Abstract
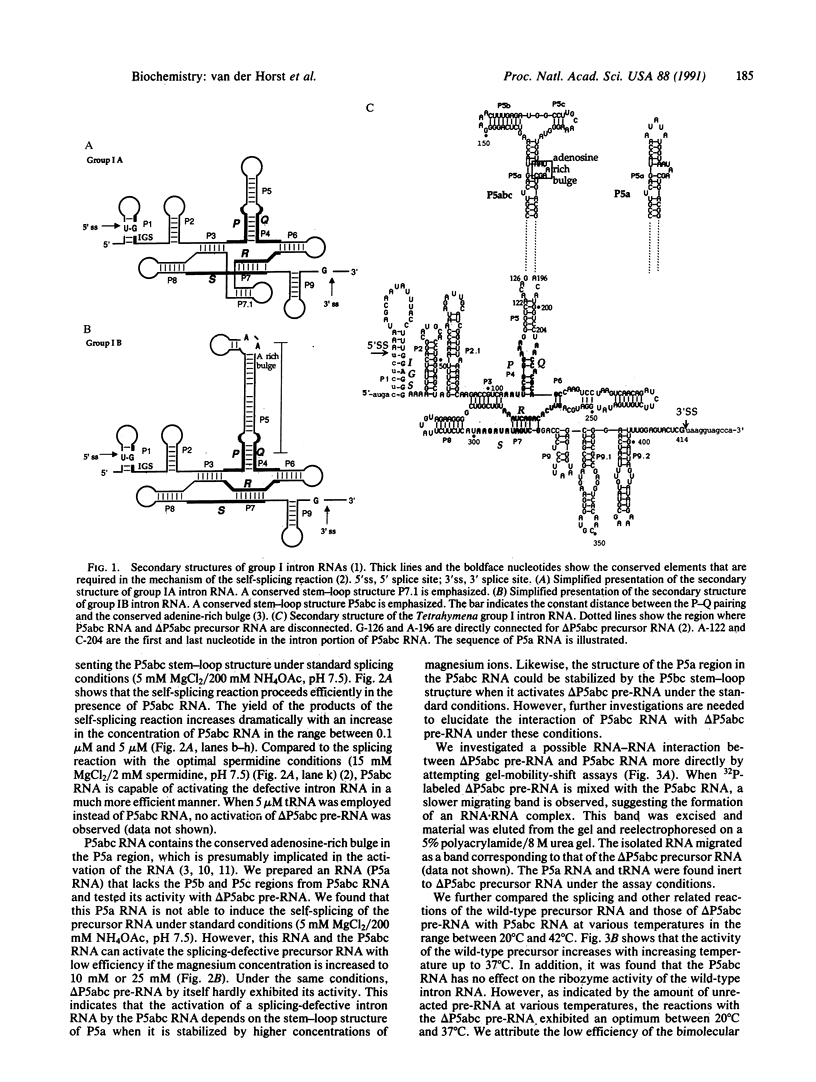
The self-splicing rRNA intron of Tetrahymena thermophila belongs to a subgroup of group I introns that contain a conserved extra stem-loop structure termed P5abc. A Tetrahymena mutant precursor RNA lacking this P5abc is splicing-defective under standard conditions (5 mM MgCl2/200 mM NH4Cl, pH 7.5) in vitro. However, the mutant precursor RNA by itself is capable of performing the self-splicing reaction without P5abc under different conditions (15 mM MgCl2/2 mM spermidine, pH 7.5). We have investigated the functional role of the P5abc in the mechanism of the self-splicing reaction. When an RNA consisting of the P5abc but lacking the rest of the Tetrahymena intron is incubated with the mutant precursor, the self-splicing reaction proceeds highly efficiently under standard conditions (5 mM MgCl2/200 mM NH4Cl, pH 7.5). Two steps of the bimolecular self-splicing reaction can be performed accurately by a shortened precursor RNA containing all essential components required in the self-splicing reaction and an activator RNA consisting of the P5abc. Gel-mobility-shift assays suggest that two molecules associate by a direct RNA-RNA interaction during the splicing reaction. The results imply that there might exist other small RNAs whose role is to activate ribozymes.
Full text
PDF
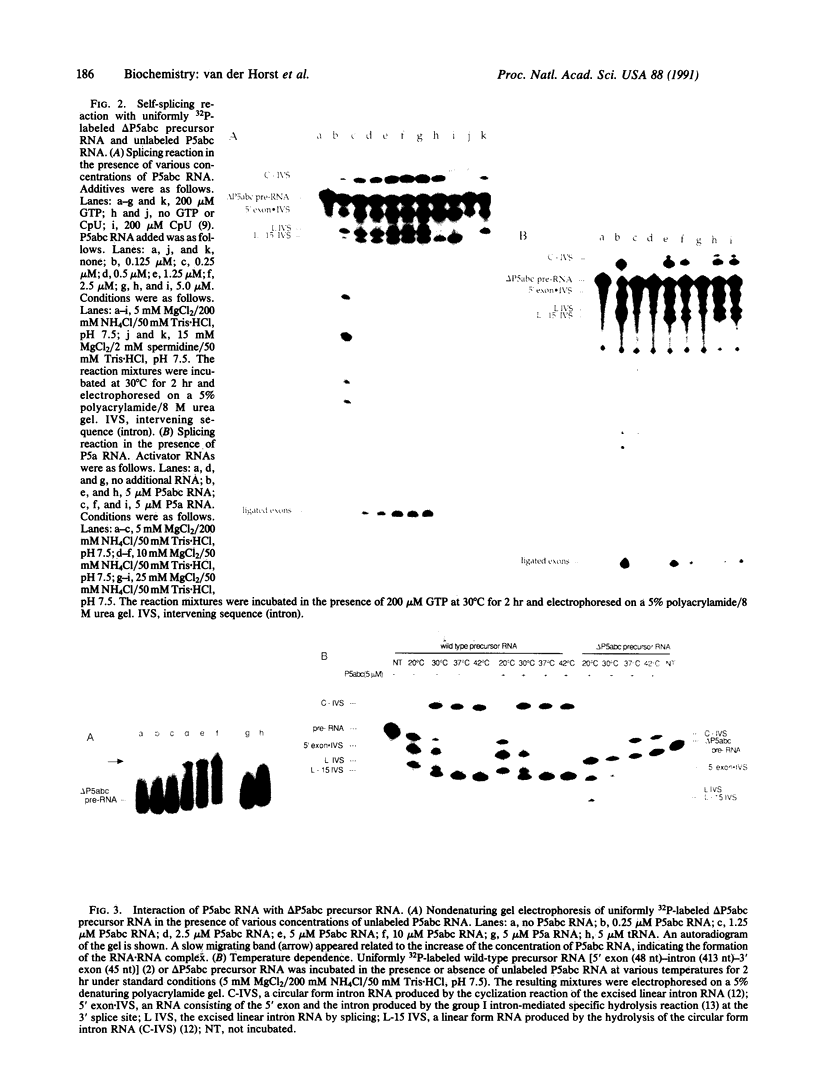
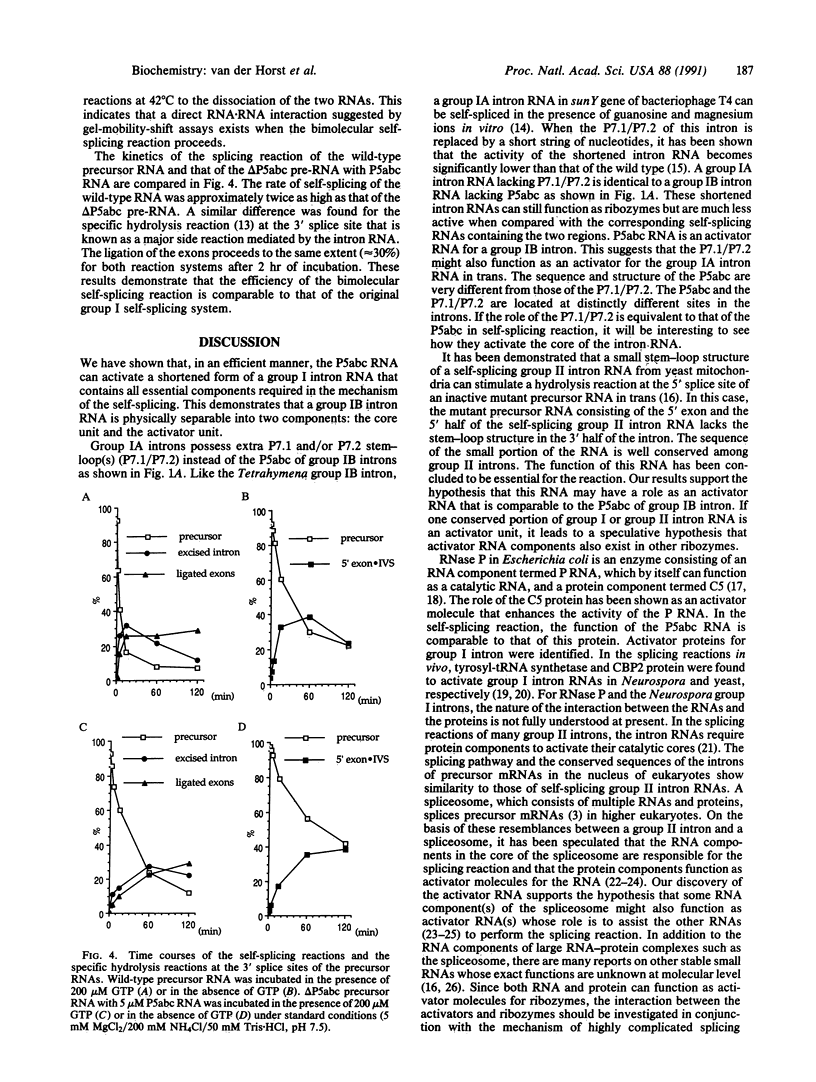

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Burke J. M., Belfort M., Cech T. R., Davies R. W., Schweyen R. J., Shub D. A., Szostak J. W., Tabak H. F. Structural conventions for group I introns. Nucleic Acids Res. 1987 Sep 25;15(18):7217–7221. doi: 10.1093/nar/15.18.7217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch H., Reddy R., Rothblum L., Choi Y. C. SnRNAs, SnRNPs, and RNA processing. Annu Rev Biochem. 1982;51:617–654. doi: 10.1146/annurev.bi.51.070182.003153. [DOI] [PubMed] [Google Scholar]
- Cech T. R. Conserved sequences and structures of group I introns: building an active site for RNA catalysis--a review. Gene. 1988 Dec 20;73(2):259–271. doi: 10.1016/0378-1119(88)90492-1. [DOI] [PubMed] [Google Scholar]
- Cech T. R. The generality of self-splicing RNA: relationship to nuclear mRNA splicing. Cell. 1986 Jan 31;44(2):207–210. doi: 10.1016/0092-8674(86)90751-8. [DOI] [PubMed] [Google Scholar]
- Cherniack A. D., Garriga G., Kittle J. D., Jr, Akins R. A., Lambowitz A. M. Function of Neurospora mitochondrial tyrosyl-tRNA synthetase in RNA splicing requires an idiosyncratic domain not found in other synthetases. Cell. 1990 Aug 24;62(4):745–755. doi: 10.1016/0092-8674(90)90119-y. [DOI] [PubMed] [Google Scholar]
- Collins R. A. Evidence of natural selection to maintain a functional domain outside of the 'core' in a large subclass of group I introns. Nucleic Acids Res. 1988 Mar 25;16(6):2705–2715. doi: 10.1093/nar/16.6.2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna J. A., Szostak J. W. Miniribozymes, small derivatives of the sunY intron, are catalytically active. Mol Cell Biol. 1989 Dec;9(12):5480–5483. doi: 10.1128/mcb.9.12.5480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedor M. J., Uhlenbeck O. C. Substrate sequence effects on "hammerhead" RNA catalytic efficiency. Proc Natl Acad Sci U S A. 1990 Mar;87(5):1668–1672. doi: 10.1073/pnas.87.5.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flor P. J., Flanegan J. B., Cech T. R. A conserved base pair within helix P4 of the Tetrahymena ribozyme helps to form the tertiary structure required for self-splicing. EMBO J. 1989 Nov;8(11):3391–3399. doi: 10.1002/j.1460-2075.1989.tb08503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich P. Biochemistry in Hungary. Trends Biochem Sci. 1990 Jul;15(7):251–252. doi: 10.1016/0968-0004(90)90046-e. [DOI] [PubMed] [Google Scholar]
- Gampel A., Nishikimi M., Tzagoloff A. CBP2 protein promotes in vitro excision of a yeast mitochondrial group I intron. Mol Cell Biol. 1989 Dec;9(12):5424–5433. doi: 10.1128/mcb.9.12.5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrier-Takada C., Gardiner K., Marsh T., Pace N., Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983 Dec;35(3 Pt 2):849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- Inoue T., Sullivan F. X., Cech T. R. New reactions of the ribosomal RNA precursor of Tetrahymena and the mechanism of self-splicing. J Mol Biol. 1986 May 5;189(1):143–165. doi: 10.1016/0022-2836(86)90387-6. [DOI] [PubMed] [Google Scholar]
- Inouye M., Delihas N. Small RNAs in the prokaryotes: a growing list of diverse roles. Cell. 1988 Apr 8;53(1):5–7. doi: 10.1016/0092-8674(88)90480-1. [DOI] [PubMed] [Google Scholar]
- Joyce G. F., van der Horst G., Inoue T. Catalytic activity is retained in the Tetrahymena group I intron despite removal of the large extension of element P5. Nucleic Acids Res. 1989 Oct 11;17(19):7879–7889. doi: 10.1093/nar/17.19.7879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger K., Grabowski P. J., Zaug A. J., Sands J., Gottschling D. E., Cech T. R. Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell. 1982 Nov;31(1):147–157. doi: 10.1016/0092-8674(82)90414-7. [DOI] [PubMed] [Google Scholar]
- Latham J. A., Cech T. R. Defining the inside and outside of a catalytic RNA molecule. Science. 1989 Jul 21;245(4915):276–282. doi: 10.1126/science.2501870. [DOI] [PubMed] [Google Scholar]
- McClain W. H., Guerrier-Takada C., Altman S. Model substrates for an RNA enzyme. Science. 1987 Oct 23;238(4826):527–530. doi: 10.1126/science.2443980. [DOI] [PubMed] [Google Scholar]
- Michel F., Jacquier A., Dujon B. Comparison of fungal mitochondrial introns reveals extensive homologies in RNA secondary structure. Biochimie. 1982 Oct;64(10):867–881. doi: 10.1016/s0300-9084(82)80349-0. [DOI] [PubMed] [Google Scholar]
- Sharp P. A. On the origin of RNA splicing and introns. Cell. 1985 Sep;42(2):397–400. doi: 10.1016/0092-8674(85)90092-3. [DOI] [PubMed] [Google Scholar]
- Sharp P. A. Splicing of messenger RNA precursors. Science. 1987 Feb 13;235(4790):766–771. doi: 10.1126/science.3544217. [DOI] [PubMed] [Google Scholar]
- Séraphin B., Simon M., Boulet A., Faye G. Mitochondrial splicing requires a protein from a novel helicase family. Nature. 1989 Jan 5;337(6202):84–87. doi: 10.1038/337084a0. [DOI] [PubMed] [Google Scholar]
- Waring R. B., Davies R. W. Assessment of a model for intron RNA secondary structure relevant to RNA self-splicing--a review. Gene. 1984 Jun;28(3):277–291. doi: 10.1016/0378-1119(84)90145-8. [DOI] [PubMed] [Google Scholar]
- Xu M. Q., Shub D. A. The catalytic core of the sunY intron of bacteriophage T4. Gene. 1989 Oct 15;82(1):77–82. doi: 10.1016/0378-1119(89)90032-2. [DOI] [PubMed] [Google Scholar]
- Zaug A. J., Grabowski P. J., Cech T. R. Autocatalytic cyclization of an excised intervening sequence RNA is a cleavage-ligation reaction. Nature. 1983 Feb 17;301(5901):578–583. doi: 10.1038/301578a0. [DOI] [PubMed] [Google Scholar]