Abstract
Recent advances in genome engineering are starting a revolution in biological research and translational applications. The CRISPR-associated RNA-guided endonuclease Cas9 and its variants enable diverse manipulations of genome function. In this review, we describe the development of Cas9 tools for a variety of applications in cell biology research, including the study of functional genomics, the creation of transgenic animal models, and genomic imaging. Novel genome engineering methods offer a new avenue to understand the causality between genome and phenotype, thus promising a fuller understanding of cell biology.
Keywords: CRISPR/Cas9, CRISPRi/a, gene editing, gene regulation, cell biology
From DNA repair pathways to CRISPR/Cas9-mediated genome editing
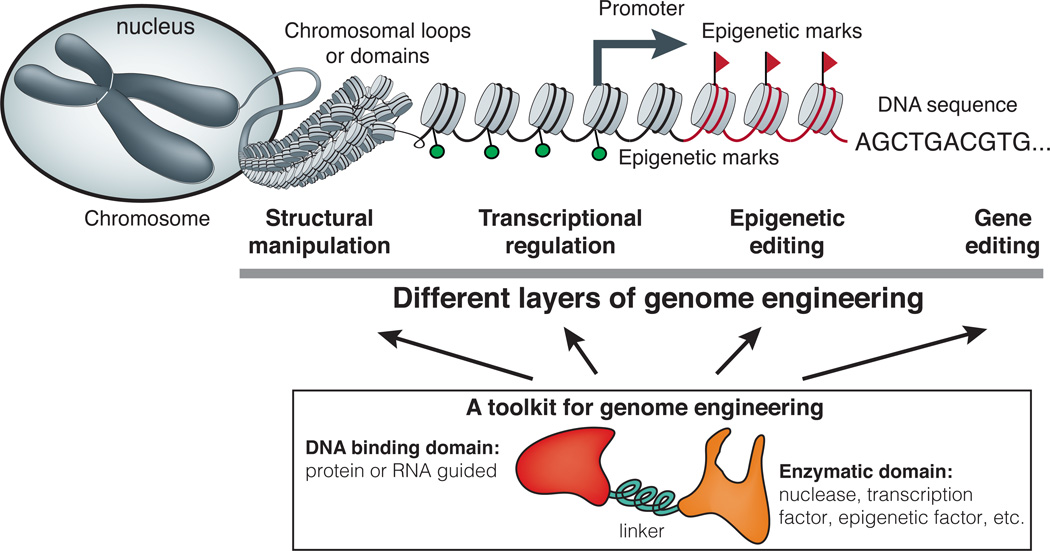
Eukaryotic cells use a sophisticated network of genes and genomic regulatory elements to carry out functions related to cell growth and death, organelle formation and organization, metabolite production, and microenvironment sensing. The ability to precisely manipulate the genome is essential to understanding complex and dynamic cellular processes. Broadly speaking, genome engineering defines methodological approaches to alter genomic DNA sequence (gene editing), modify epigenetic marks (epigenetic editing), modulate functional output (transcriptional regulation), and reorganize chromosomal structure (structural manipulation) (Figure 1). These goals require a toolkit of designer molecules that can be conveniently constructed and delivered into cells to perform one of the above functions.
Figure 1. A schematic view of the diverse goals of genome engineering.
Genome engineering defines methodological approaches to alter the DNA sequence (gene editing), modify the epigenetic marks (epigenetic editing), modulate the functional output (transcriptional regulation), and reorganize the chromosomal structure (structural manipulation).
Naturally occurring systems and pathways have provided a rich resource for tool building. The discovery of the homology-directed repair (HDR) pathway inspired a method to modify the DNA sequence at a precise genomic locus in a targeted manner. Using the HDR pathway, a designed DNA template with flanking homologous sequences could be used to precisely recombine at the target genomic locus [1]. However, this application is usually a highly inefficient process in mammalian cells and tissues. By contrast, the presence of a double stranded DNA break (DSB) can enhance efficiency [2, 3]. Furthermore, it has been shown that in the absence of a DNA template, eukaryotic cells may generate almost random deletion or insertion indels at the site of a DSB via the alternative non-homology end joining (NHEJ) pathway, offering another approach for targeted gene knockout [4].
Since then, a major question in the field of gene editing has been how to introduce site-specific DSBs to initiate the DNA repair process. Molecules that allow sequence-specific DNA binding were of primary interest. These include programmable endonucleases engineered from zinc finger proteins (ZFNs) or transcription activator-like effectors (TALENs) [5, 6]. The peptide domains of these proteins could be designed following a simple set of rules for protein-DNA recognition. However, their utility was hindered by an often costly and tedious process to construct and by a context dependence issue in the protein design [7, 8]. Nevertheless, previous work has shown these programmable DNA binding proteins could be coupled to nuclease domains, transcriptional repressors or activators, and epigenetic modifiers to enable diverse types of genomic manipulation [9–12]. However, it remained to be understood how to precisely target a specific DNA sequence of interest via an even simpler mechanism such as Watson-Crick base pairing.
The CRISPR (clustered regularly interspaced short palindromic repeats)/Cas (CRISPR associated protein) system performs such a function. Truly a gift from Nature [13, 14], the CRISPR/Cas system was discovered initially in Escherichia coli in the 1980s [15], but its function remained elusive until 2007. Working in the yogurt production bacterium Streptococcus thermophilus, earlier work demonstrated that encoding the bacteriophage sequence from the host CRISPR locus conferred acquired resistance against the same bacteriophage [16]. Later work showed that CRISPR utilized small CRISPR-associated RNAs (crRNAs) to guide the nuclease activity of Cas proteins in E. coli [17]. Together, this work uncovered a RNA-guided nuclease mechanism for the CRISPR system, which also suggested a genetic system with high specificity and efficiency for DNA binding and cleavage.
The practical use of CRISPR for gene editing began with the elucidation of the mechanism of the type II CRISPR system [18]. The type II CRISPR from Streptococcus pyogenes encodes a RNA-guided endonuclease protein Cas9, which was shown to use only two small RNAs (a mature crRNA and a trans-acting tracrRNA) for sequence-specific DNA cleavage [18–20]. Furthermore, a chimeric single guide RNA (sgRNA) fused between crRNA and tracrRNA recapitulated the structure and function of the tracrRNA-crRNA complex, which could efficiently direct Cas9 to induce DSBs in vitro [18]. The rules used by Cas9 to search for a DNA target are strikingly elegant and simple, requiring only a 20 nucleotide (nt) sequence on the sgRNA that base pairs with the target DNA and the presence of a DNA protospacer adjacent motif (PAM) adjacent to the complimentary region [18, 21].
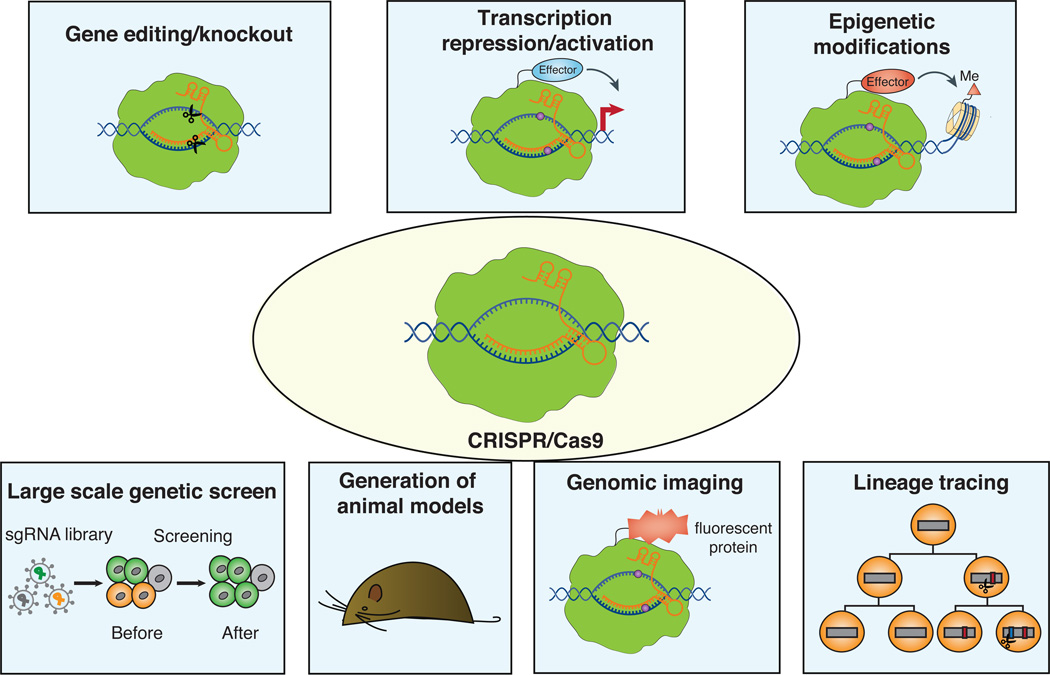
The Cas9 complex has since been developed as a remarkably useful tool of genome editing. As demonstrated by the pioneering work in a number of cell types and organisms [22–26], the Cas9/sgRNA complex can efficiently generate DSBs, which then facilitates NHEJ-mediated gene knockout or HDR-mediated recombination. This system has since gained rapid acceptance and has been used for genome editing in essentially all organisms that can be cultured in the laboratory setting. In this review, we focus on recent applications of CRISPR-Cas9 in cell biology research using mammalian cell cultures and animal models (Figure 2).
Figure 2. Applications of CRISPR/Cas9 to the cell biology research.
The CRISPR/Cas9 technology has been used for gene editing, transcriptional regulation, epigenetic regulation, large-scale genetic screens, generation of animal models, and genomic imaging.
An expanding CRISPR toolkit for RNA-guided genome editing
The different types of natural CRISPR systems encode a toolkit for genome editing. Six major types of CRISPR systems have been identified from different organisms (types I–VI), with various subtypes in each major type [27, 28].Within the type II CRISPR system, several species of Cas9 have been characterized from S. pyogenes, S. thermophilus, N. meningitidis, S. aureus, and F. novicida [18, 29–34].While these Cas9s possess a similar RNA-guided DNA binding mechanism, they often have distinct PAM recognition sequences. Similar to the toolkit of restriction enzymes for molecular cloning, a large toolkit of Cas9s expands the targetable genome sequence for gene editing and genome manipulation.
Other types of CRISPR systems may exhibit different mechanisms. For example, the Type III-B CRISPR system from Pyrococcus furiosus uses a Cas complex for RNA-directed RNA cleavage [35, 36], which is indicative of a mechanism for targeting and modulating RNAs in cells. The recent discovery of the protein Cpf1 from Prevotella and Francisella-1 type V CRISPR showed that Cpf1 uses a short crRNA without a tracrRNA for RNA-guided DNA cleavage [37–40]. Both biochemical and cell culture work showed that Cpf1-mediated genome targeting is effective and specific, comparable to the S. pyogenes Cas9. The type VI-A CRISPR effector C2c2 from the bacterium Leptotrichia shahii is a RNA-guided RNase, which can be programmed to knock down specific mRNAs in bacteria [41]. These results broaden our understanding of the diversity of the natural CRISPR-Cas systems, which also provides a functionally diverse set of tools.
Other enzymatic domains can also be harnessed for genome editing. For example, instead of using the endonuclease activity of Cas9, a mutation in one nuclease domain of Cas9 can create a nickase Cas9 (nCas9) that can cleave one strand of DNA [42].With a pair of sgRNAs, the specificity of genome editing could be enhanced by using a pair of nCas9s that target each strand of DNA at adjacent sites. Furthermore, recent work demonstrated that a Cas9-fused cytidine deaminase enzyme allowed for direct conversion of a C to T (or G to A) substitution [43]. In this work, fusing the nuclease-deactivated dCas9 or the nCas9 with a cytidine deaminase domain corrected point mutations relevant to human disease without DSBs; therefore, avoiding NHEJ-mediated indel formation.
Applications of CRISPR/Cas9 for cell biological studies
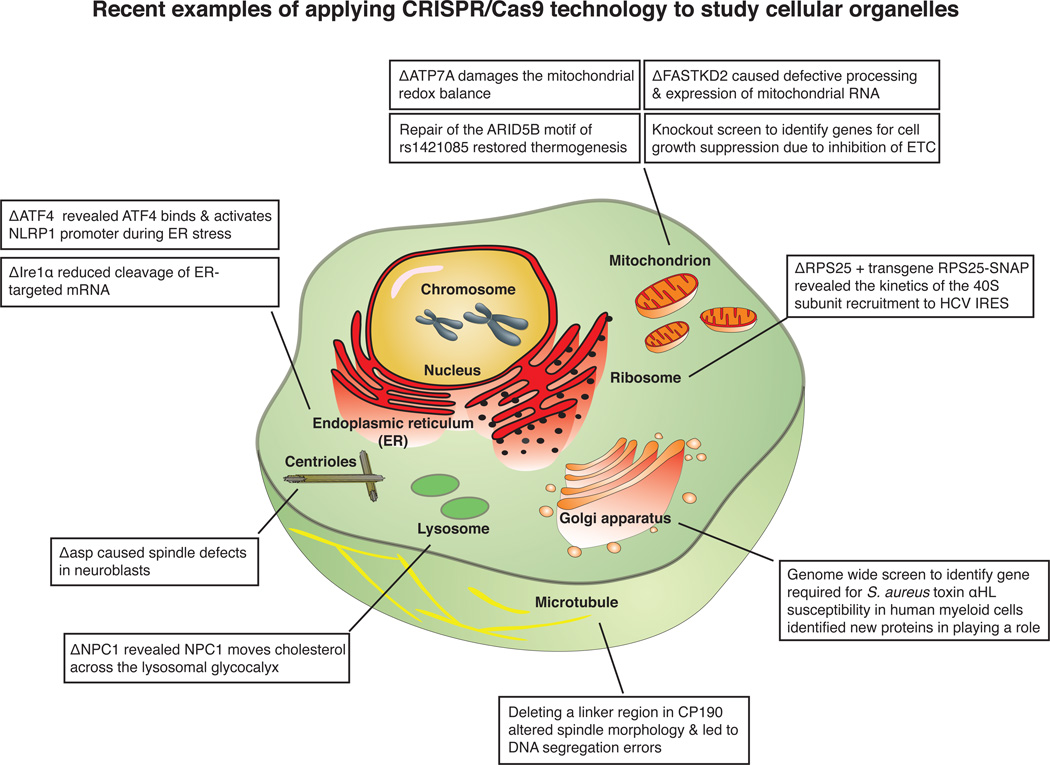
The CRISPR/Cas9 technology has accelerated the discovery and mechanistic interrogation of the genome and organelles in diverse types of cells and organisms. Some examples of utilizing CRISPR/Cas9 for studying cellular organelles are summarized in Table 1 and Figure 3. Beyond using CRISPR/Cas9 as a gene-editing tool, we describe the development of CRISPR/Cas9 as a versatile toolkit for transcriptional control and epigenetic regulation, and highlight its utilities for large-scale genetic screens, generation of animal models, genomic imaging, and lineage tracing (Figure 2).
Table 1.
Examples of CRISPR/Cas9 being used for cell biology research
| Organelle | CRISPR-Cas9 target | Finding | Ref |
|---|---|---|---|
| Microtubule | CRISPR/Cas9 generation of mutant flies by deleting a linker region in the centrosome protein CP190 |
Identified a centrosome and microtubule targeting region in CP190 for spindle localization Deletion of the linker region altered spindle morphology and led to DNA segregation errors |
[44] |
| Mitochondria | CRISPR/Cas9 knockout of the copper transporting ATPase ATP7A in mouse 3T3-L1 cells and in Menkes Disease (MD) patient fibroblasts |
ATP7A dysfunction damages the mitochondrial redox balance |
[45] |
| CRISPR/Cas9 knockout of FASTKD2, a RNA binding protein of the FAS-activated serine/threonine kinase family |
Defective processing and expression of mitochondrial RNA Cellular respiration damage with depressed activities of respiratory complexes |
[46] | |
| CRISPR/Cas9 mediated repair of the ARID5B motif of rs1421085 in primary adipocytes from a patient carrying the risk allele |
IRX3 and IRX5 repression restored Browning expression programs activated Thermogenesis restored |
[47] | |
| CRISPR/Cas9-based genetic screen to study cell proliferation suppression due to inhibition of the mitochondrial electron transport chain (ETC) |
Identified the cytosolic aspartate aminotransferase (GOT1) as the key gene GOT1 loss of function kills cells upon ETC inhibition. |
[48] | |
| Endoplasmic Reticulum | CRISPR/Cas9 knockout of ATF4 or NLRP1 |
NLRP1 is up-regulated during severe ER stress ATF4 binds and activates NLRP1 promoter during ER stress |
[49] |
| CRISPR/Cas9 mediated deletion of transmembrane endoribonuclease Ire1α in HEK293 cells |
Ire1α forms a complex with the Sec61 translocon to cleave its mRNA substrates Disruption of Ire1α complex caused reduced cleavage of ER-targeted mRNA |
[50] | |
| Centrosome | CRISPR/Cas9 dual-sgRNA to generate a null abnormal spindle (asp) allele by excising a 750-bp fragment that included the promoter, 5’ UTR, and the first exon in Drosophila neuroblasts |
Asp null mutations cause spindle defects in neuroblasts Asp is regulated by Drosophila melanogaster calmodulin (CaM) to cross-link spindle microtubules |
[51] |
| Lysosome | Generation of Niemann-Pick type C 1 (NPC1)-deficient cell line using CRISPR/Cas9 |
NPC1 moves cholesterol across the lysosomal glycocalyx |
[52] |
| Ribosome | CRISPR/Cas9 knockout of nonessential gene of ribosomal protein eS25 (RPS25) in Hap1 cell line. RPS25-SNAP (mutant O6- alkylguanine DNA alkyl- transferase) transgene was transduced into RPS25-KO Hap1 cells to be the only source of the protein. |
Demonstrated an approach to create fluorescently labeled 40S ribosomal subunits from human cells Studied kinetics of the 40S subunit recruitment to the hepatitis C virus (HCV) internal ribosome entry site (IRES) |
[53] |
| Golgi apparatus | Genome-wide CRISPR/Cas9 loss-of-function screen to identify host targets required for S. aureus toxin alpha hemolysin (αHL) susceptibility in human myeloid cells. |
Identified new proteins (SYS1, ARFRP1, TSPAN14) in regulating the presentation of ADAM10 on the plasma membrane post-translationally Cells lacking sphingomyelin synthase 1 (SGMS1) resist αHL intoxication |
[54] |
Figure 3. Examples of applying the CRISPR/Cas9 technology to study cellular organelles.
The figure illustrates exemplar studies in particular organelles, with more details listed in Table 1.
Transcriptional regulation of the genome with CRISPR/dCas9
The nuclease-dead dCas9 has provided a broad platform for programming diverse types of transcriptional or epigenetic manipulation of the genome, without altering the genome sequence. In brief, dCas9 was created by introducing point mutations into the HNH and RuvC domains to eliminate endonuclease activity [55]. This repurposed protein became a RNA-guided DNA binding protein. In bacteria, the dCas9 protein was sufficient to induce strong sequence-specific gene repression, simply by sterically hindering the transcriptional activity of RNA polymerase [55, 56]. In eukaryotic cells, fusing dCas9 to transcriptional effector proteins allowed for more efficient RNA-guided transcriptional modulation for both gene interference (CRISPRi) and activation (CRISPRa) [12, 57–59].
By fusing dCas9 to transcriptional repressors, such as the Kruppel-associated box (KRAB) domain, CRISPRi can efficiently repress coding and noncoding genes such as microRNAs and large intergenic noncoding RNAs (lincRNAs) in mammalian cells [57, 58, 60, 61]. Compared to complete loss-of-function using Cas9, CRISPRi can use different sgRNAs that bind to different genomic loci for tunable and titratable gene repression [58].While complete knockout is useful for studying gene function in many cases, tunable repression of a gene to different levels offers advantages when knocking out a gene leads to lethality of cells or an organism [56].
Earlier work using dCas9 fused to a peptide containing multiple VP16 domains (VP64 or VP128) could only activate endogenous genes mildly [57, 62, 63]; therefore, several strategies have been developed to improve CRISPRa efficiency. These include recruiting multiple copies of the VP64 domain via a multimeric peptide array (SunTag), wherein each peptide domain could bind to a single-chain variable fragment (scFv) fused to VP64 [64]; fusing dCas9 to a synergistic tripartite activator system containing VP64, the activation domain of p65 (p65AD), and Rta (Epstein-Barr virus R transactivator) [65]; and combining dCas9-VP64 with a modified sgRNA engineered with two copies of an MS2 RNA hairpin that could recruit p65AD and the HSF1 (human heat shock factor 1) activation domain via interaction with the MS2 binding protein [59]. A systematic comparison of the efficacy of these methods revealed that these systems perform comparable but are dependent on the genomic and cellular context [66], suggesting that activation efficiency may vary for different genes and in different types of cells. In the future, simpler yet more effective tools for RNA-guided gene activation should be further developed.
To repurpose more complex gene regulation, sgRNA was engineered as a class of “scaffold” RNAs (scRNAs) that directly recruit transcription effectors without protein fusion [67]. scRNAs are generated by fusing RNA hairpins to the sgRNA, which interact with the cognate protein to recruit activators or repressors. Using engineered scRNAs, multiple genes can be simultaneously activated and repressed in the same cells. In addition to using scRNAs, multiple orthogonal species of dCas9s could also provide a platform for complex transcription regulation and sophisticated manipulation of the transcriptome.
Epigenetic regulation with CRISPR/dCas9
dCas9 fused to epigenetic modifying enzymes has been used to introduce locus-specific epigenetic modifications in the genome. Examples include fusing dCas9 to the core catalytic domain of the human acetyltransferase p300 (p300core), which allows acetylation of histone H3 Lys27 (H3K27), and upregulation of genes when binding to proximal or distal enhancers [68]; fusing dCas9 to lysine demethylase 1 (LSD1) reduces the acetylation level of H3K27 [69]; fusing dCas9 to KRAB increases the H3K9me3 mark near the target site [70]; and fusing dCas9 to the DNA methyltransferase DNMT3A increases CpG methylation near the target site [71]. These works also demonstrated modified gene expression levels due to Cas9-mediated locus-specific epigenetic modifications. For example, in mouse embryonic stem cells, the enhancers of pluripotency factors, such as Oct4 and Tbx3 could be repressed by dCas9-LSD1 fusion, leading to loss of pluripotency [69, 72].
While these examples provide an approach to edit the epigenetic states of essentially any locus in the genome, a largely unexplored question is the fate of the synthetic epigenetic marks, and whether or not they can be stably inherited when cells proliferate. Furthermore, given the diverse types of epigenetic modifications and their mutual interactions, a comprehensive toolkit consisting of multiple orthogonally acting dCas9s and their cognate sgRNA that allows flexibly editing multiple epigenetic (histone or DNA) marks simultaneously is needed. The toolkit is useful for understanding the function of diverse epigenetic marks, their interactions, and their relationship to genomic and cellular functions.
Large-scale functional genomic studies using CRISPR/Cas9
One of the powerful applications of the CRISPR/Cas9 technology is the high-throughput screening of genomic functions. The oligo libraries encoding hundreds of thousands of sgRNAs can be computationally designed and chemically synthesized to target a broad set of genome sequences. Pairing with Cas9 or dCas9 fusion proteins, this provides an approach to systematically knock out, repress, or activate genes on a large-scale. The technique requires a delicate delivery method that ensures every cell only receives a single sgRNA, usually via lentiviral or retroviral delivery into mammalian cells. The screens are frequently performed in a pooled manner, as cells transduced with the lentiviral library as a mixed population are cultured together. Via deep sequencing and analysis of the sgRNA features in the pooled cells, genes causing changes in cell growth and death can be inferred with bioinformatics. Indeed, CRISPR screens can easily identify genes, their regulatory elements, and protein domains in the mammalian genome responsible for cell growth and drug resistance [73]. A genomic tiling screen using CRISPR/Cas9 precisely mapped functional domains within enhancer elements and found that a p53-bound enhancer of the p53 effector gene CDKN1Ais required for oncogene-induced senescence in immortalized human cells [74].
Using the endonuclease Cas9, loss-of-function genome-wide knockout screens have been performed in cultured or primary mammalian cells with sgRNA libraries (usually 3–10 sgRNAs per gene) to investigate a range of phenotypes including cell growth, cancer cell drug resistance, and viral susceptibility [75–77]. A genome-scale sgRNA library can also be used to manipulate cultured cells that are later introduced in vivo. Indeed a genome-scale sgRNA library was created to mutagenize a non-metastatic mouse cancer cell line for the study of metastasis in a mouse model [78]. The mutant cell pool rapidly generated metastases when transplanted into immunocompromised mice in vivo. Sequencing of the metastatic cells suggested genes that accelerate lung cancer metastases and development of late-stage primary tumors. Moreover, this screening method can be extended to use in primary cells, which can lead to novel findings that are often overlooked using cell lines. Indeed, introducing a genome-wide sgRNA library into primary dendritic cells (DCs) allowed for the identification of genes related to cell growth that induce tumor necrosis factor (TNF) in response to bacterial lipopolysaccharide (LPS), an essential host response to pathogens [79], which would otherwise be technically challenging with other genome editing tools.
Cas9-mediated loss-of-function screens have also performed to knock out pairs of genes in combination [80]. A library of 23,409 barcoded dual sgRNA combinations were created and a pooled screen was performed to identify gene pairs in human cells that inhibit ovarian cancer cell growth in the presence of small molecule drugs. While further work is needed to characterize the efficacy and accuracy of multiplex genetic screening, this work highlights the potential of more sophisticated functional screening studies using CRISPR.
Beyond Cas9-based complete loss-of-function screens, the invention of CRISPRi and CRISPRa further enables both partial loss-of-function and gain-of-function genetic screens [58, 59]. Growth-based screens using CRISPRi/a have been used to identify essential genes, tumor suppressor genes, and potential mechanisms that confer cytotoxicity induced by a cholera-diphtheria toxin [58]. Using a library consisting of ~70,000 guides targeting the human RefSeq coding isoforms, a CRISPRa-based screen identified genes that, upon activation, conferred resistance to a BRAF inhibitor [59].
In addition to the use of pooled screens, multi-well plates have been used in combination with the partial repression feature of CRISPRi to study the function of the full set of essential genes in the gram-positive bacterium Bacillus subtilis [56]. As knocking out essential genes results in lethality that prevents further assay of the phenotype, partial knockdown of essential genes becomes a powerful approach. A mutant B. subtilis library was created to include gene partial knockdowns (~3-fold) of all essential genes using CRISPRi, which was tested for the growth phenotype under 35 unique compounds. Using this chemical genomic approach, a comprehensive interconnecting essential gene network was identified, as well as targeted genes that interact with uncharacterized antibiotics. Inducible knockdown of essential genes also allowed for systematic characterization of cell morphology and terminal death phenotypes.
An important question is how these screens compare to each other and to other existing approaches. Several works compared different screens based on CRISPR, CRISPRi and RNA interference (RNAi). One work performed comparative screens of 46 essential and 47 nonessential genes, and concluded that the CRISPR/Cas9 nuclease system outperformed the shRNA- and CRISPRi/dCas9-based gene regulation systems for the sets of essential and nonessential genes [81]. From the CRISPR screening data, they observed less variation across the data, and detected more functional constructs with less off-target effects. Another work concluded that CRISPR could identify more essential gene targets compared to RNAi [82]. Since similar precision was observed between the two approaches, it was suggested that combining data from both screens could improve the predictive accuracy. The systematic comparison of different approaches may suggest that a comparative screening approach will be more powerful for studying complex cell biology phenotypes.
In addition, new methods to generate CRISPR libraries may help reduce the overall cost associated with this technique and extend its uses to screen a larger chromosomal region (e.g., the tiling along a whole chromosome). While most CRISPR libraries are generated via chemical synthesis of large pools of oligos, a new method termed CRISPR EATING (Everything Available Turned Into New Guides) can inexpensively generate large quantities of sgRNAs for whole genome targeting [83]. In this approach, PAM-proximal sequences are extracted by digesting input DNA with restriction enzymes that target immediately 5’ to an NGG or NAG (the PAM sequences for S. pyogenes Cas9, N = any nucleic acid). In this study, one library was generated and used to label the whole 3.4-mb region on X. laevis chromosome 4 in the egg extracts. The method allows for the generation of complex and customized libraries from any source of DNA via routine molecular biology methods.
CRISPR/Cas9 for generating animal models
Genetically engineered animal models are crucial for the study of complex cellular and physiological processes. While mouse models have been widely used, the CRISPR/Cas9 gene editing approach has been established in many other animal models including worm [84], fly [85], fish [86, 87], rat [88], rabbit [89, 90], goat [91], sheep [92], dog [93], pig [94], and monkeys [95]. The expansion of transgenic animal models beyond mouse is advantageous to biomedical research that accelerates the development of new therapeutic strategies.
CRISPR provides a much easier approach to establish these transgenic animal models compared to previous gene-editing tools. Traditional approaches to construct transgenic mice via insertional mutagenesis or TALEN-mediated gene editing are time-consuming, costly, and inefficient. The robustness and high efficiency of CRISPR-Cas9 simplifies the process for creating model systems [96, 97]. Moreover, nucleic acids encoding the Cas9 protein and target-specific sgRNAs can be conveniently injected into embryos to generate gene-modified mice with deletions of multiple genes, mutations in defined genes, or insertions of fluorescence reporters or other peptide tags to endogenous genes. For example, coinjection of Cas9 mRNA and sgRNAs targeting Tet1 and Tet2 into zygotes generated mice with biallelic mutations in both genes with an efficiency of 80% [96]. Furthermore, coinjection of Cas9 mRNA and sgRNAs with mutant oligos generated precise point mutations simultaneously in two target genes, while coinjecting Cas9 mRNA and sgRNAs into one-cell-stage cynomolgus monkey embryos generated founder animals harboring two gene modifications [95].
The establishment of a Cre-conditional Cas9 knockin mouse has broadened the applications of Cas9 in vivo [98]. The Cas9 knockin mouse is a great resource to rapidly generate mutations in a subpopulation of cells in vivo, and test how mutations cause disease phenotypes. Different methods based on adeno-associated virus (AAV), lentivirus, or nanoparticles can be used to deliver sgRNAs into multiple cell types such as neurons, immune cells, and endothelial cells in a Cas9 knockin mouse to model the dynamics of significantly mutated genes in lung adenocarcinoma [98]. Another work demonstrated that the Cre-conditional Cas9 knockin mouse phenocopied Cre-mediated genetic deletion of genes in Cre/LoxP mouse models in studying pancreatic ductal adenocarcinoma [99]. Via retrograde pancreatic ductal injection of lentiviral vectors expressing Cre and an sgRNA into Cre-conditional Cas9 knockin mice, the authors showed knockout of Lkb1 together with manipulated expression of oncogenic Kras. However, due to the heterogeneity of delivery and Cas9-mediated gene editing, caution is required when interpreting results.
In addition to using a Cas9 knockin mouse model, viral vectors encoding Cas9 and an sgRNA can be directly delivered into wildtype mice or Cre/loxP mouse models to probeg gene function. One work used adeno-associated viral (AAV) vectors encoding Cas9 and sgRNAs to target a single gene or multiple genes in the normal adult mouse brain in vivo [100]. Characterizing the effects of gene modifications in postmitotic neurons revealed similar phenotypes as observed in gene knockout mice. Another work used a lentiviral system that delivers both the CRISPR system and Cre recombination to examine CRISPR-induced mutation of genes in the context of well-studied conditional Cre/loxP mouse models of lung cancer and other cancer types [101]. In another work to study cancer genes in the mouse liver, a hydrodynamic injection delivered a plasmid DNA expressing Cas9 and sgRNAs that directly targeted the tumor suppressor genes (p53 or PTEN) alone and in combination into the liver. The authors demonstrated the feasibility of Cas9-mediated mutation of tumor suppressor genes in the liver as an avenue for rapid development of liver cancer models [102]. However, similar to the Cas9 knockin mouse, the virally delivered Cas9 may only edit genes in a fraction of cells, and the approach may be most effective for studying the effects of loss-of-function mutations on cell autonomous properties.
Genome imaging using CRISPR/Cas9
Imaging offers a direct approach for studying the spatial and temporal behavior of the genome in living cells [103]. The ability of Cas9 to target specific sequences in the genome makes it a promising imaging tool for directly observing genomic organization and dynamics in cells. The first proof-of-concept work fused the S. pyogenes dCas9 to EGFP and used the fusion protein to visualize the dynamics of coding or non-coding sequences in living human cell lines [104]. The authors tracked the dynamics of telomeres, and the repetitive and nonrepetitive sequences of coding genes (MUC4, MUC1) in a short time frame (~minutes) and throughout the whole cell cycle. In addition, dCas9 fused to EGFP has been used to label endogenous centromeres and telomeres loci in live mouse embryonic stem cells [105]. The development of the SunTag system, a repeating peptide array that can recruit multiple copies of an antibody-fusion protein, enhanced the sensitivity to amplify the dCas9 fluorescent signal in the genome [64]. Using dCas9 orthologs tagged with different fluorescent proteins, it was shown that the dynamics of multiple repetitive genomic loci could be tracked in living cells [106]. A method termed CASFISH (Cas9-mediated fluorescence in situ hybridization) further combined dCas9 with FISH (fluorescence in situ hybridization) [107]. Due to the specific DNA targeting and unwinding activity of dCas9, CASFISH allowed a fast and convenient process for labeling DNA elements while avoiding treatment of heat and disruptive chemicals that distort the natural organization of the nucleus, which is normally seen in FISH. Thus the approach preserves the spatial relationships of the genetic elements that are important for studying gene expression.
Recent work also established a CRISPR approach to facilitate super-resolution imaging in living mammalian cells [108]. Current live cell super-resolution imaging normally relies on the overexpression of a host protein fused to a fluorescent protein, which results in artifacts that may obscure the interpretation of imaging results. Using CRISPR/Cas9 to fluorescently tag the endogenous genes that are expressed from their native genomic loci could allow genes to be expressed at close to endogenous levels, thus avoiding artifacts. Based on the idea, a method termed RESOLFT (reversible saturable optical fluorescence transitions) was developed, wherein heterozygous and homozygous Cas9-edited human knockin cell lines were generated that expressed the reversibly switchable fluorescent protein rsEGFP2 from their respective native genomic, which prevent the appearance of typical overexpression-induced artifacts in these cells.
To enhance signals for endogenous proteins imaging, one work adapted self-complementing split fluorescent proteins, GFP11 and sfCherry11, derived from the sfGFP and sfCherry [109]. The small sizes of these split fluorescent domains (16–18 amino acids) allow easily inserting them into endogenous genomic loci via CRISPR gene editing. Tandem arrays of these domains further amplify fluorescence signals in imaging, such as for tracking intraflagellar transport particles.
In addition to DNA imaging, S. pyogenes dCas9 can also allow for endogenous RNA imaging in living cells [110]. In the presence of sgRNAs targeting mRNA and a stabilized PAMmer oligonucleotide that contains the PAM domain for dCas9 binding, specifically targeted RNA can be visualized. Indeed, it was observed that nuclear localized dCas9 could be exported to the cytoplasm. Furthermore dCas9 allowed for tracking of RNA during induced RNA/protein accumulation in the presence of oxidative stress.
Lineage tracing using CRISPR/Cas9
It is worth noting that gene editing has been used as tools for cell lineage tracing. One recent work demonstrated a lineage tracing method termed genome editing of synthetic target arrays for lineage tracing (GESTALT) [111]. This method uses CRISPR/Cas9 gene editing to generate a combinatorial diversity of mutations that accumulate over cell divisions within a series of DNA barcodes. Via deep sequencing, lineage relationship between many cells can be inferred using patterns of the edited barcodes. The approach was developed in both cell culture and zebrafish, by editing synthetic arrays of approximately a dozen CRISPR/Cas9 target sites. The approach generated thousands of unique edited barcodes in cell lines, which could be sequenced from either DNA or RNA. By injecting fertilized eggs with editing reagents that targeted a genomic barcode with 10 target sites, the authors observed the accumulation of hundreds to thousands of uniquely edited barcodes per animal, and further inferred the lineage relationship between ancestral progenitors and organs based on mutation patterns. This proof-of-principle work showed that combinatorial and cumulative genome editing is a powerful approach to record lineage information in multicellular systems.
In another work, the type I-E CRISPR/Cas system of Escherichia coli was harnessed to generate records of specific DNA sequences in bacterial genomes [112]. Unlike gene editing, the work is based on the native adaptive immunity acquisition ability of CRISPR, as new spacer sequences can be acquired and integrated stably into the CRISPR crRNA array. Using this feature, it was demonstrated that the Cas1–Cas2 complex enables recording of defined sequences over many days and in multiple modalities. The work elucidated fundamental aspects of the CRISPR acquisition process. The recording system developed could be useful for applications that require tracing long histories of in vivo cellular activity.
While optimization of these methods is required for more robust performance, genome editing and the unique features (i.e., adaptation) of the CRISPR system provide promising approaches to record biological information and history in living cells and tissues. One can envision that these tools may enable mapping of the complete cell lineage in multicellular organisms as well as linking cell lineage information to molecular profiles (e.g., transcription, epigenetics, and proteomics) such as those in single cells.
Concluding remarks
CRISPR/Cas9 technology has revolutionized cell biology research. The system is versatile, enabling diverse types of genome engineering approaches. While most of the work has used Cas9- mediated knockout or dCas9-mediated repression and activation to study gene function, we expect expansion of these tools to study the epigenome and 3-dimensional chromosomal organization in greater detail in the future. Furthermore, studies have used CRISPR to model complex genomic rearrangements in vitro and in vivo, which brought breakthroughs in studying chromosomal translocations [113, 114]. Most research has been performed in cell lines, and future work related to the interrogation of cellular functions should be carried out in primary cells derived from animals or human patients or in vivo using relevant animal models.
CRISPR/Cas9 is emerging as a major genome-manipulation tool for research and therapeutics, yet there are remaining challenges to improve its specificity, efficiency, and utility (see Outstanding Questions). One major concern is the off-target effects, since Cas9 can tolerate mismatches between sgRNA and target DNA [115–117]. Methods have been developed to profile the off-target effects such as GUIDE-seq [118]. To improve specificity, several strategies have been developed including using paired nickase variants of Cas9 [32, 42], paired dCas9-FokI nucleases [119, 120], truncated sgRNAs ((17 to 18 bp) that are more sensitive to mismatches [121], and controlling acting concentration of the Cas9/sgRNA complex [122]. Using structure-guided protein engineering approaches, a few work recently created S. pyogenes Cas9 variants with improved specificity [123, 124]. For example, a high fidelity variant of Cas9 harboring designed alterations showed reduced non-specific DNA contacts, while retaining robust on-target activities comparable to wild-type Cas9 [124]. Combinations of these methods could provide a route to its ultimate use for gene therapy.
Outstanding questions.
How can the off-target effects of CRISPR/Cas9 be avoided in mammalian cells and whole organisms?
Can CRISPR/Cas9 technology be developed to insert a large gene fragment into the mammalian genome for gene knockin studies with similar efficiency to that of gene knockout studies?
Will CRISPR/Cas9 technology be able to efficiently modulate different types of epigenetic modifications? Can it control the fate of synthetic epigenetic marks, and whether or not they can be stably inherited when cells proliferate?
Can CRISPR/Cas9-mediated genetic screens be performed on non-proliferation based phenotypes such as differentiation?
Can CRISPR/Cas9 technology enable more robust transgenic animal generation by deleting, mutating, and inserting any gene of interest?
Beyond gene editing, how can CRISPR/Cas9 be used to help advance cell biology research?
As a powerful yet versatile gene editing and regulation tool, the CRISPR-Cas9 technology is already accelerating both research and therapeutics. We believe its broad applications in genomics research and cell biology research will greatly advance our knowledge of both basic biology and diseases in the years to come.
Trends.
The RNA-guided CRISPR/Cas9 endonuclease and the endonuclease-dead dCas9 protein are powerful genomic manipulations tools for gene editing, transcriptional regulation, and epigenetic modifications.
Both Cas9 and dCas9 enable diverse types of high-throughput screenings of gene functions in cell lines and in vivo
The CRISPR/Cas9 accelerates the establishment of many useful transgenic animal models for biomedical research.
The CRISPR/Cas9 is repurposed for genomic imaging and lineage tracing in living cells and tissues.
Acknowledgments
F.W. acknowledges support from the Center of Synthetic Biology and Shanghai Institute of Rheumatology in Renji Hospital affiliated to Shanghai Jiaotong University School of Medicine, National Natural Science Foundation of China (No. 81502233), the Science and Technology Commission of Shanghai Municiplality (No. 134119a8100) and the Young Scientific Research Project of Shanghai Municipal Health Bureau (No. 20134Y176). L.S.Q. acknowledges support from the NIH Office of the Director (OD), National Institute of Dental & Craniofacial Research (NIDCR), and NIH Director’s Early Independence Award DP5 OD017887.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Capecchi MR. Altering the genome by homologous recombination. Science. 1989;244:1288–1292. doi: 10.1126/science.2660260. [DOI] [PubMed] [Google Scholar]
- 2.Rudin N, et al. Genetic and physical analysis of double-strand break repair and recombination in Saccharomyces cerevisiae. Genetics. 1989;122:519–534. doi: 10.1093/genetics/122.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bibikova M, et al. Stimulation of homologous recombination through targeted cleavage by chimeric nucleases. Mol Cell Biol. 2001;21:289–297. doi: 10.1128/MCB.21.1.289-297.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bibikova M, et al. Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics. 2002;161:1169–1175. doi: 10.1093/genetics/161.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Urnov FD, et al. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- 6.Christian M, et al. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010;186:757–761. doi: 10.1534/genetics.110.120717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaj T, et al. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wolfe SA, et al. DNA recognition by Cys2His2 zinc finger proteins. Annu Rev Biophys Biomol Struct. 2000;29:183–212. doi: 10.1146/annurev.biophys.29.1.183. [DOI] [PubMed] [Google Scholar]
- 9.Beerli RR, Barbas CF., 3rd Engineering polydactyl zinc-finger transcription factors. Nat Biotechnol. 2002;20:135–141. doi: 10.1038/nbt0202-135. [DOI] [PubMed] [Google Scholar]
- 10.Konermann S, et al. Optical control of mammalian endogenous transcription and epigenetic states. Nature. 2013;500:472–476. doi: 10.1038/nature12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang F, et al. Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nat Biotechnol. 2011;29:149–153. doi: 10.1038/nbt.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maeder ML, et al. CRISPR RNA-guided activation of endogenous human genes. Nat Methods. 2013;10:977–979. doi: 10.1038/nmeth.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wiedenheft B, et al. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–338. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- 14.Marraffini LA, Sontheimer EJ. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat Rev Genet. 2010;11:181–190. doi: 10.1038/nrg2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakata A, et al. Cloning of alkaline phosphatase isozyme gene (iap) of Escherichia coli. Gene. 1982;19:313–319. doi: 10.1016/0378-1119(82)90021-x. [DOI] [PubMed] [Google Scholar]
- 16.Barrangou R, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 17.Brouns SJ, et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deltcheva E, et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sapranauskas R, et al. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res. 2011;39:9275–9282. doi: 10.1093/nar/gkr606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marraffini LA, Sontheimer EJ. Self versus non-self discrimination during CRISPR RNA-directed immunity. Nature. 2010;463:568–571. doi: 10.1038/nature08703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jinek M, et al. RNA-programmed genome editing in human cells. Elife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hwang WY, et al. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cho SW, et al. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31:230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- 27.Makarova KS, et al. An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol. 2015;13:722–736. doi: 10.1038/nrmicro3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chylinski K, et al. Classification and evolution of type II CRISPR-Cas systems. Nucleic Acids Res. 2014;42:6091–6105. doi: 10.1093/nar/gku241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gasiunas G, et al. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci U S A. 2012;109:E2579–E2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ran FA, et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y, et al. Processing-independent CRISPR RNAs limit natural transformation in Neisseria meningitidis. Mol Cell. 2013;50:488–503. doi: 10.1016/j.molcel.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mali P, et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sampson TR, et al. A CRISPR/Cas system mediates bacterial innate immune evasion and virulence. Nature. 2013;497:254–257. doi: 10.1038/nature12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hirano H, et al. Structure and Engineering of Francisella novicida Cas9. Cell. 2016;164:950–961. doi: 10.1016/j.cell.2016.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hale CR, et al. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009;139:945–956. doi: 10.1016/j.cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hale CR, et al. Essential features and rational design of CRISPR RNAs that function with the Cas RAMP module complex to cleave RNAs. Mol Cell. 2012;45:292–302. doi: 10.1016/j.molcel.2011.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zetsche B, et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamano T, et al. Crystal Structure of Cpf1 in Complex with Guide RNA and Target DNA. Cell. 2016;165:949–962. doi: 10.1016/j.cell.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dong D, et al. The crystal structure of Cpf1 in complex with CRISPR RNA. Nature. 2016;532:522–526. doi: 10.1038/nature17944. [DOI] [PubMed] [Google Scholar]
- 40.Fonfara I, et al. The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature. 2016;532:517–521. doi: 10.1038/nature17945. [DOI] [PubMed] [Google Scholar]
- 41.Abudayyeh OO, et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016;353:aaf5573. doi: 10.1126/science.aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ran FA, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Komor AC, et al. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533:420–424. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Plevock KM, et al. Newly Characterized Region of CP190 Associates with Microtubules and Mediates Proper Spindle Morphology in Drosophila Stem Cells. PLoS One. 2015;10:e0144174. doi: 10.1371/journal.pone.0144174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bhattacharjee A, et al. Activity of Menkes Disease Protein ATP7A is Essential for Redox Balance in Mitochondria. J Biol Chem. 2016;291:16644–16658. doi: 10.1074/jbc.M116.727248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Popow J, et al. FASTKD2 is an RNA-binding protein required for mitochondrial RNA processing and translation. RNA. 2015;21:1873–1884. doi: 10.1261/rna.052365.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Claussnitzer M, et al. FTO Obesity Variant Circuitry and Adipocyte Browning in Humans. N Engl J Med. 2015;373:895–907. doi: 10.1056/NEJMoa1502214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Birsoy K, et al. An Essential Role of the Mitochondrial Electron Transport Chain in Cell Proliferation Is to Enable Aspartate Synthesis. Cell. 2015;162:540–551. doi: 10.1016/j.cell.2015.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.D'Osualdo A, et al. Transcription Factor ATF4 Induces NLRP1 Inflammasome Expression during Endoplasmic Reticulum Stress. PLoS One. 2015;10:e0130635. doi: 10.1371/journal.pone.0130635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Plumb R, et al. A functional link between the co-translational protein translocation pathway and the UPR. Elife. 2015;4:e07426. doi: 10.7554/eLife.07426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schoborg T, et al. An Asp-CaM complex is required for centrosome-pole cohesion and centrosome inheritance in neural stem cells. J Cell Biol. 2015;211:987–998. doi: 10.1083/jcb.201509054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li J. Glycosylation inhibition reduces cholesterol accumulation in NPC1 protein-deficient cells. Proc Natl Acad Sci U S A. 2015;112:14876–14881. doi: 10.1073/pnas.1520490112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fuchs G, et al. Kinetic pathway of 40S ribosomal subunit recruitment to hepatitis C virus internal ribosome entry site. Proc Natl Acad Sci U S A. 2015;112:319–325. doi: 10.1073/pnas.1421328111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Virreira Winter S, et al. Genome-wide CRISPR screen reveals novel host factors required for Staphylococcus aureus alpha-hemolysin-mediated toxicity. Sci Rep. 2016;6:24242. doi: 10.1038/srep24242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Qi LS, et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Peters JM, et al. A Comprehensive, CRISPR-based Functional Analysis of Essential Genes in Bacteria. Cell. 2016;165:1493–1506. doi: 10.1016/j.cell.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gilbert LA, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gilbert LA, et al. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell. 2014;159:647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Konermann S, et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517:583–588. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Larson MH, et al. CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat Protoc. 2013;8:2180–2196. doi: 10.1038/nprot.2013.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhao Y, et al. Sequence-specific inhibition of microRNA via CRISPR/CRISPRi system. Sci Rep. 2014;4:3943. doi: 10.1038/srep03943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Perez-Pinera P, et al. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat Methods. 2013;10:973–976. doi: 10.1038/nmeth.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cheng AW, et al. Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res. 2013;23:1163–1171. doi: 10.1038/cr.2013.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tanenbaum ME, et al. A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell. 2014;159:635–646. doi: 10.1016/j.cell.2014.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chavez A, et al. Highly efficient Cas9-mediated transcriptional programming. Nat Methods. 2015;12:326–328. doi: 10.1038/nmeth.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chavez A, et al. Comparison of Cas9 activators in multiple species. Nat Methods. 2016;13:563–567. doi: 10.1038/nmeth.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zalatan JG, et al. Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds. Cell. 2015;160:339–350. doi: 10.1016/j.cell.2014.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hilton IB, et al. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol. 2015;33:510–517. doi: 10.1038/nbt.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kearns NA, et al. Functional annotation of native enhancers with a Cas9-histone demethylase fusion. Nat Methods. 2015;12:401–403. doi: 10.1038/nmeth.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Thakore PI, et al. Highly specific epigenome editing by CRISPR-Cas9 repressors for silencing of distal regulatory elements. Nat Methods. 2015;12:1143–1149. doi: 10.1038/nmeth.3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vojta A, et al. Repurposing the CRISPR-Cas9 system for targeted DNA methylation. Nucleic Acids Res. 2016;44:5615–5628. doi: 10.1093/nar/gkw159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kearns NA, et al. Cas9 effector-mediated regulation of transcription and differentiation in human pluripotent stem cells. Development. 2014;141:219–223. doi: 10.1242/dev.103341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shi J, et al. Discovery of cancer drug targets by CRISPR-Cas9 screening of protein domains. Nat Biotechnol. 2015;33:661–667. doi: 10.1038/nbt.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Korkmaz G, et al. Functional genetic screens for enhancer elements in the human genome using CRISPR-Cas9. Nat Biotechnol. 2016;34:192–198. doi: 10.1038/nbt.3450. [DOI] [PubMed] [Google Scholar]
- 75.Shalem O, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 77.Zhou Y, et al. High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells. Nature. 2014;509:487–491. doi: 10.1038/nature13166. [DOI] [PubMed] [Google Scholar]
- 78.Chen S, et al. Genome-wide CRISPR screen in a mouse model of tumor growth and metastasis. Cell. 2015;160:1246–1260. doi: 10.1016/j.cell.2015.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Parnas O, et al. A Genome-wide CRISPR Screen in Primary Immune Cells to Dissect Regulatory Networks. Cell. 2015;162:675–686. doi: 10.1016/j.cell.2015.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wong AS, et al. Multiplexed barcoded CRISPR-Cas9 screening enabled by CombiGEM. Proc Natl Acad Sci U S A. 2016;113:2544–2549. doi: 10.1073/pnas.1517883113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Evers B, et al. CRISPR knockout screening outperforms shRNA and CRISPRi in identifying essential genes. Nat Biotechnol. 2016;34:631–633. doi: 10.1038/nbt.3536. [DOI] [PubMed] [Google Scholar]
- 82.Morgens DW, et al. Systematic comparison of CRISPR/Cas9 and RNAi screens for essential genes. Nat Biotechnol. 2016;34:634–636. doi: 10.1038/nbt.3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lane AB, et al. Enzymatically Generated CRISPR Libraries for Genome Labeling and Screening. Dev Cell. 2015;34:373–378. doi: 10.1016/j.devcel.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Friedland AE, et al. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat Methods. 2013;10:741–743. doi: 10.1038/nmeth.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bassett AR, et al. Highly efficient targeted mutagenesis of Drosophila with the CRISPR/Cas9 system. Cell Rep. 2013;4:220–228. doi: 10.1016/j.celrep.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jao LE, et al. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc Natl Acad Sci U S A. 2013;110:13904–13909. doi: 10.1073/pnas.1308335110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chang N, et al. Genome editing with RNA-guided Cas9 nuclease in zebrafish embryos. Cell Res. 2013;23:465–472. doi: 10.1038/cr.2013.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li W. Simultaneous generation and germline transmission of multiple gene mutations in rat using CRISPR-Cas systems. Nat Biotechnol. 2013;31:684–686. doi: 10.1038/nbt.2652. [DOI] [PubMed] [Google Scholar]
- 89.Lv Q. Efficient Generation of Myostatin Gene Mutated Rabbit by CRISPR/Cas9. Sci Rep. 2016;6:25029. doi: 10.1038/srep25029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yan Q, et al. Generation of multi-gene knockout rabbits using the Cas9/gRNA system. Cell Regen (Lond) 2014;3:12. doi: 10.1186/2045-9769-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang X, et al. Generation of gene-modified goats targeting MSTN and FGF5 via zygote injection of CRISPR/Cas9 system. Sci Rep. 2015;5:13878. doi: 10.1038/srep13878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Crispo M, et al. Efficient Generation of Myostatin Knock-Out Sheep Using CRISPR/Cas9 Technology and Microinjection into Zygotes. PLoS One. 2015;10:e0136690. doi: 10.1371/journal.pone.0136690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zou Q, et al. Generation of gene-target dogs using CRISPR/Cas9 system. J Mol Cell Biol. 2015;7:580–583. doi: 10.1093/jmcb/mjv061. [DOI] [PubMed] [Google Scholar]
- 94.Wang K, et al. Efficient Generation of Myostatin Mutations in Pigs Using the CRISPR/Cas9 System. Sci Rep. 2015;5:16623. doi: 10.1038/srep16623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Niu Y, et al. Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell. 2014;156:836–843. doi: 10.1016/j.cell.2014.01.027. [DOI] [PubMed] [Google Scholar]
- 96.Wang H, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yang H, et al. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell. 2013;154:1370–1379. doi: 10.1016/j.cell.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Platt RJ, et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014;159:440–455. doi: 10.1016/j.cell.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chiou SH, et al. Pancreatic cancer modeling using retrograde viral vector delivery and in vivo CRISPR/Cas9-mediated somatic genome editing. Genes Dev. 2015;29:1576–1585. doi: 10.1101/gad.264861.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Swiech L, et al. In vivo interrogation of gene function in the mammalian brain using CRISPR-Cas9. Nat Biotechnol. 2015;33:102–106. doi: 10.1038/nbt.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sanchez-Rivera FJ, et al. Rapid modelling of cooperating genetic events in cancer through somatic genome editing. Nature. 2014;516:428–431. doi: 10.1038/nature13906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Xue W. CRISPR-mediated direct mutation of cancer genes in the mouse liver. Nature. 2014;514:380–384. doi: 10.1038/nature13589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lanctot C, et al. Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions. Nat Rev Genet. 2007;8:104–115. doi: 10.1038/nrg2041. [DOI] [PubMed] [Google Scholar]
- 104.Chen B, et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell. 2013;155:1479–1491. doi: 10.1016/j.cell.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Anton T, et al. Visualization of specific DNA sequences in living mouse embryonic stem cells with a programmable fluorescent CRISPR/Cas system. Nucleus. 2014;5:163–172. doi: 10.4161/nucl.28488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ma H. Multicolor CRISPR labeling of chromosomal loci in human cells. Proc Natl Acad Sci U S A. 2015;112:3002–3007. doi: 10.1073/pnas.1420024112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Deng W. CASFISH: CRISPR/Cas9-mediated in situ labeling of genomic loci in fixed cells. Proc Natl Acad Sci U S A. 2015;112:11870–11875. doi: 10.1073/pnas.1515692112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ratz M, et al. CRISPR/Cas9-mediated endogenous protein tagging for RESOLFT super-resolution microscopy of living human cells. Sci Rep. 2015;5:9592. doi: 10.1038/srep09592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kamiyama D, et al. Versatile protein tagging in cells with split fluorescent protein. Nat Commun. 2016;7:11046. doi: 10.1038/ncomms11046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Nelles DA, et al. Programmable RNA Tracking in Live Cells with CRISPR/Cas9. Cell. 2016;165:488–496. doi: 10.1016/j.cell.2016.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.McKenna A, et al. Whole-organism lineage tracing by combinatorial and cumulative genome editing. Science. 2016;353:aaf7907. doi: 10.1126/science.aaf7907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Shipman SL, et al. Molecular recordings by directed CRISPR spacer acquisition. Science. 2016;353:aaf1175. doi: 10.1126/science.aaf1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Maddalo D, et al. In vivo engineering of oncogenic chromosomal rearrangements with the CRISPR/Cas9 system. Nature. 2014;516:423–427. doi: 10.1038/nature13902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Choi PS, Meyerson M. Targeted genomic rearrangements using CRISPR/Cas technology. Nat Commun. 2014;5:3728. doi: 10.1038/ncomms4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hsu PD, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Fu Y. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Crosetto N, et al. Nucleotide-resolution DNA double-strand break mapping by next-generation sequencing. Nat Methods. 2013;10:361–365. doi: 10.1038/nmeth.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Tsai SQ, et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol. 2015;33:187–197. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Guilinger JP, et al. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat Biotechnol. 2014;32:577–582. doi: 10.1038/nbt.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tsai SQ, et al. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat Biotechnol. 2014;32:569–576. doi: 10.1038/nbt.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Fu Y. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol. 2014;32:279–284. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Davis KM, et al. Small molecule-triggered Cas9 protein with improved genome-editing specificity. Nat Chem Biol. 2015;11:316–318. doi: 10.1038/nchembio.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Slaymaker IM, et al. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351:84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kleinstiver BP, et al. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529:490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]