Abstract
Ribose can be used for energy or as a component of several important biomolecules, but for it to be used in either capacity it must first be phosphorylated by ribokinase (RBSK). RBSK proteins are part of the phosphofructokinase-B (pfkB) family of carbohydrate kinases. Sequence comparisons of pfkB proteins from the model plant Arabidopsis thaliana with the human and Escherichia coli RBSK identified a single candidate RBSK, At1g17160 (AtRBSK). AtRBSK is more similar to predicted RBSKs from other plant species and known mammalian and prokaryotic RBSK than to all other PfkB proteins in Arabidopsis. AtRBSK contains a predicted chloroplast transit peptide, and we confirmed plastid localization using AtRBSK fused to YFP. Structure prediction software verified that the AtRBSK sequence mapped onto a known RBSK structure. Kinetic parameters of purified recombinant AtRBSK were determined to be Kmribose = 150 μm ± 17 μm, KmATP = 45 μm ± 5.6 μm, and kcat = 2.0 s−1. Substrate inhibition was observed for AtRBSK (KiATP = 2.44 mm ± 0.36 mm), as has been demonstrated for other RBSK proteins. Ribose accumulated in Arabidopsis plants lacking AtRBSK. Such plants grew normally unless media was supplemented with ribose, which led to chlorosis and growth inhibition. Both chlorosis and ribose accumulation were abolished upon the introduction of a transgene expressing AtRBSK-MYC, demonstrating that the loss of protein is responsible for ribose hypersensitivity. Ribose accumulation in plants lacking AtRBSK was reduced in plants also deficient in the nucleoside ribohydrolase NSH1, linking AtRBSK activity to nucleoside metabolism.
Keywords: carbohydrate metabolism, carbohydrate processing, enzyme, nucleoside/nucleotide metabolism, phosphorylation, plant biochemistry, carbohydrate kinase
Introduction
Ribose is a constituent of many critical biologically active molecules, including ATP and other energy information molecules, signaling molecules such as plant hormone cytokinins, metabolites such as NAD and S-AdoMet, and most notably nucleotides and their polymers RNA and DNA. Ribose can also be metabolized via the non-oxidative phase of the pentose phosphate pathway and glycolysis/TCA cycle to produce energy. In both prokaryotes and eukaryotes ribose is incorporated into these larger molecules or catabolized by initial conversion into ribose 5-phosphate via a phosphoryl transfer from ATP to the O5′ position on ribose by ribokinase (RBSK)2 (EC 2.7.1.15). No known major pathway utilizes unphosphorylated ribose. Indeed, the main source of ribose 5-phosphate is the isomerization of glucose 6-phosphate in the pentose phosphate pathway. As such, little is known about the significance of unphosphorylated ribose in cells or about the function of RBSK in plants.
RBSK is a member of the ribokinase family of proteins, also known as the pfkB family (reviewed in Ref. 1), which includes several carbohydrate and nucleoside kinases. Although RBSK is likely conserved throughout evolution, as there are similar predicted proteins in genomes ranging from prokaryotes, to fungi, plants, and mammals, only bacterial and human RBSK proteins have been characterized biochemically (2–4). The structures of a handful of bacterial and human RBSKs have been solved, and the RBSK reaction mechanism is well understood (5, 6).
The phosphorylation of ribose by RBSK requires the presence of both divalent and monovalent cations for catalysis and enzyme activation, respectively. Divalent cations, thought to be magnesium physiologically (7), aid in catalysis and form the metal-ATP chelate, which is the form bound by the enzyme. Monovalent cations, potassium physiologically, instead activate the Escherichia coli and human enzyme via an induced conformational change that forms an anion hole (4, 7, 8). Inorganic phosphate has been shown to activate both E. coli and human RBSK and other pfkB family proteins (4, 9, 10).
The in vivo roles for RBSK activity vary dramatically by organism. Prokaryotes contain a ribose operon, encoding ribose-binding protein, PM-localized transporter, and RBSK (11), indicating a major function of this operon in utilization of extracellular ribose. Similarly for heterotrophic organisms, RSBK produces phosphorylated ribose for anabolic and catabolic reactions. In photosynthetic organisms, extracellular ribose might be a rare occurrence; hence the role of RSBK activity in these organisms is more obscure. In contrast to animals that primarily degrade nucleosides by phosphorolysis (12), plants begin nucleotide salvage and degradation by hydrolysis, releasing the nucleobase and free ribose (13, 14). Although the metabolic fates of the freed bases have been elucidated (13), the fate of this ribose is unknown.
Here we report characterization of the protein encoded by the Arabidopsis gene At1g17160 as a ribokinase (AtRBSK) and demonstrate its in vivo role in nucleotide metabolism. The protein encoded by At1g17160 is an active RBSK with high specificity for ribose. Its activity is dependent on both mono- and divalent cations. Surprisingly, in contrast to other RBSKs, addition of inorganic phosphate was not required and did not affect its kinase activity. Sequence analysis of AtRBSK indicates that it is a member of the pfkB family, which includes RBSK proteins from other species. AtRBSK is predicted to have a chloroplast transit peptide (cTP) and has been identified as plastid localized in large scale plastid proteomic studies (15). We confirm this prediction and untargeted proteomic studies via microscopy showing that YFP fusions are plastid-localized in puncta, similar to the nucleoid protein marker PLASTID ENVELOPE DNA BINDING PROTEIN (PEND) (16). Arabidopsis plants lacking AtRBSK grow much more poorly on growth media supplemented with ribose compared with growth on sucrose or xylose. Ribose accumulates in plants lacking AtRBSK when grown on media not supplemented with ribose. Both the inhibition of growth and the accumulation of ribose in mutant plants are lost upon the introduction of a transgene encoding AtRBSK. We also provide evidence that the breakdown of nucleosides by the nucleoside ribohydrolase, NSH1 (13, 14, 17), contributes to the accumulation of ribose in plants lacking functional AtRBSK. This finding provides one example of a role for AtRBSK, in plant nucleoside metabolism.
Results
At1g17160.1 Is Member of the RBSK Clade
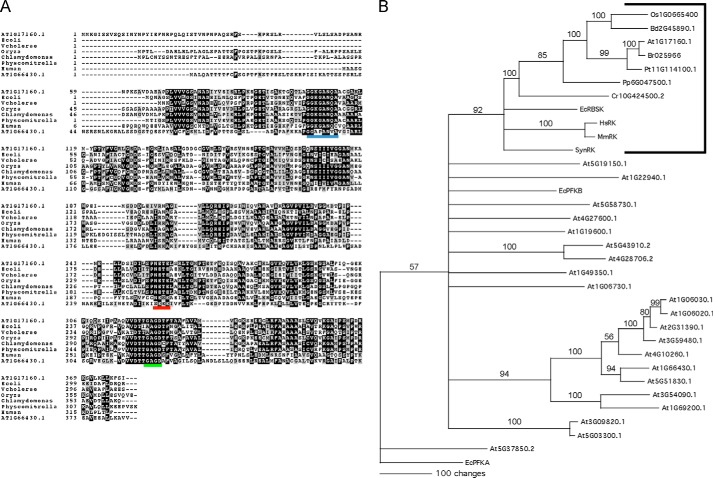
To identify the putative Arabidopsis RBSK we used the NCBI BLAST search tool (18) with the E. coli RBSK sequence as a query against predicted Arabidopsis protein sequences. The most significant hit was At1g17160.1 (35% identity, 53% similarity, E = 3 × 10−47), hereafter referred to as At1g17160. An alignment of the protein sequence of At1g17160 with known and predicted RBSK sequences from various other species showed a high degree of identity, especially in the C-terminal region (Fig. 1A). Interestingly, At1g66430, At1g17160, and proteins with highest sequence similarity to At1g17160 from Chlamydomonas reinhartii and Oryza sativa contain N-terminal extensions with predicted cTPs (19), the sequences of which are known to be highly divergent and account for the diverged N-terminal regions in those sequences. The At1g17160 peptide sequence contains the NXXE motif (residues 255–258, Fig. 1A underlined in red) and the (G/A)XGD motif (residues 322–325, Fig. 1A, underlined in green), which are both signature components of proteins in the pfkB family (6, 10, 20). We also observed a region (residues 105–111, Fig. 1A, underlined in blue) that is conserved in all RBSK sequences we queried, including At1g17160, but not other pfkB proteins such as an Arabidopsis fructokinase, At1g66430, included in the alignment (Fig. 1A). A subset of those RBSK unique residues interact with ribose in the active site of the Vibrio cholerae RBSK (5).
FIGURE 1.
Analyses of At1g17160.1 predict that it is the A. thaliana RBSK. A, alignment of characterized RBSK protein sequences from human and E. coli with predicted RBSKs from the plant lineage indicate that RBSK-like sequences are conserved and share a substrate-specific motif; from rice O. sativa, the moss P. patens, green algal C. reinhardtii, along with a non-RBSK Arabidopsis pfkB protein, At1g66430.1, for comparison. Black and gray boxes indicate amino acids identical or with conservative substitutions in the majority of proteins, respectively. The conserved Gly-Gly (Di-gly) ribose binding motif, the NXXE motif, and the (G/A)XGD motifs are underlined in blue, red, and green, respectively. B, consensus bootstrap tree of 22 Arabidopsis pfkB-type proteins with known RBSKs from A and predicted RBSKs from A with additional predicted RBSKs from B. distachyon, B. rapa, P. trichocarpa, Synechosistis sp. PCC 7509, and mouse (M. musculus). E. coli PFKA was included as an outgroup as it is not related to E. coli PFKB (36). The tree shows the results of 1000 bootstrap replicates of heuristic searches using maximum parsimony using PAUP. Bootstrap values are shown for clades with >50% support and clades with less than 50% support were collapsed.
To ascertain whether At1g17160 was the only predicted Arabidopsis RBSK, a phylogenetic tree was made that included all predicted Arabidopsis pfkB family protein sequences, several characterized bacterial and mammalian RBSKs, and plant proteins from other species with signature RBSK sequences (Fig. 1B, bracketed). All RBSK protein sequences formed a clade that included At1g17160, which was the only Arabidopsis pfkB sequence included in this clade. Within the RBSK clade, the predicted RBSK sequences from plants formed their own subbranch. Within the plant subbranch, the eudicots grouped together. At1g17160 was most similar to the sequence from Brassica rapa, and the Populus trichocarpa sequence was also included in that branch. The monocots O. sativa and Brachypodium distachyon formed their own subbranch. The sequence from Physcomitrella patens was further removed and the sequence from C. reinhartii was sister to the plant RBSK subbranch. The human and mouse RBSK proteins were very similar to each other and were also more similar to bacterial RBSK sequences than to the predicted plant RBSK sequences. There was one other large clade that was apparent in our phylogram that included predicted and demonstrated Arabidopsis fructokinases, two fructokinase-like proteins, and all other pfkB proteins.
Homology Modeling of AtRBSK
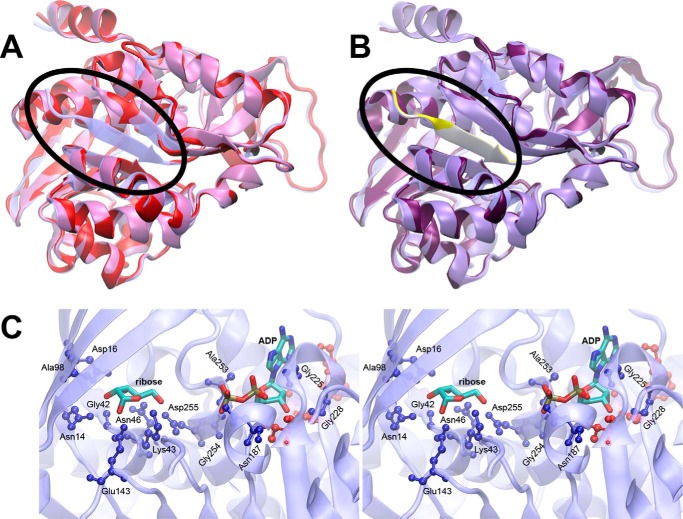
The genomic sequence of open reading frame At1g17160 encodes a 379-amino acid protein with a 74-amino acid chloroplast transit peptide predicted by the chloroP software (19). To assess possible ribokinase activity of this protein, we first aligned the predicted mature sequence, lacking the 74-amino acid cTP, with E. coli ribokinase and used the crystal structure of the E. coli protein with bound ribose and ADP (PDB accession 1RKD (6)) to prepare a homology model using MODELLER (21). The resulting model lacked a central strand in one of two β sheets in the ribokinase-fold (Fig. 2A). Inspection of the full sequence revealed that additional amino acids at the N terminus of At1g17160 were conserved relative to the E. coli ribokinase sequence resolved in the crystal structure (Fig. 1A). An equivalent homology model confirmed that the re-addition of seven residues upstream of the predicted cTP cleavage site would be sufficient to provide the missing structural element (Fig. 2B). We also projected sequence identity between At1g17160 and 1RKD onto 1RKD using homolmapper (22), confirming that interactions between 1RKD and bound ribose would be conserved in At1g17160 (Fig. 2C). This analysis thus implicated At1g17160 as a functional ribokinase enzyme but indicated that the predicted transit-peptide cleavage site was unlikely to be correct.
FIGURE 2.
Structure modeling of At1g17160 using E. coli RBSK structure. A, a homology model of At1g17160.1 (red) based on the predicted transit peptide cleavage site is shown superimposed on the template structure of E. coli ribokinase (transparent blue). The N terminus of the E. coli RBSK containing the β-sheet (blue) missing from the At1g17160 model (red) is circled. B, a similar superposition is shown for the homology model with a longer N-terminal sequence (purple), with the added N-terminal sequence highlighted (yellow). C, a stereo view of the E. coli ribokinase active site is shown. Bound ADP and ribose are shown as sticks, colored by atom type. Protein residues within 3.5 Å of ADP or ribose (ball-and-stick) are color-coded by sequence identity to At1g17160 (blue, conserved; red, not conserved).
At1g17160 Is an Active Ribokinase
To assess its biochemical properties we first cloned the At1g17160 cDNA without the predicted transit peptide and purified it after recombinant expression in bacteria. Consistent with the prediction made by homology modeling, the protein lacking the 74-amino acid predicted cTP was inactive. We then expressed At1g17160 without most of its predicted cTP but with seven additional residues upstream of the predicted cTP cleavage site restored to reconstruct the N-terminal β sheet discussed above (Fig. 2B). We then tested the activity of At1g17160 in concentrations of ribose and ATP similar to what has been published for other RBSK enzymes (4). The At1g17160 protein was active, phosphorylating ribose with a Km of 150 μm ± 17 μm and kcat of 2 s−1 (Fig. 3A), and hence will be referred to as AtRBSK. Activity was 5% or less of the ribose stimulated activity when xylose, another aldopentose, or fructose, a hexose used as a substrate by other family members, was used as the carbohydrate substrate. Substrate inhibition was observed at higher concentrations of ATP (Km 45 μm ± 5.6 μm, Ki 2.4 mm ± 0.36 mm, Fig. 3B).
FIGURE 3.
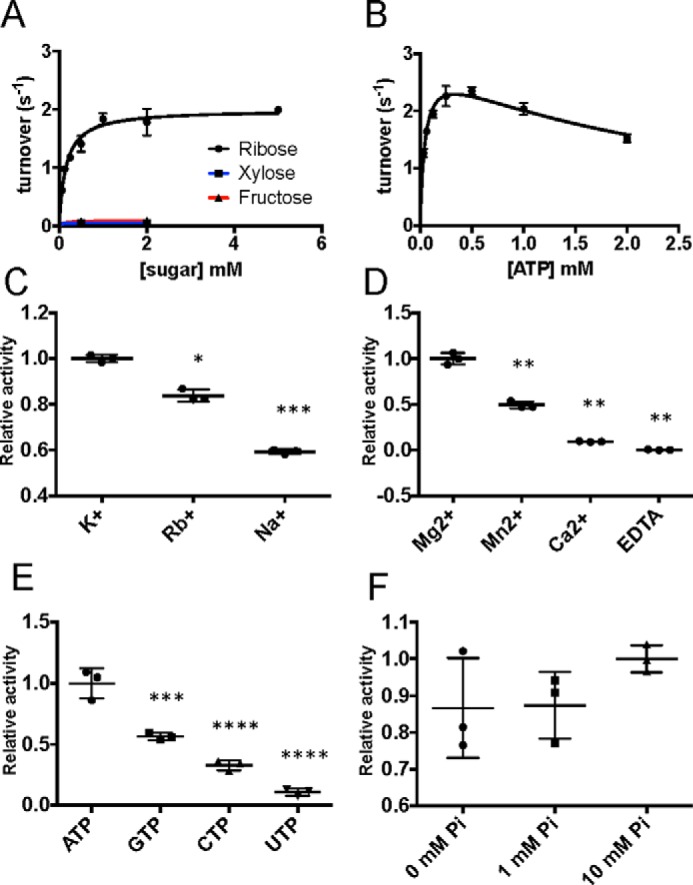
At1g17160 is an active and specific RBSK. A, plot of initial reaction rates at increasing concentrations of ribose (black line), xylose (blue line), and fructose (red line). B, plot of initial reaction rates at increasing concentrations of ATP. C, scatter plot comparison of enzyme activity in the presence of 100 mm monovalent K+, Rb+, and Na+, normalized to K+. Asterisks indicate statistical difference from K+. D, comparison of enzyme activity in the presence of 10 mm Mg2+, Mn2+, Ca2+, or EDTA, normalized to MgCl2. Asterisks indicate statistical difference from Mg2+. E, comparison of enzyme activity in the presence of 0.5 mm nucleotide phosphate donors, normalized to ATP. Asterisks indicate statistical difference from ATP. F, comparison of enzyme activity at varying concentrations of inorganic phosphate. All experiments were performed in triplicate. Scatter plots show all data points with mean ± S.D. indicated. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001.
RBSK proteins are known to require both mono- and divalent cations as cofactors (7, 8), so we tested the effect of various ions on the activity of AtRBSK. Potassium ions most strongly activate other known RBSK proteins and AtRBSK followed that pattern. AtRBSK activity dropped to 83 and 59% of full stimulation when rubidium or sodium, respectively, were substituted (Fig. 3C). Many ATP hydrolyzing enzymes use magnesium as a cofactor, including those of the pfkB family. The substitution of manganese for magnesium in the reaction mixture reduced AtRBSK activity to 49% of that observed in the presence of magnesium, and substitution of calcium in the reaction further reduced activity to 9% (Fig. 3D). No activity was observed upon the addition of EDTA to the reaction (Fig. 3D).
We then tested the preference of AtRBSK for different phosphoryl donors. As expected, activity was highest in the presence of ATP. GTP resulted in the next highest activity, which was 56% of the ATP value (Fig. 3E). CTP and UTP could be used as phosphate donors but resulted in only 32 and 10%, respectively, of activity stimulated by ATP (Fig. 3E). Another hallmark of RBSK proteins that have been studied to date is their requirement for inorganic phosphate. Surprisingly, we did not observe significant stimulation of RBSK activity in the presence of 1 or 10 mm inorganic phosphate (Fig. 3F).
AtRbsK Localizes to Plastids
AtRBSK has been identified as plastid localized in mass spectrometry analysis of chloroplast fractions (15). To confirm whether the prediction of an N-terminal cTP was correct, albeit with a probably incorrect cleavage site, and to obtain independent verification of published proteomics studies, we examined the intracellular localization of C terminally YFP-tagged AtRBSK. We cloned the full-length AtRbsK cDNA into a binary vector carrying an in-frame C-terminal YFP tag under the constitutive CaMV35S promoter. We transiently expressed AtRBSK-YFP along with a CFP-tagged plastid nucleoid marker PEND-CFP as a positive control for plastids via Agrobacterium infiltration of tobacco leaves. We then isolated chloroplasts, treated them with Hoescht dye to label plastid DNA, and imaged the isolated chloroplasts. AtRBSK-YFP formed puncta in the isolated chloroplasts that overlapped with both PEND-CFP signal and DNA staining, suggesting a possible association of AtRBSK with plastid nucleoids (Fig. 4, A–E). There was also a faint YFP signal around the outer membrane of the chloroplasts that was somewhat irregular.
FIGURE 4.
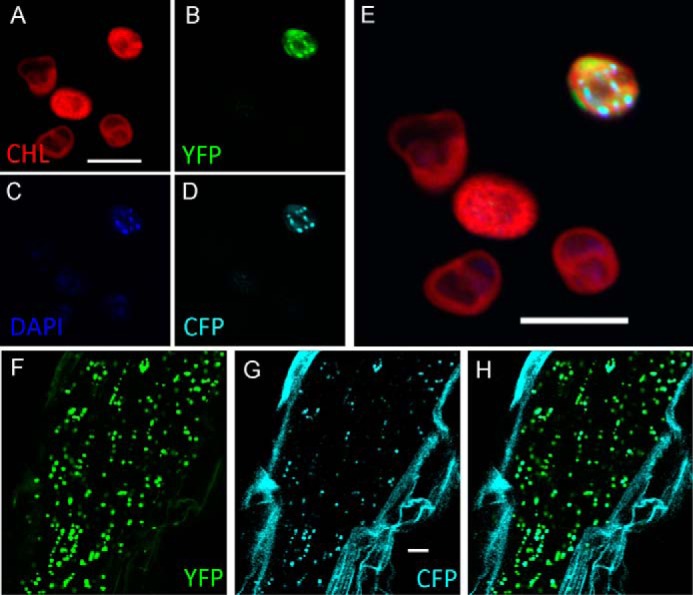
AtRBSK localizes to plastids. Confocal microscopy of isolated chloroplasts shows localization of A, chlorophyll; B, AtRBSK-YFP; C, DAPI; D, PEND-CFP; E, overlay of all channels. Not all leaf cells are transformed in transient assays and hence not chloroplasts fluoresce for CFP and YFP. F-H, expression of AtRBSK-YFP and PEND-CFP in roots of stably transformed Arabidopsis thaliana ecotype Col roots. F, AtRBSK-YFP; G, PEND-CFP; H, overlay. Scale bars = 10 μm.
A publicly available microarray data set (genevestivator.com) suggests that expression of AtRBSK is highest in roots. Because root plastids are developmentally different from chloroplasts in leaves we examined localization of AtRBSK in roots. We generated Arabidopsis that stably expressed both previously mentioned proteins and imaged the roots of 7-day-old seedlings. In contrast to leaf chloroplasts, root plastids contain a single large nucleoid that contains PEND (16). AtRBSK-YFP localized to large puncta in Arabidopsis roots (Fig. 4F) that coincided with the large puncta formed by PEND-CFP in the same plants (Fig. 4G).
Ribose Inhibits Growth of Arabidopsis Lacking Functional RBSK
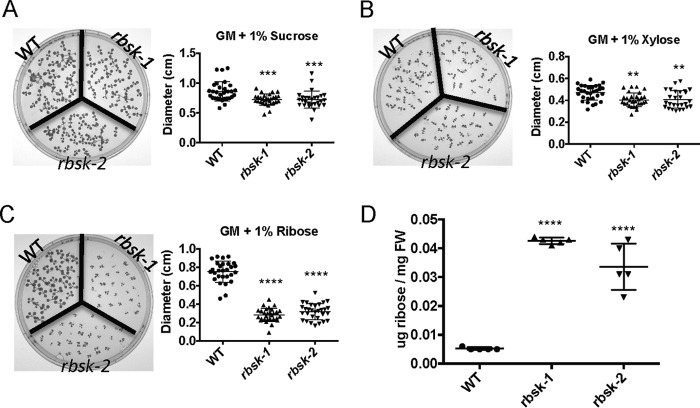
To study the in vivo function of AtRBSK, we obtained and characterized two independent plant lines harboring T-DNA insertions in AtRBSK (Fig. 5A): SALK_005371 (rbsk-1) and SAIL_815_E08 (rbsk-2), both in the Columbia (Col) ecotype background that was used as WT in comparison studies. We confirmed that individual plants were homozygous for the T-DNA insertion in their genomic DNA (Fig. 5B) and that no functional mRNA was present (Fig. 5C). We observed no overt phenotype in plants grown on either germination media (GM) or on soil. Because sucrose is commonly added to GM as a carbon source for seedlings before they become autotrophic, we tested the ability of rbsk-1 and rbsk-2 seedlings to use carbohydrates other than sucrose as a carbon source. We replaced the sucrose in GM with either ribose or xylose and compared the growth of rbsk-1 and rbsk-2 with WT by measuring leaf diameter. After 10 days of growth on GM with sucrose, both rbsk-1 and rbsk-2 were slightly smaller than WT (Fig. 6A), although the difference was statistically significant. All seedlings grown on GM with xylose were smaller than those grown on GM with sucrose but again, rbsk-1 and rbsk-2 seedlings were only slightly smaller than WT (Fig. 6B). WT plants grown on GM supplemented with ribose were roughly 90% of the size of WT seedlings grown on GM with sucrose, but rbsk-1 and rbsk-2 seedlings were reduced to 37 and 42%, respectively, of the size of the WT line on ribose (Fig. 6C), indicating a stronger growth repression on ribose as a carbon source.
FIGURE 5.
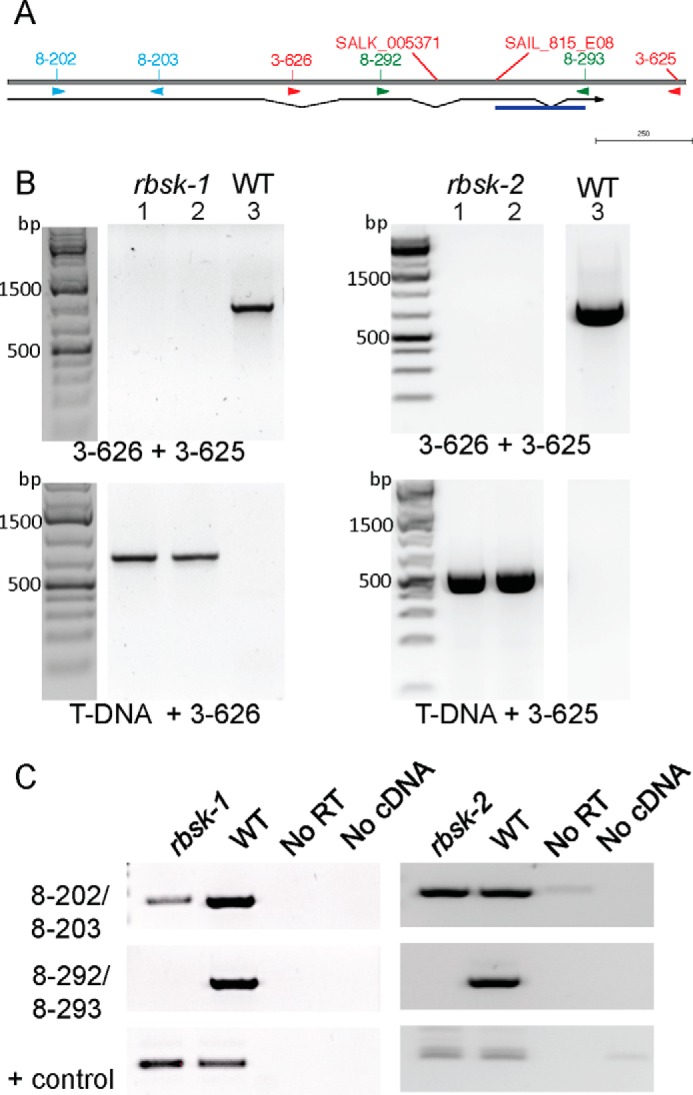
rbsk-1 and rbsk-2 are T-DNA insertional loss of function alleles of AtRBSK. A, schematic of AtRBSK gene architecture with T-DNA insertion positions and primer binding sites shown. Thin horizontal line below indicates position of coding region with introns connecting exons, and arrow indicates direction of open reading frame. DNA sequencing of the T-DNA PCR product revealed a sequence inversion in rbsk-2 (underlined in blue). B, lanes 1 and 2 are PCR products from 2 independent pools of genomic DNA from 4–6 seedlings each, lane 3 is a PCR product from a WT control, using primers flanking the T-DNA insertion sites (top panels) or primers to produce a DNA product spanning the T-DNA-genomic DNA junctions (lower panels) to verify the genotypes of progeny from a homozygous individual for each insertion. Primers used are indicated below their respective ethidium bromide-stained gel image. bp denotes base pairs in molecular weight markers. C, RT-PCR on cDNA from homozygous seedlings or WT (Col ecotype) using primers upstream (top panel) or flanking (middle panel) the T-DNA insertion. Primers specific for an unrelated pfkB family gene, At1g49350, were used as a positive control for the cDNA synthesis and RT-PCR (lower panels). Primer pairs used are indicated. RT, reverse transcriptase; No RT and No cDNA are negative control samples to indicate products from rbsk and WT cDNA are derived from cDNA added, that there is no or low levels of contaminating genomic DNA.
FIGURE 6.
Ribose inhibits growth of rbsk-1 and rbsk-2. 10-Day-old Arabidopsis seedlings grown in constant light on solid GM supplemented with A, 1% sucrose; B, 1% xylose; C, 1% ribose and quantification of seedling diameter. Experiments were performed at least three times each. Two rbsk mutant biological replicates were compared with one biological replicate of WT as a control group as shown. Scatter plots show diameter measurements of >25 seedlings from one representative experiment. D, graphical representation of μg of ribose/mg of fresh weight of 10-day-old Arabidopsis seedlings grown in liquid GM supplemented with 1% sucrose determined by mass spectrometry as compared with ribose standards run at the same time and normalized to fresh weight of the seedlings. 5 technical replicates were analyzed for each of two rbsk mutant biological replicates and one biological replicate of WT as a control group as shown. Scatter plots show all data points with mean ± S.D. indicated. All asterisks indicate statistical difference from WT. **, p < 0.01; ***, p < 0.001; ****, p < 0.0001.
We analyzed the metabolome of WT, rbsk-1, and rbsk-2 lines grown for 10 days in liquid GM containing sucrose and found that, although it had not been added to the growth media, ribose had accumulated in rbsk-1 and rbsk-2 seedlings to roughly 10-fold higher than what was present in the WT seedlings (Fig. 6D).
To verify whether the phenotypes observed were caused by the loss of function of AtRBSK and not by off-target effects of the T-DNA insertional mutagenesis processes, we generated vectors carrying the AtRBSK full-length cDNA under the control of the 35S-promoter and harboring a C-terminal 6×MYC tag (35S:RBSK-MYC) and introduced the gene into rbsk-2. Four independent transformants were selected and brought to homozygosity of the introduced transgene, although protein expression was detectable in only three of those lines. The three lines with detectable protein are referred to as RKOE-1, RKOE-2, and RKOE-3 (Fig. 7A). In addition to the growth inhibition on ribose (Fig. 6), rbsk-1 and rbsk-2 lines become chlorotic when grown on GM supplemented with ribose (Fig. 7B). To quantify complementation in RKOE-1–3 lines, we compared the percent of germinated seedlings that were green after 11 days of growth. All seedlings from all genotypes were green on GM supplemented with sucrose (Fig. 7C). WT seedlings were also all green when plated on GM supplemented with ribose as opposed to rbsk-2, of which roughly only 10% were green (Fig. 7D). RKOE-1–3 seedlings were all greater than 90% green at 11 days after germination (Fig. 7D) indicating successful complementation of the rbsk-2 ribose hypersensitive phenotype. Thus, the selective reduction of growth and ultimately lack of viability on ribose observed for rbsk-1 and rbsk-2 grown in light results from lack of RBSK.
FIGURE 7.
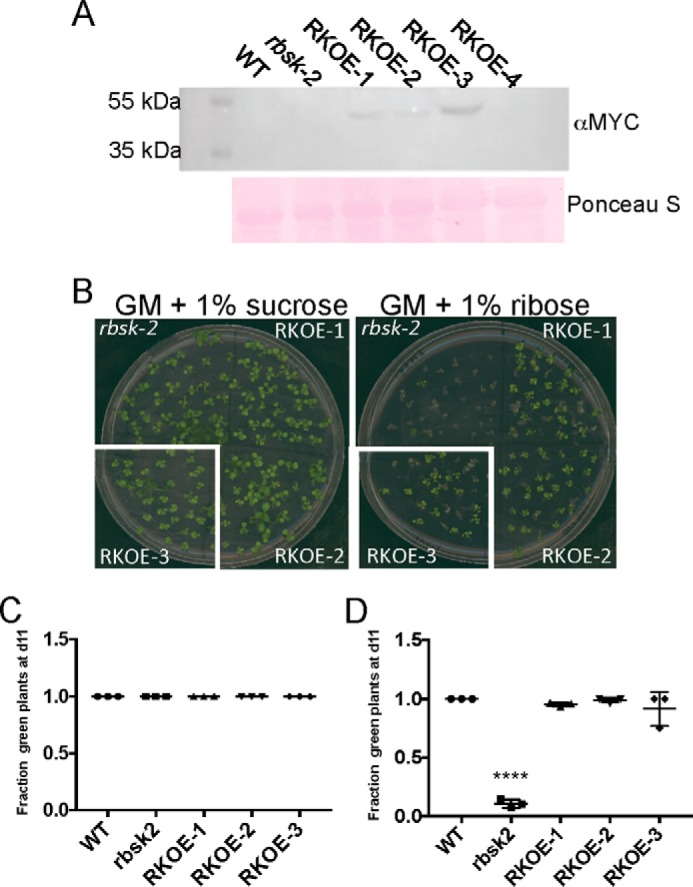
Complementation of rbsk-2 with a transgene expressing epitope-tagged RBSK. A, immunoblot analysis of 35S:RBSK-MYC expression in seedlings. 50 μg of total protein from each indicated plant line was loaded per lane. Top panel is anti-MYC immunoblot. Bottom panel is Ponceau S-stained membrane as a loading control. B, representative images of 11-day-old seedlings of the indicated genotypes on either GM containing either 1% sucrose (left) or 1% ribose (right). C and D, quantification of the fraction of green seedlings out of germinated seedlings from B, grown on GM containing either C, 1% sucrose, or D, 1% ribose. Three technical replicates were analyzed for each condition. Analysis included three independent biological replicates of seedlings expressing 35S:RBSK-MYC in the rbsk-2 background and one biological replicate of the parental rbsk-2 for comparison. Counts included >20 seedlings per replica. Scatter plots show the fraction of green seedlings on each of the three technical replicates with mean ± S.D. indicated. The fraction of green seedlings was not different from Col for the 3 35S:RBSK-MYC expressing lines and much greater than observed for the rbsk-2 progenitor line. All asterisks indicate statistical difference from WT. ****, p < 0.0001.
Because ribose hyper-accumulated in rbsk-1 and rbsk-2, we sought to determine whether this hyper-accumulation was reduced in the AtRBSK complementation lines. To this end we obtained a set of previously characterized protein-based ribose sensors for rapid detection of ribose in plant extracts (23). These sensors consist of cyan fluorescent protein (CFP) and yellow fluorescent protein (YFP) fused to the N and C terminus, respectively, of a ribose-binding protein. CFP and YFP constitute a FRET pair and, when fused to a single protein, can detect ligand-induced changes in protein conformation. In the case of the particular sensor used in our experiments, FRET signal, as measured by the ratio of YFP fluorescence emission to CFP fluorescence emission when excited by wavelengths of light in the CFP range, is strong in the unbound form and is weaker upon binding of ribose to the protein (Fig. 8A). FRET signal was high in extracts from WT plants, indicating a level of ribose at or below the detection limit of the sensor. However, when exposed to extracts from rbsk-1 or rbsk-2 seedlings the FRET signal was weaker (Fig. 8B), suggesting an accumulation of ribose. We then tested whether or not ribose accumulated in our complementation plant lines. Extracts from RKOE-1 and RKOE-3 both resulted in high FRET signal that was comparable with WT (Fig. 8C), indicating a lack of ribose accumulation due to the overexpression of AtRBSK.
FIGURE 8.
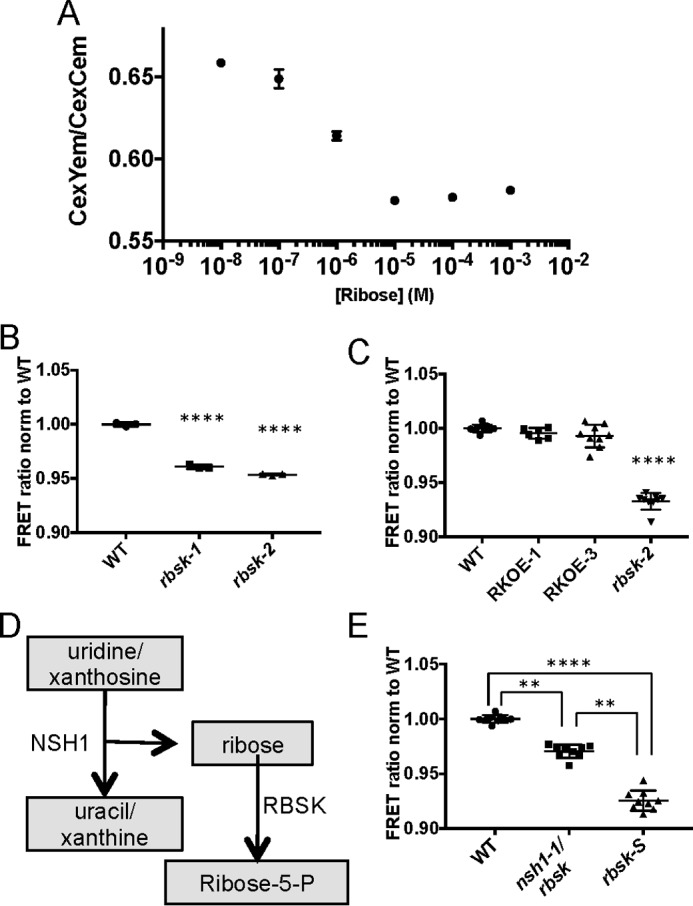
Protein-based ribose sensors confirm ribose accumulation in rbsk-2. A, standard curve of ratio of YFP emission to CFP emission when excited by CFP excitation in ribose FRET sensors at indicated by ribose concentrations. Mean ± S.D. are shown of three technical replicates from a representative experiment. The experiment was repeated three times. B, comparison of FRET ratios, normalized to WT, in extracts from WT, rbsk-1, and rbsk-2 seedlings diluted 100-fold in FRET lysis buffer and added to crude FRET sensor lysate. This experiment was performed once and included three technical replicates each from two biological replicates of rbsk mutant seedlings and one biological replicate of a WT control. The mean ± S.D. of three technical replicates are shown. C, comparison of FRET ratios, normalized to WT, in extracts from WT, RKOE-1, and RKOE-3 diluted 100-fold in FRET lysis buffer and added to crude FRET sensor lysate. Experiments were performed three times, or two times in the case of RKOE-1. Each experiment included three technical replicates from each of two biological replicates from seedlings expressing 35S:RBSK-MYC in the rbsk-2 background and one biological replicate each of the parental rbsk-2 and WT control. All data points from all experiments are shown with mean ± S.D. indicated. D, pathway diagram of generation of ribose from NSH1 activity. E, comparison of FRET ratios, normalized to WT, in extracts from WT, rbsk nsh-1, and rbsk-S, diluted 100-fold in FRET lysis buffer and added to crude FRET sensor lysate. Experiments were performed three times each. Each experiment included three technical replicates from one biological replicate each of rbsk nsh-1 double mutant, rbsk-s single mutant, and WT control seedlings. All data points from each of the three experiments are shown with mean ± S.D. indicated. All asterisks indicate statistical difference from WT unless otherwise noted. **, p < 0.01; ****, p < 0.0001.
Ribose Accumulation in rbsk Results in Part from Nucleotide Metabolism
Ribose is an intermediate in several biochemical pathways. However, in those pathways the phosphorylated form, ribose 5-phosphate, is synthesized via isomerization reactions and not through the direct phosphorylation of ribose. Because our data provide evidence that unphosphorylated ribose accumulates in plants lacking active AtRBSK, we wanted to determine which biochemical pathway generates unphosphorylated ribose to better understand the biological context of AtRBSK. One attractive pathway is nucleotide salvage. This pathway includes nucleoside hydrolase enzymes that catalyze hydrolysis of the nucleosides, releasing the base and an unphosphorylated ribose (Fig. 8D). The major nucleoside hydrolase in Arabidopsis, NSH1, has been identified and characterized (14, 17). We obtained a previously characterized NSH1 T-DNA insertion line, nsh1-1, and crossed it to rbsk-2 to generate double KO plants in the F2 generation. Seed from double KO (rbsk-2 nsh1) and an F2 sibling homozygous for only the rbsk T-DNA insertion (rbsk-S, for single) were germinated on GM, and extracts were analyzed for ribose content. We used the ribose-binding FRET sensor to determine whether the lack of NSH1 affected ribose content in rbsk-2 nsh1 plants. In extracts from rbsk-S and WT plants we again observed low and high FRET signal, respectively (Fig. 8E), indicating ribose hyperaccumulation in rbsk-S and below detection in WT plants. In extracts from rbsk-2 nsh1 plants, the FRET signal was higher than what was observed in extracts from rbsk-S but lower than that from WT plants (Fig. 8E), indicating intermediate ribose levels. These data support our hypothesis that nucleoside hydrolysis generates unphosphorylated ribose in vivo and that AtRBSK acts to phosphorylate that ribose and return it to a metabolically available form.
Discussion
Here we provide evidence that At1g17160.1 encodes the major protein with RBSK activity in Arabidopsis. Sequence analysis suggests that AtRBSK is a member of the pfkB family of carbohydrate kinases, which includes all previously described RBSK proteins. AtRBSK contains both the NXXE and (G/A)XGD motifs, both of which are conserved among pfkB family proteins. It also has strong sequence identity to other RBSK proteins, but not to other pfkB proteins at a region known to contact the sugar itself. These residues likely confer specificity of these proteins for ribose. Finally, AtRBSK was the only member of the 22 predicted pfkB family proteins in Arabidopsis to clade with known RBSKs from other species. The hyperaccumulation of ribose in plants lacking AtRBSK also suggests that AtRBSK is the major protein responsible for phosphorylating the pool of ribose and thus for maintaining low free ribose levels. Although other related proteins that are either temporally regulated or expressed only in a tissue-specific manner may also possess RBSK activity, our data supports the hypothesis that AtRBSK is the sole RBSK protein in Arabidopsis.
Most residues associated with ATP binding were conserved between RBSK proteins and the non-RBSK Arabidopsis protein At1g66430 (Figs. 1−2), specifically Gly-225, Gly-228, Ala-253, Gly-254, and Asp-255, with the latter three residues being part of the signature (G/A)XGD motif. Asn-187 is part of the NXXE motif and also contacts ATP. Asn-187 was conserved within RBSK proteins but not conserved in non-RBSK proteins. Residues contacting ribose were not as well conserved with the non-RBSK protein, although many were shared. Asn-14 and Lys-43 were conserved between all RBSK proteins but not by a non-RBSK protein. Asp-16, Gly-42, and Asn-46 were all conserved between both RBSK and non-RBSK proteins. One of the residues unique to RBSK proteins, Lys-43, is in the region previously hypothesized to establish specificity of these enzymes for their carbohydrate substrate.
Biochemically, AtRBSK behaves similarly to other RBSKs that have been studied. Human and E. coli RBSK bind ribose with slightly different affinities with a Km of 279 μm and 2.17 mm, respectively (2, 4). We determined the Km for ribose of AtRBSK to be 150 μm, indicating that the affinity of AtRBSK for ribose is approximately within the same order of magnitude as the human RBSK. A similar pattern was seen for the affinity of the enzymes for ATP. Human and E. coli RBSKs Km for ATP was 70 and 213 μm, respectively (2, 4), as compared with the Km of 46 μm that we determined for AtRBSK. AtRBSK was less sensitive to substitutions in monovalent cations as its activity only dropped to 80 and 60% of maximal activity in the presence of rubidium and sodium, respectively. E. coli RBSK was not assayed in the presence of rubidium but dropped to less than 10% of maximal activity in the presence of sodium (2). E. coli RBSK has been shown to favor K+ ions (8) and the same appears to be true for AtRBSK. AtRBSK also behaves similarly to known RBSKs in the presence of divalent cations. Like the E. coli RBSK, AtRBSK has maximal activity in the presence of magnesium. Replacing magnesium with manganese resulted in reduced activity, 80% of maximum in EcRBSK and 50% of maximum in AtRBSK, and less than 10% of maximum in both EcRBSK and AtRBSK with either calcium or the absence of a divalent cation (2). Interestingly, AtRBSK was active in the absence of inorganic phosphate, which has been shown to strongly activate both human and E. coli RBSK (4). The assay developed by Andersson and colleagues (8) and used by us does not include inorganic phosphate so there is at least one other example in the literature of a RBSK enzyme active in the absence of inorganic phosphate in addition to ours.
The clade formed by RBSK protein sequences within the pfkB family suggests that specific RBSK activity has been maintained across time. One inference that can be drawn from this observation is that RBSK proteins play an important role in biology. RBSK activity in prokaryotes likely served to sequester ribose imported from the environment and activate it for use in metabolism or for synthesis of biomolecules. The presence of prokaryotic RBSK genes in operons with ribose import machinery is consistent with this hypothesis, and putative protein-protein interactions between bacterial RBSK proteins and such machinery could mitigate the intrinsically lower ribose affinity of the E. coli RBSK relative to eukaryotic RBSK proteins. Animals consume other organisms, which likely results in a significant amount of dietary ribose from the breakdown of biomolecules in food that must be phosphorylated for further use. The accumulation of ribose in plants grown on media not supplemented with ribose suggests that plants generate unphosphorylated ribose internally. Because the reduction of nucleoside hydrolysis lowers ribose levels in rbsk plants, these data support the model that unphosphorylated ribose normally arises in the NSH1-dependent branches of nucleotide salvage pathways and RBSK is responsible for phosphorylating ribose produced in these pathways for further utilization.
The ribose-induced growth inhibition and chlorosis of plants lacking AtRBSK suggests that plants are also able to take in ribose from their environment and that extracellular ribose is toxic for rbsk plants by unknown mechanisms. A natural source of extracellular ribose could be from NSH3 activity. NSH3, an extracellular protein related to NSH1, degrades extracellular purine nucleosides (17), which would generate extracellular ribose that could then be taken up by plants. Ribose would then require intracellular phosphorylation for use in intracellular metabolism as described above.
Our data suggest that NSH1, a cytosolic protein (13), generates some of the intercellular ribose that then requires phosphorylation by RBSK, making the plastidic localization of AtRBSK interesting. The import of ribose by chloroplasts has been demonstrated (24) suggesting that cytosolically produced ribose can enter plastids for phosphorylation. Several reactions in nucleotide metabolism, including de novo synthesis take place in plastids (25) and ribose 5-phosphate is a precursor for many of those reactions. The presence of AtRBSK in plastids may thus act to provide a precursor, ribose-5-P, for conversion to phosphoribosyl pyrophosphate for nucleotide synthesis.
Ribose levels are not reduced to WT levels in the rbsk-2 nsh1 double mutant, suggesting that other pathways/enzymes may make a contribution to production of unphosphorylated ribose. Ribose may also be generated via epimerase activities on pentoses such as arabinose or xylose from cell wall remodeling or breakdown, although there is currently no experimental evidence to support such a hypothesis. The breakdown of extracellular pathogens or detritus in the environment may also provide a source of epimerase substrates, nucleosides for NSH3 breakdown, or ribose itself. Although outside the scope of our experiments it would prove interesting to determine other sources of unphosphorylated ribose in plant cells. Knowledge of the effects caused by the hyperaccumulation of ribose, and of sources of unphosphorylated ribose would help us better understand the in vivo roles for AtRBSK in plant biology.
Materials and Methods
Plant Material and Growth Conditions
We obtained Arabidopsis seeds from the Arabidopsis Resource Center (ABRC) carrying T-DNA insertions in At1g17160: SALK_005371 (rbsk-1) and SAIL_815_E08 (rbsk-2); and in At2g36310 (NSH1, URH1): SALK_083120C, all in the Columbia (Col-0) ecotype. Col-0 plants (referred to as WT and serves as wild type control line for most experiments). Col-0 and homozygous rbsk-1 and rbsk-2 plants were grown at the same time, in the same trays, and seeds were harvested at the same time. Seeds from these “age and growth condition matched” plants were used for phenotypic analyses. For growth on various sugars, seeds from Col, rbsk-1, and rbsk-2 were surface sterilized in a solution of 30% bleach and 0.1% Triton X-100 (Sigma), stratified for 48 h at 4 °C and grown for 10 days on solid (0.8% bacto-agar) GM (4.3 g/liter of Murashige and Skoog (MS) basal salts (Sigma), 2.5 mm MES, 1× B vitamins (0.5 μg/ml of nicotinic acid, 1.0 μg/ml of thiamine·HCl, 0.5 μg/ml of pyroxidine·Cl, 0.1 μg/ml of myo-inositol), pH 5.7) supplemented with either 1% (w/v) sucrose, 1% xylose, or 1% ribose. All plate-grown seedlings were grown at 20 °C under constant white light at 40–50 μmol s−1 m−2.
For FRET sensor analysis, rbsk-2 was crossed with nsh1, a previously characterized T-DNA insertion line in NSH1 re-verified in the laboratory (13, 14) and brought to homozygosity for T-DNA insertions in both genes, creating a double KO plant line referred to as rbsk nsh1. Genotypes were verified by PCR (see below). A sibling from the cross that was homozygous for the T-DNA insertion in RBSK and wild type for NSH1 was also selected as a rbsk alone mutant control line from as similar a genetic background as possible for comparison and referred to as rbsk-S (for single). Both of these plant lines as well as others indicated in the text were surface sterilized, stratified for 48 h, and grown in liquid GM for 7 days prior to carbohydrate extraction and analysis.
Preparation of Arabidopsis Seedlings for Metabolomics Analysis
For preparation of seedlings for metabolomic analyses, Arabidopsis seedlings were grown in liquid GM with 1% sucrose for 10 days, washed three times in several milliliters of PBS (10 mm sodium phosphate, 137 mm NaCl, 2.7 mm KCl, pH 7.5), blotted dry, flash frozen, and submitted to the West Coast Metabolomics Center for processing and mass spectrometry analysis via their standard procedures (26, 27). Ribose content in seedlings was determined by comparing the mass spectrometry signal of ribose against a set of ribose standards of known concentrations and then normalized to fresh weight of the seedlings.
Protein Sequence Analysis
Full-length protein sequences of RBSK from human (GenBank ref seq NP_071411.1) and E. coli (GenBank ref seq NP_418208.1), and V. cholerae (GenBank ref seq number AKB06975.1), and predicted RBSK proteins from C. reinhartii (GenBank ref seq number XP_001702739.1), P. patens (GenBank ref seq number XP_001774463.1), and O. sativa (GenBank ref seq number NP_001043796.1) as well as At1g17160 and At1g66430 were aligned using CLUSTAL (28) and visualized using Boxshade. Phylogenetic relationships between the previously mentioned sequences and additionally, 22 Arabidopsis pfkB-type proteins with predicted RBSKs from B. distachyon (GenBank ref seq number XP_003569514.1), B. rapa (GenBank ref seq number XP_009149161.1), P. trichocarpa (GenBank ref seq number XP_002317476.1), Synechosistis sp. PCC 7509 (GenBank ref seq number WP_009633544.1), and mouse Mus musculus RBSK (GenBank ref seq number NP_071411.1). E. coli PFKA (GenBank ref seq number NP 418351.1) was used as an outgroup were evaluated by maximum parsimony in PAUP*4.0b10 (29) using an alignment of pfkB protein sequences constructed with Clustal Omega and manually adjusted to maximize alignment and to remove phylogenetically uninformative sequences. Statistical support for relationships was evaluated by 1000 bootstrap replicates, with 20 random addition heuristic searches using the branch and bound algorithm for each bootstrap replicate using PAUP*4.0b10 (29).
Cloning
RNA was extracted from 7-day-old Arabidopsis seedlings with the RNeasy Plant Mini Kit (Qiagen) and used to generate cDNA using the SuperScript III First-Strand Supermix (Invitrogen). Clones for transient expression in tobacco leaves and for Arabidopsis transformation had their stop codon mutated and gateway (Invitrogen) recombination sites added via PCR using the forward primer, 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTGATGATGAAAGGGATCTCTTC-3′ and reverse primer, 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCTGAGATACTAAACTTCAGGAGCTTCAAAAC-3′. For the generation of clones for bacterial expression, the codons for the N-terminal 67 amino acids were not included and gateway (Invitrogen) recombination sites were added using the forward primer, 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTGGCGCCGCCACTGGTGGTTGTGGG-3′ and the reverse primer, 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCTTAGATACTAAATTTAAGGAG-3′. All PCR for cloning was carried out using Phusion high fidelity DNA polymerase (Fisher) according to the manufacturer's instructions. PCR products for all clones were recombined into pDONR201 or pDONR207 (Invitrogen) via the BP clonase (Invitrogen) reaction. Sequences were verified at the University of California, Davis DNA sequencing facility. Clones were then recombined into pEAK2 (30) for bacterial expression; or pEARLEYGATE101 (31) or pGWB17 (32) via the LR clonase (Invitrogen) reaction for transient expression in tobacco leaves or floral dipping in Arabidopsis, respectively.
Chloroplast Isolation
Chloroplasts were isolated from tobacco leaves 48 h after infiltration of Agrobacterium carrying vectors to overexpress our genes of interest. Tobacco leaves were chopped using scissors in grinding buffer (0.05 m HEPES, 0.33 m sorbitol, 2 mm EDTA, 1% (w/v) BSA, pH 8.0) and then blended two times for 5 s each in a Waring blender with a 2-s pause. The buffer containing the plant material was then passed through two layers of Miracloth (Millipore) and centrifuged over a 50% Percoll (GE Healthcare) gradient. Isolated chloroplasts were re-suspended to 0.5 mg/ml of chlorophyll in buffer (0.05 m HEPES, 0.33 m sorbitol, pH 8.0) containing 1 μg/ml of Hoescht dye 33258 (Polysciences) and mounted on slides.
Homology Modeling
Protein sequence alignments were performed with MUSCLE (33), and sequence identity was projected onto protein structures using homolmapper (22). Homology modeling was performed using MODELLER (21). Structure figures were prepared using VMD and Tachyon (34, 35).
Protein Expression Analysis
14-day-old seedlings from Col-0 (WT), rbsk-2, and RKOE-1–4 were ground in RIPA buffer (10 mm Tris, 140 mm NaCl, 1 mm EDTA, 0.5 mm EGTA, 1% (v/v) Triton X-100, 0.1% (w/v) deoxycholate, 0.1% (w/v) SDS, pH 8.0). Cellular debris was cleared by centrifugation at maximum speed on a tabletop microcentrifuge for 10 min. Protein concentration in the supernatant was determined using the Protein Assay Dye Reagent (Bio-Rad). 50 μg of total protein from each plant line was loaded and run on a 10% SDS-PAGE gel and then transferred to a PVDF membrane (Millipore). The membrane was blocked with blotto (50 mm Tris, 200 mm NaCl, 5% (w/v) nonfat dry milk) and Western blotting analysis was done using an anti-MYC antibody conjugated to horseradish peroxidase (1:2000 dilution, Roche catalog number 11814-150001), visualized using Amersham Biosciences ECL Western blotting Detection Reagent (GE Healthcare). HRP chemiluminescence was imaged using the CCD on an ImageQuant LAS4000 (GE Healthcare).
Microscopy
Chloroplasts were prepared for microscopy as described above. Arabidopsis seeds were surface sterilized, stratified for 48 h at 4 °C, and grown for 7 days on solid GM and mounted in mounting medium (50 mm Tris, 20% glycerol, pH 8.0). All images were captured on an Olympus FV1000 microscope using the appropriate filter sets for YFP, CFP, DAPI, or chlorophyll autofluorescence.
Preparation of Recombinant Proteins
Proteins were expressed in E. coli strain BL21-pLys-S and purified via nickel-Sepharose (GE Healthcare) affinity chromatography. Bacterial cells were lysed by sonication in lysis buffer (50 mm Tris, 100 mm NaCl, 50 mm imidazole, pH 7.5, Complete mini protease inhibitor mixture (Roche)) washed with at least 300 column volumes of wash buffer (50 mm Tris, 1.5 m NaCl, 50 mm imidazole, pH 7.5) and eluted in elution buffer (50 mm Tris, 100 mm NaCl, 1.0 m imidazole, pH 7.5). Eluate was dialyzed exhaustively against storage buffer (50 mm Tris, 100 mm NaCl, 20% glycerol, pH 7.5), concentrated in an Amicon Ultra 15 concentrator (Millipore) with 10-kDa MWCO, flash frozen, and stored at −80 °C. Concentration of recombinant proteins was determined using Protein Assay Reagent (Bio-Rad). Purity was determined by Coomassie Blue staining of an SDS-PAGE gel.
Enzymatic Assays
Enzymatic assays were carried out using the indicator dye phenol red to visualize the generation of protons in the kinase reaction by measuring changes in absorbance at 430 nm (8). The reaction mixture consisted of 10 mm MgCl2, 140 mm KCl, 0.003% phenol red, 1 mm ribose, and 0.1 mm ATP, pH 8.0. For determination of Km values of ribose and ATP, initial reaction rates were measured in concentrations of ribose or ATP, respectively, that were varied between 2 and 0.0625 mm with all other parameters held constant. Data were plotted and biochemical parameters were determined via non-linear curve fitting to either the Michaelis-Menten equation or substrate inhibition equation as appropriate in Prism 7 (GraphPad). To determine the effect of monovalent cations, initial reaction rates were determined in the presence of 100 mm KCl, NaCl, or RbCl, and normalized to that of KCl. To determine the effect of divalent cations, initial reaction rates were determined in the presence of 10 mm MgCl2, MnCl2, CaCl2, or EDTA, and normalized to that of MgCl2. Inorganic phosphate dependence was determined using a NADH coupled assay in the presence of 50 mm Tris, pH 7.5, 100 mm KCl, 10 mm MgCl2, 0.3 mm ATP, 1 mm phosphoenolpyruvate, 0.2 mm NADH, 1 mm ribose, 2 units/ml of lactate dehydrogenase (Sigma), 2 units/ml of pyruvate kinase (Sigma), and the indicated concentrations of inorganic phosphate. All assays were performed in triplicate.
Genotyping and Expression Analysis
For genotyping rbsk-1 and rbsk-2 alleles, the gene-specific primers were used: 3-625, 5′-CTCGTTAGTTGCTTAGGCCAC-3′ and 3-626, 5′-GAATGGATGCAGCTGCTTTAG-3′. The T-DNA primers, 5′-TGGTTCACGTAGTGGGCCATCG-3′ and 5′-TAGCATCTGAATTTCATAACCAATCTC-3′, were used with 3-626 for rbsk1 and with 3-625 for rbsk-2. Previously published primers were used for nsh1 analysis (14). To analyze mRNA expression, RNA was extracted from 7-day-old Arabidopsis seedlings with the RNeasy Plant Mini Kit (Qiagen) and used to generate cDNA using the SuperScript III First-Strand Supermix (Invitrogen). The forward primer 8-202 5′-TCGATCCCTCCGTGTTCTCT-3′ and reverse primer 8-203 5′-CTCTCCCAAACGACCAACGA-3′ were used to produce an upstream product on the At1g17160 mRNA. The forward primer 8-292 5′-TGAGCTCAGTCGCTTGACAG-3′ and reverse primer 8-293 5′-TGCTCGGTATTGCACCCTTT-3′ were chosen to flank the T-DNA insertion site. The control primers forward 5′-GCGTGGATGCCGTTGAAAAT-3′ and reverse 5′-TCGTCTTGGTTTGGGGACAC-3′ produce a product from another Arabidopsis pfkB family member, At1g49350, and were used to confirm the presence of cDNA in the PCR. GoTaq (Promega) was used in all genotyping and reverse transcriptase PCR according to the manufacturer's instructions. PCR products were visualized by ethidium bromide staining after agarose gel electrophoresis. O'GeneRuler 1 kb Plus (Thermo) was used as molecular weight markers.
Ribose FRET Sensors
7-Day-old liquid grown seedlings were rinsed with dH2O three times, then blotted dry and weighed to determine fresh weight. They were then flash frozen in liquid nitrogen and ground to a powder with a mortar and pestle. Extraction buffer (10 mm Tris, 100 mm NaCl, 1 mm EDTA, 0.05% Nonidet P-40, 50% ethanol, pH 8.0) was added to achieve a concentration of 100 mg of fresh weight/ml and grinding continued. The extracts were mixed with chloroform to remove chlorophyll and spun at maximum velocity on a tabletop microcentrifuge (Beckman Coulter Microfuge 22R) to remove debris. Serial 1:10 dilutions of plant extracts were made in FRET lysis buffer (20 mm Tris, 200 mm NaCl, 2 mm EDTA, 0.1% Nonidet P-40, pH 8.0). E. coli strain BL21(DE3)gold carrying pFLIPrib-250n (23) were grown overnight in 50 ml of LB supplemented with 100 μg/ml of ampicillin at 37 °C, washed in 20 ml of 25 mm Tris, pH 8.5, and resuspended in 10 ml of FRET lysis buffer. 20 mg of lysozyme (Sigma) and 10 units of DNase (Promega) were added to the FRET lysis solution and lysis proceeded for 30 min at room temperature. Cellular debris was cleared via centrifugation at 4,000 × g for 5 min. 100 μl of bacterial lysate and 50 μl of each plant extract dilution were loaded in triplicate to a 96-well plate (Packard) and scanned in a Fusion α-FP fluorescent plate reader (PerkinElmer Life Sciences) using a CFP (434 nm, bandwidth 17 nm) excitation filter and both YFP (535 nm, bandwith 22 nm), and CFP (479 nm, bandwidth 40 nm) emission filters (Thorlabs Inc.). FRET signal is expressed as a ratio of YFP to CFP fluorescence under CFP excitation.
Statistical Analysis
Analysis of variance with Tukey's post hoc multiple comparison test was applied to the means of all scatter plot graphs in Prism 7 (GraphPad).
Author Contributions
J. R., N. C. R., and J. C. designed the work; J. R., P. C., N. C. R., and J. C. performed the research; J. R., N. C. R., and J. C. analyzed the data; J. R. wrote the initial manuscript draft; and J. R., N. C. R., and J. C. edited the manuscript.
Acknowledgments
We thank Alexander Jones and Wolf Frommer (Carnegie Institution Stanford) for the gifts of the ribose sensors, Torsten Mohlmann (Universitat Kaiserslautern) for gift of NSH1 over-expression lines, and Jessica Floret and Sonia Chapiro for technical assistance. We thank Professors Kentaro Inoue, Li Tian, and Oliver Fiehn for helpful suggestions.
This work was supported by the Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences of the United States Department of Energy Grants DE-FG02-09ER16077 and DE-SC0002175 and a Eric and Louise Conn Graduate Student Fellowship in Plant Biochemistry (to J. R.). The authors declare they have no conflict of interest with the contents of this article.
We dedicate this paper to the memory of Kentaro Inoue, who unexpectedly passed away on August 31, 2016.
- RBSK
- ribokinase
- pfkB
- phosphofructokinase-B
- cTP
- chloroplast transit peptide
- NSH1
- nucleoside ribohydrolase-1
- GM
- germination media
- PEND
- plastid envelope DNA binding protein.
References
- 1. Park J., and Gupta R. S. (2008) Adenosine kinase and ribokinase: the RK family of proteins. Cell Mol. Life Sci. 65, 2875–2896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chuvikovsky D. V., Esipov R. S., Skoblov Y. S., Chupova L. A., Muravyova T. I., Miroshnikov A. I., Lapinjoki S., and Mikhailopulo I. A. (2006) Ribokinase from E. coli: expression, purification, and substrate specificity. Bioorg. Med. Chem. 14, 6327–6332 [DOI] [PubMed] [Google Scholar]
- 3. Hope J. N., Bell A. W., Hermodson M. A., and Groarke J. M. (1986) Ribokinase from Escherichia coli K12: nucleotide sequence and overexpression of the rbsK gene and purification of ribokinase. J. Biol. Chem. 261, 7663–7668 [PubMed] [Google Scholar]
- 4. Park J., van Koeverden P., Singh B., and Gupta R. S. (2007) Identification and characterization of human ribokinase and comparison of its properties with E. coli ribokinase and human adenosine kinase. FEBS Lett. 581, 3211–3216 [DOI] [PubMed] [Google Scholar]
- 5. Paul R., Patra M. D., and Sen U. (2015) Crystal structure of apo and ligand bound Vibrio cholerae ribokinase (Vc-RK): role of monovalent cation induced activation and structural flexibility in sugar phosphorylation. Adv. Exp. Med. Biol. 842, 293–307 [DOI] [PubMed] [Google Scholar]
- 6. Sigrell J. A., Cameron A. D., Jones T. A., and Mowbray S. L. (1998) Structure of Escherichia coli ribokinase in complex with ribose and dinucleotide determined to 1.8-Å resolution: insights into a new family of kinase structures. Structure 6, 183–193 [DOI] [PubMed] [Google Scholar]
- 7. Quiroga-Roger D., Babul J., and Guixé V. (2015) Role of monovalent and divalent metal cations in human ribokinase catalysis and regulation. Biometals 28, 401–413 [DOI] [PubMed] [Google Scholar]
- 8. Andersson C. E., and Mowbray S. L. (2002) Activation of ribokinase by monovalent cations. J. Mol. Biol. 315, 409–419 [DOI] [PubMed] [Google Scholar]
- 9. Maj M. C., Singh B., and Gupta R. S. (2002) Pentavalent ions dependency is a conserved property of adenosine kinase from diverse sources: identification of a novel motif implicated in phosphate and magnesium ion binding and substrate inhibition. Biochemistry 41, 4059–4069 [DOI] [PubMed] [Google Scholar]
- 10. Parducci R. E., Cabrera R., Baez M., and Guixé V. (2006) Evidence for a catalytic Mg2+ ion and effect of phosphate on the activity of Escherichia coli phosphofructokinase-2: regulatory properties of a ribokinase family member. Biochemistry 45, 9291–9299 [DOI] [PubMed] [Google Scholar]
- 11. Iida A., Harayama S., Iino T., and Hazelbauer G. L. (1984) Molecular cloning and characterization of genes required for ribose transport and utilization in Escherichia coli K-12. J. Bacteriol. 158, 674–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pérignon J. L., Bories D. M., Houllier A. M., Thuillier L., and Cartier P. H. (1987) Metabolism of pyrimidine bases and nucleosides by pyrimidine-nucleoside phosphorylases in cultured human lymphoid cells. Biochim. Biophys. Acta 928, 130–136 [DOI] [PubMed] [Google Scholar]
- 13. Jung B., Flörchinger M., Kunz H. H., Traub M., Wartenberg R., Jeblick W., Neuhaus H. E., and Möhlmann T. (2009) Uridine-ribohydrolase is a key regulator in the uridine degradation pathway of Arabidopsis. Plant Cell 21, 876–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Riegler H., Geserick C., and Zrenner R. (2011) Arabidopsis thaliana nucleosidase mutants provide new insights into nucleoside degradation. New Phytol. 191, 349–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zybailov B., Rutschow H., Friso G., Rudella A., Emanuelsson O., Sun Q., and van Wijk K. J. (2008) Sorting signals, N-terminal modifications and abundance of the chloroplast proteome. PLoS ONE 3, e1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Terasawa K., and Sato N. (2005) Visualization of plastid nucleoids in situ using the PEND-GFP fusion protein. Plant Cell Physiol. 46, 649–660 [DOI] [PubMed] [Google Scholar]
- 17. Jung B., Hoffmann C., and Möhlmann T. (2011) Arabidopsis nucleoside hydrolases involved in intracellular and extracellular degradation of purines. Plant J. 65, 703–711 [DOI] [PubMed] [Google Scholar]
- 18. Altschul S. F., Gish W., Miller W., Myers E. W., and Lipman D. J. (1990) Basic local alignment search tool. J. Mol. Biol. 215, 403–410 [DOI] [PubMed] [Google Scholar]
- 19. Emanuelsson O., Nielsen H., and von Heijne G. (1999) ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 8, 978–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cabrera R., Babul J., and Guixé V. (2010) Ribokinase family evolution and the role of conserved residues at the active site of the PfkB subfamily representative, Pfk-2 from Escherichia coli. Arch. Biochem. Biophys. 502, 23–30 [DOI] [PubMed] [Google Scholar]
- 21. Fiser A., and Sali A. (2003) Modeller: generation and refinement of homology-based protein structure models. Methods Enzymol. 374, 461–491 [DOI] [PubMed] [Google Scholar]
- 22. Rockwell N. C., and Lagarias J. C. (2007) Flexible mapping of homology onto structure with homolmapper. BMC Bioinformatics 8, 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lager I., Fehr M., Frommer W. B., and Lalonde S. (2003) Development of a fluorescent nanosensor for ribose. FEBS Lett. 553, 85–89 [DOI] [PubMed] [Google Scholar]
- 24. Schäfer G., and Heber U. (1977) Glucose transport into spinach chloroplasts. Plant Physiol. 60, 286–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zrenner R., Stitt M., Sonnewald U., and Boldt R. (2006) Pyrimidine and purine biosynthesis and degradation in plants. Annu. Rev. Plant Biol. 57, 805–836 [DOI] [PubMed] [Google Scholar]
- 26. Fiehn O. (2006) Metabolite profiling in Arabidopsis. Methods Mol. Biol. 323, 439–447 [DOI] [PubMed] [Google Scholar]
- 27. Weckwerth W., Wenzel K., and Fiehn O. (2004) Process for the integrated extraction, identification and quantification of metabolites, proteins and RNA to reveal their co-regulation in biochemical networks. Proteomics 4, 78–83 [DOI] [PubMed] [Google Scholar]
- 28. Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., Thompson J. D., and Higgins D. G. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rogers J. S., and Swofford D. L. (1999) Multiple local maxima for likelihoods of phylogenetic trees: a simulation study. Mol. Biol. Evol. 16, 1079–1085 [DOI] [PubMed] [Google Scholar]
- 30. Kraft E. (2007) An investigation of the ubiquitin conjugating enzymes and RING E3 ligases in Arabidopsis thaliana. Ph.D. thesis, University of California, Davis [Google Scholar]
- 31. Earley K. W., Haag J. R., Pontes O., Opper K., Juehne T., Song K., and Pikaard C. S. (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45, 616–629 [DOI] [PubMed] [Google Scholar]
- 32. Nakagawa T., Kurose T., Hino T., Tanaka K., Kawamukai M., Niwa Y., Toyooka K., Matsuoka K., Jinbo T., and Kimura T. (2007) Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104, 34–41 [DOI] [PubMed] [Google Scholar]
- 33. Edgar R. C. (2004) MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5, 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Humphrey W., Dalke A., and Schulten K. (1996) VMD: visual molecular dynamics. J. Mol. Graph. 14, 33–38, 27–8 [DOI] [PubMed] [Google Scholar]
- 35. Stone J. (1998) An efficient library for parallel ray tracing and animation. University of Missouri, Rolla, MO [Google Scholar]
- 36. Babul J. (1978) Phosphofructokinases from Escherichia coli: purification and characterization of the nonallosteric isozyme. J. Biol. Chem. 253, 4350–4355 [PubMed] [Google Scholar]