Abstract
Nonribosomal peptide synthetases (NRPSs) produce a wide variety of peptide natural products. During synthesis, the multidomain NRPSs act as an assembly line, passing the growing product from one module to the next. Each module generally consists of an integrated peptidyl carrier protein, an amino acid-loading adenylation domain, and a condensation domain that catalyzes peptide bond formation. Some adenylation domains interact with small partner proteins called MbtH-like proteins (MLPs) that enhance solubility or activity. A structure of an MLP bound to an adenylation domain has been previously reported using a truncated adenylation domain, precluding any insight that might be derived from understanding the influence of the MLP on the intact adenylation domain or on the dynamics of the entire NRPS module. Here, we present the structures of the full-length NRPS EntF bound to the MLPs from Escherichia coli and Pseudomonas aeruginosa. These new structures, along with biochemical and bioinformatics support, further elaborate the residues that define the MLP-adenylation domain interface. Additionally, the structures highlight the dynamic behavior of NRPS modules, including the module core formed by the adenylation and condensation domains as well as the orientation of the mobile thioesterase domain.
Keywords: acyl carrier protein (ACP), bacterial metabolism, enzyme mechanism, enzyme mutation, multifunctional protein, natural product biosynthesis, siderophore, structural biology
Introduction
Nonribosomal peptide synthetases (NRPSs)2 are fascinating modular enzymes that use an assembly line architecture to produce important peptide natural products (1–3). During synthesis, the amino acid building blocks are bound to peptidyl carrier protein (PCP) domains that migrate between catalytic active sites for the requisite steps in the biosynthetic pathway. Most NRPS modules contain an adenylation domain that activates the correct amino acid and loads the PCP domain. Internal modules contain a condensation domain that transfers the upstream amino acid or peptide to the newly loaded amino acid, extending the peptide length by one residue. Freed from the constraints of standard ribosomal synthesis, NRPS products display a wide range of chemical structures.
The chemical diversity of NRPS products is further enhanced by the presence of additional internal domains or external proteins that modify the nascent peptide (4–6). In addition to these tailoring enzymes, some NRPS biosynthetic clusters contain genes encoding small (∼70-residue) proteins. Named after the MbtH protein from the Mycobacterium tuberculosis mycobactin operon (7), these MbtH-like proteins (MLPs) were shown simultaneously by Thomas and co-workers (8) and Walsh and co-workers (9) to function as activators of acyladenylate formation. The MLP-NRPS interactions exhibit several interesting features. Some adenylation domains also require MLPs as chaperones and cannot be expressed without their MLP partner (10). Furthermore, genetic and biochemical studies have shown that MLPs can activate NRPS proteins in different biosynthetic clusters within a species and can be substituted heterologously in different species (11).
Structural studies of MLPs and MLP-adenylation domain complexes have been performed to aid in explaining the role of MLPs in NRPS biochemistry. MLPs have a consensus sequence identified as NXEXQXSXWPX5PXGWX13LX7WTDXRP (12) where X represents any amino acid. Particularly striking are the highly conserved proline and tryptophan residues. The crystal structure of an MLP from Pseudomonas aeruginosa called PA2412 (13) and the solution structures of MLPs from M. tuberculosis (14) and three other organisms (Mycobacterium marinum (Protein Data Bank code 2MYY), Mycobacterium avium (Protein Data Bank code 2N6G), and Burkholderia pseudomallei (Protein Data Bank code 2LPD)) that have not been published all show a flat architecture with three β-strands and an α-helix that lies across the β-sheet. Both the N and C termini form flexible coils or, in two structures, a short helix at the C terminus. On one face of the MLP, two of the three conserved tryptophan residues are oriented parallel to each other, forming a small cavity (13).
NRPS adenylation domains have two subdomains, a large N-terminal subdomain and a smaller (∼110-residue) C-terminal subdomain, that adopt multiple orientations to catalyze the adenylation and thioester-forming partial reactions (15). The crystal structure of SlgN1, an unusual NRPS adenylation domain with an MLP fused to the N terminus in a single protein chain, offered the first view of the interaction between the MLP and adenylation domains (16). To crystallize SlgN1, the C-terminal subdomain of the adenylation domain was truncated. The MLP domain of SlgN1 bound to the N-terminal subdomain distal to the active site, with the tryptophan cavity coordinating the side chain of Ala-433 (16). Mutation of this alanine to glutamate abolished acyladenylate formation, confirming that the observed binding interface was required for MLP activation of SlgN1.
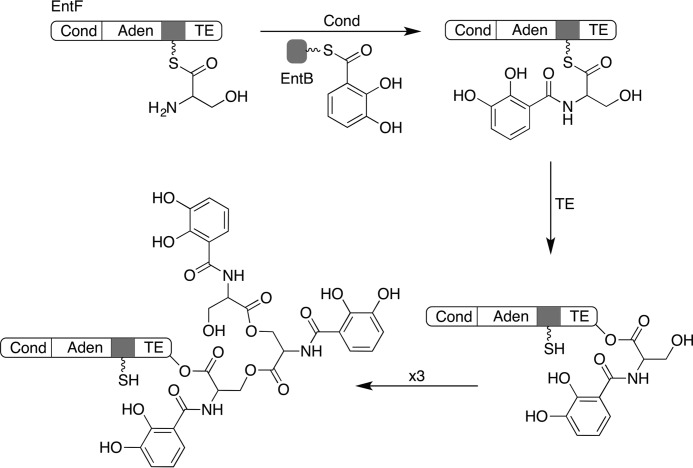
The NRPS pathway for the biosynthesis of enterobactin in Escherichia coli has served as a model system for understanding the assembly line architecture (17). Enterobactin is produced by three NRPS proteins, EntE, EntB, and EntF, that convert three molecules of serine and three molecules of 2,3-dihydroxybenzoic acid (DHB) into the trilactone product (Fig. 1). In enterobactin biosynthesis, EntE first loads a molecule of DHB onto the aryl carrier protein EntB (18). Similarly, a molecule of serine is loaded on the EntF PCP domain by the activity of the upstream adenylation domain. The EntF condensation domain then binds to loaded EntB protein as well as the downstream PCP to catalyze amide formation. The terminal thioesterase domain transfers the DHB-Ser amide to a catalytic serine within its active site. Three cycles allow for the formation of linear enterobactin that is released through lactone formation catalyzed by the thioesterase domain. Key chemical steps in enterobactin synthesis, namely the thiolation of EntB catalyzed by EntE (19, 20), the thiolation of the internal PCP by the EntF adenylation domain (21), and the interaction of the EntF PCP with the downstream thioesterase domain (22, 23), have all been structurally characterized.
FIGURE 1.
Chemical reactions catalyzed by the EntF proteins. The EntF condensation (Cond) domain catalyzes transfer of a molecule of DHB, which has been loaded on the EntB carrier protein domain, onto the serine that has been loaded onto the EntF PCP domain. The EntF thioesterase (TE) domain catalyzes transfer of the DHB-Ser amide to the catalytic serine within the thioesterase domain. Three cycles of this results in the formation of linear enterobactin, which is released through formation of the final trilactone. Aden, adenylation domain.
The adenylation activity by EntF is enhanced in the presence of the MLP YbdZ, which is encoded within the enterobactin operon. In contrast, activity of the freestanding adenylation domain EntE is not influenced by YbdZ (8).
We have recently determined the structures of EntF and AB3403, a terminal NRPS module from Acinetobacter baumannii with the same domain architecture (21). These structures showed how the large conformational change in the adenylation domain (15) results in two conformations that transport the PCP between the adenylation and condensation domains. Comparison of the crystal structures of EntF, AB3403, and SrfA-C, the terminal module from surfactin NRPS cluster, showed that NRPS modules are highly dynamic (21, 24). Additionally, the thioesterase domains of SrfA-C and AB3403 adopt different positions, and this domain was disordered in the structure of EntF. Negative stain electron microscopy corroborated the dynamic NRPS module observation and showed multiple locations for the thioesterase domain. A second recent study presented multiple structures of LgrA, the initiation module of the linear gramicidin NRPS (25), which contains a formyltransferase domain upstream of the adenylation and PCP domains. The LgrA structures showed an additional subdomain movement within the adenylation domain that delivers the PCP to the formyltransferase domain.
We present here new crystal structures of EntF obtained via co-crystallization of EntF with the E. coli MLP YbdZ and the MLP from P. aeruginosa PA2412, which can also activate EntF. Although the MLP-adenylation domain interaction is similar to SlgN1, the interaction can now be analyzed in the context of not only an intact adenylation domain but also a complete NRPS module. The structure also allows us to compare an adenylation domain in the presence and absence of an MLP. The structures show that MLP binding has no effect on the structure of the EntF adenylation domain.
The EntF structures also show a new position for the thioesterase domain compared with the earlier characterized NRPS modules. The downstream thioesterase domain makes very limited contacts to the core of the module, supporting the highly dynamic architecture for NRPS modules. Finally, the condensation domain structure both supports the dynamic module hypothesis and depicts a new potential opening mechanism for the downstream PCP.
Results
Structures of EntF Bound to MLPs
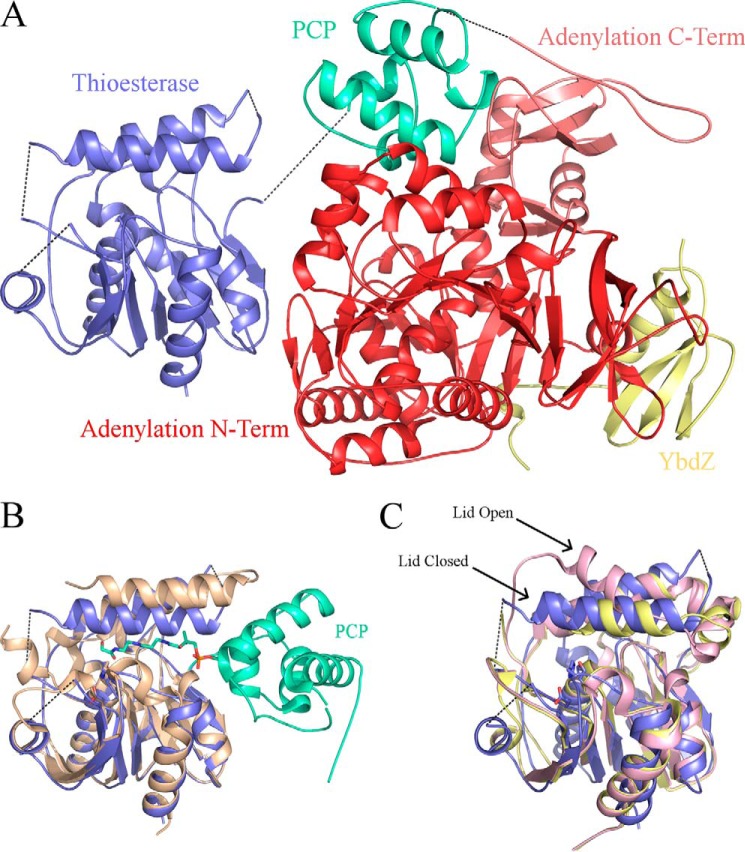
We determined the crystal structures of EntF in complex with two MLPs in a new crystal form. Crystals of EntF bound to the E. coli YbdZ diffracted to 3.0 Å. The structure was solved by combing partial data sets from three crystals (Table 1). Crystals were obtained by incubating EntF with the serine adenosine vinylsulfonamide (Ser-AVS) mechanism-based inhibitor used in the original EntF structure (21). This resulted in covalent trapping of the PCP in an interaction with the adenylation domain in the thioester-forming conformation. In contrast to the previous EntF structure, the thioesterase domain was also ordered in the new crystal form (Fig. 2).
TABLE 1.
Crystallization data
r.m.s.d., root mean square deviation; Avg, average; Aden, adenylation domain; Cond, condensation domain; TE, thioesterase domain. Correlation coefficients CC1/2 and CC* are from iMOSFLM/Scala.
| EntF-YbdZ complex | EntF-PA2412 complex | |
|---|---|---|
| Diffraction data | ||
| Protein Data Bank code | 5JA1 | 5JA2 |
| Beamline | APS 23-ID-B | SSRL12-2 |
| Wavelength (Å) | 1.0332 | 0.9795 |
| Number of crystals | 3 | 1 |
| Space group | I2 | I2 |
| Unit cell a, b, c; β (Å; °) | 161.2, 57.7, 184.0; 97.9 | 159.2, 57.7, 182.4; 97.6 |
| Resolution range (Å)a | 56.3–3.0 (3.1–3.0) | 59.0–3.0 (3.1–3.0) |
| Total reflections | 106,064 (10,391) | 96,975 (8,389) |
| Unique reflections | 31,781 (3,163) | 31,760 (2,973) |
| Multiplicity | 3.3 (3.3) | 3.1 (2.8) |
| Completeness (%) | 93.1 (93.7) | 94.8 (90.5) |
| Mean I/σ(I) | 7.1 (3.1) | 6.7 (1.4) |
| Rmerge | 0.1426 (0.397) | 0.1025 (0.6214) |
| Rmeas | 0.1681 | 0.1253 |
| CC1/2 | 0.971 (0.749) | 0.914 (0.669) |
| CC* | 0.993 (0.926) | 0.977 (0.896) |
| Structure refinement | ||
| R-factor | 0.1811 (0.2566) | 0.2047 (0.2927) |
| Rfree | 0.2496 (0.3227) | 0.2426 (0.3204) |
| No. protein/ligand atoms | 9,795/50 | 9,824/61 |
| r.m.s.d. bonds (Å) | 0.003 | 0.002 |
| r.m.s.d. angles (°) | 0.58 | 0.55 |
| Wilson B-factor (Å2) | 36.3 | 71.8 |
| Avg B-factor (Å2), overall | 46.7 | 71.8 |
| Cond N (residues 1–180) | 44.1 | 73.9 |
| Cond C (residues 181–440) | 45.6 | 71.6 |
| Aden N (residues 450–857) | 34.6 | 55.2 |
| Aden C (residues 858–963) | 51.4 | 74.7 |
| PCP (residues 969–1042) | 70.4 | 108.3 |
| TE (residues 1052–1292) | 75.6 | 110.4 |
| MLP | 43.1 | 84.0 |
| Ramachandran analysis | ||
| Favored (%) | 95.6 | 96.1 |
| Allowed (%) | 3.4 | 3.4 |
| Outliers (%) | 1.0 | 0.5 |
| MolProbity Clashscore | 5.38 | 5.8 |
a Values in parentheses reflect the highest resolution shell.
FIGURE 2.
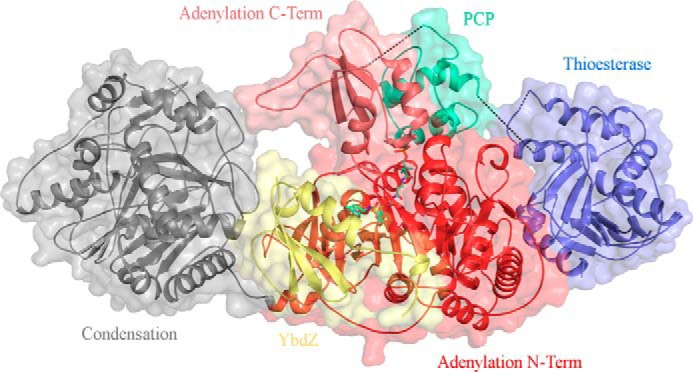
Crystal structure of EntF bound to the MLP YbdZ. The Ser-AVS inhibitor was used to covalently trap the PCP in an interaction with the adenylation domain in the thioester-forming conformation. Shown is the complete terminal module with condensation domain (gray), adenylation N-terminal subdomain (red), adenylation C-terminal subdomain (salmon), PCP (green-cyan), thioesterase domain (blue), and YbdZ (yellow). Black dotted lines represent unresolved coils, including the adenylation domain-PCP linker, the PCP-thioesterase linker, and a loop connecting the two α-helices of the lid on the thioesterase domain.
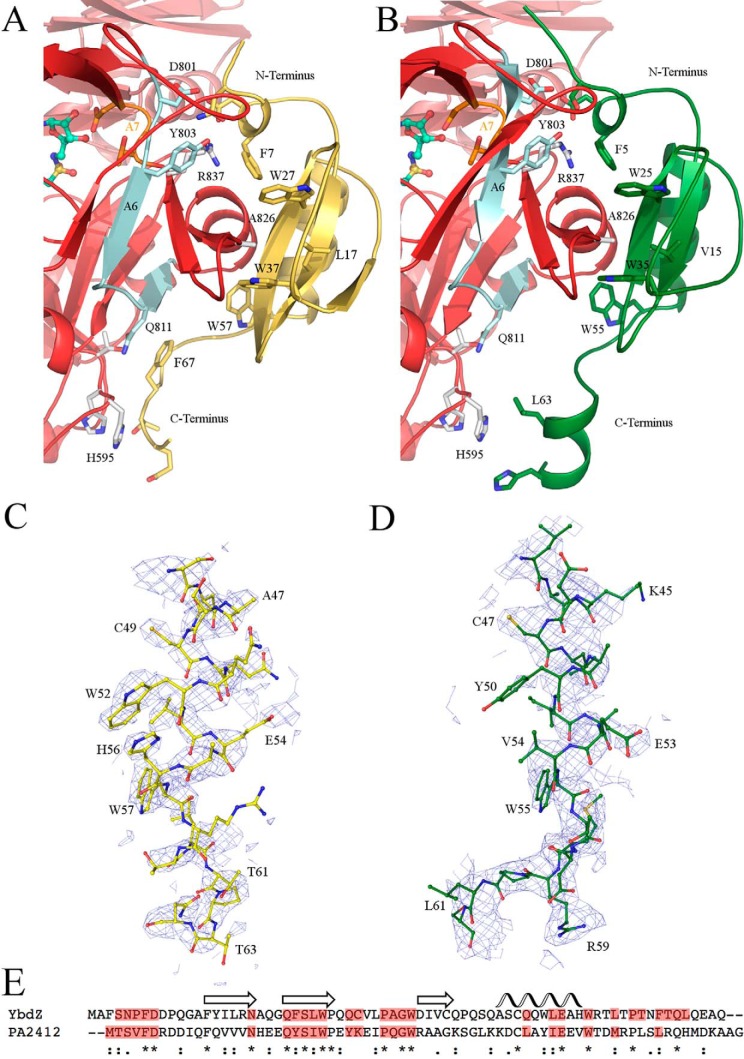
PA2412, the MLP from the P. aeruginosa pyoverdine NRPS cluster, activates EntF acyladenylate formation similarly to YbdZ (see below). Crystals of EntF bound to PA2412 were isomorphous with the EntF-YbdZ crystals and also diffracted to 3.0 Å. PA2412 was bound in the same location as YbdZ with similar contacts (Fig. 3).
FIGURE 3.
Interactions of EntF and MLPs. Interactions between the N-terminal subdomain of the adenylation domain (red) and YbdZ (yellow) (A) or PA2412 (green) (B) are shown. Simulated annealing 2Fo − Fc composite omit maps, contoured at 1 σ, are shown for the helix of YbdZ residues 42–61 (C) and PA2412 residues 43–62 (D). E, YbdZ and PA2412 share 25% sequence identity. Conserved secondary structural elements of MLPs are shown above. Residues for each MLP that interact with the adenylation N-terminal subdomain are highlighted in red. Strongly (:) or weakly (.) homologous, or identical (*) residues are indicated.
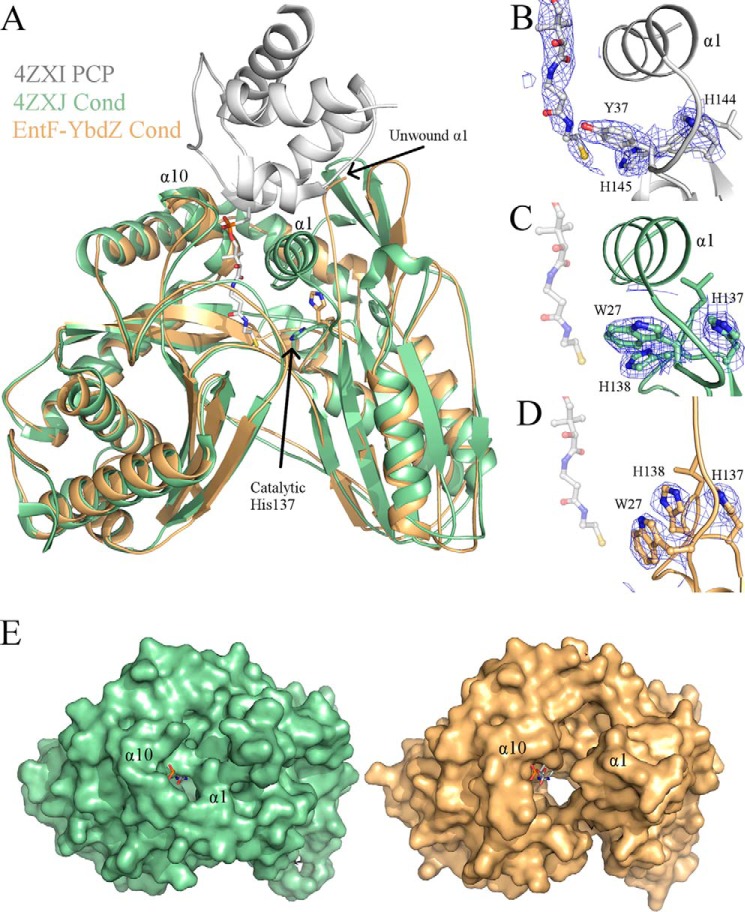
In both structures, there are several short unresolved loops. EntF from the YbdZ complex starts at residue 21, whereas EntF of the PA2412 complex starts at residue 16. This is accompanied by an unwinding of α1 in the condensation domain. There are four or five unresolved residues starting at residue 63 as well as two or three unresolved residues starting at residue 335 in the condensation domain. The entire adenylation domains of both structures are intact. The adenylation-PCP and the PCP-thioesterase domain linkers are disordered in both structures. The adenylation-PCP linker becomes unresolved at residue Leu-963, two residues after the important LPXP motif, which is critical for anchoring of the A10 motif (26). Finally, within the thioesterase domain, residues 1172–1181 are unresolved in both structures; residues 1245–1249 are only unresolved in the YbdZ complex. Both YbdZ and PA2412 are completely resolved with the exception of a few C-terminal residues. The overall average B-factors as well as the average B-factors for each individual domain are higher in EntF-PA2412 compared with EntF-YbdZ (Table 1). Simulated annealing 2Fo − Fc composite omit maps show unambiguous electron density for the presence of both YbdZ and PA2412 (Fig. 3, C and D). The refined electron density is clear; however, the less biased omit map density does show some disorder, perhaps reflecting the low resolution or possibly substoichiometric binding of the MLP or dynamics at the solvent-facing side of the MLP.
The Interaction of MLPs with the EntF Module
Both MLPs bound to the EntF adenylation domain in the same location and orientation as seen in SlgN1, ∼15 Å from the active site of the adenylation domain. They are oriented in such a way that only one of the three β-strands (β2), the α-helix, and termini make contact with the N-terminal subdomain of the adenylation domain, forming a 1437- and 1224-Å2 interface for EntF-YbdZ and EntF-PA2412, respectively. No contacts are made either between MLP and the C-terminal subdomain of the adenylation domain or the thioesterase domain. Minimal contacts are made between the MLPs and the condensation domain. Most notably Gln-303, Leu-427, and Asp-430 of the condensation domain interact with residues found on the loop between β2 and β3 of the MLPs.
Similar to the structure of SlgN1, two of the three conserved tryptophan residues of YbdZ (Trp-27 and Trp-37) and PA2412 (Trp-25 and Trp-35) form a cavity that surrounds the side chain of Ala-826 of the adenylation domain (Fig. 3). Leu-17 is located in the back of this pocket in YbdZ, and Val-15 is located in the back of this pocket in PA2412. The third conserved tryptophan for YbdZ and PA2412 (Trp-57 and Trp-55, respectively), located immediately after the longer α-helix, interacts with a pocket on the adenylation domain formed by Pro-817, Thr-820, and Ala-821. Interestingly, although the Trp residues have been shown to be important for MLP function, the equally well conserved proline residues on the MLP TioT can be substituted with alanine alone or in combination with no impact on MLP activity (27).
Based on an interface analysis by the Proteins, Interfaces, Structures, Assemblies software (PISA) server (28), there are 12 potential hydrogen bonds and two salt bridge between YbdZ and the EntF N-terminal subdomain of the adenylation domain. There are nine potential hydrogen bonds and three salt bridges between PA2412 and the EntF adenylation N-terminal subdomain. YbdZ and PA2412 share 25% sequence identity and 47% sequence similarity (Fig. 3E). Several of these similar residues interact with the adenylation domain.
The main difference between the interaction of YbdZ and PA2412 with EntF is the C termini of the MLPs (Fig. 3, A and C). The C terminus of PA2412 (13) adopts a two-turn α-helix that was not seen in the solution structure of MbtH, which instead contains an extended coil (14). SlgN1 is unique because the MLP is directly tethered to the N terminus of the adenylation domain. The linker between the MLP and the adenylation domain of SlgN1 is only resolved in two of the four monomers of the asymmetric unit in the unliganded Protein Data Bank structure 4GR4 and one of the four monomers in the AMPCPP-bound Protein Data Bank structure 4GR5. This linker forms a coil containing a single turn helix, which does not make contact with the adenylation domain.
The C termini of YbdZ and PA2412 illustrate different interactions with the adenylation domain. The C terminus of YbdZ forms an extended coil that is cradled by His-595, His-596, Thr-597, and Gln-811 of the adenylation domain (Fig. 3A). By contrast, the analogous adenylation domain residues of SlgN1 cradle a single arginine residue located in the C-terminal coil. The C terminus of PA2412 forms a two-turn helix as it did in Protein Data Bank structure 2PST (13). This helix is pulled away from the adenylation domain with Leu-63 oriented toward His-595 (Fig. 3B), reducing the size of the surface interface for PA2412 compared with YbdZ. Both N termini of YbdZ and PA2412 form a single turn helix and interact with Asp-801, Tyr-803, and Arg-837 in a similar manner (Fig. 3, A and B).
MLP Binding Does Not Alter the Structure of the Adenylation Domain
We examined the EntF-MLP complexes to determine whether MLP binding affects either the adenylation domain structure or influences the position of the adenylation C-terminal subdomain. The adenylation domain is trapped in the thioester-forming conformation and interacts with the PCP domain. The complexes show that MLPs can bind adenylation domains that are in the thioester-forming conformation. Published work showed that YbdZ can activate the adenylate-forming reaction and can co-purify with recombinant adenylation domains that bind MLPs (8). These data, along with the large interaction surface, suggest that MLPs bind to adenylation domains throughout the entire reaction cycle of the adenylation domain and not just during acyladenylate formation.
We next compared EntF structures in the presence and absence of the MLP. Alignments of our original structure with the current EntF-YbdZ complex show that steric conflicts preclude any trace YbdZ contaminants (shown to have the ability to co-purify with EntF from wild-type cells (8)) from interacting with the adenylation domain in the crystal lattice. The prior structure therefore represents an MLP-free EntF protein.
A comparison of the MLP-free and -bound states shows no changes to the protein structure that result from MLP binding. The r.m.s. displacement of the Cα atoms of the adenylation domains for the MLP-free structure to either the YbdZ- and PA2412-bound structures is 0.3 Å. Aligning the entire adenylation domain of EntF to the two EntF-MLP complexes results in an r.m.s. displacement of 0.4–0.6 Å. In all three structures, the residues that interact with an MLP, as well as active site residues that interact with the adenylate or serine moiety, are in nearly identical positions.
Biochemical Analysis of MLP Activation
To assess biochemically the MLP activation, the pyrophosphate exchange assay (29) was utilized to monitor acyladenylate formation (Table 2) with three adenylation domains and their substrates: EntF adenylation of serine, the excised adenylation domain from PvdL module 2 (PvdL-M2A) adenylation of glutamate, and NikP1 adenylation of histidine. PvdL is a four-module NRPS protein from the pyoverdine cluster of P. aeruginosa (30), which is sensitive to activation by PA2412.3 NikP1 is present in the nikkomycin biosynthetic operon from Streptomyces tendae. Like SlgN1, NikP1 contains the MLP fused to the N terminus of the adenylation domain. NikP1 also contains a PCP immediately downstream of the adenylation domain (31).
TABLE 2.
Apparent kinetic constants for proteins in presence and absence of MLPs
ND, not detected (see “Experimental Procedures”).
| Protein | Substrate | Temperature | kcat | Km | kcat/Km |
|---|---|---|---|---|---|
| °C | s−1 | μm | s−1 m−1 | ||
| EntF | Serine | 37 | 0.30 ± 0.02 | 27.7 ± 7.4 | 1.09 × 104 |
| EntF + YbdZ | Serine | 37 | 0.28 ± 0.01 | 6.9 ± 1.0 | 4.02 × 104 |
| EntF | Serine | 0 | 0.07 ± 0.01 | 503 ± 57 | 1.35 × 102 |
| EntF + YbdZ | Serine | 0 | 0.14 ± 0.01 | 13.8 ± 4.4 | 9.84 × 103 |
| EntF + PA2412 | Serine | 0 | 0.32 ± 0.01 | 59.6 ± 9.6 | 5.28 × 103 |
| EntF | ATP | 37 | 0.25 ± 0.02 | 267 ± 32 | 9.48 × 102 |
| EntF + YbdZ | ATP | 37 | 0.23 ± 0.01 | 279 ± 15 | 8.17 × 102 |
| EntF | ATP | 0 | 0.15 ± 0.01 | 251 ± 20 | 6.06 × 102 |
| EntF + YbdZ | ATP | 0 | 0.32 ± 0.01 | 271 ± 21 | 1.17 × 103 |
| NikP1 WT | Histidine | 37 | 0.28 ± 0.02 | 170 ± 49 | 1.62 × 103 |
| NikP1 (A-PCP) | Histidine | 37 | ND | ND | ND |
| NikP1 (A-PCP) + PA2412 | Histidine | 37 | 0.36 ± 0.01 | 202 ± 15 | 1.78 × 103 |
| NikP1 (P449A) | Histidine | 37 | 0.23 ± 0.01 | 509 ± 26 | 4.42 × 102 |
| PvdL-M2A | Glutamate | 37 | ND | ND | ND |
| PvdL-M2A + PA2412 | Glutamate | 37 | 0.12 ± 0.01 | 11,800 ± 3,000 | 10 |
All three proteins were purified from a strain lacking the endogenous E. coli MLP. To assess the MLP dependence of NikP1, the tethered MLP was removed, and the truncated NikP1, containing the adenylation domain and PCP, was expressed and purified. The MLP domain from NikP1 could not be expressed solubly. Therefore, PA2412 was used as a substitute to demonstrate whether the truncated adenylation domain-PCP construct remained functional. Both NikP1 and PvdL-M2A were inactive in the absence of MLP, and activity was observed in the presence of an MLP. Addition of PA2412 was able to restore activity to the truncated NikP1 adenylation domain-PCP protein.
Unlike PvdL-M2A and NikP1, EntF is active in the absence of an MLP. This was reported previously by Felnagle et al. (8), who concluded that MLP binding is required to lower the Km for serine to concentrations found within the cytosol of E. coli. Because EntF retained some activity in the absence of YbdZ, apparent kinetic rates for both serine and ATP were measured at 37 and 0 °C in the presence and absence of YbdZ.
At 37 °C, there was no appreciable difference in the kcat for serine between YbdZ-bound and unbound EntF. There was, however, a modest 4-fold increase in the Km for serine when YbdZ was not bound. At 0 °C, addition of YbdZ led to a 35-fold decrease in Km and a 2-fold increase in kcat for serine, resulting in a ∼70-fold increase in enzymatic efficiency (kcat/Km). The addition of PA2412 results in a 39-fold increase in kcat/Km.
The differences in apparent kinetic efficiency for ATP were not as significant as those for serine. At 37 °C, there was no difference in either kcat or Km. At 0 °C, addition of YbdZ results in a 2-fold increase in enzymatic efficiency. These data suggest that MLP binding more significantly influences the kinetic efficiency for amino acid utilization.
The adenylation domains of both EntF and SlgN1 (16) contain an alanine residues that inserts into the tryptophan pocket of the MLP. The importance of the alanine (16) and tryptophan residues (9) have been confirmed biochemically. Several adenylation domains known to be MLP-activated, for example PacL (9), VbsS (10), NovH (11), and Cgc18 (32), all have a proline in place of the alanine. NikP1 also contains a proline at this position. A mutation of Pro-449 to alanine in full-length NikP1 retained MLP-dependent activity (Table 2). Therefore, alanine and proline are interchangeable residues, both capable of inserting into the tryptophan cavity of a single MLP.
The EntF Thioesterase Domain Adopts a New Orientation
The overall structure of the thioesterase domain of EntF is similar to other structures. The core of the domain contains six parallel β-strands and one antiparallel β-strand, which form one continuous central β-sheet. This β-sheet is surrounded by five α-helices. Within this α,β-fold is the active site that consists of Ser-1138, His-1271, and Asp-1165. This catalytic triad aligns well with the active sites of other thioesterase domain structures, including SrfA-C (24, 33), AB3404 (21), and the excised EntF thioesterase domain interacting with the PCP in a catalytic manner (22, 23) (Fig. 4B).
FIGURE 4.
EntF thioesterase domain. A, EntF-YbdZ ribbon diagram with adenylation domain (red), PCP (green-cyan), and YbdZ (yellow). Unresolved coils are represented by dotted lines. The condensation domain is not shown for clarity. B, alignment of the thioesterase domain of EntF-YbdZ (blue) with the crystal structure of the EntF PCP-thioesterase domain (tan and green-cyan) that depicts a catalytic interaction between the two domains. C, alignment of the thioesterase domain of EntF-YbdZ (blue) with the closed (yellow) and open (pink) thioesterase domains of SrfTE (Protein Data Bank code 1JMK, chains C and O, respectively).
Atop the thioesterase α,β-fold housing the catalytic triad sits a lid composed of two α-helices. This region is known to adopt both an opened and closed conformation (33). In the open conformation, the first α-helix of the lid is angled upward, forming an opening on the face opposite of where the PCP interacts. In the closed conformation, both α-helices of the lid are parallel relative to each other. In the MLP-bound structures of EntF, the lid region adopts a closed conformation with overall good electron density for both α-helices (Fig. 4C).
In the previous EntF crystal structure, the thioesterase domain was completely unresolved (21). In the new crystal form, the thioesterase domains of the MLP-EntF structures are resolved and located alongside the adenylation domain, forming a 565-Å2 interface with the N-terminal subdomain (Fig. 4A). This results in an overall linear architecture of the condensation, adenylation, and thioesterase domains (Fig. 2). This striking new position of the thioesterase domain represents an 83- and 92-Å movement of the center of mass of this domain relative to the comparable positions in the SrfA-C and AB3403 structures, respectively. Our structures further support a dynamic system in which the thioesterase domain is free to adopt several different conformations and locations while the module is in the thioester-forming conformation. The lack of interactions between the thioesterase domain and the core of the protein suggests it may be loosely tethered to the PCP and relatively free to move in solution.
Changes to the Condensation-Adenylation Didomain Core of the Module
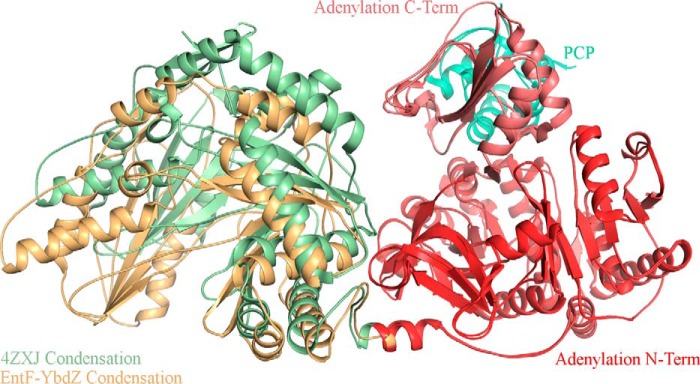
The interactions between the condensation domain and the adenylation domain of modular NRPSs are quite extensive. The condensation-adenylation domain interface is 1023, 1097, and 780 Å2 for AB3403, SrfA-C, and EntF, respectively. In each of these three structures, the orientation of the condensation domain relative to the adenylation domain differs slightly. The condensation domains of AB3403 and EntF are rotated by ∼25° compared with SrfA-C. Furthermore, the condensation domain of EntF is kinked upward toward the adenylation domain. This orientation appears to be incompatible with the adenylate-forming conformation as the C-terminal subdomain of the adenylation domain would clash with the C-terminal lobe of the condensation domain. We therefore hypothesized that to adopt the adenylate-forming conformation the condensation domain of EntF in the original structure (21) would need to shift away from the adenylation domain to a similar position as seen in AB3403. In both EntF-MLP complexes, this is precisely the case. Despite being in the thioester-forming conformation, the condensation domain moves away from the adenylation domain (Fig. 5). This change in the condensation-adenylation domain interface confirms that the condensation domain can move relative to the adenylation domain within a single protein.
FIGURE 5.
Orientation of the condensation and adenylation didomain platform in EntF structures. Superposition of the previous and current EntF structures shows that in the structure of MLP-free EntF the condensation domain (green) is shifted upward toward the adenylation domain (red) and PCP (green-cyan).
Conformational Changes at the PCP Binding Site within the Condensation Domain
To accommodate both the upstream and downstream PCPs, it was hypothesized that the bilobed condensation domain undergoes an opening and closing (24, 34). Both AB3403 and EntF are in a closed conformation despite the modules being in different stages of the NRPS catalytic cycle. Although the condensation domains of the EntF-MLP complexes demonstrate mobility relative to the adenylation domain, the condensation domains from all three EntF structures are in the same closed conformations with an overall r.m.s. displacement of 1.2 Å. The largest difference between the condensation domains, besides their orientation, is an unraveling of α1 in the MLP complexes.
The PCP of AB3403 sits on top of α1 and α10 of the condensation domain to allow the 4′-phosphopantetheine (PPant) arm of the PCP to extend into the active site (21). In the new structures, α1, consisting of residues 10–20, is unwound and largely unresolved (Fig. 6A); however, α10 is in the same location as seen previously in other condensation domains. As a result of this unwinding, the tunnel in which the downstream PCP PPant enters is much larger than seen in any other condensation domain structure (Fig. 6E).
FIGURE 6.
Condensation domains of EntF structures. A, comparison of the downstream PPant tunnel of the condensation domain (Cond) between the MLP-free (green) and YbdZ-bound (orange) EntF proteins. Superimposed is the PCP from AB3403 (Protein Data Bank code 4ZXI; gray) with PPant, represented in stick, included. In all current structures of condensation domains, α1 and α10 are located at the entrance of the tunnel where the downstream PCP sits. This results in the catalytic histidine pointing into the tunnel toward the thiol of the PPant. Electron density around the catalytic histidine with coefficients from 2Fo − Fc contoured at 1.5 σ is shown for AB3403 (B), MLP-free EntF (C), and YbdZ-EntF (D). The pantetheine arm is shown in C and D for reference only. E, this results in a larger opening into the active site tunnel as demonstrated by surface representation of Protein Data Bank structures 4ZXJ (green) and 5JA1 (orange). The pantetheine moiety is again modeled in the tunnel from alignment with AB3403.
This unwinding of α1 also alters the active site. The condensation domain active sites possess a conserved HHXXXDG motif. The second histidine of this motif is crucial for peptide bond formation (35). In all current crystal structures of condensation domains, this histidine is orientated down, away from α1 and α10 and into the center of the condensation domain tunnel (Fig. 6). The new EntF structures show that unwinding of α1 allows this catalytic histidine to change rotamers and rotate upward toward the opening formed by the unwinding of α1 (Fig. 6, A and D). Trp-27 is now shifted down, impeding His-127 from pointing into the active site (Fig. 6D). Based on these structural changes, it is possible that this unwinding of α1 may be an opening mechanism specific for the downstream PCP and PPant.
Discussion
We present herein the structures of EntF bound to two MLPs, its true partner and a homolog from P. aeruginosa. In this structure, EntF is trapped in the thioester-forming conformation with the PCP directed into the adenylation domain active site bound to the mechanism-based inhibitor Ser-AVS. The MLP-adenylation domain interaction is similar to that seen with SlgN1, and the availability of multiple structures allows us to test several structural hypotheses regarding the nature of the MLP-adenylation domain interaction.
Adenylation domains catalyze two partial reactions, the combination of ATP and the amino acid to form an aminoacyladenylate and pyrophosphate and the subsequent thioester-forming reaction where the pantetheine thiol displaces the AMP to load the pantetheine with the amino acid. These two steps are catalyzed by two conformations of the adenylation domain, a process referred to as domain alternation (15). The MLP could enhance or activate the adenylation domain by altering the structure of the adenylation domain to adopt a catalytically competent conformation. Comparison of the structures of EntF in the presence and absence of MLPs does not show any difference to support, for example, a movement of an important catalytic loop into position. Alternately, enhancement of adenylation domain activity could occur if MLP binding promoted the formation of the adenylate-forming conformation by preventing formation of the alternate catalytic orientation. This also does not seem to be the case. It appears therefore that the activation of the adenylation domain by an MLP is not the result of a structural change.
An important caveat is that EntF is, as noted, only enhanced by the MLP and not fully activated. The other complete NRPS modules that have been structurally characterized appear not to be MLP-dependent. The organisms encoding AB3403 and LgrA do not contain any MLPs in the genome, and SrfA-C contains a glutamate residue at the site of the tryptophan insertion residue that is an alanine in EntF and SlgN1. Therefore, additional structural studies of MLP interactions with complete NRPS modules, particularly those that are fully MLP-dependent, may offer different views into the impact of MLP binding on the adenylation domain.
Adenylation domains can require MLPs for either solubility or activity, and some, such as EntF and NovH (11), are unusual as they function without MLPs but show increased activity upon MLP binding. We asked whether these structural and biochemical data could allow one to determine from an amino acid sequence if an adenylation domain will accommodate an MLP. Both alanine and proline are compatible with the stacked tryptophans of MLPs. The MLP of NikP1 can coordinate both residues potentiating adenylation domain activity (Table 2). PA1221, an adenylation-PCP didomain protein from P. aeruginosa, has a glutamate in this position and is not activated by the only MLP in P. aeruginosa (36); similarly, inserting a glutamate in place of the alanine in SlgN1 abolished MLP binding (16). Therefore, a larger polar residue in this position is likely incompatible with MLP binding.
This crucial tryptophan-coordinated residue is located 13 residues after the A6 motif (15, 37) and two residues after a highly conserved hydrophobic residue, most often phenylalanine. Using this fingerprint, GEX10GYX10FX(A/P) with GEX10GY representing the structural A6 motif and A/P representing the tryptophan-coordinated residue, a bioinformatics search was carried out to determine the variation of the tryptophan-coordinated residue among NRPS adenylation domains. To do this, the same adenylation domain database used to assess adenylation-PCP domain linkers (26) was again analyzed. To eliminate potential shifts due to gaps in the sequence, any sequence that did not match the fingerprint was not included in the analysis. Some degree of freedom was given to GE, GY, and Phe as long as it was clear the substitute residue was comparable and that no more than two of the five residues varied. Of the 6,375 sequences in the original adenylation domain database, 5,237 (82%) fit these criteria. Of these, 38% contained an alanine at the tryptophan-coordinated site, and 26% contained a proline. Aspartate and glutamate occupy this position in 15% of adenylation domain sequences. The remaining 21% of sequences contain the other 16 amino acids of which threonine is most common at 4.5% (supplemental Fig. S1).
This analysis indicates that the majority of NRPS adenylation domains can potentially accommodate MLPs to some extent because alanine and proline were heavily favored at this position. However, it is known that some adenylation domains, for example EntE, contain an alanine at the tryptophan-coordinated site and yet do not show MLP binding or enhancement (8). Consequently, it may be easier to predict that an adenylation domain is MLP-independent rather than MLP-dependent. Sequence analysis alone is insufficient to classify an adenylation domain as MLP-dependent, and definitive support will continue to require biochemical verification.
Listed in supplemental Table S1 are adenylation domains that to our knowledge have been tested for MLP dependence. Of the 27 listed, 13 require MLPs for acyladenylate formation, six require MLPs for solubility, two are enhanced by MLPs, four are not affected by MLPs, and two do not require MLPs for in vivo activity because the organism lacks an endogenous MLP. Interestingly TioK, which requires MLP binding for solubility (27), has a leucine at the tryptophan-coordinated site (Fig. 7). This suggests that larger non-polar residues, such as leucine, isoleucine, and valine, which make up 3.8% of all possible residues at this location, may also insert into the tryptophans of MLPs.
FIGURE 7.
Sequences of adenylation domains that have been tested for MLP dependence. Alignment of all adenylation domains tested for MLP dependence from supplemental Table S1 is shown. Highlighted regions include the A6 motif (cyan), the tryptophan-coordinated residues (pink), and the A7 motif (orange). Asterisks above indicate the residues that interact with the MLPs from the crystal structures of SlgN1 (green), EntF-PA2412 (red), and EntF-YbdZ (blue). MLP dependence is noted on the left.
Our structural analysis identified other binding interactions between the MLP and adenylation domain. For example, the third tryptophan of the MLP interacts with a pocket formed by Pro-817, Thr-820, and Ala-821. These three residues are well conserved in the adenylation domains that require an MLP interaction (Fig. 7).
The EntF structures presented here provide additional views that expand our understanding of the conformations adopted by NRPSs. The EntF crystal structures are in the thioester-forming conformation with the PCP covalently trapped in an interaction with the adenylation domain due to the serine adenosine vinylsulfonamide inhibitor. The three NRPS modules with the same architecture, SrfA-C, AB3403, and EntF, show very different locations for the terminal thioesterase domain. In none of these structures does the thioesterase domain make significant contacts with the remainder of the protein. Therefore, it appears that the location of the thioesterase domain within the module is highly variable and is loosely based on the location of the PCP as it cycles through the NRPS catalytic cycle.
The unwinding of α1 and the alternate rotamer for the catalytic histidine in the condensation domain of the new EntF structures are the only condensation domain structures in which this exists (Fig. 6). Because α1 is one of two helices that coordinate the binding of the downstream PCP, it seems plausible that the unwound structure would be unable to facilitate a downstream PCP interaction. This unwinding also creates a larger active site tunnel in the condensation domain. It is possible that this unwinding may be a mechanism to allow for binding and release of the downstream PPant and bound nascent natural product. Further biochemical work will need to be done to confirm this hypothesis. Together with a recent structure of an upstream PCP with an epimerization domain, a structural homolog of NRPS condensation domains (38), our understanding of the functional complexes with condensation domains is becoming much clearer.
Both the adenylation-PCP and PCP-thioesterase domain linkers of the EntF structures are disordered. This suggests that these regions do not have a single conformation in the thioester-forming conformation. The adenylation-PCP linker in EntF is ordered through the LPXP motif, located at Leu-958 through Pro-961, but becomes disordered at residue Leu-963. This supports our prior hypothesis that the LPXP motif of the adenylation-PCP linker anchors it to the C-terminal subdomain of the adenylation domain (26). This motif secures the adenylation A10 loop, harboring the catalytic lysine, and assists in coordinating the movement between the C-terminal subdomain of the adenylation domain and the PCP.
The recent NRPS modular structures of AB3403, LgrA, and EntF will assist in understanding the required intradomain interactions for natural product biosynthesis. These insights could lead to new development in combinatorial biosynthesis and production of novel peptide natural products. The structures of EntF presented here now allow analysis of MLP binding in the context of a full module and shed light on the dynamics of the thioesterase domain and the movement of the condensation domain relative to the adenylation domain.
Experimental Procedures
Cloning of NRPS Genes
The entF, entE, and entB genes were amplified from the genomic DNA of E. coli strain JM109 (26, 39). Accession numbers and domain boundaries are reported in supplemental Table S2. The adenylation-PCP domains from pvdL module 2 and the MLP pa2412 were amplified from P. aeruginosa strain PAO1 (13). The adenylation domain from the amplified pvdL gene (PvdL-M2A) was created by placing a stop codon after the adenylation domain and before the PCP. The nikP1 gene was generously provided by Walsh and co-workers (31). To remove the tethered MLP from NikP1, an NdeI-cut site was introduced after the attached MLP and before the adenylation domain. After removal, T4 ligase was used to ligate the NikP1 adenylation-PCP domain construct into a pET15b plasmid. Finally, the ybdZ gene was chemically synthesized (GenScript) and provided in a pUC57 plasmid. All genes were cloned into a modified pET15b vector encoding an N-terminal His5 tag sequence and TEV protease recognition site (40). GenBankTM accession numbers and domain boundaries for protein fragments are listed in supplemental Table S2.
Expression of EntF, EntE, EntB, and YbdZ
Expression of EntE, EntB, and YbdZ was carried out in BL21-DE3 cell line. EntF was expressed in BL21-DE3 ΔybdZ cells kindly provided by Dr. Michael G. Thomas, University of Wisconsin. This eliminated the possibility of co-purifying the endogenous E. coli MLP, YbdZ, with EntF. Cells grew at 37 °C in LB medium until an A600 of 0.6 was reached. Expression was induced with 1 mm IPTG, and the cells were incubated at 16 °C overnight (∼18 h). Cells were harvested via centrifugation and flash frozen in liquid nitrogen. The cells were lysed via sonication in lysis buffer containing 50 mm Tris, pH 7.5, 400 mm NaCl, 0.2 mm TCEP, 10% glycerol (v/v), and 10 mm imidazole. After centrifugation to remove cell debris, the lysate was passed over a 5-ml Ni2+·HiTrap Chelating HP column (GE Healthcare), and bound proteins were eluted with lysis buffer containing 300 mm imidazole. The eluate was dialyzed overnight in cleavage buffer containing 50 mm Tris, pH 7.5, 400 mm NaCl, 0.2 mm TCEP, 10% glycerol (v/v), and 0.5 mm EDTA. During the overnight dialysis, TEV protease was incubated with the protein to remove the His tag. For EntF and EntB, which contain a PCP, 200 nm Sfp (the promiscuous phosphopantetheinyltransferase from Bacillus subtilis), 100 μm CoA, and 1 mm MgCl2 were added to the dialyzing proteins. The next morning the proteins were passed over a nickel affinity column once more to remove cleaved His tag, Sfp, and TEV. The flow-through was collected and concentrated to 3 ml. The concentrated flow-throughs for EntF, EntE, and EntB were run over a Superdex 200 16/600 gel filtration column in 50 mm EPPS, pH 8.0, 150 mm NaCl, 0.2 mm TCEP, and 10% glycerol (v/v). All proteins were concentrated to the desired concentration via centrifugal filters and flash frozen in liquid nitrogen.
Expression and Purification of PvdL-M2A and PA2412
PvdL-M2A and PA2412 were purified similarly as EntE, EntB, and EntF. Briefly, BL21-DE3 ΔybdZ for PvdL-M2A and BL21-DE3 cells for PA2412 were grown at 37 °C to an A600 of 0.6, and protein expression was induced with the addition of 750 μm IPTG. The induced cells were incubated overnight at 16 °C. Cells were lysed via sonication with 50 mm Tris, pH 7.5, 150 mm NaCl, 20 mm imidazole, and 0.2 mm TCEP. After centrifugation, the proteins were eluted from a 5-ml Ni2+·HiTrap Chelating HP column with lysis buffer with 300 mm imidazole. TEV protease was added to the eluted proteins and dialyzed overnight at 4 °C in 50 mm Tris, pH 7.5, 150 mm NaCl, 0.2 mm TCEP, and 0.5 mm EDTA. The next day a second nickel column was run to separate the cleaved proteins from the His tags and protease. PA2412 was then flash frozen in liquid nitrogen and stored. PvdL-M2A was run over a Superdex 200 16/600 gel filtration column in 20 mm Tris, pH 7.5, 40 mm NaCl, and 0.2 mm TCEP. Protein was flash frozen in liquid nitrogen and stored at −80 °C.
Expression and Purification of NikP1 Constructs
BL21-DE3 ΔybdZ cells containing the NikP1 and truncated NikP1 plasmids were grown at 37 °C until an A600 of 0.6 was reached. Protein expression was induced with 500 μm IPTG, and cells were incubated overnight at above for the other proteins. The lysis buffer contained 25 mm Tris, pH 8.0, 400 mm NaCl, 0.2 mm TCEP, 10% glycerol (v/v), and 10 mm imidazole. Protein was eluted from the nickel column in lysis buffer that contained 300 mm imidazole. TEV protease, Sfp, CoA, and MgCl2 were added, and the protein was dialyzed overnight at 4 °C in 25 mm Tris, pH 8.0, 400 mm NaCl, 0.2 mm TCEP, 10% glycerol (v/v), and 0.5 mm EDTA. Dialyzed protein eluted from the second nickel affinity column was further purified with the gel filtration column in 25 mm Tris, pH 7.5, 50 mm NaCl, 0.2 mm TCEP, and 10% glycerol (v/v).
Pyrophosphate Exchange Assay
To assess acyladenylate formation for EntF, PvdL-M2A, and NikP1, the pyrophosphate exchange assay was used to monitor the reverse formation of radiolabeled ATP (29). 100-μl reactions were set up containing 1 μm enzyme, 200 μm Na4PPi, and 0.15 μCi of Na4[32P]PPi in 50 mm HEPES, pH 8.0, 100 mm NaCl, 10 mm MgCl2, and 1 mm EDTA. For measuring apparent amino acid kinetics (EntF-serine, PvdL-M2A-glutamate (pH 7.5), and NikP1-histidine), saturating concentrations of ATP were added. For EntF and NikP1, this was 2 mm ATP, and for PvdL-M2A, this was 5 mm ATP. Amino acid concentrations varied from 1 μm to 1 mm. For measuring apparent ATP kinetics, amino acids were kept at a saturating concentration of 5 mm. The ATP concentration varied from 1 μm to 1 mm. Reactions were carried out at 37 °C or on ice (0 °C) for 10 min and then quenched with 500 μl of 1.2% (w/v) activated charcoal, 0.1 m Na4PPi, and 0.35 m perchloric acid. Samples were centrifuged, and the pelleted charcoal was washed with 1 ml of distilled H2O twice. After the final wash, the charcoal was resuspended and transferred to 10 ml of liquid scintillation fluid. Radiolabeled nucleotide was quantified using a Packard Tri-Carb 1900 TR liquid scintillation counter. Apparent kinetic values were calculated using non-linear regression curve fitting and the Michaelis-Menten equation. Activity for the EntF + YbdZ data yields a specific activity ranging from 2 nmol/min/mg with 1 μm serine to 108 nmol/min/mg with 1 mm serine; activity below 0.5 nmol/min/mg was considered below the limits of detection.
EntF-YbdZ Crystallization and Structure Determination
Prior to crystallization of EntF, a Ser-AVS inhibitor (21) was added to the protein at a concentration 4× that of EntF and incubated at room temperature (∼22 °C) for 2–4 h. Also added at that time was YbdZ in equal molar amounts to EntF. Crystal conditions were first identified using the Hauptman-Woodward high throughput screen (41). Long thin needles grew from a single nucleation point in a mixture containing 100 mm Bistris propane, pH 7.5, 125–150 mm MgCl2, and 22–28% PEG 4000 (w/v). Crystals were replicated using hanging drop vapor diffusion at 20 °C. EntF (30 mg/ml) was used with a protein to mixture volume ratio of 1:1. A batch mimic approach (19, 21) was used in which the mixture was diluted in half with EntF dialysis buffer in the reservoir. Although the diluted mixture was used in the reservoir, the undiluted mixture was used in the 1:1 protein drop. Because these needle-like crystals were too thin to work with, they were used as seeds for larger crystals. A Seed Bead (42) was used to crush the original needle crystals. These microcrystals were used for seeding with 20 mg/ml EntF in the same mixtures and reservoirs as above with a protein-mixture-seed ratio of 0.8:0.6:0.2. Larger single crystals grew at 14 and 20 °C.
The larger crystals grown from seeds were harvested and cryoprotected with the original undiluted mixture supplemented with 10% 2,3-butanediol (v/v). Diffraction data were collected on Advanced Photon Source (APS) beamline 23-ID-B using the rastering option to find the optimal spots on the crystals. Diffraction data from several partial data sets from three crystals were indexed, merged, and scaled using iMOSFLM (43) in space group I2. Combining these data sets resulted in overall good statistics (Table 1) with a completeness of 93.1% overall and 93.7% in the outer resolution shell. Structure determination was carried out in PHENIX (44). Molecular replacement was performed using the previous EntF structure (21) as a model. Due to slight differences in overall architecture, the EntF molecular replacement model was split into individual domains with each domain being a separate model. Because the thioesterase domain was unresolved in the EntF model, the thioesterase domain from the excised EntF PCP-thioesterase crystal structure was used (23). The crystal structure of the MLP PA2412 (13) was used as a search model for YbdZ. However, due to the small size of MLPs (8 kDa), PhaserEP was unable to find an appropriate position for the YbdZ domain. Therefore, a homology model of the EntF adenylation domain interaction with PA2412, created by comparison with SlgN1 (16), was used. The EntF-YbdZ model was built and refined iteratively using Coot (45) and PHENIX. Translation-libration-screw (TLS) refinement was used with refinement groups defined along the functional NRPS domains or subdomains for the condensation and adenylation domains; individual isotropic B-factors were used.
EntF-PA2412 Crystallization and Structure Determination
EntF-PA2412 crystals were grown using the same technique as for the EntF-YbdZ crystals. Prior to mixing, PA2412 was dialyzed into the same final buffer as EntF. The large crystals grown from EntF-PA2412 seeds and cryoprotected with 10% 2,3-butandiol (v/v) were shipped to Stanford Synchrotron Radiation Lightsource (SSRL) beamline 12-2 for data collection. A complete data set was collected from a single crystal. A molecular replacement solution was found using the protein atoms from the EntF-YbdZ structure as a model. The structure was modeled and refined using the same procedure used for the EntF-YbdZ structure.
Author Contributions
B. R. M. performed the structural and biochemical analysis. E. J. D. cloned and expressed the proteins from P. aeruginosa. C. S. and C. C. A. designed and synthesized the Ser-AVS inhibitor. B. R. M. and A. M. G designed the experiments, analyzed the results, and wrote the manuscript.
Supplementary Material
Acknowledgments
We thank Alyssa Aldridge for technical assistance with the NikP1 mutagenesis experiments. We also thank Professors Christopher Walsh and Michael Thomas for generously providing bacterial strains. The General Medical Sciences and Cancer Institutes Structural Biology Facility at the Advanced Photon Source (GM/CA@APS) has been funded in whole or in part with federal funds from the National Cancer Institute (Grant ACB-12002) and the National Institute of General Medical Sciences (Grant AGM-12006). This research used resources of the Advanced Photon Source, a United States Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract DE-AC02-06CH11357. Use of the Stanford Synchrotron Radiation Lightsource (SSRL) is supported by the United States Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research and by the National Institutes of Health, National Institute of General Medical Sciences, including Grant P41GM103393.
This work was supported by National Institutes of Health Grant R01 GM116957 (to A. M. G.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The atomic coordinates and structure factors (codes 5JA1 and 5JA2) have been deposited in the Protein Data Bank (http://wwpdb.org/).

This article contains supplemental Fig. S1 and Tables S1 and S2.
M. G. Thomas, personal communication.
- NRPS
- nonribosomal peptide synthetase
- AMPCPP
- α,β-methyleneadenosine 5′-triphosphate
- DHB
- 2,3-dihydroxybenzoic acid
- EPPS
- 3-[4-(2-hydroxyethyl)-1-piperazinyl]propanesulfonic acid
- IPTG
- isopropyl β-d-1-thiogalactopyranoside
- MLP
- MbtH-like protein
- PPant
- 4′-phosphopantetheine
- PCP
- peptidyl carrier protein
- PvdL-M2A
- adenylation domain from PvdL module 2
- r.m.s.
- root mean square
- Ser-AVS
- serine adenosine vinylsulfonamide
- TCEP
- tris(2-carboxyethyl)phosphine
- TEV
- tobacco etch virus
- Bistris propane
- 1,3-bis[tris(hydroxymethyl)methylamino]propane.
References
- 1. Miller B. R., and Gulick A. M. (2016) Structural biology of nonribosomal peptide synthetases. Methods Mol. Biol. 1401, 3–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Walsh C. T. (2016) Insights into the chemical logic and enzymatic machinery of NRPS assembly lines. Nat. Prod. Rep. 33, 127–135 [DOI] [PubMed] [Google Scholar]
- 3. Weissman K. J. (2015) The structural biology of biosynthetic megaenzymes. Nat. Chem. Biol. 11, 660–670 [DOI] [PubMed] [Google Scholar]
- 4. Labby K. J., Watsula S. G., and Garneau-Tsodikova S. (2015) Interrupted adenylation domains: unique bifunctional enzymes involved in nonribosomal peptide biosynthesis. Nat. Prod. Rep. 32, 641–653 [DOI] [PubMed] [Google Scholar]
- 5. Walsh C. T., Chen H., Keating T. A., Hubbard B. K., Losey H. C., Luo L., Marshall C. G., Miller D. A., and Patel H. M. (2001) Tailoring enzymes that modify nonribosomal peptides during and after chain elongation on NRPS assembly lines. Curr. Opin. Chem. Biol. 5, 525–534 [DOI] [PubMed] [Google Scholar]
- 6. Samel S. A., Marahiel M. A., and Essen L. O. (2008) How to tailor non-ribosomal peptide products—new clues about the structures and mechanisms of modifying enzymes. Mol. Biosyst. 4, 387–393 [DOI] [PubMed] [Google Scholar]
- 7. Quadri L. E., Sello J., Keating T. A., Weinreb P. H., and Walsh C. T. (1998) Identification of a Mycobacterium tuberculosis gene cluster encoding the biosynthetic enzymes for assembly of the virulence-conferring siderophore mycobactin. Chem. Biol. 5, 631–645 [DOI] [PubMed] [Google Scholar]
- 8. Felnagle E. A., Barkei J. J., Park H., Podevels A. M., McMahon M. D., Drott D. W., and Thomas M. G. (2010) MbtH-like proteins as integral components of bacterial nonribosomal peptide synthetases. Biochemistry 49, 8815–8817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang W., Heemstra J. R. Jr., Walsh C. T., and Imker H. J. (2010) Activation of the pacidamycin PacL adenylation domain by MbtH-like proteins. Biochemistry 49, 9946–9947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Heemstra J. R. Jr., Walsh C. T., and Sattely E. S. (2009) Enzymatic tailoring of ornithine in the biosynthesis of the Rhizobium cyclic trihydroxamate siderophore vicibactin. J. Am. Chem. Soc. 131, 15317–15329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Boll B., Taubitz T., and Heide L. (2011) Role of MbtH-like proteins in the adenylation of tyrosine during aminocoumarin and vancomycin biosynthesis. J. Biol. Chem. 286, 36281–36290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Baltz R. H. (2011) Function of MbtH homologs in nonribosomal peptide biosynthesis and applications in secondary metabolite discovery. J. Ind. Microbiol. Biotechnol. 38, 1747–1760 [DOI] [PubMed] [Google Scholar]
- 13. Drake E. J., Cao J., Qu J., Shah M. B., Straubinger R. M., and Gulick A. M. (2007) The 1.8 Å crystal structure of PA2412, an MbtH-like protein from the pyoverdine cluster of Pseudomonas aeruginosa. J. Biol. Chem. 282, 20425–20434 [DOI] [PubMed] [Google Scholar]
- 14. Buchko G. W., Kim C. Y., Terwilliger T. C., and Myler P. J. (2010) Solution structure of Rv2377c-founding member of the MbtH-like protein family. Tuberculosis 90, 245–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gulick A. M. (2009) Conformational dynamics in the acyl-CoA synthetases, adenylation domains of non-ribosomal peptide synthetases, and firefly luciferase. ACS Chem. Biol. 4, 811–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Herbst D. A., Boll B., Zocher G., Stehle T., and Heide L. (2013) Structural basis of the interaction of MbtH-like proteins, putative regulators of nonribosomal peptide biosynthesis, with adenylating enzymes. J. Biol. Chem. 288, 1991–2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gehring A. M., Mori I., and Walsh C. T. (1998) Reconstitution and characterization of the Escherichia coli enterobactin synthetase from EntB, EntE, and EntF. Biochemistry 37, 2648–2659 [DOI] [PubMed] [Google Scholar]
- 18. Gehring A. M., Bradley K. A., and Walsh C. T. (1997) Enterobactin biosynthesis in Escherichia coli: isochorismate lyase (EntB) is a bifunctional enzyme that is phosphopantetheinylated by EntD and then acylated by EntE using ATP and 2,3-dihydroxybenzoate. Biochemistry 36, 8495–8503 [DOI] [PubMed] [Google Scholar]
- 19. Sundlov J. A., and Gulick A. M. (2013) Structure determination of the functional domain interaction of a chimeric nonribosomal peptide synthetase from a challenging crystal with noncrystallographic translational symmetry. Acta Crystallogr. D Biol. Crystallogr. 69, 1482–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sundlov J. A., Shi C., Wilson D. J., Aldrich C. C., and Gulick A. M. (2012) Structural and functional investigation of the intermolecular interaction between NRPS adenylation and carrier protein domains. Chem. Biol. 19, 188–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Drake E. J., Miller B. R., Shi C., Tarrasch J. T., Sundlov J. A., Allen C. L., Skiniotis G., Aldrich C. C., and Gulick A. M. (2016) Structures of two distinct conformations of holo-non-ribosomal peptide synthetases. Nature 529, 235–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Frueh D. P., Arthanari H., Koglin A., Vosburg D. A., Bennett A. E., Walsh C. T., and Wagner G. (2008) Dynamic thiolation-thioesterase structure of a non-ribosomal peptide synthetase. Nature 454, 903–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu Y., Zheng T., and Bruner S. D. (2011) Structural basis for phosphopantetheinyl carrier domain interactions in the terminal module of nonribosomal peptide synthetases. Chem. Biol. 18, 1482–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tanovic A., Samel S. A., Essen L. O., and Marahiel M. A. (2008) Crystal structure of the termination module of a nonribosomal peptide synthetase. Science 321, 659–663 [DOI] [PubMed] [Google Scholar]
- 25. Reimer J. M., Aloise M. N., Harrison P. M., and Schmeing T. M. (2016) Synthetic cycle of the initiation module of a formylating nonribosomal peptide synthetase. Nature 529, 239–242 [DOI] [PubMed] [Google Scholar]
- 26. Miller B. R., Sundlov J. A., Drake E. J., Makin T. A., and Gulick A. M. (2014) Analysis of the linker region joining the adenylation and carrier protein domains of the modular nonribosomal peptide synthetases. Proteins 82, 2691–2702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zolova O. E., and Garneau-Tsodikova S. (2012) Importance of the MbtH-like protein TioT for production and activation of the thiocoraline adenylation domain of TioK. Med. Chem. Commun. 3, 950–955 [Google Scholar]
- 28. Krissinel E., and Henrick K. (2007) Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 29. Rusnak F., Faraci W. S., and Walsh C. T. (1989) Subcloning, expression, and purification of the enterobactin biosynthetic enzyme 2,3-dihydroxybenzoate-AMP ligase: demonstration of enzyme-bound (2,3-dihydroxybenzoyl)adenylate product. Biochemistry 28, 6827–6835 [DOI] [PubMed] [Google Scholar]
- 30. Mossialos D., Ochsner U., Baysse C., Chablain P., Pirnay J. P., Koedam N., Budzikiewicz H., Fernández D. U., Schäfer M., Ravel J., and Cornelis P. (2002) Identification of new, conserved, non-ribosomal peptide synthetases from fluorescent pseudomonads involved in the biosynthesis of the siderophore pyoverdine. Mol. Microbiol. 45, 1673–1685 [DOI] [PubMed] [Google Scholar]
- 31. Chen H., Hubbard B. K., O'Connor S. E., and Walsh C. T. (2002) Formation of β-hydroxy histidine in the biosynthesis of nikkomycin antibiotics. Chem. Biol. 9, 103–112 [DOI] [PubMed] [Google Scholar]
- 32. Al-Mestarihi A. H., Garzan A., Kim J. M., and Garneau-Tsodikova S. (2015) Enzymatic evidence for a revised congocidine biosynthetic pathway. Chembiochem 16, 1307–1313 [DOI] [PubMed] [Google Scholar]
- 33. Bruner S. D., Weber T., Kohli R. M., Schwarzer D., Marahiel M. A., Walsh C. T., and Stubbs M. T. (2002) Structural basis for the cyclization of the lipopeptide antibiotic surfactin by the thioesterase domain SrfTE. Structure 10, 301–310 [DOI] [PubMed] [Google Scholar]
- 34. Bloudoff K., Rodionov D., and Schmeing T. M. (2013) Crystal structures of the first condensation domain of CDA synthetase suggest conformational changes during the synthetic cycle of nonribosomal peptide synthetases. J. Mol. Biol. 425, 3137–3150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Roche E. D., and Walsh C. T. (2003) Dissection of the EntF condensation domain boundary and active site residues in nonribosomal peptide synthesis. Biochemistry 42, 1334–1344 [DOI] [PubMed] [Google Scholar]
- 36. Mitchell C. A., Shi C., Aldrich C. C., and Gulick A. M. (2012) Structure of PA1221, a nonribosomal peptide synthetase containing adenylation and peptidyl carrier protein domains. Biochemistry 51, 3252–3263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Marahiel M. A., Stachelhaus T., and Mootz H. D. (1997) Modular peptide synthetases involved in nonribosomal peptide synthesis. Chem. Rev. 97, 2651–2674 [DOI] [PubMed] [Google Scholar]
- 38. Chen W. H., Li K., Guntaka N. S., and Bruner S. D. (2016) Interdomain and intermodule organization in epimerization domain containing nonribosomal peptide synthetases. ACS Chem. Biol. 11, 2293–2303 [DOI] [PubMed] [Google Scholar]
- 39. Drake E. J., Nicolai D. A., and Gulick A. M. (2006) Structure of the EntB multidomain nonribosomal peptide synthetase and functional analysis of its interaction with the EntE adenylation domain. Chem. Biol. 13, 409–419 [DOI] [PubMed] [Google Scholar]
- 40. Kapust R. B., Tözsér J., Fox J. D., Anderson D. E., Cherry S., Copeland T. D., and Waugh D. S. (2001) Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Protein Eng. 14, 993–1000 [DOI] [PubMed] [Google Scholar]
- 41. Luft J. R., Collins R. J., Fehrman N. A., Lauricella A. M., Veatch C. K., and DeTitta G. T. (2003) A deliberate approach to screening for initial crystallization conditions of biological macromolecules. J. Struct. Biol. 142, 170–179 [DOI] [PubMed] [Google Scholar]
- 42. Luft J. R., and DeTitta G. T. (1999) A method to produce microseed stock for use in the crystallization of biological macromolecules. Acta Crystallogr. D Biol. Crystallogr. 55, 988–993 [DOI] [PubMed] [Google Scholar]
- 43. Battye T. G., Kontogiannis L., Johnson O., Powell H. R., and Leslie A. G. (2011) iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D Biol. Crystallogr 67, 271–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.