Abstract
Stress granules (SGs) are cytoplasmic condensates of stalled messenger ribonucleoprotein complexes (mRNPs) that form when eukaryotic cells encounter environmental stress. RNA-binding proteins are enriched for arginine methylation and facilitate SG assembly through interactions involving regions of low amino acid complexity. How methylation of specific RNA-binding proteins regulates RNA granule assembly has not been characterized. Here, we examined the potent SG-nucleating protein Ras-GAP SH3-binding protein 1 (G3BP1), and found that G3BP1 is differentially methylated on specific arginine residues by protein arginine methyltransferase (PRMT) 1 and PRMT5 in its RGG domain. Several genetic and biochemical interventions that increased methylation repressed SG assembly, whereas interventions that decreased methylation promoted SG assembly. Arsenite stress quickly and reversibly decreased asymmetric arginine methylation on G3BP1. These data indicate that arginine methylation in the RGG domain prevents large SG assembly and rapid demethylation is a novel signal that regulates SG formation.
Keywords: mRNA, post-translational modification (PTM), protein arginine N-methyltransferase 5 (PRMT5), RNA binding protein, stress granule, G3BP1, protein arginine N-methyltransferase (PRMT)
Introduction
Eukaryotic cells exposed to environmental stress rapidly form cytoplasmic stress granules (SGs),2 RNA granules that contain stalled translation initiation complexes. SG formation typically follows translational inhibition due to stress-induced eIF2α phosphorylation, and SGs are viewed as extensions of translation control mechanisms (1–4). SGs are composed of messenger RNAs, small ribosomal subunits, eukaryotic translation initiation factors, and key RNA-binding proteins such as G3BP1, Tia1, HuR, TDP-43, and FMRP, several of which are proposed to play roles in nucleating SG assembly (1, 3, 5–8). The architecture of SGs likely involves stable cores enriched for G3BP1 surrounded by a dynamic shell that rapidly undergoes assembly and disassembly (9). G3BP1 is thus considered a potent SG-nucleating protein. G3BP1 is targeted by many viruses to block SG assembly (10, 11). SGs are thought to function as short-term repositories for mRNAs to prevent degradation (12), and may also trigger the innate immune system by providing a platform for activation of double-stranded RNA-dependent protein kinase PKR (13) and recruit factors to regulate signaling cascades (14–17). Dysregulation of RNA granule components has also been identified in several human diseases (18–20). Although SGs have been under intense investigation for over a decade, the mechanisms promoting SG assembly are poorly understood.
SG assembly is a complex process resulting from the net consequences of multiple RNA-RNA, RNA-protein, and protein-protein interactions (PPI) (21). However, the mechanisms that turn these interactions on and off are not understood. Currently, a liquid-liquid phase separation model has been proposed to explain aspects of RNP granule assembly. There are twenty-one proteins that have been described as having SG nucleating functions, and all of them contain multiple low complexity (LC) and intrinsically disordered (ID) regions (21, 22). Tia1 and FUS were the first RNA-binding proteins shown to regulate SGs and be regulated by interactions of LC/ID regions (23, 24), and similar types of interactions have been shown to regulate P-bodies in yeast (25, 26). High concentrations of LC/ID-containing proteins spontaneously assemble liquid-like droplets through weak multivalent interactions, and these droplets display surface tension and fuse with adjacent droplets in vitro (24, 27–31). These liquid droplets are of a fairly uniform shape and size. Although SGs exhibit similar fusion and fission features, this model does not entirely explain the biophysical features of SGs because SGs are amorphous and regulatory mechanisms are lacking in vitro.
Various post-translational modifications (PTMs) in LC/ID regions of SG-nucleating proteins have been proposed to regulate SG assembly. This includes phosphorylation (32, 33), and dephosphorylation (5), poly(ADP)ribosylation (34), deacetylation (35), glycosylation (36), and methylation (37). However, only phosphorylation at Ser-149 (5) and deacetylation by HDAC6, both within a LC/ID region of G3BP1, have been linked to changes in SG assembly (35).
Protein arginine methylation is a PTM mostly studied in nuclear proteins, and regulates maturation of heterogenous ribonucleoproteins, PPI, and protein distribution in eukaryotic cells (38, 39). Quantification and identification of methylation sites in mammalian cells via SILAC labeling revealed that ∼75% of RNA-binding proteins, including G3BP1, are methylated, predominantly in RGG domains (40). The RGG domain of Cold-inducible RNA-binding protein controls its localization and migration into cold-induced SGs but a linkage to methylation is unknown (37). In addition, TDRD3, a methyl-arginine-binding protein, is a SG constituent protein (41, 42). Moreover, cells treated with MTA, a relatively selective inhibitor of protein arginine methyltransferase (PRMT) 5 (38, 43), displayed altered SG disassembly kinetics (41). Together these studies indicate methylation may be an important PTM that regulates SG condensation, but the key SG components that are methylated are unclear.
PRMTs methylate arginine residues in glycine and arginine-rich motifs. Nine mammalian PRMTs have been identified, and they are divided into three types (39, 44, 45). Type I PRMTs (PRMT1–4, PRMT6, and PRMT8) catalyze formation of ω-NG,NG-asymmetric dimethylarginines (ADMA). Type II PRMTs (PRMT5, and PRMT9) catalyze the formation of ω-NG,N'G-symmetric dimethylarginines (SDMA). Both Type I and II PRMTs catalyze the formation of ω-NG-monomethylarginines (MMA) before catalyzing their respective dimethyl arginine products. The only type III PRMT (PRMT7) only generates a MMA product. PRMTs are functionally linked to transcriptional regulation, mRNA metabolism, signal transduction, DNA repair, and have roles in human diseases involving modulation of protein localization (46–49). PRMT1 can regulate nuclear-cytoplasmic shuttling of FUS and ATXN2L but has not been linked to SG dynamics; however, PRMT1 affects RAP55 localization to P-bodies (50–52). G3BP1 is methylated by PRMT1 at several arginine residues in mice in conjunction with β-catenin gene regulation (40). Methylation inhibitors can also modulate SGs (41, 53), but methylation of specific SG-nucleating protein targets and methylation-dependent changes in SG dynamics has not been established.
Here we describe a new regulatory mechanism for SG assembly that involves arginine methylation of G3BP1 by PRMT1 and PRMT5. We demonstrate that the RGG domain of G3BP1 is crucial for SG assembly, and it is methylated by both PRMT1 and PRMT5. Surprisingly, methylation of the RGG domain was not associated with SG assembly, rather rapid demethylation of G3BP1 in response to oxidative stress was linked to SG assembly. Collectively, these data indicate that arginine demethylation in the RGG domain of G3BP1 is a novel signal to trigger SG formation during oxidative stress.
Results
The RGG Domain of G3BP1 Is Required for SG Assembly
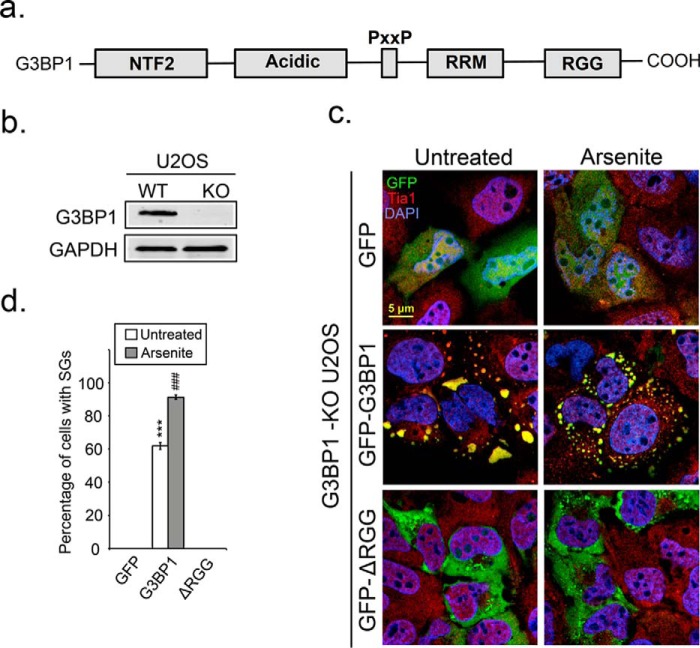
Certain PTMs on RNA-binding proteins can modulate RNA granule assembly. SILAC mass spectrometry indicates that G3BP1 is methylated in the RGG domain (40, 54). Since G3BP1 is a potent SG-nucleating protein we investigated whether the C-terminal RGG domain is important for SG assembly. We generated a GFP-tagged G3BP1-ΔRGG construct expressing truncated G3BP1 lacking the RGG domain (Fig. 1a). The G3BP1-ΔRGG construct was transfected into U2OS cells that were genetically modified, using CRISPR/Cas9 technology, to abrogate G3BP1 expressing (G3BP1 KO cells; Fig. 1b). This allowed us to examine whether the RGG domain of G3BP1 is required for SG formation in cells where there are no complications resulting from coexpression of wild type endogenous protein. Expression of GFP in cells did not generate SGs even under arsenite stress conditions, demonstrating the strong dependence of SG assembly on G3BP1 (Fig. 1c, top panel and d). Expression of G3BP1 led to formation of SGs spontaneously (Fig. 1c, middle panel) in ∼60% of transfected cells (Fig. 1d) as expected, and 90% of arsenite-treated transfected cells (Fig. 1c, middle panel and d). However, expression of G3BP1-ΔRGG was severely impaired SG assembly in unstressed and arsenite-treated cells (Fig. 1, c, bottom and d), suggesting that the RGG domain of G3BP1 is critical for SG formation.
FIGURE 1.
Deletion of the RGG domain of G3BP1 impairs SG assembly. a, diagram depicting the domain structure of G3BP1. b, endogenous G3BP1 in U2OS cells was depleted by CRISPR/Cas9 knock-out. A single G3BP1 KO clone was selected and verified by Western blot analysis. c, G3BP1 KO U2OS cells were transfected with GFP, GFP-G3BP1, or GFP-G3BP1-ΔRGG constructs and SG responses following arsenite stress (0.2 mm for 30 min) monitored by IFA. Cells were counterstained with the SG marker Tia1 in red, and DAPI for nuclei in blue. d, bar graph shows the percentage of cells that produced SGs in each condition. White bars indicate untreated cells, and gray bars refer to arsenite-stressed cells. The results shown are representative of three independent experiments counting >100 cells. ***, p < 0.001 versus untreated GFP control; ###, p < 0.001 against arsenite-treated GFP control. Original magnification: 63×.
G3BP1 Is a Substrate of PRMT1 and PRMT5
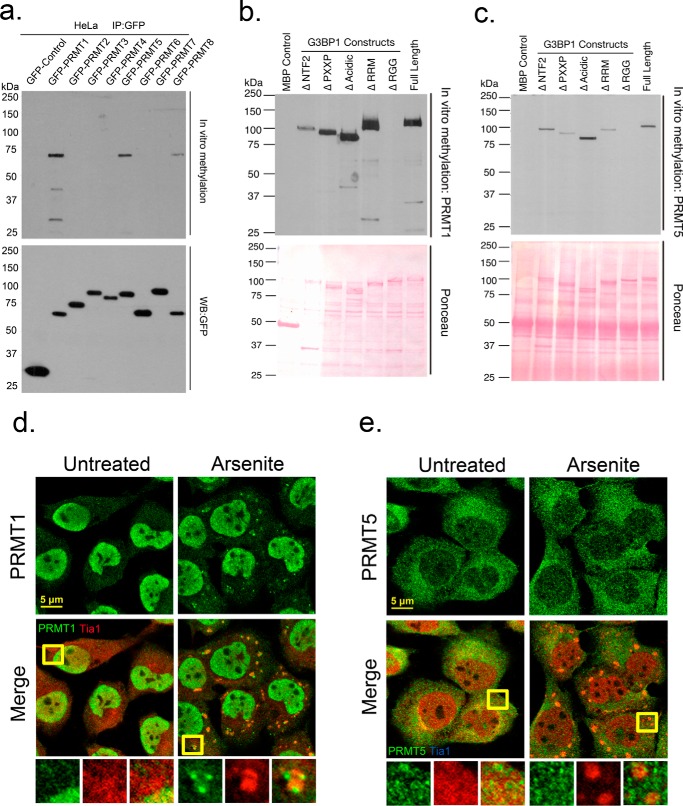
PRMT1 is the only PRMT family member known to methylate G3BP1 in mice but which PRMTs modify human G3BP1 is not known (54). To investigate whether the RGG domain of G3BP1 is methylated, we performed in vitro methylation assays with purified recombinant MBP-G3BP1 protein and GFP-tagged PRMTs (Fig. 2a). We identified three PRMTs that methylated G3BP1 in vitro: PRMT1, PRMT5, and PRMT8 (Fig. 2a). The most efficient methylation of G3BP1 was catalyzed by PRMT1 and 5, which were studied further with a panel of G3BP1 deletions (Fig. 2, b or c, respectively). We found that the RGG domain completely abolished in vitro methylation on G3BP1, indicating that PRMT1 and PRMT5 methylated G3BP1 in the RGG domain (Fig. 2, b and c, top panel). Deletion of other domains affected methylation efficiency, but all could be modified in vitro. We also visualized the distribution of PRMT1 and PRMT5 in arsenite stressed U2OS cells via immunofluorescence microscopy (IF). Although both enzymes methylated G3BP1 in the RGG domain, only PRMT1 colocalized with SGs (Fig. 2, d and e), suggesting PRMT1 may play a more dominant role in SG dynamics.
FIGURE 2.
G3BP1 is methylated by PRMT1 and PRMT5 in vitro. Recombinant MBP-G3BP1 was incubated with a panel of purified GFP-tagged PRMTs in in vitro methylation reactions as described under “Experimental Procedures.” b and c, recombinant proteins bearing deletions of the indicated domains were incubated with GFP-PRMT1 or GFP-PRMT5, respectively. Loading controls of PRMTs are shown as Western blots against GFP (a, lower panel) or for G3BP constructs via ponceau staining (b, c, lower panel). U2OS cells were arsenite-treated and processed for IFA. Cells were stained for endogenous PRMT1 (d) or PRMT5 (e) in green, and counterstained with Tia1 in red. Yellow squares indicate regions depicted in vignettes. Original magnification: 63×.
The Enzymatic Activity of PRMT1 and PRMT5 Suppress SG Formation
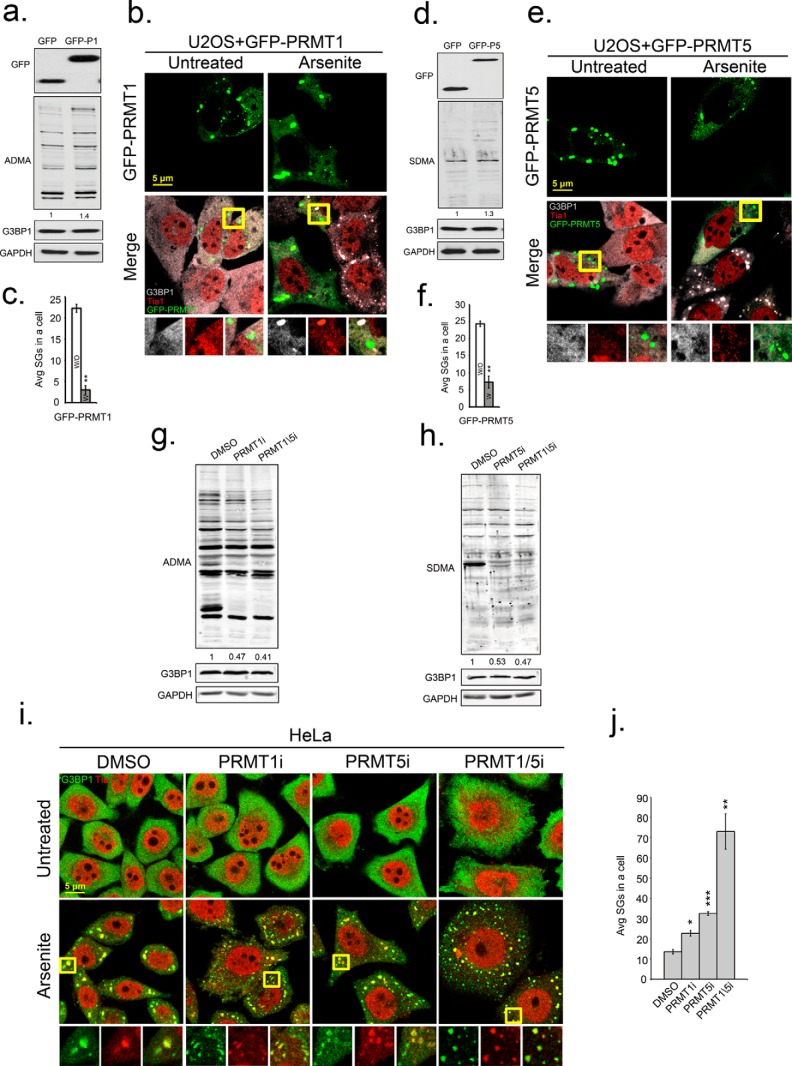
To determine if PRMT1 and PRMT5 modulate SG formation, we used genetic and chemical approaches to manipulate PRMT1 and PRMT5 activity. Overexpression of GFP-PRMT1 or GFP-PRMT5 in U2OS cells slightly increased ADMA (30%), and SDMA (25%) signals and did not change the expression of G3BP1 (Fig. 3, a and d). Ectopic expression of each PRMT led to formation of PRMT-containing foci in the cytoplasm; however, only GFP-PRMT1 foci contained SG constituent proteins including G3BP1, Tia1 (Fig. 3, b and e), and HuR (data not shown). To our surprise, there was an approximate 5-fold reduction in SG formation in cells that expressed either GFP-PRMT1 or GFP-PRMT5 (Fig. 3, c and f). Next, we used small molecule inhibitors of PRMT1 or PRMT5 in HeLa cells, and observed a 2-fold loss of ADMA and SDMA signal in endogenous proteins, but no alteration of G3BP1 protein levels (Fig. 3, g and h). Treatment of cells with inhibitors of PRMT1 or PRMT5 increased average numbers of arsenite-induced SGs per cell (22.7 ± 1.42 or 32.5 ± 0.89, respectively) compared with controls cells (13.6 ± 1.11). However, the average size of SGs in inhibitor treated cells was smaller than in controls. Simultaneous treatment of cells with both PRMT1 and 5 inhibitors strongly promoted SG formation during arsenite stress (73.1 ± 8.7) as compared with DMSO controls (13.6 ± 1.11) (Fig. 3, i and j). This increase in SGs upon PRMT inhibition correlates with reduced translation rates as measured by puromycin incorporation into nascent polypeptides (supplemental Fig. S3). These data indicate that a loss of methylation on G3BP1 promoted SG assembly marked by the canonical SG marker Tia1 (Fig. 3i). Inhibition of PRMT1 and 5 similarly promoted SGs marked by HuR and eIF3b (supplemental Fig. S1).
FIGURE 3.
Enzymatic activity of PRMT1 and PRMT5 suppresses SG formation. U2OS cells were transfected with GFP, GFP-PRMT1, or GFP-PRMT5. Antibodies specific for GFP (top panels in a and d), asymmetric (ADMA) methyl modifications (a and g), and symmetric methyl modifications (SDMA) (b and h) were used in Western blot analysis. b and e, to measure effects of PRMT overexpression on SGs, cells expressing PRMT1 or PRMT5 were stressed with arsenite and processed for IFA with antibodies against G3BP1 in gray and Tia1 in red. c and f, bar graphs represent average number of SGs/cell in cells that did express (white bars) or did not express transgenes (gray bars), **, p < 0.01. i, enzymatic activities of PRMTs in HeLa cells were inhibited by PRMT1 inhibitor (PRMT1i), PRMT5 inhibitor (PRMT5i) or both together (PRMT1/5i). HeLa cells were fixed after arsenite treatment and stained for SG markers G3BP1 in green and Tia1 in red. Yellow squares indicate regions depicted in vignettes, and the merged channel vignette is on the right of each series of three panels. j, quantification of average SGs/cell after inhibitor treatments. *, p < 0.05; **, p < 0.01; and ***, p < 0.001 versus DMSO-treated cells. The results shown are representative of three independent experiments in which 100 cells were counted in each. Original magnification: 63×.
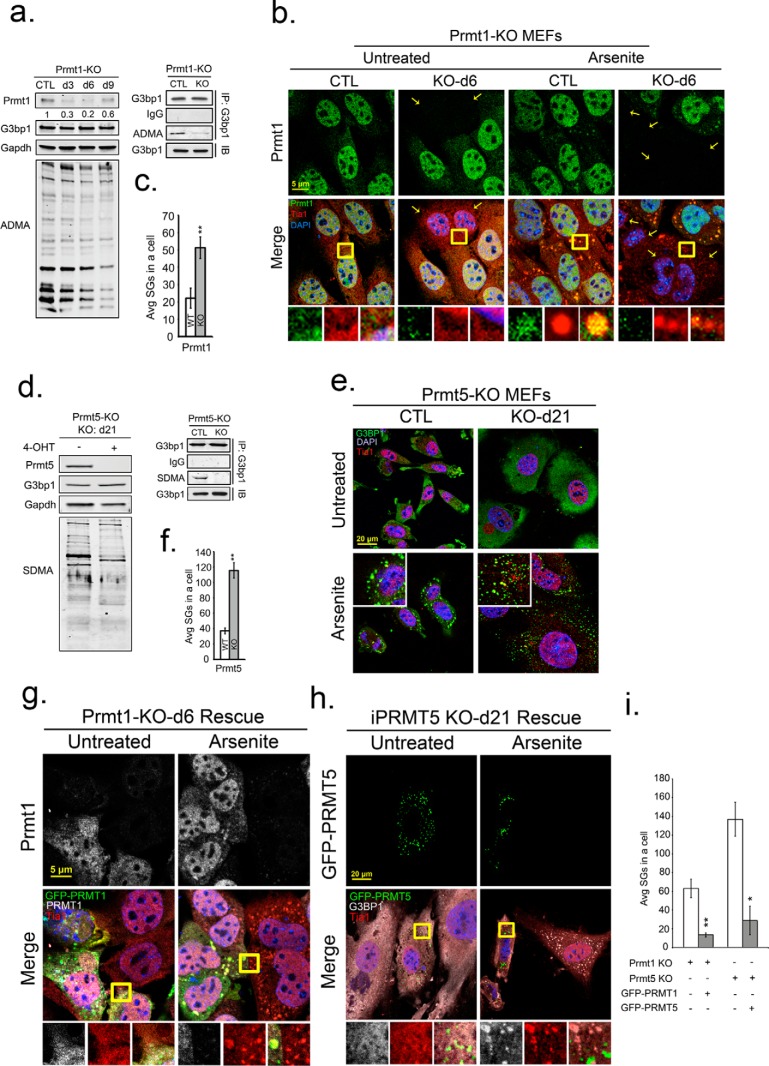
To further investigate the role of PRMTs in regulating SG assembly, we used tamoxifen-inducible PRMT1 (PRMT1-KO) and PRMT5 (PRMT5-KO) knock-out MEFs. PRMT KO MEFs displayed reduced ADMA and SDMA signals on total endogenous proteins similar to chemical inhibition of these PRMTs. G3BP1 protein levels were unaffected (Fig. 4, a and d), but immunoprecipitated G3BP1 had a large reduction of ADMA and SDMA (Fig. 4, a and d, right panels) confirming that KO of PRMT1 and PRMT5 affected G3BP1 methylation status. Assessment of G3BP1 and Tia1-containing SGs in response to arsenite stress in PRMT KO cells revealed a 2–3-fold increase in SGs with both PRMT1 KO and PRMT5 KO (Fig. 4, b, c, e, and f). SGs containing HuR and eIF3b were also elevated in arsenite-stressed PRMT KO MEFs (supplemental Fig. S1, b and c). While KO of both PRMT1 and 5 elevated SGs, only PRMT5 KO significantly reduced basal translation rates under unstressed conditions as monitored by puromycin incorporation into nascent polypeptides (4, 59) supplemental Fig. S4). Knock-out of either PRMT1 or 5 did not alter translation inhibition during arsenite stress. These findings suggest that G3BP1 methylation regulates the physical condensation of SGs rather than indirect effects resulting from overt changes in protein synthesis.
FIGURE 4.
Knockdown of PRMT1 or PRMT5 promotes SG assembly. Inducible PRMT1 (PRMT1-KO) and PRMT5 (PRMT5-KO) knock-out MEFs were treated with 4-OHT for 6 and 21 days, respectively, to deplete PRMT1 and PRMT5. a and d, endogenous PRMT1, PRMT5, G3BP1 protein levels, and methylation status of total proteins was determined by Western blot with antibodies against PRMT1, PRMT5, G3BP1, asymmetric (ADMA), or symmetric (SDMA) dimethylarginine as indicated. GAPDH protein levels serve as loading controls (a, d). b, PRMT1-KO MEFs were assessed for knock-out efficiency and effect on methylation (small panel shown in a), arsenite treated and processed for IFA with antibodies against PRMT1 in green, Tia1 in red, and DAPI in blue. Average SGs/cell were quantified before or after PRMT1 KO (c; white versus gray bars). **, p < 0.01. e, similarly, PRMT5-null MEFs (shown by small panel in d, were arsenite treated and processed for IFA. Cells were counterstained for G3BP1, Tia1, and DAPI, and the average SGs/cell were quantified with or without PRMT5 KO (f), **, p < 0.01 versus untreated PRMT5-KO MEFs. (g) and (h), PRMT1 and PRMT5 expression was rescued in PRMT-KO MEFs by transfection of GFP-tagged PRMT expression constructs. Cells were arsenite treated, fixed, and stained in IFA for PRMT1 in gray (g, top panels), G3BP1 in gray (h), Tia1 in red, and DAPI. Yellow squares indicate regions depicted in vignettes. Prmt1 KO was always incomplete, thus staining for both endogenous and transgene Prmt1 is shown (i). Quantification of average SGs/cell in rescue experiments with PRMT KO MEFs (white bar), and PRMT KO rescued MEFs (gray bar). *, p < 0.05; **, p < 0.01 versus PRMT-KO MEFs. The results shown are representative of three independent experiments counting >100 cells in each. Original magnification: 63×.
In agreement with PRMT overexpression results (Fig. 3), only PRMT1 colocalized with SGs in stressed PRMT1-KO MEF control cells (Fig. 4b). Further, depletion of PRMT1 or PRMT5 strongly increased the number of arsenite-induced SGs in cells compared with wild type cells. Finally we attempted to reverse the phenotype of the smaller, more numerous SG in PRMT KO MEFs by ectopic expression of PRMT1 and PRMT5. Expression of PRMTs in PRMT KO MEFs by transfection of GFP-tagged PRMT constructs reduced the number of SGs in arsenite-stressed cells to levels observed in wild type control cells (Fig. 4, g, h, and i). Collectively, depletion of PRMT enzymatic activity by chemical or genetic approaches promoted SG assembly, suggesting that methylation of G3BP1 (and/or other key SG-nucleating factors) may serve as a SG off-switch.
Methyl-deficient G3BP1 Mutants Promote SGs
To better link the methylation phenotype observed when PRMT activity is manipulated to G3BP1 methylation and function in SGs, two different types of GFP-tagged G3BP1 methylation mutants were generated. Arginine residues at position 429, 435, 443, 447, or 460 were changed to lysine (methyl-deficient) or phenylalanine (charge-neutral) (Fig. 5a). GFP-G3BP1 mutants were expressed in U2OS G3BP1 KO cells to analyze the behavior of G3BP1-induced SGs in the absence of endogenous G3BP1. As shown above, overexpression of GFP-G3BP1 induced SGs in 60 and 90% of unstressed transfected and arsenite-stressed transfected cells, respectively, and deletion of the RGG domain of G3BP1 abolished SG assembly (Fig. 5, b and c). Overexpression of G3BP1 charge-neutral mutants repressed SGs whereas methyl-null mutants generally promoted SGs marked by Tia1 (Fig. 5, b–d). These SGs also contained other canonical SG components (i.e. eIF3b and HuR; supplemental Fig. S2). Only the R435K methyl-deficient mutant was defective in SG formation, but only under unstressed conditions (Fig. 5, b and d). G3BP1 containing R to F mutations at all five arginine sites was very defective in SG formation under untreated conditions (Fig. 5, d, right panel, and b). Under arsenite-stressed conditions, most of the G3BP1 single point mutants responded similar to wild type regardless of whether they were charged-neutral or methyl-null mutations (Fig. 5b). Strikingly, cells that expressed the R447F mutant strongly suppressed SG assembly during stress, suggesting that Arg-447 might be targeted by PRMT1 or PRMT5 to regulate SGs.
FIGURE 5.
Mutation of putative methylation sites in the G3BP1 RGG domain modulates SG nucleation. a, a schematic showing potential arginine methylation sites in the RGG domain of G3BP1. Two types of GFP-tagged G3BP1 mutants were created at each site, charge-neutral mutants substituted Arg to Phe and methyl-deficient mutants replaced Arg to Lys. c and d, G3BP1 KO U2OS cells were transfected with GFP-tagged G3BP1 methylation mutants and left untreated or stressed, processed for IFA with staining for SGs with Tia1 (red). b, percentages of cells with SGs were quantified in charge-neutral (white bars) transfected cells and methyl-deficient (dark gray bars) transfected cells by analyzing 100 cells. *, p < 0.05; **, p < 0.01; and ***, p < 0.001 versus GFP-G3BP1-transfected cells (light gray bars). The results shown are representative of three independent experiments. e, in vitro methylation reactions (top panels) using recombinant MBP-G3BP1 variants bearing individual Arg point mutations were incubated with purified GFP-tagged PRMT 1 or PRMT5 as described under “Experimental Procedures.” Protein levels are indicated by Ponceau stain of the blots (in the lower panels).
To locate specific arginine residues in the RGG domain targeted by PRMT1 and PRMT5, we performed in vitro methylation assays with a panel of recombinant G3BP1 Arg to Lys mutant proteins. The quintuple methyl-deficient G3BP1 (All K) mutant was completely refractory to in vitro methylation with either PRMT. Mutation of individual residues Arg-447 and Arg-435 affected the efficiency of G3BP1 methylation by PRMT1. The Arg-460 G3BP1 mutant was not methylated by PRMT5 (Fig. 5e, top panel). Collectivity, these data indicate PRMT1 and PRMT5 differentially methylate G3BP1, with PRMT1 methylating Arg-435 and Arg-447, and PRMT5 preferentially targeting Arg-460. Together with SG formation assays in cells, these results suggest Arg-447 is an important residue in regulation of SGs, likely through methylation by PRMT1.
G3BP1 Is Demethylated in Arsenite-induced SGs
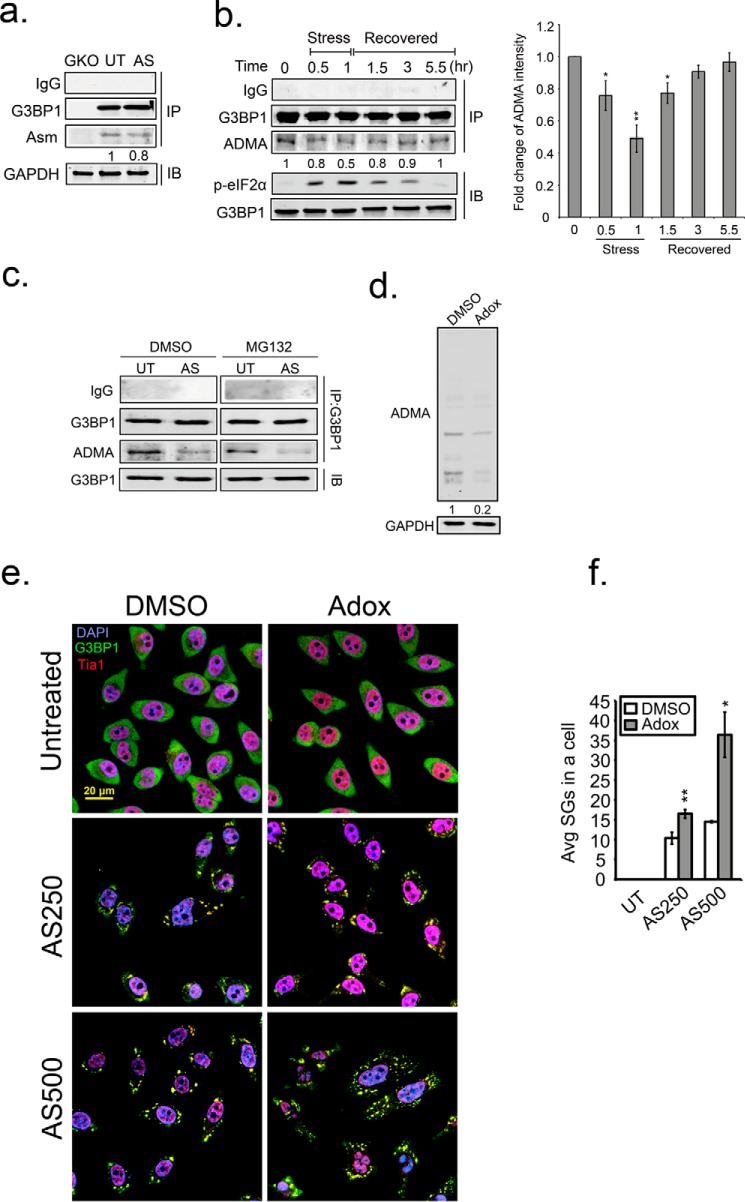
Based on the results above we speculated that G3BP1 arginines are demethylated to allow SGs assembly. Therefore, we compared ADMA of G3BP1 immunoprecipitated from unstressed and arsenite stressed cells. Indeed, we observed 20% loss of ADMA on G3BP1 after 30 min of arsenite treatment in U2OS cells (Fig. 6a). To validate the result, we performed a stress-recovery experiment to monitor the methylation status of G3BP1 through the recovery period in HeLa cells. Arsenite stress induced strong eIF2α phosphorylation that persisted for 3.5 h after release from stress. This is a proxy for translation inhibition during stress. In parallel to eIF2α phosphorylation, we also detected a loss of ADMA on G3BP1 from 20% at 30 min of stress to 50% by 1 h of arsenite stress. G3BP1 was progressively methylated with ADMA during stress recovery (Fig. 6b). This indicates that stress induces demethylation of G3BP1, and G3BP1 is methylated again during stress recovery. Interestingly, inhibition of proteasomal degradation by MG132 did not block the reduction of G3BP1 ADMA signal (Fig. 6c), indicating that the loss of ADMA signal on G3BP1 is not due to proteasomal G3BP1 turnover. This is also supported by constant G3BP1 levels at all timepoints (Fig. 6, a–c).
FIGURE 6.
Demethylation of G3BP1 promotes SG formation. a, pulldown of endogenous G3BP1 in G3BP1 KO (GKO), unstressed (UT), and arsenite-treated (AS) HeLa cells. The methylation status of G3BP1 was detected with the asymmetric dimethylation (ADMA) antibody in Western blot analysis. b, asymmetric dimethylation was detected on G3BP1 (ADMA) through stress and recovery periods. b, right panel, quantification of ADMA intensity is shown. The results are representative of three independent experiments. *, p < 0.05; **, p < 0.01 against the 0 h time point. Arsenite stress was applied for 60 min, removed, then cells were washed and incubated for the indicated recovery period. The level of phospho-eIF2α (p-eIF2α) was monitored by immunoblot analysis as a proxy for translation inhibition during the recovery period. G3BP1 asymmetric dimethyl (ADMA) status was determined by immunoblot analysis of immunoprecipitated G3BP1. c, HeLa cells were treated with either DMSO or MG132 simultaneously with either vehicle or arsenite for 1.5 h. The methylation status (ADMA) of G3PB1 was detected as described above. d, HeLa cells were treated with Adox for 48 h, and endogenous ADMA levels were determined by immunoblot. e, Adox-treated cells were stressed with 250 mm (AS250) or 500 mm (AS500) arsenite, and processed for IFA with G3BP1 in green, Tia1 in red, and DAPI. f, quantification of average SGs/cell were calculated in >100 cells for each condition. White bar indicates DMSO control, and gray bars indicates Adox treatment. *, p < 0.05; **, p < 0.01 against DMSO control. Original magnification: 63×.
To further address whether demethylation promotes SG formation, we treated HeLa cells with adenosine dialdehyde (AdOx), which is a pan methyltransferase inhibitor. There was an 80% loss of ADMA signal on total cellular proteins after 48 h of AdOX treatment (Fig. 6d). AdOX treatment did not induce SG formation in unstressed cells, whereas stressed, AdOX-treated cells displayed more SGs similar to genetic inhibition of PRMTs described above. We found that the number of SGs was arsenite concentration dependent under AdOX treatment (Fig. 6, e and f). These findings support the above data indicating inhibition of PRMT1 and PRMT5 enzymatic activity stimulates SG formation (Fig. 3, i and j), and are consistent with the hypothesis that SG regulation occurs through G3BP1 demethylation.
Demethylation of G3BP1 on Three Arginine Residues in Response to Stress
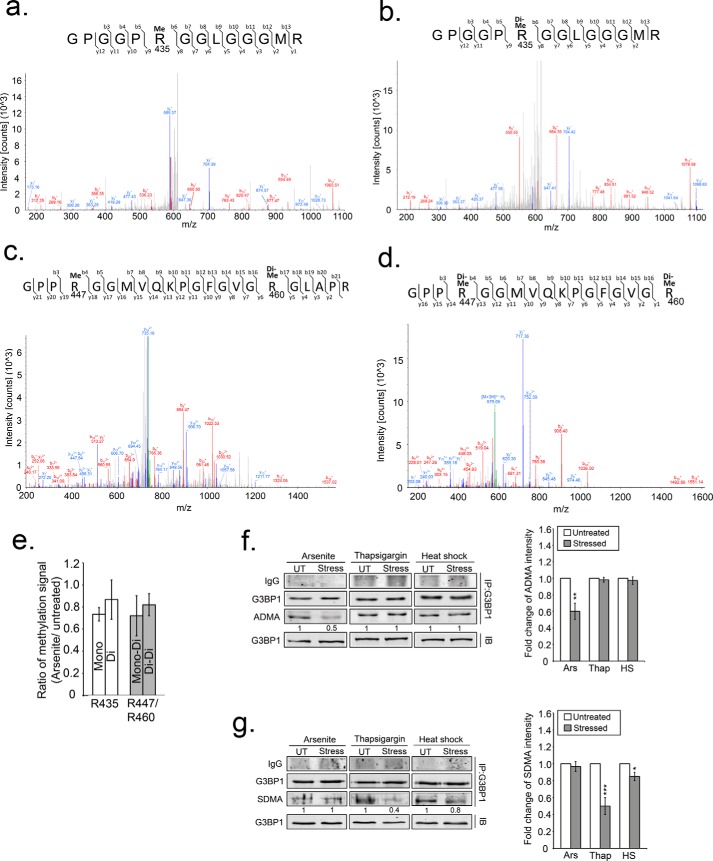
To confirm changes in G3BP1 methylation reported with the ADMA antibody, we performed liquid chromatography-mass spectrometry (LC-MS) analysis of changes in G3BP1 methylation after oxidative stress. Two methylated peptides from the RGG domain were detected, with sequences GPGGPRGGLGGGMR (amino acid 430–443 of G3BP1) (Fig. 7, a and b), and GPPRGGMVQKPGFGVGRGLAPR (amino acid 444–465 of G3BP1) (Fig. 7, c and d). Arg-435 and Arg-447 was either monomethylated (Fig. 7, a and c) or dimethylated (Fig. 7, b and d), and Arg-460 was only dimethylated (Fig. 7, c and d) in unstressed cells. After 30 min of arsenite treatment, a reduction of arginine methylation in G3BP1 was detected in both these peptides. Arg-435 lost about 27% (0.73 ± 0.06) of monomethylation and 14% (0.86 ± 0.18) of dimethylation, whereas R447/R460 lost about 28% of mono/dimethylation (0.72 ± 0.18) and 18% (0.82 ± 0.1) of dimethylation/dimethylation (Fig. 7e). These results confirmed that G3BP1 undergoes rapid demethylation during oxidative stress.
FIGURE 7.
Demethylation of G3PB1 confirmed by MS analysis and detected in ER stress and heat shock. a–d, fragment ion (MS/MS) spectrums isolated from immunoprecipitated G3BP1 analysis. The first arginine residue (R435) within the peptide GPGGPRGGLGGGMR was identified as either monomethylated (a) or dimethylated (b). Two methylated arginine residues (Arg-447 and Arg-460) were identified in the peptide GPPRGGMVQKPGFGVGR in two different forms; either (c) monomethylated Arg-447 and dimethylated Arg-460, or (d) dimethylation of Arg-447 and dimethylation of Arg-460. e, bar graph indicates triplicate MS data of methylation status of G3BP1 under oxidative stress comparing the ratio of methylated over non-methylated peptides. White bars represent two methylated forms of Arg-435. Gray bars represent either monomethylated (mono) Arg-447 and dimethylated (di) Arg-460 or dimethylated Arg-447 and Arg-460 (di-di). Immunoblot for ADMA (f) or SDMA (g) of G3BP1 that was immunoprecipitated from either arsenite-treated, thapsigargin-treated or heat shocked cells. UT indicates untreated, and Stress indicates the different stressors. The quantification is the intensity of ADMA (f, right panel) and SDMA (g, right panel) shown as bar graphs and normalized against untreated conditions. The results shown are representative of three independent experiments. White bars indicate untreated, and gray bars indicate different stressors. *, p < 0.05; **, p < 0.01; and ***, p < 0.01 against untreated controls.
Finally, we tested if demethylation of G3BP1 is a general stress response or only specific to arsenite stress. We used antibodies to monitor ADMA and SDMA signals on G3BP1 after arsenite, thapsigargin (ER stress) and heat shock. Interestingly, G3BP1 methylation status was differentially dependent on the stressors. Unlike arsenite treatment, the ADMA signal in G3BP1 was unchanged during ER stress and heat shock, but SDMA signal was reduced by these stressors (Fig. 7, f and g). Together this indicates that demethylation of G3PB1 might be a common mechanism to trigger SG nucleation, and PRMT1 may play an important role in oxidative stress, whereas PRMT5 may regulate G3BP1 during ER stress and heat shock.
Discussion
SGs help regulate gene expression and cell survival (55) (56) (57). As a consequence of these functions, defects in SG assembly and dynamics are implicated in several human diseases (58) (20). Although SGs have been studied for over a decade, the fundamental mechanisms governing how they assemble remain unresolved. PTMs are common endpoints of signaling cascades that coordinate protein functions within the context of whole cell gene expression and responses. Fine tuning of protein function is gained from multiple PTMs on the same protein. G3BP1 is known to be phosphorylated, methylated, and acetylated, but only regulation of phosphorylation at serine 149 has been linked to SG formation (5, 40). Here we found that the methylation status of G3BP1 is altered during cellular stress, and G3BP1 is demethylated in concert with SG assembly in response to oxidative stress with arsenite. Arginine methylation is a common PTM on nuclear proteins that is generally considered a permanent modification due to the lack of definitive arginine demethylases. Methylation of RNA-binding proteins ATAXIN2L and SERBP1 promotes nuclear localization but does not influence their entry into SGs (52, 59). Methylation of the cold shock SG component CIRP promotes its entry into RNA granules (37). In contrast, PRMT1 overexpression reduced SGs promoted by FUS overexpression and extensive methylation of Ddx4 destabilized Ddx4 liquid droplets (29, 51). The work reported here provides a mechanistic framework for understanding the role of methylation of these other RNA-binding proteins in RNA granule biology. Our work also suggests the presence of a potent demethylase that catalyzes rapid arginine demethylation of a key SG assembly factor in response to several stressors. We detected a cycle of loss and gain of ADMA signal on G3PB1 during the arsenite stress-recovery period, which was confirmed by MS analysis. The cycle of arginine demethylation and methylation in stress-recovery experiments is not a result of G3BP1 protein turnover and new protein synthesis because 1) arsenite rapidly shuts down bulk translation because of eIF2α phosphorylation, 2) G3BP1 is a stable protein with a half-life of over six hours (data not shown), and 3) no reduction of ADMA signal on G3PB1 resulted from treatment of cells with MG132 during arsenite stress. We also show PRMT1, 5 and 8 are responsible for decorating G3BP1 with methyl groups. PRMTs 1 and 5 recognize different arginine residues on G3BP1, and expression of methyl-null G3BP1 point mutants at R429, R447, and R460F repressed SG formation in cells. Although previous work indicates PRMT1 methylates murine G3BP1, they did not show effects on SGs (55).
We focused on PRMTs 1 and 5 in this study since PRMT8 methylation was weak and it is expressed only in the brain (60). PRMT methylation of G3BP1 occurs in the RGG domain, which is critical for SG assembly under oxidative stress conditions. Previous work suggested the RGG domain of G3BP1 is involved in PPI and protein-RNA interactions (21) and interactions with 40S ribosomes (61). We elevated arginine methylation levels by ectopic expression of PRMT1 and PRMT5 and found reduced numbers of SGs during oxidative stress, whereas chemical and genetic inhibition of PRMT enzymatic activity promoted SG assembly. We also discovered differential methylation of G3BP1 by PRMTs. Arg-435 and Arg-447 are sites methylated by PRMT1, and R460 is a methylation site for PRMT5. Though Arg-447 methylation by PRMT1 strongly represses the SG phenotype, a sophisticated methylation regulatory system for G3BP1 function may exist. Arg-435 remained arginine methylated during stress, and potentially cooperates with PRMT5 methylation of Arg-460. PRMT5 does not penetrate SGs, as we show, but might function to promote G3BP1 signaling, thus modifying G3BP1 outside of SGs as it undergoes rapid exchange in and out of SGs. Conversely, consistent with another report (51), PRMT1 enters SGs and may fine-tune Arg-447 methylation in situ, trigger G3BP1 egress and promote SG disassembly. The detailed molecular mechanisms still require further investigation.
Collectively, our data indicate that arginine methylation of G3BP1 inhibits SG assembly; and loss of PRMT1 and PRMT5 enzymatic activity might result in failure of secondary assembly of SGs (small SG phenotype). An emerging hypothesis in the field encompasses a two stage assembly of SGs, nucleated in the first stage by G3BP1-formation of many small granules that then merge into larger granules (9, 62). The size of SGs seems to be independent of translational repression in other studies, but disturbs SG-PB interactions (63). SG have also been proposed to modulate signal transduction cascades by recruiting signaling molecules into SGs or providing a platform for these molecules to work (21) (64) (65). Recent MS analysis of the composition of crosslinked G3BP1-enriched SGs showed some enrichment of both PRMT1 and PRMT5 with G3BP1 (9), and either could play roles in these events.
In conclusion, our findings indicate stress-induced, rapid demethylation of G3BP1, a critical SG nucleating factor, is a key requirement for SG assembly. Our work suggests the presence of an arginine demethylase involved in SG accumulation. Jumonji domain containing domain 6 (JMJD6) is the only enzyme that has been reported to have arginine demethylase activity (66, 67), although lysine demethylases with activity on histones are known (68) (69). Although some conflicting reports challenge whether JMJD6 has true arginine demethylase activity (70), several reports link JMJD6 to demethylation of mitochondrial PAX, Hsp70, estrogen receptor, and RNA helicase A (67), and JMJD6 is activated by hypoxic stress (71). Further work will determine if JMJD6 or yet unknown demethylases are involved in SG accumulation through G3BP1.
Experimental Procedures
Cell Culture and Transfections
Cells were cultured under standard conditions of 10% FBS in DMEM. CRISPR G3BP1 knock-out U2OS cells are a single clone generated by sgRNA targeting to exon1 of G3BP1. After transfection of CRISPR/Cas9, cells were serially diluted and propagated to obtain single cell clones. Several clones were compared before selecting the one used in this study. All expression constructs were transfected into cells by Lipofectamine 3000 (Thermo Fisher Scientific) in accordance with the manufacturer's instructions.
PRMT1 and PRMT5 inducible knock-out MEFs were treated with 2 μm 4-hydroxytamoxifen (4-OHT; Sigma) for 6 days and 21 days, respectively, to induce PRMT KO. We verified PRMT KO efficiency by Western blotting analysis with primary antibodies against PRMT1 (Cell Signaling), PRMT5 (Abcam), ADMA, and SDMA (EpiCypher).
PRMT1 and PRMT5 Inhibitors
The PRMT5 inhibitor was developed by EpiZyme (EPZ015666) (72). The PRMT1 inhibitor (MS023) was developed in the Jin laboratory, and blocks Type I PRMTs, but does not prevent SDMA addition by PRMT5 (73).
Plasmid Constructs
A panel of G3BP1 methylation mutants was generated from a GFP-G3BP1 construct, generously provided by N. Kedersha (Brigham and Women's Hospital, Boston, MA). Two sets of site-directed mutagenesis primers were used to sequentially mutate Arg residues. Methyl-deficient primers switched arginine residues (Arg-429, Arg-435, Arg-443, Arg-447, and Arg-460) in the RGG domain of G3BP1 to lysine, whereas charge-neutral primer sets mutated arginine residues at the same positions to phenylalanine. Site-directed mutagenesis was performed with Herculase II fusion DNA Polymerase (Agilent technologies) as described by the manufacturer. The GFP-tagged PRMT plasmid set was described previously (74).
Protein Purification
MS2-MBP, MBP-G3BP1, and MBP-G3BP1 deletion mutant expression was induced in Rosetta cells (Novagen) for 3 h at 30 °C with 0.5 mm isopropyl-β-d-1-thiogalactopyranosode (IPTG). Proteins were purified by passing over amylose resin at 4 °C (New England Biolabs). Proteins were then washed and eluted from the resin with 10 mm maltose prior to dialysis against 0.1 m phosphate-buffered saline (PBS) and 50% glycerol. MBPs were analyzed by SDS-PAGE for protein integrity, and protein concentrations were quantified by comparison to bovine serum albumin (BSA) standards.
In Vitro Methylation Assay
In vitro methylation was conducted essentially as described previously (75). Briefly, PRMTs were purified from HeLa cells on protein A/G beads then reacted with 1 μg of recombinant G3BP1, 1 μl of S-adenosyl-l-[methyl-3H]methionine (85 Ci/mmol; Perkin-Elmer), 3 μl of 10× PBS, and H2O in a reaction volume of 30 μl. Samples were incubated at 30 °C for 1.5 h followed by separation with SDS-PAGE, transfer to a PVDF membrane, and then sprayed with EN3HANCE (Perkin-Elmer). Finally, modified proteins were detected with autoradiography.
Mass Spectrometry Analysis
G3BP1 was immunoprecipitated from untreated and arsenite-treated cells, gel purified, and excised for trypsin digestion. The resulting peptide fragments were analyzed by liquid chromatography and mass spectrometry (LC-MS) using an LTQ ion-trap mass spectrometer equipped with a nano LC electrospray ionization (ESI) source to determine arginine methylation of G3BP1 peptides. Approximately 85% of total G3BP1 peptide coverage was achieved in each independent experiment.
Immunofluorescence Assay
Microscopy was performed essentially as described previously (4). Primary antibodies were incubated overnight at 4 °C. Primary antibodies used were: anti-PRMT1 (Cell Signaling, 2449), anti-PRMT5 (Abcam, ab109451), anti-G3BP1 (10), anti-Tia1 (Santa Cruz Biotechnology SC11386), anti-HuR (Santa Cruz Biotechnology SC-5261), anti-TDRD3 (Santa Cruz Biotechnology, SC-84626). All secondary antibodies (Molecular Probes) were used at 1:1,000 for 30 min at 25 °C. All images were taken with a Zeiss LSM 710 or 880 with 63× oil immersion objective equipped with Zen software, and processed with Image J and Adobe Photoshop CS4.
Immunoblots
Protein lysates (50 μg) were separated and transferred according to standard procedures. Membranes were blocked with Sea Block (Thermo Fisher Scientific) for 30 min at 25 °C then incubated with primary antibodies overnight at 4 °C or 3 h at 25 °C. Fluorescent conjugated secondary antibodies anti-mouse IgG (Dylight 680, 5470) and anti-rabbit IgG (Dylight 800, 5151) (Cell Signaling) were incubated for 30 min. Primary antibodies used herein not listed under the immunofluorescence heading above were: anti-G3BP1 from Abcam (ab181149 and ab181150), anti-phospho-eIF2α (Cell Signaling, 3597), anti-GFP (Santa Cruz Biotechnology SC-9996), anti-GAPDH (Millipore MAB374), anti-ADMA (EpiCypher, 12-0011), and anti-SDMA (EpiCypher, 13-0012). Signals were detected using an Odyssey CLx (LI-COR). The relative levels of protein expression were normalized against GAPDH.
Immunoprecipitation
U2OS or HeLa cells were harvested in RIPA buffer then quantified by a bicinchoninic acid (BCA) assay (Thermo Fisher Scientific). 1 mg of cleared protein lysates from each condition were diluted in 60% RIPA, 40% NETN buffer, then incubated with monoclonal primary antibody against G3BP1 (Abcam, 1:100) for 18 h at 4 °C. Protein A-Sepharose beads (Thermo Fisher Scientific) were equilibrated with NETN buffer and 40 μl of beads were added into each reaction for 1 h at 4 °C. Samples were washed with 1 ml of NETN buffer 5 times and eluted with 2× Laemmli sample buffer (Bio-Rad).
Statistical Analysis
All data are expressed as mean ± S.D. and compared between groups using the Student's t test. p value <0.05 was considered statistically significant. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Author Contributions
Conceptualization, WC. T., M. T. B., and R. E. L.; Conducting experiments and design, WC. T, S. G, M. T. B, and R. E. L; Reagents and discussions, L. C. R.; Compound synthesis, G. S.; Writing and Editing, WC. T, G. S, M. T. B., and R. E. L.; Funding Acquisition, M. T. B. and R. E. L.
Supplementary Material
Acknowledgments
We thank Sung Yun Jung and Antrix Jain of the BCM Mass Spectrometry-Proteomic Core for technical support. We further thank Phil Jones for the kind gift of MS023 ADMA inhibitor.
This work was funded by National Institutes of Health Public Health Service Grant AI50237 and NCI Cancer Center Support Grant (P30CA125123). Additional support was provided by the Integrated Microscopy Core at Baylor College of Medicine with funding from the National Institutes of Health (HD007495, DK56338, and CA125123), the Dan L. Duncan Cancer Center, and the John S. Dunn Gulf Coast Consortium for Chemical Genomics. Work in the laboratory of Mark T. Bedford is supported by National Institutes of Health funding (DK062248). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.

This article contains supplemental Figs. S1–4.
- SG
- stress granule
- G3BP1
- RasGAP-SH3-binding protein1
- PRMT
- protein arginine methyltransferase
- ADMA
- asymmetric dimethylarginine
- SDMA
- symmetric diemethylarginine.
References
- 1. Kedersha N. L., Gupta M., Li W., Miller I., and Anderson P. (1999) RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2α to the assembly of mammalian stress granules. J. Cell Biol. 147, 1431–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kedersha N., Chen S., Gilks N., Li W., Miller I. J., Stahl J., and Anderson P. (2002) Evidence that ternary complex (eIF2-GTP-tRNA(i)(Met))-deficient preinitiation complexes are core constituents of mammalian stress granules. Mol. Biol. Cell. 13, 195–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kedersha N., Stoecklin G., Ayodele M., Yacono P., Lykke-Andersen J., Fritzler M. J., Scheuner D., Kaufman R. J., Golan D. E., and Anderson P. (2005) Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 169, 871–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Reineke L. C., Dougherty J. D., Pierre P., and Lloyd R. E. (2012) Large G3BP-induced granules trigger eIF2α phosphorylation. Mol. Biol. Cell. 23, 3499–3510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tourrière H., Chebli K., Zekri L., Courselaud B., Blanchard J. M., Bertrand E., and Tazi J. (2003) The RasGAP-associated endoribonuclease G3BP assembles stress granules. J. Cell Biol. 160, 823–831 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 6. Mcdonald K. K., Aulas A., Destroismaisons L., Pickles S., Beleac E., Camu W., Rouleau G. A., and Vande Velde C. (2011) TAR DNA-binding protein 43 (TDP-43) regulates stress granule dynamics via differential regulation of G3BP and TIA-1. Hum. Mol. Genet. 20, 1400–1410 [DOI] [PubMed] [Google Scholar]
- 7. Kimball S. R., Horetsky R. L., Ron D., Jefferson L. S., and Harding H. P. (2003) Mammalian stress granules represent sites of accumulation of stalled translation initiation complexes. Am. J. Physiol., Cell Physiol. 284, C273—C284 [DOI] [PubMed] [Google Scholar]
- 8. Anderson P., and Kedersha N. (2006) RNA granules. J. Cell Biol. 172, 803–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jain S., Wheeler J. R., Walters R. W., Agrawal A., Barsic A., and Parker R. (2016) ATPase-modulated stress granules contain a diverse proteome and substructure. Cell 164, 487–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. White J. P., Cardenas A. M., Marissen W. E., and Lloyd R. E. (2007) Inhibition of cytoplasmic mRNA stress granule formation by a viral proteinase. Cell Host Microbe 2, 295–305 [DOI] [PubMed] [Google Scholar]
- 11. Reineke L. C., and Lloyd R. E. (2013) Diversion of stress granules and P-bodies during viral infection. Virology 436, 255–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kedersha N., and Anderson P. (2002) Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem. Soc. Trans. 30, 963–969 [DOI] [PubMed] [Google Scholar]
- 13. Reineke L. C., Kedersha N., Langereis M. A., van Kuppeveld F. J. M., and Lloyd R. E. (2015) Stress granules regulate double-stranded RNA-dependent protein kinase activation through a complex containing G3BP1 and caprin1. MBio. 6, e02486–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Eisinger-Mathason T. S. K., Andrade J., Groehler A. L., Clark D. E., Muratore-Schroeder T. L., Pasic L., Smith J. A., Shabanowitz J., Hunt D. F., Macara I. G., and Lannigan D. A. (2008) Codependent functions of RSK2 and the apoptosis-promoting factor TIA-1 in stress granule assembly and cell survival. Mol Cell. 31, 722–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Takahashi M., Higuchi M., Matsuki H., Yoshita M., Ohsawa T., Oie M., and Fujii M. (2013) Stress granules inhibit apoptosis by reducing reactive oxygen species production. Mol. Cell. Biol. 33, 815–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kobayashi T., Winslow S., Sunesson L., Hellman U., and Larsson C. (2012) PKCα binds G3BP2 and regulates stress granule formation following cellular stress. PLoS ONE 7, e35820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Brown J. A. L., Roberts T. L., Richards R., Woods R., Birrell G., Lim Y. C., Ohno S., Yamashita A., Abraham R. T., Gueven N., and Lavin M. F. (2011) A novel role for hSMG-1 in stress granule formation. Mol. Cell. Biol. 31, 4417–4429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Johnson M. E., Grassetti A. V., Taroni J. N., Lyons S. M., Schweppe D., Gordon J. K., Spiera R. F., Lafyatis R., Anderson P. J., Gerber S. A., and Whitfield M. L. (2016) Stress granules and RNA processing bodies are novel autoantibody targets in systemic sclerosis. Arthritis Res. Ther. 18, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bosco D. A., Lemay N., Ko H. K., Zhou H., Burke C., Kwiatkowski T. J., Sapp P., McKenna-Yasek D., Brown R. H., and Hayward L. J. (2010) Mutant FUS proteins that cause amyotrophic lateral sclerosis incorporate into stress granules. Hum. Mol. Genet. 19, 4160–4175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gal J., Zhang J., Kwinter D. M., Zhai J., Jia H., Jia J., and Zhu H. (2010) Nuclear localization sequence of FUS and induction of stress granules by ALS mutants. Neurobiology of Aging 32, e27–e40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kedersha N., Ivanov P., and Anderson P. (2013) Stress granules and cell signaling: more than just a passing phase? Trends Biochem. Sci. 38, 494–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Anderson P., and Kedersha N. (2008) Stress granules: the Tao of RNA triage. Trends Biochem. Sci. 33, 141–150 [DOI] [PubMed] [Google Scholar]
- 23. Gilks N., Kedersha N., Ayodele M., Shen L., Stoecklin G., Dember L. M., and Anderson P. (2004) Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol. Biol. Cell. 15, 5383–5398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kato M., Han T. W., Xie S., Shi K., Du X., Wu L. C., Mirzaei H., Goldsmith E. J., Longgood J., Pei J., Grishin N. V., Frantz D. E., Schneider J. W., Chen S., Li L., Sawaya M. R., Eisenberg D., Tycko R., and McKnight S. L. (2012) Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell 149, 753–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Decker C. J., Teixeira D., and Parker R. (2007) Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J. Cell Biol. 179, 437–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Reijns M. A. M., Alexander R. D., Spiller M. P., and Beggs J. D. (2008) A role for Q/N-rich aggregation-prone regions in P-body localization. J. Cell Sci. 121, 2463–2472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Han T. W., Kato M., Xie S., Wu L. C., Mirzaei H., Pei J., Chen M., Xie Y., Allen J., Xiao G., and McKnight S. L. (2012) Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell 149, 768–779 [DOI] [PubMed] [Google Scholar]
- 28. Elbaum-Garfinkle S., Kim Y., Szczepaniak K., Chen C. C.-H., Eckmann C. R., Myong S., and Brangwynne C. P. (2015) The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc. Natl. Acad. Sci. U.S.A. 112, 7189–7194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nott T. J., Petsalaki E., Farber P., Jervis D., Fussner E., Plochowietz A., Craggs T. D., Bazett-Jones D. P., Pawson T., Forman-Kay J. D., and Baldwin A. J. (2015) Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol. Cell 57, 936–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lin Y., Protter D. S., Rosen M. K., and Parker R. (2015) Formation and maturation of phase-separated liquid droplets by RNA-binding proteins. Mol. Cell 60, 208–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Molliex A., Temirov J., Lee J., Coughlin M., Kanagaraj A. P., Kim H. J., Mittag T., and Taylor J. P. (2015) Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163, 123–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Stoecklin G., Stubbs T., Kedersha N., Wax S., Rigby W. F. C., Blackwell T. K., and Anderson P. (2004) MK2-induced tristetraprolin:14–3-3 complexes prevent stress granule association and ARE-mRNA decay. EMBO J. 23, 1313–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Courchet J., Buchet-Poyau K., Potemski A., Brès A., Jariel-Encontre I., and Billaud M. (2008) Interaction with 14–3-3 adaptors regulates the sorting of hMex-3B RNA-binding protein to distinct classes of RNA granules. J. Biol. Chem. 283, 32131–32142 [DOI] [PubMed] [Google Scholar]
- 34. Leung A. K. L., Vyas S., Rood J. E., Bhutkar A., Sharp P. A., and Chang P. (2011) Poly(ADP-Ribose) regulates stress responses and microRNA activity in the cytoplasm. Mol. Cell 42, 489–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kwon S., Zhang Y., and Matthias P. (2007) The deacetylase HDAC6 is a novel critical component of stress granules involved in the stress response. Genes Dev. 21, 3381–3394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ohn T., Kedersha N., Hickman T., Tisdale S., and Anderson P. (2008) A functional RNAi screen links O-GlcNAc modification of ribosomal proteins to stress granule and processing body assembly. Nat. Cell Biol. 10, 1224–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. De Leeuw F., Zhang T., Wauquier C., Huez G., Kruys V., and Gueydan C. (2007) The cold-inducible RNA-binding protein migrates from the nucleus to cytoplasmic stress granules by a methylation-dependent mechanism and acts as a translational repressor. Exp. Cell Res. 313, 4130–4144 [DOI] [PubMed] [Google Scholar]
- 38. Kryukov G. V., Wilson F. H., Ruth J. R., Paulk J., Tsherniak A., Marlow S. E., Vazquez F., Weir B. A., Fitzgerald M. E., Tanaka M., Bielski C. M., Scott J. M., Dennis C., Cowley G. S., Boehm J. S., Root D. E., Golub T. R., Clish C. B., Bradner J. E., Hahn W. C., and Garraway L. A. (2016) MTAP deletion confers enhanced dependency on the PRMT5 arginine methyltransferase in cancer cells. Science 351, 1214–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bedford M. T., and Clarke S. G. (2009) Protein arginine methylation in mammals: who, what, and why. Mol. Cell 33, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ong S.-E., Mittler G., and Mann M. (2004) Identifying and quantifying in vivo methylation sites by heavy methyl SILAC. Nature Methods 1, 119–126 [DOI] [PubMed] [Google Scholar]
- 41. Goulet I., Boisvenue S., Mokas S., Mazroui R., and Côté J. (2008) TDRD3, a novel Tudor domain-containing protein, localizes to cytoplasmic stress granules. Hum. Mol. Genet. 17, 3055–3074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Linder B., Plöttner O., Kroiss M., Hartmann E., Laggerbauer B., Meister G., Keidel E., and Fischer U. (2008) Tdrd3 is a novel stress granule-associated protein interacting with the Fragile-X syndrome protein FMRP. Hum. Mol. Genet. 17, 3236–3246 [DOI] [PubMed] [Google Scholar]
- 43. Mavrakis K. J., McDonald E. R. 3rd, Schlabach M. R., Billy E., Hoffman G. R., deWeck A., Ruddy D. A., Venkatesan K., Yu J., McAllister G., Stump M., deBeaumont R., Ho S., Yue Y., Liu Y., Yan-Neale Y., Yang G., Lin F., Yin H., Gao H., Kipp D. R., Zhao S., McNamara J. T., Sprague E. R., Zheng B., Lin Y., Cho Y. S., Gu J., Crawford K., Ciccone D., Vitari A. C., Lai A., Capka V., Hurov K., Porter J. A., Tallarico J., Mickanin C., Lees E., Pagliarini R., Keen N., Schmelzle T., Hofmann F., Stegmeier F., and Sellers W. R. (2016) Disordered methionine metabolism in MTAP/CDKN2A-deleted cancers leads to dependence on PRMT5. Science 351, 1208–1213 [DOI] [PubMed] [Google Scholar]
- 44. Yang Y., and Bedford M. T. (2013) Protein arginine methyltransferases and cancer. Nat. Rev. Cancer 13, 37–50 [DOI] [PubMed] [Google Scholar]
- 45. Fuhrmann J., Clancy K. W., and Thompson P. R. (2015) Chemical biology of protein arginine modifications in epigenetic regulation. Chem. Rev. 115, 5413–5461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Morales Y., Cáceres T., May K., and Hevel J. M. (2016) Biochemistry and regulation of the protein arginine methyltransferases (PRMTs). Arch. Biochem. Biophys. 590, 138–152 [DOI] [PubMed] [Google Scholar]
- 47. Chen C., Nott T. J., Jin J., and Pawson T. (2011) Deciphering arginine methylation: Tudor tells the tale. Nat. Rev. Mol. Cell Biol. 12, 629–642 [DOI] [PubMed] [Google Scholar]
- 48. Mowen K. A., Schurter B. T., Fathman J. W., David M., and Glimcher L. H. (2004) Arginine methylation of NIP45 modulates cytokine gene expression in effector T lymphocytes. Mol. Cell 15, 559–571 [DOI] [PubMed] [Google Scholar]
- 49. Lee Y.-H., and Stallcup M. R. (2011) Roles of protein arginine methylation in DNA damage signaling pathways is CARM1 a life-or-death decision point? Cell Cycle 10, 1343–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tradewell M. L., Yu Z., Tibshirani M., Boulanger M.-C., Durham H. D., and Richard S. (2012) Arginine methylation by PRMT1 regulates nuclear-cytoplasmic localization and toxicity of FUS/TLS harbouring ALS-linked mutations. Hum. Mol. Genet. 21, 136–149 [DOI] [PubMed] [Google Scholar]
- 51. Yamaguchi A., and Kitajo K. (2012) The effect of PRMT1-mediated arginine methylation on the subcellular localization, stress granules, and detergent-insoluble aggregates of FUS/TLS. PLoS ONE 7, e49267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kaehler C., Guenther A., Uhlich A., and Krobitsch S. (2015) PRMT1-mediated arginine methylation controls ATXN2L localization. Exp. Cell Res. 334, 114–125 [DOI] [PubMed] [Google Scholar]
- 53. Dolzhanskaya N. (2006) Methylation regulates the intracellular protein-protein and protein-RNA interactions of FMRP. J. Cell Sci. 119, 1933–1946 [DOI] [PubMed] [Google Scholar]
- 54. Bikkavilli R. K., and Malbon C. C. (2011) Arginine methylation of G3BP1 in response to Wnt3a regulates β-catenin mRNA. J. Cell Sci. 124, 2310–2320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Arimoto-Matsuzaki K., Saito H., and Takekawa M. (2016) TIA1 oxidation inhibits stress granule assembly and sensitizes cells to stress-induced apoptosis. Nat. Commun. 7, 10252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Thedieck K., Holzwarth B., Prentzell M. T., Boehlke C., Kläsener K., Ruf S., Sonntag A. G., Maerz L., Grellscheid S.-N., Kremmer E., Nitschke R., Kuehn E. W., Jonker J. W., Groen A. K., Reth M., Hall M. N., and Baumeister R. (2013) Inhibition of mTORC1 by astrin and stress granules prevents apoptosis in cancer cells. Cell 154, 859–874 [DOI] [PubMed] [Google Scholar]
- 57. Arimoto K., Fukuda H., Imajoh-Ohmi S., Saito H., and Takekawa M. (2008) Formation of stress granules inhibits apoptosis by suppressing stress-responsive MAPK pathways. Nat. Cell Biol. 10, 1324–1332 [DOI] [PubMed] [Google Scholar]
- 58. Li Y. R., King O. D., Shorter J., and Gitler A. D. (2013) Stress granules as crucibles of ALS pathogenesis. J. Cell Biol. 201, 361–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lee Y.-J., Wei H.-M., Chen L.-Y., and Li C. (2014) Localization of SERBP1 in stress granules and nucleoli. FEBS J. 281, 352–364 [DOI] [PubMed] [Google Scholar]
- 60. Lee J., Sayegh J., Daniel J., Clarke S., and Bedford M. T. (2005) PRMT8, a new membrane-bound tissue-specific member of the protein arginine methyltransferase family. J. Biol. Chem. 280, 32890–32896 [DOI] [PubMed] [Google Scholar]
- 61. Kedersha N., Panas M. D., Achorn C. A., Lyons S., Tisdale S., Hickman T., Thomas M., Lieberman J., McInerney G. M., Ivanov P., and Anderson P. (2016) G3BP-Caprin1-USP10 complexes mediate stress granule condensation and associate with 40S subunits. J. Cell Biol. 212, 845–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Aulas A., Stabile S., and Vande Velde C. (2012) Endogenous TDP-43, but not FUS, contributes to stress granule assembly via G3BP. Mol. Neurodegener. 7, 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Aulas A., Caron G., Gkogkas C. G., Mohamed N.-V., Destroismaisons L., Sonenberg N., Leclerc N., Parker J. A., and Vande Velde C. (2015) G3BP1 promotes stress-induced RNA granule interactions to preserve polyadenylated mRNA. J. Cell Biol. 209, 73–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sahoo P. K., Murawala P., Sawale P. T., Sahoo M. R., Tripathi M. M., Gaikwad S. R., Seshadri V., and Joseph J. (2012) Wnt signalling antagonizes stress granule assembly through a Dishevelled-dependent mechanism. Biol. Open. 1, 109–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kozubowski L., Aboobakar E. F., Cardenas M. E., and Heitman J. (2011) Calcineurin colocalizes with P-bodies and stress granules during thermal stress in Cryptococcus neoformans. Eukaryotic Cell 10, 1396–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Chang B., Chen Y., Zhao Y., and Bruick R. K. (2007) JMJD6 is a histone arginine demethylase. Science 318, 444–447 [DOI] [PubMed] [Google Scholar]
- 67. Poulard C., Rambaud J., Hussein N., Corbo L., and Le Romancer M. (2014) JMJD6 regulates ERα methylation on arginine. PLoS ONE 9, e87982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Shi Y., Lan F., Matson C., Mulligan P., Whetstine J. R., Cole P. A., Casero R. A., and Shi Y. (2004) Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119, 941–953 [DOI] [PubMed] [Google Scholar]
- 69. Tsukada Y.-I., Fang J., Erdjument-Bromage H., Warren M. E., Borchers C. H., Tempst P., and Zhang Y. (2006) Histone demethylation by a family of JmjC domain-containing proteins. Nature 439, 811–816 [DOI] [PubMed] [Google Scholar]
- 70. Böttger A., Islam M. S., Chowdhury R., Schofield C. J., and Wolf A. (2015) The oxygenase Jmjd6–a case study in conflicting assignments. Biochem. J. 468, 191–202 [DOI] [PubMed] [Google Scholar]
- 71. Alahari S., Post M., and Caniggia I. (2015) Jumonji domain containing protein 6: A novel oxygen sensor in the human placenta. Endocrinology 156, 3012–3025 [DOI] [PubMed] [Google Scholar]
- 72. Chan-Penebre E., Kuplast K. G., Majer C. R., Boriack-Sjodin P. A., Wigle T. J., Johnston L. D., Rioux N., Munchhof M. J., Jin L., Jacques S. L., West K. A., Lingaraj T., Stickland K., Ribich S. A., Raimondi A., Scott M. P., Waters N. J., Pollock R. M., Smith J. J., Barbash O., Pappalardi M., Ho T. F., Nurse K., Oza K. P., Gallagher K. T., Kruger R., Moyer M. P., Copeland R. A., Chesworth R., and Duncan K. W. (2015) A selective inhibitor of PRMT5 with in vivo and in vitro potency in MCL models. Nat. Chem. Biol. 11, 432–437 [DOI] [PubMed] [Google Scholar]
- 73. Eram M. S., Shen Y., Szewczyk M. M., Wu H., Senisterra G., Li F., Butler K. V., Kaniskan H. Ü., Speed B. A., dela Seña C., Dong A., Zeng H., Schapira M., Brown P. J., Arrowsmith C. H., Barsyte-Lovejoy D., Liu J., Vedadi M., and Jin J. (2016) A potent, selective, and cell-active inhibitor of human Type I protein arginine methyltransferases. ACS Chem. Biol. 11, 772–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Yang Y., Hadjikyriacou A., Xia Z., Gayatri S., Kim D., Zurita-Lopez C., Kelly R., Guo A., Li W., Clarke S. G., and Bedford M. T. (2015) PRMT9 is a type II methyltransferase that methylates the splicing factor SAP145. Nat. Commun. 6, 6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Cheng D., Vemulapalli V., and Bedford M. T. (2012) Methods applied to the study of protein arginine methylation. Methods Enzymol. 512, 71–92 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.