Abstract
We describe here the identification and functional characterization of the enzyme O-GlcNAcase (OGA) as an RNA polymerase II elongation factor. Using in vitro transcription elongation assays, we show that OGA activity is required for elongation in a crude nuclear extract system, whereas in a purified system devoid of OGA the addition of rOGA inhibited elongation. Furthermore, OGA is physically associated with the known RNA polymerase II (pol II) pausing/elongation factors SPT5 and TRIM28-KAP1-TIF1β, and a purified OGA-SPT5-TIF1β complex has elongation properties. Lastly, ChIP-seq experiments show that OGA maps to the transcriptional start site/5′ ends of genes, showing considerable overlap with RNA pol II, SPT5, TRIM28-KAP1-TIF1β, and O-GlcNAc itself. These data all point to OGA as a component of the RNA pol II elongation machinery regulating elongation genome-wide. Our results add a novel and unexpected dimension to the regulation of elongation by the insertion of O-GlcNAc cycling into the pol II elongation regulatory dynamics.
Keywords: metabolism, O-GlcNAcylation, RNA polymerase II, transcription, transcription elongation factor
Introduction
RNA polymerase II is the species of polymerase that transcribes the protein-coding genes in the cell. Largely based on in vitro transcription data and bacterial transcriptional regulation, it was thought that the pathway of transcription initiation was the de novo assembly and recruitment of general factors and pol II4 into a preinitiation complex at promoters. In other words, transcription was solely controlled by whether initiation occurred or not. However, in the late 1980s, a transcriptionally engaged pol II was found on several genes, located roughly at +50 relative to the transcriptional start site (TSS) (1–3). More recently, genome-wide approaches have shown that paused pol II exists on at least 40% of promoters in the genome (4–9). These data show that the regulation of elongation is a significant and widespread method by which cells regulate gene expression.
Biochemically, the establishment of a paused polymerase requires the recruitment of DSIF and NELF to pol II early in elongation to prevent pol II from productive elongation (10–13). Release of the paused polymerase is achieved with P-TEFb phosphorylation of DSIF and NELF, ejecting NELF from pol II and converting DSIF into a positive elongation factor (14, 15). The more recent discoveries of additional factors likely involved in pause establishment and release include human capping enzyme (16), the TFIIH-associated kinase, CDK7 (17, 18), the TFIIH ERCC3 helicase (19), Mediator (20–22), Integrator (23, 24), ELL (25), TFIIS (26), TRIM28-KAP1-TIF1β (27, 28), Top1 (29), SEC (30), PAF complex (31, 32), and Gdown1 (33). The plethora of factors suggests that more complicated dynamics are at play in regulating pausing and elongation. Additionally, it is not clear, and indeed difficult to know, whether all of the factors involved in pol II pausing and elongation have been identified. This difficulty is due mostly to the absence of a human cell-based in vitro transcription system that recapitulates a paused polymerase and with which the full complement of factors can be functionally identified. Lastly, it is not clear how all of these factors integrate into a unified mechanistic and regulatory model of pol II pausing.
The requirements for serine/threonine phosphorylation in pausing release imply that O-GlcNAcylation might also be a participatory factor; the O-GlcNAcylation of proteins is a very common post-translational modification, occurring on serine and threonine residues, and is often mutually exclusive with the phosphorylation of those same residues (34). O-GlcNAc transferase (OGT) utilizes UDP-GlcNAc as a high energy donor substrate and the β-N-acetylglucosaminidase or O-GlcNAcase (OGA) removes GlcNAc from protein substrates (35–37). Physiologically, O-GlcNAcylation likely plays significant causal roles in type 2 diabetes and cancer. OGA is essential for tumor cell viability and metabolic homeostasis (38–40). Additionally, OGT knock-out mice are embryonic lethal, and brain-specific OGT knock-out disrupts eating behavior in mice (41, 42), underscoring both the essential nature of OGT and the nutrient-sensing properties of the O-GlcNAcylation regulatory space. Lastly, ∼2–5% of glucose flux into the cell is diverted to the hexosamine biosynthetic pathway for the synthesis of UDP-GlcNAc, and it has been hypothesized then that UDP-GlcNAc levels represent a nutrient sensor and reflect the intracellular glucose concentration (35).
We describe here the unexpected involvement of O-GlcNAcylation in RNA pol II pausing and elongation. We had previously found requirements for both OGT and OGA in transcription in vitro (43), although the point at which the enzymes acted was not determined. Here we examined OGA function further using in vitro transcription assays. In vitro elongation assays showed a requirement for OGA during elongation in crude nuclear extracts. Secondly, we identified an ∼600-kDa complex that contains OGA and the pausing/elongation factors SPT5 and TRIM28-KAP1-TIF1β (27, 28). In vivo ChIP-seq analysis shows that OGA localizes to TSS and OGA peaks overlap considerably with pol II, SPT5, and TRIM28-KAP1-TIF1β peaks. These data suggest a novel role for OGA activity in the regulation of RNA pol II pausing and elongation.
Results
OGA Activity Is Required for Efficient Elongation in Vitro
Previous results showed that chemical inhibition of OGA blocked RNA pol II-dependent transcription in vitro (43). However, it was not clear exactly when in the transcription process OGA activity was required. A hint came from our recent study of the effects of OGT and OGA inhibitors on PIC formation and pol II promoter recruitment (44). The absence of an effect of the OGA inhibitor PUGNAc suggested that the inhibition of transcription might occur after PIC formation, thus implicating early elongation steps as being OGA-dependent.
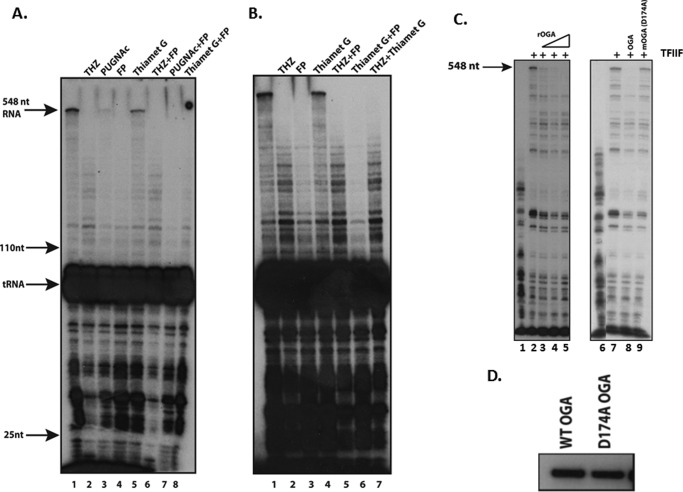
To address a possible OGA function during elongation, we adopted the elongation assay established by Price and co-workers (46). We used nuclear extracts made from HeLa cells (45) and assayed transcription activity using the CMV promoter and a pulse-chase assay (46). We used this assay to test various kinase and OGA inhibitors specifically for effects on elongation by adding the inhibitor(s) after allowing PIC formation to occur for 30 min. Inhibitors were then added for 10 min followed by NTPs to initiate transcription. We found as expected a full-length 548-nucleotide run-off transcript using untreated HeLa nuclear extracts (Fig. 1A). We next tested inhibitors of TFIIH CDK7 kinase activity (THZ) and the P-TEFb CDK9 inhibitor flavopiridol (FP) in our assay (47, 48). Both THZ and flavopiridol have been shown previously to block transcription elongation in vitro (48, 49). Indeed, both inhibitors blocked elongation in our assay (Fig. 1A). We next tested two OGA inhibitors: PUGNAc and Thiamet G. PUGNAc is considered a more general hexosaminidase inhibitor and can inhibit, for example, lysosomal hexosaminidases as well as OGA (however, due to the use of a nuclear extract and the neutral pH of the transcription assay, we do not expect any lysosomal hexosaminidase activity), whereas Thiamet-G is specific for OGA (50–52). Both inhibited transcription elongation of the full-length product but not smaller (<75 nucleotides) RNAs (Fig. 1A). These data show that OGA activity in vitro is required for efficient elongation in crude nuclear extracts.
FIGURE 1.
OGA is a positive elongation factor in crude nuclear extracts and is required for efficient elongation. A, lanes 1–5, the addition of hexosaminidase inhibitor PUGNAc and OGA-specific inhibitor Thiamet G (after PIC formation) both block efficient elongation in crude nuclear extract elongation assays. Shown are pulse-chase elongation assays using a CMV promoter template. Transcription was initiated by the addition of [32P]CTP/GAT for 30 s and chased for 5′ with 1.2 mm cold CTP (46). Labeled RNA was isolated and separated on an 8% urea, Tris borate-EDTA polyacrylamide gel. The full-length labeled RNA run-off product of 548 nucleotides is indicated by the arrow. THZ is an inhibitor of the CDK7 kinase subunit of TFIIH, and FP is a P-TEFb inhibitor (47, 48). PUGNAc and Thiamet G were added after a 30 h PIC formation step as our previous work suggested that OGA might be required for PIC formation (43). THZ and FP were added either concomitantly with PIC formation or afterward (as with PUGNAc and OGA). Lanes 6–8, epistatic assays indicate that OGA acts after P-TEFb and CDK7/TFIIH. In vitro transcriptions were done as in A, but in addition to assaying THZ, FP, PUGNAc, or Thiamet G separately, inhibitors were paired to assay for a temporal defect. nt, nucleotides. B, in a separate experiment, done as in panel A, we also assessed the epistatic relationship between THZ and Thiamet G (in addition to a THZ/FP and Thiamet G/FP assay). Note the identical effects of THZ, whether assayed by itself or in combination with FP or Thiamet G (compare lanes 2 (THZ), 5 (THZ+FP), and 7 (THZ+Thiamet G) to the effect of FP or Thiamet G in lanes 3 and 4, respectively). C, rOGA inhibits RNA pol II elongation on immobilized templates. PICs were formed on immobilized templates followed by a 30-s pulse of [32P]CTP and 0.5 mm GTP/ATP/UTP. Beads were washed and then incubated with or without rTFIIF and wild type or mutant OGA (mOGA; D174A) as indicated. This incubation step was followed by a 7-min incubation with 0.5 mm NTPs. Labeled RNA was isolated and run on an 8% polyacrylamide/urea/Tris borate-EDTA gel. D, Western blots of recombinant wild type or mutant Asp-174 catalytically inactive OGA showing that equal amounts were added to the transcription assays in panel C.
Because TFIIH, P-TEFb, and OGA activities are required for elongation (Fig. 1A), we asked whether one acted before the other or whether their activities were additive or synergistic. To do this, we compared the P-TEFb or OGA inhibitors separately (as in Fig. 1A) to the effects of mixing both inhibitors together. If the effects of the drugs are temporally distinct, then mixing inhibitors A and B together would have the same effect as either A or B by itself. For example, if the mixing of A and B has the same effect as A, then we would conclude that A acts before B. Conversely, an additive/synergistic effect from mixing A and B would appear as an elongation defect more severe than observed for either A or B by itself. Our results are shown in Fig. 1A, lanes 6–8. The combination of THZ and FP showed a THZ effect, indicating that CDK7 acted before P-TEFb (49) (compare lanes 2 and 4 to lane 6 in Fig. 1A; see similar results in Fig. 1B). Both OGA inhibitor (PUGNAc and Thiamet G) combinations with FP showed FP effects and not PUGNAc or Thiamet G effects, indicating that P-TEFb acts before OGA in the elongation process (Fig. 1A, compare lanes 3, 4, and 5 to lanes 7 and 8). We also assayed the combination of THZ and Thiamet G (Fig. 1B). Strikingly, THZ effects are predominant to those of Thiamet G (compare lanes 2 and 3 to lanes 5 and 7, respectively). Overall, these experiments indicate that TFIIH/CDK7 acts first, P-TEFb acts second, and OGA acts third during the elongation process, and equally importantly, they do not appear to be acting in any sort of additive or synergistic way, suggesting each enzyme represents a distinct step in the elongation process.
We next asked whether OGA protein could affect elongation. To that end, we purified recombinant OGA from bacterial extracts via a His tag and titrated rOGA into an elongation assay designed to specifically measure later elongation events. PICs were formed on immobilized CMV templates and given a 30-s pulse of [32P]CTP/cold GTP/UTP/ATP. Elongation was stopped with EDTA, and complexes were washed. The elongation was then chased with cold NTPs. A representative elongation product after the cold NTP chase is shown in Fig. 1C, lane 1. TFIIF is an established initiation and elongation factor (54–56), and we exploited its elongation properties by adding it to the chase step where it efficiently promoted elongation (compare lanes 1 and 2, Fig. 1C) (46). To this TFIIF-dependent elongation assay we titrated rOGA, and we found a clear reduction in elongation (lanes 3–5, Fig. 1C).
We then asked whether OGA catalytic activity was required for the block to elongation. We purified a catalytically inactive rOGA (D174A) (57) from bacteria and added identical amounts (Fig. 1D) of the wild type or mutant rOGA to the TFIIF-dependent elongation assay and found that the Asp-174 mutant had no effect on elongation (Fig. 1C, lanes 6–9). The data in Fig. 1 show that OGA can have both positive and negative elongation functions, depending on the assay used.
OGA Is Associated with the Pausing and Elongation Factor SPT5 and TRIM28-KAP1-TIF1β
The requirement for OGA activity in transcription elongation processes in vitro and in vivo suggested that OGA might interact with members of the elongation machinery. The now classic purification scheme of nuclear extracts for the components of the transcription machinery was done by first fractionating a nuclear extract over a P11 resin (58). This scheme fractionates TFIIA in the flow-through, DSIF and NELF in the 0.1 and 0.3 m fraction, RNA pol II, TFIIH, TFIIB, TFIIE, TFIIF, and Mediator in the 0.5 m fraction, and TFIID and some Mediator in the 1 m fraction (59). To determine OGT and OGA chromatographic behavior relative to the other transcriptional machinery, we ran a HeLa nuclear extract over a P11 resin and collected fractions from the flow-through, 0.1 m KCl elution, the 0.3 m, 0.5 m, and 1 m KCl step elutions. We then analyzed each step of elution for the presence of OGT and OGA by Western blot. We found that OGA eluted solely in the flow-through/0.1 m fraction (Fig. 2A). In contrast, OGT was distributed in all four of the KCl step elutions (Fig. 2A and Ref. (43). As evidence of the proper fractionation of the extract, RNA pol II was found predominantly in the 0.5 m KCl elution as expected (58).
FIGURE 2.
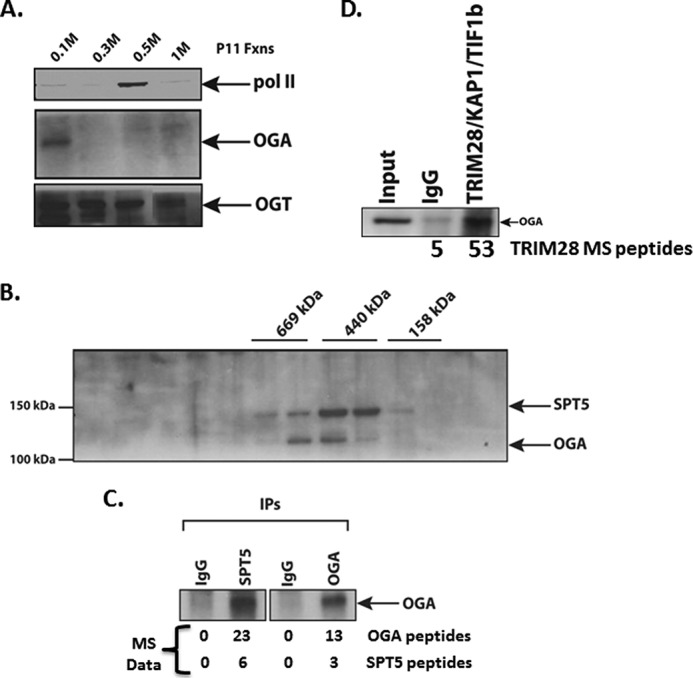
OGA is associated with the pausing and elongation factor SPT5. A, fractionation of HeLa nuclear extract over P11 resin showing the distribution of OGA, OGT, and RNA pol II in each of the 0.1 m flow-through, 0.3 m, 0.5 m, and 1 m KCl step elutions. Westerns blots of pol II, OGA, and OGT are shown. B, Superose 6 gel filtration analysis of P11 0.1 m fraction showing co-elution of the SPT5 subunit of DSIF and OGA. Molecular weight marker elutions (done separately) are indicated above the Western blot of SPT5 and OGA. C, co-immunoprecipitation (IPs) of either SPT5 or OGA from the 600-kDa Superose 6 fraction shows a direct interaction of SPT5 and OGA. Underneath are listed the numbers of OGA and SPT5 peptides identified by mass spectrometry analysis from each respective immunoprecipitation. D, TRIM28-KAP1-TIF1β is associated with OGA. Anti-TRIM28 antibody or a control rabbit IgG was incubated with a 0.1 m P11 fraction, and immunoprecipitates were collected on protein A-agarose beads, washed, and eluted in sample buffer, run on a 4–12% gradient SDS-PAGE gel, transferred overnight to nitrocellulose, and probed with an anti-OGA antibody. Listed below the Western blot are the number of TRIM28 peptides obtained by mass spectroscopy of an IgG control or OGA immunoprecipitation of a 0.1 m P11 fraction.
We next looked for the coelution of OGA with elongation factors that are present in the P11 flow-through fraction. The most obvious factor was DSIF, which was mentioned by Handa and co-workers (10, 12) as existing in both the 0.1 and 0.3 m P11 elutions. We separated our 0.1 m material over a Superose 6 gel filtration column and probed for both OGA and the SPT5 subunit of DSIF by Western blot. We found that these two factors coeluted at ∼500–600 kDa (Fig. 2B). We then assayed for an interaction between the 2 by immunoprecipitating the 500-kDa Superose 6 fraction with anti-SPT5 and anti-OGA antibodies and probing both immunoprecipitates for OGA. The anti-OGA immunoprecipitation precipitated OGA, as expected, whereas the SPT5 immunoprecipitation also precipitated OGA (Fig. 2C), demonstrating that they are found in the same complex, interacting either directly or indirectly. To confirm this interaction, we immunoprecipitated the Superose 6 fractions containing OGA and SPT5 and analyzed them by mass spectrometry. We found that the SPT5 antibody coimmunoprecipitated OGA and vice versa as indicated by the peptide numbers in Fig. 2C. These experiments show that OGA and SPT5 are physically interacting, thus connecting OGA to the elongation machinery.
In addition to confirming the interaction between OGA and SPT5, mass spectroscopy analysis indicated that the pausing/elongation factor TRIM28-KAP1-TIF1β (27, 28) also immunoprecipitated with OGA (Fig. 2D). To confirm this interaction, we were able to detect OGA by immunoprecipitating TRIM28-KAP1-TIF1β (hereafter referred to as TIF1β) from the P11 0.1 m fraction (Fig. 2D). Thus, we have multiple confirmations of the interaction of OGA with two RNA pol II factors that are known participants in elongation, SPT5 and TIF1β, consistent with an OGA-dependent elongation function.
An OGA-SPT5-TIF1β Complex Has Elongation Properties
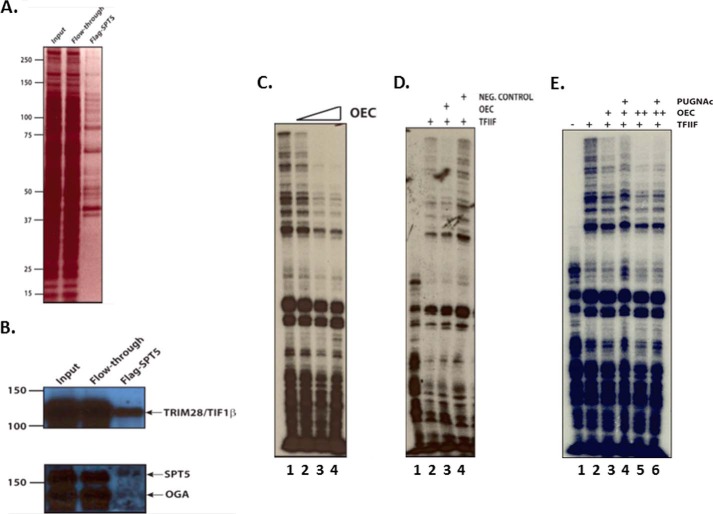
To further examine the relationship between OGA, SPT5, and TIF1β, we attempted to construct various FLAG-tagged cell lines. Although we were unable to establish tagged OGT and OGA cells lines, we were successful in creating a FLAG-SPT5 HeLa cell line. We grew ∼10 liters of the FLAG-SPT5 cell line, made a nuclear extract, and fractionated the extract over a P11 column. We then isolated FLAG-tagged SPT5 from the P11 0.1 m fraction using M2-agarose beads and eluted the complex from the resin with FLAG peptide (Fig. 3A). To confirm the content of the F-SPT5 complex, we immunoblotted the FLAG-tagged material with OGA, SPT5, and TIF1β antibodies and found that all three were present in the complex (Fig. 3B). Hereafter, we refer to this complex as the OGA elongation complex (OEC).
FIGURE 3.
OEC has elongation factor properties. A, purification of OEC. Shown is a silver-stained PAGE of FLAG-tagged SPT5/OEC as well as input and flow-through of the M2-agarose resin used to capture the FLAG epitope. B, OEC contains OGA, SPT5, and TIF1β. F-SPT5/OEC was run on SDS-PAGE and subjected to immunoblotting with the indicated antibodies on the right. C, OEC inhibits transcription elongation. F-SPT5/OEC was titrated into TFIIF-dependent elongation assays (see Fig. 1C and lanes 1 and 2 of panel D for examples of the assay). D, a non-FLAG eluate from M2-agarose does not inhibit elongation. An untagged P11 0.1 m fraction was incubated with M2-agarose and subsequently eluted with FLAG peptide. The resulting eluate was used as a control in the TFIIF elongation assays (compare lanes 3 and 4; lane 3 contains the F-SPT5/OEC activity). Equal amounts of protein input onto the M2 resin, and equal volumes of each eluate were used. E, OEC activity is not dependent on OGA catalytic activity. The OEC inhibition in the TFIIF-dependent elongation assay was assessed in the presence of the OGA inhibitor PUGNAc. Two different OEC concentrations were used (indicated by + and ++) and were incubated with PUGNAc before adding to the elongation assay, as in Fig. 3D.
We examined the OEC for elongation properties by titrating the OEC into the TFIIF-dependent elongation assay (Fig. 1C). We found that the OEC inhibited elongation of longer RNAs (>200 nucleotides) while having little effect on shorter RNAs (Fig. 3C). To confirm the specificity of this effect, we incubated a P11 0.1 m fraction that did not contain FLAG-SPT5 with M2-agarose and eluted bound material from that resin. The addition of that material (neg. control lane, Fig. 3D) had no effect on TFIIF-dependent elongation. These results indicate that the OEC has properties characteristic of a complex regulating pol II elongation.
Lastly, we asked whether OGA catalytic activity was required for OEC activity. We again used the TFIIF-dependent elongation assay and assayed two different OEC amounts, both of which inhibited elongation (compare lanes 2 and 3 and lanes 2 and 5, Fig. 3E). We then incubated each OEC amount with PUGNAc before adding it to the elongation assay (compare lanes 3 and 4 and lanes 5 and 6, Fig. 3E). In both cases, the PUGNAc did not change the level of elongation relative to the untreated OEC, indicating that OGA activity is not required for OEC activity in this assay.
OGA and O-GlcNAc Map to TSS/5′ Ends of genes in Vivo
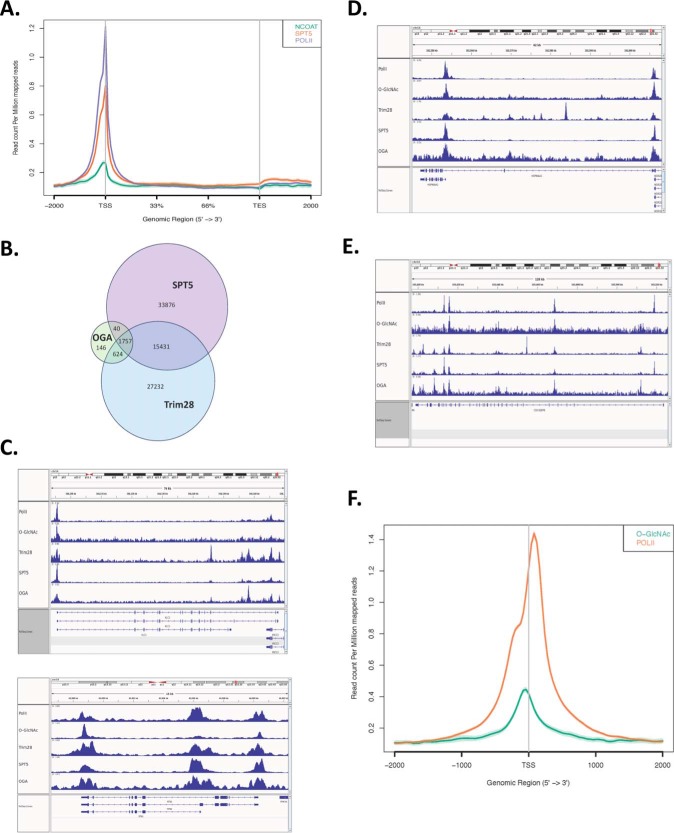
Using the ChIP-seq assay we looked in vivo at the distribution of OGA across the genome and its overlap with pol II, SPT5, and TIF1β. Remarkably, we found discrete peaks of OGA that mapped to TSSs in metagene analysis (Fig. 4A): 46% of OGA peaks overlapped with pol II (Table 1), 70% of OGA peaks overlapped with SPT5, and 93% of OGA peaks overlapped with TIF1β (Fig. 4B and Table 2). OGA overlapped with only 6% of promoter pol II (Table 3); however, of those overlaps, 100% also contained SPT5, and 91% contained SPT5 and TIF1β (Table 4). In conclusion, there is a clear partitioning of OGA, SPT5, and TIF1β to pol II promoter peaks. Visual inspection of individual genes, randomly selected, showed that OGA, pol II, SPT5, and TIF1β peaks all tightly correlated with each other (Fig. 4, C–F), including prominent peaks on paused genes such as the heat shock gene HSP90AA1 (Fig. 4C). These tight co-localizations between the three proteins are consistent with their biochemical interactions in Figs. 2 and 3.
FIGURE 4.
ChIP-seq analysis of OGA distribution genome-wide shows localization to promoters and sites of RNA pol II pausing. A, OGA occupies transcription start sties and the 5′ proximal ends of genes. Metagene analysis was performed on the OGA, pol II, and SPT5 ChIP-seq data. TES, transcription end site. B, peaks from MACS peak calling were aligned against each other, and the numbers of peak overlaps are indicated in the Venn diagram. Percent overlaps indicated in Tables 1–4 were then calculated. C, ChIP-seq peak distribution of pol II, O-GlcNAc, OGA, SPT5, and TRIM28/TIF1β on the hsp90AA1 gene. Note the overlaps of the three proteins at the two promoters. D, ChIP-seq peak distribution of pol II, O-GlcNAc, OGA, SPT5, and TRIM28/TIF1β on the KLC1 gene. Note the peak overlaps on the exon at the center of the diagram. E, ChIP-seq peak distribution of pol II, O-GlcNAc, OGA, SPT5, and TRIM28/TIF1β on the RTN2 gene. F, ChIP-seq peak distribution of pol II, O-GlcNAc, OGA, SPT5, and TRIM28/TIF1β on the CDC42BPB gene. As with panel C, note the OGA, SPT5, and pol II peaks in the gene body. G, Metagene analysis of pol II and O-GlcNAc ChIP-seq peak distributions relative to TSSs.
TABLE 1.
Peak overlaps of RNA pol II, O-GlcNAc, TIF1β, and OGA
X refers to the peaks listed in the left column. The number of overlaps are listed in the middle column, and the % of overlaps of each X species relative to the OGA peaks are in parentheses in the middle column. The right column lists the % of OGA peaks that overlap with the total peaks in left column.
| Peaks | X overlaps/total OGA peaks | OGA overlaps/X total |
|---|---|---|
| Total pol II (18,990) | 1183 (46.1%) | 6.2% |
| Total O-GlcNAc (12,801) | 621 (24.2%) | 4.9% |
| Total TIF1β (45,044) | 2306 (89.8%) | 5.1% |
| Total OGA (2,567) |
TABLE 2.
Overlaps between OGA, SPT5, and TIF1β peaks
Overlap % are listed as indicated on the top row. The absolute numbers are indicated in each cell, and total peaks are indicated in the bottom row.
| Peak overlaps | OGA/X total peaks | X/OGA total peaks | SPT5/X total peaks | X/SPT5 total peaks | TIF1β/X total peaks | X/TIF1β total peaks | SPT5+ TIF1β/OGA total peaks |
|---|---|---|---|---|---|---|---|
| OGA overlaps | 1797 (70%) | 1797 (3%) | 2381 (93%) | 2381 (5%) | 1757 (68%) | ||
| SPT5 overlaps | 1797 (3.2%) | 1797 (70%) | 17188 (30%) | 17188 (38%) | |||
| TIF1βoverlaps | 2381 (5%) | 2381 (93%) | 17188 (38%) | 17188 (30%) | |||
| Total OGA 2567 | Total SPT5 56104 | Total TIF1β 45044 |
TABLE 3.
Overlaps of pol II peaks with OGA, SPT5, TIF1β, O-GlcNAc, and TIF1β +O-GlcNAc
Overlaps of the indicated factors are indicated on the top row relative to the total number of pol II peaks listed in the left-most column. All percents are the % of pol II peaks either at promoters or in gene bodies.
| Pol II peaks | OGA peaks/pol II | SPT5 peaks/pol II | TIF1β /pol II | TIF1β +O-GlcNAc/pol II | O-GlcNAc/pol II peaks |
|---|---|---|---|---|---|
| Total pol II promoter (5332) | 335 (6.2%) | 5306 (99%) | 3749 (65%) | 2643 (49%) | 3734 (70%) |
| Total pol II gene body (1221) | 112 (9%) | 1207 (99%) | 874 (72%) | 520 (43%) | 717 (59%) |
TABLE 4.
Overlaps of the factors indicated in the top row relative to the total number of overlaps of OGA and promoter pol II in Table 3
| OGA+SPT5 | OGA+SPT5+TIF1β | OGA+SPT5+TIF1β+O-GlcNAc |
|---|---|---|
| 334 (100% of OGA pol II peaks) | 305 (91%of OGA pol II peaks) | 250 (75% of OGA pol II peaks) |
| 112 (100% of OGA pol II peaks) | 104 (93% of OGA pol II peaks) | 76 (68% of OGA pol II peaks) |
Our analysis also showed a significant number of pol II peaks within gene bodies. These peaks are not at all well understood, and it not clear whether they represent paused or arrested/stalled pol II. Our mapping shows that at gene-body pol II peaks, there are coincident peaks of OGA, SPT5, and TIF1β (Fig. 4, D and F, and Table 4). 99% of pol II peaks had an overlapping SPT5 peak, and 72% had an overlapping TIF1β peak. Of those, 100% had both OGA and SPT5, and 93% had OGA, SPT5, and TIF1β. As with promoter pol II, the overlap of gene body pol II and OGA also shows enormous overlaps with SPT5 and TIF1β. Although there are many more SPT5 and TIF1β peaks, virtually all of the OGA peaks co-localize with these two elongation factors (Table 3). Because of the presence of SPT5 and TIF1β (as well as OGA), these data suggest that these pol II peaks are likely paused in some manner and should be considered legitimate concentrations of SPT5-dependent paused pol II within gene bodies genome-wide due to the prior assignment of both SPT5 and TIF1β as pausing factors (27, 28).
Lastly, we investigated the genome-wide distribution of O-GlcNAc itself by ChIP-seq using the anti-GlcNAc antibody RL2. Previous work in Caenorhabditis elegans showed that O-GlcNAc was considerably enriched at TSSs (60). We found similar and striking enrichments of O-GlcNAc at TSSs in human BJAB cells (Fig. 4G and Table 3). 70% of pol II promoter peaks overlapped with an O-GlcNAc peak, and ∼30% ofok;1 O-GlcNAc peaks overlapped with promoter pol II. Thus, all components of the O-GlcNAcylation regulatory space are localized at promoters: O-GlcNAc (Fig. 4G), OGA (Fig. 4A), and OGT (61).
Discussion
We describe here the unexpected discovery of a novel elongation factor, O-GlcNAcase, which is the enzyme responsible for O-GlcNAc removal from a protein substrate, as a regulator of RNA pol II pausing and elongation. Thus, the O-GlcNAcylation system is part of a novel elongation regulatory phenomenon, one that is catalytic and adds an entirely new regulatory space governing both pausing and elongation by inserting O-GlcNAcylation regulatory steps into the elongation process. We show for the first time that OGA inhibition blocks elongation, that OGA is in physical contact with the elongation machinery, and that OGA maps to promoters genome-wide.
OGA Is an Elongation Factor
Inhibition of OGA in a crude nuclear extract (Fig. 1) results in an inhibition of elongation that is qualitatively very similar to the well known inhibition of P-TEFb-dependent elongation by flavopiridol and 5,6-Dichlorobenzimidazole riboside (62). Epistatic assays also show that OGA activity is part of a sequence of enzymatic activities in vitro: CDK7/TFIIH activity, followed by P-TEFb activity, followed by OGA activity (Fig. 1). In contrast to the crude nuclear extract system, in a partially purified elongation system where OGA has been removed by washes of the DNA template, elongation is inhibited by the addition of rOGA (Fig. 1). All of these experiments also argue for a direct effect of OGA on transcription elongation. But here, when OGA is assayed without other components of a nuclear extract (which were by definition removed during template washing), the assay revealed a possible positive function for OGA in elongation under different biochemical conditions. In any case it is clear that OGA and OGA-containing complexes can regulate pol II elongation (Figs. 1 and 3).
OGA, SPT5, and TIF1β
Our finding of a physical interaction between OGA and SPT5 (Fig. 2), a component of the elongation machinery, further suggests that the relationship between the regulation of O-GlcNAcylation and elongation is a bone fide one. SPT5 is a well known elongation factor required for establishing a paused pol II and, after phosphorylation, promoting productive elongation (62). To our knowledge, SPT5 has only been documented in a complex with SPT4, and we have yet to find any indication of SPT4 in the OEC. This suggests that SPT5 then has elongation functions other than those as part of DSIF.
Our finding of TIF1β in the OEC is also quite significant. TIF1β is also known as TRIM28 and KAP1 and was recently described as a factor participating in pol II pausing on the human HSPA1B promoter (27). A subsequent analysis suggested similar functions (28, 63). We have elected to use the TIF1β designation to emphasize the relationship to elongation, as TIF1γ also has elongation properties, including recruiting P-TEFb to promoters (64, 65). Additionally, TIF1α, -β, and -γ, all, are quite similar in primary structure (66).
We also assayed whether OGA catalytic activity was required for the OEC's ability to inhibit elongation. Somewhat surprisingly, we found that PUGNAc did not affect OEC function. This suggests that the OGA is inactive in the complex and that the OEC inhibitory functions are due to the activity of SPT5 and TIF1β. It is thus possible that OGA activity is regulated by the other factors in the OEC and that it is then activated at some point in the elongation process. To examine this further, a reconstituted elongation assay, based on the nuclear extract system, will be needed.
ChIP-seq Analysis
The cross-linking of OGA to promoters and its overlap with pol II positions across the genome is a remarkable result, further supporting the functional data that OGA is an elongation factor. Approximately half of the OGA peaks overlap with pol II, suggesting that OGA significantly partitions with pol II. Additionally, there are considerable overlaps of both SPT5 and TIF1β with OGA. These correlations are consistent with the physical isolation of both SPT5 and TIF1β with OGA. Furthermore, the overlaps of these three factors with pol II in the individual gene tracks further implicates OGA with pol II pausing and elongation. Finally, the localization of all of the components of the O-GlcNAcylation system at promoters, OGT (61), OGA (Fig. 4), and O-GlcNAc itself is quite striking (Fig. 4). Thus, promoters (and to a similar extent pol II peaks) appear to be focal points of O-GlcNAc information and regulation. We suggest that the promoter itself is acting, via the presence of the O-GlcNAcylation system, as a nutrient sensor, where activities of both OGT and OGA dynamically sample the levels of intracellular UDP-GlcNAc, and thus indirectly, the flux of glucose into the cell. This information would then be transmitted to various steps in PIC formation and pol II recruitment (44) and the regulation of elongation (this work). Others have proposed that UDP-GlcNAc concentrations are governed by nutrient flux, as glucose, glutamine, acetyl CoA, and UTP are all necessary for UDP-GlcNAc biosynthesis (35, 67). It is likely though that the UDP-GlcNAc is not the only component of the nutrient-sensing mechanism. OGT obviously must participate too, but more importantly, it is the set of O-GlcNAcylated proteins that are undergoing a dynamic cycling of O-GlcNAcylation that are the nutrient sensors. By constantly adding and removing O-GlcNAc from a protein(s), cycling allows the OGT and the target protein to continuously sample the nutrient state of the cell and respond to any changes in the supply and concentration of those nutrients.
Two-step Model of Pausing/Pause Release
The general implication of these data suggest that the removal of O-GlcNAc is necessary for proper elongation. It is well known that O-GlcNAc and phosphorylation are often found in a mutually exclusive relationship. It follows then that removal of O-GlcNAc might be a necessary prior event to the subsequent phosphorylation of elongation factors (for example, DSIF and NELF) by various kinases (such as P-TEFb). In that regard we have noted that virtually all of the known elongation machinery is O-GlcNAcylated, as determined by mass spectroscopy (68). Nevertheless, we do not have a good grasp on the functional targets of either OGT or OGA in elongation. However, it is clear that OGT is required for elongation events just as much as OGA is, as OGA needs O-GlcNAcylated substrates. As such, we predict that OGT is also functioning as an elongation factor and is coupled to other elongation factors. Determining the mechanistic function and regulation of OGA in the elongation processes will likely require the establishment of a more highly purified O-GlcNAc-dependent transcription system. Additionally, the elucidation of these processes will likely require the identification of OGT and OGA substrate(s) in the elongation machinery.
The OGA requirement may also impinge on the phosphorylation state of RNA pol II, as we have shown that both serine 2 and serine 5 residues of the pol II CTD can be O-GlcNAcylated (44). It is likely that OGA inhibition leads to altered phosphorylation states of the CTD, which would then influence pausing (in the case of serine 5) and various post-transcriptional events (in the case of serine 2). Again, it might be possible to test some of these predictions with highly purified pol II added into an O-GlcNAc-dependent reconstituted in vitro transcription system.
A Possible Connection between Cancer and Diabetes Phenotypes and OGA Activity Genome-wide
We suggest that the dysregulation of O-GlcNAcylation in cancer cells (69) results in the concomitant dysregulation of gene expression genome-wide via altered pol II elongation. The same idea can also be invoked to explain O-GlcNAcylation functions in type 2 diabetes (35), thus reframing both disease phenotypes as a defect in pol II elongation genome-wide. Indeed, increased O-GlcNAcylation and glucose flux resulted in increased expression of a subset of genes in beta cells of the pancreas (70).
Additional Functions of O-GlcNAcylation during Pausing and Elongation
One major question that arises from this work is why would a cell employ an apparently continuous cycling of O-GlcNAc on and off of a set of protein targets? When looked at in isolation, this appears to be nothing more than a waste of energy and resources, a classic futile cycle. However, the UDP-GlcNAc substrate for OGT is a high energy donor, contributing a ΔG (−8.8 kcal/mol; Ref. 71) similar to that of ATP hydrolysis (standard ΔG = −7.3 kcal/mol; Ref. 72), which may be a part of an energy requirement for pol II elongation and pausing.
Summary
Our studies on the regulation of O-GlcNAcylation in RNA pol II-dependent transcription provide novel, unforeseen, and fascinating insights into the regulation of initiation and elongation. Our results have described a novel regulatory space that regulates RNA pol II transcription at promoters and during elongation. Furthermore, these studies also connect the nutrient state of the cell to the transcriptional machinery and all that is implied by the metabolic dysregulation seen in human disease, from diabetes to cancer (73). Lastly, these studies underscore the immense power of transcription biochemistry and in vivo approaches that together provide avenues of investigation that neither one can supply on its own.
Experimental Procedures
ChIP-seq
Approximately 30–50 million cells were cross-linked in 1% formaldehyde for 5 min and quenched with 1 m Tris, pH 8.0. Cells were washed twice with cold PBS and placed in 1 ml of low salt buffer (150 mm NaCl, 50 mm Tris-HCl, pH 7.5, 5 mm EDTA, 0.5% Nonidet P-40, 1% Triton X-100) with complete protease inhibitor mixture (Roche Applied Science). Cells were sonicated for 15 min (20 s on, 40 s off) with a Sonicator Ultrasonic Processor XL (Misonix Inc.). Lysate was spun down at 13,200 rpm for 20 min, and immunoprecipitation was performed with 10 μg of 8WG16 (Covance, MMS-126R), normal mouse IgG (Santa Cruz, sc-2025), normal rabbit IgG (Santa Cruz, sc-2027), SPT5 (Santa Cruz sc-28878), TRIM28/TIF1b (Abcam ab622553), OGA/NCOAT (Santa Cruz sc-376429), or O-GlcNAc (RL2; Santa Cruz sc-59624) overnight at 4 °C. 50 μl of protein G beads (Roche Applied Science), pre-blocked with 0.5% BSA, were incubated with the lysate for 3 h at 4 °C. Beads were washed for 5 min 4 times with radioimmune precipitation assay buffer (10 mm Tris, pH 7.6, 150 mm NaCl, 1 mm EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS) and once with rinse buffer (10 mm Tris, pH 7.6, 50 mm NaCl, 1 mm EDTA). The cross-link was reversed in 100 μl of elution buffer 1 (10 mm Tris, pH 7.6, 1 mm EDTA, 1% SDS) for 10 min at 65 °C. 150 μl of elution buffer 2 (10 mm Tris, pH 7.6, 1 mm EDTA, 0.67% SDS) was added and treated with RNase A (Roche Applied Science) for 30 min at 37 °C and then with Proteinase K (Roche Applied Science) overnight at 65 °C. DNA was recovered using a MinElute PCR Purification kit (Qiagen). DNA was quantified by Qubit dsDNA High Sensitivity Quantification. Libraries were constructed with 25 ng of DNA using the KAPA Hyper Prep kit (KAPA Biosystems) with NEXTflex DNA Barcodes (BIOO Scientific).
ChIP-seq Bioinformatics Analysis
ChIP-seq reads were mapped to human genome (hg19) using BWA (PMID: 19451168). Low quality reads and adaptor sequences were removed by Trim-Galore. Peaks regions were identified by Macs2 using default options. Overlaps between peaks were calculated using Bedtools (PMID: 20110278.) Promoter regions were defined as −30 and +300 in reference to the TSS, and the gene bodies were defined as +301 of the TSS to the transcription termination site defined in Gencode release 19 (PMID: 22955987). Elongation rates were calculated by the ratio between the read density of 5′ promoter region (between −30 and +300 from TSS) and the read density of gene body (between +301 and +2000 from TSS). Genes with <2000 nucleotides and overlapping with other genes were omitted from the analyses.
In Vitro Transcriptions
In vitro transcriptions were done essentially as described (46). Nuclear extracts were prepared essentially as described (45). All drug inhibitors were added after PIC formation for 15 min before nucleotide addition. rOGA was made as described (43). Additions of flavopiridol and THZ were done as described (49). rTFIIF was expressed in Escherichia coli and purified as described previously (58).
Chromatography
Nuclear extract was separated by P11 on an AKTA Purifier using the indicated step elutions. 0.1 m fraction was loaded onto a Superose 6 column and developed in BC100. FLAG-SPT5 was purified from Flag-SPT5 expressing HeLa cells (10L) nuclear extract that was fractionated over a P11 column. The 0.1 m fraction was then subjected to an affinity purification step using M2-agarose beads (Sigma). FLAG-SPT5 was eluted using 0.5 mg/ml FLAG peptide (Sigma) in BC100 buffer.
Mass Spectrometry
On Bead Trypsin Digestion
The beads were resuspended in 25 mm NH4HCO3, pH 8.4, and heated at 95 °C for 5 min to denature the proteins. The samples were digested overnight with 2 μg of trypsin at 37 °C. After centrifugation the supernatant containing the peptides was collected, the beads washed twitch with 25 mm NH4HCO3, pH 8.4, and the supernatant was collected for maximal recovery of peptides. The tryptic digest was lyophilized and then reconstituted in 25% acetonitrile, 0.1% FA for fractionation using strong cation exchange liquid chromatography (LC). The strong cation exchange fractionation was performed as described (53).
LC-MS/MS Analysis
All LC-MS/MS analysis was performed on an LTQ Velos Pro ion trap mass spectrometer (Thermo Scientific, San Jose, CA). Dried tryptic digest were resuspended in 12 μl of 0.1% TFA, and 6 μl was loaded onto AcclaimTM PepMapTM 100 C18 LC column (Thermo Scientific) using a Thermo Easy nLC 1000 liquid chromatography system (Thermo Scientific) connected online to the mass spectrometer. After sample injection, the column was washed, and peptides were eluted using a linear gradient. The mass spectrometer was operated in a data-dependent mode with each full MS scan followed by 15 data-dependent MS/MS acquisitions.
Protein Identification
Acquired MS/MS spectra were searched against a human UniProt protein database, 2014, using a SEQUEST and Fixed Value PSM validator algorithms in the Proteome Discoverer 1.4 software (Thermo Scientific). The precursor ion tolerance was set at 1.5 Da, and the fragment ions tolerance was set at 0.6 Da along with methionine oxidation included as dynamic modification. Only fully tryptic peptides with up to two miscleavages with charge state-dependent cross-correlation Xcorr ≥ 2.1 for [M+H]1+, ≥ 2.5 for [M+2H]2+, and ≥ 3.2 for [M+3H]3+ and delta correlation (ΔCn) ≥ 0.08 were considered as positive identification.
Reagents
RNA pol II antibody (8WG16, Covance), SPT5 antibody (Santa Cruz), OGA antibody (Santa Cruz), PUGNAc (Sigma), flavopiridol (Sigma), Thiamet-G (Sigma), P11 (Whatman), [32P]CTP (PerkinElmer Life Sciences).
Author Contributions
M. R. made the recombinant OGA protein and performed the ChIP-seq experiments. A. G. F. identified OGA antibodies for ChIP-seq, B.-H. K. analyzed the ChIP-seq data for O-GlcNAc, OGA, TRIM28, pol II, and SPT5. B. J. A. and K. Z. analyzed ChIP-seq data for O-GlcNAc and pol II. B. A. L. performed the experiments in Figs. 1–3, helped analyze all ChIP-seq data, designed all of the experiments, and wrote the manuscript.
Acknowledgments
We thank John Hanover for the rOGA expression vector, Jerry Hart for anti-OGT antibodies, David Price for the CMV DNA template, Yoki Yamaguchi for the FLAG-SPT5 expression vector, Nathanial Gray for THZ, and Thorkell Andresson and Sudipto Das at the NCI Laboratory of Proteomics and Analytical Technologies for the MS analysis. Illumina sequencing was performed by the NCI Sequencing Facility. We also thank Jason Piotrowski for expert technical assistance. Mass spectrometry Excel files are available upon request.
Note Added in Proof
Preinitiation complexes were formed as described (43) using the CMV promoter. The elongation assays were performed as described (46, 74). The TFIIF-dependent assays were done as described (74), using the CMV promoter conjugated to Dynabeads (ThermoFisher) via a biotin-streptavidin linkage (75). Pulsed elongation complexes were stopped by adding EDTA to 20 mm final concentration and then were washed twice in high salt buffer (1.6 m KCl, 20 mm Hepes, pH 7.5) and twice in low-salt buffer (60 mm KCl, 20 mm Hepes, pH 7.5, 200 μg/ml BSA) (74). Magnetic beads were suspended in elongation buffer (20 mm Hepes, pH 7.5, 60 mm KCl, 200 μg/ml BSA, 3 mm MgCl2) (74). Recombinant TFIIF, rOGA, PUGNAc, and OEC were added, as indicated, for 5 min before chasing the elongation complexes with 0.5 mm NTP for 3 min. Assays were then stopped with stop buffer (74) and processed as described (74) on 8% polyacrylamide/urea/TBE gels, followed by autoradiography.
This work was supported by the NCI Intramural Program (to B. A. L.) and the NHLBI Intramural Program (to K. Z.), National Institutes of Health. The authors declare they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
ChIP-seq data have been deposited in the NCBI GEO database under GSE86154.
- pol
- polymerase
- TSS
- transcriptional start site
- OGT
- O-GlcNAc transferase
- OGA
- O-GlcNAcase
- rOGA
- recombinant OGA
- FP
- flavopiridol
- OEC
- OGA elongation complex
- PIC
- preinitiation complex and THZ, (E)-N-(3-((5-chloro-4-(1H-indol-3-yl)pyrimidin-2-yl)amino)phenyl)-4-(4-(dimethylamino)but-2-enamido)benzamide.
References
- 1. Rougvie A. E., and Lis J. T. (1988) The RNA polymerase II molecule at the 5′ end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell 54, 795–804 [DOI] [PubMed] [Google Scholar]
- 2. Krumm A., Meulia T., Brunvand M., and Groudine M. (1992) The block to transcriptional elongation within the human c-myc gene is determined in the promoter-proximal region. Genes Dev. 6, 2201–2213 [DOI] [PubMed] [Google Scholar]
- 3. Strobl L. J., and Eick D. (1992) Hold back of RNA polymerase II at the transcription start site mediates down-regulation of c-myc in vivo. EMBO J. 11, 3307–3314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rahl P. B., Lin C. Y., Seila A. C., Flynn R. A., McCuine S., Burge C. B., Sharp P. A., and Young R. A. (2010) c-Myc regulates transcriptional pause release. Cell 141, 432–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Core L. J., Waterfall J. J., Gilchrist D. A., Fargo D. C., Kwak H., Adelman K., and Lis J. T. (2012) Defining the status of RNA polymerase at promoters. Cell Rep. 2, 1025–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Core L. J., Waterfall J. J., and Lis J. T. (2008) Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322, 1845–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Muse G. W., Gilchrist D. A., Nechaev S., Shah R., Parker J. S., Grissom S. F., Zeitlinger J., and Adelman K. (2007) RNA polymerase is poised for activation across the genome. Nat. Genet. 39, 1507–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nechaev S., Fargo D. C., dos Santos G., Liu L., Gao Y., and Adelman K. (2010) Global analysis of short RNAs reveals widespread promoter-proximal stalling and arrest of Pol II in Drosophila. Science 327, 335–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zeitlinger J., Stark A., Kellis M., Hong J. W., Nechaev S., Adelman K., Levine M., and Young R. A. (2007) RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat. Genet. 39, 1512–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wada T., Takagi T., Yamaguchi Y., Ferdous A., Imai T., Hirose S., Sugimoto S., Yano K., Hartzog G. A., Winston F., Buratowski S., and Handa H. (1998) DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 12, 343–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yamaguchi Y., Inukai N., Narita T., Wada T., and Handa H. (2002) Evidence that negative elongation factor represses transcription elongation through binding to a DRB sensitivity-inducing factor/RNA polymerase II complex and RNA. Mol. Cell. Biol. 22, 2918–2927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yamaguchi Y., Takagi T., Wada T., Yano K., Furuya A., Sugimoto S., Hasegawa J., and Handa H. (1999) NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell 97, 41–51 [DOI] [PubMed] [Google Scholar]
- 13. Wu C.-H, Yamaguchi Y., Benjamin L. R., Horvat-Gordon M., Washinsky J., Enerly E., Larsson J., Lambertsson A., Handa H., and Gilmour D. (2003) NELF and DSIF cause promoter proximal pausing on the hsp70 promoter in Drosophila. Genes Dev. 17, 1402–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wada T., Takagi T., Yamaguchi Y., Watanabe D., and Handa H. (1998) Evidence that P-TEFb alleviates the negative effect of DSIF on RNA polymerase II-dependent transcription in vitro. EMBO J. 17, 7395–7403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yamada T., Yamaguchi Y., Inukai N., Okamoto S., Mura T., and Handa H. (2006) P-TEFb-mediated phosphorylation of hSpt5 C-terminal repeats is critical for processive transcription elongation. Mol. Cell 21, 227–237 [DOI] [PubMed] [Google Scholar]
- 16. Mandal S. S., Chu C., Wada T., Handa H., Shatkin A. J., and Reinberg D. (2004) Functional interactions of RNA-capping enzyme with factors that positively and negatively regulate promoter escape by RNA polymerase II. Proc. Natl. Acad. Sci. U.S.A. 101, 7572–7577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Glover-Cutter K., Larochelle S., Erickson B., Zhang C., Shokat K., Fisher R. P., and Bentley D. L. (2009) TFIIH-associated Cdk7 kinase functions in phosphorylation of C-terminal domain Ser-7 residues, promoter-proximal pausing, and termination by RNA polymerase II. Mol. Cell. Biol. 29, 5455–5464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wen Y., and Shatkin A. J. (1999) Transcription elongation factor hSPT5 stimulates mRNA capping. Genes Dev. 13, 1774–1779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Weber A., Liu J., Collins I., and Levens D. (2005) TFIIH operates through an expanded proximal promoter to fine-tune c-myc expression. Mol. Cell. Biol. 25, 147–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Takahashi H., Parmely T. J., Sato S., Tomomori-Sato C., Banks C. A., Kong S. E., Szutorisz H., Swanson S. K., Martin-Brown S., Washburn M. P., Florens L., Seidel C. W., Lin C., Smith E. R., Shilatifard A., Conaway R. C., and Conaway J. W. (2011) Human mediator subunit MED26 functions as a docking site for transcription elongation factors. Cell 146, 92–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Donner A. J., Ebmeier C. C., Taatjes D. J., and Espinosa J. M. (2010) CDK8 is a positive regulator of transcriptional elongation within the serum response network. Nat. Struct. Mol. Biol. 17, 194–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Park J. M., Werner J., Kim J. M., Lis J. T., and Kim Y. J. (2001) Mediator, not holoenzyme, is directly recruited to the heat shock promoter by HSF upon heat shock. Mol. Cell 8, 9–19 [DOI] [PubMed] [Google Scholar]
- 23. Stadelmayer B., Micas G., Gamot A., Martin P., Malirat N., Koval S., Raffel R., Sobhian B., Severac D., Rialle S., Parrinello H., Cuvier O., and Benkirane M. (2014) Integrator complex regulates NELF-mediated RNA polymerase II pause/release and processivity at coding genes. Nat. Commun. 5, 5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gardini A., Baillat D., Cesaroni M., Hu D., Marinis J. M., Wagner E. J., Lazar M. A., Shilatifard A., and Shiekhattar R. (2014) Integrator regulates transcriptional initiation and pause release following activation. Mol. Cell 56, 128–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Smith E. R., Winter B., Eissenberg J. C., and Shilatifard A. (2008) Regulation of the transcriptional activity of poised RNA polymerase II by the elongation factor ELL. Proc. Natl. Acad. Sci. U.S.A. 105, 8575–8579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Adelman K., Marr M. T., Werner J., Saunders A., Ni Z., Andrulis E. D., and Lis J. T. (2005) Efficient release from promoter-proximal stall sites requires transcript cleavage factor TFIIS. Mol. Cell 17, 103–112 [DOI] [PubMed] [Google Scholar]
- 27. Bunch H., Zheng X., Burkholder A., Dillon S. T., Motola S., Birrane G., Ebmeier C. C., Levine S., Fargo D., Hu G., Taatjes D. J., and Calderwood S. K. (2014) TRIM28 regulates RNA polymerase II promoter-proximal pausing and pause release. Nat. Struct. Mol. Biol. 21, 876–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. McNamara R. P., Reeder J. E., McMillan E. A., Bacon C. W., McCann J. L., and D'Orso I. (2016) KAP1 Recruitment of the 7SK snRNP complex to promoters enables transcription elongation by RNA polymerase II. Mol. Cell 61, 39–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Baranello L., Wojtowicz D., Cui K., Devaiah B. N., Chung H. J., Chan-Salis K. Y., Guha R., Wilson K., Zhang X., Zhang H., Piotrowski J., Thomas C. J., Singer D. S., Pugh B. F., Pommier Y., et al. (2016) RNA polymerase II regulates topoisomerase 1 activity to favor efficient transcription. Cell 165, 357–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Luo Z., Lin C., and Shilatifard A. (2012) The super elongation complex (SEC) family in transcriptional control. Nat. Rev. Mol. Cell Biol. 13, 543–547 [DOI] [PubMed] [Google Scholar]
- 31. Chen F. X., Woodfin A. R., Gardini A., Rickels R. A., Marshall S. A., Smith E. R., Shiekhattar R., and Shilatifard A. (2015) PAF1, a molecular regulator of promoter-proximal pausing by RNA polymerase II. Cell 162, 1003–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yu M., Yang W., Ni T., Tang Z., Nakadai T., Zhu J., and Roeder R. G. (2015) RNA polymerase II-associated factor 1 regulates the release and phosphorylation of paused RNA polymerase II. Science 350, 1383–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cheng B., Li T., Rahl P. B., Adamson T. E., Loudas N. B., Guo J., Varzavand K., Cooper J. J., Hu X., Gnatt A., Young R. A., and Price D. H. (2012) Functional association of Gdown1 with RNA Polymerase II poised on human genes. Mol. Cell 45, 38–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hart G. W., Slawson C., Ramirez-Correa G., and Lagerlof O. (2011) Cross talk between O-GlcNAcylation and phosphorylation: roles in signaling, transcription, and chronic disease. Annu. Rev. Biochem. 80, 825–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bond M. R., and Hanover J. A. (2013) O-GlcNAc cycling: a link between metabolism and chronic disease. Annu. Rev. Nutr. 33, 205–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lewis B. A. (2013) O-GlcNAcylation at promoters, nutrient sensors, and transcriptional regulation. Biochim. Biophys. Acta 1829, 1202–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lewis B. A., and Hanover J. A. (2014) O-GlcNAc and the epigenetic regulation of gene expression. J. Biol. Chem. 289, 34440–34448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Keembiyehetty C., Love D. C., Harwood K. R., Gavrilova O., Comly M. E., and Hanover J. A. (2015) Conditional knock-out reveals a requirement for O-linked N-acetylglucosaminase (O-GlcNAcase) in metabolic homeostasis. J. Biol. Chem. 290, 7097–7113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hart T., Chandrashekhar M., Aregger M., Steinhart Z., Brown K. R., MacLeod G., Mis M., Zimmermann M., Fradet-Turcotte A., Sun S., Mero P., Dirks P., Sidhu S., Roth F. P., Rissland O. S., Durocher D., Angers S., and Moffat J. (2015) High-resolution CRISPR screens reveal fitness genes and genotype-specific cancer liabilities. Cell 163, 1515–1526 [DOI] [PubMed] [Google Scholar]
- 40. Wang T., Birsoy K., Hughes N. W., Krupczak K. M., Post Y., Wei J. J., Lander E. S., and Sabatini D. M. (2015) Identification and characterization of essential genes in the human genome. Science 350, 1096–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lagerlöf O., Slocomb J. E., Hong I., Aponte Y., Blackshaw S., Hart G. W., and Huganir R. L. (2016) The nutrient sensor OGT in PVN neurons regulates feeding. Science 351, 1293–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. O'Donnell N., Zachara N. E., Hart G. W., and Marth J. D. (2004) Ogt-dependent X-chromosome-linked protein glycosylation is a requisite modification in somatic cell function and embryo viability. Mol. Cell. Biol. 24, 1680–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ranuncolo S. M., Ghosh S., Hanover J. A., Hart G. W., and Lewis B. A. (2012) Evidence of the involvement of O-GlcNAc-modified human RNA polymerase II CTD in transcription in vitro and in vivo. J. Biol. Chem. 287, 23549–23561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lewis B. A., Burlingame A. L., and Myers S. A. (2016) Human RNA polymerase II promoter recruitment in vitro is regulated by O-linked N-Acetylglucosaminyltransferase (OGT). J. Biol. Chem. 291, 14056–14061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dignam J. D., Lebovitz R. M., and Roeder R. G. (1983) Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11, 1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Adamson T. E., Shore S. M., and Price D. H. (2003) Analysis of RNA polymerase II elongation in vitro. Methods Enzymol. 371, 264–275 [DOI] [PubMed] [Google Scholar]
- 47. Kwiatkowski N., Zhang T., Rahl P. B., Abraham B. J., Reddy J., Ficarro S. B., Dastur A., Amzallag A., Ramaswamy S., Tesar B., Jenkins C. E., Hannett N. M., McMillin D., Sanda T., Sim T., Kim N. D., et al. (2014) Targeting transcription regulation in cancer with a covalent CDK7 inhibitor. Nature 511, 616–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chao S. H., Fujinaga K., Marion J. E., Taube R., Sausville E. A., Senderowicz A. M., Peterlin B. M., and Price D. H. (2000) Flavopiridol inhibits P-TEFb and blocks HIV-1 replication. J. Biol. Chem. 275, 28345–28348 [DOI] [PubMed] [Google Scholar]
- 49. Nilson K. A., Guo J., Turek M. E., Brogie J. E., Delaney E., Luse D. S., and Price D. H. (2015) THZ1 reveals roles for Cdk7 in co-transcriptional capping and pausing. Mol. Cell 59, 576–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yuzwa S. A., Macauley M. S., Heinonen J. E., Shan X., Dennis R. J., He Y., Whitworth G. E., Stubbs K. A., McEachern E. J., Davies G. J., and Vocadlo D. J. (2008) A potent mechanism-inspired O-GlcNAcase inhibitor that blocks phosphorylation of tau in vivo. Nat. Chem. Biol. 4, 483–490 [DOI] [PubMed] [Google Scholar]
- 51. Haltiwanger R. S., Grove K., and Philipsberg G. A. (1998) Modulation of O-linked N-acetylglucosamine levels on nuclear and cytoplasmic proteins in vivo using the peptide O-GlcNAc-β-N-acetylglucosaminidase inhibitor O-(2-acetamido-2-deoxy-d-glucopyranosylidene)amino-N-phenylcarbamate. J. Biol. Chem. 273, 3611–3617 [DOI] [PubMed] [Google Scholar]
- 52. Teo C. F., El-Karim E. G., and Wells L. (2016) Dissecting PUGNAc-mediated inhibition of the pro-survival action of insulin. Glycobiology 10.1093/glycob/cww043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Das S., Bosley A. D., Ye X., Chan K. C., Chu I., Green J. E., Issaq H. J., Veenstra T. D., and Andresson T. (2010) Comparison of strong cation exchange and SDS-PAGE fractionation for analysis of multiprotein complexes. J. Proteome Res. 9, 6696–6704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bengal E., Flores O., Krauskopf A., Reinberg D., and Aloni Y. (1991) Role of the mammalian transcription factors IIF, IIS, and IIX during elongation by RNA polymerase II. Mol. Cell. Biol. 11, 1195–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Flores O., Maldonado E., and Reinberg D. (1989) Factors involved in specific transcription by mammalian RNA polymerase II. Factors IIE and IIF independently interact with RNA polymerase II. J. Biol. Chem. 264, 8913–8921 [PubMed] [Google Scholar]
- 56. Price D. H., Sluder A. E., and Greenleaf A. L. (1989) Dynamic interaction between a Drosophila transcription factor and RNA polymerase II. Mol. Cell. Biol. 9, 1465–1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Cetinba N., Macauley M. S., Stubbs K. A., Drapala R., and Vocadlo D. J. (2006) Identification of Asp174 and Asp175 as the key catalytic residues of human O-GlcNAcase by functional analysis of site-directed mutants. Biochemistry 45, 3835–3844 [DOI] [PubMed] [Google Scholar]
- 58. Maldonado E., Drapkin R., and Reinberg D. (1996) Purification of human RNA polymerase II and general transcription factors. Methods Enzymol. 274, 72–100 [DOI] [PubMed] [Google Scholar]
- 59. Thomas M. C., and Chiang C. M. (2006) The general transcription machinery and general cofactors. Crit. Rev. Biochem. Mol. Biol. 41, 105–178 [DOI] [PubMed] [Google Scholar]
- 60. Love D. C., Ghosh S., Mondoux M. A., Fukushige T., Wang P., Wilson M. A., Iser W. B., Wolkow C. A., Krause M. W., and Hanover J. A. (2010) Dynamic O-GlcNAc cycling at promoters of Caenorhabditis elegans genes regulating longevity, stress, and immunity. Proc. Natl. Acad. Sci. U.S.A. 107, 7413–7418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Deplus R., Delatte B., Schwinn M. K., Defrance M., Méndez J., Murphy N., Dawson M. A., Volkmar M., Putmans P., Calonne E., Shih A. H., Levine R. L., Bernard O., Mercher T., Solary E., Urh M., Daniels D. L., and Fuks F. (2013) TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. EMBO J. 32, 645–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Guo J., and Price D. H. (2013) RNA polymerase II transcription elongation control. Chem. Rev. 113, 8583–8603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. McNamara R. P., Guzman C., Reeder J. E., and D'Orso I. (2016) Genome-wide analysis of KAP1, the 7SK snRNP complex, and RNA polymerase II. Genom Data 7, 250–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Bai X., Kim J., Yang Z., Jurynec M. J., Akie T. E., Lee J., LeBlanc J., Sessa A., Jiang H., DiBiase A., Zhou Y., Grunwald D. J., Lin S., Cantor A. B., Orkin S. H., and Zon L. I. (2010) TIF1γ controls erythroid cell fate by regulating transcription elongation. Cell 142, 133–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Bai X., Trowbridge J. J., Riley E., Lee J. A., DiBiase A., Kaartinen V. M., Orkin S. H., and Zon L. I. (2013) TiF1-γ plays an essential role in murine hematopoiesis and regulates transcriptional elongation of erythroid genes. Dev. Biol. 373, 422–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Iyengar S., and Farnham P. J. (2011) KAP1 protein: an enigmatic master regulator of the genome. J. Biol. Chem. 286, 26267–26276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Harwood K. R., and Hanover J. (2014) Nutrient-driven O-GlcNAc cycling: think globally but act locally. J. Cell Sci. 127, 1857–1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hahne H., Sobotzki N., Nyberg T., Helm D., Borodkin V. S., van Aalten D. M., Agnew B., and Kuster B. (2013) Proteome wide purification and identification of O-GlcNAc-modified proteins using click chemistry and mass spectrometry. J. Proteome Res. 12, 927–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Slawson C., and Hart G. W. (2011) O-GlcNAc signalling: implications for cancer cell biology. Nat. Rev. Cancer 11, 678–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Durning S. P., Flanagan-Steet H., Prasad N., and Wells L. (2016) O-linked β-N-acetylglucosamine (O-GlcNAc) acts as a glucose sensor to epigenetically regulate the insulin gene in pancreatic β cells. J. Biol. Chem. 291, 2107–2118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Okada T., Ihara H., Ito R., Taniguchi N., and Ikeda Y. (2009) Bidirectional N-acetylglucosamine transfer mediated by β-1,4-N-acetylglucosaminyltransferase III. Glycobiology 19, 368–374 [DOI] [PubMed] [Google Scholar]
- 72. Voet D., and Voet J. G. (2011) Biochemistry, 4th Ed., p. 566, John Wiley & Sons, Hoboken, NJ [Google Scholar]
- 73. Vaidyanathan K., and Wells L. (2014) Multiple Tissue-specific roles for the O-GlcNAc post-translational modification in the induction of and complications arising from type II diabetes. J. Biol. Chem. 289, 34466–34471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Cheng B., and Price D. H. (2007) Properties of RNA polymerase II elongation complexes before and after the P-TEFb-mediated transition into productive elongation. J. Biol. Chem. 282, 21901–21912 [DOI] [PubMed] [Google Scholar]
- 75. Cheng B., and Price D. H. (2009) Isolation and functional analysis of RNA polymerase II elongation complexes. Methods 48, 346–352 [DOI] [PMC free article] [PubMed] [Google Scholar]