Abstract
SKN-1/Nrf are the primary antioxidant/detoxification response transcription factors in animals and they promote health and longevity in many contexts. SKN-1/Nrf are activated by a remarkably broad-range of natural and synthetic compounds and physiological conditions. Defining the signaling mechanisms that regulate SKN-1/Nrf activation provides insights into how cells coordinate responses to stress. Nrf2 in mammals is regulated in part by the redox sensor repressor protein named Keap1. In C. elegans, the p38 MAPK cascade in the intestine activates SKN-1 during oxidative stress by promoting its nuclear accumulation. Interestingly, we find variation in the kinetics of p38 MAPK activation and tissues with SKN-1 nuclear accumulation among different pro-oxidants that all trigger strong induction of SKN-1 target genes. Using genome-wide RNAi screening, we identify new genes that are required for activation of the core SKN-1 target gene gst-4 during exposure to the natural pro-oxidant juglone. Among 10 putative activators identified in this screen was skr-1/2, highly conserved homologs of yeast and mammalian Skp1, which function to assemble protein complexes. Silencing of skr-1/2 inhibits induction of SKN-1 dependent detoxification genes and reduces resistance to pro-oxidants without decreasing p38 MAPK activation. Global transcriptomics revealed strong correlation between genes that are regulated by SKR-1/2 and SKN-1 indicating a high degree of specificity. We also show that SKR-1/2 functions upstream of the WD40 repeat protein WDR-23, which binds to and inhibits SKN-1. Together, these results identify a novel p38 MAPK independent signaling mechanism that activates SKN-1 via SKR-1/2 and involves WDR-23.
Author Summary
Oxidative stress is the result of imbalanced control of reactive oxidative species in cells, is a common occurrence during aerobic metabolism, and must be managed to limit cellular damage and disease. Many details about the signaling mechanisms utilized by animal cells in response to pro-oxidants remain to be discovered. We provide evidence that the signaling mechanisms that activate SKN-1, a master regulator of detoxification genes and aging in the model organism C. elegans, may vary in response to different oxidative stressors. From a genome-wide genetic screen, we identify highly conserved genes skr-1/2 as central regulators of the gene response to pro-oxidants. During exposure to oxidants, SKR-1/2 function upstream from SKN-1 and a direct SKN-1 repressor named WDR-23. These results provide new insights into our understanding of SKN-1 regulation and lay the foundation for future studies to define in detail novel signaling pathways that respond to pro-oxidants.
Introduction
Reactive small molecules are common in natural environments and are produced as byproducts of oxygen metabolism. Reactive small molecules in excess can cause oxidative damage with widespread detrimental effects, but also function as signaling molecules for normal physiological processes [1]. Appropriate response to and regulation of these compounds is crucial as aberrant accumulation has been implicated in early onset of aging along with many pathological states that include metabolic syndromes, neurological disorders, and cancer [2,3,4]. In C. elegans, the cap ‘n’ collar transcription factor family member SKN-1 is homologous to mammalian Nrf2 and functions to promote longevity and resistance to a wide range of environmental stressors [5].
In response to a wide-range of reactive small molecules, SKN-1/Nrf transcription factors translocate into the nucleus and bind to response elements in target genes to activate a conserved detoxification response [6,7,8]. In mammals, Keap1 represses basal Nrf2 activity through a direct interaction that promotes ubiquitylation and degradation, and there is strong support for a model in which small molecules directly react with Keap1 releasing Nrf2 from repression [6,7,8]. Additional Keap1-independent signaling mechanisms exist that are less-defined [9].
Genetic tractability and the conserved nature of the SKN-1/Nrf response have made C. elegans an important model for regulation of this pathway [5]. C. elegans has also been instrumental for defining SKN-1 as a central determinant of aging and longevity [10,11,12] and is being used to understand the role of SKN-1 in antiparasitic drug resistance [13,14,15]. Although C. elegans lacks a close Keap1 homolog, it is repressed under basal conditions by an analogous mechanism via the WD40 repeat protein WDR-23, which binds to and inhibits SKN-1 [15,16]. The protein kinases AKT-1/2, SGK-1, and GSK-3 also function to inhibit SKN-1 under basal conditions [11,17]. A number of protein kinases have been implicated in activation of SKN-1 (MKK-4, IKKɛ-1, NEKL-2, and PHDK-2), although it is not known if any of these regulate SKN-1 directly [18]. During oxidative stress, the p38 MAPK signaling cascade directly phosphorylates and promotes nuclear accumulation of SKN-1 in cells of the intestine [19], a tissue thought to be a primary site for detoxification; p38 MAPK is also required for activation of SKN-1 in the intestine during infection [20,21]. A recent study demonstrated that TIR-1, Toll/interleukin-1 receptor domain protein, functions upstream from p38 MAPK during exposure to an oxidant [22]. Although protein kinases, particularly p38 MAPK, are clearly important, it is not known if this one mechanism is responsible for activation of SKN-1 by all the diverse reactive small molecules known to strongly activate the pathway.
We demonstrate here that the kinetics of p38 MAPK activation and tissues with SKN-1::GFP accumulation vary with different pro-oxidants that all elicit a strong SKN-1 dependent detoxification response. Using genome-wide RNAi screening, we identified SKR-1/2 as required for the core SKN-1 transcriptional response to diverse pro-oxidant compounds. SKR-1/2 are highly conserved orthologues of Skp1, a component of many protein complexes including the Skp-Cullin-F box ubiquitin ligase (SCF) that regulates cell cycle progression and differentiation [23,24]. Loss of skr-1/2 strongly and specifically attenuates induction of SKN-1 dependent genes independent of p38 MAPK signaling and reduces survival of pro-oxidants. SKR-1/2 functions upstream of WDR-23 and influences the accumulation of a WDR-23::GFP fusion protein in nuclei. We hypothesize that this newly identified pathway regulates SKN-1 activity by modulating WDR-23 function.
Results
Kinetics of p38 MAPK activation varies among pro-oxidants that induce a SKN-1 dependent detoxification response
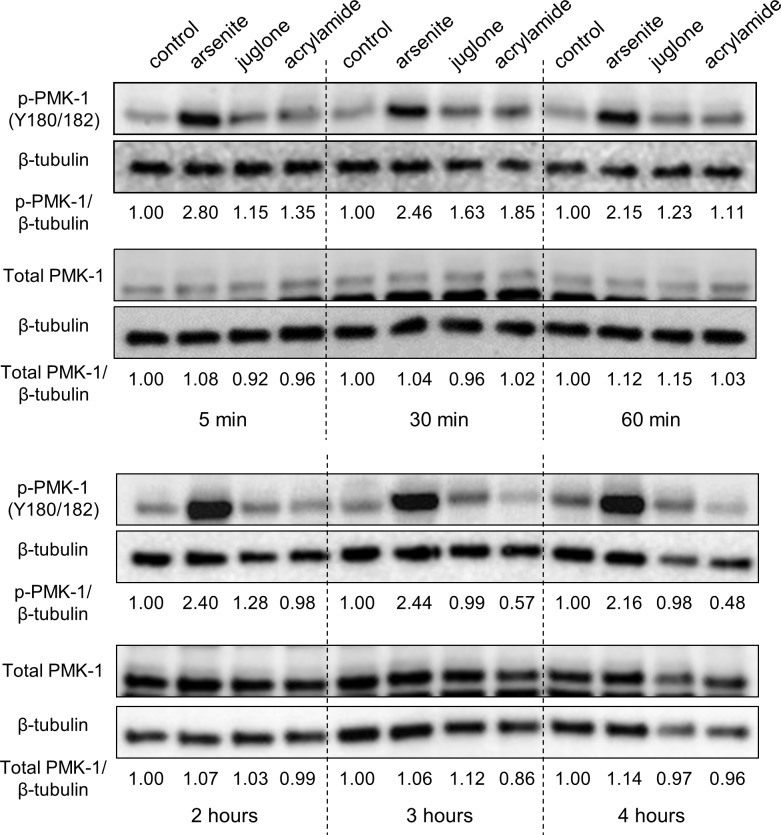
SKN-1 activation has been shown to be mediated through direct phosphorylation by PMK-1 (p38 MAPK) in response to oxidative stress induced by arsenite and during pathogen infection (Pseudomonas aeruginosa) [19,20,21]. SKN-1 residues phosphorylated by PMK-1 are required for nuclear accumulation, a step sometimes correlated with induction of SKN-1 dependent detoxification genes. Arsenite, paraquat, juglone, and acrylamide are a diverse set of small molecules that all strongly activate SKN-1 dependent detoxification genes [16,19,25]. To test if PMK-1 responded similarly to these different compounds, we measured phosphorylation of PMK-1 at its kinase activation residue (Y180/182) over a four hour time course following exposure to arsenite, juglone, and acrylamide (Fig 1). Arsenite increased PMK-1 phosphorylation levels strongly at all time points, and juglone and acrylamide caused smaller transient increases during short term exposure (5–60 min). Interestingly, PMK-1 phosphorylation levels decreased with acrylamide after 3 and 4 h (Fig 1).
Fig 1. Kinetics of PMK-1 activation.
Relative level of PMK-1 phosphorylation at the kinase activating residues (Y180/182) and total PMK-1 protein levels in L4/YA worms treated with 5 mM sodium arsenite, 38 μM juglone, or 7 mM acrylamide for up to four hours. Band intensities relative to β-tubulin are given below each pair of blots with control arbitrarily set to 1.
We also measured phosphorylation of PMK-1 with replicates to confirm effects at specific time points and to test paraquat; doses and durations used were all sub-lethal (S1 Fig) yet strongly activate SKN-1 dependent genes (5 mM arsenite for 1 h, 35 mM paraquat for 2 h, 38 μM juglone for 3 h, and 7 mM acrylamide for 4 h). An increase in phosphorylation of PMK-1 was observed with arsenite and paraquat and a decrease was confirmed for 4 h of acrylamide exposure (Fig 2A). Total levels of PMK-1 were not altered with any treatment or duration (Figs 1 and 2A), indicating that changes to p-PMK-1 were at the posttranslational level. Given that we used whole animal lysates, these results may not reflect PMK-1 phosphorylation kinetics in all tissues. However, our results do demonstrate that the overall patterns of PMK-1 phosphorylation can vary between conditions that all strongly activate SKN-1.
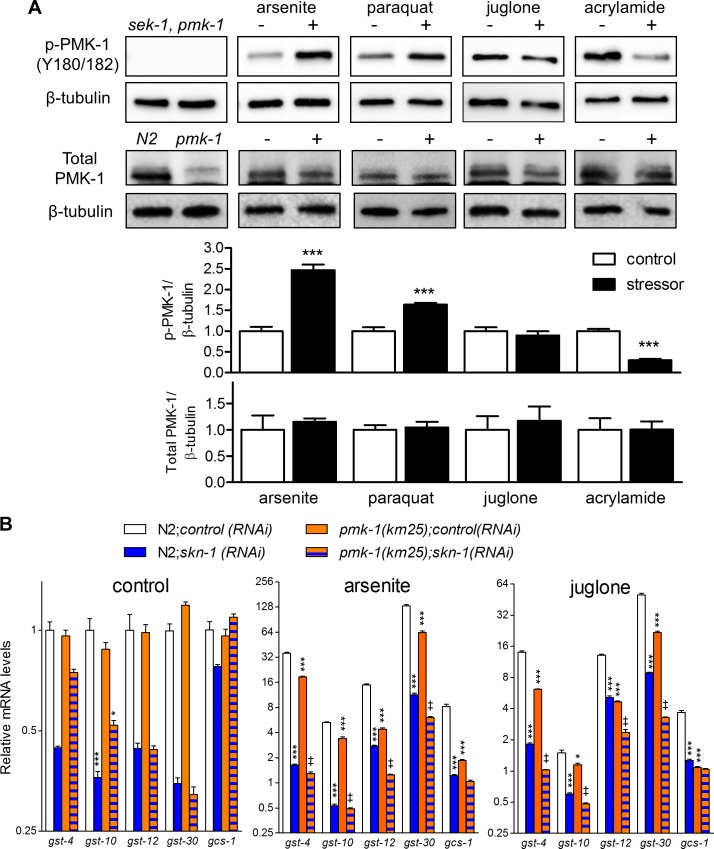
Fig 2. PMK-1 activation and its requirement for SKN-1 transcriptional responses.
(A) Relative level of PMK-1 phosphorylation at the kinase activating residues (Y180/182) and total PMK-1 protein levels in L4/YA worms treated with 5 mM sodium arsenite for 1 h, 35 mM paraquat for 2 h, 38 μM juglone for 3 h, or 7 mM acrylamide for 4 h. PMK-1 phosphorylation and protein levels were normalized to β-tubulin, which was detected on the same blot after stripping; histograms represent mean plus standard error of densitometry from n = 4 replicates of ~1,000 worms. ***P<0.001 compared to corresponding control as determined by Student’s T-test. Representative Western blot images are shown. (B) Fold changes in gst-4, gst-10, gst-12, gst-30, and gcs-1 mRNA levels relative to N2 control in L4/YA N2 wildtype and pmk-1(km25) mutant worms treated with 5 mM sodium arsenite or 38 μM juglone for 1 h, worms were fed either control RNAi or skn-1 RNAi from L1. Histograms represent mean plus standard error of n = 4 replicates of 200–300 worms. All genes were induced significantly by arsenite or juglone (P<0.001); *P<0.05, *** P<0.001 compared to N2;control(RNAi), ‡ P<0.001 compared to pmk-1(km25);control(RNAi).
We next used real-time RT-PCR (qPCR) in deletion mutants of pmk-1 with and without skn-1(RNAi) to assess the requirement of the p38 MAPK pathway after exposure to 5 mM arsenite or 38 μM juglone for 1 h. As expected, five detoxification genes directly regulated by SKN-1 were strongly activated by both compounds in N2 wild-type worms and this was partially dependent on pmk-1 and largely dependent on skn-1 (Fig 2B). Although all four gst detoxification genes were partially dependent on pmk-1, they were still induced. Given that the pmk-1(km25) allele we used is a deletion considered to be null, these results suggest that there are mechanisms that can compensate for loss of PMK-1 in these contexts (e.g., PMK-1 paralogs or other pathways).
SKN-1 accumulation in nuclei can be decoupled from induction of SKN-1 dependent detoxification genes
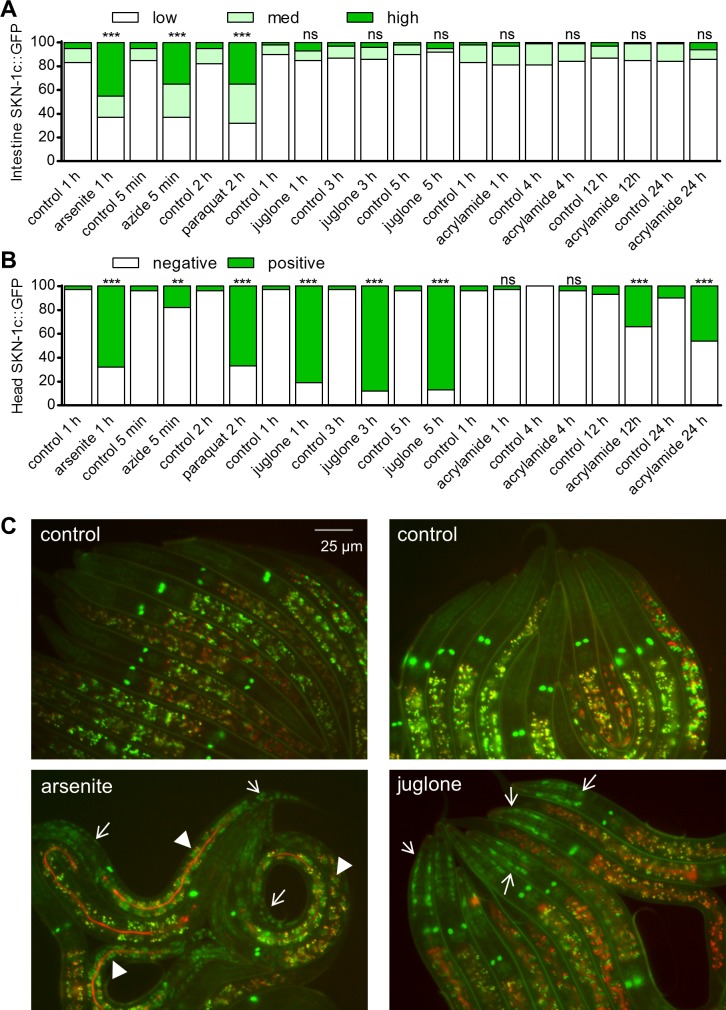
Arsenite induces strong nuclear accumulation of SKN-1b/c::GFP fusion proteins in the intestine, which is dependent on the p38 MAPK pathway [19]. Although SKN-1 protein accumulation in nuclei is thought to be one mechanism of pathway activation [19], genetic and environmental conditions have been identified that activate SKN-1 target genes without causing detectable SKN-1b/c::GFP accumulation implicating other yet-to-be defined regulatory mechanisms [26,27,28]. We scored the percentage of worms with three levels of intestinal SKN-1b/c::GFP nuclear accumulation with arsenite, azide, paraquat, juglone, and acrylamide exposure to determine if SKN-1 accumulation varies with SKN-1 inducer. The LD001 strain we used expresses fusion proteins of SKN-1c and b variants, but not the longer SKN-1a variant [6]. Consistent with previous reports, we found that 5 mM arsenite, 5 mM azide, and 35 mM paraquat induced high levels of nuclear SKN-1b/c::GFP localization in the intestine (Fig 3A). Conversely, nuclear SKN-1b/c::GFP was not detected in the intestine with short (5–15 min, S2 Fig) or long-term (up to 5 h of 38 μM juglone or 24 h of 7 mM acrylamide, Fig 3A) exposure to juglone or acrylamide even though these treatments strongly activate SKN-1 dependent detoxification genes in the same tissue [16,25] (S3 Fig).
Fig 3. SKN-1 is localized to multiple tissues in a stress dependent manner.
Animals integrated with a SKN-1b/c::GFP transgene were treated with 5 mM sodium arsenite, 5 mM sodium azide, 35 mM paraquat, 38 μM juglone, or 7 mM acrylamide. Accumulation of intestinal (A) and head (B) SKN-1b/c::GFP were scored separately. n = 60–100 worms from 3 independent trials. ***P<0.001 from corresponding controls as determined by Chi-Square tests. For intestinal SKN-1b/c::GFP, low refers to little to no SKN-1b/c::GFP, medium refers to SKN-1b/c::GFP observed only at the anterior or posterior of the intestine, and high refers to SKN-1b/c::GFP observed throughout the intestine. For head SKN-1b/c::GFP, negative refers to no observation of GFP signals in the head region, SKN-1b/c::GFP positive refers to GFP signals observed throughout the head region. (C) Representative fluorescence micrographs are shown for arsenite and juglone. Arrows mark head GFP and arrowheads mark intestinal nuclei.
Scoring of stress-inducible SKN-1b/c::GFP localization is typically limited to the intestine, which is credited with being the primary site of detoxification and has the largest nuclei that are readily visible. However, we and others [25] observe strong induction of SKN-1 dependent detoxification gene reporters in other tissues, particularly the hypodermis (S3 Fig). Careful observation revealed SKN-1b/c::GFP accumulation in nuclei throughout worms exposed to arsenite (Fig 3B and 3C and S4A Fig); these other nuclei are most obvious in the head and tail regions, which have less autofluorescence than areas around the intestine. No accumulation was observed in intestine, head, or tail regions of worms fed skn-1 dsRNA (S4A Fig) verifying specificity of the signal. We also scored nuclear accumulation with a transgene that covers all forms of SKN-1 including the long SKN-1a variant, skn-1op::GFP [11]. As shown in S4B Fig, SKN-1op::GFP generally responded similar to SKN-1b/c::GFP with no intestinal accumulation with juglone.
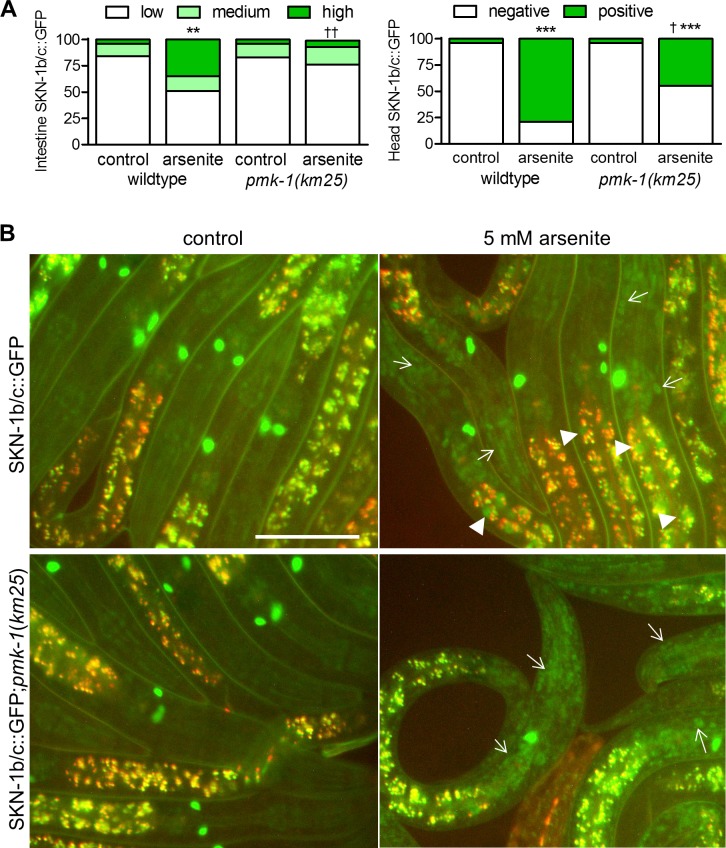
The head nuclei SKN-1b/c::GFP signals were also consistently induced by azide, paraquat, and juglone (Fig 3B and 3C); head nuclei SKN-1b/c::GFP signals were also induced by acrylamide, but this took several hours. Although identification of all tissues with SKN-1b/c::GFP is difficult, at least some of the signal appears to be in hypodermal cell nuclei based on location and morphology (S4C Fig) when compared to images of hypodermal specific markers [29]. The pmk-1(km25) allele was crossed into the SKN-1b/c::GFP strain to test the requirement of p38 MAPK for the accumulation of hypodermal SKN-1. As expected, SKN-1b/c::GFP failed to accumulate in the intestine of pmk-1 worms under arsenite exposure (Fig 4). Alternatively, loss of pmk-1 only slightly reduced the number of worms with SKN-1b/c::GFP accumulation in the head region (Fig 4).
Fig 4. SKN-1b/c::GFP can accumulate in the head without pmk-1.
(A) Scoring of SKN-1b/c::GFP was conducted as in Fig 2. **P<0.01 and ***P<0.001 relative to the corresponding controls without arsenite as determined by Chi-Square tests, †P<0.05 and ††P<0.01 relative to the arsenite exposed wildtype worms as determined by Chi-Square tests, n = 51–92. (B) Representative images of SKN-1b/c::GFP. Arrows mark head GFP and arrowheads mark intestinal nuclei.
To determine the physiological role of SKN-1 in the hypodermis and intestine, we next measured the effects of hypodermis and intestine-specific skn-1(RNAi) on juglone survival. As shown in S5 Fig, loss of skn-1 from either tissue reduced survival consistent with skn-1 mediating resistance in both tissues even though SKN-1::GFP accumulation is not obvious in the intestine during exposure to juglone. Therefore, juglone and acrylamide are able to activate a robust SKN-1 dependent detoxification gene response without detectable SKN-1::GFP nuclear accumulation in the intestine, and SKN-1b/c::GFP accumulation during stress can be partially decoupled from pmk-1 in tissues other than the intestine.
Genome-wide screen for new regulators of SKN-1 target gene activation
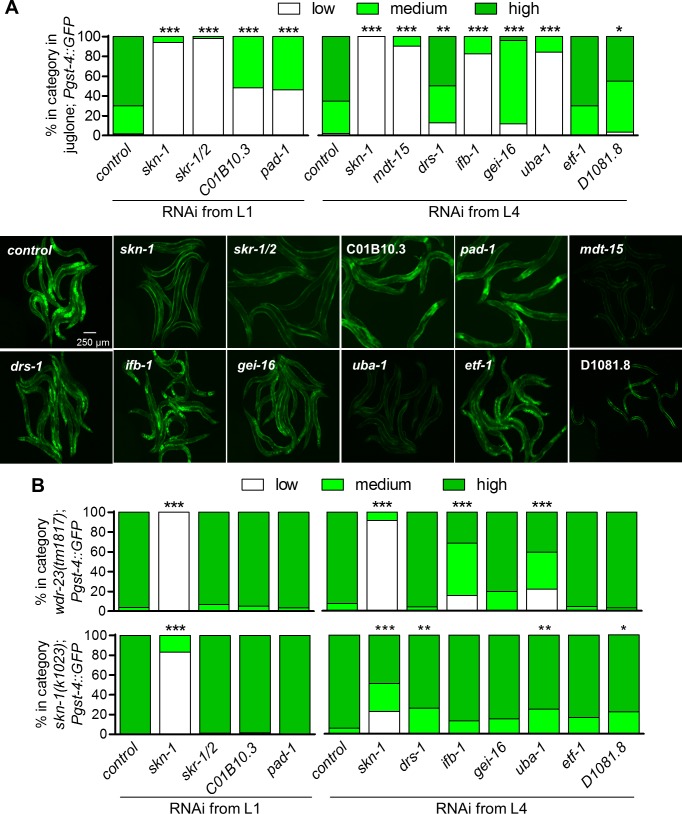
To identify new regulators of the SKN-1 detoxification response, we utilized a C. elegans strain harboring an integrated Pgst-4::GFP transcriptional reporter in a genome-wide RNAi screen for genes that regulate gst-4 expression during exposure to juglone. gst-4 is a phase 2 detoxification gene regulated directly by SKN-1 under numerous conditions, and it has been shown to be a reliable reporter for SKN-1 transcriptional activity [25,27,30,31]. We screened approximately 19,000 dsRNA clones and identified 10 genes that when silenced consistently reduced juglone-induced Pgst-4::GFP fluorescence. RNAi of seven of these genes caused developmental defects or general sickness. After retesting these by initiating silencing at the L4 larval stage rather than L1 to bypass developmental requirements, there were a total of six RNAi clones that strongly reduced Pgst-4::GFP fluorescence. These were skr-1/2, C01B10.3, pad-1, mdt-15, ifb-1, and uba-1 (Fig 5A); mdt-15 has previously been reported to function with SKN-1 [32]. We tested the candidate novel regulators with two other stressors to assess specificity. None of the novel gst-4 regulators had a significant effect on induction of a heat shock reporter, Phsp-16.2::GFP (S6A Fig), but ifb-1, uba-1, etf-1, and D1081.8 were required for induction of an osmotic stress reporter, Pgpdh-1::RFP (S6B Fig).
Fig 5. New gst-4 regulators identified through genome wide RNAi screening function upstream of wdr-23 and skn-1.
Animals integrated with Pgst-4::GFP were fed bacteria producing dsRNA to positive hits obtained from the RNAi screen and exposed to juglone. (A) Pgst-4::GFP fluorescence was scored and representative images are shown. For Pgst-4::GFP scoring, low refers to little to no GFP signals observed throughout the worm, medium refers to GFP signals observed at the anterior and posterior ends of the worm, and high refers to GFP signals observed throughout the body. n = 50–118 worms. (B) Pgst-4::GFP reporter scoring in either a wdr-23 loss of function (tm1817) or a skn-1 gain of function allele (k1023) mutant background. n = 54–75 worms. *P<0.05, **P<0.01 and ***P<0.001 relative to corresponding control as determined by Chi-Square tests; note that pad-1(RNAi) in skn-1(k1023) could not be tested for significance because it had zero worms with low or medium fluorescence.
We next tested the candidate genes for genetic interactions with wdr-23 and skn-1 using strains carrying the Pgst-4::GFP reporter and a wdr-23(tm1817) loss of function allele or a skn-1(k1023) gain of function allele [10]. Both strains have constitutive activation of Pgst-4::GFP that is suppressed by skn-1(RNAi). dsRNA targeting the candidates had little or no effect on Pgst-4::GFP with the exception of ifb-1 and uba-1 in wdr-23(tm1817) (Fig 5B); although not confirmed in these experiments, skr-1/2(RNAi) consistently causes a partial embryonic lethality phenotype. These findings suggest that skr-1/2, C01B10.3, and pad-1 likely function upstream of skn-1 and wdr-23.
skr-1/2 is required for induction of skn-1 dependent detoxification genes by diverse compounds
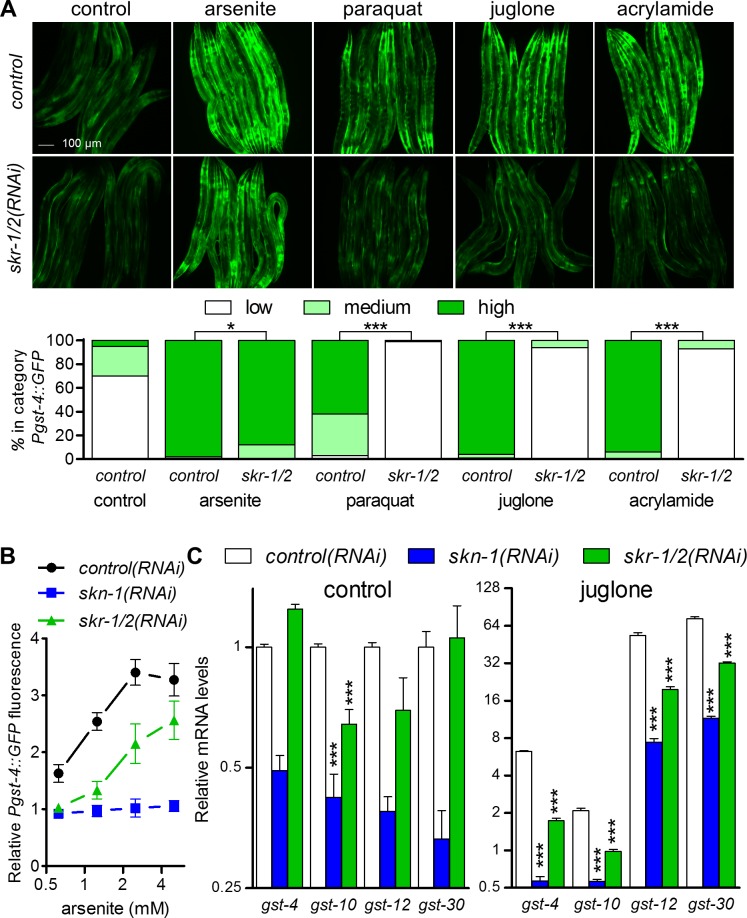
skr-1/2(RNAi) (Skp-related) caused the strongest inhibition of Pgst-4::GFP with juglone, and most closely resembled the pattern observed for skn-1(RNAi) (Fig 5A). SKR-1/2 was previously shown to be required for longevity extension in daf-2 insulin and IGF-1-like receptor mutants [33], but has not previously been reported to function as a regulator of stress responses. Because the neighboring skr-1 and skr-2 genes are recent duplications that are 83% identical at the nucleotide level, skr-1 dsRNA likely silences both skr-1 and skr-2 [24]. Consistent with this, skr-1 dsRNA reduces both skr-1 and skr-2 mRNA (S1 Table) and skr-2 dsRNA has the same effect on Pgst-4::GFP as skr-1 dsRNA (S7A Fig). We therefore refer to ‘skr-1/2(RNAi)’ when using skr-1 dsRNA.
C. elegans has a total of 21 SKR proteins that are orthologous to the single yeast and mammalian Skp1 proteins, which are one of the four core components of the highly conserved SCF ubiquitin-ligase complex. Skp1 interacts directly with cullin and F-box proteins, with the latter functioning to selectively recruit substrate targets for ubiquitin ligation [23]. To address whether other skr genes or components of the SCF complex are also required for gst-4 activation in response to juglone, we performed an RNAi screen against a sub-library of available skr dsRNA clones (skr-3, 5, 7, 8, 9, 10, 11, 12, 13, 15, 17, 18, 19, 20, and 21) and cul-1, which is the only cullin that is known to interact with SKR-1 and 2 [23,24]. Unlike skr-1/2, none of the other skr clones or cul-1 had a significant effect on Pgst-4::GFP induction after juglone exposure (S7B Fig). Due to a general sickness and larval arrest of cul-1(e1756) null mutants [34], we were unable to confirm the cul-1(RNAi) results using a mutant. To enhance RNAi, we also tested eri-1 RNAi hypersensitive worms fed cul-1 dsRNA for two generations. Embryonic lethality (a well-established cul-1 phenotype [35]) was observed in second generation worms fed cul-1 dsRNA, but there was still no effect on Pgst-4::GFP when induced with juglone in second generation worms that were able to develop to the L4 stage (S7C and S7D Fig).
To investigate the role of skr-1/2 in response to reactive compounds, we tested its requirement for Pgst-4::GFP induction during exposure to arsenite, paraquat, juglone, and acrylamide. skr-1/2(RNAi) strongly inhibited induction of Pgst-4::GFP with juglone, paraquat, and acrylamide and had a smaller effect with arsenite (Fig 6A); a role for skr-1/2 in Pgst-4::GFP induction by arsenite was confirmed in a more sensitive plate reading assay at four doses (Fig 6B). The requirement for skr-1/2 was also tested with qPCR of four gst mRNAs directly controlled by SKN-1 under control conditions and after exposure to 38 μM juglone for 3 h (Fig 6C). Loss of skr-1/2 only reduced expression of one of the gst genes under control conditions. Juglone activated all four gst mRNAs (P<0.001, Fig 6C), and as expected, skn-1(RNAi) strongly reduced induction of all four gst genes in juglone. Silencing of skr-1/2 also strongly inhibited induction of all four gst genes by juglone by 57–74%. Therefore, skr-1/2 is partially required for induction of core SKN-1 dependent detoxification genes by diverse reactive compounds.
Fig 6. skr-1/2 is required for induction of SKN-1 dependent detoxification genes.
(A) Pgst-4::GFP fluorescence scoring and representative fluorescence micrographs of worms fed with control or skr-1/2 dsRNA after exposure to 5 mM sodium arsenite for 1 h (recovered for 3 h on NGM agar to induce GFP), 35 mM paraquat for 2 h (recovered for 2 h), 38 μM juglone for 3 h (recovered for 1 h), or 7 mM acrylamide for 4 h (no recovery). n = 70–89 worms from 3 independent trials, *P<0.05, ***P<0.001 as determined by Chi-Square test. (B) Relative Pgst-4::GFP fluorescence measured by a plate reader after a 6 h exposure to a range of arsenite concentrations; all values are normalized to control(RNAi) with no arsenite. n = 4–8 wells of worms in a 384 well plate; P<0.001 for skr-1/2(RNAi) and skn-1(RNAi) versus control(RNAi) at all concentrations except for the lowest. (C) Fold changes in mRNA of gst-4, gst-10, gst-12, and gst-30 relative to control (no stressors) in worms with control, skn-1, or skr-1/2(RNAi) after exposure to 38 μM juglone for 3 h. mRNA levels were normalized to rpl-2; values are means plus standard error of n = 4 replicates of 200–400 worms. All genes were induced significantly by juglone (P<0.001); ***P<0.001 compared to control (RNAi).
Global RNA-seq analysis demonstrates that skr-1/2 is specifically required for skn-1 transcriptional responses
To obtain a global view of gene regulation by skr-1/2, we extracted RNA from worms grown on control, skr-1/2, and skn-1(RNAi) after exposure to juglone and identified differentially expressed genes (DEG) using unbiased whole-genome RNA sequencing (RNA-seq). The FOXO transcription factor daf-16 is also widely involved in detoxification responses [36] and a previous study implicated skr-1/2 in regulation of a DAF-16 dependent gene and in promoting longevity in daf-2 mutant worms, which have elevated DAF-16 activity [33]. Therefore, we also included daf-16(RNAi) to evaluate the specificity of gene regulation by skr-1/2. We identified 309 (174 up, 135 down) DEG by juglone, 273 (142 up, 131 down) DEG by skr-1/2(RNAi), 1241 (517 up, 737 down) DEG by skn-1(RNAi), and 562 (281 up, 281 down) DEG by daf-16(RNAi) (Fig 7A and 7B, S8A and S8B Fig and S1 Table).
Fig 7. Whole transcriptome profiling reveals correlation between loss of skn-1 and skr-1/2.
(A) Heat map of log2 fold-change of 174 genes found to be significantly up-regulated by juglone (38 μM for 3 h) with their corresponding fold-changes in worms fed with skn-1, skr-1/2, or daf-16 dsRNA compared to control dsRNA. Fold-change analysis for RNA-seq data were carried out using the CuffDiff application in Galaxy with q-value <0.05 determined by a false discovery rate of <5%. n = 3 replicates of 1,000–2,000 worms. Coefficients of correlation are listed below the map and 95% confidence intervals are as follows: skn-1 and skr-1/2 (0.693–0.818), skn-1 and daf-16 (0.533–0.712), and skr-1/2 and daf-16 (0.090–0.370). (B) Venn diagrams showing numbers of genes overlapping. (C) DAVID functional enrichment analysis of skr-1/2(RNAi) down-regulated genes. (D) Linear regression analysis of all genes down-regulated by either skn-1 or daf-16(RNAi) plotted against their fold change with skr-1/2(RNAi). ***P<0.001 as determined by linear regression F-test.
A heat map of all genes up-regulated by juglone is shown in Fig 7A demonstrating that many of the strongest up-regulated genes were dependent on both skn-1 and skr-1/2. Correlation coefficients are shown below the heat map for all comparisons demonstrating a much higher correlation of skr-1/2 with skn-1 (0.763) than with daf-16 (0.235) for the genes up-regulated by juglone. Of the 174 genes up-regulated by juglone, 78 were skn-1 dependent and 35 were skr-1/2 dependent (Fig 7B). Of the 35 skr-1/2 dependent juglone induced genes, almost all (32, or 91%) were also skn-1 dependent. Furthermore, 110 of the 131 (84%) total skr-1/2 dependent genes were also skn-1 dependent; 14.7% of all skn-1 dependent genes were skr-1/2 dependent. Compared to skn-1(RNAi), daf-16(RNAi) affected fewer overall genes but had a similar proportional overlap with skr-1/2 (13.5%, Fig 7B). Using DAVID analysis, skr-1/2 dependent genes were primarily enriched for glutathione mediated detoxification and metabolism, glucosyltransferase activities, and collagen/cuticle development (Fig 7C), while the genes up-regulated by skr-1/2(RNAi) were enriched for regulation of growth rate, collagen, and metal ion binding (S8C Fig). Genes up-regulated by skr-1/2(RNAi) had little overlap with genes influenced by juglone (S8B Fig).
To gain deeper insights into the gene expression effects of skr-1/2 relative to skn-1 and daf-16, we performed linear regression analysis on all genes that were down-regulated by skn-1(RNAi) or daf-16(RNAi), regardless of how they were affected by juglone, by plotting fold change with skr-1/2(RNAi) versus fold change by skn-1 or daf-16(RNAi) (Fig 7D). Fold-changes for skr-1/2(RNAi) were significantly correlated with fold-changes for all genes significantly downregulated by skn-1(RNAi) (slope = 0.265±0.02, R2 = 0.24, P < 0.0001) (Fig 7D). No correlation was observed between skr-1/2 and daf-16(RNAi) for all genes significantly downregulated by daf-16(RNAi) (slope = 0.08±0.05, R2 = 0.01, P = 0.07) (Fig 7D). A significant, but very small, correlation was found between skr-1/2 and skn-1(RNAi) for genes up-regulated by skn-1(RNAi), not for genes up-regulated by daf-16(RNAi) (S8D Fig). Taken together, these results reveal that skr-1/2 is required for expression of a subset of detoxification and extracellular matrix genes that are largely (84%) nested within a set of skn-1 dependent genes and not correlated with daf-16 dependent genes.
Loss of skr-1/2 reduces oxidative stress resistance
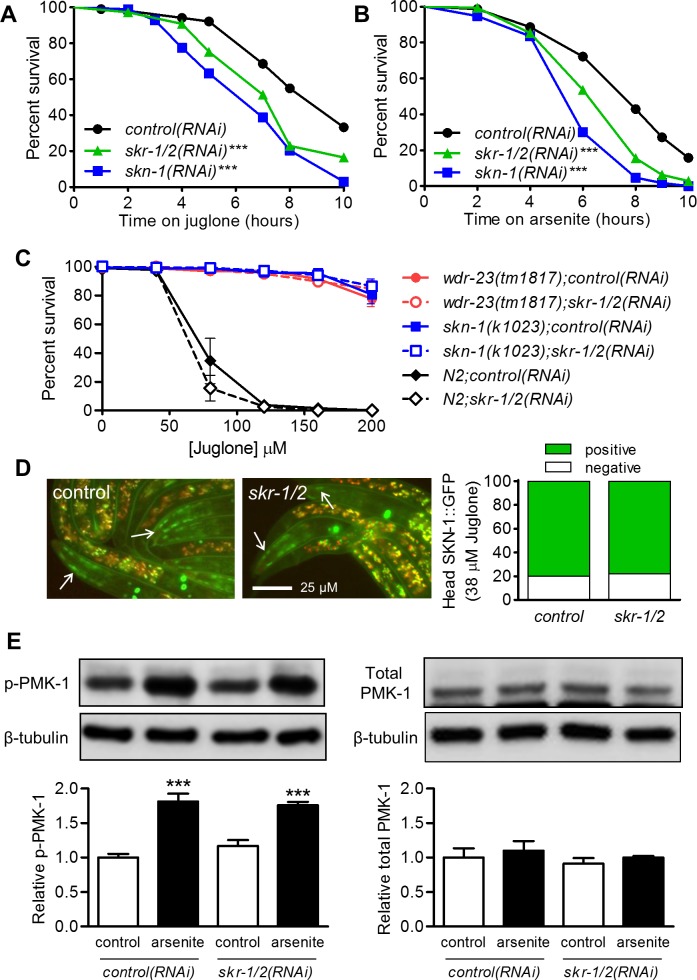
We next examined the requirement of skr-1/2 for juglone and arsenite resistance. In our hands, skn-1(RNAi) often does not have a reproducible effect on survival of juglone in worms that have not previously been exposed suggesting that basal activity may not play a large role in survival of an acute lethal juglone dose. We then pre-conditioned worms to a low and hormetic level of juglone (38 μM for 2 h), which strongly activates SKN-1 dependent genes (Fig 2B) and dramatically increases resistance [37], before measuring survival in a much higher lethal dose (125 μM). In these experiments, skn-1(RNAi) significantly decreased juglone survival compared to control worms in all three trials and skr-1/2(RNAi) significantly decreased survival in two of three trials (Fig 8A and S9 Fig). In 10 mM arsenite, either skn-1(RNAi) or skr-1/2(RNAi) significantly decreased survival compared to the control worms (Fig 8B and S9 Fig). These results show that skr-1/2 is required for oxidative stress resistance.
Fig 8. skr-1/2 is required for juglone and arsenite resistance but not for SKN-1 accumulation or PMK-1 phosphorylation.
(A) Survival of eri-1 worms fed control, skn-1, or skr-1/2 RNAi on 125 μM juglone after a 2 h pre-treatment at 38 μM. ***P<0.001 compared to control RNAi as determined by Log-rank (Mantel-Cox) test. n = 98–109 worms from a single trial; statistics and results from two other trials are shown in S9 Fig. (B) Survival of N2 worms fed control, skn-1, or skr-1/2 RNAi on 10 mM arsenite. ***P<0.001 compared to control RNAi as determined by Log-rank (Mantel-Cox). n = 140–170 worms from a single trial; statistics and results from two other trials are shown in S9 Fig. (C) Survival of worms with enhanced SKN-1 activity exposed to a range of juglone concentrations for 16 h. skr-1/2(RNAi) did not significantly decrease survival at any concentration in any strain. n = 4 independent trials of 12–78 worms per condition and trial. (D) Animals with integrated SKN-1b/c::GFP were fed with either control or skr-1/2(RNAi) and treated with 38 μM juglone for 3 h and accumulation of head SKN-1b/c::GFP was scored. Representative fluorescence micrographs of head SKN-1b/c::GFP are shown with arrows marking GFP. n = 45 to 54 worms. (E) Phosphorylation of PMK-1 was measured in worms fed control or skr-1/2 dsRNA and exposed to 5 mM sodium arsenite for 1 h. PMK-1 phosphorylation levels were normalized to β-tubulin and then to control (without stressor); β-tubulin was detected on the same blots after stripping. Total PMK-1 was measured on the same lysates in a different blot and also normalized to β-tubulin. Values are mean plus standard error for densitometry of protein bands from n = 3 replicates of >1,000 worms. Representative immunoblot bands are shown, ***P<0.001 relative to control. There was no significant effect of skr-1/2(RNAi).
We next tested the effects of skr-1/2(RNAi) on juglone survival in two strains with greatly increased SKN-1 activity, wdr-23(tm1817) loss of function and skn-1(k1023) gain of function [10]. As expected, both of these strains were highly resistant over a range of juglone concentrations compared to N2, and skr-1/2(RNAi) did not have a reproducible effect on juglone resistance in N2 worms not previously exposed to juglone (Fig 8C). As expected given that skr-1/2 appears to regulate gst-4 upstream from wdr-23 (Fig 5B), skr-1/2(RNAi) did not reduce resistance of either wdr-23(tm1817) or skn-1(k1023) worms (Fig 8C).
skr-1/2(RNAi) does not affect nuclear accumulation of SKN-1::GFP or PMK-1 phosphorylation
Having established skr-1/2 as a requirement for SKN-1 dependent detoxification responses, we conducted additional experiments to gain insight into potential mechanisms. We demonstrated above that skr-1/2 likely regulates detoxification gene expression upstream from WDR-23 and SKN-1 (Fig 5B). To test if skr-1/2 is required for nuclear localization of SKN-1, we performed skr-1/2(RNAi) in a strain carrying the SKN-1b/c::GFP transgene and scored accumulation after exposure to juglone. Because we did not observe an increase of SKN-1b/c::GFP accumulation in the intestine nuclei by juglone (Fig 3A), we scored the effects of skr-1/2(RNAi) on SKN-1b/c::GFP accumulation in the head region. RNAi against skr-1/2 failed to prevent accumulation of SKN-1b/c::GFP in the head after juglone exposure (Fig 8D).
We next determined if skr-1/2 loss influences phosphorylation of PMK-1. RNAi against skr-1/2 had no effect on total or phosphorylated PMK-1 levels under basal conditions or when increased by arsenite (Fig 8E), a condition that strongly increased phosphorylated PMK-1 levels (Fig 1). Taken together, these data suggest that although skr-1/2 is required for activation of SKN-1 target genes by stress, it does so without observable changes to SKN-1 nuclear accumulation or p38 MAPK phosphorylation.
An skr-1 deletion mutant decreases SKN-1 target gene activation
As mentioned earlier, skr-1 and skr-2 are very similar even at the nucleotide level (83% identical). In order to investigate if one or both are required for gst induction, we conducted qPCR experiments with skr-1(tm2391) and skr-2(ok1938) deletion mutants. skr-1(tm2391) and skr-2(ok1938) homozygotes are each maternal effect embryonic lethal and skr-1 mutants also display late larval arrest [34]; RNA was isolated by picking healthy homozygote larvae at the L3 and early L4 stages. The skr-1(tm2391) allele reduced mRNA levels of four juglone induced gst genes (S10A Fig). The skr-2(ok1938) allele only slightly reduced mRNA levels for two genes (gst-4 and 12) and increased induction of gst-10 and gst-30. Interestingly, skr-2(ok1938) had a ~40% reduction in skr-1 mRNA levels (S10B Fig), likely due to a partial deletion of the skr-1 promoter in this mutant; however, this reduction in skr-1 mRNA appeared to have minimal effect on the activation of skn-1 dependent genes in response to juglone. These results help confirm a role of skr-1 and are consistent with SKR-2 being less important than SKR-1 for SKN-1 pathway activation.
To test if overexpression of skr-1/2 alone is sufficient to increase expression of SKN-1 transcriptional targets, we generated a transgenic worm overexpressing a 4 kb DNA fragment that contained full genomic sequences of both skr-1 and skr-2 and an integrated Pgst-4::GFP reporter. Transgenic worms carrying the 4 kb DNA fragment overexpressed (oe) both skr-1 and skr-2 mRNA, but had wild-type levels of Pgst-4::GFP fluorescence (S11A and S11B Fig); qPCR confirmed that the mRNA levels of SKN-1 transcriptional targets gst-4 and gcs-1 were unaffected by skr-1/2 overexpression at basal conditions, and the overexpression also did not further activate SKN-1 target transcription during juglone exposure (S11C Fig). Therefore, overexpression of skr-1/2 is not sufficient to activate the SKN-1 stress response.
SKR-1 is expressed broadly and interacts with and influences WDR-23
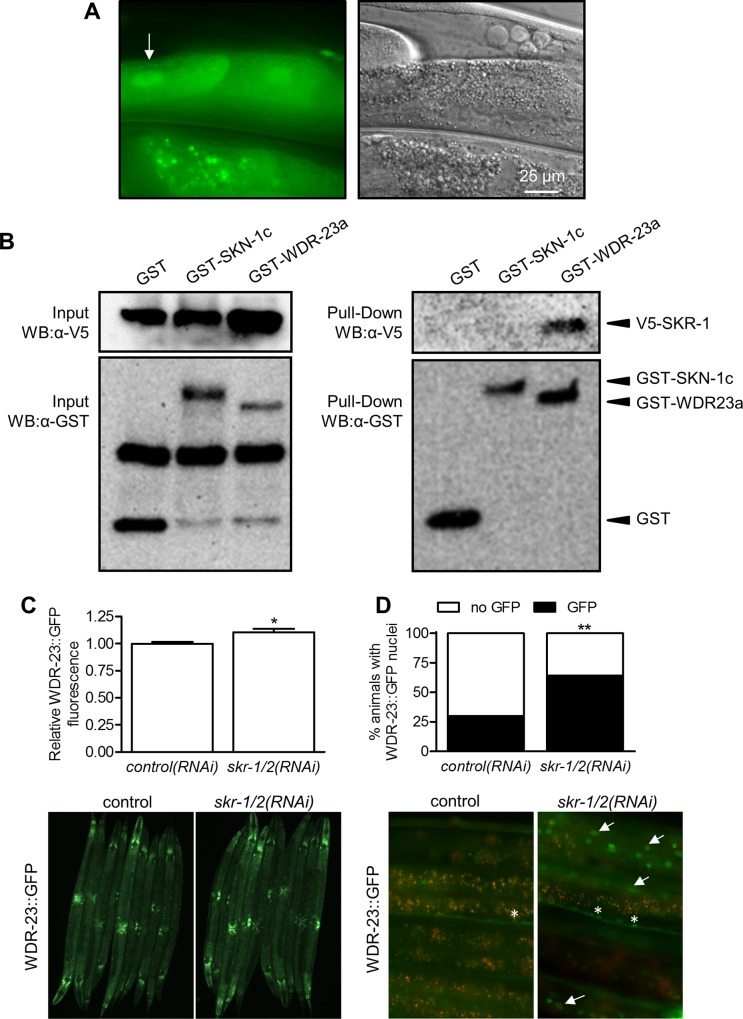
To determine the expression pattern of SKR-1, we generated transgenic worms expressing a SKR-1::GFP fusion protein. Consistent with a previous study [24], SKR-1::GFP was expressed throughout the worm, with SKR-1::GFP highly expressed in the intestine, pharynx, neurons, and spermatheca (S12 Fig); intracellular distribution of SKR-1 was not previously determined. Within the large cells of the intestine, we observed SKR-1::GFP in the cytosol and nuclei suggesting that the protein is broadly distributed within cells (Fig 9A). To test if SKR-1 is regulated under stress, we treated this transgenic worm to conditions that activate SKN-1. Exposure to juglone and arsenite did not result in obvious changes in expression or localization of SKR-1::GFP (S11D Fig).
Fig 9. SKR-1 is expressed broadly and interacts with WDR-23.
(A) Fluorescence and differential interference contrast micrographs showing cytoplasmic and nuclear (arrow) expression of SKR-1::GFP in the intestine (QV254). (B) SKR-1 interacts with WDR-23. HEK293 cells were co-transfected with V5-SKR-1 fusion protein along with either GST only vector (pDEST27), GST-SKN-1c fusion protein, or GST-WDR23a fusion protein. Complexes were captured with GSH beads and interactions with SKR-1 were determined by immunoblotting with anti-V5 mAb. Co-pulldown of V5-SKR-1 with GST-WDR-23a was also detected in a separate independent trial. (C-D) Total WDR-23::GFP fluorescence (normalized to RFP) and percentage of worms with visible nuclear WDR-23::GFP fluorescence with and without skr-1/2(RNAi). *P<0.05 and **P<0.01. Arrows point to hypodermal nuclei and asterisks mark neuronal cells.
To test whether SKR-1 might interact with key members of the SKN-1 pathway, we co-transfected either full length SKN-1c or WDR-23a fused to an N-terminal GST tag together with SKR-1 fused to an N-terminal V5 fusion tag in HEK293 cells and performed GST pull-downs. Pulldown of GST-SKN-1c failed to capture SKR-1 as determined by Western blot using a V5 monoclonal antibody (Fig 9B). Alternatively, SKR-1 was captured by pull-down of GST-WDR-23a. We also used a strain expressing an integrated transgene of wdr-23a cDNA fused to GFP [38], which rescues misexpression of a gst-4 transgene in wdr-23 mutants, to test if SKR-1/2 might regulate WDR-23 in vivo. As shown in Fig 9C, skr-1/2(RNAi) had a significant but very small effect on WDR-23::GFP fluorescence levels. Alternatively, skr-1/2(RNAi) doubled the proportion of worms with obvious nuclear localization in the hypodermis (Fig 9D). Obvious changes to intestinal WDR-23::GFP were not observed.
Discussion
In C. elegans, the p38 MAPK signaling cascade activates SKN-1 in the intestine during oxidative stress induced by arsenite exposure [19] and during pathogen infection [20,21]. Here, we demonstrate striking variation in PMK-1 activation kinetics among diverse pro-oxidant and electrophilic compounds that all strongly induce SKN-1 dependent transcriptional responses. SKN-1 can also accumulate in the nuclei of tissues other than the intestine partially independent of PMK-1. To begin defining other regulatory pathways that may function parallel to PMK-1, we leveraged the genetic tractability of C. elegans to identify a novel role for the highly conserved Skp1 homologs skr-1/2 in the SKN-1 mediated stress response. SKR-1 interacts with and influences the localization of the SKN-1 repressor WDR-23, and loss of skr-1/2 inhibits the expression of skn-1 dependent detoxification genes and impairs survival during exposure to pro-oxidants. A revised working model for SKN-1 regulation by reactive small molecules that incorporates our findings is presented in Fig 10 and discussed below.
Fig 10. Model of SKR-1 activation by pro-oxidants and reactive small molecules.
In response to pro-oxidants, PMK-1 is activated and contributes to SKN-1 activation. Additional parallel or compensatory mechanisms likely exist given the ability of detoxification genes to be induced strongly without PMK-1 (Fig 2B). In response to diverse pro-oxidants and electrophiles, SKN-1 dependent gene expression is induced by a mechanism that requires SKR-1/2 (Figs 5 and 6). SKR-1/2 is not required for phosphorylation of p38 MAPK (Fig 8) and likely functions independently. SKR-1 acts upstream of WDR-23 (Fig 5), is able to interact with WDR-23 (Fig 9), and influences its nuclear levels suggesting a mechanism through this direct SKN-1 repressor.
PMK-1 phosphorylation and intestinal SKN-1::GFP accumulation vary with different pro-oxidants that activate SKN-1 dependent stress responses
Phosphorylation of PMK-1 at activating residues by arsenite and pathogen exposure is associated with intestinal accumulation of nuclear SKN-1 consistent with the prevailing model in which PMK-1 phosphorylates SKN-1 in the intestine during oxidative stress and causes nuclear accumulation [19]. Interestingly, PMK-1 phosphorylation kinetics varied greatly between different sub-lethal pro-oxidant and electrophile exposures that all strongly activate SKN-1 dependent detoxification gene expression (Fig 1). Acrylamide even decreased PMK-1 phosphorylation levels during chronic exposure, but caused strong and sustained Pgst-4::GFP induction (Figs 1 and 2 and S3 Fig). We also observed strong SKN-1 dependent detoxification gene activation in pmk-1 deletion mutants during sub-lethal exposure to arsenite and juglone (Fig 2) and SKN-1 nuclear accumulation in tissues other than the intestine (Fig 3 and S4 Fig) and in pmk-1 deletion mutants (Fig 4).
Importantly, SKN-1 dependent gene induction was partially dependent on pmk-1 (Fig 2B), which is consistent with a recent study showing that sek-1 and pmk-1 are required for induction of a downstream target of SKN-1 in neurons (nlg-1) that promotes survival of juglone [39]. We also cannot rule out the possibility that SKN-1 accumulates in the intestine with juglone at levels below what we are able to detect. Our data establish that pro-oxidants and electrophiles are capable of activating SKN-1 and its downstream detoxification genes outside the intestine and with varying degrees of SKN-1::GFP nuclear accumulation and PMK-1 activation suggesting the presence of parallel or compensatory mechanisms; e.g., other kinases downstream from NSY-1/SEK-1 (such as PMK-2 or PMK-3) or other unknown pathways. It is also possible that other transcription factors could help compensate.
C. elegans skr-1/2 is required for detoxification gene induction
SKR homologs function in SCF multi-subunit E3 ubiquitin ligases that are conserved from yeast to humans. In yeast and humans, a single SKR named Skp1 forms the SCF complex with Rbx, Cul1, and various F-box proteins to promote protein ubiquitination [40,41]. Skp1 also functions as a scaffold in protein complexes independently of SCF [42,43,44,45]. In C. elegans, 21 skr genes have been identified [23,24]. Loss of function of either skr-1 or 2 results in embryonic arrest that is characterized by excessive cell numbers and hyperplasia [23,24]. Interestingly, skr-1/2(RNAi) delays degradation of SKN-1 during embryonic development leading to a delay in development [46]. SKR-1/2 was previously shown to be required for longevity in daf-2 mutants and not wildtype worms [33], but a role in stress responses has not been reported. In this study, we identified a new role for skr-1/2 in permitting skn-1 dependent stress responses in larval and adult stages of C. elegans.
skr-1/2 were required for transcriptional induction of a SKN-1 dependent gst-4 reporter by diverse pro-oxidants at sub-lethal doses (Fig 6) and for resistance to lethal doses of juglone and arsenite (Fig 8A and 8B). We found that skr-1/2(RNAi) did not reduce levels of total or phosphorylated PMK-1 (Fig 8E), and in our RNA-seq data, knockdown of skr-1/2 did not alter mRNA levels of core MAPK genes (nsy-1, sek-1, or pmk-1) (S1 Table). Although it remains possible that skr-1/2 could affect protein levels of NSY-1 or SEK-1, these data raise the possibility of SKR-1/2 functioning by a different mechanism.
skr-1/2 specifically regulates skn-1 dependent responses
The function of skr-1/2 in our study was shown to be tightly linked to SKN-1. Knockdown of skr-1/2 did not block heat-shock or osmotic stress transcriptional response reporters (S6 Fig) and skr-1/2 dependent genes were enriched for functions in detoxification and cuticle collagen (Fig 7), which are both also enriched among skn-1 dependent genes [12,30,47]. Furthermore, our RNA-seq data demonstrated that a strikingly large majority (84%) of skr-1/2 dependent genes were also skn-1 dependent and that expression changes caused by skr-1/2 and skn-1 RNAi were correlated (Fig 7). skr-1/2 had no correlation with genome-wide daf-16 dependent expression (Fig 7).
How might SKR-1/2 regulate SKN-1 activity?
The SCF ubiquitin ligase complex has previously been reported to negatively regulate Nrf2 protein levels in human cells; phosphorylation of Nrf2 by glycogen synthase kinase 3 promotes ubiquitin mediated Nrf2 degradation by the SCF complex via its interaction with the β-transducin repeat-containing protein (β-TrCP) F-box [48]. Our results show that skr-1/2 Skp1 homologs in C. elegans positively regulate skn-1 mediated stress responses. RNAi against the gene encoding the central scaffold of the C. elegans SCF complex, cul-1, failed to mimic the effects of skr-1/2(RNAi) on regulation of the gst-4 promoter during exposure to juglone (S7 Fig). However, we cannot rule out a role for cul-1 because of the possibility of residual CUL-1 function after RNAi treatment.
C. elegans and Drosophila melanogaster each contain expanded families of Skp1 homologs whereas yeast and vertebrates, including humans, have only one member [23,24]. Having multiple Skp1 homologs has been hypothesized to permit the evolution of more flexible and variable functions. Even in yeast and human cells where single homologs are present, evidence has been provided for Skp1 functioning independently of the SCF complex to regulate membrane protein recycling, kinetochore function, ion transport, and protein degradation [42,43,44,45,49]. In all these cases, the molecular role of Skp1 appears to be as a scaffold to assist in protein complex assembly.
In C. elegans, SKN-1 is regulated directly by WDR-23 and PMK-1 [16,19]. Our data do not support a positive association between SKR-1/2 and PMK-1 function as skr-1/2(RNAi) did not reduce PMK-1 protein levels or its stress activated phosphorylation (Fig 8E). Instead, SKR-1 has the potential to interact with WDR-23 and the requirement of skr-1/2 for skn-1 dependent gene induction was abolished in a wdr-23 null mutant (Fig 5B). WDR-23 is a direct repressor of SKN-1 that is present in many cells [16,25,50], and loss of wdr-23 promotes stress resistance and a long-lived phenotype that is skn-1 dependent [10]. SKR-1 was present throughout cells including within some nuclei (Fig 9A) where it could interact with WDR-23. Our genetic interaction and WDR-23::GFP results (Figs 5B, 9C and 9D) are consistent with SKR-1/2 functioning to negatively regulate WDR-23 accumulation in nuclei likely by serving as a scaffold to influence protein complex assembly (Fig 10). SKN-1 dependent genes are extremely sensitive to changes in WDR-23 function [16] and one possible mechanism is for SKR-1/2 to negatively regulate WDR-23 protein levels via ubiquitination. In this model, loss of skr-1/2 would increase WDR-23 levels and repression of SKN-1.
Importantly, we did not observe accumulation of either SKN-1b/c or SKN-1op(a/b/c) reporters in the intestine after juglone exposure and skr-1/2 RNAi did not impair SKN-1b/c::GFP accumulation in head nuclei (Fig 8D). Although SKR-1/2 could influence SKN-1 accumulation at levels that we were not able to detect, it is also possible that SKR-1/2 and WDR-23 may be able to regulate SKN-1 by alternative mechanisms such as DNA binding or transactivation activity. Future experiments to investigate these alternatives and to identify other SKR-1 and WDR-23 interacting proteins required for the detoxification response could help clarify the molecular mechanisms of WDR-23 and SKN-1 regulation. Induction of SKN-1 detoxification genes can also be decoupled from nuclear accumulation in the intestine during genetic impairment of translation and fatty acid metabolism pathways [51,52]. It remains to be seen if SKR-1/2 plays a role under these conditions.
Summary
Our study provides evidence that PMK-1 phosphorylation and SKN-1 nuclear localization are differentially regulated in response to different reactive small molecule exposures that activate SKN-1 dependent detoxification responses. We also identify SKR-1/2 as required for SKN-1 dependent detoxification gene induction in response to diverse pro-oxidants and electrophiles that may interact with and regulate WDR-23. SKN-1 is activated by a broad range of different conditions and our study highlights the fact that distinct upstream regulatory pathways are present that may permit tailored responses to oxidative and reactive small molecule stressors. These findings and the identification of SKR-1/2 as a key regulator lay the foundation for defining a novel SKN-1 regulatory mechanism that involves WDR-23.
Materials And Methods
C. elegans strains
C. elegans strains were grown and maintained at 20°C using standard methods [53], unless noted otherwise. The following strains were used: wild-type N2 Bristol, VP596 dvIs19 [pAF15(Pgst-4::GFP::NLS)]; vsIs33[Pdop-3::RFP]), QV65 gpIs [Phsp-16.2::GFP]; vsIs33, CF1580 daf-2(e1370) III; muIs84 [(pAD76) sod-3p::GFP + rol-6], QV25 wdr-23(tm1817); eri-1(mg666)IV; dvIs19, QV130 skn-1(k1023); dvIs19, KU25 pmk-1(km25)IV, KU4 sek-1(km4)X, LD1 ldIs7 [skn-1B/C::GFP + pRF4(rol-6(su1006))], LD1008 [skn-1(operon)::GFP, rol-69su1006)], VP537 eri-1(mg666)IV; dvIs19, GR1373 eri-1(mg366)IV, VP604 kbIs24 [Pgpdh-1::dsRed2;Pmyo-2::GFP;unc-119] X, QV254 zjEx114 [SKR-1::GFP; Pmyo-2::tdTomato], QV256 zjEx115 [skr-1/2 gDNA; Pmyo-2::tdTomato; Pmyo-3::dsRed], QV288 pmk-1(km25)IV; ldIs7, VC1439 skr-2(ok1938) I/hT2 [bli-4(e937) let-?(q782) qIs48] (I;III), CU6110 skr-1(tm2391) I/hT2 I;III; +/hT2 I;II, and KHA116 unc-119(ed3); chuIs116 [wdr-23p::wdr-23a(cDNA)::GFP, unc-119].
RNAi and genome-wide RNAi screening
RNAi was performed as described previously [16] by feeding worms strains of E. coli [HT115(DE3)] that are engineered to transcribe double stranded RNA (dsRNA) homologous to a target gene. For screening, L1 larvae of VP537 worms were grown in liquid medium with dsRNA-producing bacteria for 3 days, and subsequently exposed to 38 μM juglone for 4 h and screened for Pgst-4::GFP expression with a Zeiss Stemi SV11 microscope. The entire ORFeome RNAi feeding library (Open Biosystems, Huntsville, AL) was screened with additional missing clones supplemented from the original genomic RNAi feeding library (Geneservice, Cambridge, United Kingdom). Clones that resulted in reduced Pgst-4::GFP were rescreened three additional times, clones with positive scores in all three trials were considered novel regulators of gst-4 and subsequently sequenced for identification. With the exception of the genome-wide RNAi screen, all subsequent RNAi experiments were performed on nematode growth media (NGM) plates that were made with the addition of 50 μg mL-1 carbenicillin, 0.2% lactose, and seeded with appropriate HT115 RNAi bacteria and grown overnight before use. RNAi feeding was initiated at either synchronized L1 larvae stage, or at L4 to young adult stage for RNAi clones that displayed developmental effects when fed from L1. Bacteria with plasmid pPD129.36 were used as a control for non-specific RNAi effects. This control plasmid expresses 202 bases of dsRNA that are not homologous to any predicted C. elegans gene.
Fluorescence microscopy with GFP reporters
To visualize SKN-1b/c::GFP, LD1 worms at L1/L2 stages were incubated with 5 mM sodium arsenite for 1 h, 5 mM sodium azide for 5 min, 35 mM paraquat for 2h, 38 μM juglone for 5–15 min, 1, 3, and 5 h, or 7 mM acrylamide for 5–15 min, 1, 4, 12, or 24 h. Worms were then washed and anesthetized with 5 mM levamisole. Anesthetized worms were mounted on 2% agarose pads and visualized and imaged using an Olympus BX60 microscope and Zeiss AxioCam MRm camera. Differential interference contrast and fluorescent images of SKN-1b/c:GFP and SKN-1op::GFP were taken. Both grayscale images taken by the GFP and RFP channels were merged into green and red channels respectively to produce a composite image using ImageJ (NIH). When needed for clarity, brightness and contrast adjustments were made equally to images from the same color channel and within the same experiment. Accumulation of SKN-1b/c::GFP and SKN-1op::GFP expression in intestinal nuclei was scored as in previous studies [19], while accumulation of SKN-1b/c::GFP and SKN-1op::GFP in the head regions were scored as either negative, referring to no observation of GFP signals in the head region, or positive, referring to GFP signals observed throughout the head region.
To visualize Pgst-4::GFP, young adult worms were treated with 5 mM sodium arsenite for 1 h in liquid NGM (and were washed 3x with NGM buffer and allowed to recover for 3 h on NGM agar), 35 mM paraquat for 2 h in liquid NGM (recovery for 2 h), 38 μM juglone in liquid NGM for 3 h (recovery for 1 h), or 7 mM acrylamide in liquid NGM for 4 h (no recovery). These conditions were all sub-lethal (S1A Fig) but strongly induce Pgst-4::GFP fluorescence (Fig 5A). For Pgst-4::GFP scoring, low refers to little to no GFP observed throughout the worm, medium refers to GFP signals observed only at the anterior and posterior ends of the worm, and high refers to GFP signals observed throughout the body. To visualize Phsp-16.2::GFP, young adult worms were exposed to 33°C for 1 h followed by 5 h recovery at 20°C; for Pgpdh-1::RFP, young adult worms were exposed to 250 mM NaCl for 24 h. To visualize effects of oxidants on SKR-1::GFP expression and localization, young adult worms were exposed to 38 μM of juglone for 3 h or 5 mM arsenite for 1 h.
Real-time PCR
Quantitative real-time RT-PCR was used to measure mRNA levels in L4 to young adult stage worms fed with appropriate RNAi as described previously [16]. Worms were incubated with 5 mM sodium arsenite or 38 μM juglone in liquid NGM for 1 or 3 h at 20°C with gentle shaking. Total RNA from 200–300 worms was isolated with a Quick-RNA MicroPrep Kit (Zymo Research, Irvine, USA), and cDNA was synthesized using 1 μg of RNA with GoTaq 2-Step RT-qPCR System (Promega, Madison, WI, USA) following the manufacturer’s protocol. Quantitative real-time PCR was performed in 10 μL reactions in a Realplex ep gradient S Mastercycler (Eppendorf AG, Hamburg, Germany) with GoTaq Green Master Mix (Promega, Madison, WI) according to the manufacturer’s protocol. Data was analyzed by the standard curve method, with the housekeeping genes rpl-2 and cdc-42 used as internal reference controls. Primer sequences are available upon request.
Western blotting
Synchronized L4 to young adult stage N2 worms were treated with compounds in liquid NGM while gently shaking at 20°C. Each treatment was accompanied by a control group of worms incubated in NGM buffer for the same duration. After incubation with each stressor, worms were washed three times with NGM buffer, and approximately 1,000 L4 to young adult stage worms were lysed in homogenization buffer for each replicate (homogenization buffer content: 50 mM Tris Base pH 7.5, 150 mM NaCl, 0.1% SDS, 0.5% NaDeoxycholate, 1x Halt™ protease inhibitor cocktail, and 1x Halt phosphatase inhibitor cocktail (LifeTechnologies, Cat# 78430, #78420, Rockford, IL). Worms were sonicated for complete lysis by ultrasonication (Misonix XL 2000, Farmingdale, NY). Worm lysates were centrifuged at 13,000 x g for 10 min at 4°C and supernatants were normalized for protein concentration with BCA protein assay (Pierce, Cat#23227) and collected for SDS-PAGE electrophoresis. Equal volume of lysates totaling 30 μg of proteins were loaded and separated by SDS-PAGE, and detected by immunoblotting with phosphorylated PMK-1 (1:2000; Promega, Cat# V1211x), total PMK-1 antibody (1:1000; gift from K. Matsumoto [54]), and β-tubulin antibody (1:100; Developmental Studies Hybridoma Bank, Cat # E7) with methods as previously described [16].
Co-transfection and GST pulldown
HEK293 cells were cultured in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum, 4.5 g/l glucose, 584 mg/l L-glutamine, 100 mg/l sodium pyruvate, and 1 U/ml penicillin. Co-transfections were performed with Lipofectamine LTX with PLUS reagents (Life Technologies, Cat#1533810) using full length SKN-1c or WDR-23a cloned into pDEST27 vector and full length SKR-1 cloned into V5-DEST vector. Two days after transfection, cells were lysed with immunoprecipitation lysis buffer (Life Technologies, Cat # 87787) and pulldown was conducted using glutathione-Sepharose 4B at room temperature for 1 h (GE Healthcare, Cat#17-0756-01 Little Chalfont, United Kingdom). Beads from pulldowns were washed extensively with PBS+0.1% Triton-X and eluted with an equal volume of 2X SDS loading buffer by heating at 90°C for 5 minutes. Western blots were carried out as described above, with mouse anti-GST MAb (1:1,000; Santa Cruz Biotech, Cat #B-14 Dallas, TX), and mouse anti-V5 tag MAb (1:1000; Invitrogen, Cat #R960-25).
Stress resistance assays
Juglone stress resistance assays were performed on synchronized worms at L4 stages fed HT115 bacteria. Worms were pretreated with 38 μM of juglone in liquid NGM for 2 h and subsequently transferred to 125 μM juglone for survival analysis similar to [37]. In the arsenite stress resistance assay, worms were incubated with 10 mM of arsenite in liquid NGM and analyzed for survival. Worms were considered dead if they did not display any movement in response to prodding with a thin wire. A total of three independent trials were performed for each survival assay.
Whole transcriptome RNA sequencing
N2 worms were synchronized via by hypochlorite treatment and grown on RNAi for two days. Worms were then incubated in either NGM buffer (control) or 38 μM juglone for 3 h. RNA was extracted from ~1,000–2,000 worms per sample using the RNAqueous-Micro Total RNA Isolation Kit (ThermoFisher Scientific, Cat#AM1931). Total RNA was sent to The Yale Center for Genome Analysis for 75 nucleotide single-end sequencing in an Illumina HiSeq 2000. Raw sequencing data was processed using the public Galaxy server and mapped to the C. elegans genome (ce10). Using the Cufflink package and CuffDiff application from Galaxy [55], FPKM (Fragments Per Kilobase of transcript per Million mapped reads) were calculated and tested for differential expression with a FDR score of 5%. Differentially expressed genes were clustered for GO (Gene Ontology) analysis by DAVID for gene functional classification [56].
Statistical analyses
Statistical significance was determined using Student’s T-test when two means were compared and a one-way analysis of variance (ANOVA) with Tukey’s or Bonferonni multiple comparison tests when three or more means were compared. Log-Rank tests in the OASIS online tool were used when survival curves were compared [57]. Chi-square tests were used to evaluate categorical data, and linear regression was used for testing correlations in gene expression. P values of <0.05 were taken to indicate statistical significance except for comparing more than two survival curves, in which Bonferonni adjustments were made to P values to account for repeated comparisons. Statistical significance is indicated in figures as *P<0.05, **P<0.01, ***P<0.001, and ns = not significant. Data is available at: https://figshare.com/s/fb874c6dc1aa9b77375c
Supporting Information
Fragments Per Kilobase of transcript per Million mapped reads (FPKM) values are provided for triplicates.
(XLSX)
Percentage survival of worms after treatment with each oxidant were scored after 24 h recovery on NGM agar plate seeded with OP50 E. coli. n = 3 trials of 274–402 worms total.
(TIF)
Accumulation of intestinal and head SKN-1b/c::GFP after 5 or 15 min exposure to 2% azide, 5 mM arsenite, 38 μM juglone or 7 mM acrylamide, n = 63–84 worms per condition. ** P<0.01, ***P<0.001 from corresponding controls as determined by the Chi-Square test (bottom). Representative images of SKN-1b/c::GFP. Arrows mark head GFP and arrowheads mark intestinal nuclei.
(TIF)
Arrows mark Pgst-4::GFP in hypodermal nuclei, arrowheads mark Pgst-4::GFP in the intestine.
(TIF)
(A) Representative fluorescence micrographs of worms expressing SKN-1b/c::GFP treated with NGM buffer (control) or 5 mM sodium arsenite for 1h fed with control or skn-1(RNAi). (B) Scoring of intestinal and head SKN-1op::GFP and representative fluorescence micrographs. n = 52–71 worms. ***P<0.001 from corresponding controls as determined by Chi-Square tests. Scoring is the same as in Fig 2. (C) Representative fluorescence micrographs and differential interference contrast (DIC) images showing worms expressing SKN-1b/c::GFP in the hypodermis after exposure to 5 mM sodium arsenite for 1 h. Arrows mark head GFP and arrowheads mark ASI neurons.
(TIF)
Wildtype and tissue-specific RNAi worms fed with control and skn-1(RNAi) were exposed to 38 μM juglone for 2 h and then the concentration was raised to a total of 125 μM and survival was measured for up to 10 h. Graphs for trial 2 are shown above and a summary of the results for all three trials is show below. skn-1(RNAi) reduced survival in all three strains.
(TIF)
(A-B) Animals integrated with Phsp16.2::GFP or Pgpdh-1::RFP were fed bacteria producing dsRNA and exposed to either heat shock (transfer from 20 to 33°C for 1 h and 5 h recovery for Phsp16.2::GFP) or osmotic stress (transfer from 51 to 250 mM NaCl for 24 h for Pgpdh-1::RFP). Fluorescence was scored as in Fig 4. ***P<0.001 compared to control as determined by Chi-Square tests.
(TIF)
(A) skr-1 and skr-2 RNAi have similar effects on Pgst-4::GFP induced by juglone. (B) RNAi screen against a sub-library of genes functioning within the SCF complex in animals carrying the Pgst-4::GFP reporter after juglone exposure (38 μM for 3 h). ***P<0.001 compared to control as determined by Chi-Square tests. (C) Pgst-4::GFP scoring and representative fluorescence micrographs of eri-1 worms fed with control or cul-1(RNAi) for two generations and exposed to 38 μM juglone for 3 h. (A-C) n = 53–139 worms. (D) High penetrance of embryonic lethal and larval arrest phenotypes are observed in F2 generation of cul-1 RNAi fed eri-1 worms. F1 mothers were allowed to lay eggs and then removed for 24 h before taking an image; note a high number of dead eggs and sick L1 larvae.
(TIF)
(A) Heat map of fold-changes in 135 genes down-regulated by juglone with their corresponding fold-changes in worms fed with dsRNA for skn-1, skr-1/2 or daf-16. n = 3 replicates of 1,000–2,000 worms. Coefficients of correlation are listed below the heat map and 95% confidence intervals are as follows: skn-1 and skr-1/2 (0.442–0.649), skn-1 and daf-16 (0.299–0.542), and skr-1/2 and daf-16 (0.238–0.494). (B) Venn diagrams showing numbers of genes overlapping. (C) DAVID functional enrichment analysis of skr-1/2(RNAi) up-regulated genes. (D) Linear regression analysis of all genes up-regulated by either skn-1 or daf-16(RNAi) plotted against their fold change with skr-1/2(RNAi); ***P < 0.001 linear regression F-test.
(TIF)
Data from three individual trials of survival assays are provided. Representative trials are plotted in Fig 7A and 7B.
(TIF)
(A) Fold changes in mRNA level of gst-4, gst-10, gst-12, and gst-30 relative to control in N2 wild-type, skr-1(tm2391), and skr-2(ok1938) mutants after exposure to 38 μM juglone for 3 h. ***P<0.001 compared to N2 treated with juglone. (B) mRNA levels of skr-1 in the skr-2(ok1938) mutant and skr-2 in the skr-1(tm2391) mutant. Values are means plus standard error, n = 4 replicates of 200–400 worms.
(TIF)
(A) Relative Pgst-4::GFP fluorescence and (B) representative micrographs of worms (QV256) with and without skr-1/2(oe); note that the extrachromosomal array carries a genomic fragment that includes both skr-1 and 2, which are adjacent to each other. n = 33 to 56 worms. (C) Fold changes in mRNA level of skr-1, skr-2, gst-4, and gcs-1 in skr-1/2(oe) animals relative to control and when treated with or without 38 μM juglone for 3 h. mRNA levels were normalized to cdc-42, values are mean plus standard error. n = 4 replicates of ~50 worms. ***P<0.001 compared to control. (D) Expression and localization patterns of SKR-1::GFP (QV254) were not obviously affected when treated with 38 μM of juglone for 3h or 5 mM arsenite for 1 h.
(TIF)
Animals carrying an extrachromosomal array containing SKR-1::GFP and a Pmyo-2::tdTomato marker (QV254) were fed with either control or skr-1/2 dsRNA. (A) Shown are DIC images along with fluorescence micrographs taken with GFP and RFP filters. (B) Paired high magnification micrographs of SKR-1::GFP fluorescence and DIC (QV254); the upper right pair is a merger of red and green channels (compare to only GFP in the image immediately below).
(TIF)
Acknowledgments
C. elegans strains were provided by the Caenorhabditis Genetics Center (University of Minnesota, Minneapolis, MN) supported by the National Institutes of Health Office of Research Infrastructure Programs (P40 OD010440). We thank Dr. Ding Xue (University of Colorado Boulders) for providing the CU6110 strain, Dr. Koichi Hasegawa (Chubu University) for providing the KHA116 strain, and Dr. Kunihiro Matsumoto for the total PMK-1 antibody. This work was supported by NSF grants IOS-1120130 and IOS-1452948 to KPC, a National Sciences and Engineering Research Council of Canada (NSERC) postdoctoral award to CWW, National Institutes of Health grant R01 DK61168 to KS from the National Institute of Diabetes, Digestive and Kidney Diseases, by Institutional Development Award (IDeA) grant P20 GM103423 and Center of Biomedical Research Excellence (COBRE) grant P20 GM104318 from the National Institute of General Medical Sciences. AP performed the genome-wide RNAi screen; other experiments described in this paper were conducted by CWW and AD. CWW and KPC analyzed data and wrote the first draft of the manuscript; all authors participated in interpretation of data and revision and approval of the final version of the manuscript.
Data Availability
Data is available at: https://figshare.com/s/63e2e8045a83bac665dc.
Funding Statement
This work was supported by NSF grants IOS-1120130 and IOS-1452948 to KPC, a National Sciences and Engineering Research Council of Canada postdoctoral award to CWW, National Institutes of Health grant R01 DK61168 to KS from the National Institute of Diabetes, Digestive and Kidney Diseases, by Institutional Development Award (IDeA) grant P20 GM103423 and Center of Biomedical Research Excellence (COBRE) grant P20 GM104318 from the National Institute of General Medical Sciences. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Finkel T (2011) Signal transduction by reactive oxygen species. J Cell Biol 194: 7–15. 10.1083/jcb.201102095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Finkel T, Holbrook NJ (2000) Oxidants, oxidative stress and the biology of ageing. Nature 408: 239–247. 10.1038/35041687 [DOI] [PubMed] [Google Scholar]
- 3.Waris G, Ahsan H (2006) Reactive oxygen species: role in the development of cancer and various chronic conditions. J Carcinog 5: 14 10.1186/1477-3163-5-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Uttara B, Singh AV, Zamboni P, Mahajan RT (2009) Oxidative stress and neurodegenerative diseases: a review of upstream and downstream antioxidant therapeutic options. Curr Neuropharmacol 7: 65–74. 10.2174/157015909787602823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blackwell TK, Steinbaugh MJ, Hourihan JM, Ewald CY, Isik M (2015) SKN-1/Nrf, stress responses, and aging in Caenorhabditis elegans. Free Radic Biol Med 88: 290–301. 10.1016/j.freeradbiomed.2015.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.An JH, Blackwell TK (2003) SKN-1 links C. elegans mesendodermal specification to a conserved oxidative stress response. Genes Dev 17: 1882–1893. 10.1101/gad.1107803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giudice A, Arra C, Turco MC (2010) Review of molecular mechanisms involved in the activation of the Nrf2-ARE signaling pathway by chemopreventive agents. Methods Mol Biol 647: 37–74. 10.1007/978-1-60761-738-9_3 [DOI] [PubMed] [Google Scholar]
- 8.Niture SK, Khatri R, Jaiswal AK (2013) Regulation of Nrf2—An update. Free Radic Biol Med 66: 36–44. 10.1016/j.freeradbiomed.2013.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bryan HK, Olayanju A, Goldring CE, Park BK (2013) The Nrf2 cell defence pathway: Keap1-dependent and -independent mechanisms of regulation. Biochem Pharmacol 85: 705–717. 10.1016/j.bcp.2012.11.016 [DOI] [PubMed] [Google Scholar]
- 10.Tang L, Choe KP (2015) Characterization of skn-1/wdr-23 phenotypes in Caenorhabditis elegans; pleitrophy, aging, glutathione, and interactions with other longevity pathways. Mech Ageing Dev 149: 88–98. 10.1016/j.mad.2015.06.001 [DOI] [PubMed] [Google Scholar]
- 11.Tullet JM, Hertweck M, An JH, Baker J, Hwang JY, et al. (2008) Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in C. elegans. Cell 132: 1025–1038. 10.1016/j.cell.2008.01.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ewald CY, Landis JN, Abate JP, Murphy CT, Blackwell TK (2014) Dauer-independent insulin/IGF-1-signalling implicates collagen remodelling in longevity. Nature: 10.1038/nature14021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peddibhotla S, Fontaine P, Leung CK, Maloney P, Hershberger PM, et al. (2015) Discovery of ML358, a selective small molecule inhibitor of the SKN-1 pathway involved in drug detoxification and resistance in nematodes. ACS Chem Biol 10: 1871–1879. 10.1021/acschembio.5b00304 [DOI] [PubMed] [Google Scholar]
- 14.Peddibhotla S, Leung CK, Maloney P, Hershberger PM, Nguyen K, et al. (2013) A high throughput screen for inhibitors of nematode detoxification genes Probe Reports from the NIH Molecular Libraries Program [Internet] Bethesda (MD): National Center for Biotechnology Information (US); 2010–2013 April 11. [PubMed] [Google Scholar]
- 15.Choe KP, Leung CK, Miyamoto MM (2012) Unique structure and regulation of the nematode detoxification gene regulator, SKN-1: implications to understanding and controlling drug resistance. Drug Metab Rev 44: 209–223. 10.3109/03602532.2012.684799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choe KP, Przybysz AJ, Strange K (2009) The WD40 repeat protein WDR-23 functions with the CUL4/DDB1 ubiquitin ligase to regulate nuclear abundance and activity of SKN-1 in Caenorhabditis elegans. Mol Cell Biol 29: 2704–2715. 10.1128/MCB.01811-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.An JH, Vranas K, Lucke M, Inoue H, Hisamoto N, et al. (2005) Regulation of the Caenorhabditis elegans oxidative stress defense protein SKN-1 by glycogen synthase kinase-3. PNAS 102: 16275–16280. 10.1073/pnas.0508105102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kell A, Ventura N, Kahn N, Johnson TE (2007) Activation of SKN-1 by novel kinases in Caenorhabditis elegans. Free Radical Biology and Medicine 43: 1560–1566. 10.1016/j.freeradbiomed.2007.08.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inoue H, Hisamoto N, An JH, Oliveira RP, Nishida E, et al. (2005) The C. elegans p38 MAPK pathway regulates nuclear localization of the transcription factor SKN-1 in oxidative stress response. Genes Dev 19: 2278–2283. 10.1101/gad.1324805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Papp D, Csermely P, Soti C (2012) A role for SKN-1/Nrf in pathogen resistance and immunosenescence in Caenorhabditis elegans. PLoS Pathog 8: e1002673 10.1371/journal.ppat.1002673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoeven R, McCallum KC, Cruz MR, Garsin DA (2011) Ce-Duox1/BLI-3 generated reactive oxygen species trigger protective SKN-1 activity via p38 MAPK signaling during infection in C. elegans. PLoS Pathog 7: e1002453 10.1371/journal.ppat.1002453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crook-McMahon HM, Olahova M, Button EL, Winter JJ, Veal EA (2014) Genome-wide screening identifies new genes required for stress-induced phase 2 detoxification gene expression in animals. BMC Biol 12: 64 10.1186/s12915-014-0064-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nayak S, Santiago FE, Jin H, Lin D, Schedl T, et al. (2002) The Caenorhabditis elegans Skp1-related gene family: diverse functions in cell proliferation, morphogenesis, and meiosis. Curr Biol 12: 277–287. [DOI] [PubMed] [Google Scholar]
- 24.Yamanaka A, Yada M, Imaki H, Koga M, Ohshima Y, et al. (2002) Multiple Skp1-related proteins in Caenorhabditis elegans: diverse patterns of interaction with Cullins and F-box proteins. Curr Biol 12: 267–275. [DOI] [PubMed] [Google Scholar]
- 25.Hasegawa K, Miwa S, Isomura K, Tsutsumiuchi K, Taniguchi H, et al. (2008) Acrylamide-responsive genes in the nematode Caenorhabditis elegans. Toxicol Sci 101: 215–225. 10.1093/toxsci/kfm276 [DOI] [PubMed] [Google Scholar]
- 26.Glover-Cutter KM, Lin S, Blackwell TK (2013) Integration of the unfolded protein and oxidative stress responses through SKN-1/Nrf. PLoS Genet 9: e1003701 10.1371/journal.pgen.1003701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kahn NW, Rea SL, Moyle S, Kell A, Johnson TE (2008) Proteasomal dysfunction activates the transcription factor SKN-1 and produces a selective oxidative-stress response in Caenorhabditis elegans. Biochem J 409: 205–213. 10.1042/BJ20070521 [DOI] [PubMed] [Google Scholar]
- 28.Paek J, Lo JY, Narasimhan SD, Nguyen TN, Glover-Cutter K, et al. (2012) Mitochondrial SKN-1/Nrf mediates a conserved starvation response. Cell Metab 16: 526–537. 10.1016/j.cmet.2012.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Altun ZF, Hall DH (2005) Handbook of C. elegans anatomy. WormAtlas: http://www.wormatlas.org.
- 30.Oliveira RP, Abate JP, Dilks K, Landis J, Ashraf J, et al. (2009) Condition-adapted stress and longevity gene regulation by Caenorhabditis elegans SKN-1/Nrf. Aging Cell 8: 524–541. 10.1111/j.1474-9726.2009.00501.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leung CK, Deonarine A, Strange K, Choe KP (2011) High-throughput screening and biosensing with fluorescent C. elegans strains. J Vis Exp: e2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goh GY, Martelli KL, Parhar KS, Kwong AW, Wong MA, et al. (2013) The conserved Mediator subunit MDT-15 is required for oxidative stress responses in C. elegans. Aging Cell 13: 70–79. 10.1111/acel.12154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ghazi A, Henis-Korenblit S, Kenyon C (2007) Regulation of Caenorhabditis elegans lifespan by a proteasomal E3 ligase complex. Proceedings of the National Academy of Sciences 104: 5947–5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Killian DJ, Harvey E, Johnson P, Otori M, Mitani S, et al. (2008) SKR-1, a homolog of Skp1 and a member of the SCF(SEL-10) complex, regulates sex-determination and LIN-12/Notch signaling in C. elegans. Dev Biol 322: 322–331. 10.1016/j.ydbio.2008.07.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen N, Harris TW, Antoshechkin I, Bastiani C, Bieri T, et al. (2005) WormBase: a comprehensive data resource for Caenorhabditis biology and genomics. Nucleic Acids Res 33: D383–389. 10.1093/nar/gki066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, et al. (2003) Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature 424: 277–283. 10.1038/nature01789 [DOI] [PubMed] [Google Scholar]
- 37.Przybysz AJ, Choe KP, Roberts LJ, Strange K (2009) Increased age reduces DAF-16 and SKN-1 signaling and the hormetic response of Caenorhabditis elegans to the xenobiotic juglone. Mechanisms of Ageing and Development 130: 357–369. 10.1016/j.mad.2009.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hasegawa K, Miwa J (2010) Genetic and cellular characterization of Caenorhabditis elegans mutants abnormal in the regulation of many phase II enzymes. PLoS ONE 5: e11194 10.1371/journal.pone.0011194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Staab TA, Evgrafov O, Knowles JA, Sieburth D (2014) Regulation of synaptic nlg-1/neuroligin abundance by the skn-1/Nrf stress response pathway protects against oxidative stress. PLoS Genet 10: e1004100 10.1371/journal.pgen.1004100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Skowyra D, Craig KL, Tyers M, Elledge SJ, Harper JW (1997) F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell 91: 209–219. [DOI] [PubMed] [Google Scholar]
- 41.Lyapina SA, Correll CC, Kipreos ET, Deshaies RJ (1998) Human CUL1 forms an evolutionarily conserved ubiquitin ligase complex (SCF) with SKP1 and an F-box protein. Proc Natl Acad Sci U S A 95: 7451–7456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu M, Zhu C, Zhao X, Chen C, Zhang H, et al. (2015) Atypical ubiquitin E3 ligase complex Skp1-Pam-Fbxo45 controls the core epithelial-to-mesenchymal transition-inducing transcription factors. Oncotarget 6: 979–994. 10.18632/oncotarget.2825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matsuzawa SI, Reed JC (2001) Siah-1, SIP, and Ebi collaborate in a novel pathway for beta-catenin degradation linked to p53 responses. Mol Cell 7: 915–926. [DOI] [PubMed] [Google Scholar]
- 44.Galan JM, Wiederkehr A, Seol JH, Haguenauer-Tsapis R, Deshaies RJ, et al. (2001) Skp1p and the F-box protein Rcy1p form a non-SCF complex involved in recycling of the SNARE Snc1p in yeast. Mol Cell Biol 21: 3105–3117. 10.1128/MCB.21.9.3105-3117.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seol JH, Shevchenko A, Shevchenko A, Deshaies RJ (2001) Skp1 forms multiple protein complexes, including RAVE, a regulator of V-ATPase assembly. Nat Cell Biol 3: 384–391. 10.1038/35070067 [DOI] [PubMed] [Google Scholar]
- 46.Du Z, Santella A, He F, Tiongson M, Bao Z (2014) De novo inference of systems-level mechanistic models of development from live-imaging-based phenotype analysis. Cell 156: 359–372. 10.1016/j.cell.2013.11.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Park S-K, Tedesco PM, Johnson TE (2009) Oxidative stress and longevity in Caenorhabditis elegans as mediated by SKN-1. Aging Cell 8: 258–269. 10.1111/j.1474-9726.2009.00473.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chowdhry S, Zhang Y, McMahon M, Sutherland C, Cuadrado A, et al. (2013) Nrf2 is controlled by two distinct beta-TrCP recognition motifs in its Neh6 domain, one of which can be modulated by GSK-3 activity. Oncogene 32: 3765–3781. 10.1038/onc.2012.388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Russell ID, Grancell AS, Sorger PK (1999) The unstable F-box protein p58-Ctf13 forms the structural core of the CBF3 kinetochore complex. J Cell Biol 145: 933–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Staab TA, Griffen TC, Corcoran C, Evgrafov O, Knowles JA, et al. (2013) The conserved SKN-1/Nrf2 stress response pathway regulates synaptic function in Caenorhabditis elegans. PLoS Genet 9: e1003354 10.1371/journal.pgen.1003354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Steinbaugh MJ, Narasimhan SD, Robida-Stubbs S, Moronetti Mazzeo LE, Dreyfuss JM, et al. (2015) Lipid-mediated regulation of SKN-1/Nrf in response to germ cell absence. Elife 4: 10.7554/eLife.07836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang J, Robida-Stubbs S, Tullet JM, Rual JF, Vidal M, et al. (2010) RNAi screening implicates a SKN-1-dependent transcriptional response in stress resistance and longevity deriving from translation inhibition. PLoS Genet 6: e1001048 10.1371/journal.pgen.1001048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brenner S (1974) The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim DH, Feinbaum R, Alloing G, Emerson FE, Garsin DA, et al. (2002) A conserved p38 MAP kinase pathway in Caenorhabditis elegans innate immunity. Science 297: 623–626. 10.1126/science.1073759 [DOI] [PubMed] [Google Scholar]
- 55.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, et al. (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28: 511–515. 10.1038/nbt.1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang DW, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- 57.Yang JS, Nam HJ, Seo M, Han SK, Choi Y, et al. (2011) OASIS: online application for the survival analysis of lifespan assays performed in aging research. PLoS One 6: e23525 10.1371/journal.pone.0023525 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fragments Per Kilobase of transcript per Million mapped reads (FPKM) values are provided for triplicates.
(XLSX)
Percentage survival of worms after treatment with each oxidant were scored after 24 h recovery on NGM agar plate seeded with OP50 E. coli. n = 3 trials of 274–402 worms total.
(TIF)
Accumulation of intestinal and head SKN-1b/c::GFP after 5 or 15 min exposure to 2% azide, 5 mM arsenite, 38 μM juglone or 7 mM acrylamide, n = 63–84 worms per condition. ** P<0.01, ***P<0.001 from corresponding controls as determined by the Chi-Square test (bottom). Representative images of SKN-1b/c::GFP. Arrows mark head GFP and arrowheads mark intestinal nuclei.
(TIF)
Arrows mark Pgst-4::GFP in hypodermal nuclei, arrowheads mark Pgst-4::GFP in the intestine.
(TIF)
(A) Representative fluorescence micrographs of worms expressing SKN-1b/c::GFP treated with NGM buffer (control) or 5 mM sodium arsenite for 1h fed with control or skn-1(RNAi). (B) Scoring of intestinal and head SKN-1op::GFP and representative fluorescence micrographs. n = 52–71 worms. ***P<0.001 from corresponding controls as determined by Chi-Square tests. Scoring is the same as in Fig 2. (C) Representative fluorescence micrographs and differential interference contrast (DIC) images showing worms expressing SKN-1b/c::GFP in the hypodermis after exposure to 5 mM sodium arsenite for 1 h. Arrows mark head GFP and arrowheads mark ASI neurons.
(TIF)
Wildtype and tissue-specific RNAi worms fed with control and skn-1(RNAi) were exposed to 38 μM juglone for 2 h and then the concentration was raised to a total of 125 μM and survival was measured for up to 10 h. Graphs for trial 2 are shown above and a summary of the results for all three trials is show below. skn-1(RNAi) reduced survival in all three strains.
(TIF)
(A-B) Animals integrated with Phsp16.2::GFP or Pgpdh-1::RFP were fed bacteria producing dsRNA and exposed to either heat shock (transfer from 20 to 33°C for 1 h and 5 h recovery for Phsp16.2::GFP) or osmotic stress (transfer from 51 to 250 mM NaCl for 24 h for Pgpdh-1::RFP). Fluorescence was scored as in Fig 4. ***P<0.001 compared to control as determined by Chi-Square tests.
(TIF)
(A) skr-1 and skr-2 RNAi have similar effects on Pgst-4::GFP induced by juglone. (B) RNAi screen against a sub-library of genes functioning within the SCF complex in animals carrying the Pgst-4::GFP reporter after juglone exposure (38 μM for 3 h). ***P<0.001 compared to control as determined by Chi-Square tests. (C) Pgst-4::GFP scoring and representative fluorescence micrographs of eri-1 worms fed with control or cul-1(RNAi) for two generations and exposed to 38 μM juglone for 3 h. (A-C) n = 53–139 worms. (D) High penetrance of embryonic lethal and larval arrest phenotypes are observed in F2 generation of cul-1 RNAi fed eri-1 worms. F1 mothers were allowed to lay eggs and then removed for 24 h before taking an image; note a high number of dead eggs and sick L1 larvae.
(TIF)
(A) Heat map of fold-changes in 135 genes down-regulated by juglone with their corresponding fold-changes in worms fed with dsRNA for skn-1, skr-1/2 or daf-16. n = 3 replicates of 1,000–2,000 worms. Coefficients of correlation are listed below the heat map and 95% confidence intervals are as follows: skn-1 and skr-1/2 (0.442–0.649), skn-1 and daf-16 (0.299–0.542), and skr-1/2 and daf-16 (0.238–0.494). (B) Venn diagrams showing numbers of genes overlapping. (C) DAVID functional enrichment analysis of skr-1/2(RNAi) up-regulated genes. (D) Linear regression analysis of all genes up-regulated by either skn-1 or daf-16(RNAi) plotted against their fold change with skr-1/2(RNAi); ***P < 0.001 linear regression F-test.
(TIF)
Data from three individual trials of survival assays are provided. Representative trials are plotted in Fig 7A and 7B.
(TIF)
(A) Fold changes in mRNA level of gst-4, gst-10, gst-12, and gst-30 relative to control in N2 wild-type, skr-1(tm2391), and skr-2(ok1938) mutants after exposure to 38 μM juglone for 3 h. ***P<0.001 compared to N2 treated with juglone. (B) mRNA levels of skr-1 in the skr-2(ok1938) mutant and skr-2 in the skr-1(tm2391) mutant. Values are means plus standard error, n = 4 replicates of 200–400 worms.
(TIF)
(A) Relative Pgst-4::GFP fluorescence and (B) representative micrographs of worms (QV256) with and without skr-1/2(oe); note that the extrachromosomal array carries a genomic fragment that includes both skr-1 and 2, which are adjacent to each other. n = 33 to 56 worms. (C) Fold changes in mRNA level of skr-1, skr-2, gst-4, and gcs-1 in skr-1/2(oe) animals relative to control and when treated with or without 38 μM juglone for 3 h. mRNA levels were normalized to cdc-42, values are mean plus standard error. n = 4 replicates of ~50 worms. ***P<0.001 compared to control. (D) Expression and localization patterns of SKR-1::GFP (QV254) were not obviously affected when treated with 38 μM of juglone for 3h or 5 mM arsenite for 1 h.
(TIF)
Animals carrying an extrachromosomal array containing SKR-1::GFP and a Pmyo-2::tdTomato marker (QV254) were fed with either control or skr-1/2 dsRNA. (A) Shown are DIC images along with fluorescence micrographs taken with GFP and RFP filters. (B) Paired high magnification micrographs of SKR-1::GFP fluorescence and DIC (QV254); the upper right pair is a merger of red and green channels (compare to only GFP in the image immediately below).
(TIF)
Data Availability Statement
Data is available at: https://figshare.com/s/63e2e8045a83bac665dc.