Abstract
Trichoderma reesei Cel6A (TrCel6A) is a cellobiohydrolase that hydrolyzes crystalline cellulose into cellobiose. Here we directly observed the reaction cycle (binding, surface movement, and dissociation) of single-molecule intact TrCel6A, isolated catalytic domain (CD), cellulose-binding module (CBM), and CBM and linker (CBM-linker) on crystalline cellulose Iα. The CBM-linker showed a binding rate constant almost half that of intact TrCel6A, whereas those of the CD and CBM were only one-tenth of intact TrCel6A. These results indicate that the glycosylated linker region largely contributes to initial binding on crystalline cellulose. After binding, all samples showed slow and fast dissociations, likely caused by the two different bound states due to the heterogeneity of cellulose surface. The CBM showed much higher specificity to the high affinity site than to the low affinity site, whereas the CD did not, suggesting that the CBM leads the CD to the hydrophobic surface of crystalline cellulose. On the cellulose surface, intact molecules showed slow processive movements (8.8 ± 5.5 nm/s) and fast diffusional movements (30–40 nm/s), whereas the CBM-Linker, CD, and a catalytically inactive full-length mutant showed only fast diffusional movements. These results suggest that both direct binding and surface diffusion contribute to searching of the hydrolysable point of cellulose chains. The duration time constant for the processive movement was 7.7 s, and processivity was estimated as 68 ± 42. Our results reveal the role of each domain in the elementary steps of the reaction cycle and provide the first direct evidence of the processive movement of TrCel6A on crystalline cellulose.
Keywords: cellulase, cellulose, enzyme kinetics, enzyme mechanism, microscopic imaging, molecular motor, processivity, protein domain, single-molecule biophysics
Introduction
Cellulose, a major component of plant cell walls, is the most abundant biopolymer on earth and has potential as a regeneratable source of bioenergy and chemicals (1, 2). Cellulases from microorganisms are used to degrade physically and chemically stable crystalline celluloses into oligosaccharides under mild conditions (3, 4). Among cellulases, cellobiohydrolase (CBH)2 directly hydrolyzes crystalline cellulose into cellobiose. The fungal CBHs Trichoderma reesei Cel7A (TrCel7A) and Cel6A (TrCel6A) are the most studied CBHs.
The previous studies proposed the processive hydrolysis of TrCel7A and TrCel6A, a continuous reaction without dissociation from cellulose surface. Electron microscopic observations revealed that the edges of crystalline cellulose were sharpened after treatment with TrCel7A and TrCel6A, suggesting the processive hydrolysis from the chain ends (5, 6). Furthermore, it has been shown that the unique tunnel-shaped catalytic site keeps the enzyme bound on the substrate after each hydrolytic event, which enables processive hydrolysis and movement on crystalline cellulose (7). The processive reaction has been thought to be a key for the efficient cellulose degradation. However, it is not easy to directly prove the processive hydrolysis and movement of TrCel7A and TrCel6A by biochemical assays and electron microscopy observations.
Single-molecule imaging of biomolecules is a powerful method to visualize the elementary steps of reactions and mechanical motions of enzymes and motor proteins (8–10). The processive movement of TrCel7A on crystalline cellulose has been directly proved by single-molecule imaging with high speed atomic force microscopy (HS-AFM), and the translational rate constant (ktr) has been successfully estimated (11, 12). Very recently, even the step size (∼1 nm) of the movement was analyzed with single-molecule measurements using optical tweezers (13). However, the processive movement of TrCel6A has not been demonstrated (11), and it is unclear whether TrCel6A can processively move on crystalline cellulose. Because external force applied by the cantilever of the HS-AFM is one possible factor preventing the movement of TrCel6A, single-molecule methods without perturbation by external force are required to verify this issue.
In addition to the processive movement (hydrolysis), the overall cycle of the reaction of CBH includes binding and dissociation on crystalline cellulose. The binding rate constant (kon) and dissociation rate constant (koff) for intact TrCel7A against crystalline cellulose were successfully estimated using single-molecule fluorescence imaging (14–16). However, the kon and koff for TrCel6A have not been estimated at the single-molecule level. Furthermore, although both TrCel7A and TrCel6A contain the cellulose binding module (CBM), catalytic domain (CD), and glycosylated linker region connecting the CBM and CD, the role of each domain on binding and dissociation is not fully understood yet (17, 18). Previous biochemical studies showed that the CBM increases the amount of enzyme bound on the cellulose, indicating that the main role of the CBM is to increase the affinity to cellulose (19). However, most previous studies only estimated the dissociation constant (Kd) in equilibrium, and the kon and koff are not reported, except for the study by Carrand and Linder in 1999 (20). In addition, although high affinity binding of the glycosylated linker region of TrCel6A to the cellulose surface has been predicted by a computational analysis (21), experimental evidence has not been reported yet. To understand how CBHs efficiently hydrolyze crystalline cellulose on the liquid-solid interface, the rate constants of binding and dissociation for each domain should be analyzed independently and quantitatively.
Here, we directly observed the binding and dissociation of the full-length TrCel6A (Intact) and the CD, CBM and linker (CBM-linker), and CBM of TrCel6A (Fig. 1) on crystalline cellulose Iα using single-molecule fluorescence imaging. The kon and koff values were quantitatively estimated and compared to reveal the contribution of each domain to the binding and dissociation events. Furthermore, we directly verified the processive movement of the full-length TrCel6A on crystalline cellulose Iα using single-molecule fluorescence imaging with improved localization precision (22) and appropriate control experiments including an catalytically inactive mutant. The ktr, duration of the movement, and the processivity of TrCel6A were estimated quantitatively at the single-molecule level for the first time.
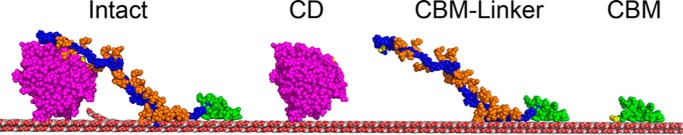
FIGURE 1.
Domain structures of Intact (left), CD (center left), CBM-linker (center right), and CBM (right) of TrCel6A. Pink spheres represent CD, blue spheres represent the linker, and green spheres represent CBM. Yellow spheres in CBM represent added free cysteine at position 43. Yellow spheres in CD represent the S386C mutation, and yellow spheres in the linker represent the S83C mutation. Orange spheres represent estimated sugar modifications on the linker.
Results
Hydrolysis Activities of Cy3-TrCel6A, Inactive Cy3-TrCel6A, and Cy3-CD
For single-molecule fluorescence imaging analysis, mutants of the full-length molecule (447aa), TrCel6A (S386C) and TrCel6A (D221A/S386C), isolated CD (S386C), CBM-linker (S83C), and CBM (S43C) were generated (Fig. 2A). These mutants were conjugated with the thiol-reactive fluorescent dye Cy3-maleimide and termed Intact, Inactive, CD, CBM-linker, and CBM, respectively (Fig. 2B). The hydrolysis activities of Intact against crystalline cellulose Iα were similar to those of wild-type TrCel6A (WT) at various concentrations of the substrate (Fig. 3 and Table 1). As the substrate concentration increased from 0.03% (w/v) to 0.5% (w/v), the hydrolysis activity of Intact increased, as did the WT. The plots were fitted with the Michaelis-Menten equation, and values of kcat = 3.1 s−1 and Km = 0.26% (w/v) for the WT and kcat = 2.8 s−1 and Km = 0.27% (w/v) for Intact were obtained. These results indicate that cysteine mutation and Cy3 conjugation do not affect enzyme function.
FIGURE 2.

A, domain constructions and mutated amino acid residues of the samples. B, SDS-PAGE of the samples used in this study. Lane 1, WT; lane 2, Intact; lane 3, Inactive; lane 4, CD; lane 5, CBM-linker; lane 6, CBM. Samples (10 pmol) were loaded in lanes 1–4, and 30 pmol and 100 pmol of sample was loaded in lanes 5 and 6, respectively. Samples were separated in a 15–20% gradient gel with Tris-Tricine buffer.
FIGURE 3.
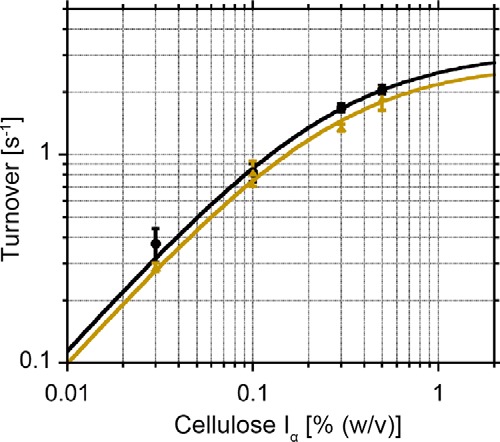
Hydrolysis activities of WT and Intact at various substrate concentrations. Hydrolysis activities of WT (black circles) and Intact (yellow triangles) at various substrate concentrations (from 0.03 to 0.5% (w/v)) and the Michaelis-Menten fit are shown. kcat values of 3.1 s−1 and 2.8 s−1 and Km values of 0.26% (w/v) and 0.27% (w/v) were obtained for WT and Intact, respectively.
TABLE 1.
Turnovers for 0.1% (w/v) of substrates at 25 °C in 50 mm sodium acetate buffer, pH 5.0
Enzymes (0.1 μm) were incubated with crystalline cellulose Iα prepared from Cladophora sp. and PASC for 2 and 30 min, respectively. Products from crystalline cellulose Iα were quantified by HPLC, and those from PASC were quantified by the p-hydroxybenzaldehyde method.
| Substrate | Turnover |
|||
|---|---|---|---|---|
| WT | Intact | Inactive | CD | |
| s−1 | ||||
| Cellulose Iα | 0.79 ± 0.04 | 0.76 ± 0.03 | Not detected | 0.031 ± 0.002 |
| PASC | 0.42 ± 0.03 | 0.41 ± 0.07 | 0.017 ± 0.022 | 0.43 ± 0.09 |
The CD showed lower activity (0.031 ± 0.002 s−1, n = 3) than the WT (0.79 ± 0.04 s−1) and Intact (0.76 ± 0.03 s−1) against crystalline cellulose (measured at 0.1% (w/v)) (Table 1). However, hydrolysis activities against amorphous cellulose (phosphoric acid swollen cellulose (PASC)) were similar among the WT, Intact, and CD (0.42 ± 0.03, 0.41 ± 0.07, and 0.43 ± 0.09 s−1, respectively) (Table 1). These results indicate that the CBM-linker is important for crystalline cellulose degradation.
Inactive showed no detectable activity against crystalline cellulose (product concentration was lower than the detection limit, 0.1 μm) and very little activity against PASC within the standard deviation (0.017 ± 0.022 s−1). Thus, the D221A mutation significantly impairs the catalytic activity of TrCel6A, as reported previously (23).
The Binding Rate Constants for Intact, CD, CBM-Linker, and CBM
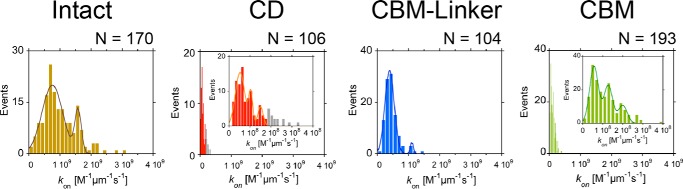
The kon values for Intact, CD, CBM-linker, and CBM were determined according to a previous report (Fig. 4) (14). Here, the kon value was defined as the number of cellulase molecules bound on the cellulose in a unit of cellulase concentration, cellulose length, and time (m−1 μm−1 s−1). The concentrations of each sample were set at 25, 250, 50, and 200 pm for Intact, CD, CBM-linker, and CBM, respectively, to observe more than two molecules binding on all of the cellulose microfibrils in the image field. Distributions of the kon for Intact, CD, CBM-linker, and CBM are shown in Fig. 4. All samples showed multiple peaks as in previous measurements of TrCel7A (14), corresponding to the number of cellulose Iα microfibrils in a bundle. The minimum peaks, which represent the kon to a single microfibril, were 7.5 × 108, 5.2 × 107, 3.9 × 108, and 5.6 × 107 m−1 μm−1 s−1 for Intact, CD, CBM-linker, and CBM, respectively (Table 2). The kon of the CBM-linker was more than half that of Intact, whereas those of the CD and CBM were less than one-tenth that of Intact.
FIGURE 4.
Distributions of the binding rate constant (kon) for Intact (left), CD (center left), CBM-linker (center right), and CBM (right). The distributions of Intact, CD, CBM-linker, and CBM were fitted with two or three Gaussians. The concentrations of the samples were 25, 250, 50, and 200 pm for Intact, CD, CBM-linker, and CBM, respectively. The number of cellulose microfibrils analyzed was 170, 106, 104, and 193 for Intact, CD, CBM-linker, and CBM, respectively. The bin widths were 1.25 × 108 m−1 μm−1 s−1 for Intact and CBM-linker, 1.4 × 107 m−1 μm−1 s−1 for CD, and 2.0 × 107 m−1 μm−1 s−1 for CBM.
TABLE 2.
Values of kon and koff
| Sample |
kona |
koffb |
|||
|---|---|---|---|---|---|
| 1st peak | 2nd peak | 3rd peak | Fast (fraction) | Slow (fraction) | |
| m−1 μm−1 s−1 | s−1 | ||||
| Intact | 7.5 × 108 ± 2.7 × 107 | 1.6 × 109 ± 2.4 × 107 | 1.1 ± 0.03 (70%) | 0.10 ± 0.02 (30%) | |
| CD | 5.2 × 107 ± 4.2 × 106 | 1.0 × 108 ± 4.9 × 106 | 1.5 × 108 ± 7.7 × 106 | 1.5 ± 0.03 (72%) | 0.16 ± 0.02 (28%) |
| CBM-linker | 3.9 × 108 ± 4.5 × 106 | 1.1 × 109 ± 3.6 × 108 | 2.6 ± 0.05 (65%) | 0.14 ± 0.02 (35%) | |
| CBM | 5.6 × 107 ± 2.3 × 106 | 1.3 × 108 ± 3.5 × 106 | 2.0 × 108 ± 6.9 × 106 | 2.3 ± 0.05 (70%) | 0.083 ± 0.02 (30%) |
a The peak values ± S.E. of the Gaussian fitting.
b The means ± S.E. of the fitting with the double exponential decay functions.
The Dissociation Rate Constants for Intact, CD, CBM-linker, and CBM
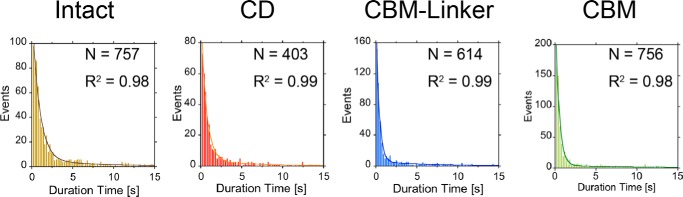
The koff values for Intact, CD, CBM-linker, and CBM were determined (Fig. 5). For all samples, the distributions of the duration time on cellulose were better fitted by double exponential decay than single exponential decay, indicating that two components exist: slow (koffslow) and fast (kofffast) (Table 2). For Intact, koffslow and kofffast were 0.10 s−1 (30%) and 1.1 s−1 (70%), respectively. For the CD, koffslow and kofffast were 0.16 s−1 (28%) and 1.5 s−1 (72%), and those for the CBM-linker were 0.14 s−1 (35%) and 2.6 s−1 (65%), respectively. For the CBM, koffslowand kofffast were 0.083 s−1 (30%) and 2.3 s−1 (70%), respectively. The koffslow values for Intact and the CBM were similar but less than those for the CD and CBM-linker. The kofffast values for the CBM-linker and CBM and those for Intact and the CD were similar. Our results also indicate that the time resolution of the observation (0.2 s, reciprocal of 5 fps) was sufficient for kinetic analysis of koff because the time constant (reciprocal of koff) for the fast component of all samples was longer than 0.2 s.
FIGURE 5.
Distributions of the duration time on cellulose for Intact (left), CD (center left), CBM-linker (center right), and CBM (right) fitted with double exponential decay functions. For Intact, kofffast was 1.1 s−1 (70%), and koffslow was 0.10 s−1 (30%). For CD, kofffast was 1.5 s−1 (72%), and koffslow was 0.16 s−1 (28%). For CBM-linker, kofffast was 2.6 s−1 (65%), and koffslow was 0.14 s−1 (35%). For CBM, kofffast was 2.3 s−1 (70%), and koffslow was 0.083 s−1 (30%). The number of events was 757, 403, 614, and 756 for Intact, CD, CBM-linker, and CBM, respectively. The bin widths of the distributions were 0.2 s.
Processive Movement and Translational Rate Constants for Intact and Other Samples
Next, we determined whether Intact undergoes processive movement on crystalline cellulose Iα using the improved localization precision (see “Experimental Procedures”) (22). As shown in Fig. 6, “apparent” unidirectional movements were observed in all samples, i.e. Intact, Inactive, CD, and CBM-linker. However, the movements of Inactive and the CBM-linker are not processive because they are deficient in hydrolysis activity. When the ktr distributions for Intact, Inactive, and CBM-linker were compared (Fig. 7, left panels), only Intact showed a slow component (peak: 8.8 ± 5.5 nm/s) in addition to a fast one (34.9 ± 16.3 nm/s), which was seen for Inactive (35.5 ± 17.4 nm/s) and the CBM-linker (42.6 ± 28.0 nm/s). Furthermore, when correlations between ktr and moving time on cellulose were plotted, only the slow component had a moving time longer than 10 s (Fig. 7, right panels). From these results, we concluded that the slow component seen only in Intact corresponds to the processive movement accompanying the hydrolysis of crystalline cellulose, whereas the fast component is the diffusive movement on the cellulose surface without hydrolysis. In addition to Inactive and the CBM-linker, the CD also showed only the fast component (peak: 39.2 ± 28.3 nm/s), and most of their moving times were shorter than 10 s (Fig. 7).
FIGURE 6.
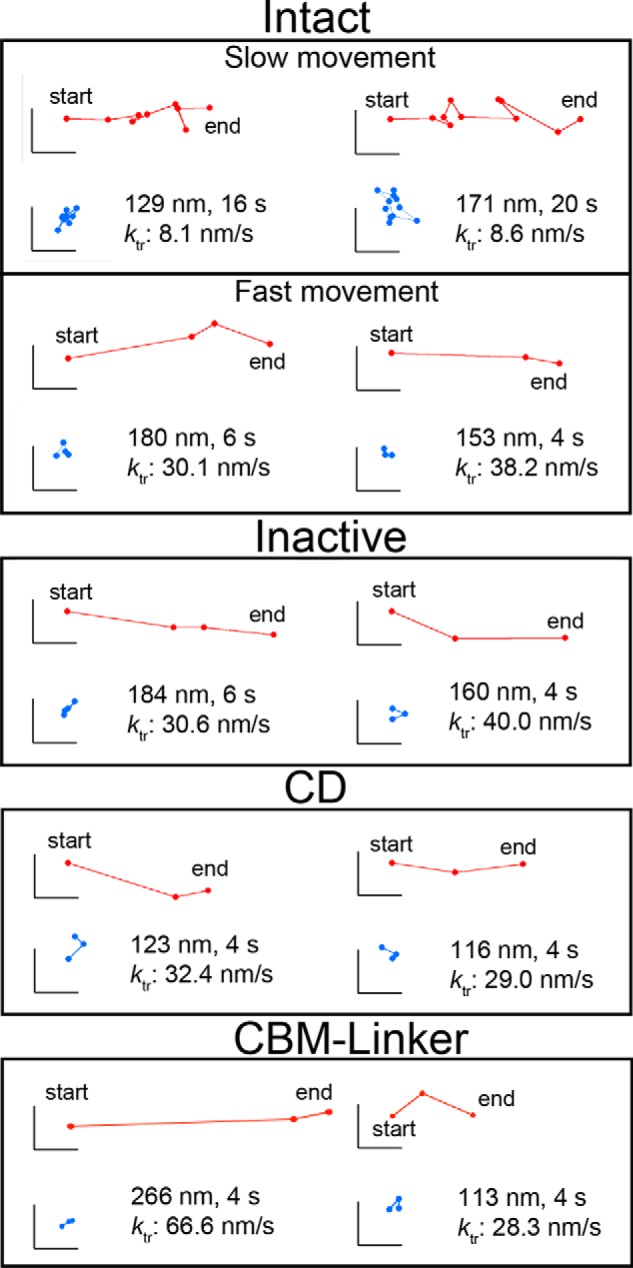
Trajectories of the movement for Intact, Inactive, CD, and CBM-linker. The red plots show the centroid trajectory of the movement. The blue plots show the centroid trajectory of a non-moving molecule in the same field of view and demonstrate that the coverslip did not drift. The frame rate of observation was 0.5 fps. The scale bars are 40 nm for both the vertical and horizontal axes.
FIGURE 7.
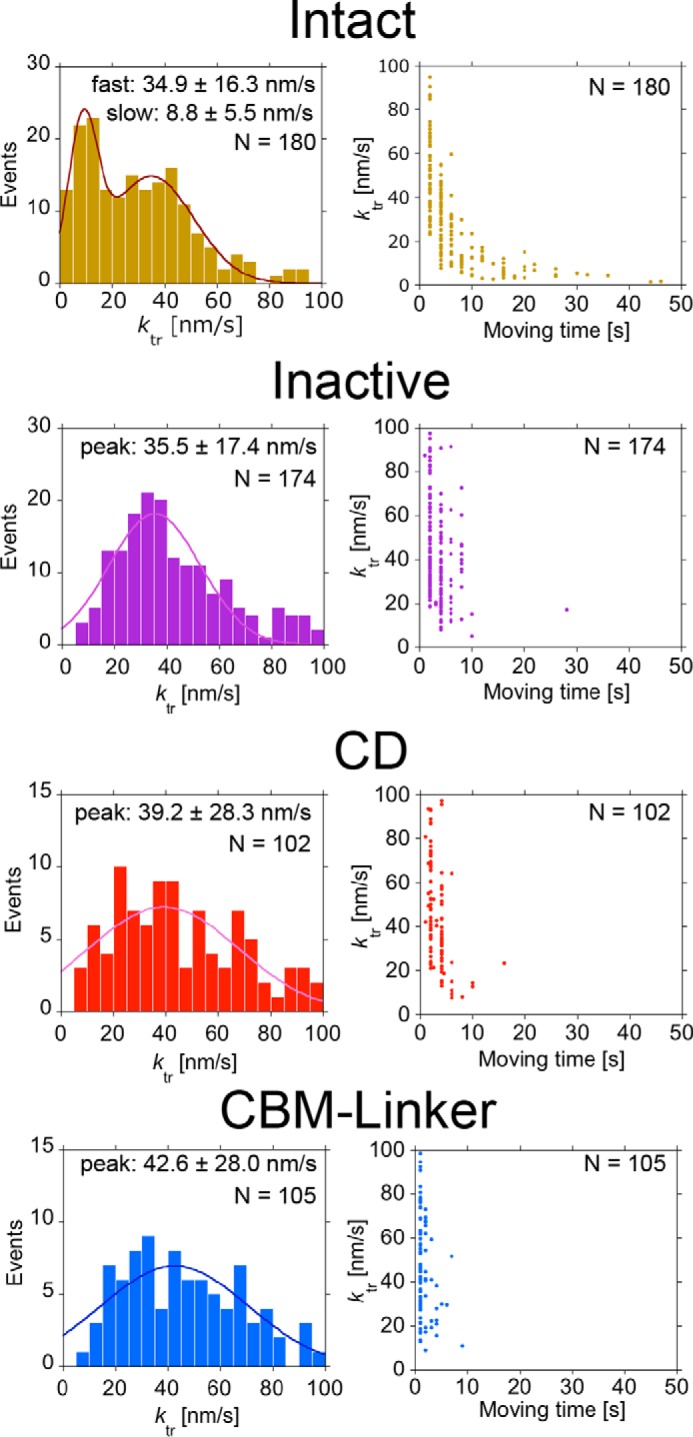
Distribution of translational rate constant (ktr) for Intact, Inactive, CD, and CBM-linker and plots of ktrversus moving time on crystalline cellulose. Distribution of the ktr for Intact was fitted by two Gaussians; the peak ± S.D. values were 8.8 ± 5.5 and 34.9 ± 16.3 nm/s (n = 180), respectively. Distributions of the ktr for Inactive, CD, and CBM-linker were fitted by a single Gaussian. The peak ± S.D. values were 35.5 ± 17.4 nm/s (n = 174), 39.2 ± 28.3 nm/s (n = 102), and 42.6 ± 28.0 nm/s (n = 105), respectively.
Next, the moving time distribution for Intact, which showed slow movement (0–20 nm/s), was plotted (Fig. 8). The molecules that showed moving times longer than 10 s, which is unique for Intact, are highlighted (Fig. 8, pink bars). The distributions for both the entire range and longer than 10 s were fitted with single exponential decay functions with the same time constant of 7.7 s.
FIGURE 8.

Distribution of moving time for Intact showing slow (0–20 nm/s), processive movement. Molecules that moved for longer than 10 s are highlighted in pink. The distributions were fitted by single exponential decay functions: y = 31.9 × exp(−0.13 × t) (all range) and y = 36.4 × exp(−0.13 × t) (>10 s). The bin width was 2 s. The numbers of molecules were 100 (all range) and 40 (>10 s).
Discussion
TrCel6A, together with TrCel7A, is one of the most well studied cellulases, and the first cellulase for which the x-ray crystal structure was determined (24). In addition, the processive reaction of CBH was first proposed based on the comparison between the tunnel-like structure of TrCel6A and cleft-like structure of an endo-glucanase Thermononospora fusca Cel6A (25). However, the processive movement of TrCel6A has not been directly observed, and the reaction cycle of TrCel6A was still unclear. Moreover, the role of each domain was not clear because quantitative measurements of the binding and dissociation rate constants are difficult with biochemical assays. In this report, we verified the processive movement of TrCel6A and clarified the kinetic role of the CBM and glycosylated linker region using single-molecule fluorescence imaging.
The role of the CBM and linker region was clearly shown by the comparison of the kon and koff values among Intact, CD, and CBM-linker (Table 2). Focusing on the binding event, the CBM-linker is highly important because the kon for the CD (5.2 × 107 m−1 μm−1 s−1) was less than one-tenth that of Intact (7.5 × 108 m−1 μm−1 s−1) (Fig. 4). However, the kon for the CBM-linker was only half (3.9 × 108 m−1 μm−1 s−1) that of Intact. This difference suggests that half of the binding events of Intact are caused by the CBM-linker and half are caused by the synergistic effect of the CBM-linker and CD. Weak and transient bindings of the CBM-linker, which could not be detected as clear fluorescence signals in our observations, may help binding of the CD and increase the kon for Intact. For dissociation, both the kofffast (1.1 s−1) and koffslow (0.10 s−1) for Intact were the lowest among Intact, CD, and CBM-linker (Fig. 5). If the CBM-linker and CD of the intact molecule cannot bind to cellulose simultaneously, the koff for Intact should be same as that for the CD or CBM-linker. Therefore, the low koff values for Intact indicate that both the CD and CBM-linker simultaneously interact with the crystalline cellulose surface. Comparing the kon and koff values between Intact and the CD (with and without CBM-linker), CBM-linker caused the kon to increase 14 times and the koff to decrease to two-thirds that of the CD. Thus, the CBM-linker mainly contributes to the binding of TrCel6A by increasing the kon directly and assisting the binding of the CD. The binding of the glycosylated linker region to the cellulose surface has been demonstrated using molecular dynamics simulation (21). The synergistic binding of the CD and CBM-linker is reasonable because the freedom of the orientation of the CD will be restricted after the binding of the CBM-linker.
After comparing kon and koff between the CBM and CBM-linker, the contribution of the glycosylated linker region becomes clear. The kon of the CBM was one-seventh that of the CBM-linker. Thus, the linker region is more important than the CBM for the initial interaction with crystalline cellulose. The main role of the glycosylation has been thought to protect cellulase from proteolysis, and information on the effect of glycosylation on the binding to crystalline cellulose is limited, especially for TrCel6A (26). One good example is the comparison of WT TrCel7A and recombinant TrCel7A produced by Aspergillus niger (27). The mass of recombinant TrCel7A is only few hundred Daltons larger compared with the WT because of more glycosylation sites in the linker and CBM, but it binds more than twice as much as the WT. Moreover, the association constants of the CBM and CBM-linker of TrCel7A were compared, and the affinity of the CBM was reported to be only one-tenth that of the CBM-linker (21). Taken together, our results strongly suggest that the interaction between glycans on the linker region of TrCel6A and the crystalline cellulose surface causes the initial binding events. However, the koff values for the CBM were smaller than those for the CBM-linker, indicating that the CBM-linker binds crystalline cellulose mainly by the CBM and that the linker region somehow disturbs the binding of the CBM. Considering the binding mechanism of type A CBMs such as CBM1 and CBM2a with a flat hydrophobic surface, binding of the CBM-linker should be more entropically unfavorable than the isolated CBM because the binding of the CBM-linker decreases the conformational entropy of the flexible linker region (28).
The heterogeneity of the binding modes of CBH to crystalline cellulose has long been discussed. First, bindings with high and low affinities were reported for TrCel7A and attributed to its multidomain structure (19). Recently, the reason for the complicated binding isotherm was explained by the steric effect using a CBM-linker connected with red fluorescent protein (29). In our previous single-molecule fluorescence imaging of intact TrCel7A, we also observed two components for the koff and attributed the slow and fast components to productive and non-productive binding, respectively (14). However, in the present study on TrCel6A, not only Intact and CD but also CBM-linker and CBM showed the slow and fast components (Fig. 5). In addition, two components were observed at very low sample concentrations (∼pm). These results indicate that the slow and fast components of dissociation are caused by other effects of the multidomain structure, the steric effect, and productive/non-productive bindings. The ratios between the slow and fast components for the CBM-linker and CD were 35 and 65% and 28 and 72%, respectively. If the fast component is caused by structurally unstable bindings (such as only one tryptophan residue on the CBM binds the cellulose surface), the ratio of the fast component of Intact should be smaller than that of the truncated domains. However, Intact showed ratios of 30% (slow) and 70% (fast), which were between the values for the CBM-linker and CD. Thus, this is not plausible.
Other reasons for the two components are structural heterogeneities of cellulose. First, a single cellulose chain has polarity, i.e. reducing and non-reducing ends. Second, the ends and middle of the cellulose chain can have different interactions with CBH. Third, cellulose can form crystalline and amorphous structures. Furthermore, crystalline cellulose has flat hydrophobic and solvated hydrophilic surfaces (30). If the two bound states were caused by the binding orientation of the enzyme, the probabilities of the fast and slow components would be similar. However, all of the samples showed higher fractions of the fast component (∼70%). In addition, in our single-molecule observations, the positions at which the samples strongly bound were random and not localized to the ends or middle of the crystalline cellulose microfibrils (Fig. 9). These results rule out the first two possibilities. In this study, we used crystalline cellulose Iα prepared from Cladophora sp., and the ratio of the amorphous region was very low and less than the detection limit of XRD measurement (Fig. 10). In addition, if the surface of crystalline cellulose is highly disordered and amorphous-like, CD will show similar kon to Intact because hydrolysis activities of Intact and CD against amorphous cellulose are similar (Table 1). Therefore, the most likely reason for the two bound states is the two different surfaces of the crystalline cellulose. The CBM has a flat surface with hydrophobic, aromatic amino acid residues. It has been reported that the CBM preferentially binds to the hydrophobic surface of crystalline cellulose (31), although the area ratio of the hydrophobic surface is much smaller than that of the hydrophilic surface in crystalline cellulose Iα (26). Thus, it is more likely that the slow and fast components correspond to the binding to the hydrophobic (high affinity) and hydrophilic (low affinity) surfaces, respectively. The dissociation constants (Kd = koff/kon) for the two binding modes are summarized in Table 3. The kon for short and long bindings were calculated from the ratio of the fast and slow components of the koff. The affinity of Intact to the hydrophobic surface was 25 and 11 times higher than that of the CD and CBM, respectively, and that to the hydrophilic surface was 19 and 28 times higher, respectively. In a previous report, the partition coefficients of the intact TrCel6A, CD, and CBM to bacterial microcrystalline cellulose were compared at nanomolar order enzyme concentrations, and the values for the intact form were 34 and 3.4 times higher than those for the CD and CBM (32). Thus, our results are similar to those from this previous study, although the CBM showed much lower affinity than the Intact in our study.
FIGURE 9.
Binding specificity and distribution of Intact, Inactive, CD, CBM-linker, and CBM to crystalline cellulose. Bright field images of cellulose microfibrils (top). Single-molecule fluorescence images of each sample (middle). Fluorescence images of each sample obtained by accumulating 150 consecutive images (bottom). The scale bars are 3 μm.
FIGURE 10.
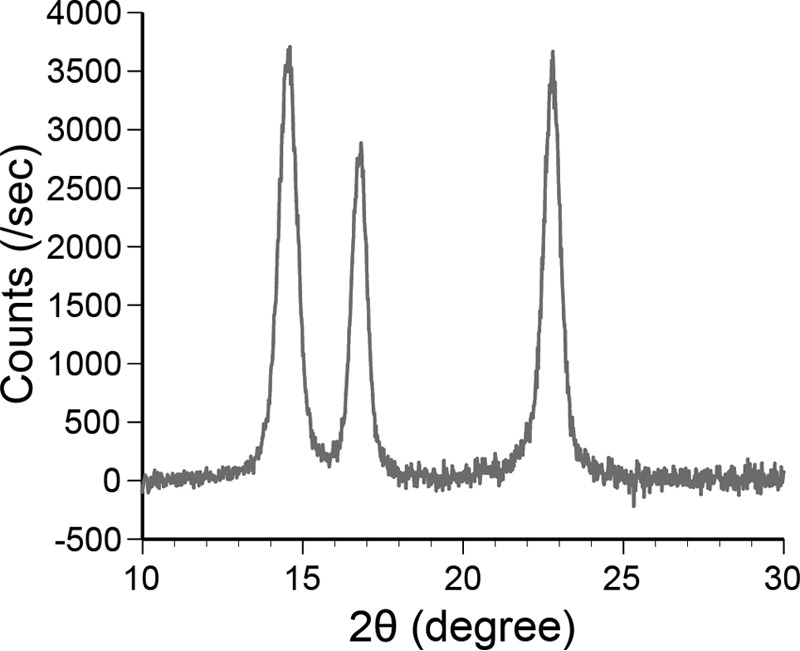
XRD profile of crystalline cellulose Iα prepared from Cladophora sp. used in this study. Voltage and current were 40 kV and 40 mA. Scattering from the glass holder was subtracted.
TABLE 3.
Values of Kd calculated from kon and koff
| Sample | kona | koff | Kdb |
|---|---|---|---|
| m−1 μm−1 s−1 | s−1 | m μm | |
| Intact | |||
| Low affinity site | 5.3 × 108 | 1.1 | 0.21 × 10−8 |
| High affinity site | 2.3 × 108 | 0.10 | 0.044 × 10−8 |
| CD | |||
| Low affinity site | 3.7 × 107 | 1.5 | 4.0 × 10−8 |
| High affinity site | 1.5 × 107 | 0.16 | 1.1 × 10−8 |
| CBM-linker | |||
| Low affinity site | 2.5 × 108 | 2.6 | 1.0 × 10−8 |
| High affinity site | 1.4 × 108 | 0.14 | 0.10 × 10−8 |
| CBM | |||
| Low affinity site | 3.9 × 107 | 2.3 | 5.9 × 10−8 |
| High affinity site | 1.7 × 107 | 0.083 | 0.49 × 10−8 |
a The kon for short and long bindings was calculated from the ratio of the fast and slow components of the koff.
b The unit of μm represents the length of a crystalline cellulose microfibril.
Between the hydrophobic and hydrophilic surfaces, the CBM and CBM-linker showed 12 and 10 times higher affinities to the hydrophobic surface than to the hydrophilic surface, respectively. Therefore, the CBM is necessary to determine the surface specificity of binding, as shown by a molecular dynamics simulation of the CBM of TrCel7A (33). The glycosylated linker region increases the kon and affinity to both surfaces but decreases the binding specificity to the hydrophobic surface (Table 3). It has been reported that TrCel7A hydrolyzes Valonia crystalline cellulose from the hydrophobic surface (34). The cellulose polymer chains accessible to the catalytic site of CBH exist mainly on the hydrophobic surface, because the cellulose chains forming the hydrophilic surface stack each other (35). Thus, our result can explain the increase in non-productive binding of the recombinant TrCel7A with extra O-glycosylation in the linker and CBM (27). Furthermore, the affinity of the CD to the hydrophobic surface is only 3.6 times higher than to the hydrophilic surface. This result suggests that the CBM leads the CD to the hydrophobic surface and increases the probability of catching the hydrolysable chain of cellulose.
One of the most important issues regarding TrCel6A is whether it actually undergoes processive movement. With single-molecule fluorescence imaging with improved localization precision, we addressed this issue and verified that intact TrCel6A shows slow, processive movements. Thus, TrCel6A can act as a linear molecular motor moving on crystalline cellulose. The estimated value of the ktr (8.8 ± 5.5 nm/s) for TrCel6A was comparable with or slightly higher than those for TrCel7A (5–7 nm/s) determined by HS-AFM observations (11, 14). Because TrCel6A has a shorter catalytic tunnel structure compared with that of TrCel7A, TrCel6A would be more easily detached from crystalline cellulose by the tapping force of the cantilever, preventing the movement under the HS-AFM. The distribution of the moving time for the slow, processive movement of TrCel6A can be fitted by a single exponential decay function with time constants of 7.7 s (Fig. 8). Considering the length (∼1 nm) of the product cellobiose (13), the processivity of TrCel6A was calculated to be 68 ± 42 (8.8 ± 5.5 nm/s × 7.7 s × 1 nm−1) from the ktr and the time constant. This value is three times larger than that of TrCel7A estimated by HS-AFM observation (16). By the electron microscopy observation, the processivity of TrCel7A was expected to be higher than that of TrCel6A, because the edge of cellulose microfibril hydrolyzed by TrCel7A was more sharpened than that by TrCel6A (5, 6). To compare the value of processivity of TrCel6A estimated in our study with that of TrCel7A quantitatively, the TrCel7A also need to be analyzed with single-molecule fluorescence imaging.
It has been reported that fungal glycoside hydrolase 6 (GH6) enzymes has mixed modes of endo- and exo-initiations (36). In our analysis, however, we could not distinguish endo- and exo-initiations of TrCel6A. Furthermore, we cannot exclude the possibility that TrCel6A also acts non-processively, because a significant fraction of Intact showed dissociation without translational movement.
The CD of TrCel6A has shorter loops covering the catalytic tunnel than that of a bacterial GH6 cellulase from Thermobifida fusca Cel6B (37), and structures of the linker region and CBM are also different between fungal and bacterial GH6 cellulases. Therefore, the correlation between structure and function such as between loop length and degree of processivity is an interesting question that can be assessed by single-molecule fluorescence imaging. By quantitatively analyzing the role of each domain of not only fungal but also bacterial GH6 cellulases, valuable insights to engineer more efficient hybrid cellulases will be gained.
Previous single-molecule studies have reported processive movements of the isolated CD of TrCel7A (12, 13). However, in our study, we did not observe the CD of TrCel6A undergoing slow and long movement (Fig. 7). One possible explanation is that the ratio of productive binding decreased in the absence of the CBM-linker, as we described above. Alternatively, the absence of the CBM-linker may significantly decrease the processivity. Regarding this second possibility, Bu et al. (38) predicted that the CBM has important roles in not only binding but also introducing a single cellulose chain to the catalytic site and increasing the processivity of the reaction. Their molecular simulation showed that the CBM exerts a driving force and assists processive movement by biased sliding when the CBM binds near the cellulose chain end. However, the effect of the CBM-linker on the processivity of TrCel6A is still open for discussion.
In addition to the slow and long processive movements of Intact, fast (30–40 nm/s) and short (<10 s) movements were observed for all samples. We attributed these fast movements to diffusion on the surface of crystalline cellulose. TrCel6A may employ multiple strategies to catch the hydrolysable point of the cellulose chain efficiently: direct binding from solution and diffusional searching on the crystalline cellulose surface. In the near future, high resolution and long-term single-molecule imaging with much brighter optical probes such as quantum dots (39) and gold nanoparticles (40–42) will be applied to TrCel6A and other CBHs. These studies will resolve the detail of the entire cycle of the processive reaction including the decrystallization, chain threading, bond cleavage, and product release and will give valuable insights for the rational design and engineering of better, non-natural cellulases.
Experimental Procedures
Construction of the TrCel6A Mutant Gene and Transformation into Pichia pastoris
The wild-type TrCel6A (WT) gene in the pPICZα plasmid was used as a template, and the S386C point mutation was introduced by PCR. The TrCel6A S386C mutant (Intact) gene and a signal sequence of the vector were transferred into a new pPICZα plasmid with MfeI and NotI. The Asp221 to Ala mutation (Inactive) was introduced into the gene of the TrCel6A S386C mutant by the same method. The CD gene of TrCel6A S386C was amplified by PCR with forward and reverse primers containing the 15 bp of the homologous sequence to the end of the α-factor and the 5′-end of the NotI recognition site, respectively. The region from the 3′-end of the α-factor to the 5′-end of the NotI site of pPICZa was also amplified by PCR and ligated with the gene of the CD of TrCel6A S386C by an In-fusion kit (Clontech). The gene was also transferred into a new pPICZα by MfeI and NotI. The CD region of TrCel6A was determined according to the x-ray crystal structure (Protein Data Bank code 1QK2). The gene encoding the CBM and linker region (CBM-linker) was amplified from the MfeI site of the pPICZα plasmid to the end of the linker region. The sequence of the recognition site of Factor Xa protease, the His6 tag, and the NotI site was included in the reverse primer. The amplified product was ligated into pPICZa by MfeI and NotI. The gene encoding the CBM, the recognition site of Factor Xa protease, and EGFP were connected by PCR. The His6 tag and NotI site were included in the reverse primer for EGFP. The amplified product was ligated into pPICZa by MfeI and NotI. All vectors with the WT and mutant genes were linearized by BglII and transformed into P. pastoris strain KM71H (Invitrogen) by electroporation.
Production and Purification of WT and Mutant TrCel6A
WT and mutants were produced using a jar fermenter (Takasugi Seisakusho) or flask. WT, Intact, Inactive, and CD were purified by passing through a hydrophobic interaction column and an anion exchange column. The detailed conditions were described previously (43). CBM-linker and CBM were purified by a nickel-nitrilotriacetic acid-agarose column (Qiagen). The culture medium was loaded into the column and washed with 50 mm imidazole in 20 mm sodium phosphate buffer (pH 7.0) and 100 mm NaCl. The proteins were eluted with 100 mm imidazole in the same buffer.
Conjugation with Cy3
Purified Intact and Inactive were reduced by 10 mm 2-mercaptoethanol for 1 h at 4 °C, and 2-mercaptoethanol was removed by HPLC with a size exclusion column (YMC-Pack Diol-200G; YMC). Intact (262 μm, 68 μl and 28.1 μm, 120 μl in two trials) and Inactive (40.8 μm, 100 μl) in 20 mm sodium phosphate buffer (pH 7.0) containing 100 mm NaCl was mixed with Cy3-maleimide monoreactive dye (GE Healthcare) at a molecular ratio of enzyme:Cy3-maleimide = 1:20 and reacted for 4 days at room temperature. Unreacted Cy3-maleimide was removed by ultrafiltration (Vivaspin 500 5 K PES membrane; Sartorius, Germany). CD (30 μm, 50 μl) in 20 mm sodium phosphate buffer (pH 7.0) containing 100 mm NaCl was reduced with 10 mm DTT for 16 h at 4 °C and then mixed with Cy3-maleimide monoreactive dye. DTT was removed by HPLC with a size exclusion column (YMC-Pack Diol-200G). The reaction with Cy3-maleimide was conducted at a molecular ratio of enzyme:Cy3-maleimide = 1:20 for 4 days at room temperature. Unreacted Cy3-maleimide was removed by HPLC equipped with a size exclusion column. CBM-linker was reduced by 10 mm 2-mercaptoethanol for 1 h at room temperature, and 2-mercaptoethanol was removed by HPLC with a size exclusion column (YMC-Pack Diol-120G; YMC). Then 500 μl of 66.5 μm CBM-linker was mixed with three times the amount of Cy3-maleimide and incubated for 16 h at room temperature. Unreacted Cy3 was removed by an EconoPack DE10 column (Bio-Rad), and the sample was eluted with 20 mm Tris-HCl (pH 8.0) containing 100 mm NaCl. Cy3-labeled CBM-linker (2.8 μm, 500 μl) was incubated with 2.5 μg of Factor Xa (NEB, England) and 2 mm CaCl2 for 16 h at 23 °C. Factor Xa was removed by HPLC with a size exclusion column (YMC-Pack Diol-120G), and the sample was eluted with 20 mm sodium acetate buffer (pH 5.0) containing 100 mm NaCl. CBM was reduced by 10 mm 2-mercaptoethanol for 1 h at room temperature, and 2-mercaptoethanol was removed by HPLC with a size exclusion column (YMC-Pack Diol-120G; YMC). CBM (40.0 μm, 1500 μl) was mixed with 3 times the amount of Cy3-maleimide and incubated 16 h at room temperature. Unreacted Cy3 was removed by an EconoPack DE10 column (Bio-Rad), and the sample was eluted with 20 mm Tris-HCl (pH 8.0) containing 100 mm NaCl. Cy3-labeled CBM (288 μm, 200 μl) was incubated with 65 μg of Factor Xa (NEB, England) and 2 mm CaCl2 for 16 h at 23 °C. Factor Xa and EGFP were removed by HPLC with a size exclusion column (YMC-Pack Diol-120G), and the sample was eluted with 20 mm sodium acetate buffer (pH 5.0) containing 100 mm NaCl. The labeling ratios of Intact was 85 and 91% for 2 trials, and the labeling ratios of Inactive, CD, CBM-linker, and CBM were 74, 64, 69, and 85%, respectively. The molecular extinction coefficient used for Cy3 was 150,000 m−1 cm−1 at 550 nm, and those for WT, Intact, and Inactive were 95,880 m−1 cm−1; that for CD was 80,300 m−1 cm−1; and those for CBM-linker and CBM were 15,580 m−1 cm−1 at 280 nm.
Cellulose Preparation
Crystalline cellulose Iα was prepared from Cladophora sp. The plant body was incubated at room temperature in 1 m NaOH for 16 h. NaOH was removed by washing with deionized water, and the sample was incubated at 70 °C for 3 h in 1.0 liter of 100 mm sodium acetate buffer (pH 4.9) containing 0.3% NaClO2. The sample was washed with water and incubated in 1 m NaOH at room temperature for 16 h. Hard tips were removed, and fibers were cut into short pieces with scissors after washing with water. The fiber was incubated in 0.1 m HCl at 110 °C for 20 min and washed with water. The fiber was suspended in water for 16 h at room temperature and homogenized by Physcotron (Microtec). The suspension was centrifuged at 8,000 × g for 10 min, and cellulose was collected. Cellulose (30 g), 30 ml of water, and 120 ml of 6 n HCl were mixed and incubated at 80 °C for 9 h with agitation at 300 rpm. Cellulose crystals were collected by centrifugation at 10,000 × g and 15,000 × g and washed with milliQ water until the pH became neutral. The aggregated crystals and large fibers were removed by centrifugation at 6,000 × g. Crystallinity was checked by XRD, and scattering intensity from amorphous regions was not observed (Fig. 10).
Hydrolysis Activity Measurement
WT, Intact, CD, and Inactive were mixed with cellulose Iα (final concentration, 0.1% w/v) for 2 min at 25 °C in 50 mm sodium acetate buffer (pH 5.0). The final concentration of enzymes was 0.1 μm. The hydrolysis activities were determined by HPLC analysis. Glucose (Sigma-Aldrich); cellobiose, cellotetraose, and cellopentaose (Serva); and cellotriose (Seikagaku) were used as standards. For PASC hydrolysis, 0.1 μm enzyme was mixed with 0.1% substrate for 30 min at 25 °C in 50 mm sodium acetate buffer (pH 5.0). Product concentrations were quantified by the p-hydroxybenzaldehyde method with a glucose standard.
Single-molecule Fluorescence Imaging Analysis
Single-molecule fluorescence imaging was performed as described previously (14). A glass coverslip (thickness, 0.12–0.17 mm; Matsunami Glass) was cleaned with 10 m KOH for 1 day. A sample chamber was constructed with the coverslip. A cellulose Iα solution (0.02% w/v, 20 μl) was spin-coated (3,000 rpm, 10 s) on the coverslip. Then Cy3-conjugated enzymes (25–250 pm, 20 μl) in 50 mm sodium acetate (pH 5.0) were dropped on the coverslip and observed.
For the kon and koff analysis, image sequences were obtained at 5 fps with a laser power of 0.14 μW/μm2 before entering the objective lens. The photo-breaching time constant for Cy3 conjugated to TrCel6A under these conditions was 18.8 ± 0.8 s. The binding specificity of the samples to crystalline cellulose Iα was confirmed by comparing the bright field image of crystalline cellulose Iα microfibrils and single-molecule fluorescence image of the samples in the same field of view obtained at a frame rate of 0.5 fps (Fig. 9).
For the analysis of movement, the localization precision was improved to 6–8 nm by increasing the laser power to 0.28 μW/μm2 and adjusting the frame rate to 0.5 or 1 fps. The photo-breaching time constant under these conditions was 15.0 ± 0.7 s. The images were analyzed using ImageJ software (National Institutes of Health).
Author Contributions
A. N. conducted all experiments and data analysis and wrote the manuscript. T. T., D. I., A. V., and M. M. conducted single-molecule experiments and analysis. M. Y. and Y. O. prepared samples. T. T. and T. U. conducted hydrolysis activity measurements. H. N., M. S., K. I., and R. I. organized the experiments. R. I. conceived the project and wrote the manuscript with A. N.
Acknowledgments
We thank Dr. Takayuki Uchihashi and Dr. Kentaro Ishii for helpful discussion and Kaori Nakane for administrative assistance.
This work is supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology 16H00789, 16H00858, and 15H04366 (to R. I.) and 15H06898 (to A. N.); Imaging Science Project of the Center for Novel Science Initiatives, National Institutes of Natural Sciences Grants IS271006 and IS281005 (to R. I.), and ORION project of Okazaki Institute for Integrative Bioscience, National Institutes of Natural Sciences (to R. I.). The authors declare that they have no conflicts of interest with the contents of this article.
- CBH
- cellobiohydrolase
- TrCel7A
- T. reesei Cel7A
- TrCel6A
- T. reesei Cel6A
- CBM
- cellulose binding module
- CD
- catalytic domain
- GH
- glycoside hydrolase
- HS-AFM
- high speed atomic force microscopy
- PASC
- phosphoric acid-swollen cellulose.
References
- 1. Himmel M. E., Ding S. Y., Johnson D. K., Adney W. S., Nimlos M. R., Brady J. W., and Foust T. D. (2007) Biomass recalcitrance: engineering plants and enzymes for biofuels production. Science 315, 804–807 [DOI] [PubMed] [Google Scholar]
- 2. Chundawat S. P., Beckham G. T., Himmel M. E., and Dale B. E. (2011) Deconstruction of lignocellulosic biomass to fuels and chemicals. Annu. Rev. Chem. Biomol. Eng. 2, 121–145 [DOI] [PubMed] [Google Scholar]
- 3. Payne C. M., Knott B. C., Mayes H. B., Hansson H., Himmel M. E., Sandgren M., Ståhlberg J., and Beckham G. T. (2015) Fungal cellulases. Chem. Rev. 115, 1308–1448 [DOI] [PubMed] [Google Scholar]
- 4. Wilson D. B. (2011) Microbial diversity of cellulose hydrolysis. Curr. Opin. Microbiol. 14, 259–263 [DOI] [PubMed] [Google Scholar]
- 5. Imai T., Boisset C., Samejima M., Igarashi K., and Sugiyama J. (1998) Unidirectional processive action of cellobiohydrolase Cel7A on Valonia cellulose microcrystals. FEBS Lett. 432, 113–116 [DOI] [PubMed] [Google Scholar]
- 6. Chanzy H., and Henrissat B. (1985) Undirectional degradation of valonia cellulose microcrystals subjected to cellulase action. FEBS Lett. 184, 285–288 [Google Scholar]
- 7. Kipper K., Väljamäe P., and Johansson G. (2005) Processive action of cellobiohydrolase Cel7A from Trichoderma reesei is revealed as “burst” kinetics on fluorescent polymeric model substrates. Biochem. J. 385, 527–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bustamante C., Chemla Y. R., Forde N. R., and Izhaky D. (2004) Mechanical processes in biochemistry. Annu. Rev. Biochem. 73, 705–748 [DOI] [PubMed] [Google Scholar]
- 9. Joo C., Balci H., Ishitsuka Y., Buranachai C., and Ha T. (2008) Advances in single-molecule fluorescence methods for molecular biology. Annu. Rev. Biochem. 77, 51–76 [DOI] [PubMed] [Google Scholar]
- 10. Greenleaf W. J., Woodside M. T., and Block S. M. (2007) High-resolution, single-molecule measurements of biomolecular motion. Annu Rev. Biophys. Biomol. Struct. 36, 171–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Igarashi K., Uchihashi T., Koivula A., Wada M., Kimura S., Okamoto T., Penttilä M., Ando T., and Samejima M. (2011) Traffic jams reduce hydrolytic efficiency of cellulase on cellulose surface. Science 333, 1279–1282 [DOI] [PubMed] [Google Scholar]
- 12. Igarashi K., Koivula A., Wada M., Kimura S., Penttilä M., and Samejima M. (2009) High speed atomic force microscopy visualizes processive movement of Trichoderma reesei cellobiohydrolase I on crystalline cellulose. J. Biol. Chem. 284, 36186–36190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Brady S. K., Sreelatha S., Feng Y., Chundawat S. P., and Lang M. J. (2015) Cellobiohydrolase 1 from Trichoderma reesei degrades cellulose in single cellobiose steps. Nat. Commun. 6, 10149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shibafuji Y., Nakamura A., Uchihashi T., Sugimoto N., Fukuda S., Watanabe H., Samejima M., Ando T., Noji H., Koivula A., Igarashi K., and Iino R. (2014) Single-molecule imaging analysis of elementary reaction steps of Trichoderma reesei cellobiohydrolase I (Cel7A) hydrolyzing crystalline cellulose Iα and IIII. J. Biol. Chem. 289, 14056–14065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jung J., Sethi A., Gaiotto T., Han J. J., Jeoh T., Gnanakaran S., and Goodwin P. M. (2013) Binding and movement of individual Cel7A cellobiohydrolases on crystalline cellulose surfaces revealed by single-molecule fluorescence imaging. J. Biol. Chem. 288, 24164–24172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nakamura A., Watanabe H., Ishida T., Uchihashi T., Wada M., Ando T., Igarashi K., and Samejima M. (2014) Trade-off between processivity and hydrolytic velocity of cellobiohydrolases at the surface of crystalline cellulose. J. Am. Chem. Soc. 136, 4584–4592 [DOI] [PubMed] [Google Scholar]
- 17. Reinikainen T., Teleman O., and Teeri T. T. (1995) Effects of pH and high ionic strength on the adsorption and activity of native and mutated cellobiohydrolase I from Trichoderma reesei. Proteins Struct. Funct. Genet. 22, 392–403 [DOI] [PubMed] [Google Scholar]
- 18. Tomme P., Van Tilbeurgh H., Pettersson G., Van Damme J., Vandekerckhove J., Knowles J., Teeri T., and Claeyssens M. (1988) Studies of the cellulolytic system of Trichoderma reesei QM 9414. Analysis of domain function in two cellobiohydrolases by limited proteolysis. Eur. J. Biochem. 170, 575–581 [DOI] [PubMed] [Google Scholar]
- 19. Stahlberg J., Johansson G., and Pettersson G. (1991) A new model for enzymatic hydrolysis of cellulose based on the two-domain structure of cellobiohydrolase I. Nat. Biotechnol. 9, 286–290 [Google Scholar]
- 20. Carrard G., and Linder M. (1999) Widely different off rates of two closely related cellulose-binding domains from Trichoderma reesei. Eur. J. Biochem. 262, 637–643 [DOI] [PubMed] [Google Scholar]
- 21. Payne C. M., Resch M. G., Chen L., Crowley M. F., Himmel M. E., Taylor L. E. 2nd, Sandgren M., Ståhlberg J., Stals I., Tan Z., and Beckham G. T. (2013) Glycosylated linkers in multimodular lignocellulose-degrading enzymes dynamically bind to cellulose. Proc. Natl. Acad. Sci. U.S.A. 110, 14646–14651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yildiz A., Forkey J. N., McKinney S. A., Ha T., Goldman Y. E., and Selvin P. R. (2003) Myosin V walks hand-over-hand: single fluorophore imaging with 1.5-nm localization. Science 300, 2061–2065 [DOI] [PubMed] [Google Scholar]
- 23. Koivula A., Ruohonen L., Wohlfahrt G., Reinikainen T., Teeri T. T., Piens K., Claeyssens M., Weber M., Vasella A., Becker D., Sinnott M. L., Zou J. Y., Kleywegt G. J., Szardenings M., Ståhlberg J., et al. (2002) The active site of cellobiohydrolase Cel6A from Trichoderma reesei: the roles of aspartic acids D221 and D175. J. Am. Chem. Soc. 124, 10015–10024 [DOI] [PubMed] [Google Scholar]
- 24. Rouvinen J., Bergfors T., Teeri T., Knowles J. K., and Jones T. A. (1990) Three-dimensional structure of cellobiohydrolase II from Trichoderma reesei. Science 249, 380–386 [DOI] [PubMed] [Google Scholar]
- 25. Davies G., and Henrissat B. (1995) Structures and mechanisms of glycosyl hydrolases. Structure 3, 853–859 [DOI] [PubMed] [Google Scholar]
- 26. Beckham G. T., Dai Z., Matthews J. F., Momany M., Payne C. M., Adney W. S., Baker S. E., and Himmel M. E. (2012) Harnessing glycosylation to improve cellulase activity. Curr. Opin. Biotech. 23, 338–345 [DOI] [PubMed] [Google Scholar]
- 27. Jeoh T., Michener W., Himmel M. E., Decker S. R., and Adney W. S. (2008) Implications of cellobiohydrolase glycosylation for use in biomass conversion. Biotechnol. Biofuels 1, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Creagh A. L., Ong E., Jervis E., Kilburn D. G., and Haynes C. A. (1996) Binding of the cellulose-binding domain of exoglucanase Cex from Cellulomonas fimi to insoluble microcrystalline cellulose is entropically driven. Proc. Natl. Acad. Sci. U.S.A. 93, 12229–12234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sugimoto N., Igarashi K., Wada M., and Samejima M. (2012) Adsorption characteristics of fungal family 1 cellulose-binding domain from Trichoderma reesei cellobiohydrolase I on crystalline cellulose: negative cooperative adsorption via a steric exclusion effect. Langmuir 28, 14323–14329 [DOI] [PubMed] [Google Scholar]
- 30. Nishiyama Y., Sugiyama J., Chanzy H., and Langan P. (2003) Crystal structure and hydrogen bonding system in cellulose 1α, from synchrotron x-ray and neutron fiber diffraction. J. Am. Chem. Soc. 125, 14300–14306 [DOI] [PubMed] [Google Scholar]
- 31. Boraston A. B., Bolam D. N., Gilbert H. J., and Davies G. J. (2004) Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem. J. 382, 769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Palonen H., Tenkanen M., and Linder M. (1999) Dynamic interaction of Trichoderma reesei cellobiohydrolases Ce16A and Ce17A and cellulose at equilibrium and during hydrolysis. Appl. Environ. Microbiol. 65, 5229–5233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nimlos M. R., Beckham G. T., Matthews J. F., Bu L., Himmel M. E., and Crowley M. F. (2012) Binding preferences, surface attachment, diffusivity, and orientation of a family 1 carbohydrate-binding module on cellulose. J. Biol. Chem. 287, 20603–20612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liu Y.-S., Baker J. O., Zeng Y., Himmel M. E., Haas T., and Ding S.-Y. (2011) Cellobiohydrolase hydrolyzes crystalline cellulose on hydrophobic faces. J. Biol. Chem. 286, 11195–11201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Beckham G. T., Matthews J. F., Peters B., Bomble Y. J., Himmel M. E., and Crowley M. F. (2011) Molecular-level origins of biomass recalcitrance: decrystallization free energies for four common cellulose polymorphs. J. Phys. Chem. B 115, 4118–4127 [DOI] [PubMed] [Google Scholar]
- 36. Boisset C., Fraschini C., Schülein M., Henrissat B., and Chanzy H. (2000) Imaging the enzymatic digestion of bacterial cellulose ribbons reveals the endo character of the cellobiohydrolase Cel6A from Humicola insolens and its mode of synergy with cellobiohydrolase Cel7A. Appl. Environ. Microbiol. 66, 1444–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wu M., Bu L., Vuong T. V., Wilson D. B., Crowley M. F., Sandgren M., Ståhlberg J., Beckham G. T., and Hansson H. (2013) Loop motions important to product expulsion in the Thermobifida fusca glycoside hydrolase family 6 cellobiohydrolase from structural and computational studies. J. Biol. Chem. 288, 33107–33117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bu L., Beckham G. T., Crowley M. F., Chang C. H., Matthews J. F., Bomble Y. J., Adney W. S., Himmel M. E., and Nimlos M. R. (2009) The energy landscape for the interaction of the family 1 carbohydrate-binding module and the cellulose surface is altered by hydrolyzed glycosidic bonds. J. Phys. Chem. B 113, 10994–11002 [DOI] [PubMed] [Google Scholar]
- 39. Kairdolf B. A., Smith A. M., Stokes T. H., Wang M. D., Young A. N., and Nie S. (2013) Semiconductor quantum dots for bioimaging and biodiagnostic applications. Annu. Rev. Anal. Chem. 6, 143–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Isojima H., Iino R., Niitani Y., Noji H., and Tomishige M. (2016) Direct observation of intermediate states during the stepping motion of kinesin-1. Nat. Chem. Biol. 12, 290–297 [DOI] [PubMed] [Google Scholar]
- 41. Ueno H., Minagawa Y., Hara M., Rahman S., Yamato I., Muneyuki E., Noji H., Murata T., and Iino R. (2014) Torque generation of Enterococcus hirae V-ATPase. J. Biol. Chem. 289, 31212–31223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Minagawa Y., Ueno H., Hara M., Ishizuka-Katsura Y., Ohsawa N., Terada T., Shirouzu M., Yokoyama S., Yamato I., Muneyuki E., Noji H., Murata T., and Iino R. (2013) Basic properties of rotary dynamics of the molecular motor Enterococcus hirae V1-ATPase. J. Biol. Chem. 288, 32700–32707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Igarashi K., Maruyama M., Nakamura A., Ishida T., Wada M., and Samejima M. (2012) Degradation of crystalline celluloses by Phanerochaete chrysosporium cellobiohydrolase II (Cel6A) heterologously expressed in methylotrophic yeast Pichia pastoris. J. Appl. Glycosci. 59, 105–110 [Google Scholar]