changes with those of lipids (data not shown). These observations indicate that mice lacking ATF3 developed spontaneous hepatosteatosis and were refractory to Dex treatment, suggesting ATF3 deficiency may be important for Dex-induced hepatic steatosis.
FIGURE 1.
GC-induced hepatic steatosis is dependent on ATF3 deficiency. WT and ATF3−/− mice (n = 6) were injected with Dex (1 mg/kg weight/day) or DMSO (Veh) for 5 days. A, ratio of liver to body weight and lipid contents in the serum and liver were measured. TG, triglyceride; Chol, cholesterol; FFA, free fatty acids. Data represent the mean ± S.E. of all mice analyzed. *, p < 0.05; **, p < 0.01, unpaired t tests. B, representative results of Oil-Red O staining and hematoxylin and eosin (HE) staining of liver section. Original magnification, ×200.
GC-induced MDSC Accumulation Is Dependent on ATF3 Deficiency
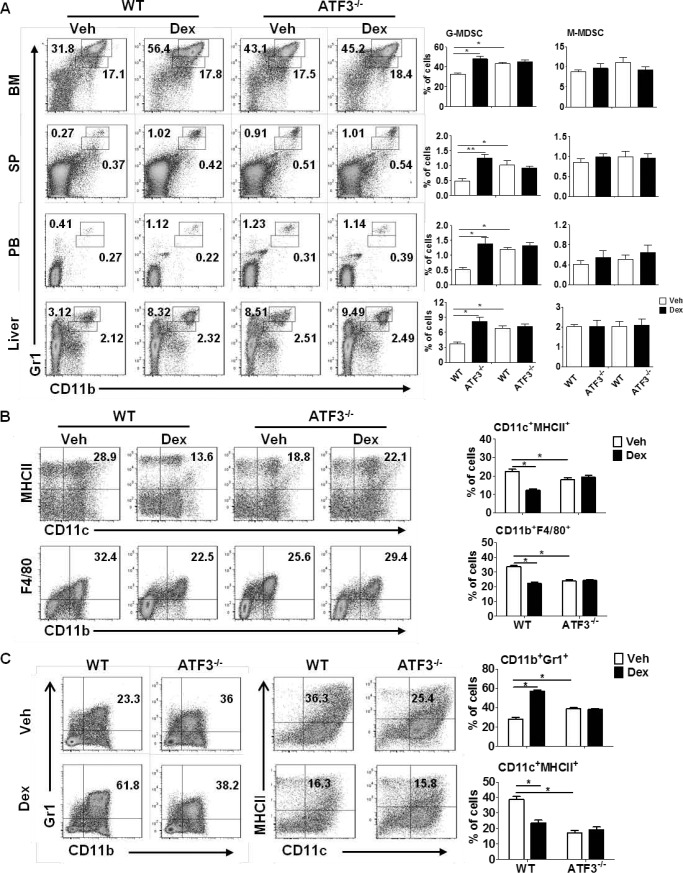
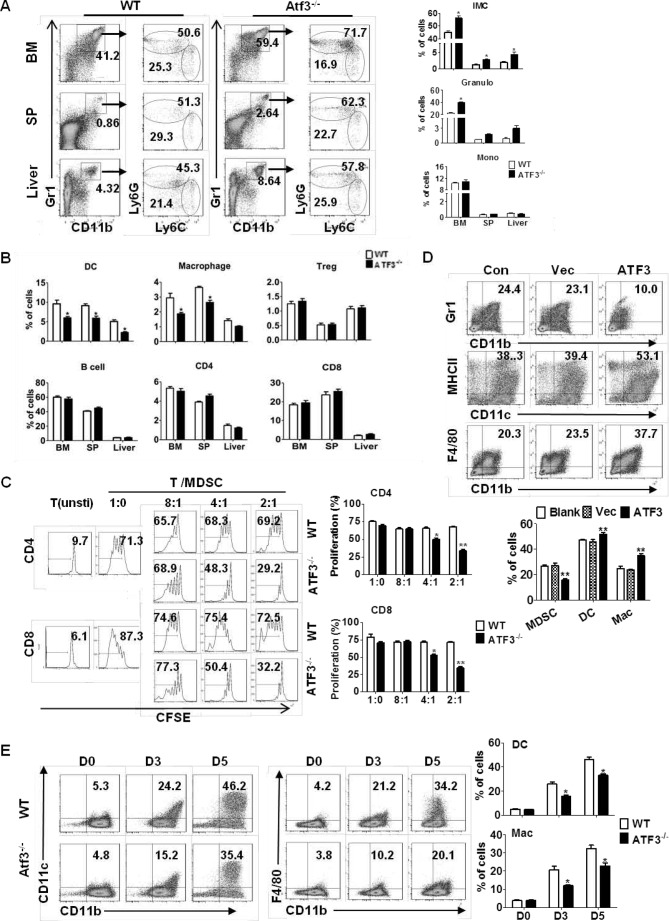
To investigate whether MDSCs contributed to the development of hepatic steatosis, flow cytometric analysis was performed in WT and ATF3−/− mice before and after Dex treatment. The CD11b+Gr1high and CD11b+Gr1inter populations corresponded to granulocytic (G)-MDSCs (CD11b+Ly6G+Ly6C−) and monocytic (M)-MDSCs (CD11b+Ly6G−Ly6C+) subsets, respectively (data not shown). Consistent with a previous report (16), Dex treatment caused significant increase in the frequency of MDSCs in WT mice, including liver and multiple immune organs. Further analysis showed that this increase was due to G-MDSCs but not M-MDSCs (Fig. 2A). ATF3 deficiency caused a 2–3-fold increase in the proportion of G-MDSCs in multiple tissues, reaching the levels of Dex-treated WT animals; however, no further increase was observed upon Dex administration in ATF3−/− mice. The levels of M-MDSCs did not display any noticeable difference among distinct groups (Fig. 2A). In line with the changes in MDSCs, the proportions of mature myeloid cells, including conventional dendritic cells (DCs, CD11c+MHCII+) and macrophages (CD11b+F4/80+), were lower in Dex-treated WT and Veh-ATF3−/− mice, compared with those in Veh-treated WT control (Fig. 2B). In line with these observations, the levels of mature myeloid cells upon Dex treatment was observed only in WT BM cells and not in ATF3−/− mice (Fig. 2B). As a further confirmation, BM cells lacking ATF3 displayed stronger capability in the generation of MDSCs than WT control in vitro assays. Dex promoted MDSC accumulation from WT BM as expected, but no effect was observed for cells from ATF3−/− mice (Fig. 2C), which were accompanied by a reduction in the mature CD11c+MHCII+ DC cells upon Dex treatment or ATF3 deletion (Fig. 2C). These results indicate that ATF3 deficiency may be required for GC-induced MDSC accumulation.
FIGURE 2.
GC-induced MDSC accumulation is dependent on ATF3 deficiency. A and B, WT and ATF3−/− mice were injected with dexamethasone (Dex) or DMSO (Veh) for 5 days (n = 6). A, levels of MDSC subsets in BM, spleen (SP), peripheral blood (PB), and liver were evaluated by flow cytometric analysis. B, population of DC (CD11c+MHCII+) and macrophage (CD11b+F4/80+) subsets in BM were evaluated by flow cytometric analysis. A and B, left flow panels are representative results, and the right graphs were mean ± S.E. from all mice analyzed. C, BM cells were cultured in medium containing GM-CSF, in the presence of Dex or Veh. The frequencies of MDSCs (CD11b+Gr1+) and DCs (CD11c+MHCII+) were analyzed by flow cytometry on day 5. Both representative results (left) and mean ± S.E. from three independent experiments (right) are shown. *, p < 0.05, unpaired t tests.
ATF3 Mediates the Effect of G-MDSCs on GC-induced Hepatic Steatosis
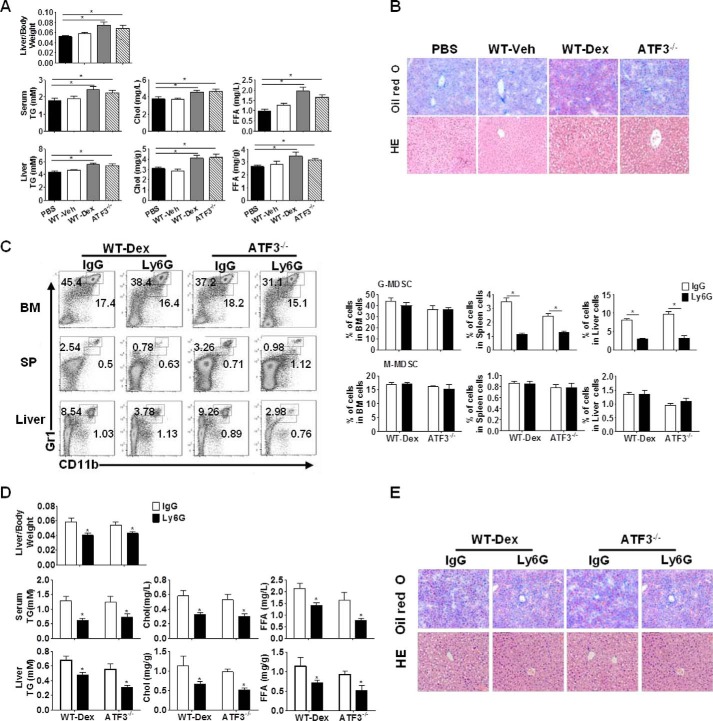
To determine whether a causal relationship exists between MDSC and hepatic lipid accumulation in response to GC administration, adoptive transfer of G-MDSCs was performed. Results showed that G-MDSCs from Dex-treated WT or Veh-treated ATF3−/− mice caused a significant increase in liver weight, as well as lipid contents in WT recipient mice. The corresponding control from Veh-treated WT mice, however, did not elicit any effects (Fig. 3A). Purified M-MDSCs from these groups failed to display any effects (data not shown). The hepatic lipid accumulation was further confirmed by Oil-Red O and hematoxylin/eosin staining (Fig. 3B). We also tested the survival of MDSCs in the adoptive transfer assay. Results showed that G-MDSCs from donor mice (CD45.2+) proliferate at the same rate as those from recipient mice (CD45.1+), indicating that they survived normally in vivo (data not shown). The role of G-MDSCs in the development of hepatic steatosis was further corroborated when G-MDSCs were depleted using anti-Ly6G antibody. The 2–3-fold reduction in the G-MDSC population in liver clearly alleviated steatosis symptoms in both Dex-treated WT and ATF3−/− mice (Fig. 3, C–E). Therefore, these results further determined the important role of G-MDSCs in the development of GC-induced hepatic steatosis and represent a novel mechanism of GC-induced fatty liver, in which ATF3 deficiency may be required.
FIGURE 3.
ATF3 mediates the effect of G-MDSCs on GC-induced hepatic steatosis. A and B, G-MDSC adoptive transfer: G-MDSCs (3 × 106) from WT, Dex-treated WT (WT-Dex), or ATF3−/− mice were adoptively transferred into WT receipts (n = 6); mice were sacrificed at day 6. PBS was used as negative control. A, ratio of liver to body weight and the lipid contents. B, representative Oil-Red O and hematoxylin/eosin (HE) staining of liver sections were shown. C–E, G-MDSCs depletion assay: anti-Ly6G or anti-IgG antibodies were injected intravenously into Dex-treated WT or Veh-treated ATF3−/− mice, as described under “Experimental Procedures.” C, proportions of MDSCs were evaluated by flow cytometric analysis. SP, spleen. D, ratio of liver to the body weight, and lipid contents in the serum and liver tissue. E, representative Oil-Red O staining and hematoxylin and eosin (HE) staining of liver tissue. B and E, original magnification, ×200. A, C, and D, data represent the mean ± S.E. of six mice per group of four independent experiments. *, p < 0.05, unpaired t tests.
ATF3 Is Transrepressed by GR in MDSCs
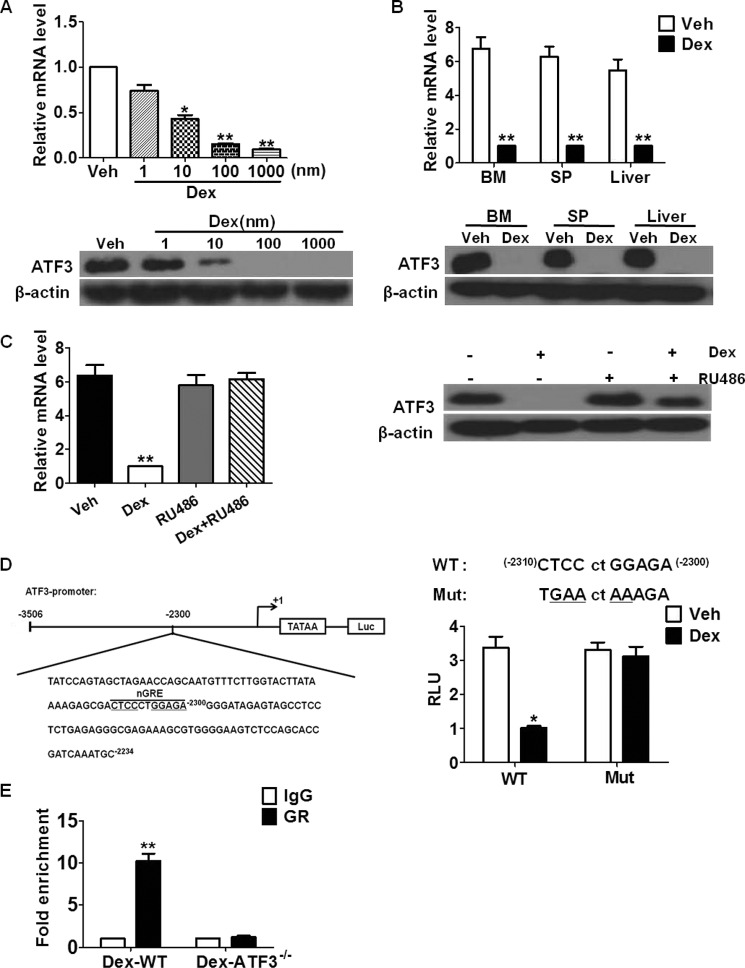
Based on the observations that ATF3 deficiency may be a prerequisite for GC-induced MDSC expansion and fatty liver, we hypothesized that GC might transrepress ATF3 in myeloid cells. It was found that Dex inhibited ATF3 expression in cultured BM cells in a concentration-dependent manner, both at mRNA and protein levels (Fig. 4A). This observation was confirmed in purified MDSCs from BM, spleen, and liver (Fig. 4B), whereas no effects were detected in T cells from these tissues (data not shown). This indicates that suppression of ATF3 by GC was limited to myeloid cells. Pretreatment with the GR antagonist RU486 completely abrogated this effect (Fig. 4C), supporting that the suppression is GR-dependent. Binding with negative GR-response elements (nGREs, with core sequence CTCCnnGGAG) mediates the transrepression of targets by GR (17). Detailed analysis of the ATF3 regulatory region revealed a potential nGRE located between −2300 and −2310 bp from the transcription start site (Fig. 4D). Transfection with ATF3 luciferase reporter plasmid containing the predicted nGRE showed that GC down-regulated the activity of the ATF3 promoter in 32D myeloid cells, whereas site-directed mutation of nGRE completely abolished this repression (Fig. 4D). As expectedly, RU486 abrogated the effect of Dex on the ATF3 reporter (data not shown). Further chromatin immunoprecipitation (ChIP) assay confirmed the binding of GR protein to nGRE located within the ATF3 regulatory region, and no signals were detected in BM cells from ATF3−/− mice, as expected (Fig. 4E). Overall, these results revealed that GR transrepresses ATF3 in myeloid cells.
FIGURE 4.
ATF3 is transrepressed by GR in MDSCs. A, BM cells were treated with Dex at indicated concentrations, and ATF3 expression was measured by qRT-PCR (upper) and WB (lower). B, ATF3 expression in sorted MDSCs from BM, spleen (SP), and liver was determined by qRT-PCR (upper) and Western blotting (lower). C, BM cells were treated as indicated, ATF3 expression was measured by qRT-PCR (left) and Western blotting (right). RU486, GR antagonist. D, left, scheme of ATF3 promoter sequence with potential nGRE indicated. Right, site-directed mutant (Mut): ATF3-Luc activity, 32D cells transfected with pATF3/Luc plasmid were treated with Dex, and luciferase activity was measured using the Dual-Luciferase Reporter Assay (Promega). RLU, relative luciferase unit. E, ChIP assay was performed on BM cells from Dex-treated WT or ATF3−/− mice; using anti-GR or anti-IgG antibody, the presence of ATF3 promoter harboring the nGRE was determined by qRT-PCR. Data were normalized against input and presented as fold increase over IgG control. A–E, results were graphed as mean ± S.E. from three independent experiments; Western blottings in A–C were representative data from three independent experiments, each sample was pooled from three mice. *, p < 0.05; **, p < 0.01, unpaired t tests were used.
ATF3 Regulates the Development of G-MDSCs
The participation of ATF3 in MDSC-mediated hepatic steatosis in response to GC suggests a potential role of ATF3 in the differentiation of myeloid cells. Flow cytometric analysis showed that the level of CD11b+Gr1+ immature myeloid cells (iMCs) was elevated in the tissues from ATF3−/− mice, with a more than 2-fold increase in the liver and spleen. The increase was mostly provided by CD11b+Ly6G+Ly6C− granulocytes and not CD11b+Ly6G−Ly6C+ monocytes (Fig. 5A). In line with the increase in iMCs, the proportion of mature myeloid cells, including CD11c+MHCII+ dendritic cells and CD11b+F4/80+ macrophages, was clearly lower in ATF3−/− mice than in their WT littermates. In contrast, the composition of lymphocyte population, including total T and B cells, as well as regulatory T cells, was not impacted by ATF3 deficiency (Fig. 5B). The major characteristic of MDSCs is their suppressive activity (18). To further prove the elevated CD11b+Ly6G+Ly6C− cells were G-MDSCs, we co-cultured CD11b+Ly6G+Ly6C− cells with autologous T cells to evaluate their suppressive activity. Results showed that splenic CD11b+Ly6G+Ly6C− cells from ATF3−/−mice were suppressive toward T cells, indicating that they were G-MDSCs. Consistent with a previous report (19), the corresponding control iMCs from WT mice had no effect (Fig. 5C). The reactive oxygen species (ROS) content, and the expression of p47phox (component of the NADPH oxidase complex responsible for ROS production), was significantly higher in G-MDSCs from ATF3−/− mice in both mRNA and protein levels, when compared with WT control (data not shown). Administration of ROS inhibitor N-acetyl-l-cysteine completely abrogated the suppressive activity of MDSCs in ATF3−/− mice (data not shown). These observations indicate that ATF3 deficiency facilitated the activation of MDSC in an ROS-dependent manner.
FIGURE 5.
ATF3 regulates the development of G-MDSCs. A, proportions of the indicated myeloid cell populations in WT and ATF3-KO mice were analyzed by flow cytometry. iMC (CD11b+Gr1+), Granulo (granulocytes, CD11b+Ly6G+Ly6C−), and Mono (monocytes, CD11b+Ly6G−Ly6C+) are shown. B, proportions of indicated immune cells in WT and ATF3-KO mice were determined by flow cytometric analysis: DC (CD11c+MHCII+), macrophage (CD11b+Gr1−F4/80+), Treg (CD4+CD25+), B cell (B220+), CD4 T (CD3+CD4+), and CD8 T (CD3+CD8+). Con, control. C, allogeneic mixed lymphocytes reaction (MLR). CD3+T cells were stimulated with ConA and cocultured with allogeneic splenic CD11b+Ly6G+ at different ratios for 3 days. T cell proliferation was evaluated by carboxyfluorescein diacetate, succinimidyl ester (CFSE) dilution, and unstimulated T cells were used as negative control. D, BM cells were infected with lentivirus expressing ATF3 or vector (Vec) and cultured in medium containing GM-CSF and IL6. The frequencies of MDSCs (Gr1+CD11b+), DCs (CD11c+MHCII+), and macrophages (CD11b+F4/80+) among GFP + cells were analyzed by flow cytometry at day 6. Medium alone was used as control. E, splenic MDSCs were cultured with GM-CSF and IL-4 for 3 and 5 days, and the proportions of DCs (CD11c+CD11b+) and macrophages (CD11b+F4/80+) were determined. A–D, both representative data and mean ± S.E. from six mice (A) or three independent experiments (B–D) are shown. *, p < 0.05; **, p < 0.01, unpaired t tests. SP, spleen.
The apoptosis and proliferation of MDSC, as revealed by annexin-V or Ki67 staining, remained unchanged in the presence or absence of ATF3 (data not shown), indicating that the elevation of G-MDSCs was not caused by cell survival. We next evaluated whether ATF3 regulates the development of G-MDSCs. Lentiviral overexpression of ATF3 caused a reduction in the percentage of MDSCs generated from cultured bone marrow cells (Fig. 5D). MDSCs could be induced to differentiating into mature myeloid cells under proper conditions (20). Splenic CD11b+Gr1+ cells with ATF3 deletion clearly had a lower ability to differentiate into mature myeloid cells when cultured with granulocyte-macrophage colony-stimulating factor (GM-CSF) and IL-4 (Fig. 5E). These results collectively indicate that ATF3 represent a novel regulator in the development and activation of MDSCs.
S100A9 Is a Novel Target of ATF3 in G-MDSCs
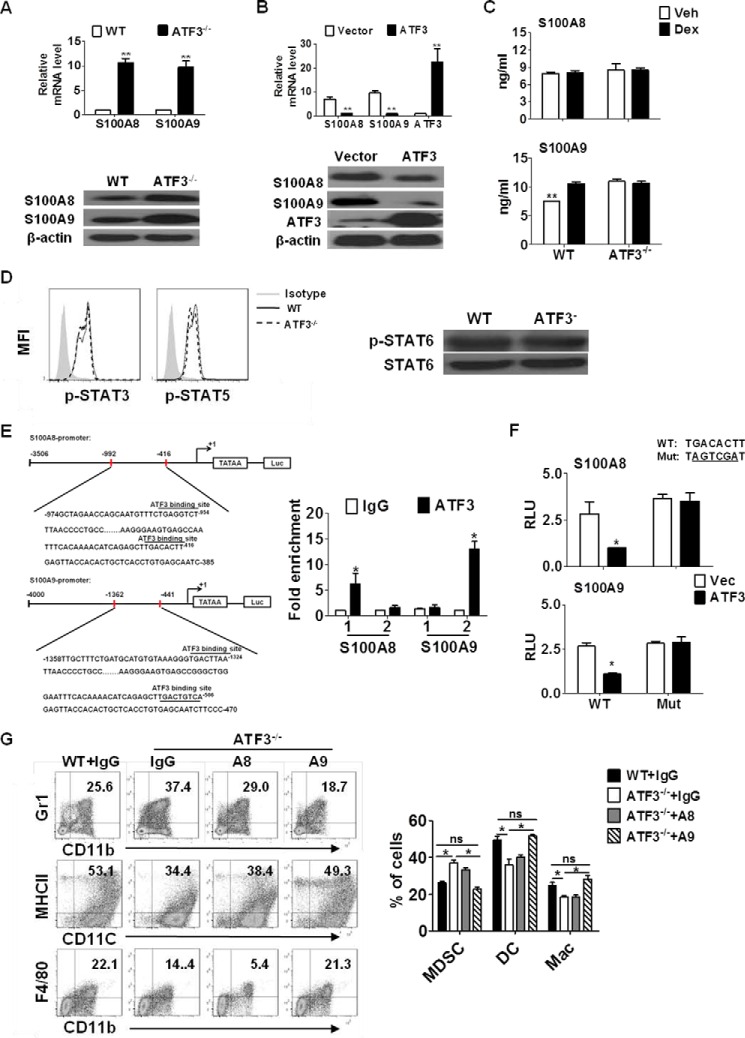
To investigate the mechanism underlying the role of ATF3 in MDSCs, gene expression analysis was performed to screen the potential targets of ATF3 by qRT-PCR. S100A8 and S100A9, known to be involved in myeloid cell differentiation (21), were significantly up-regulated in ATF3-deficient MDSCs, both at the mRNA and protein levels (Fig. 6A). Other MDSC-related genes, including GM-CSF and IL-6, did not show noticeable differences in the presence or absence of ATF3 (data not shown). The effect was further confirmed when lentiviral overexpression of ATF3 led to down-regulation of S100A8 and S100A9 in MDSCs (Fig. 6B). The amounts of S100A8/9 in serum from Dex-treated WT and ATF3−/− mice were elevated, as compared with WT controls (Fig. 6C). Heterodimer formed by S100A8 and S100A9, functioning as transcriptional targets of STAT3, has been demonstrated to be important in MDSC accumulation and migration in cancer (22). The phosphorylation of STAT3 and other STAT proteins, however, remained largely unchanged in MDSCs, regardless of ATF3 expression (Fig. 6D), indicating that the regulation of S100A8 and S100A9 by ATF3 is independent of STAT3.
FIGURE 6.
S100A9 mediates the effect of ATF3 on G-MDSCs. A, expression of S100A8/A9 in splenic MDSCs was determined by qRT-PCR (upper) and Western blotting (below). B, BM cells were infected with lentivirus expressing ATF3 or vector (with GFP tag); gene expression in GFP+ cells was evaluated by qRT-PCR (upper) and Western blotting (below). C, levels of S100A8/A9 protein in the serum from the indicated animals were measured by ELISA. D, phosphorylation of STAT3 and STAT5 in splenic MDSCs was measured by flow cytometry (left), the STAT6 phosphorylation was evaluated by Western blotting (right). MFI, mean fluorescence intensity. E, scheme of S100A8/A9 promoters with potential ATF3-binding sites as indicated. S100A8: site 1, −416 to −425 bp, and site 2, −954 to −961 bp; S100A9: site 1, −506 to −515 bp, and site 2, −1324 to −1331 bp. Overexpressed ATF3 lentivirus were infected with WT BM cells for 3 days, and then cells were fixed in 1% paraformaldehyde for ChIP assay. The presence of S100A8/A9 promoters harboring the ATF3-binding sites was determined by qRT-PCR. Data were normalized against input and presented as fold increase over IgG control. F, S100A8/A9-Luc activity, 32D cell transfected with pS100A8/Luc or pS100A9/Luc plasmid and then were infected with overexpressed ATF3 lentivirus. Luciferase activity was measured using the Dual-Luciferase Reporter Assay (Promega). G, BM cells were cultured with GM-CSF and IL6 in presence or absence of neutralizing antibodies against S100A8 or S100A9, and anti-IgG was used as control. The percentages of the indicated populations were analyzed by flow cytometry. A–C and E–G, results were mean ± S.E. from three independent experiments; Western blots in A, B, and D are representative data from three independent experiments, each sample was pooled from three mice. *, p < 0.05; **, p < 0.01, unpaired t tests were used. ns, no significance; RLU, relative luciferase units; Mac, macrophage.
Further sequence analysis revealed two potential ATF/cAMP-response element consensus sites within the promoters of S100A8 and S100A9 (Fig. 6E). Chromatin immunoprecipitation (ChIP) assay showed that ATF3 bound to canonical ATF3-binding sites located in the promoters of S100A8 (−416 to −425 bp) and S100A9 (−1324 to −1331 bp) (Fig. 6E). Further luciferase reporter assays using 32D cell line confirmed that the corresponding binding sites mediate the regulation of S100A8/A9 by ATF3, whereas site-directed mutation completely abolished this effect (Fig. 6F). Neutralizing antibodies against S100A8 or S100A9 were administered to cultured BM cells. Results showed that blocking S100A9 reversed the effect of ATF3 on MDSC levels, whereas neutralization of S100A8 produced a limited effect (Fig. 6G). These results demonstrate that S100A8 and S100A9 are transcriptional targets of ATF3, and S100A9 play a major role in mediating the effect of ATF3 on MDSCs.
S100A9 Silencing Alleviates GC-induced Hepatosteatosis
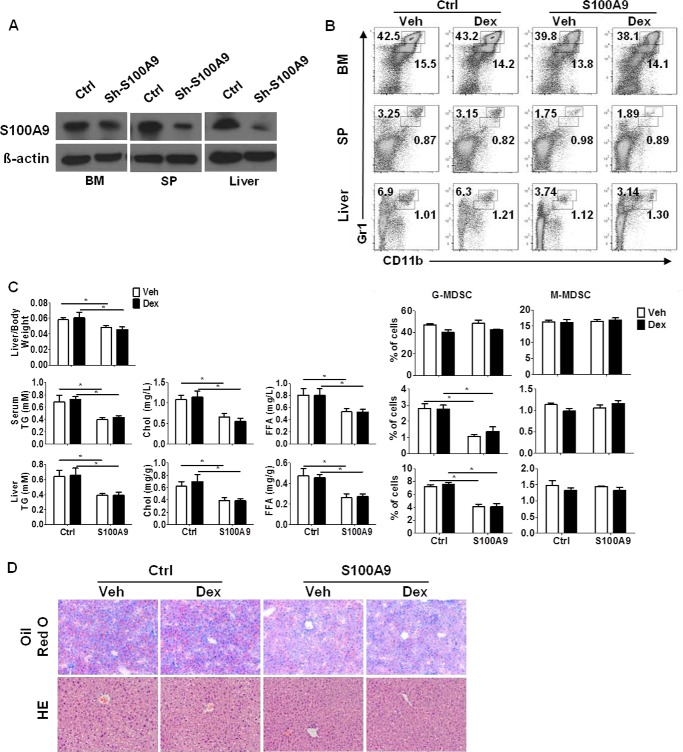
We next determined whether silencing S100A9 could alleviate hepatic steatosis under either Dex treatment or ATF3 deficiency conditions. For this purpose, lentiviral particles carrying S100A9-specific short hairpin (sh) RNA or scrambled shRNA were intravenously injected into ATF3−/− or Dex-treated WT mice. Knockdown of S100A9 in MDSCs was confirmed by Western blotting (Fig. 7A). A clear reduction of G-MDSCs, in both the liver and immune organs, was observed upon S100A9 silencing in ATF3−/− mice (Fig. 7B) and Dex-treated WT mice (data not shown). The lipid accumulation in the liver and serum was alleviated as expected (Fig. 7, C and D). Meanwhile, Dex failed to display any effect on hepatic steatosis after silencing S100A9 in WT animals (data not shown). These results demonstrate that ATF3/S100A9 mediates the effect of MDSCs on hepatic steatosis caused by GC administration.
FIGURE 7.
S100A9 silencing alleviates GC-induced hepatosteatosis. Lentiviral particles carrying S100A9-shRNA or scrambled control were intravenously injected into ATF3−/− mice, and then mice were injected with Dex or DMSO (Veh). A, knockdown of S100A9 expression in sorted MDSCs was determined by Western blotting. B, proportions of MDSC subsets were determined by flow cytometric analysis; both representative data (left) and mean ± S.E. from 6 mice (right) were included. C, ratio of liver to the body weight and the lipid contents in the serum and liver tissue were measured. Data were presented as mean ± S.E. from six mice. D, representative Oil-Red O and hematoxylin and eosin (HE) staining of liver sections. Original magnification, ×200. *, p < 0.05, unpaired t tests were used. SP, spleen; Ctrl, control.
TLR4−/− Mice Is Sensitive to GC-induced Hepatosteatosis
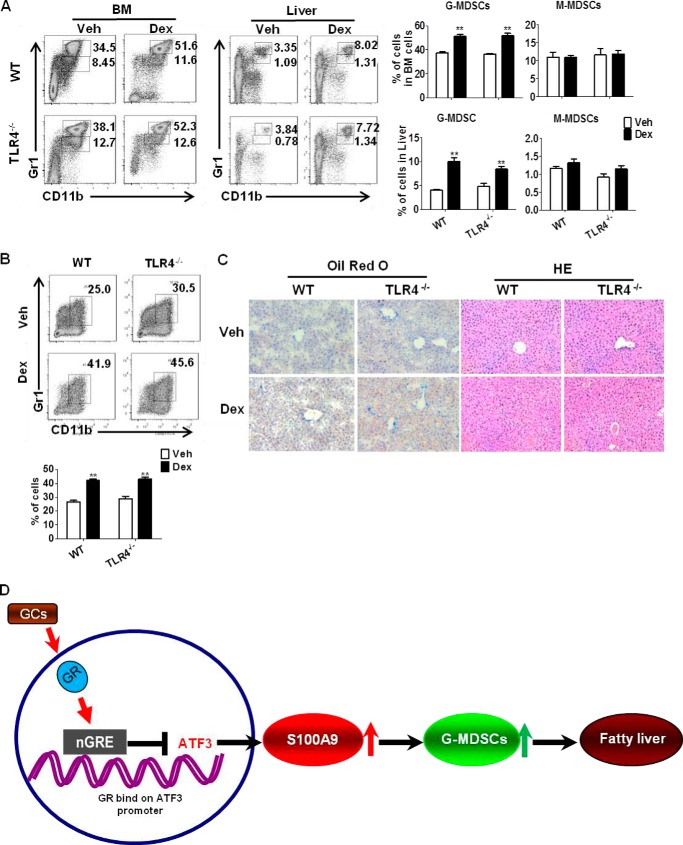
Toll-like receptor (TLR) signaling, especially TLR4, plays an important role in mediating the anti-inflammatory and immunosuppressive effects of glucocorticoids (23). Activation of TLRs is associated with liver diseases, including alcoholic liver injury, liver fibrosis, and liver cancer (24, 25). We therefore investigated the potential role of TLRs in GC-induced hepatosteatosis. TLR4 was chosen due to its recognition in inflammation and liver diseases (26). Flow cytometric analysis showed that G-MDSC (CD11b+Gr1high) populations were significantly increased in TLR4−/− mice, upon Dex treatment (Fig. 8A), which is consistent with the results from in vitro culture (Fig. 8B). No difference was observed between TLR4−/− and WT controls mice (Fig. 8, A and B). The development of steatosis liver in Dex-treated TLR4−/− mice was further confirmed by staining with lipid-specific Oil-Red O and hematoxylin/eosin (Fig. 8C). These observations showed that deficiency of TLR4 failed to impair the effects of Dex on G-MDSCs and fatty liver, indicating that TLR4 is dispensable for the GC-induced hepatosteatosis. However, we have to point out that the possibility of other TLRs in GC-induced hepatosteatosis could not be excluded at this time.
FIGURE 8.
TLR4−/− mice are sensitive to GC-induced hepatosteatosis. A, WT, TLR4−/− mice were injected with Dex or DMSO (Veh) for 5 days (n = 6). The levels of MDSCs subsets in BM and liver were evaluated by flow cytometric analysis. The left flow panels are representative results; the right graphs show mean ± S.E. from all mice analyzed. B, BM cells were cultured in medium containing GM-CSF, in the presence of Dex or Veh. The frequencies of MDSCs were analyzed by flow cytometry on day 5. C, representative results of Oil-Red O staining and hematoxylin and eosin (HE) staining of liver section. Original magnification, ×200. *, p < 0.05, unpaired t tests were used. D, scheme depicting ATF3 mediated GC-induced hepatic steatosis. The mechanism underlying the role of MDSCs in GC induced hepatic steatosis. Activation of GR by GC represses ATF3 transcription by binding to nGRE, which leads to transactivation of S100A9. The induction of S100A9 causes the expansion and activation of G-MDSCs and hepatic lipid accumulation.
In summary, we uncovered the mechanism underlying the role of MDSCs in GC-induced hepatic steatosis. Upon activation by GC, GR represses ATF3 transcription by binding to nGRE, which leads to transactivation of S100A9, the important pro-inflammatory molecule and regulator of myeloid cell differentiation. The induction of S100A9 causes expansion and activation of G-MDSCs and hepatic lipid accumulation. Therefore, our study uncovers an important role of G-MDSCs in GC-induced hepatic steatosis, in which ATF3 may have potential therapeutic implications (Fig. 8D).
Discussion
GC is a widely prescribed anti-inflammatory and immunosuppressive medicine in clinics. The side effects of GC, mostly dysregulated lipid metabolism and fatty liver, remain the major concern in patients (27). Understanding the mechanism of GC-induced hepatic steatosis is expected to provide an intervention target to avoid this side effect. In this study, we demonstrated that G-MDSCs represent a novel player in promoting GC-induced hepatic steatosis, in which ATF3/S100A9 is the dominant signaling event.
MDSCs, characterized by their immunosuppressive capability, have been well studied in a variety of diseases, especially in cancer (7). Studies have shown that liver is one of the major sites of MDSCs, in addition to the immune organs (28). Accumulation of MDSC in liver has been observed in tumor-bearing hosts, hepatitis B virus-infected individuals, and high fat diet-fed animals (4, 29, 30). Also, hepatic stellate cells were reported to support the expansion of MDSCs in vitro (31). Our study reveals the importance of MDSCs in the modulation of lipid metabolism in liver. These studies collectively support that MDSCs are an active player in hepatic immunity.
The origin of hepatic MDSCs remains to be elucidated. It is possible that MDSCs could migrate to the liver from bone marrow in response to chemokines released by hepatocytes or liver stroma cells. Alternatively, MDSCs could differentiate from hematopoietic stem or progenitor cells residing in the liver. In this study, we found that adoptive transfer of MDSCs resulted in their homing to the liver and spleen, which supports the first possibility. However, MDSC differentiation within the liver cannot be excluded.
The exact role of MDSCs in liver is context-dependent. MDSC could facilitate tumor escape by down-regulation of anti-tumor immunity in both the liver and circulation (32). In another aspect, MDSCs in liver could protect against ConA-induced T cell-mediated hepatitis (33). Accumulated immature myeloid cells in mouse liver caused by a high fat diet could induce apoptosis of natural killer cells by releasing some pro-inflammatory cytokines (5). In this study, we found that G-MDSCs in liver could promote lipid accumulation and fatty liver in the GC treatment. The mechanism underlying MDSC-promoted lipid dysregulation deserves further investigation. It is plausible that MDSCs could secrete some pro-inflammatory cytokines, such as S100A9, and cause inflammatory responses and the dysfunction of hepatocytes, which finally leads to lipid accumulation in liver. Cumulatively, these findings indicate that the beneficial or detrimental activities of MDSCs in the liver may be context-dependent.
S100A9 belongs to the S100 family of calcium-binding proteins and functions as a pro-inflammatory danger signal transducer in the form of a homodimer or heterodimer with S100A8. The S100A9 protein is released by myeloid cells in response to cell damage, infection, or inflammation (34). Functioning as transcriptional target of STAT3, S100A9 could facilitate MDSC accumulation and migration in cancer (21). Our study shows that S100A9 can be transrepressed by ATF3, a well known stress-responsive protein. This reveals a novel mechanism of S100A9 regulation in myeloid cells, suggesting that expression of S100A9 may be responsive to stress.
ATF3 has been demonstrated as a key intersection point in linking lipid metabolism, inflammation, and immune responses. ATF3 functions as a negative regulator of the inflammatory responses in macrophages by antagonizing TLR-induced NF-κB signaling (35), and ATF3 deletion increases lipid accumulation and foam cell formation through epigenetic repression of the cholesterol 25-hydroxylase gene Ch25h (36). ATF3 has been demonstrated to mediate the anti-inflammatory and metabolic activities of high density lipoprotein (HDL) by down-regulating TLR-induced proinflammatory cytokines (37). Here, we provide novel evidence that ATF3 plays an important role in linking lipid metabolism and immunity through regulation of MDSC development and activation.
The role of ATF3 in neutrophils biology has recently been reported. ATF3 was shown to be important for neutrophil migration in the lungs via transrepression of CXCL1 chemokine (38). Another study reported that ATF3 functions as a novel negative regulator of neutrophil differentiation by modulating Ly6G expression (39). Consistently, we found that ATF3 deletion promoted the increase in CD11b+Ly6G+ cells and their acquisition of immunosuppressive functions, which characterizes this cell population as G-MDSCs and not neutrophils. The accumulation of G-MDSCs in ATF3-deficient mice was also confirmed in a tumor-bearing mouse model (data not shown). Our study provides further insights into the role of ATF3 in the differentiation of myeloid cells.
The clinical significance of our data remains to be investigated. The evaluation of the MDSC population in liver biopsies of patients with GC-induced hepatic steatosis would be helpful; however, given that GC-treated patients usually suffer from complicated disorders and may take other medications, the results of clinical studies should be carefully analyzed in view of confounding factors.
Experimental Procedures
Mouse Strain
All experiments performed on mice were approved by the Institutional Animal Care and Use Committee of Sun Yat-Sen University. ATF3−/− mice (background C57BL/6) were kindly provided by Dr. Tsonwin Hai (Ohio State University, Columbus, OH). Female TLR4−/− mice (background C57BL/6) and the congenic female C57BL/6SJL strain (C57BL/6, CD45.1 alloantigen) were obtained from Nanjing Biomedical Research Institute of Nanjing University and The Jackson Laboratory (Bar Harbor, ME), respectively. All mice were housed in a specific pathogen-free facility, and age-matched wild-type littermates (6–8 weeks) were used as controls. All mice were given normal feed diet.
Reagents
Dexamethasone, mifepristone (RU486), N-acetyl-l-cysteine, l-arginine, ConA, and dimethyl sulfoxide were purchased from Sigma. GM-CSF was obtained from PeproTech (Rocky Hill, NJ). The antibodies against ATF3, S100A8, S100A9, p47phox, Arg1, STAT6(Tyr-641, catalog no. 9361), p-STAT6, GR (Cell Signaling Technology, catalog no. 12041), β-actin, and HRP-conjugated secondary antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA) or Cell Signaling Technology. NW-hydroxy-nor-arginine and l-NG-monomethyl-arginine were obtained from BioVision (Milpitas, CA). The following fluorescein-conjugated anti-mouse antibodies were obtained from eBioscience (San Diego): CD11c-PE-Cy5, MHC-II-PE, Gr-1-PE-Cy7, Gr-1-PE, Ly-6C-PerCP-Cyanine5.5, Ly6G-PE, B220-PE, CD11b-FITC, CD11b-PE-Cy7, F4/80-PerCP-Cyanine5.5, CD3e-FITC, CD4-PE, CD25-PE-Cy7, CD8a-PE-Cy5, CD8a-PE-Cy7, FITC-p-STAT3 (Tyr-705(D3A7), catalog no. 9145), PE-cy7-p-STAT5(Tyr-694(D47E7), catalog no. 4322), and the corresponding IgG isotype antibodies.
5-Bromo-2-deoxyuridine (BrdU) was from BD Biosciences. S100A8 and S100A9 neutralizing antibodies were purchased from R&D Systems (Minneapolis, MN). The shRNAs for S100A9 and the control shRNA were purchased from Cell Signaling Technology (Beverly, MA). In vivo-jetPEITM was from PolyPlus-Transfection, Illkirch, France. Lipofectamine 2000, 5,6-carboxyfluorescein diacetate succinimidyl ester, and the reagents for cell culture were from Invitrogen. Anti-Ly6G antibody (IA8) or anti-IgG antibody was from eBioscience (San Diego).
Flow Cytometric Analysis and Sorting
Flow cytometric analysis and sorting were performed as described previously (40). Briefly, single-cell suspensions were prepared and stained with fluorochrome-conjugated antibodies. Data were collected on an LSRII flow cytometer (BD Biosciences, San Jose, CA) and analyzed with FlowJo software (Tree Star, Ashland, OR). Data were acquired as the fraction of labeled cells within a live-cell gate set for 50,000 events. For flow cytometric sorting, cells were stained with specific antibodies and isolated on a FACS Aria cell sorter (BD Bioscience). The purity after sorting was greater than 95%.
In Vivo Dex Treatment
Female 8-week-old mice were intraperitoneally injected with Dex (1 mg/kg body weight/daily, 5 days) (41). The symptoms of hepatic steatosis and the levels of immune cells were analyzed on day 6. For blocking of T cell apoptosis, IL-7 (5 μg/kg body weight) was intraperitoneally injected with or without Dex (42).
MDSC Adoptive Transfer
Adoptive transfer of G-MDSCs or M-MDSCs was performed as described previously (43). Briefly, MDSC subsets were isolated from spleen of ATF3-KO or Dex-treated WT mice by flow cytometric sorting, followed by tail vein injection (3 × 106) into WT recipients on days 1 and 3. The symptoms of hepatic steatosis were evaluated on day 6. G-MDSCs (3 × 106) from Dex-treated WT (WT-Dex) or Veh-treated ATF3-KO mice were adoptively transferred into WT CD45.1 recipient mice, and recipient mice were injected with BrdU at days 5. The levels of CD45.2+ cell, CD11b+Gr1+ cell, and BrdU+ cell were evaluated by flow cytometry.
MDSC Depletion
G-MDSC depletion was performed as described previously (44). Briefly, anti-Ly6G antibody (1A8) or anti-IgG antibody (rat IgG2b) was injected (120 μg per injection) through the tail vein on days 0 and 3 at post-Dex injection. The levels of immune cells and hepatic steatosis were evaluated on day 6.
In Vivo S100A9 shRNA Infection
Lentiviral particles carrying S100A9-specific short hairpin (sh) RNA (scrambled control shRNA as control) were prepared. A commercially available cationic polymer transfection reagent (in vivo-jetPEITM) was used to deliver lentivirus via intravenous injection. Briefly, 150 μg of lentiviral particles diluted in 200 μl of 5% glucose solution was mixed with in vivo-jetPEITM solution. The mixture was incubated for 15 min at room temperature and then was injected into mice (45). The knockdown efficiency was determined by RT-PCR.
qRT-PCR
Total RNA was extracted with TRIzol (Invitrogen) and was subjected to reverse transcription using reverse transcriptase (TAKARA) at 42 °C for 15 min, and the resulting cDNA was amplified by PCR using gene-specific primers (40). The primers were designed based on genome-wide procedures using the Primer 3 program and cross-checked by a BLAST search of the NCBI database. The qRT-PCR primers are as follows: S100A8-for (NM_009114.3), 5′-ATGCCGTCTACAGGGATGAC-3′, and S100A8-rev, 5′-ACTGAGGACACTCGGTCTCTA-3′; S100A9-for (NM_009114.3), 5′-GGACCTGGACACAAATGCAG-3′, and S100A9-rev, 5′-ACCCTCGTGCATCTTCTCG-3′; ATF3-for (NM_007498), 5′-TTTGCTAACCTGACACCCTTTG-3′, and ATF3-rev, 5′-AGAGGACATCCGATGGCAGA-3′; and β-actin-for (NM_007393.5), 5′TACCACAGGCATTGTGATGG-3′, and β-actin-rev, 5′TTTGATGTCACGCACGATTT-3′.
Luciferase Activity Analysis
Rat ATF3 promoter reporter pATF3/Luc vector, pS100A8/Luc, and pS100A9/Luc vector were constructed by pGL3/Luc plasmid. Corresponding site-mutated plasmids were constructed by the two-step PCR method. Lipofectamine 2000 (Invitrogen) was used to transfect 32D cells according to the technical manual. Twenty four hours post-transfection, cells were treated with Veh (DMSO) or Dex in DMEM only for 48 h. Cells were then harvested, and their luciferase activities were measured with the Dual-Luciferase Reporter Assay kit (Promega) according to the technical manual. Cloning primers used were as follows: ATF3-reporter-for, 5′-TCTGAGGGAGTATGGACGAA-3′, and ATF3-reporter-rev, 5′-CGCTGAGTGAGACTGTGGC-3′; S100A8-reporter-for, 5′-CGCTGAGTGAGACTGTGGC-3′, and S100A8-reporter-rev, 5′-CACCTGCACAAACTGAGGACAC-3′; and S100A9-reporter-for, 5′-GGCAGGTAGGCTGTCTTG-3′, and S100A9-reporter-rev, 5′-GGCAGGTAGGCTGTCTTG-3′.
Chromatin Immunoprecipitation (ChIP) Assay
To test whether activated GR could bind to the nGRE sequence within the ATF3 promoter, WT or ATF3−/− BM cells were treated with Dex for 3 days. To prove that ATF3 could bind to the S100A8/A9 promoter region, WT BM cells were infected with lentivirus overexpressing ATF3. After treatment, cells were fixed in 1% paraformaldehyde for 10 min at room temperature and harvested in cell lysis buffer containing protease inhibitors (Millipore). Nuclei were collected and resuspended in shearing buffer (10 mm Tris-HCl (pH 8), 1 mm EDTA, 140 mm NaCl, 1% SDS, 0.1% sodium deoxycholate, 1% Triton X-100, protease inhibitor mixture). DNA was sheared by sonication using a Branson Sonifier. Protein-DNA complexes were immunoprecipitated overnight with equivalent amounts of specific antibody or anti-IgG. Immunoprecipitated protein-DNA complexes were recovered with protein A magnetic beads (Millipore), washed, and eluted. After the cross-links were reversed, the DNA was purified using a QIAquick DNA purification kit (Qiagen). ChIP DNA samples were then subject for real time qPCR analysis with the following primers:ChIP-ATF3-for, 5′-CTGGGATAGGCGTTTCAT-3′, and ChIP-ATF3-rev, 5′-CCAAGTCGTACCTGGATAGA-3′; ChIP-A8-site1-for, 5′-TGGCAACTCTGGAAGGGAA-3′, and ChIP-A8-site1-rev, 5′-CGCTAAGGAGAGCCTACACTG-3′; ChIP-A8-site2-for, 5′-TACCCATCTAAATAGAACCCCA-3′, and ChIP-A8-site2-rev, 5′-CGCTAAGGAGAGCCTACACT-3′; ChIP-A9-site1-for, 5′-GCTGGGAATTTCACAAAACA-3′, and ChIP-A9-site1-rev, 5′-GTTAGATACATGGTTATCTCG-3′; and ChIP-A9-site2-for, 5′-GGAACCCTTGATGCCCAGCCC-3′, and ChIP-A9-site2-rev, 5′-TTCTCCTTGTACCTCTCTGA-3′.
Statistical Analysis
Statistical tests were performed using GraphPad Prism version 5.0a software and SPSS 17.0. Unpaired Student's t tests and analysis of variance were used to confirm most comparisons. Correlations between different parameters were analyzed using the Spearman rank test. *, p < 0.05; **, p < 0.01,was considered significant.
Author Contributions
J. Z. supervised the study. Y. L. and J. Z. designed the experiments and wrote the manuscript. J. W., M. S., and H. J. performed and interpreted the flow cytometry analysis and animal experiments. All the authors approved the final manuscript and contributed critical revisions to its intellectual contents.
Acknowledgments
We thank Dr. Tsonwin Hai from the Department of Molecular and Cellular Biochemistry, Ohio State University, for kindly providing the ATF3-KO mouse strain. We thank Dr. Hongliang Li from Collaborative Innovation Center of Model Animal, Wuhan University (China), for providing assistance in ATF3-KO mouse breeding.
This work was supported by National Key Basic Research Program of China Grant 2012CB524900; Guangdong Innovative Research Team Program Grant 2009010058; Guangdong Province Universities and Colleges Pearl River Scholar Funded Scheme (GDUPS, 2014); Programme for the 12th Five-year Plan 2012ZX10001003; National Natural Science Foundation of China Grants 91542112, 81571520, 31270921; Fundamental Research Funds for the Central Universities; the Provincial Talents Cultivated by “Thousand-Hundred-Ten” Programme of Guangdong Province; 111 Project B12003; China Postdoctoral Science Foundation Grant 2015M582365, and Natural Science Foundation of Guangdong Province Grant 2016A030310252. The authors declare that they have no conflicts of interest with the contents of this article.
- GC
- glucocorticoid
- MDSC
- myeloid-derived suppressor cells
- G-MDSC
- glucocorticoid MDSC
- M-MDSC
- monocytic MDSC
- Dex
- dexamethasone
- ATF3
- activating transcription factor 3
- RLU
- relative luciferase unit
- GR
- GC receptor
- nGRE
- negative glucocorticoid-response element
- DC
- dendritic cell
- BM
- bone marrow
- qRT
- quantitative reverse transcription
- ROS
- reactive oxygen species
- iMC
- immature myeloid cell
- TLR
- Toll-like receptor
- ConA
- concanavalin A
- Veh
- vehicle.
References
- 1. Cohen J. C., Horton J. D., and Hobbs H. H. (2011) Human fatty liver disease: old questions and new insights. Science 332, 1519–1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. van der Goes M. C., Jacobs J. W., and Bijlsma J. W. (2014) The value of glucocorticoid co-therapy in different rheumatic diseases–positive and adverse effects. Arthritis Res. Ther. 16, S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schäcke H., Döcke W. D., and Asadullah K. (2002) Mechanisms involved in the side effects of glucocorticoids. Pharmacol. Ther. 96, 23–43 [DOI] [PubMed] [Google Scholar]
- 4. Varga G., Ehrchen J., Tsianakas A., Tenbrock K., Rattenholl A., Seeliger S., Mack M., Roth J., and Sunderkoetter C. (2008) Glucocorticoids induce an activated, anti-inflammatory monocyte subset in mice that resembles myeloid-derived suppressor cells. J. Leukocyte Biol. 84, 644–650 [DOI] [PubMed] [Google Scholar]
- 5. Deng Z. B., Liu Y., Liu C., Xiang X., Wang J., Cheng Z., Shah S. V., Zhang S., Zhang L., Zhuang X., Michalek S., Grizzle W. E., and Zhang H. G. (2009) Immature myeloid cells induced by a high-fat diet contribute to liver inflammation. Hepatology 50, 1412–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nagaraj S., and Gabrilovich D. I. (2008) Tumor escape mechanism governed by myeloid-derived suppressor cells. Cancer Res. 68, 2561–2563 [DOI] [PubMed] [Google Scholar]
- 7. Gabrilovich D. I., and Nagaraj S. (2009) Myeloid-derived suppressor cells as regulators of the immune system. Nat. Rev. Immunol. 9, 162–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liao J., Wang X., Bi Y., Shen B., Shao K., Yang H., Lu Y., Zhang Z., Chen X., Liu H., Wang J., Chu Y., Xue L., Wang X., and Liu G. (2014) Dexamethasone potentiates myeloid-derived suppressor cell function in prolonging allograft survival through nitric oxide. J. Leukocyte Biol. 96, 675–684 [DOI] [PubMed] [Google Scholar]
- 9. Ostrand-Rosenberg S., and Sinha P. (2009) Myeloid-derived suppressor cells: linking inflammation and cancer. J. Immunol. 182, 4499–4506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Qin A., Cai W., Pan T., Wu K., Yang Q., Wang N., Liu Y., Yan D., Hu F., Guo P., Chen X., Chen L., Zhang H., Tang X., and Zhou J. (2013) Expansion of monocytic myeloid-derived suppressor cells dampens T cell function in HIV-1-seropositive individuals. J. Virol. 87, 1477–1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liao J., Wang X., Bi Y., Shen B., Shao K., Yang H., Lu Y., Zhang Z., Chen X., Liu H., Wang J., Chu Y., Xue L., Wang X., and Liu G. (2014) Dexamethasone potentiates myeloid-derived suppressor cell function in prolonging allograft survival through nitric oxide. J. Leukocyte Biol. 96, 675–684 [DOI] [PubMed] [Google Scholar]
- 12. Varga G., Ehrchen J., Brockhausen A., Weinhage T., Nippe N., Belz M., Tsianakas A., Ross M., Bettenworth D., Spieker T., Wolf M., Lippe R., Tenbrock K., Leenen P. J., Roth J., and Sunderkötter C. (2014) Immune suppression via glucocorticoid-stimulated monocytes: a novel mechanism to cope with inflammation. J. Immunol. 193, 1090–1099 [DOI] [PubMed] [Google Scholar]
- 13. Thompson M. R., Xu D., and Williams B. R. (2009) ATF3 transcription factor and its emerging roles in immunity and cancer. J. Mol. Med. 87, 1053–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hai T., Wolfgang C. D., Marsee D. K., Allen A. E., and Sivaprasad U. (1999) ATF3 and stress responses. Gene Expr. 7, 321–335 [PMC free article] [PubMed] [Google Scholar]
- 15. Hai T., Wolford C. C., and Chang Y. S. (2010) ATF3, a hub of the cellular adaptive-response network, in the pathogenesis of diseases: is modulation of inflammation a unifying component? Gene Expr. 15, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang K., Bai X., Li R., Xiao Z., Chen J., Yang F., and Li Z. (2012) Endogenous glucocorticoids promote the expansion of myeloid-derived suppressor cells in a murine model of trauma. Int. J. Mol. Med. 30, 277–282 [DOI] [PubMed] [Google Scholar]
- 17. Oakley R. H., Revollo J., and Cidlowski J. A. (2012) Glucocorticoids regulate arrestin gene expression and redirect the signaling profile of G protein-coupled receptors. Proc. Natl. Acad. Sci. U.S.A. 109, 17591–17596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Movahedi K., Guilliams M., Van den Bossche J., Van den Bergh R., Gysemans C., Beschin A., De Baetselier P., and Van Ginderachter J. A. (2008) Identification of discrete tumor-induced myeloid-derived suppressor cell subpopulations with distinct T cell-suppressive activity. Blood 111, 4233–4244 [DOI] [PubMed] [Google Scholar]
- 19. Yan D., Yang Q., Shi M., Zhong L., Wu C., Meng T., Yin H., and Zhou J. (2013) Polyunsaturated fatty acids promote the expansion of myeloid-derived suppressor cells by activating the JAK/STAT3 pathway. Eur. J. Immunol. 43, 2943–2955 [DOI] [PubMed] [Google Scholar]
- 20. Sade-Feldman M., Kanterman J., Ish-Shalom E., Elnekave M., Horwitz E., and Baniyash M. (2013) Tumor necrosis factor-α blocks differentiation and enhances suppressive activity of immature myeloid cells during chronic inflammation. Immunity 38, 541–554 [DOI] [PubMed] [Google Scholar]
- 21. Cheng P., Corzo C. A., Luetteke N., Yu B., Nagaraj S., Bui M. M., Ortiz M., Nacken W., Sorg C., Vogl T., Roth J., and Gabrilovich D. I. (2008) Inhibition of dendritic cell differentiation and accumulation of myeloid-derived suppressor cells in cancer is regulated by S100A9 protein. J. Exp. Med. 205, 2235–2249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Condamine T., and Gabrilovich D. I. (2011) Molecular mechanisms regulating myeloid-derived suppressor cell differentiation and function. Trends Immunol. 32, 19–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sternberg E. M. (2006) Neural regulation of innate immunity: a coordinated nonspecific host response to pathogens. Nat. Rev. Immunol. 6, 318–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gustot T., Lemmers A., Moreno C., Nagy N., Quertinmont E., Nicaise C., Franchimont D., Louis H., Devière J., and Le Moine O. (2006) Differential liver sensitization to toll-like receptor pathways in mice with alcoholic fatty liver. Hepatology 43, 989–1000 [DOI] [PubMed] [Google Scholar]
- 25. Miura K., Seki E., Ohnishi H., and Brenner D. A. (2010) Role of toll-like receptors and their downstream molecules in the development of nonalcoholic fatty liver disease. Gastroenterol. Res. Pract. 2010, 362847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hritz I., Mandrekar P., Velayudham A., Catalano D., Dolganiuc A., Kodys K., Kurt-Jones E., and Szabo G. (2008) The critical role of toll-like receptor (TLR) 4 in alcoholic liver disease is independent of the common TLR adapter MyD88. Hepatology 48, 1224–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vegiopoulos A., and Herzig S. (2007) Glucocorticoids, metabolism and metabolic diseases. Mol. Cell. Endocrinol. 275, 43–61 [DOI] [PubMed] [Google Scholar]
- 28. Ilkovitch D., and Lopez D. M. (2009) The liver is a site for tumor-induced myeloid-derived suppressor cell accumulation and immunosuppression. Cancer Res. 69, 5514–5521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Huang A., Zhang B., Yan W., Wang B., Wei H., Zhang F., Wu L., Fan K., and Guo Y. (2014) Myeloid-derived suppressor cells regulate immune response in patients with chronic hepatitis B virus infection through PD-1-induced IL-10. J. Immunol. 193, 5461–5469 [DOI] [PubMed] [Google Scholar]
- 30. Hu C. E., Gan J., Zhang R. D., Cheng Y. R., and Huang G. J. (2011) Up-regulated myeloid-derived suppressor cell contributes to hepatocellular carcinoma development by impairing dendritic cell function. Scand. J. Gastroenterol. 46, 156–164 [DOI] [PubMed] [Google Scholar]
- 31. Chou H. S., Hsieh C. C., Yang H. R., Wang L., Arakawa Y., Brown K., Wu Q., Lin F., Peters M., Fung J. J., Lu L., and Qian S. (2011) Hepatic stellate cells regulate immune response by way of induction of myeloid suppressor cells in mice. Hepatology 53, 1007–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ostrand-Rosenberg S., Sinha P., Beury D. W., and Clements V. K. (2012) Cross-talk between myeloid-derived suppressor cells (MDSC), macrophages, and dendritic cells enhances tumor-induced immune suppression. Semin. Cancer Biol. 22, 275–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liu G., Bi Y., Wang R., Yang H., Zhang Y., Wang X., Liu H., Lu Y., Zhang Z., Chen W., Chu Y., and Yang R. (2014) Targeting S1P1 receptor protects against murine immunological hepatic injury through myeloid-derived suppressor cells. J. Immunol. 192, 3068–3079 [DOI] [PubMed] [Google Scholar]
- 34. Sinha P., Okoro C., Foell D., Freeze H. H., Ostrand-Rosenberg S., and Srikrishna G. (2008) Proinflammatory S100 proteins regulate the accumulation of myeloid-derived suppressor cells. J. Immunol. 181, 4666–4675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gilchrist M., Thorsson V., Li B., Rust A. G., Korb M., Roach J. C., Kennedy K., Hai T., Bolouri H., and Aderem A. (2006) Systems biology approaches identify ATF3 as a negative regulator of Toll-like receptor 4. Nature 441, 173–178 [DOI] [PubMed] [Google Scholar]
- 36. Gold E. S., Ramsey S. A., Sartain M. J., Selinummi J., Podolsky I., Rodriguez D. J., Moritz R. L., and Aderem A. (2012) ATF3 protects against atherosclerosis by suppressing 25-hydroxycholesterol-induced lipid body formation. J. Exp. Med. 209, 807–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. De Nardo D., Labzin L. I., Kono H., Seki R., Schmidt S. V., Beyer M., Xu D., Zimmer S., Lahrmann C., Schildberg F. A., Vogelhuber J., Kraut M., Ulas T., Kerksiek A., Krebs W., et al. (2014) High-density lipoprotein mediates anti-inflammatory reprogramming of macrophages via the transcriptional regulator ATF3. Nat. Immunol. 15, 152–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Boespflug N. D., Kumar S., McAlees J. W., Phelan J. D., Grimes H. L., Hoebe K., Hai T., Filippi M. D., and Karp C. L. (2014) ATF3 is a novel regulator of mouse neutrophil migration. Blood 123, 2084–2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Maruyama K., Fukasaka M., Vandenbon A., Saitoh T., Kawasaki T., Kondo T., Yokoyama K. K., Kidoya H., Takakura N., Standley D., Takeuchi O., and Akira S. (2012) The transcription factor Jdp2 controls bone homeostasis and antibacterial immunity by regulating osteoclast and neutrophil differentiation. Immunity 37, 1024–1036 [DOI] [PubMed] [Google Scholar]
- 40. Yang Q., Shi M., Shen Y., Cao Y., Zuo S., Zuo C., Zhang H., Gabrilovich D. I., Yu Y., and Zhou J. (2014) COX-1-derived thromboxane A2 plays an essential role in early B-cell development via regulation of JAK/STAT5 signaling in mouse. Blood 124, 1610–1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Patel R., Patel M., Tsai R., Lin V., Bookout A. L., Zhang Y., Magomedova L., Li T., Chan J. F., Budd C., Mangelsdorf D. J., and Cummins C. L. (2011) LXRβ is required for glucocorticoid-induced hyperglycemia and hepatosteatosis in mice. J. Clin. Invest. 121, 431–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sade H., and Sarin A. (2003) IL-7 inhibits dexamethasone-induced apoptosis via Akt/PKB in mature, peripheral T cells. Eur. J. Immunol. 33, 913–919 [DOI] [PubMed] [Google Scholar]
- 43. Shi M., Shi G., Tang J., Kong D., Bao Y., Xiao B., Zuo C., Wang T., Wang Q., Shen Y., Wang H., Funk C. D., Zhou J., and Yu Y. (2014) Myeloid-derived suppressor cell function is diminished in aspirin-triggered allergic airway hyperresponsiveness in mice. J. Allergy Clin. Immunol. 134, 1163–1174 [DOI] [PubMed] [Google Scholar]
- 44. Waight J. D., Netherby C., Hensen M. L., Miller A., Hu Q., Liu S., Bogner P. N., Farren M. R., Lee K. P., Liu K., and Abrams S. I. (2013) Myeloid-derived suppressor cell development is regulated by a STAT/IRF-8 axis. J. Clin. Invest. 123, 4464–4478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kim J. Y., Hwang J. Y., Lee D. Y., Song E. H., Park K. J., Kim G. H., Jeong E. A., Lee Y. J., Go M. J., Kim D. J., Lee S. S., Kim B. J., Song J., Roh G. S., Gao B., and Kim W. H. (2014) Chronic ethanol consumption inhibits glucokinase transcriptional activity by Atf3 and triggers metabolic syndrome in vivo. J. Biol. Chem. 289, 27065–27079 [DOI] [PMC free article] [PubMed] [Google Scholar]