Abstract
The propagation of hepatitis C virus (HCV) is highly dependent on host cellular factors. To identify the cellular factors involved in HCV propagation, we have previously performed protein microarray assays using the HCV nonstructural 5A (NS5A) protein as a probe. Of ∼9,000 host proteins immobilized in a microarray, ∼90 cellular proteins were identified as HCV NS5A interacting partners. Of these candidates, we selected Abelson interactor 1 (Abi1) for further characterization. Binding of HCV NS5A to Abi1 was verified by both in vitro pulldown and coimmunoprecipitation assays. HCV NS5A interacted with Abi1 through regions I + II of Abi1 and domain I of NS5A. We further demonstrated that Abi1 colocalized with the HCV NS5A protein in the cytoplasm. We showed that NS5A inhibited epidermal growth factor-mediated ERK and Egr1 activations and this inhibitory activity of NS5A was nullified in Abi1-knockdown cells. Moreover, silencing of Abi1 expression impaired HCV replication, whereas overexpression of Abi1 promoted HCV propagation. Collectively, these data indicate that HCV exploits host Abi1 protein via NS5A to modulate MEK/ERK signaling pathway for its own propagation.
Keywords: early growth response protein 1 (EGR1), epidermal growth factor (EGF), extracellular-signal-regulated kinase (ERK), growth factor receptor-bound protein 2 (GRB2), hepatitis C virus (HCV), signal transduction
Introduction
Hepatitis C virus (HCV)3 is a major etiologic agent of chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma worldwide. HCV is an enveloped virus with a positive-sense, single-stranded RNA genome. HCV belongs to the genus Hepacivirus within the Flaviviridae family (1). The HCV genome is ∼9.6 kb in length and encodes a 3,010-amino acid protein from a single large open reading frame. This polyprotein precursor undergoes cleavage by both host cellular and viral proteases to generate 3 structural proteins (core, E1, and E2) and 7 nonstructural proteins (p7 and NS2 to NS5B) (2). The structural proteins are the components of the virion, whereas the nonstructural proteins are involved in the replication of the viral genome. Nonstructural 5A (NS5A) is a multifunctional protein and interacts with many cellular proteins to regulate cellular signaling pathways and viral propagation. NS5A contains three consensus proline-rich PXXP motifs, which bind to SH3 domains. These motifs are commonly found in many cellular signaling molecules (3). NS5A modulates the MAPK signaling pathway to regulate cell growth and activation. NS5A specifically interacts with the two SH3 domains of growth factor receptor-bound protein 2 (Grb2) and inhibits the MAPK/ERK pathway (4). NS5A containing mutations within the C-terminal proline-rich motif neither interacts with Grb2 nor inhibits epidermal growth factor (EGF)-stimulated ERK1/2 phosphorylation (5–7). Furthermore, NS5A expressed in recombinant herpes simplex virus-infected cells also interacts with Grb2 and inhibits the Erk1/2 signaling pathway (8). These data indicate that NS5A regulates the MAPK pathway via Grb2. However, the exact molecular mechanisms by which NS5A interferes with function of Grb2 remains unclear.
Abelson interactor 1 (Abi1), also known as E3B1, is an adaptor protein in the receptor tyrosine kinase (RTK) signaling pathway. It contains three highly conserved domains: a homeo-domain homologous region, a proline-rich (PR), and a Src homology 3 (SH3) domain (9). Abi1, together with Eps8 and Sos1, is a crucial molecule in the regulation of actin reorganization via RTKs to activate Ras and initiate Rac-induced membrane ruffle formation (10–13). Abi1 is also involved in the regulation of cell proliferation, cell adhesion, and cell migration (14, 15). Abi1 is known to regulate EGF-induced ERK pathway along with cytoskeletal reorganization and lamellipodia formation during metastasis (16–18). Overexpression of Abi1 specifically inhibits EGF-induced activation of ERK but not c-Jun N-terminal kinase or Akt (14). It has been previously reported that the HCV NS5A protein interacts with Grb2 and perturbs Sos-mediated Ras activation without disrupting the Grb2-Sos interaction and further inhibits EGF-mediated ERK1/2 activation (3, 4).
Using protein microarray technology, we have previously identified ∼90 NS5A-interacting cellular proteins (19). Here we verify the protein interaction between NS5A and Abi1 by both in vitro pulldown and coimmunoprecipitation assays. Abi1 is required for HCV replication. Moreover, EGF-stimulated ERK and Egr1 activations were inhibited by NS5A and this inhibition was mediated by the Abi1 protein. Overall, our data suggest that HCV usurps cellular Abi1 to modulate the MEK/ERK signaling pathway to promote viral propagation.
Results
Identification of Abi1 as an NS5A Interactor in Protein Array
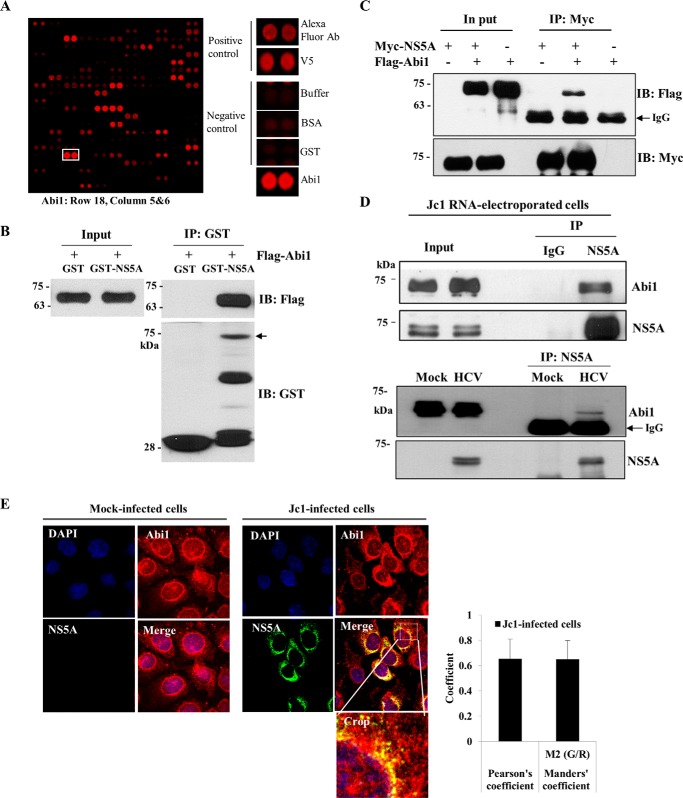
To identify cellular proteins interacting with the HCV NS5A protein, we previously performed protein microarray assays using the HCV NS5A protein as a probe. Approximately 90 cellular proteins were identified as HCV NS5A interactors (19). Abi1 was identified as one of the candidate hits, and both positive- and negative-control hits are shown in Fig. 1A. Abi1 plays a critical role in modulation of actin polymerization (16–18) and regulation of EGF signaling by transduction of the signal from Ras to Rac (10–13). Overexpression of Abi1 leads to inhibit EGF-induced ERK activation (14). It has been previously reported that NS5A also perturbs the MEK/ERK signaling pathway under the stimulation of EGF via the interaction with Grb2 (3, 4), but this mechanism still remains unclear. We therefore selected Abi1 to explore the possible involvement in modulation of MEK/ERK signaling to regulate HCV propagation. To verify the protein array data, we first performed an in vitro GST pulldown assay using GST-NS5A protein purified from Escherichia coli and cell lysates expressing FLAG-tagged Abi1. Fig. 1B shows that Abi1 selectively bound to the GST-NS5A protein but not the GST protein. To confirm the in vitro binding result, we performed a coimmunoprecipitation assay. HEK293T cells were cotransfected with Myc-tagged NS5A and FLAG-tagged Abi1. Cell lysates were immunoprecipitated with an anti-Myc antibody and then the coprecipitated protein was detected by immunoblot analysis using an anti-FLAG antibody. Coimmunoprecipitation data further confirmed that Abi1 specifically interacted with the NS5A protein (Fig. 1C). Next, we investigated whether the HCV NS5A protein interacted with endogenous Abi1 in the context of HCV replication. For this purpose, Huh7.5 cells were electroporated with Jc1 RNA. Total cell lysates harvested at 3 days after electroporation were immunoprecipitated with either IgG or an anti-NS5A antibody, and bound proteins were analyzed by immunoblotting with an anti-Abi1 antibody. As shown in Fig. 1D (upper panel), HCV NS5A interacted with endogenous Abi1 protein. To corroborate this result, we performed an immunoprecipitation assay using cell lysates harvested from either mock-infected or Jc1-infected cells with an NS5A antibody. Indeed, we verified that NS5A interacted with endogenous Abi1 in HCV-infected cells (Fig. 1D, lower panel). These data suggest that NS5A may colocalize with Abi1 in HCV-infected cells. To investigate this possibility, Huh7.5 cells were either mock-infected or infected with Jc1 for 3 days and followed by an immunofluorescence assay. As shown in Fig. 1E, both Abi1 and HCV NS5A were colocalized in the cytoplasm as yellow fluorescence in the merged image in Jc1-infected cells but not in mock-infected cells. We confirmed the colocalization of Abi1 and NS5A by determining the Pearson and Manders coefficients using ImageJ with the JaCop plugin as we reported previously (20). Collectively, these data suggest that Abi1 specifically interacts with HCV NS5A protein both in vitro and in vivo.
FIGURE 1.
Abi1 is a novel cellular factor interacting with HCV NS5A protein. A, identification of Abi1 in a protein microarray. Both positive and negative controls are shown. B, Abi1 interacts with HCV NS5A protein. HEK293T cells were transiently transfected with FLAG-tagged Abi1 expression plasmid. Total cell lysates harvested at 48 h after transfection were incubated with either GST or GST-NS5A protein. Bound proteins were precipitated with glutathione-Sepharose beads and detected by immunoblotting with an anti-FLAG monoclonal antibody. Protein expressions of GST and GST-NS5A fusion protein were verified by immunoblot analysis using an anti-GST antibody. Arrow indicates GST-NS5A. Input corresponds to 10% of total protein. C, HEK293T cells were transiently transfected with either Myc-tagged NS5A or FLAG-tagged Abi1 expression plasmid, or cotransfected with both plasmids. At 48 h after transfection, cell lysates were immunoprecipitated with an anti-Myc monoclonal antibody, and bound proteins were detected by immunoblot analysis using an anti-FLAG monoclonal antibody (upper panel). Immunoprecipitation efficiency was verified by immunoblot analysis using an anti-Myc antibody (lower panel). D, upper panel, Huh7.5 cells were electroporated with 10 μg of Jc1 RNA and incubated for 3 days. Cell lysates were immunoprecipitated with either IgG or an anti-NS5A antibody and then bound protein was detected by immunoblotting with an anti-Abi1 antibody. Immunoprecipitation efficiency was verified by immunoblot analysis using NS5A antibody. Lower panel, Huh7.5 cells were either mock-infected or infected with Jc1. Total cell lysates were immunoprecipitated with an anti-NS5A antibody. Bound proteins were immunoblotted with an anti-Abi1 antibody. Immunoprecipitation efficiency was verified by immunoblotting with an anti-NS5A antibody. E, Huh7.5 cells were either mock-infected or infected with Jc1 for 4 h. At 3 days postinfection, cells were fixed in cold methanol at −20 °C for 5 min, and immunofluorescence staining was performed using an anti-Abi1 monoclonal antibody and TRITC-conjugated goat anti-mouse IgG to detect Abi1 (red) and a rabbit anti-NS5A antibody and FITC-conjugated goat anti-rabbit IgG to detect NS5A (green). Dual staining showed colocalization of Abi1 and NS5A in the cytoplasm as yellow fluorescence in the merged images. Cells were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) to label nuclei (blue). The enlarged selection marked by a white square is shown as a crop image. Colocalization of Abi1 and HCV NS5A was quantified by both Pearson's and Manders' overlap coefficients. More than 10 cells were applied to ImageJ for quantification of overlap coefficient, and error bars indicate the mean ± S.D. Experiments were performed in duplicate.
HCV NS5A Interacts with Abi1 through Regions I + II of Abi1 and Domain I of NS5A
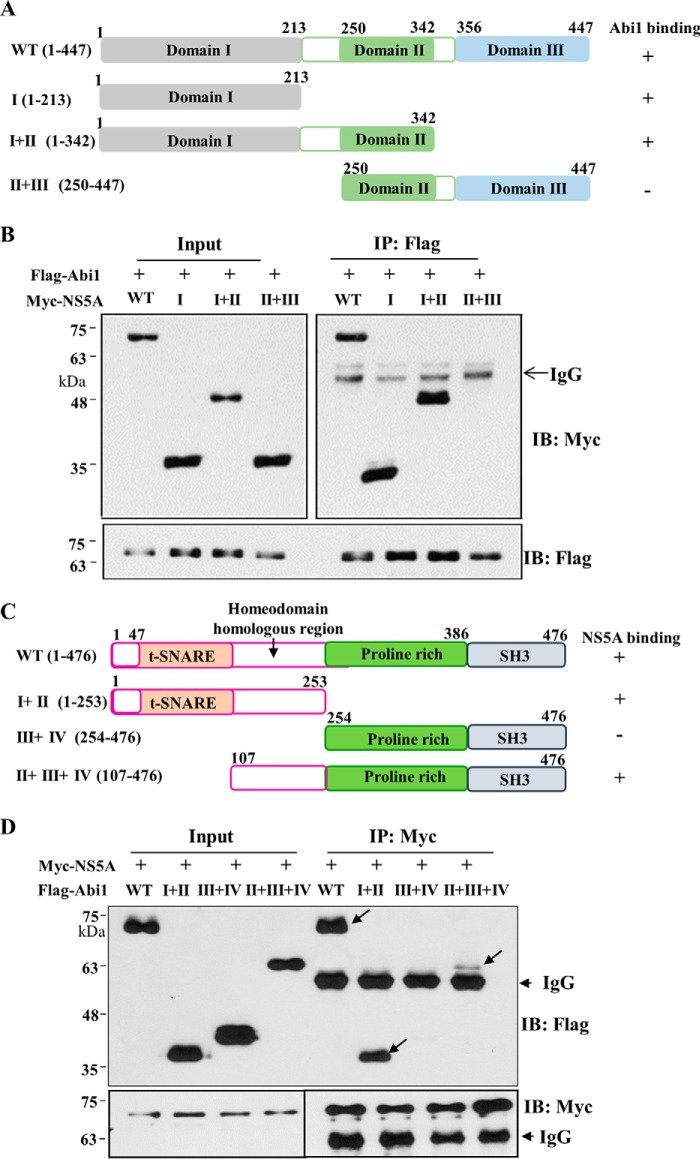
To determine the domain in NS5A responsible for Abi1 binding, the interactions between Abi1 and various deletion mutants of NS5A (Fig. 2A) were analyzed by a transfection-based coimmunoprecipitation assay. As shown in Fig. 2B, Abi1 interacted with the domain I mutant but not with a mutant harboring domains II and III of NS5A. This result clearly indicated that domain I of NS5A was responsible for binding with Abi1. Next, we determined the region in Abi1 for NS5A binding. For this purpose, we constructed various truncated mutants of Abi1 (Fig. 2C). Region I contains a syntaxin-binding region (SNARE) and a WAVE binding domain. Region II contains an homeodomain homologous region rich in serine and threonine. Region III consists of a proline-rich region. Region IV contains an Src homology 3 domain. Using these various truncated mutants of Abi1, the binding region was determined as described above. As shown in Fig. 2D, NS5A interacted with mutants containing both regions I + II and regions II + III + IV. However, NS5A no longer interacted with a mutant containing regions III + IV, indicating that NS5A interacted with regions I + II of Abi1 (Fig. 2D). It was noteworthy that regions II + III + IV of Abi1 showed weaker interaction with NS5A as compared with regions I + II. Although region II of Abi1 is required for protein interaction, these data suggest that region I of Abi1 may play an additional role in protein interaction with NS5A.
FIGURE 2.
HCV NS5A interacts with Abi1 through the domain I of NS5A and regions I + II of Abi1. A, schematic illustration of both wild type and mutant constructs of the NS5A expression plasmid. B, Abi1 interacts with domain I of NS5A. HEK293T cells were cotransfected with FLAG-tagged Abi1 and Myc-tagged NS5A expression plasmids. Total cell lysates harvested at 48 h after transfection were immunoprecipitated (IP) with an anti-FLAG antibody, and bound proteins were immunoblotted (IB) with an anti-Myc antibody. The 10% input proteins are shown. C, schematic illustration of both wild type and mutant Abi1 expression plasmids. D, NS5A interacts with regions I + II of Abi1. HEK293T cells were cotransfected with Myc-tagged NS5A and FLAG-tagged Abi1 expression plasmids. At 36 h after transfection, cell lysates were immunoprecipitated with an anti-Myc antibody, and bound proteins were immunoblotted with an anti-FLAG antibody. Protein expressions of Myc-tagged NS5A and FLAG-tagged Abi1 were verified by immunoblotting with an anti-Myc or anti-FLAG antibody using the same cell lysates. The 10% input proteins are shown. Experiments were carried out in duplicate.
NS5A Down-regulates EGF Signaling via Abi1
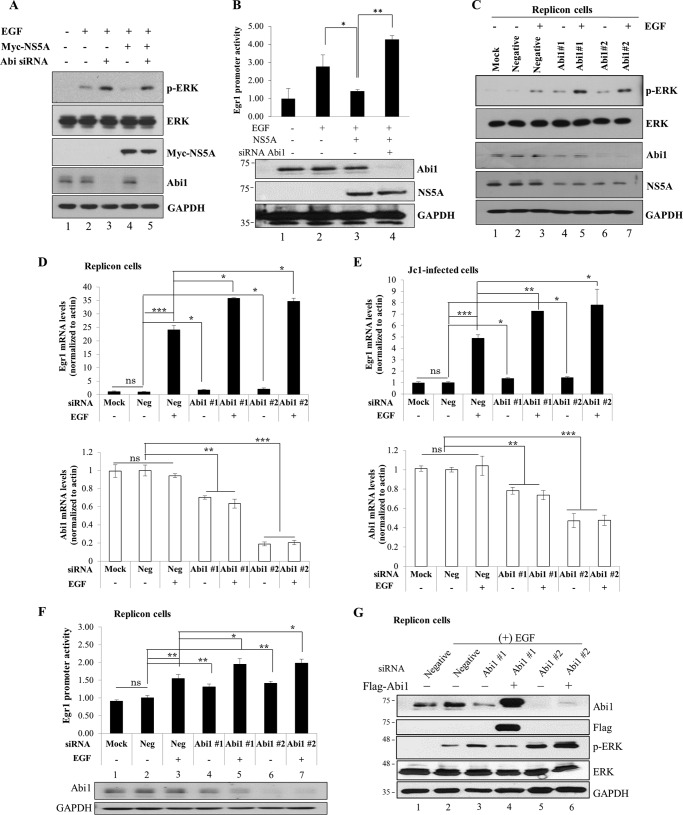
It has been previously reported that the HCV NS5A protein inhibits EGF-stimulated activation of the Ras-ERK mitogen-activated protein kinase pathway (6). Because NS5A interacts with the Abi1 protein, we selected Abi1 to explore the possible involvement in modulation of the EGF signaling pathway. We showed that overexpression of NS5A inhibited EGF-stimulated ERK activation (Fig. 3A, lane 4). However, NS5A was unable to inhibit EGF-stimulated ERK activation in Abi1 knockdown cells (Fig. 3A, lane 5). Of note, silencing of Abi1 resulted in an increase in ERK activation under EGF stimulation in vector-transfected cells (Fig. 3A, lane 3). In the MEK/ERK signaling pathway, EGF-stimulated phospho-ERK is translocated to the nucleus and then activates early growth response protein 1 (Egr1) promoter activity. To further confirm the involvement of Abi1 in the inhibitory activity of NS5A on EGF-stimulated ERK activation, we analyzed Egr1 promoter activity in Abi1 knockdown cells. Indeed, EGF-mediated Egr1 promoter activation was inhibited by NS5A (Fig. 3B, lane 3) and the inhibitory activity of NS5A was nullified in the absence of Abi1 (Fig. 3B, lane 4). We further confirmed that negative regulation of EGF-stimulated ERK activation in HCV replicon cells was also impaired in Abi1-knockdown cells (Fig. 3C, lanes 5 and 7 versus lane 3). It was noteworthy that ERK was basally activated in the absence of EGF stimulation in Abi1-knockdown cells, where NS5A expression was also impaired (Fig. 3C, lanes 4 and 6 versus lane 2). In HCV replicon cells, the Egr1 mRNA level was prominently higher in EGF-stimulated cells than control cells, and this EGF-stimulated Egr1 mRNA level was significantly increased in Abi1-knockdown cells (Fig. 3D). We verified the same results using Jc1-infected cells (Fig. 3E). Likewise, EGF-mediated Egr1 promoter activation in HCV replicon cells was significantly increased in the absence of Abi1, indicating that Abi1 was required for the negative regulation of EGF signaling pathway in HCV replicating cells (Fig. 3F, lanes 5 and 7 versus lane 3). To rule out the off-target effect of Abi1 siRNA, we used siRNA targeting of either the 3′ UTR of Abi1 (#1) or coding region of Abi1 (#2) to estimate the recovery ability of EGF-mediated ERK activation by overexpression of Abi1. For this purpose, we used FLAG-tagged Abi1 plasmid, which contains only the coding region of Abi1 but lacks 3′ UTR of Abi1. Fig. 3G shows that EGF-stimulated ERK activation in the HCV replicon cells was increased by knockdown of Abi1 (lanes 3 and 5). However, exogenous expression of Abi1 suppressed EGF-mediated ERK activation in Abi #1 siRNA knockdown cells (Fig. 3G, lane 4) but not in Abi #2 siRNA knockdown cells (Fig. 3G, lane 6). Collectively, these data suggest that the protein interplay between Abi1 and NS5A may be necessary for the modulation of EGF signaling pathway.
FIGURE 3.
NS5A down-regulates EGF signaling via modulation of Abi1. A, Huh7 cells were transfected with either negative control siRNA (−) or Abi1-specific siRNA (+). At 24 h after transfection, cells were transfected with 2 μg of Myc-tagged NS5A for 24 h. Cells were starved overnight and then treated with 100 ng/ml of EGF for 1 h. Total cell lysates were immunoblotted with the indicated antibodies. B, upper panel, Huh7 cells were transfected with either negative control siRNA or Abi1-specific siRNA. At 24 h after transfection, cells were further cotransfected with 2 μg of Myc-tagged NS5A and 1 ng of Egr1-luc promoter construct and 0.5 μg of pCH110. Cells were starved overnight and then treated with 100 ng/ml of EGF. Egr1 promoter activity was determined at 5 h after treatment. Experiments were carried out in triplicate. Error bars indicate the mean ± S.D. Lower panel, protein expressions in the same cell lysates were verified with the indicated antibodies. C, Huh7 cells harboring HCV subgenomic replicon were either mock-transfected or transfected with either negative control siRNA or 2 different Abi1-specific siRNA. At 48 h after transfection, cells were starved overnight and then either left untreated or treated with 100 ng/ml of EGF. Following 1 h incubation, cell lysates were immunoblotted with the indicated antibodies. D, Huh7 cells harboring HCV subgenomic replicon were transfected with siRNAs as described in C. At 48 h after transfection, cells were starved overnight and then either left untreated or treated with 100 ng/ml of EGF for 2 h. Both intracellular Abi1 and Egr1 mRNA levels were quantified by qRT-PCR. E, Huh7 cells were either mock-transfected or transfected with the indicated siRNAs. At 24 h after transfection, cells were infected with Jc1 for 4 h. At 48 post-infection, cells were starved for 24 h and then treated with EGF as described above. At 2 h after EGF treatment, both intracellular Abi1 and Egr1 mRNA levels were quantified by qRT-PCR. The asterisks indicate significant differences (ns, non-significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001) from the value for the negative control. Experiments were carried out in duplicate. Error bars indicate the mean ± S.D. F, upper panel, Huh7 cells harboring HCV subgenomic replicon were either mock-transfected or transfected with either negative control siRNA or 2 different Abi1-specific siRNA. At 24 h after transfection, cells were further cotransfected with Egr1-luc promoter and pCH110 plasmids for 24 h. Cells were starved overnight and then treated with 100 ng/ml of EGF. Egr1 promoter activity was determined at 5 h after treatment. Lower panel, protein expressions in the same cell lysates were verified with the indicated antibodies. Experiments were carried out in duplicate. Error bars indicate the mean ± S.D. G, Huh7 cells harboring HCV subgenomic replicon were transfected with the indicated siRNAs. At 24 h after transfection, cells were further transfected with either empty vector (−) or FLAG-tagged Abi1 plasmid. Cells were starved overnight and then treated with 100 ng/ml of EGF. Total cell lysates harvested at 48 h after plasmid transfection were immunoblotted with the indicated antibodies. Abi1 siRNA #1 targets to the 3′ UTR of Abi1, whereas Abi1 siRNA #2 binds to the sequence inside of coding region of Abi1. Experiments were carried out in duplicate.
HCV NS5A Forms a Ternary Complex with Abi1 and Sos1
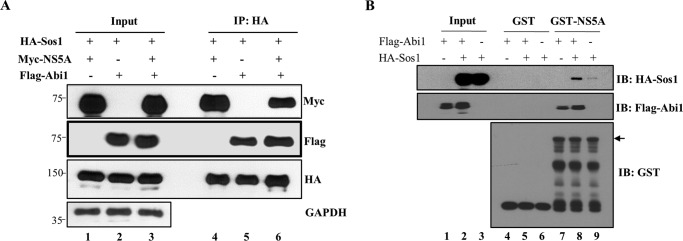
To further investigate how NS5A could down-regulate MEK/ERK signaling, we examined whether NS5A and Abi1 binding might interfere with the Abi1-Sos1 interaction. To demonstrate this, HEK293T cells were cotransfected with FLAG-tagged Abi1, Myc-tagged NS5A, and HA-tagged Sos1. At 48 h after transfection, cell lysates were immunoprecipitated with an anti-HA antibody and coprecipitated proteins were detected by immunoblotting with either an anti-Myc antibody or an anti-FLAG antibody. As shown in Fig. 4A, Abi1 was coprecipitated with NS5A and Sos1 (lane 6). Our binding data suggest that NS5A and Abi1 may form a ternary complex with Sos1. To further verify this result, an in vitro binding assay was performed using either GST or GST-NS5A fusion protein and cell lysates expressing either FLAG-tagged Abi1 or Ha-tagged Sos1. As shown in Fig. 4B, GST-NS5A weakly interacted with Sos1 in the absence of Abi1 (lane 9). Of note, GST-NS5A strongly pulled down Sos1 in the presence of Abi1 (Fig. 4B, lane 8), indicating that Abi1 interacted with both NS5A and Sos1. These data further suggest that a ternary complex may form among Sos1, NS5A, and Abi1. It is clear that NS5A perturbs the MEK/ERK signal transduction without disrupting Abi1-Sos1 interaction. These results indicate that NS5A modulates EGF signaling via Abi1 and it may contribute to HCV-induced pathogenesis.
FIGURE 4.
HCV NS5A forms a ternary complex with Abi1 and Sos1. A, HEK293T cells were cotransfected with FLAG-tagged Abi1, Myc-tagged NS5A, and HA-tagged Sos1. At 48 h after transfection, cell lysates were immunoprecipitated (IP) with an anti-HA antibody and bound proteins were immunoblotted with an anti-Myc antibody or an anti-FLAG antibody. Input corresponds to 10% of total proteins as indicated. B, HEK293T cells were transfected with either FLAG-tagged Abi1 or HA-tagged Sos1, or cotransfected with both constructs as indicated. Total cell lysates harvested at 24 h after transfection were incubated with either purified GST- or GST-NS5A-conjugated glutathione beads. Bound proteins were detected by immunoblot analysis with either anti-HA or anti-FLAG antibodies. Protein expression levels of GST and GST-NS5A were verified by using an anti-GST antibody. The arrow indicates GST-NS5A fusion protein. Experiments were performed in duplicate.
Knockdown of Abi1 Impairs HCV Propagation
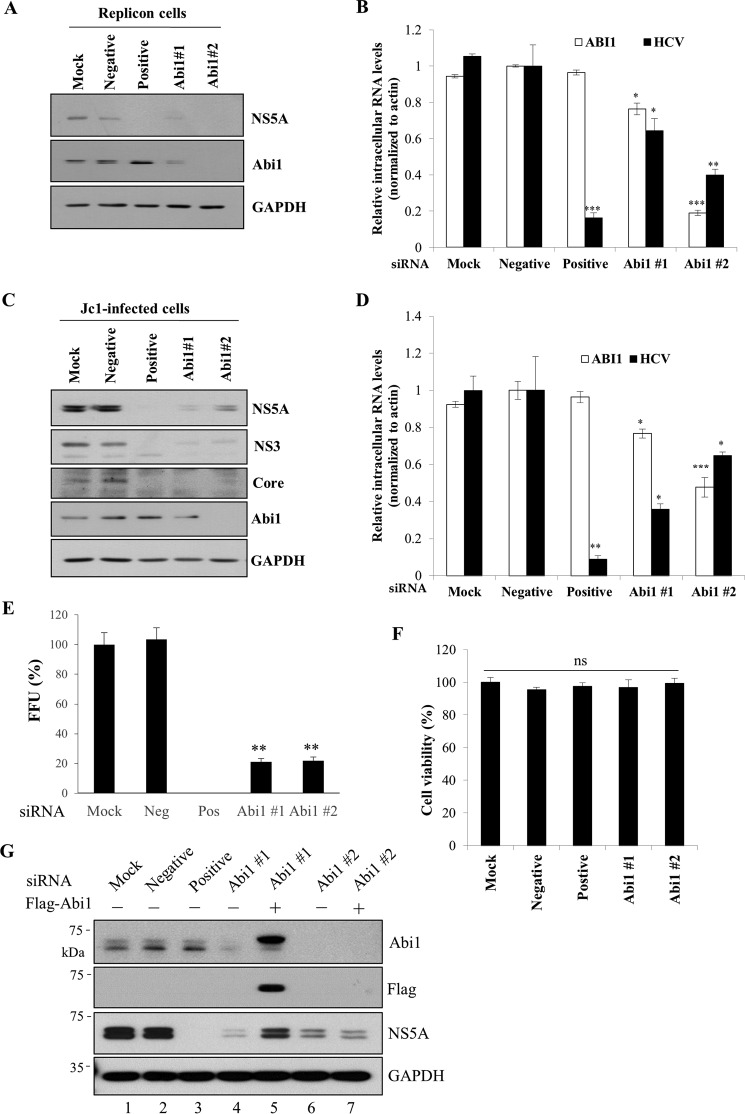
We showed that knockdown of Abi1 resulted in reduced HCV replication as well as the ability of HCV to down-regulate EGF-stimulated signal transduction (Fig. 3C). To further explore the involvement of Abi1 in HCV replication, HCV subgenomic replicon cells were either mock-transfected or transfected with either control siRNA constructs or siRNA targeting two different regions of Abi1 and then both HCV RNA and protein levels were determined. Fig. 5A demonstrated that knockdown of Abi1 decreased the HCV protein level in replicon cells. Likewise, knockdown of Abi1 significantly reduced the HCV RNA levels as determined by qRT-PCR (Fig. 5B). These data suggest that Abi1 may be involved in HCV replication. To further investigate the effect of Abi1 on HCV propagation, Huh7.5 cells were either mock-transfected or transfected with either control siRNAs or siRNA targeting Abi1 and then infected with Jc1. At 48 h post-infection, HCV protein levels were determined. Silencing of Abi1 expression led to a strong reduction of HCV protein levels in Jc1-infected cells (Fig. 5C). We further showed that knockdown of Abi1 resulted in a dramatic reduction in intracellular HCV RNA level (Fig. 5D) and HCV infectivity (Fig. 5E) without causing cellular toxicity (Fig. 5F). To rule out the off-target effect of Abi1 siRNA, we performed recovery experiments by overexpressing FLAG-tagged Abi1 in Abi1-knockdown cells using either siRNA targeting 3′ UTR of Abi1 (#1) or siRNA targeting coding region of Abi1 (#2). As shown in Fig. 5G, exogenous expression of Abi1 rescued HCV protein expression in Abi #1 siRNA-knockdown cells (lane 5) but not in Abi #2 siRNA-knockdown cells (lane 7) because FLAG-tagged Abi1 contains only the coding region of Abi1. Collectively, these data indicate that Abi1 is required for HCV propagation.
FIGURE 5.
Knockdown of Abi1 suppresses HCV propagation. A, Huh7 cells harboring HCV subgenomic replicon (genotype 1b) were either mock-transfected or transfected with 60 nm of the indicated siRNA constructs. Total cell lysates harvested at 96 h after transfection were immunoblotted with the indicated antibodies. Negative, scrambled siRNA; positive, HCV-specific siRNA; suffixes #1 and #2 refer to the siRNA sequences targeting two different regions of Abi1. B, total RNAs were extracted from siRNA-transfected HCV replicon cells and both intracellular HCV RNA and Abi1 mRNA levels were quantified by qRT-PCR. The asterisks indicate significant differences (*, p < 0.05; **, p < 0.01; ***, p < 0.001) from the value for the negative control. Experiments were carried out in triplicate. Error bars indicate the mean ± S.D. C, Huh7.5 cells were either mock-transfected or transfected with 60 nm of the indicated siRNA constructs. At 48 h after transfection, cells were infected with Jc1 for 4 h. Total cell lysates harvested at 48 h after HCV infection were immunoblotted with the indicated antibodies. D, Huh7.5 cells were either mock-transfected or transfected with the indicated siRNAs and infected with Jc1 at 48 h after transfection. At 96 h after siRNA transfection, both intracellular HCV RNA and Abi1 mRNA levels were quantified by qRT-PCR. The asterisks indicate significant differences (*, p < 0.05; **, p < 0.01; ***, p < 0.001) from the value for the negative control. Experiments were carried out in triplicate. Error bars indicate the mean ± S.D. E, Huh7.5 cells were either mock-transfected or transfected with two different Abi1-specific siRNAs for 48 h and then infected with Jc1 for 4 h. At 2 days postinfection, culture supernatant was harvested and then used to infect naive Huh7.5 cells. HCV infectivity was determined by FFU/ml. Experiments were performed in triplicate. Error bars indicate the mean ± S.D. F, Huh7.5 cells were either mock-transfected or transfected with 60 nm of the indicated siRNAs. At 96 h after transfection, cell viability was determined by WST assay. G, Huh7.5 cells were either mock-transfected or transfected with the indicated siRNAs for 24 h. Cells were further transfected with either empty vector (−) or FLAG-tagged Abi1 plasmid for 24 h and then infected with Jc1. Total cell lysates harvested at 48 h postinfection were immunoblotted with the indicated antibodies. siRNA #1 targets to the 3′ UTR of Abi1, whereas siRNA #2 binds to the coding region of Abi1. Experiments were carried out in triplicate.
Overexpression of Abi1 Promotes HCV Propagation
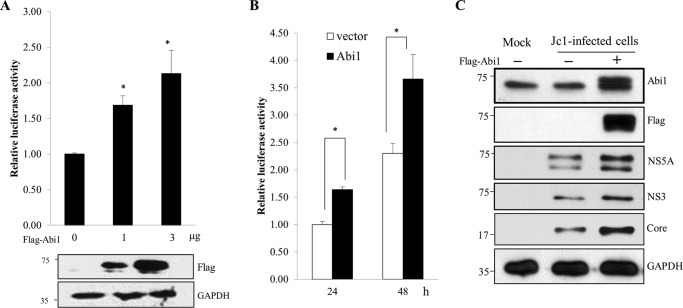
Because silencing of Abi1 suppressed HCV replication, we hypothesized that overexpression of Abi1 might have a positive effect on HCV replication. To determine this possibility, Huh7.5 cells were electroporated with JFH1-luc RNA for 48 h and then transiently transfected with FLAG-tagged Abi1. We showed that overexpression of Abi1 significantly increased the luciferase activity in a dose-dependent manner (Fig. 6A) as compared with vector-transfected cells. Similarly, overexpression of Abi1 significantly elevated the luciferase activity in a time-dependent manner (Fig. 6B). We further demonstrated that overexpression of Abi1 resulted in an increase of HCV protein levels in Jc1-infected cells as compared with vector-transfected cells (Fig. 6C). All these data indicate that Abi1 is required for HCV propagation.
FIGURE 6.
Overexpression of Abi1 increases HCV propagation. A, Huh7.5 cells were electroporated with 10 μg of JFH1-luc RNA. At 48 h after electroporation, cells were transfected with increasing amounts of FLAG-tagged Abi1. Luciferase activities were determined at 48 h after transfection. Protein expressions in the same cell lysates were verified with the indicated antibodies (lower panels). Experiments were carried out in triplicate. Error bars indicate the mean ± S.D. B, Huh7.5 cells were electroporated with JFH1-luc RNA and cotransfected with the same expression plasmids as described in A. Luciferase activities were determined at the indicated time points. Experiments were carried out in triplicate. Error bars indicate the mean ± S.D. C, Huh7.5 cells were transfected with 2 μg of either control vector (−) or FLAG-tagged Abi1 expression plasmid for 24 h, and then either mock infected or infected with Jc1 for 4 h. At 2 days postinfection, total cells lysates were immunoblotted with the indicated antibodies. Experiments were carried out in triplicate.
Discussion
HCV NS5A protein is a membrane-associated, essential component of the viral replication complex. NS5A has been shown to interact with various host proteins to modulate cell growth and cellular signaling pathways. To identify the cellular factors necessary for HCV propagation, we employed protein microarray screening using HCV NS5A protein as a probe. Of ∼9,000 human proteins, ∼90 cellular proteins were identified as the novel HCV NS5A interactors. Because Abi1 is an adaptor protein in the RTK signaling pathway and plays a crucial role in the regulation of actin reorganization and cellular proliferation (10, 14, 15), we explored the possible involvement of NS5A protein in EGF-mediated Abi1 signaling pathway.
We first verified the protein interaction between NS5A and Abi1 by both in vitro binding and coimmunoprecipitation assays. HCV NS5A interacted with endogenous Abi1 in the context of HCV replication. We further verified that both NS5A and Abi1 proteins were colocalized in the cytoplasm of Jc1-infected cells. We showed that NS5A interacted with Abi1 through regions I + II of Abi1 and domain I of NS5A harboring the N-terminal amphipathic α helix. Domain I of NS5A includes a membrane-anchoring region that is not only necessary for membrane localization but also for HCV replication (21). It has been previously reported that membrane targeting of NS5A is required for its ability to inhibit the EGF-mediated Ras-ERK signaling pathway (7). Deletion of the N-terminal 32 residues in the NS5A failed to block EGF-stimulated AP-1-driven luciferase expression (7). Meanwhile, region II of Abi1 is rich in serine and threonine residues and thus a target for serine/threonine kinases (22). There are three putative phosphorylation sites ((S/T)PX(K/R)) for cyclin-dependent kinases (23) and seven putative sites ((S/T)P) for MAP kinases (24). Therefore, we postulated that protein interplay between Abi1 and NS5A may be involved in MAPK/ERK signaling pathway.
Signaling from activated EGFR activates multiple downstream targets including Grb2 and Eps8. It has been previously reported that the HCV NS5A protein interacts with Grb2 and inhibits the EGF-stimulated Ras-ERK pathway without disrupting the Grb2-Sos1 interaction (6). In the present study, we show that NS5A interacted with Abi1 and inhibited EGF-stimulated Egr1 promoter activity. This inhibition was nullified in the absence of Abi1, implying that NS5A modulates EGF signaling via Abi1 protein. It was noteworthy that NS5A inhibited EGF-mediated signaling without disrupting Abi1-Sos1 interaction. We further showed that EGF-mediated signal transduction was modulated by the NS5A protein by forming a ternary complex with Abi1 and Sos1.
How could NS5A perturb the EGF signaling pathway in two different ways? In fact, Abi1 and Grb2 compete for the same binding site of VPVPPPVPPRRR on Sos1 (12). Sos1 can serve as a guanine nucleotide exchange factor (GEF) for both Ras (by binding Grb2) and Rac (as part of the Eps8/Abi1/PI3K complex). When Abi1 is overexpressed, Sos1 cannot bind Grb2 to activate Ras, and hence the GEF-Ras-stimulated Ras/Raf/MAPK/ERK signaling pathway involved in cell proliferation is inhibited. Instead, Sos1 forms a complex with Eps8/Abi1, and thus GEF-Rac could activate Rac to promote actin polymerization (12, 15). Therefore, NS5A, as a component of the Abi1 quadruple complex, may be able to down-regulate EGF-induced ERK activation. We speculate that NS5A may differentially regulate the EGF signaling pathway by exploiting either Grb2 or Abi1 at different stages of the HCV life cycle. However, the detailed mechanism of differential regulation of NS5A in EGF signaling needs further investigation.
Perturbation of the Ras-ERK signaling pathway may be a mechanism to regulate levels of HCV RNA replication. It has been previously reported that a low level of Ras-ERK signaling activity is required for HCV RNA replication, but complete inhibition of the Ras-ERK signaling is inhibitory (25). Other groups also reported that inhibition of MEK/ERK signaling enhanced HCV propagation (26). We showed that silencing of Abi1 resulted in enhancement of ERK activation. HCV NS5A could not inhibit the EGF signaling pathway in the absence of Abi1. This indicates that protein interplay between Abi1 and NS5A is required to suppress the MEK/ERK signaling. Finally, we explored the possible involvement of Abi1 in HCV propagation. We demonstrated that siRNA-mediated knockdown of Abi1 resulted in reduction of the intracellular HCV RNA level, HCV protein level, and HCV infectivity. Furthermore, exogenous expression of Abi1 rescued the HCV protein level in Abi1-knockdown cells. Indeed, overexpression of Abi1 significantly increased JFH1-luc luciferase activity and HCV protein levels in HCV-infected cells. Collectively, HCV usurps cellular Abi1 to favor its own replication and thus modulation of MEK/ERK signaling by the NS5A protein may contribute to HCV pathogenesis.
Experimental Procedures
Plasmid Constructions
Plasmids expressing Myc-tagged wild type NS5A and mutants have been described previously (27, 28). The coding region of Abi1 was amplified by PCR using the total RNAs extracted from HEK293T cells. PCR products were inserted into the EcoRI and KpnI sites of the p3xFLAG-CMV10 vector (Sigma). The Abi1 mutants were constructed using the same restriction enzyme sites of the p3xFLAG-CMV10 plasmid. Egr1 promoter (−688 to +1) linked to the luciferase gene was kindly provided by Dr. Sang Wook Son (Korea University) and pMT2-HA-hSOS1 was given by Dr. Pier Paolo Di Fiore (Fondazione Istituto FIRC di Oncologia Molecolare, Milan, Italy).
Cell Culture
All cell lines were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin in 5% CO2 at 37 °C. Both IFN-cured and HCV subgenomic replicon cells were grown as previously reported (28).
Antibodies
Antibodies were purchased from the following sources: Abi1 antibody was kindly provided by Dr. Giorgio Scita (Fondazione Istituto FIRC di Oncologia Molecolare, Milan, Italy); FLAG antibody from Sigma; GAPDH, ERK1, and Myc antibodies from Santa Cruz; p-ERK1 antibody from Cell Signaling. HCV core, NS3, and NS5A antibodies have been described elsewhere (29).
Preparation for a Microarray Probe
HCV NS5A protein expressed in E. coli was purified using Invitrogen nickel-nitrilotriacetic acid-agarose beads according to the manufacturer's instructions. The protein concentration was determined by the Bradford assay (Bio-Rad) and protein aliquots were stored at −70 °C.
Protein Array Screening
Protein microarray screening was performed as we reported previously (29). Briefly, protein array was incubated with blocking buffer (50 mm HEPES, pH 7.5, 25% glycerol, 0.08% Triton X-100, 200 mm NaCl, 20 mm reduced glutathione, and 0.1 mm DTT) for 1 h at 4 °C with gentle shaking and then 6 μg of purified NS5A protein diluted in 120 μl of probing buffer (PBS containing 0.1% Tween 20) was added to the array. Following incubation at 4 °C for 2 h, the array was washed five times in ice-cold buffer and then treated with anti-V5-Alexa Fluor 647 antibody (Invitrogen) for 1 h at 4 °C. Array was dried and the images were analyzed using a PerkinElmer Scanarray Ex-pressHT system and Invitrogen Prospector version 5.2 software. Significant interactions were identified based on a Z-score cutoff value of 3.0.
Immunoprecipitation
HEK293T cells were cotransfected with 2 μg of Myc-tagged NS5A and 2 μg of FLAG-tagged Abi1. Total amounts of DNA were adjusted by adding an empty vector. At 48 h after transfection, cells were harvested and the immunoprecipitation assay was performed as we reported previously (20). Endogenous interaction between Abi1 and NS5A was verified by using cell lysates from either mock- or Jc1-infected cells with an anti-NS5A antibody.
In Vitro Pulldown Assay
Approximately 1 μg of purified NS5A protein was incubated with 50 μl of glutathione-Sepharose beads for 1 h at 4 °C with gentle shaking. The beads were then washed three times in buffer (50 mm Na2HPO4 (pH 8.0), 100 mm NaCl, 1 mm PMSF, 1% protease mixture inhibitor) and were incubated with cell lysate expressing FLAG-tagged Abi1 for 2 h at 4 °C. The sample was washed five times in lysis buffer and then bound protein was detected by immunoblot analysis using an anti-FLAG monoclonal antibody.
Immunofluorescence Assay
Huh7.5 cells cultured on glass slides were either mock-infected or infected with Jc1 for 3 days. Cells were fixed in cold methanol at −20 °C for 5 min and the immunofluorescence assay was performed as reported previously (20).
Generation of HCV Jc1 and Virus Infection
Infectious HCVs were generated as we reported previously (20) with minor modifications. Briefly, the pFK-Jc1 plasmid was linearized by MluI digestion and purified by phenol-chloroform extraction. Jc1 RNAs were generated using T7 RiboMAXTM express large scale RNA production system (Promega) and were purified with Ribo EX (Gene All) according to the manufacturer's protocols. Ten micrograms of in vitro-transcribed genomic Jc1 RNA was mixed with 7.5 × 106 Huh7.5 cells in a 0.4-cm gap cuvette, and the mixture was electroporated at 270 V and 950 microfarads using a Bio-Rad GenePulser II electroporator. Cells were gently transferred to complete medium (low-glucose DMEM containing 10% fetal bovine serum, 100 units/ml of penicillin, 100 μg/ml of streptomycin, 2 mm l-glutamine, 1 mm nonessential amino acids, and 10 mm HEPES) and plated on a 150-mm dish. At 4 days after electroporation, the culture medium was collected, filtered through a 0.45-mm syringe-top filter unit (Millipore), and kept as a virus stock. For virus infection, cells were washed twice with phosphate-buffered saline (PBS) and incubated for 4 h with cell culture-grown HCV (HCVcc). HCVcc-infected cells were washed again with PBS and further cultured in complete medium.
RNA Interference
siRNAs targeting Abi1 (#1 sense, 5′-CUGUUGUACACUGGUUCAA-3′; antisense, 5′-UUGAACCAGUGUACAACAG-3′ and #2 sense, 5′-UCUCUAGCUAGUGUUGCUU-3′; antisense, 5′-AAGCAACACUAGCUAGAGA-3′) and the universal negative control siRNA were purchased from Bioneer. siRNA targeting the 5′-nontranslated region of Jc1 (5′-CCUCAAAGAAAAACCAAACUU-3′) was used as a positive control (20, 29). siRNA transfection was performed using a Lipofectamine RNAiMAX reagent (Invitrogen) according to the manufacturer's instructions. siRNA #1 targets to the 3′ UTR of Abi1 and siRNA #2 targets to the coding region of Abi1.
Quantification of HCV RNA
Both intracellular and extracellular RNAs were isolated from HCVcc-infected cells, culture media, or replicon cells using the TRIzol LS reagent (Invitrogen) and were reverse transcribed using the SuperScript Vilo cDNA synthesis kit (Invitrogen). Quantitative real-time PCR (qRT-PCR) experiments were carried out using an iQ5 multicolor real-time PCR detection system (Bio-Rad Laboratories) under the following conditions: 15 min at 95 °C, followed by 40 cycles of 95 °C for 20 s, 55 °C for 20 s, and 72 °C for 20 s. Seventy-one cycles of 10 s, with 0.5 °C temperature increments from 60 to 95 °C, were used for the melting curves.
Determination of Virus Titers
Naive Huh7.5 cells were seeded at 2 × 104/well in 8-well chamber culture slides (BD Biosciences) with 200 μl of culture medium. Following 24 h of incubation, Huh7.5 cells were inoculated with serial dilutions of cell culture medium harvested from HCVcc-infected cells. At 2 days after inoculation, indirect immunofluorescence assay was performed for the presence of intracellular core antigen to determine the number of focus-forming units (FFU). Each cluster of infected cells identified by staining for core protein was considered to be a single infectious focus. Virus titers were calculated as FFU/ml.
WST Assay
Cells seeded on 24-well plates were transfected with the indicated siRNAs. Cell viability was measured by using 30 μl of water-soluble tetrazolium salt (WST) in each well and incubated for 1 h at 37 °C. The plate was then shaken for 1 min and the aqueous layer in each well was transferred into 96-well plate and absorbance was measured at 450 nm.
Luciferase Reporter Gene Assay
Huh7.5 cells were transfected with Egr1-Luc and pCH110 plasmid for 24 h. Cells were then starved overnight and treated with 100 ng/ml of EGF. At 5 h after treatment, cells were harvested, and then luciferase assays were performed as we described previously (29). JFH-Luc reporter assay was carried out as we reported previously (20).
Statistical Analysis
Data are presented as mean ± S.D. Student's t test was used for statistical analysis. The asterisks in the figures indicate significant differences (*, p < 0.05; **, p < 0.01; ***, p < 0.001).
Author Contributions
V. T. T. H., Y. S. L., and S. C. T. performed experiments, analyzed data, and drafted the manuscript; L. N. N. and T. M. P. performed experiments; S. B. H. initiated the study, designed experiments, and wrote the manuscript. All authors reviewed and approved the final version of the manuscript.
Acknowledgment
We thank Dr. Ralf Bartenschlager (University of Heidelberg) for Jc1 and JFH1-luc constructs.
This work was supported by Basic Science Research Program Grant 2016R010886 from the Ministry of Science, ICT and Future Planning, Korea and Korean Health Technology R&D Project Grant HI13C1746 from the Ministry of Health and Welfare, Korea. The authors declare that they have no conflicts of interest with the contents of this article.
- HCV
- hepatitis C virus
- Abi1
- Abelson interactor 1
- NS5A
- nonstructural 5A
- HCVcc
- cell culture-grown HCV
- Grb2
- growth factor receptor-bound protein 2
- Egr1
- early growth response protein 1
- Sos
- son of sevenless
- qRT
- quantitative RT
- FFU
- focus-forming unit
- WST
- water-soluble tetrazolium salt
- TRITC
- tetramethylrhodamine isothiocyanate
- RTK
- receptor tyrosine kinase
- SH3
- Src homology domain 3
- GEF
- guanine nucleotide exchange factor.
References
- 1. Giannini C., and Bréchot C. (2003) Hepatitis C virus biology. Cell Death Differ. 10, S27–S38 [DOI] [PubMed] [Google Scholar]
- 2. Moradpour D., Penin F., and Rice C. M. (2007) Replication of hepatitis C virus. Nat. Rev. Microbiol. 5, 453–463 [DOI] [PubMed] [Google Scholar]
- 3. Tan S. L., Nakao H., He Y., Vijaysri S., Neddermann P., Jacobs B. L., Mayer B. J., and Katze M. G. (1999) NS5A, a nonstructural protein of hepatitis C virus, binds growth factor receptor-bound protein 2 adaptor protein in a Src homology 3 domain/ligand-dependent manner and perturbs mitogenic signaling. Proc. Natl. Acad. Sci. U.S.A. 96, 5533–5538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. He Y., Nakao H., Tan S. L., Polyak S. J., Neddermann P., Vijaysri S., Jacobs B. L., and Katze M. G. (2002) Subversion of cell signaling pathways by hepatitis C virus nonstructural 5A protein via interaction with Grb2 and P85 phosphatidylinositol 3-kinase. J. Virol. 76, 9207–9217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Macdonald A., Crowder K., Street A., McCormick C., Saksela K., and Harris M. (2003) The hepatitis C virus non-structural NS5A protein inhibits activating protein-1 function by perturbing ras-ERK pathway signaling. J. Biol. Chem. 278, 17775–17784 [DOI] [PubMed] [Google Scholar]
- 6. Macdonald A., Chan J. K., and Harris M. (2005) Perturbation of epidermal growth factor receptor complex formation and Ras signalling in cells harbouring the hepatitis C virus subgenomic replicon. J. Gen. Virol. 86, 1027–1033 [DOI] [PubMed] [Google Scholar]
- 7. Mankouri J., Griffin S., and Harris M. (2008) The hepatitis C virus non-structural protein NS5A alters the trafficking profile of the epidermal growth factor receptor. Traffic 9, 1497–1509 [DOI] [PubMed] [Google Scholar]
- 8. Georgopoulou U., Caravokiri K., and Mavromara P. (2003) Suppression of the ERK1/2 signaling pathway from HCV NS5A protein expressed by herpes simplex recombinant viruses. Arch. Virol. 148, 237–251 [DOI] [PubMed] [Google Scholar]
- 9. Jenei V., Andersson T., Jakus J., and Dib K. (2005) E3B1, a human homologue of the mouse gene product Abi-1, sensitizes activation of Rap1 in response to epidermal growth factor. Exp. Cell Res. 310, 463–473 [DOI] [PubMed] [Google Scholar]
- 10. Scita G., Nordstrom J., Carbone R., Tenca P., Giardina G., Gutkind S., Bjarnegård M., Betsholtz C., and Di Fiore P. P. (1999) EPS8 and E3B1 transduce signals from Ras to Rac. Nature 401, 290–293 [DOI] [PubMed] [Google Scholar]
- 11. Lanzetti L., Rybin V., Malabarba M. G., Christoforidis S., Scita G., Zerial M., and Di Fiore P. P. (2000) The Eps8 protein coordinates EGF receptor signalling through Rac and trafficking through Rab5. Nature 408, 374–377 [DOI] [PubMed] [Google Scholar]
- 12. Innocenti M., Tenca P., Frittoli E., Faretta M., Tocchetti A., Di Fiore P. P., and Scita G. (2002) Mechanisms through which Sos-1 coordinates the activation of Ras and Rac. J. Cell Biol. 156, 125–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen H., Wu X., Pan Z. K., and Huang S. (2010) Integrity of SOS1/EPS8/ABI1 tri-complex determines ovarian cancer metastasis. Cancer Res. 70, 9979–9990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fan P. D., and Goff S. P. (2000) Abl interactor 1 binds to sos and inhibits epidermal growth factor- and v-Abl-induced activation of extracellular signal-regulated kinases. Mol. Cell. Biol. 20, 7591–7601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Innocenti M., Frittoli E., Ponzanelli I., Falck J. R., Brachmann S. M., Di Fiore P. P., and Scita G. (2003) Phosphoinositide 3-kinase activates Rac by entering in a complex with Eps8, Abi1, and Sos-1. J. Cell Biol. 160, 17–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Innocenti M., Zucconi A., Disanza A., Frittoli E., Areces L. B., Steffen A., Stradal T. E., Di Fiore P. P., Carlier M. F., and Scita G. (2004) Abi1 is essential for the formation and activation of a WAVE2 signalling complex. Nat. Cell Biol. 6, 319–327 [DOI] [PubMed] [Google Scholar]
- 17. Cui M., Yu W., Dong J., Chen J., Zhang X., and Liu Y. (2010) Down-regulation of ABI1 expression affects the progression and prognosis of human gastric carcinoma. Med. Oncol. 27, 632–639 [DOI] [PubMed] [Google Scholar]
- 18. Kotula L. (2012) Abi1, a critical molecule coordinating actin cytoskeleton reorganization with PI-3 kinase and growth signaling. FEBS Lett. 586, 2790–2794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Park C., Min S., Park E. M., Lim Y. S., Kang S., Suzuki T., Shin E. C., and Hwang S. B. (2015) Pim kinase interacts with nonstructural 5A protein and regulates hepatitis C virus entry. J. Virol. 89, 10073–10086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lim Y. S., Tran H. T., Park S. J., Yim S. A., and Hwang S. B. (2011) Peptidyl-prolyl isomerase Pin1 is a cellular factor required for hepatitis C virus propagation. J. Virol. 85, 8777–8788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Penin F., Brass V., Appel N., Ramboarina S., Montserret R., Ficheux D., Blum H. E., Bartenschlager R., and Moradpour D. (2004) Structure and function of the membrane anchor domain of hepatitis C virus nonstructural protein 5A. J. Biol. Chem. 279, 40835–40843 [DOI] [PubMed] [Google Scholar]
- 22. Biesova Z., Piccoli C., and Wong W. T. (1997) Isolation and characterization of e3B1, an eps8 binding protein that regulates cell growth. Oncogene 14, 233–241 [DOI] [PubMed] [Google Scholar]
- 23. Nigg E. A. (1993) Cellular substrates of p34(cdc2) and its companion cyclin-dependent kinases. Trends Cell Biol. 3, 296–301 [DOI] [PubMed] [Google Scholar]
- 24. Clark-Lewis I., Sanghera J. S., and Pelech S. L. (1991) Definition of a consensus sequence for peptide substrate recognition by p44mpk, the meiosis-activated myelin basic protein kinase. J. Biol. Chem. 266, 15180–15184 [PubMed] [Google Scholar]
- 25. Gretton S., Hughes M., and Harris M. (2010) Hepatitis C virus RNA replication is regulated by Ras-Erk signalling. J. Gen. Virol. 91, 671–680 [DOI] [PubMed] [Google Scholar]
- 26. Ndjomou J., Park I. W., Liu Y., Mayo L. D., and He J. J. (2009) Up-regulation of hepatitis C virus replication and production by inhibition of MEK/ERK signaling. PLoS ONE 4, e7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Choi S. H., and Hwang S. B. (2006) Modulation of the transforming growth factor-β signal transduction pathway by hepatitis C virus nonstructural 5A protein. J. Biol. Chem. 281, 7468–7478 [DOI] [PubMed] [Google Scholar]
- 28. Park C. Y., Choi S. H., Kang S. M., Kang J. I., Ahn B. Y., Kim H., Jung G., Choi K. Y., and Hwang S. B. (2009) Nonstructural 5A protein activates β-catenin signaling cascades: implication of hepatitis C virus-induced liver pathogenesis. J. Hepatol. 51, 853–864 [DOI] [PubMed] [Google Scholar]
- 29. Ngo H. T., Pham L. V., Kim J. W., Lim Y. S., and Hwang S. B. (2013) Modulation of mitogen-activated protein kinase-activated protein kinase 3 by hepatitis C virus core protein. J. Virol. 87, 5718–5731 [DOI] [PMC free article] [PubMed] [Google Scholar]