Abstract
The degree of genetic relatedness among group members is influenced by dispersal, group formation and composition, mating systems, and other socioecological factors. Making inferences about differences between species in their socio-genetic structure is difficult because studies rarely compare multiple species. In this study, we use multilocus microsatellite genotype data to analyze intragroup genetic relatedness in two howler monkey species (Alouatta palliata and A. pigra). We test the prediction that their patterns of intragroup genetic relatedness will be distinct based on expectations derived from their distinct social systems. Alouatta palliata is expected to have low levels of intragroup relatedness, given that both males and females are reported to disperse from their natal groups, and to join groups with no close kin. Levels of relatedness among A. pigra group members are expected to be variable according to the history of group formation, with new groups formed by unrelated individuals and well-established groups having close kin due to female nepotism and sometimes by takeovers by coalitions of related males. Our results indicate that in both species, most groups contain closely related same-sex and/or inter-sex dyads. This suggests that philopatry in A. palliata may be more common than reported or that individuals are using alternative strategies to reside with close kin. We found greater variation among groups in female-female relatedness in A. palliata than in A. pigra, implying that these species have distinct socio-genetic structures. Further studies including both long-term observational and genetic data are necessary to understand the mechanisms that determine the degree of variation in intragroup genetic relatedness within and among populations for both species. Ecological and demographic data are also necessary to determine the importance of other factors, especially habitat loss and fragmentation, in determining the degree of relatedness in howler monkey groups.
Keywords: Kinship, genetic variation, microsatellites, relatedness
INTRODUCTION
In social animals, genetic relatedness and social interactions among members of a group influences the way in which genetic variation is structured within and between populations. The degree of genetic relatedness among group members is affected by the mode of dispersal and group formation, by group composition, mating systems, and can also be indirectly influenced by demographic stochasticity and historical factors [Di Fiore, 2012], habitat fragmentation [Oklander et al., 2010], and the distribution of resources [Henzi et al., 1997; Koenig et al., 1998; Sinha et al., 2005]. Within populations, patterns of inter- and intragroup relatedness are determined by complex interactions between these factors. Thus, understanding patterns of relatedness within and between social groups can inform us about both the factors that shape genetic structure in populations and the potential for individuals to attain inclusive fitness benefits.
Traditionally, researchers have relied on long-term observations (i.e., demographic records) in order to understand patterns of relatedness, dispersal and genetic structure in populations. However, long-term field studies in primates are often difficult, due to logistics and the costs associated with such projects, in addition to dealing with political obstacles [Strier & Mendes, 2009]. Furthermore, the inherent difficulties of keeping accurate immigration and emigration records in species with bisexual dispersal in which individuals are difficult to identify, makes this task even harder. When long-term demographic data are not available, genetic data can provide a way to understand the extent of genetic structure resulting from patterns of dispersal and social systems. In fact, genetic data can be used to infer patterns of intergroup movement because, in general, we can expect to find a greater degree of intergroup genetic structure in the more philopatric sex because individuals remaining in natal groups reside with same-sex kin [Goudet et al., 2002]. Over the last decade, molecular methods have been implemented to investigate genetic relatedness among many social species in order to understand the extent and implications of kin associations in relation to dispersal patterns [Parus major: Van De Casteele & Matthysen, 2006; Passer domesticus: Vangestel et al., 2011; Crocuta crocuta: Watts et al., 2011; delphinids: Möller, 2012].
Considering social animals, primate societies are ideal systems in which genetic data can be used to understand patterns of relatedness due to the extensive variation across taxa in their dispersal and social structure. However, relatively few primate studies have used genetic data to assess the degree of relatedness among group members [Silk, 2002] and the data are especially limited for New World monkeys [Di Fiore, 2009]. Analyses of genetic relatedness have been used to confirm that dispersal systems and social structure produce predictable patterns in genetic population substructure [e.g., Altmann et al., 1996], and to indirectly infer patterns of dispersal when demographic data are unavailable [e.g., Di Fiore & Fleischer, 2005]. However, studies rarely compare multiple species, so it is hard to make comparative inferences about differences in socio-genetic structure. Howler monkeys are an interesting taxon that can be used to address this problem because there are several closely related species that appear to vary in key aspects of their dispersal and social systems. Here, we use reports of dispersal and social structure in two species of howler monkeys (Alouatta palliata and A. pigra) to generate predictions on patterns of genetic relatedness that might emerge from these behaviors. We then test our predictions using multilocus genotype data to examine intragroup genetic relatedness by comparing patterns of relatedness between the two species.
In both species, individuals live in uni- or multi-male/multi-female groups, but A. palliata groups tend to be larger. In A. palliata, group size ranges from 6–20+ individuals [Chapman & Balcomb, 1998] and usually consists of 2–4 adult males, 2–10 adult females, and 1–10 immatures [Glander, 1980; Estrada, 1982; Arroyo-Rodríguez et al., 2008; Milton et al., 2009]. Alouatta pigra mean group size ranges from 4–9 individuals, with 1–2 adult males, 1–3 adult females, and 1–4 immatures [Chapman & Balcomb, 1998; Van Belle & Estrada, 2006]. Although bisexual dispersal has been reported in both species [Brockett et al., 2000; Horwich et al., 2000; Van Belle et al., 2008; Glander, 1992; Clarke & Glander, 2004], these reports also show variation around dispersal patterns within each species. Some A. pigra females have been reported to remain philopatric and immigration by dispersing females is thought to be rare [Brockett et al., 2000; Van Belle et al., 2011]. However, not all A. pigra females stay in their natal groups and those that disperse may form new groups instead of joining established groups as is reported for A. seniculus [Crockett, 1984; Pope, 1992]. On the other hand, A. palliata females are reported to disperse from their natal group and join groups that do not contain kin, but exceptions exist where individuals sometimes stay in their natal group [Glander, 1980; Clarke & Glander, 2008]. Regarding males, different group-joining strategies have been reported in A. pigra; whereas in some cases a single male can join a group without evicting any residents, in other instances a single male may take over a group by expelling the resident males [Horwich et al., 2000; Van Belle et al., 2012], or closely related males can form coalitions and take over a group together [Van Belle et al., 2012]. In contrast to the multiple strategies reported in A. pigra, the dispersal strategies of A. palliata males are reported to be similar to that of A. palliata females, with individuals dispersing before adulthood, remaining solitary for some time, and joining groups that do not contain kin [Glander, 1980; Glander, 1992; Clarke & Glander, 2008]. In addition, secondary transfer across multiple groups has been observed for some A. palliata individuals (both male and female) [Clarke & Glander, 2010].
Due to these reported differences, we predict that patterns of genetic relatedness among same-sex adults will differ between A. pigra and A. palliata groups. To test this, we calculate and compare coefficients of genetic relatedness for intragroup adults generated from multilocus microsatellite genotypes in both species. Specifically, we ask 1) what are the patterns of genetic relatedness among same-sex adults within social groups in these species? and 2) Do these patterns reflect our current understanding of their dispersal and social structure? If the dispersal strategy of A. pigra females is similar to that reported for A. seniculus, female relatedness should be greater in well-established groups as compared to new groups formed by dispersing individuals. Therefore, we predict that mean intragroup female relatedness among A. pigra groups will be highly variable due to likely random sampling of established and new groups in this study. Because solitary A. pigra males may join groups without necessarily expelling resident males, and closely related coalitions of males can join groups together, we should also see variation in mean intragroup adult male relatedness among groups in this species. On the other hand, since most A. palliata juveniles (female and male) are reported to disperse from their natal group and join established groups that do not contain close relatives, we predict that mean relatedness among intragroup adults will be low in A. palliata groups for both sexes, and we should see little variation among groups.
METHODS
Sample Collection
Blood and hair samples from 64 A. pigra individuals [26 adult females (F), 23 adult males (M), 15 immatures (IM)] and 140 A. palliata individuals (59 F, 42 M, 39 IM) were obtained from 37 wild groups from different locations (Figure 1, Table SI). Sampled individuals were captured between 1998 and 2012 following procedures described in Rodríguez-Luna & Cortés-Ortiz [1994]. Since our sample comprises wild-born individuals that had not been followed since birth we determined adult status following dental development and wear patterns of captured individuals according to the criteria developed in Pope [1966] (see details in Kelaita et al. [2011]). Briefly, we assigned adult status for individuals with fully-erupted dentition and the third molar in functional occlusion, and at least slight wear found on some of the premolars and first molar. Most individuals (groups designated with numbers in Figure 1) were tattooed with unique IDs to avoid duplicated sampling. Non-tattooed individuals (groups designated with letters in Figure 1) were either captured in locations sampled only once or were sampled during the same expedition from distinct groups. During sample collection, 2 ml of blood were extracted from the caudal vein of chemically immobilized individuals and mixed in 10 ml of lysis buffer [Seutin et al., 1991]. Samples were kept on ice in the field and stored at −20°C after they arrived in the laboratory. Hair samples were stored in paper envelopes, kept at room temperature in the field, and stored at −20°C in the lab. This research complies with the protocols approved by University of Michigan Committee on Use and Care of Animals, and adhered to American Society of Primatologists’ Principles for the Ethical Treatment of Non-Human Primates. Sample collection and transportation comply with all legal requirements in Mexico and the USA.
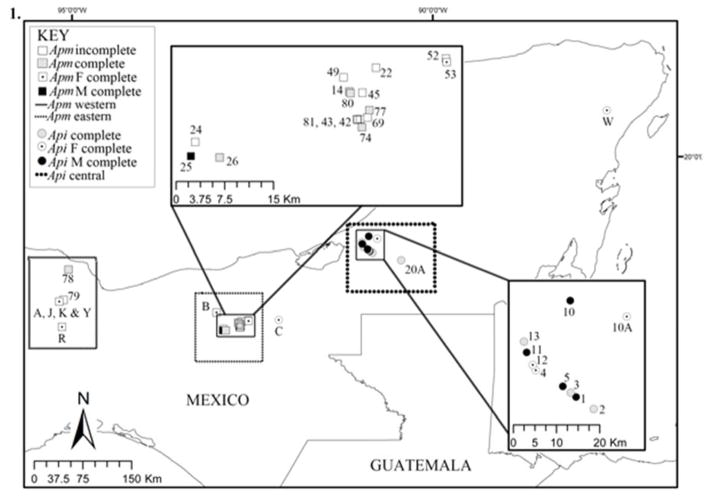
Figure 1.
Map of sampling localities. Each symbol corresponds to a group of howler monkeys, see key for details. Incomplete groups are groups in which neither all adult males nor females were sampled, but data from these groups was included in population level analyses (see methods for details). Complete groups are groups in which all adult males and females were sampled. F complete and M complete groups are those in which all adult females and all adult males, respectively, were sampled.
Because some of our samples came from geographically distant locations and relatedness estimates are sensitive to population structure [Wang 2011], we partitioned our data in three datasets: A. pigra, and Western and Eastern populations of A. palliata (Figure 1, see details in the “Analyses” section).
DNA Extraction and Microsatellite Genotyping
Genomic DNA was extracted from both blood and hair samples for all individuals (except for one infant for which we only extracted DNA from hair) using the QIAGEN DNeasy tissue kit (Qiagen Inc., Valencia, CA). We followed the manufacturer’s protocol for animal tissue extractions with the following modifications: step 1) for blood samples: starting volume of 100 μL of whole blood solution, added to 100 μL buffer ATL, for hair samples: approximately 15 hair follicles in 100 μL buffer ATL.
All A. pigra, Western A. palliata, and Eastern A. palliata individuals were genotyped at 22, 12, and 19 polymorphic microsatellite loci, respectively (28 loci analyzed in total, Table I). The presence of polymorphisms had previously been determined for each species [Cortés-Ortiz et al., 2010], but for our analyses we only used those loci that were polymorphic in each dataset, therefore different loci were analyzed for each dataset (Table I). We conducted both single and multiplex reactions to amplify these loci. Singleplex amplifications were performed in a reaction volume of 10 μL containing 1 μL 10× buffer, 1 μL dNTPs at 2μM each, 0.8 μL MgCl2 (50mM), 0.25 μL of fluorescently labeled forward primer (10 μM), 0.25 μL unlabeled reverse primer (10 μM), 5.7 μL water, 0.045 μL Platinum taq (Invitrogene), and 1 μL DNA extract. The thermal cycling profile was as follows: initial denaturation of 94 °C for 2 min, followed by 35 cycles of 94 °C for 30 s, annealing temperature (see Table I) for 30 s, 72 °C for 30 s, followed by a 72 °C for 10 min. Based on similarities in annealing temperature (Table I), we ran multiplex reactions for a number of samples using the Qiagen multiplex PCR kit (Qiagen Inc., Valencia, CA), with a total reaction volume of 10 μL. The reaction mix contained 5 μL of 2× Master Mix, 1 μL of 10× primer mix (with each primer concentrated at 2 μM), 1 μL of water, 2 μL of Q solution, and 1 μL of DNA. Multiplexed PCR reactions followed a thermal cycling profile of 95 °C for 15 min, followed by 35 cycles of 94 °C for 30 s, annealing temperature for 30 s, 72 °C for 45 s, and 60 °C extension for 30 min. A negative control was included in all PCR reactions to ensure lack of contamination. We electrophoresed PCR products on a 2% agarose gel to verify the presence and quality of amplifications in order to determine the appropriate dilutions for genotyping. PCR products were diluted with water according to the intensity of the observed band and added to a mix of fluorescent standard (GS500LIZ) and Hi-Di Formamide (Applied Biosystems) before samples were sent to the University of Michigan DNA Sequencing Core where genotyping was done on a 3730XL Applied Biosystems DNA sequencer. Allele sizes were scored using GeneMarker V 1.5 (Softgenetics, State College, PA, USA) by at least two different researchers. If researchers did not agree on a call, the sample was amplified again, genotyped and the alleles were re-scored. On average 12% of samples from individuals analyzed in this study were genotyped more than once per locus. All plates submitted for genotyping contained at least one PCR product with alleles of known size to ensure consistent migration of DNA and comparable allele sizing across runs.
Table I.
PCR conditions and variability for the microsatellite markers used for each dataset.
| Locussource | Dataset | Allele size range (bp) (Api | ApmW | ApmE) | T °C [Api|(Apm multiplex)Apm single] | Na (Api | ApmW | ApmE) | Ho:He (Api | ApmW | ApmE) |
|---|---|---|---|---|---|
| AP68a | Api, ApmE | 185–197 | na | 191–197 | 50 | (53)50 | 6 | na | 4 | 0.67:0.68 | na | 0.09:0.09 |
| AP74 a | Api | 148–152 | na | na | 52 | na | 3 | na | na | 0.38:0.44 | na | na |
| D5S111 b | Api | 169–182 | na | na | 60 | na | 3 | na | na | 0.18:0.22 | na | na |
| D6S260 b | Api, Apm | 181–187 | 177–181 | 173–187 | 53 | 53 | 4 | 3 | 7 | 0.75:67 | 0.35:0.51 | 0.45:0.44 |
| D14S51 b | Api, Apm | 143–152 | 143–147 | 141–149 | 53 | (55)60 | 3 | 3 | 5 | 0.58:0.51 | 0.06:0.12 | 0.27:034*** |
| D17S804 b | Api | 157–169 | na | na | 60 | na | 4 | na | na | 0.45:0.45 | na | na |
| PEPC8 c | Api, ApmE | 239–250 | na | 238–250 | 46 | 46 | 5 | na | 4 | 0.55:0.48 | na | 0.14:0.13 |
| AB20 d | Api, ApmE | 136–266 | na | 236–246 | 67 | na | 6 | na | 4 | 0.60:0.69 | na | 0.09:0.09 |
| APM1 e | Api, Apm | 181–208 | 208–220 | 206–210 | 64 | (64)64 | 5 | 5 | 3 | 0.75:0.70 | 0.26:0.26 | 0.45:0.49 |
| APM4 e | Api, Apm | 239–247 | 245–249 | 237–251 | 65 | (64)64 | 4 | 3 | 8 | 0.31:0.63*** | 0.19:0.52*** | 0.49:0.64*** |
| AB06 d | Api, Apm | 174–176 | 272–276 | 272–280 | 60 | (55)55 | 3 | 3 | 5 | 0.47:0.48| 0.16:0.25| 0.15:0.25*** |
| AB07 d | Api, Apm | 174–176 | 174–176 | 174–176 | 60 | 60 | 2 | 2 | 2 | 0.49:0.49 | na | na |
| AB12 d | Api | 219–234 | na | na | 65 | na | 4 | na | na | 0.51:0.60 | na | na |
| AB16 d | Api | 168–177 | na | na | 65 | na | 2 | na | na | 0.33:0.30 | na | na |
| AB17 d | Api | 224–248 | na | na | 60 | na | 7 | na | na | 0.78:0.76 | na | na |
| APM9 e | Api | 170–176 | na | na | 55 | na | 4 | na | na | 0.56:0.51 | na | na |
| API06 e | Api, Apm | 244–254 | 277–279 | 250–277 | 55 | (55)55 | 4 | 2 | 3 | 0.56:0.54 | 0.10:0.09 | 0.16:0.22*** |
| API07 e | Api, Apm | 108–121 | 115–117 | 113–117 | 50 | (55)50 | 5 | 2 | 3 | 0.56:0.60 | 0.58:0.49 | 0.44:0.52 |
| API08 e | Api | 271–279 | na | na | 55 | na | 5 | na | na | 0.62:0.70 | na | na |
| API09 e | Api | 475–471 | na | na | 60 | na | 6 | na | na | 0.40:0.57*** | na | na |
| API11 e | Api, ApmE | 253–261 | na | 253–259 | 55 | (55)55 | 2 | na | 4 | 0.16:0.15 | na | 0.14:0.16 |
| API14 e | Api, ApmE | 253–261 | na | 181–210 | 55 | (55)55 | 4 | na | 3 | 0.64:0.62 | na | 0.09:0.09 |
| 1110 f | Apm | na | 203–205 | 203–205 | na | (53)54 | na | 2 | 2 | na | 0.39:0.35 | 0.19:0.22 |
| 157 f | Apm | na | 224–226 | 222–232 | na | (53)54 | na | 7 | 6 | na | 0.58:0.76*** | 0.74:0.79 |
| AC45 g | Apm | na | 342–358 | 330–358 | na | 65 | na | 5 | 10 | na | 0.61:0.69 | 0.43:0.78*** |
| 1118 f | ApmE | na | na |132–136 | na | (53)52 | na | na | 4 | na | na | 0.14:0.21*** |
| TGMS1 h | ApmE | na | na | 304–323 | na | 60 | na | na | 3 | na | na | 0.45:0.47 |
| TGMS2 h | Apm | na | 322–328 | 312–328 | na | 60 | na | 3 | 4 | na | 0.55:0.48 | 0.42:0.49 |
Sources:
Research genetics (for all MapPairs),
Apm=both Western and Eastern A. palliata, ApmW=Western A. palliata, ApmE=Eastern A. palliata, Api=A. pigra, na=locus not amplified for dataset, T °C = annealing temp, Na = number of alleles, Ho = observed heterozygosity, He = expected heterozygosity, asterisk = test for significant deviation from HWE: * p < 0.05, ** p < 0.01, *** p < 0.001.
Analyses
Observed and expected heterozygosity, number of alleles per locus, and probability of identity (PI) were calculated in GenAlEx 6.41 [Peakall & Smouse, 2006]. We used Micro-Checker [Van Oosterhout et al., 2004] to test for evidence of null alleles, scoring errors due to stuttering, and large allele dropout. None of the loci showed evidence to suggest the presence of any of these phenomena in any of our datasets (i.e., A. pigra, Western A. palliata and Eastern A. palliata). Arlequin ver 3.5.1.3 [Excoffier & Lischer, 2010] was used to analyze linkage disequilibrium (LD) between pairs of loci and departures from Hardy-Weinberg equilibrium (HWE) in each dataset, and for each we implemented a Bonferroni correction to account for multiple comparisons. We did not find evidence for LD between any loci in A. pigra, but there was evidence for LD for several pairs in A. palliata. Since we do not know the location of these microsatellites in the genome we cannot be sure of physical linkage between any pair of loci. However, the fact that different loci show LD in each dataset (loci in LD for Western A. palliata include AB06 & AB07, and for Eastern A. palliata include PEPC8 & API11, APM4 & 157, TGMS1 & TGMS2, and D6S260 & AC45) suggests that physical proximity of loci may not be responsible for this observation. Data analysis after removal of genotype data for loci under LD did not produce results different from those of analyses utilizing the entire dataset. For each dataset, at least two loci showed evidence for deviation from HWE (Table I). For neutral loci, like microsatellites, deviations from HWE can be caused by the presence of null alleles. Based on our analyses with Micro-Checker, there is no evidence of null alleles in our datasets. Another factor that can affect HWE is the analyses of multiple populations treated as a single population, which is known as a Wahlund effect [Wahlund, 1928]. Although we partitioned our A. palliata dataset into two more evident populations (East and West) we cannot exclude the possibility that the observed deviations from HWE could have been caused by population substructure. However, excluding loci under HW disequilibrium did not affect our overall results, so we report our analyses without excluding these loci.
Relatedness
When not accounted for, population structure is known to inflate relatedness estimates [Wang 2011]. Therefore, we partitioned our genotype datasets to avoid this issue. In A. pigra, all relatedness estimates were calculated using allele frequencies in the “central population” (Figure 1). For the two groups that were geographically distant from the central location, we calculated relatedness values for each group separately by combining individuals with the central population. In A. palliata, our samples clustered in two geographically distant populations (Eastern and Western). To determine whether or not these populations are genetically distinct, we computed pairwise RST in Arlequin. Our results indicated that there is some structure between these sampling sites (RST=0.34, P<0.001). Therefore, we calculated relatedness estimates for the Eastern and Western populations of A. palliata separately using local allele frequencies within each population.
There are several relatedness estimators available and each calculates the coefficient of relatedness (r) from multilocus genotype data differently. Factors such as degree of dyadic relatedness and number of alleles per locus can influence different estimators in different ways. To determine which estimator is most appropriate for each dataset in this study, we compared r-values from several estimators using simulated genotypes [100 each of parent-offspring (r=0.5), full sibs (r=0.5), half-sibs (r=0.25), and unrelated (r=0)] against actual values using RELATED [Pew et al. 2014]. This analysis takes allele frequencies from sampled populations into account when simulating genotypes. As a result, dyadic r-values for the most appropriate estimator will match the expected value most closely. The Queller & Goodnight [1989] estimator performed best for the A. pigra, and the Eastern A. palliata dataset, while the Lynch & Ritland [1999] performed best for the Western A. palliata dataset. To be consistent in our calculations across datasets, we report here only Queller & Goodnight [1989], (QG) r-values computed in RELATED and used those estimates in statistical analyses. To confirm the appropriateness of this estimator, we compared QG r-values against others estimated by RELATED for known mother-offspring dyads in each species (N=4 dyads each). QG reliably estimated expected r-values for these dyads (r≈0.5) (see results for more details). We consider closely related dyads to be those with an r-value consistent with that of first-degree relatives. Thus, closely related dyads have an r-value greater than or equal to the mean simulated value for half-sibs in that dataset (A. pigra: r≥0.239; Western A. palliata: r≥0.213, Eastern A. palliata: r≥0.227) and unrelated dyads to be those with r-values below this threshold.
To examine patterns of intragroup relatedness in each species, we made comparisons between dyad types. We tested for significant differences in mean relatedness by permuting r-values across dyad types in R [R Core Team 2015], using the sample function to construct permuted datasets 10,000 times without replacement. We considered mean relatedness to be significantly different when the observed difference between dyad types exceeded that seen in >95% of our permuted datasets. To test if close relatives reside in the same groups, we tested for significant differences in observed mean relatedness (Di) between a) all possible adult female-female (F-F) dyads and intragroup F-F dyads, between b) all possible adult male-male (M-M) dyads and intragroup M-M dyads, and between c) all possible adult male-adult female (M-F) dyads and intragroup M-F dyads. These analyses were conducted using only within-population dyads (see above for population description). For example, for A. pigra “all F-F dyads” means all possible F-F dyads within the “central” population and for A. palliata “all F-F dyads” means all F-F dyads within the Western population plus all F-F dyads within the Eastern population, but no F-F dyads between Western and Eastern populations. For each species, comparisons involving mean relatedness among intragroup adult females were conducted using only genotype data for adult females in groups from which we collected samples from all adult females present in the group (i.e., complete groups) (A. pigra N=9 groups, A palliata N=6 groups). The same criterion was applied for comparisons involving relatedness among intragroup adult males (A. pigra=8 groups, A. palliata=7 groups; Table II). If mean relatedness is greater for intragroup dyads than for all possible dyads, this would suggest that intragroup individuals are more closely related than is any given dyad of that type at random. To evaluate possible sex-biased dispersal in each species, we also tested for significant differences in relatedness between intragroup M-M dyads vs. intragroup F-F dyads. If dispersal is not sex-biased, we should see no difference in dyadic relatedness between the sexes.
Table II.
QG estimates of the coefficient of relatedness (r) for all same-sex intragroup dyads in all complete groups sampled in this study.
| Group | M | F | r1 | r2 | r3 | r4 | r5 | r6 | Mean r |
|---|---|---|---|---|---|---|---|---|---|
| A. pigra | |||||||||
| Females | |||||||||
| 4 | 1 | 3 | 0.48* | 0.31* | 0.52* | 0.43 | |||
| 5 | 3 | 3 | 0.24 | −0.08 | −0.08 | 0.03 | |||
| 10 | 2 | 3 | 0.42* | 0.09 | 0.17 | 0.23 | |||
| 1 | 2 | 3 | 0.11 | −0.02 | 0.20 | 0.10 | |||
| 10A | 2 | 2 | 0.15 | 0.15 | |||||
| W | 1 | 2 | 0.46* | 0.46 | |||||
| 12 | 1 | 2 | −0.10 | −0.10 | |||||
| C | 1 | 2 | 0.40* | 0.40 | |||||
| 11 | 2 | 2 | 0.24 | 0.24 | |||||
| Males | |||||||||
| 5 | 3 | 3 | 0.56* | −0.21 | −0.12 | 0.08 | |||
| 1 | 2 | 3 | 0.38 | 0.38 | |||||
| 2 | 2 | 1 | 0.47* | 0.47 | |||||
| 3 | 2 | 1 | 0.11 | 0.11 | |||||
| 20A | 2 | 1 | −0.03 | −0.03 | |||||
| 10 | 2 | 3 | 0.24* | 0.24 | |||||
| 11 | 2 | 2 | 0.14 | 0.14 | |||||
| 13 | 2 | 1 | −0.19 | −0.19 | |||||
|
| |||||||||
| A. palliata | |||||||||
| Females | |||||||||
| A | 1 | 4 | 0.17 | −0.05 | 0.37* | 0.10 | 0.82* | 0.29* | 0.28 |
| 25 | 2 | 4 | 0.08 | −0.12 | −0.17 | 0.30* | 0.24* | 0.01 | 0.05 |
| B | 1 | 3 | 0.46 | 0.02 | 0.28* | 0.26 | |||
| Y | 1 | 3 | 0.33* | −0.02 | 0 | 0.10 | |||
| R | 1 | 2 | 0.74* | 0.74 | |||||
| 53 | 1 | 2 | 0.65* | 0.65 | |||||
| Males | |||||||||
| 74 | 3 | 10 | 0.58* | 0.40* | 0.53* | 0.50 | |||
| 78 | 3 | 2 | 0.42* | 0.05 | −0.09 | 0.13 | |||
| 14 | 2 | 5 | 0.56* | 0.56 | |||||
| 25 | 2 | 4 | −0.13 | −0.13 | |||||
| 26 | 2 | 2 | 0.62* | 0.62 | |||||
| 77 | 2 | 1 | 0.38* | 0.38 | |||||
| 80 | 2 | 8 | 0.47* | 0.47 | |||||
M=number of adult males, F=number of adult females, r1, r2, etc. = dyadic r-value,
denotes closely related dyad, mean r=mean intragroup relatedness of dyad type specified.
Intergroup variation
To evaluate potential differences in levels of intergroup variation in mean relatedness between species, we subtracted mean intragroup relatedness values separately for males and females for every possible combination of groups within each population (example: d1=|group1 mean r – group2 mean r|, d2=|group1 mean r – group3 mean r|, etc.), then calculated the mean difference by species (d1+d2+…+dn/n comparisons for each species) and compared this difference between species. Using this method, we will expect that populations with low variation in mean relatedness among groups will exhibit a lower mean difference than populations with a greater variation in the patterns of relatedness among groups. We then tested for significant differences in this mean difference using the permutation method in R as described above.
Because our analyses of same-sex relatedness were limited to groups in which either all males or all females were captured and due to the difficulty of capturing all females in a large group, our sample of A. palliata F-F dyads in particular is biased to groups with a low number of females (mean=3). Although smaller group size and this female composition are not uncommon for A. palliata mexicana [e.g., Estrada 1982; Arroyo-Rodríguez et al., 2008; Asensio et al., 2009; Dunn et al., 2009], we tested for an effect of group size on A. palliata F-F relatedness within groups using dyads from both complete and incomplete groups (data not shown) to determine if this bias affected our results. Female-female relatedness was not correlated with number of females sampled (Spearman’s rho=0.044, P=0.648), nor number of adults sampled (Spearman’s rho=0.034, P=0.724) per group. This suggests that group size may not affect variation in F-F relatedness between groups in A. palliata. However, we cannot be certain of the possible effect of our sampling until studies of larger groups can be completed.
RESULTS
Suitability of molecular markers
Observed heterozygosity (Ho) per locus in the central A. pigra population ranged from Ho=0.16 to Ho= 0.78 and averaged at Ho=0.51 across all 22 microsatellite markers and the mean number of alleles per locus (Na) was 4.12 (Table I). Probability of identity was very low (PI=1.7×10−13, PIsib=1.6×10−6) indicating that it is unlikely that two individuals or any two siblings, respectively, in a randomly chosen dyad share the same multilocus genotype. This supports the notion that for A. pigra our results are robust using this combination of microsatellite markers. Heterozygosity and mean number of alleles per locus were lower among the markers used for A. palliata (Western: mean Ho=0.34 [range Ho=0.06–0.61], mean Na=3.33, Eastern: mean Ho=0.30 [range Ho=0.09–0.74] mean Na=4.42). However, probability of identity was also very low (Western: PI=1.1×10−5, PIsib=4.7×10−3, Eastern: PI=4.3×10−8, PIsib=5.0×10−4), suggesting that although the markers used for A. palliata were not as polymorphic as those used for A. pigra, their combination is sufficient to distinguish among individuals.
For known mother-offspring dyads (N=4 for each species), mean QG relatedness was close to the expected value of r=0.5 (A. pigra mean r=0.43; A. palliata mean r=0.59). For A. palliata, the mean is higher than the expected value for this type of relationship, but this is not surprising given that many of the markers used for A. palliata were not highly polymorphic. Although we report QG r-values for both species since this estimator also performed well in the simulation studies, we warn that these values are probably slightly inflated for A. palliata. This should not be a problem for the purposes of this study since we are only making within-species comparisons using r-values directly.
Intragroup relatedness
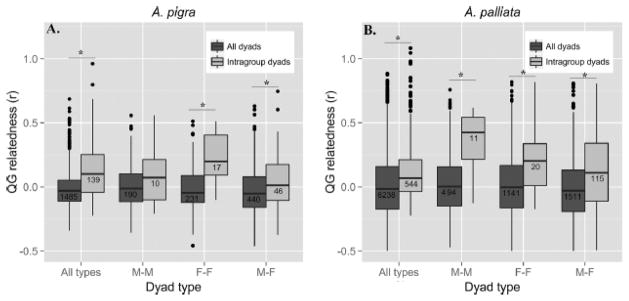
Our relatedness results for each species are summarized by dyad type in Figure 2. Within each species, mean intragroup relatedness among all dyad types was significantly greater than the general mean relatedness among all dyad types (A pigra rint=0.12 ± SE 0.017, rall=−1.7×10−2 ± SE 0.005, |Di|=0.14, P<0.001; A. palliata rint=0.17 ± SE 0.012, rall=−8.8×10−3 ± SE 0.003, |Di|=0.19, P<0.001). This result indicates that, in general, groups of both species do not contain a random sample of genotypes from the population, but actually contain close relatives. Similarly, for both species mean intragroup female-female relatedness was significantly greater than that among all female-female dyads (A. pigra rintF-F=0.21 ± SE 0.049, rallF-F=−1.8×10−2 ± SE 0.011, |Di|=0.22, P<0.001; A. palliata rintF-F=0.22 ± SE 0.063, rallF-F=8.5×10−3 ± SE 0.007, |Di|=0.22, P<0.001). For intragroup male-male dyads this difference was only significant in A. palliata (A. pigra rintM-M=0.10 ± SE 0.083, rallM-M=−7.5×10−4 ± SE 0.012, |Di|=0.10, P=0.063; A. palliata rintM-M =0.34 ± SE 0.082, rallM-M=−1.0×10−2 ± SE 0.010, |Di|=0.33, P<0.001).
Figure 2.
Box plot comparing QG relatedness by dyad type for A) A. pigra, and B) A. palliata. Samples sizes are indicated below central horizontal lines, which represent medians. Asterisks indicate significant comparisons. Dyads of “all types” include M-M, F-F, M-F, and dyads that include immatures. QG relatedness values only include within-population dyads as specified in methods.
Our analyses of relatedness between males and females show that in both species, mean intragroup male-female relatedness was significantly greater than that among all male-female dyads (A pigra rintM-F=0.04 ± SE 0.034, rallM-F=−3.9×10−2 ± SE 0.009, |Di|=0.08, P=0.005; A. palliata rintM-F=0.11 ± SE 0.028, rallM-F=−3.5×10−2 ± SE 0.006, |Di|=0.15, P<0.001), suggesting that groups also contain inter-sex relatives. Within A. pigra groups, relatedness of male-female dyads was not significantly different from relatedness of male-male dyads (|Di|=0.06, P=0.490), but was lower than mean relatedness of female-female dyads (|Di|=0.16, P=0.013). Within A. palliata groups, this trend was reversed, and relatedness of male-female dyads was not significantly different from relatedness of female-female dyads (|Di|=0.11, P=0.132), but was lower than mean relatedness of male-male dyads (|Di|=0.23, P=0.016). This implies that although there are close inter-sex relatives living in the same group in each species, there may be a larger number of closely related same-sex dyads (F-F for A. pigra and M-M for A. palliata) in the group or that levels of relatedness among same-sex dyads are higher.
Considering all dyads, mean male-male relatedness did not differ from female-female relatedness within each species (A. pigra: |Di|=1.6×10−3, P=0.902; A. palliata: |Di|=1.7×10−2, P=0.289). This trend was also true within groups; mean intragroup male-male relatedness was not significantly different from mean intragroup female-female relatedness (A. pigra: |Di|=0.11, P=0.250; A. palliata: |Di|=0.12, P=0.259), suggesting that dispersal is not sex-biased in either species.
Intergroup Variation
In A. pigra, all but one group had only two adult males and in some groups they were unrelated (e.g., groups 13 and 20A) while in other groups adult males were closely related (groups 2 and 10) (see Table II). In the only three-male group for this species (group 5), two males were closely related to each other (r=0.56), while the third appeared to be unrelated to both individuals (both dyads r<0).
There were two A. palliata groups with more than two adult males (groups 74 and 78). In group 74, all adult male dyads were closely related (r=0.40–0.58), and in group 78, two males were closely related (r=0.42), while the third seems to be unrelated to both individuals. Among the two-male A. palliata groups (N=5), there was only one in which the adult males were unrelated (r =−0.13, group 25). This dyad was one of only three intragroup M-M dyads that were unrelated for this species. However, each male was closely related to at least one of the four adult females in this group (data not shown). These results indicate that intragroup A. palliata males tend to be closely related to each other, but exceptions certainly exist.
For both species, there was variation between groups in intragroup F-F relatedness. In A. pigra, relatedness in two-female groups ranged from unrelated (groups 10A, 12, and 11) to closely related (groups C and W). In three-female groups (N=4), the degree of relatedness among intragroup adult female dyads also varied. Females in two of these groups were unrelated to each other (groups 5 and 1), while females in one of these groups were all closely related (group 4), and one group had two related, and one unrelated females (group 10). For A. palliata, there were two groups that only contained two adult females (groups R and 53). In both cases, these females were highly related to each other – on the order of mother-daughter or full siblings (r=0.65–0.74). There were four A. palliata groups in which there were more than two adult females. Each of these groups contained a mixture of unrelated and closely related dyads.
Next, we compared the degree of intergroup variation in mean same-sex relatedness between the species. The mean difference (dF) in intragroup female relatedness among groups was significantly lower in A. pigra than in A. palliata (A. pigra dF=0.21; A. palliata dF=0.41, |Di|=0.21, P=0.006), suggesting greater variation between A. palliata groups in levels of female relatedness. For males, mean difference (dM) in intragroup relatedness among groups was not different between species (A. pigra dM=0.23; A. palliata dM=0.29, |Di|=0.06, P=0.364). In both species, mean difference in group relatedness did not differ between males versus females (A. pigra |Di|=2.2×10−2, P=0.602; A. palliata |Di|=0.12, P=0.305).
DISCUSSION
In this study, we used genetic data to compare patterns of relatedness between two species of howler monkeys, A. pigra and A. palliata. We found that in both species most groups contained pairs of close relatives, which was unexpected for A. palliata. Although direct statistical comparisons of intragroup genetic relatedness between species would be inappropriate given the different number and levels of variation of the microsatellite markers used for each species, the general patterns of relatedness observed show that for both species intragroup F-F dyads are more closely related than they would be at random in the populations. This was expected for groups of A. pigra, as it is believed that mothers recruit their daughters to remain in their natal group while impeding the immigration of unrelated females [Horwich et al. 2000]. However, this was not expected for A. palliata, where it is reported that groups are predominately composed of females that emigrate from groups elsewhere (and thus would be unrelated) [Glander, 1992; Clarke & Glander, 2008].
A recent comparative behavioral study by Ho et al. [2014] analyzing groups of both species near locations sampled in this study found that A. pigra females were in closer proximity to one another and had higher rates of affiliative behavior than A. palliata females; which ratifies the expected differences based on reports made by numerous behavioral studies on each species [Ho et al., 2014 and references therein]. These differences in social interactions would be expected if within-group A. pigra females were more closely related than A. palliata within-group females, but our study does not provide support for this inference, suggesting that kinship may not always be a strong determinant of social interactions among females. The combination of genetic analyses and behavioral observations for the same groups would provide a strong approach to study the effect of kinship in determining social interactions between group members in each species.
Despite this general similarity between species in the pattern of intragroup female relatedness, variation among groups in mean female relatedness was significantly greater in A. palliata, which was also unexpected. This result implies that female social structure may be quite variable among groups within A. palliata, and perhaps even more variable than in A. pigra. For males, A. palliata groups had a higher prevalence of closely related dyads, but analyses indicate that intergroup variation in mean male relatedness is similar between species. In general, our results support the notion that some of the patterns in genetic relatedness among same-sex intragroup adults in A. pigra and A. palliata are different, but understanding the factors that contribute to these differences and similarities requires a deeper understanding of the social systems and dispersal patterns for each species.
The variable levels of relatedness within and between A. pigra groups found here for males and females are consistent with previous findings at Palenque National Park by Van Belle et al. [2012]. These authors found closely related same-sex dyads in many, but not all A. pigra groups, and they also observed immigration by coalitions of related A. pigra males. However, the presence of more than one male in a group may not always be the result of a group takeover by a coalition. For example, Horwich et al. [2000] observed solitary A. pigra males joining established groups and living with other (possibly unrelated) resident males. Additionally, multi-male groups may also be formed when juvenile males stay in their natal group until adulthood, although there is no information available on the proportion of male A. pigra juveniles that do not disperse from their natal group. Our genetic results suggest that in both species, there may be multiple strategies for males to become group residents, as groups were uni- or multimale and males in multimale groups were sometimes related and sometimes unrelated. Based on long-term census data, Crockett [1985] reported similar results for A. seniculus in that males can remain in their natal group, take over other groups, or join established groups.
Horwich et al. [2000] suggested that A. pigra female dispersal might be similar to that of A. seniculus, in which dispersing females tend to form new groups with non-relatives and over time reproductively dominant females recruit their daughters as group members [Crockett, 1984; Pope, 2000]. In a population containing both long-established and new groups, this phenomenon would be manifested in high variance in intragroup adult female relatedness. Thus, we predicted to find variation between groups in levels of intragroup female relatedness among our random sample of groups. Our results do not conflict with this idea as we observed variation between A. pigra groups in mean intragroup adult female relatedness (Table II). However, we do not have long-term demographic data for these groups and we do not know when these groups were formed. In order to determine if the pattern of female dispersal indeed produces differing levels of intragroup relatedness between new and well-established A. pigra groups, future studies should include analyses comparing female relatedness between groups of known origins.
Considering the observation that most A. palliata juveniles at La Pacifica, Costa Rica (LP) disperse from their natal group and join groups that do not contain kin [Glander, 1992; Clarke & Glander, 2008], we predicted that relatedness among same-sex adults in our A. palliata groups would be low (as they would be a mix of individuals that likely immigrated from different groups). However, our results do not support this prediction as all but one group contained closely related same-sex dyads. This may mean that either rates of male and female philopatry for the species are greater in the Mexican population than reported in the LP population [Clarke & Glander, 2008] or that individuals are using alternative strategies to reside with close relatives (e.g., dispersing from one’s natal group and later joining a group that contains full siblings). Contrary to our findings, Ellsworth [2000] revealed mean r-values within A. palliata groups at LP that did not suggest close kinship among intragroup adult dyads for both males and females. Milton et al. [2009] found closely related adult A. palliata males in some groups on BCI in Panama, but only a single pair of closely related intragroup adult females. Such mixed results in A. palliata relatedness among groups studied at different locations are consistent with the idea that dispersal strategies and social structure may vary across populations in this species. This has important implications for studies of social behavior, as social interactions may be affected by kin selection and the interactions of adults within or between groups in different populations may vary depending on the particular relatedness patterns of that population. Therefore, we urge researchers studying social behavior of howler monkeys to incorporate genetic analyses of their study groups, rather than relying on assumptions about patterns of relatedness among group members that are derived from observations alone.
Within-species differences in social structure (i.e., the pattern of social interactions and the resulting relationships among members in a population [Kappeler & van Schaik 2002]) may be attributed to variation in ecological and demographic factors between habitats [Chapman & Rothman, 2009]. Likewise, dispersal patterns may vary between populations in relation to the distribution of food resources [Henzi et al., 1997; Koenig et al., 1998; Sinha et al., 2005] and to habitat fragmentation [Oklander et al., 2010]. In particular, habitat fragmentation has been demonstrated to affect social organization and dispersal in howler monkeys [reviewed in Arroyo-Rodríguez & Dias, 2010]. Movement between forest fragments is risky since monkeys have to travel across the ground. Therefore, one might hypothesize that philopatry may be more common in fragmented forests than in continuous forests. Oklander et al. [2010] compared intragroup genetic relatedness in A. caraya between continuous and fragmented forests and found differences between habitat types. In continuous forest, intragroup adults were not closely related, but in fragmented forest, intragroup adult females were more closely related than adult males, suggesting that females tend to be more philopatric in the fragmented forest than in the continuous forest. Many of the A. palliata groups sampled in the current study live in very small forest fragments often isolated by pasturelands for cattle. In contrast, although usable habitat in the LP population is also fragmented, many fragments are connected via forest corridors [see map in Glander, 1992] and BCI has not been altered much by humans since the early 1900’s when it was deemed a nature reserve. Differences in the degree of forest connectivity between habitats at LP, BCI, and our sampling sites may be partially responsible for the greater prevalence of closely related A. palliata dyads in this study due to higher rates of philopatry as a response to the fragmented nature of their habitat. Molecular studies that compare intragroup relatedness between fragmented and continuous forest in replicate populations would be desirable to test this hypothesis.
It is now apparent that high levels of genetic relatedness among at least some intragroup adults may be a common feature in howler monkey social systems [A. seniculus: Pope, 1998; A. caraya: Oklander et al., 2010; A. pigra: Van Belle et al., 2012, present study; A. palliata: Milton et al., 2009, present study]. However, the degree of intraspecific variation in patterns of relatedness within and between howler monkey populations, and the factors that determine this variation have not been identified. Differences among groups in each species, along with the prevalence of closely related intragroup dyads in species with distinct social systems demonstrate the complexity of interactions between habitat, demography, social interactions and dispersal patterns that shape patterns of genetic relatedness in howler monkey populations. Our findings here invoke the need for deeper investigation of the ecological factors affecting dispersal patterns and social interactions in both species, and the role of these factors in shaping intragroup genetic relatedness in howler monkeys.
Supplementary Material
Acknowledgments
We thank Tony Di Fiore and two anonymous reviewers for their insightful suggestions that helped to improve this manuscript. Thore Bergman and Lucy Ho provided useful comments to earlier versions of this manuscript, Mary Kelaita and Brittney Napier helped to collect and organize the genotype data, and Paul Glaum provided helpful advice about statistical analyses. Samples analyzed in this study were part of several collaborative projects with Domingo Canales Espinosa, Ernesto Rodríguez Luna, Pedro A. Dias Duarte, Javier Hermida Lagunes and Francisco García Orduña, and the authors are thankful for their unconditional support that made possible the extensive sampling. Ma. del Socorro Aguilar, Antonio Jauregui, Antonio Jauregui Jr., Ariadna Rangel, Alba Rodas, María de Jesús Rovirosa, and Paulo Quintana provided invaluable assistance during field work. Official collecting and exportation permits were obtained from SEMARNAT Mexico, and importation permits from USFWS and CDC. Molecular methods were conducted at the Genomic Diversity Laboratory, EEB, University of Michigan. This study was supported by the National Science Foundation (DEB-0640519 and BCS-0962807) to LCO, PROMEP (103.5/03/1154EXB-9) to LCO, and a block grant from EEB, University of Michigan to MDN. MDN was supported in part, by the following NIH training grant: “Michigan Predoctoral Training in Genetics (T32GM007544). The authors declare no conflict of interest.
References
- Altman J, Alberts SC, Haines SA, et al. Behavior predicts genetic structure in a wild primate group. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:5797–5801. doi: 10.1073/pnas.93.12.5797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arroyo-Rodríguez V, Asensio N, Cristóbal-Azkarate J. Demography, life history and migrations in a Mexican mantled howler group in a rainforest fragment. American Journal of Primatology. 2008;70:114–118. doi: 10.1002/ajp.20463. [DOI] [PubMed] [Google Scholar]
- Arroyo-Rodríguez V, Dias PAD. Effects of habitat fragmentation and disturbance on howler monkeys: A review. American Journal of Primatology. 2010;72:1–16. doi: 10.1002/ajp.20753. [DOI] [PubMed] [Google Scholar]
- Asensio N, Arroyo-Rodríguez V, Dunn JC, Cristóbal-Azkarate J. Conservation value of landscape supplementation for howler monkeys living in forest patches. Biotropica. 2009;41:768–773. [Google Scholar]
- Brockett RC, Horwich RH, Jones CB. Female dispersal in the Belizean black howling monkey Alouatta pigra. Neotropical Primates. 2000;8:32–34. [Google Scholar]
- Chapman CA, Balcomb SR. Population charachteristics of howlers: Ecological conditions or group history. International Journal of Primatology. 1998;19:385–403. [Google Scholar]
- Chapman CA, Rothman JM. Within-species differences in primate social structure: evolution of plasticity and phylogenetic constraints. Primates. 2009;50:12–22. doi: 10.1007/s10329-008-0123-0. [DOI] [PubMed] [Google Scholar]
- Clarke MR, Glander KE. Adult migration patterns of the mantled howlers of La Pacifica. American Journal of Primatology. 2004;62(suppl 1):87. [Google Scholar]
- Clarke MR, Glander KE. Natal emigration by both sexes in the La Pacifica population of mantled howlers: When do some stay? American Journal of Primatology. 2008;70:195–200. doi: 10.1002/ajp.20473. [DOI] [PubMed] [Google Scholar]
- Clarke MR, Glander KE. Secondary transfer of adult mantled howlers (Alouatta palliata) on Hacienda La Pacifica, Costa Rica: 1975–2009. Primates. 2010;51:241–249. doi: 10.1007/s10329-010-0195-5. [DOI] [PubMed] [Google Scholar]
- Cortés-Ortiz L, Mondragon E, Cabotage J. Isolation and characterization of microsatellite loci for the study of Mexican howler monkeys, their natural hybrids, and other Neotroptical primates. Conservation Genetics Resources. 2010;2:21–26. [Google Scholar]
- Crockett CM. Emigration by female red howler monkeys and the case for female competition. In: Small MF, editor. Female primates: studies by women primatologists. New York: Alan R. Liss; 1984. pp. 159–173. [Google Scholar]
- Crockett CM. Population studies of red howler monkeys (Alouatta seniculus) National geographic research. 1985;1:264–273. [Google Scholar]
- Di Fiore A, Fleischer RC. Social behavior, reproductive strategies, and population genetic structure of Lagothrix poeppigii. International Journal of Primatology. 2005;26:1137–1173. [Google Scholar]
- Di Fiore A. Genetic approaches to the study of dispersal and kinship in New World primates. In: Garber PA, Estrada A, Bicca-Marques JC, Heymann EW, Strier KB, editors. South American Primates. Comparative Perspectives in the Study of Behavior, Ecology, and Conservation. New York: Springer; 2009. pp. 211–250. [Google Scholar]
- Di Fiore A. Genetic consequences of primate social organization. In: Mitani J, Call J, Kappeler P, Palombit R, Silk J, editors. The evolution of primate societies. Chicago, IL: University of Chicago Press; 2012. pp. 269–292. [Google Scholar]
- Dunn JC, Cristóbal-Azkarate J, Veà J. Differences in diet and activity pattern between two groups of Alouatta palliata associated with the availability of big trees and fruit of top food species. American Journal of Primatology. 2009;71:654–662. doi: 10.1002/ajp.20700. [DOI] [PubMed] [Google Scholar]
- Ellsworth JA. Dissertation. Reno (NV): University of Nevada, Reno; University Microfilms; Ann Arbor, MI: 2000. Molecular evolution, social structure and phylogeography of the mantled howler monkey (Alouatta palliata) p. 179. [Google Scholar]
- Ellsworth JA, Hoelzer GA. Charachterization of microsatellite loci in a New World primate, the mantled howler monkey (Alouatta palliata) Molecular Ecology. 1998;7:657–666. doi: 10.1046/j.1365-294x.1998.00340.x. [DOI] [PubMed] [Google Scholar]
- Escobar-Páramo P. Microsatellite primers for the wild brown capuchin monkey Cebus apella. Molecular Ecology. 2000;9:107–118. doi: 10.1046/j.1365-294x.2000.00764.x. [DOI] [PubMed] [Google Scholar]
- Estrada A. Survey and census of howler monkeys (Alouatta palliata) in the rain forest of “Los Tuxtlas,” Veracruz, Mexico. American Journal of Primatology. 1982;2:363–372. doi: 10.1002/ajp.1350020405. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Lischer HEL. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- Glander KE. Reproduction and populations growth in free-ranging mantled howling monkeys. American Journal of Physical Anthropology. 1980;53:25–36. doi: 10.1002/ajpa.1330530106. [DOI] [PubMed] [Google Scholar]
- Glander KE. Dispersal patterns in Costa Rican mantled howling monkeys. International Journal of Primatology. 1992;13:415–436. [Google Scholar]
- Goncalves EC, Silva A, Barbosa MSR, Schneider PC. Isolation and characterization of microsatellite loci in Amazonian red-handed howlers Alouatta belzebul (Primates, Plathyrrini) Molecular Ecology Notes. 2004;4:406–408. [Google Scholar]
- Goudet J, Perrin N, Wasser P. Tests for sex-biased dispersal using bi-parentally inherited genetic markers. Molecular Ecology. 2002;11:1103–1114. doi: 10.1046/j.1365-294x.2002.01496.x. [DOI] [PubMed] [Google Scholar]
- Henzi SP, Lycett JE, Piper SE. Fission and troop size in a mountain baboon population. Animal Behaviour. 1997;53:525–535. [Google Scholar]
- Ho L, Cortés-Ortiz L, Dias PAD, et al. Effect of ancestry on behavioral variation in two species of howler monkeys (Alouatta pigra and A. palliata) and their hybrids. American Journal of Primatology. 2014;76:855–867. doi: 10.1002/ajp.22273. [DOI] [PubMed] [Google Scholar]
- Horwich RH, Brockett RC, Jones CB. Alternative male reproductive behaviors in the Belizean black howler monkey (Alouatta pigra) Neotropical Primates. 2000;8:95–98. [Google Scholar]
- Kappeler PM, van Schaik CP. Evolution of primate social systems. Internatioal Journal of Primatology. 2002;23:707–740. [Google Scholar]
- Kelaita M, Dias PAD, Aguilar-Cucurachi M, Canales-Espinosa D, Cortés-Ortiz L. Impact of intrasexual selection on sexual dimorphism and testes size in the Mexican howler monkeys Alouatta palliata and A. pigra. American Journal of Physical Anthropology. 2011;146:179–187. doi: 10.1002/ajpa.21559. [DOI] [PubMed] [Google Scholar]
- Koenig A, Beise J, Chalise MK, Ganzhorn JU. When females should contest for food–testing hypotheses about resource density, distribution, size, and quality with Hanuman langurs (Presbytis entellus) Behavioral Ecology and Sociobiology. 1998;42:225–237. [Google Scholar]
- Lynch M, Ritland K. Estimation of pairwise relatedness with molecular markers. Genetics. 1999;152:1753–1766. doi: 10.1093/genetics/152.4.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milton K, Lozier JD, Lacey EA. Genetic structure of an isolated population of mantled howler monkeys (Alouatta palliata) on Barro Colorado Island, Panama. Conservation Genetics. 2009;10:347–358. [Google Scholar]
- Möller LM. Sociogenetic structure, kin associations and bonding in delphinids. Molecular Ecology. 2012;21:745–764. doi: 10.1111/j.1365-294X.2011.05405.x. [DOI] [PubMed] [Google Scholar]
- Oklander LI, Zunino GE, Di Fiore A, Corach D. Isolation, charachterization, and evaluation of 11 autosomal STRs sutable for population studies in black and gold howler monkeys Alouatta caraya. Molecular Ecology Notes. 2007;7:117–120. [Google Scholar]
- Oklander LI, Kowalewski MM, Corach D. Genetic consequences of habitat fragmentation in black-and-gold howler (Alouatta caraya) populations from northern Argentina. International Journal of Primatology. 2010;31:813–832. [Google Scholar]
- Peakall R, Smouse PE. GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes. 2006;6:288–295. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pew J, Muir PH, Wang J, Frasier TR. Related: an R package for analyzing pairwise relatedness from codominant molecular markers. Molecular Ecology Resources. 2014;15:557–561. doi: 10.1111/1755-0998.12323. [DOI] [PubMed] [Google Scholar]
- Pope BL. The population characteristics of howler monkeys (Alouatta caraya) in northern Argentina. American Journal of Physical Anthropology. 1966;24:361–370. doi: 10.1002/ajpa.1330240308. [DOI] [PubMed] [Google Scholar]
- Pope TR. The influence of dispersal patterns and mating systems on genetic differentiation within and between populations of the red howler monkey (Alouatta seniculus) Evolution. 1992;46:1112–1128. doi: 10.1111/j.1558-5646.1992.tb00623.x. [DOI] [PubMed] [Google Scholar]
- Pope TR. Effects of demographic change on group kin structure and gene dynamics of populations of red howling monkeys. Journal of Mammalogy. 1998;79:692–719. [Google Scholar]
- Pope TR. Reproductive success increases with degree of kinship in cooperative coalitions of female red howler monkeys (Alouatta seniculus) Behavioral Ecology and Sociobiology. 2000;48:253–267. [Google Scholar]
- Queller DC, Goodnight KF. Estimating relatedness using genetic markers. Evolution. 1989;43:258–275. doi: 10.1111/j.1558-5646.1989.tb04226.x. [DOI] [PubMed] [Google Scholar]
- R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2015. http://www.R-project.org. [Google Scholar]
- Rodríguez-Luna E, Cortés-Ortiz L. Translocación y seguimiento de un grupo de monos Alouatta palliata liberado en una isla (1988–1994) Neotropical Primates. 1994;2:1–5. [Google Scholar]
- Seutin G, White BN, Boag PT. Preservation of avian blood and tissue samples for DNA analyses. Canadian Journal of Zoology. 1991;69:82–90. [Google Scholar]
- Sinha A, Mukhopadhyay K, Datta-Roy A, Ram S. Ecology proposes, behaviour disposes: ecological variability in social organization and male behavioural strategies among wild bonnet macaques. Current Science. 2005;89:1166–1179. [Google Scholar]
- Silk JB. Kin selection in primate groups. International Journal of Primatology. 2002;23:849–875. [Google Scholar]
- Strier KB, Mendes SL. Long-term field studies of South American primates. In: Garber PA, Estrada A, Bicca-Marques JC, Heymann EW, Strier KB, editors. South American Primates. Comparative Perspectives in the Study of Behavior, Ecology, and Conservation. New York: Springer; 2009. pp. 139–155. [Google Scholar]
- Tomer Y, Greenberg DA, Concepcion E, Ban Y, Davies TF. Thyroglobulin is a thyroid specific gene for the familial autoimmune thyroid diseases. The Journal of Endocinology & Metabolism. 2002;87:404–407. doi: 10.1210/jcem.87.1.8291. [DOI] [PubMed] [Google Scholar]
- Van Belle S, Estrada A. Demographic features of Alouatta pigra populations in extensive and fragmented forests. In: Alejandro E, Garber PA, Pavelka MSM, Luecke L, editors. New Perspectives in the Study of Mesoamerican Primates. New York: Springer; 2006. pp. 121–142. [Google Scholar]
- Van Belle S, Estrada A, Strier KB. Social relationships among male Alouatta pigra. International Journal of Primatology. 2008;29:1481–1498. [Google Scholar]
- Van Belle S, Estrada A, Strier KB. Insights into social relationships among female black howler monkeys Alouatta pigra at Palenque National Park, Mexico. Current Zoology. 2011;57:1–7. [Google Scholar]
- Van Belle S, Estrada A, Strier KB, Di Fiore A. Genetic structure and kinship patterns in a populations of black howler monkeys, Alouatta pigra, at Palenque National Park, Mexico. American Journal of Primatology. 2012;74:948–957. doi: 10.1002/ajp.22047. [DOI] [PubMed] [Google Scholar]
- Van De Casteele T, Matthysen E. Natal dispersal and parental escorting predict relatedness between mates in a passerine bird. Molecular Ecology. 2006;15:2557–2565. doi: 10.1111/j.1365-294X.2006.02946.x. [DOI] [PubMed] [Google Scholar]
- Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P. MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Molecular Ecology Notes. 2004;4:535–538. [Google Scholar]
- Vangestel C, Mergeay J, Dawson DA, Vandomme V, Lens L. Spatial heterogeneity in genetic relatedness among house sparrows along an urban-rural gradient as revealed by individual-based analysis. Molecular Ecology. 2011;20:4643–4653. doi: 10.1111/j.1365-294X.2011.05316.x. [DOI] [PubMed] [Google Scholar]
- Wahlund S. The combination of populations and the appearance of correlation examined from the standpoint of the study of heredity. Hereditas. 1928;11:65–106. [Google Scholar]
- Wang J. Unbiased relatedness estimation in structured populations. Genetics. 2011;187:887–901. doi: 10.1534/genetics.110.124438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts HE, Scribner KT, Garcia HA, Holekamp KE. Genetic diversity and structure in two spotted hyena populations reflects social organization and male dispersal. Journal of Zoology. 2011;285:281–291. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.