Abstract
In this paper, we present an automated method for taxonomy learning, focusing on concept formation and hierarchical relation learning. To infer such relations, we partition the extracted concepts and group them into closely-related clusters using Hierarchical Agglomerative Clustering, informed by syntactic matching and semantic relatedness functions. We introduce a novel, unsupervised method for cluster detection based on automated dendrogram pruning, which is dynamic to each partition. We evaluate our approach with two different types of textual corpora, clinical trials descriptions and MEDLINE publication abstracts. The results of several experiments indicate that our method is superior to existing dynamic pruning and the state-of-art taxonomy learning methods. It yields higher concept coverage (95.75%) and higher accuracy of learned taxonomic relations (up to 0.71 average precision and 0.96 average recall).
Keywords: ontology learning, taxonomy extraction from text, semantic relation acquisition, term recognition, concept discovery
1. Introduction
Ontologies are formal representations of knowledge resources that describe and share a common understanding of a particular domain. They are foundational for knowledge-based systems or intelligent systems and serve a wide range of applications such as Natural Language Processing [1], Information Retrieval [2], text clustering and classification, to name a few. Machine reading [3, 4], which aims to extract structured knowledge from text with little human effort, has been a major goal of Artificial Intelligence since its early days and an important application area for ontologies. However, ontology development is a time and cost consuming task, requiring the knowledge of specialists from multiple disciplines who may have difficulties reaching consensus [5]. Current works in the field of automatic or semi-automatic ontology acquisition largely aim at overcoming this barrier.
Within this line of works, we present Ontofier, a novel framework to unsupervised ontology learning from text. In this work, we focus on the tasks of extracting domain concepts and their taxonomic relations. Concept hierarchies based on the taxonomic relations enable structuring information into categories, hence fostering efficient search, reuse, and formulation of relations.
In the wide spectrum of approaches to ontology classifications, as introduced in Uschold et al. [6], at one end are the formal, heavyweight ontologies that make intensive use of axioms for specification, and at the other end are ontologies that use little or no axioms, referred to as lightweight ontologies. Taxonomies reside somewhere in the middle of this spectrum. Our contribution is a novel, fully-automated method for taxonomic relation learning from text. We present an extensive evaluation of our approach involving several medical experts, focusing on text from the biomedical domain, which is particularly challenging and lagging behind in ontology learning techniques. We used clinical trial eligibility criteria to illustrate our methodology, which promises to generalize beyond eligibility criteria text.
Potential applications of our approach include enrichment of current ontologies with new concepts and parent-child relations, improving text understandability for machines to allow better knowledge inference and search capabilities, and automated grouping of domain concepts for better engineering of classification features (e.g. Yu et al. [7] utilize a semi-automated approach for grouping of drug concepts to improve the classification features on drugs in phenotyping algorithms).
2. Related Work
Ontology learning from text is the process of identifying terms, concepts, relations, and optionally axioms (for formal ontologies), from textual information and using them to construct and maintain an ontology [8]. For our review, we consulted numerous surveys on ontology learning methods [8, 9, 10, 11, 12]. The learning techniques are generally categorized as symbolic, statistical, and hybrid. Symbolic methods rely on static linguistic patterns (rules) that can provide high accuracy, but require extensive domain expertise and are hard to generalize to other domains. Whereas statistical methods usually exploit corpora to learn structured knowledge, requiring minimal prior knowledge but providing better generalizability.
Our focus is on unsupervised statistical methods that do not require large amounts of labeled data. The most relevant works are based on clustering, which is useful for two purposes. First, similarity measures can provide information about the hierarchical relations of concepts. Second, the discovery of distinct clusters of similar terms can help to identify concepts and their synonyms. The works of [13, 14, 15] propose methods for unsupervised concept formation, whereas [16, 17, 18, 19] introduce relation extraction techniques. These methods mainly make use of static, rare background knowledge.
In view of the shortcomings of conventional techniques, an interesting line of works is emerging. They explore the rich, heterogeneous resources of structured Web data for ontology learning. The intertwining of the Web with ontology learning enables us to harvest consensus (hence shared conceptualization) and access to large quantities of information. Among the few works [20, 21, 22, 23, 24] that explore structured Web data for relation extraction, Liu et al. [22] make use of Wikipedias categorical system to deduce relations between concepts. They apply sentence parsers and syntactic rules to extract the explicit properties and values from the category names. Wong et al. [24] use Wikipedia and search engine page count to acquire coarse-grained relations between ambiguous concepts, using lexical simplification, and association inference. Mintz et al. [23] use Freebase as lookup dictionary to provide distant supervision for extracting relations between entity pairs. Fan and Friedman [25] introduced a distributional similarity approach for the semantic classification of concepts in the Unified Medical Language System (UMLS), the biggest repository of biomedical vocabularies.
As such, we observe an increasing trend in exploring structured web data for relation extraction. Boosheri et al. [26] also proposed an approach for ontology enrichment by using DBpedia. In contrast to our work, their approach extracts the relations (predicates) that DBPedia offers, relying first on a pre-defined similarity threshold to prune the predicates and then on ontology engineers to refine the recommended relations. Our work lies in the intersection between this framework of methods that use semantic knowledge bases in the Web, i.e. semantic-based techniques, and unsupervised statistical methods. This hybrid approach is relatively new and has not been well tested.
The novelty of our work lies in its exploitation of external knowledge bases in a fully-automated approach for concept formation and unsupervised taxonomical relation learning. In contrast to purely statistical methods, Ontofier employs not only text-based similarity measures but also concept semantic relatedness by using rich information of Web knowledge bases. Moreover, unlike symbolic methods, Ontofier does not rely on lexical patterns or rules manually crafted upon analysis of datasets/domain text at hand. Compared to existing clustering approaches, ours has unique advantages in dimensionality reduction and automatic clustering within each partition, not requiring pre-defined clustering parameters, which can be nontrivial and usually require costly fine-tuning procedures or prior expert knowledge. 4
3. Lightweight Ontology Learning
The commonality in various definitions is that an ontology is a representation of entities and their relations in a particular domain [9]. A key requirement is that each entity has one unique reference, an identifier, which is linked to one or more natural language terms to capture the synonymy inherent in human language. We adhere to this definition, using the following data structure for a domain concept.
Definition 1
A domain concept, extracted from a set 𝒮 of natural language sentences of a particular domain, is defined as the tuple ci = (cid, cname,𝒜), where cid is a unique concept identifier, cname is the concept name represented as a string, and 𝒜 is the set of atoms composed of natural language phrases in the sentences 𝒮 to which the concept is linked. Each atom is defined as ai = (aphrase, sl), s.t. aphrase is the phrase (sentence fragment) linked to cid, and sl ∈ 𝒮 the sentence where the phrase occurs.
Let us illustrate this definition with an example from real-life data on biomedical text in the domain of clinical trial patient recruitment1.
Example 1
The following text describes criteria of patients eligible in clinical trials for Alzheimer’s disease:
“Exclude patients with a current diagnosis of hepatic or renal disease. Exclude patients with severe liver disorder or kidney disease.”
We identify, among others, the domain concepts:
(c1, “liver disease”, {a1, a2}) with atoms:
a1=(“hepatic disease”, s1), a2=(“liver disorder”, s2);
(c2, ’kidney disease’, {a3, a4}) with atoms:
a3=(“renal disease”, s1), a4=(“kidney disease”, s2);
The atoms represent the natural language terms to which a new concept is linked, also capturing the inherent synonymy. For brevity, we also use the concept notation ci = (cid, cname), excluding atoms set.
An important piece of semantic information in an ontology is captured by the hierarchical relations among the concepts. According to formal, logic-based semantics, we are able to structure the ontology in the form of a hierarchy by determining subconcept/superconcept relations (also referred to as subsumption relations) between the concepts [27].
According to the principles of subsumption theory, “to subsume is to incorporate new material into one’s cognitive structures. When information is subsumed into the learner’s cognitive structure it is organized hierarchically” [28]. Adhering to this theory, our learning process makes use of the derivative subsumption, which allows one to completely derive new concepts (as superconcepts) from an existing cognitive structure of known concepts. We use the following notation of the subsumption relation:
A subsumption relation, denoted as ⊑(ci,cj), is a binary relation of generic hierarchical nature between concept ci ∈ 𝒞 and concept cj ∈ 𝒞, where ci ⊑ cj states that the broader concept (or superconcept) cj subsumes the more specific concept (or subconcept) ci (i.e. ci ⊑ cj ). We can also state that ci is subsumed by cj.
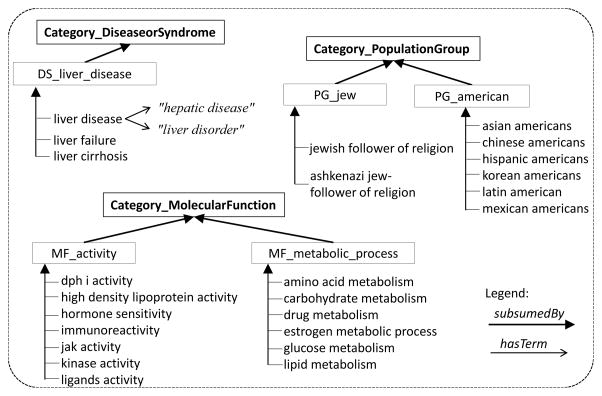
Hence, the subsumption relation is used to create a hierarchy between general concepts and specific concepts. Referring to Example 1, by grouping the discovered concepts (c1, liver disease) and (c2, liver failure), we can derive a new concept (p1, DS liver disease) introducing the subsumption relations ⊑ (c1, p1) and ⊑ (c2, p1). Figure 1 illustrates an excerpt of an automatically learned taxonomy in the biomedical domain.
Figure 1.
Sample lightweight ontology learned by Ontofier from plain biomedical text of clinical trials eligibility criteria description. Note: words in quotation marks define terms.
3.1. Ontofier Framework
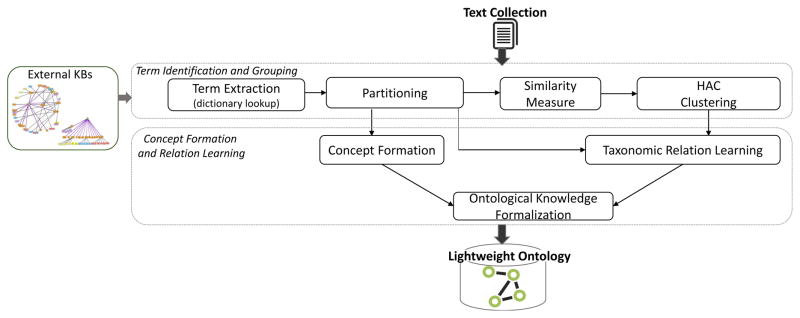
The architecture of the proposed framework is illustrated in Figure 2. The components are explained below.
Figure 2.
Ontofier architecture
Term extraction
The first tasks consist of text preprocessing, e.g. extract and parse the text from clinical trial eligibility criteria, and sentence segmentation. For each sentence, we perform term extraction by looking up the longest phrases in the background knowledge base for biomedical text, the Unified Medical Language System (UMLS). [29]2. As a result, each term is mapped to one or more entities in UMLS.
For this task, we apply two existing tools: ELIXR [30] for clinical trial descriptions, andMedEx for annotation of drugs in publication abstracts [31]. The term lookup functionality in ELIXR identifies the longest single-word or multiple-word strings that match those in the MRCONSO table of the UMLS Metathesaurus, which contains a range of lexical variants [32]. A Metathesaurus concept unique identifier (CUI) is extracted for each word string. MedEx deploys a proprietary sequential, semantic tagger that looks up terms in RxNorm [33] as a predefined semantic lexicon for drugs that contains terms and their variants. Interested readers may refer to the previously published works for more details on these tools.
Partitioning
We assemble into one group the set of terms mapped to the same entity in UMLS. For example, terms “hepatic disease” and “liver disorder” are mapped to the same entity (concept) in UMLS. We then retrieve the categories to which the entities belong in the external KB. One partition is created for each category. In the case of UMLS, these are the semantic types. An example of such category is “DiseaseOrSyndrome” in Fig. 1. The terms are then assigned to the partitions corresponding with the categories to which they belong in the KB.
Partitioning is an important step that helps with the disambiguation of a concept based on its context. In a general example, if the term “jaguar” is mapped to an entity with two categories Cars and Animals, then we will form two concepts with distinct identifiers, one for each partition. For semantic type selection, we use the feature of EliXR that applies a set of semantic preference rules [32], which extend Johnson’s approach [34] to resolve the ambiguity in UMLS semantic type assignment by recommending the most likely semantic type in the context.
Concept Formation
In each partition, one concept is distinctively formed by one group of terms that map to the same KB entity, where as concept name the label of that entity is used. For example, the concept “liver disease” is created from the terms “hepatic disease” and “liver disorder” in Fig. 1.
Concept Similarity Estimation
In this step, we compute a similarity measure for each distinct pair of concepts within a partition. This measure is designed as a combination of syntactic-based similarity and semantic-based relatedness functions. Given a pair of concepts ci and cj, the final similarity value is the weighted, linearly normalized mean of a set of similarity function scores:
| (1) |
We introduce below the set of similarity functions ft used in our approach (n=5).
f1:syntactic-based similarity function between concept names is computed as the Jaccard index of the sets of string tokens T1 and T2 extracted from the concepts names of c1 and c2, respectively:
| (2) |
Two tokens are considered to match if their Levenshtein distance [35] is smaller than a predefined parameter α (typically set to 3).
f2, f3:syntactic-based similarity function between concept name and their atoms: for this measure, we use again the Jaccard index in equation 2, but here the sets T1 and T2 are composed of the tokens extracted from the name of concept c1 and text of atoms of concepts c2 (and vice-versa for function f3).
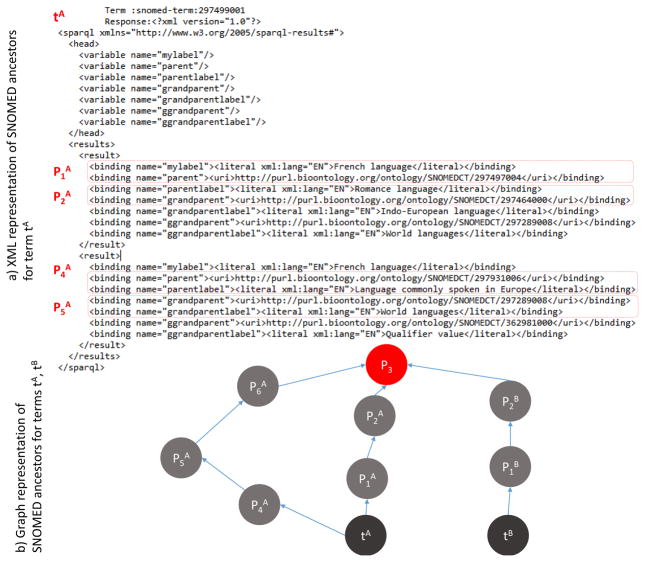
f4: besides syntactic matching, we apply semantic relatedness function using well-known knowledge bases. Since we are primarily focused on biomedical text, we use SNOMED CT, which is considered to be the most comprehensive, multilingual clinical healthcare terminology in the world3. Together with many other BioMedical ontologies, SNOMED CT is transformed into RDF triples from the original formats, asserted into triple store as an RDF graph, and made accessible via BioPortal SPARQL4 query service5.
We match each concept ci to SNOMED CT using dictionary matching approach. For those concepts that can be found in SNOMED CT, we map each of them to one or more SNOMED CT IDs. Using SPARQL service, we extract for each of the IDs si its SNOMED CT subgraph gi consisting of all its ancestor nodes up to the root by performing arbitrary depth graph traversal. For two concepts ci and cj, if they share one or more common ancestors in the respective extracted graphs gi and gj, their semantic similarity f4 is computed as the normalized, closest distance to the common ancestor node. Otherwise, f4 = 0. Figure 3 illustrates an example of of the term tA with label “French language” and the XML representation of its ancestors extracted from SNOMED CT via the SPARQL Endpoint. Whereas, part b) of Figure 3 illustrates the graph representation of the respective ancestors and of this term and another term tB. The semantic similarity is computed as the normalized, closest distance to their shared ancestor node P3 using arbitrary depth graph traversal.
Figure 3.
Semantic relatedness function f4 of two terms computed based on graph traversal of ancestors in SNOMED CT. Part a) is the XML representation of the ancestors of the term ta with label “French Language” extracted from SNOMED CT using BioPortal SPARQL Endpoint. Part b) is the graph representation of ancestors for two terms tA and tB; semantic similarity is computed as the normalized, closest distance to the common ancestor node P3.
f5: this is another semantic-based function based on DBpedia knowledge base6, which is structured knowledge extracted from gigantic source of information in Wikipedia. We implemented a method to perform DBpedia URIs look up by related keywords, which consists of tokens in the concept names. Upon resolving for each concept ci one or more URIs of DBpedia instances, we retrieve the set ui of categories (comprising URIs of instances) it belongs. The f5 value for any two concepts ci and cj is the Jaccard index of the two sets ui and uj of categories extracted from DBpedia.
Given the set of concepts Cp in each partition, the final output of the concept similarity estimation method is a |Cp| × |Cp| similarity matrix Mp for each partition. We apply these matrices to detect taxonomies of closely-related concepts.
Taxonomic Relation Learning
In order to identify informative taxonomical structures in a given set of concepts, we apply hierarchical agglomerative clustering (HAC) technique [36]. HAC organizes objects into a hierarchical cluster tree (dendrogram) whose branches represent the desired groups of closely-related concepts.
For each partition p, we apply the calculated similarity matrix Mp to generate one respective dendrogram. Hierarchical clustering methods differ primarily in the similarity measures they employ. In this work, we concentrate on complete-linkage clustering because it considers non-local criterion of cluster merging, i.e. the entire structure of the clustering can influence merge decisions.
However, the dendrogram generated by HAC is 1) often composed of many branches, especially for the case of large partitions of objects, and 2) needs to be used to detect disjoint groups (clusters) of concepts for our domain ontology. Hence, we need a method to extract optimal branches for cluster detection. The process of identifying individual branches is variously referred to as branch or tree cutting, or dendrogram pruning. We apply a novel method for unsupervised dendrogram pruning, adaptive to each partition.
We derive from each cluster in every partition one new parent concept cp and infer the subsumption relation ⊑ (cl, cp) between cp and each concept cl in that cluster.
For example, when we group with HAC under the same cluster the concepts “jak activity” and “kinase activity” illustrated in Fig. 1, and then create a parent concept “MF activity” (or likewise, “jew follower of religion” and “ashkenazi jew” under another cluster with parent “PG jew”), we are inferring a subsumption relation that is used to create a hierarchy between general concepts and specific concepts. This is where the taxonomic organization of concepts is performed, involving precisely the two steps HAC Clustering and Taxonomic Relation Learning illustrated in the architecture of Figure 2. The name of cp, e.g. “MF activity”, is inferred using cluster labeling methods [2], whose details are outside the scope of this paper.
3.2. Dynamic Dendrogram Pruning Method
The most widely used tree-cut method is the fixed height h branch cut, where each contiguous branch of objects below h is considered a separate cluster. However, this method has a major limitation for our setting. The generated dendrograms exhibit distinct branches corresponding to the data in each partition, so no single absolute fixed height can identify them correctly.
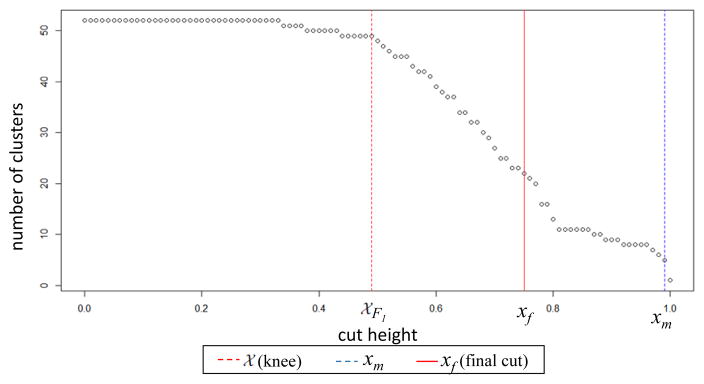
We developed a method that dynamically estimates the cut height in an unsupervised way, adapting to each partition’s individual dendrogram shape and not on an absolute global height. To find the cut height hp for dendrogram Dp of partition p, we construct the graph of function f : X → Y, where values x ∈ [0, 1] reflect the range of possible height values, and y values are the respective number of clusters generated for each value of x. This results in a concave curve, which we illustrate in Figure 4 for the dendrogram of partition “Population Group”.
Figure 4.
Illustration of true knee point and final dendrogram height cut estimation
An optimal value of the final cut height would be one that does not generate the maximal number of clusters, but does not return a low number of clusters either. In function f, the number of clusters increases for decreasing values of x (i.e. cut height), and below a particular value of x it remains constants. This reflects a knee point of the curve which we need detect first. Furthermore, we need to avoid a very low cluster number yielded by high cutoff values.
We introduce the following approach to estimate the final cut height for dendrogram pruning. In the first step, we compute the approximation of the knee point with a method that, based on our knowledge, has not been used previously for purposes of cluster detection. We can find an approximation of knee point by applying the Extremum Distance Estimator (EDE) method, as is defined in [37]. EDE method identifies the inflection point of any given concave or convex curve based on the definition and its geometrical properties. The relevant EDE approximations can theoretically be computed using Lemma 1.4 of [37] as points close to knee points. We present the modified lemma 7 and definition of estimated knee point for a strictly concave function:
Lemma 1
Let a function f : [a, b] → ℛ, f ∈ C(n), n ≥ 2 which is concave for x ∈ [a, b]. Given an arbitrary x ∈ [a, b], the knee point of f is estimated as
| (3) |
with δ taken small enough for 𝒳 to be unique unconstrained extreme in the corresponding interval8.
A visual interpretation of 𝒳 point is that it represents a slant extremum, i.e. local minimum, relative to the (slant) total chord, which connects initial and ending points of the curve. We illustrate the knee point estimation 𝒳 for our projection in Figure 4.
In the second step, we find the minimum number of clusters generated for x < 1. The rationale of this step is to avoid a height that produces only one cluster. In the graph of Figure 4, this is the point (xm, f(xm)) where xm=0.99.
In the third step, we compute final cut height hfinal as:
| (4) |
We use the average number of clusters θ estimated for the height values between the knee point cutoff xk and minimal xm to find the closest value maximal yf of a point in the curve of function f, i.e. the closest, highest value of θ in Y (performed by function findclosest : ℛ → Y ). The xf value of that point is our final height cut, which is also illustrated in Fig. 4.
4. Evaluation in Targeted Application Domains
4.1. Experimental Setting
4.1.1. Evaluation cases and datasets
We performed a series of experiments in order to evaluate our approach, using two diverse evaluation cases:
Case 1
Real-world text collections of clinical trial eligibility criteria descriptions:
D1: dataset of trials on Cardiovascular Disease from ClinicalTrials.gov. We processed 313,273 sentences and extracted 8579 concepts distributed in 67 partitions. We evaluated 3 partitions with 484 concepts.
D2: clinical trial protocols on Alzheimers disease provided by a medical institute. We processed 281 sentences and extracted 436 concepts distributed in 42 partitions. We evaluated 12 partitions with 112 concepts.
D3: protocols of clinical trial on Breast Cancer disease from ClinicalTrials.gov portal. We processed 142,219 sentences, and extracted 4456 concepts distributed in 69 partitions. We evaluated 3 partitions with 526 concepts.
In this work, we have performed a careful manual evaluation, which is in itself a very challenging and time-consuming process. It involves several experts from different fields of expertise. Dataset D1 contains many partitions with a very large number of concepts, so we left it to the experts to select the partitions from an area in which they were more competent to evaluate (e.g. “Disease or Syndrome”, or “Diagnostic Procedure”), while still keeping a big number of evaluated concepts. The proportion of partitions evaluated in datasets D1, D2 and D3 varies, but the number of concepts across the datasets remains comparable (i.e. 484, 112, and 526 respectively).
Case 2
Semantically-annotated corpus of documents describing drug-drug interactions from the MEDLINE abstracts corpus of SemEval-2013 challenge, Task 9.1 (Recognition and classification of drug names)9. The corpus had been manually annotated with pharmacological substances (drugs) for the Sem-Eval-2013-Task9.1.
D4: this dataset consists of 142 abstracts composed of 1301 sentences containing 246 drugs annotated by trained human experts.
4.2. Experimental Protocols
All the experiments of our evaluation run the approach end-to-end, but different parts/metrics are evaluated in the first two. Table 1 gives an overview of the evaluation design, specifying the experiments performed, respective datasets used for each experiment, the gold standard and other methods used as reference for comparison, and the applied metrics.
Table 1.
Overview of the performed experiments. Note: NDF-RT is the National Drug File - Reference Terminology; Expert‘s taxonomy defines the expert-defined classes of concepts.
| Experiment | Cases | Datasets | Compared Method | Gold Standard | Metrics |
|---|---|---|---|---|---|
| Experiment 1. Pruning Performance | Case 1. Clinical Trials | D1, D2, D3 | Dynamic Tree; Dynamic Hybrid | Silhouette Coefficient | |
| Experiment 2. Quality of Taxons | Case 1. Clinical Trials | D2, D3 | Expert’s taxonomy | Purity; Precision; Recall; F-measure | |
| Case 2. MEDLINE abstracts | D4 | Ontolearn; Subsumption Method; ADTCT-HAC Method | NDF-RT |
This evaluation is focused on assessing the main contribution of this work, the relational learning task, in terms of the performance of the novel pruning method and the quality of the inferred relations. The evaluation does not cover the quality of the new parent concepts, since their naming is outside the scope of this work. However, we evaluate the quality of the grouping of the siblings concepts under each newly formed parent concept.
In the meantime, we have performed a comprehensive evaluation in all four datasets of the coverage of the concepts extracted from text (presented in the Supplementary Material). The experiments demonstrate very high coverage of concepts aided by the use of dictionary lookup tools in our Ontofier framework.
Below is a detailed explanation of the protocol for each experiment:
Experiment 1: Pruning Performance
In this experiment, we compare the performance of the dendrogram pruning method with two state-of-the-art dynamic tree cut techniques proposed in [38]: dynamic tree and dynamic hybrid. The shortcoming of these methods is that one has to pre-define a set of parameters. For these methods, we experiment with different variations of the parameters deepSplit, and minimum cluster size minCl.10
We focus on the case of clinical trial descriptions, using all datasets D1, D2, D3. We apply the Silhouette Coefficient (SC) [39] to the dendrogram in each partition to evaluate clustering results, reporting the averaged SC over all partitions. Silhouette Coefficient is a measure that helps to validate the consistency with clusters, as a mean of interpreting how well each object lies within its cluster. A larger SC value indicates better quality of clustering.
Experiment 2. Quality of taxons
The goal of this experiment is to evaluate the performance of the relation learning task in terms of different aspects of the quality of clusters from which we infer the subsumption relations. This analysis focuses on the quality of the groups of sibling concepts (taxons). For example, if we group the concept “asian american” and concept “chinese americans” as siblings in one cluster under the same concept parent, we aim now to evaluate how good this grouping (i.e. subsumption) is. We conduct this experiment for both cases: clinical trial descriptions and MEDLINE abstracts.
Case 1: to evaluate the quality of the groups of sibling concepts, we randomly selected a number of concept clusters in the taxonomies generated from the datasets of clinical trial descriptions (datasets D2 and D3). To create a quasi-gold standard of taxonomies, we involved three medical experts, who were given the generated clusters and asked to assess the cohesiveness of concepts in each cluster. They were asked to change the assignment of concepts in clusters when needed. This way, we obtain expert-driven classes of concepts used as standard for comparison.
Case 2: we assess the cohesiveness of clusters (containing more than 2 concepts) in the taxonomies generated from MEDLINE corpus, focusing on the set of concepts that match MEDLINE manual annotations (dataset D4). To assess if any two concepts in one cluster are indeed siblings of the same parent class, we check if they share the common ancestor class in the hierarchy provided by the National Drug File - Reference Terminology (NDF-RT)11, used here as the gold standard. We developed our own proprietary Java tool for transitive querying, using the NDF-RT RESTful Web API 12, and results processing. From the set of common ancestors, we exclude the root and general classes such as “Chemical Ingredients”, “Drug Products by Generic Ingredient Combinations”, “Pharmaceutical Preparations”.
We compare our method to the following taxonomy learning methods:
-
Ontolearn [40]: graph-based algorithm for learning a taxonomy from the ground up. After initial term extraction, textual definitions extracted from a corpus and the Web are used to automatically create a highly dense, potentially disconnected hypernym graph. An optimal branching algorithm is then used to induce a treelike taxonomy.
For our experiments, we use the taxonomy13 (and respective terminology14) extracted with this approach from the same MEDLINE corpus.
ADTCT-HAC [41]: This is a hierarchical clustering approach to taxonomy learning, part of the ADTCT framework, which uses various text-based window and document scopes for concept co-occurrences. The terms are extracted from text documents using part-of-speech parser. For the experiments, we use an implementation provided by the authors with default parameter window = 6, and their corpus of publication abstracts in the economics domain [41] as contrastive set. This set, needed for their approach, should contain documents outside the domain of the target taxonomy.
Subsumption method [42]: statistics-based approach for deriving a hierarchy of the concepts discovered in text using a type of co-occurrence (window-or document-based) known as subsumption. In this approach, a concept x subsumes y if the documents/windows which y occurs in are a subset of the documents which x occurs in. We used the implementation of this method in the ADTCT framework with default parameters (t – value = 0:2, window = 10).
Regarding the evaluation metrics, for a given dataset D with a set of partitions P where each partition consists of C clusters, we apply the following measures of Purity, Precision, Recall, and F-measure [43] to assess the quality of the clusters in each partition:
Purity
Measures the extent to which a cluster contains concepts of a single class. Given nij the number of concepts of class i in cluster j, ni the number of data points in cluster i, and l the number of classes, the purity for a cluster i in partition p is given by
| (5) |
whereas purity Purp of partition p with k clusters is estimated as
| (6) |
where n is the total number of concepts in p.
Precision
Measures the fraction of a cluster that consists of concepts of a specified class. Using the previous notations and given the total number of classes l, the precision of cluster i with respect to class j and the overall precision of cluster i are given by
| (7) |
whereas precision Prep of partition p with k clusters is given by
| (8) |
Recall
Measures the extent to which a cluster contains all objects of a specified class. Using the previous notations, the recall of cluster i with respect to class j, and recall of cluster i are given by
| (9) |
whereas recall Recp of partition p with k clusters is given by
| (10) |
F-measure
Combines precision and recall to measure the extent to which cluster contains only concepts of a particular class and all concepts of that class. The F-measure Fp of partition p with k clusters is given by
| (11) |
For each of the given metricsM (i.e.Pur, Pre, Rec and F-measure), we compute its overall final value for the entire dataset as the weighted average of Mps in all partitions
| (12) |
4.3. Results
4.3.1. Experiment 1. Pruning Performance
The results of the evaluation of dendrogram pruning performance and its comparison with two state-of-the-art dynamic tree cut techniques (dynamic tree and dynamic hybrid [38]) are shown in Table 2.
Table 2.
Dendrogram pruning performance using average Silhouette Coefficient (SC) and its standard deviation for different variations of the parameters deepSplit and minCl (minimum number of clusters).
| D1 | D2 | D3 | ||
|---|---|---|---|---|
|
| ||||
| Dynamic Tree | minCl=3, deepSplit=true | 0.07 ± 0.04 | 0.001 ± 0.01 | 0.03 ± 0.001 |
| minCl=3, deepSplit=false | −0.01 ± 0.02 | 0.05 ± 0.02 | 0.01 ± 0.03 | |
| minCl=2, deepSplit=true | 0.06 ± 0.02 | 0.09 ± 0.03 | 0.08 ± 0.06 | |
| minCl=2, deepSplit=false | 0.06 ± 0.02 | 0.09 ± 0.03 | 0.08 ± 0.06 | |
|
| ||||
| Dynamic Hybrid | minCl=3, deepSplit=true | 0.08 ± 0.06 | −0.01 ± 0.02 | 0.01 ± 0.002 |
| minCl=3, deepSplit=false | −0.03 ± 0.03 | 0.03 ± 0.01 | −0.01 ± 0.02 | |
| minCl=2, deepSplit=true | 0.02 ± 0.04 | 0.07 ± 0.03 | 0.03 ± 0.01 | |
| minCl=2, deepSplit=false | −0.03 ± 0.03 | 0.06 ± 0.02 | 0.02 ± 0.04 | |
|
| ||||
| Our method | 0.11 ± 0.04 | 0.13 ± 0.01 | 0.13 ± 0.02 | |
As stated above, Silhouette Coefficient (SC) is a mean for interpreting how well each object lies within its cluster, so that larger SC values indicates better quality of clustering. Results show that our approach clearly outperforms the other two methods, providing higher SC values in all datasets and for different variations of parameters deepSplit and minCl (minimum number of clusters). This is an important finding, considering furthermore the fact that, unlike other methods, our approach is automated and does not require parameter setting.
4.3.2. Experiment 2. Quality of taxonomic relations
Case 1
The results of this experiment for the case of clinical trial descriptions are presented in Table 3 for datasets D2. We observe for D2 high values of cluster quality for the majority of partitions. We also see lower values in a few partitions (p6, p12), where Ontofier had generated bigger clusters, while the experts separated them in smaller classes of concepts, leading to lower precision. The overall high purity in partitions shows that the generated clusters contain in most of the cases sibling concepts correctly belonging to a single class.
Table 3.
Quality of the taxons learned by Ontofier for each partition in dataset D2
| P artitions | No. concepts | Purity | Precision | Recall | F-measure |
|---|---|---|---|---|---|
| p1-Mental or Behavioral Dysfunction | 29 | 0.95 | 0.90 | 0.90 | 0.90 |
| p2-Diagnostic Procedure | 14 | 0.93 | 0.86 | 0.57 | 0.69 |
| p3-Immunologic Factor | 7 | 1.0 | 1.0 | 1.0 | 1.0 |
| p4-Laboratory Procedure | 13 | 0.90 | 0.80 | 0.60 | 0.69 |
| p5-Disease or Syndrome | 75 | 0.75 | 0.66 | 0.46 | 0.54 |
| p6-Therapeutic or Preventive Procedure | 12 | 0.50 | 0.46 | 0.91 | 0.61 |
| p7-AminoAcid, Peptide or Protein | 13 | 0.75 | 0.57 | 0.43 | 0.49 |
| p8-Injury or Poisoning | 4 | 0.75 | 0.50 | 1.0 | 0.67 |
| p9-Population Group | 8 | 0.63 | 0.60 | 0.60 | 0.60 |
| p10-Neoplastic Process | 12 | 1.0 | 1.0 | 1.0 | 1.0 |
| p11-Organic Chemical | 26 | 0.88 | 0.77 | 0.92 | 0.84 |
| p12-Pathologic Function | 10 | 0.67 | 0.40 | 0.80 | 0.53 |
| Avg | Total:112 | 0.74 | 0.62 | 0.77 | 0.66 |
We also note that partitions p3 and p1 have perfect scores along all metrics. The clusters in these partitions were well-defined by our method, and the human expert did not change the automatically-performed assignment. Results for dataset D3 in Table 4 show high values of purity and recall. There are mixed values of precision, varying between 0.51 for p3 and 0.82 for p1. Again, in p3 the bigger clusters where divided by the expert into smaller groups.
Table 4.
Quality of the taxonomy learned by Ontofier for each partition in dataset D3
| Partitions | No. concepts | Purity | Precision | Recall | F-measure |
|---|---|---|---|---|---|
| p1-Disease or Syndrome | 194 | 0.92 | 0.82 | 0.96 | 0.88 |
| p2-Diagnostic Procedure | 116 | 0.84 | 0.62 | 0.95 | 0.75 |
| p3-Organic Chemical | 216 | 0.75 | 0.51 | 0.97 | 0.67 |
| Avg | Total: 526 | 0.84 | 0.65 | 0.96 | 0.77 |
Medical experts evaluating different partitions reported difficulties distributing the concepts into clusters. There were uncertainties about the different levels of granularity, and sometimes there is uncertainty from which (medical) perspective to separate concepts into classes. Taxonomy construction is a nontrivial task, and is usually difficult to have consistent expert agreement. Providing an automated way to generate these taxonomies from text is a promising direction to facilitate knowledge acquisition.
Case 2
Results of the evaluation in the case of the MEDLINE corpus (dataset D4) are also illustrated in Table 5. We show the results for 7 partitions out of the total of 19 partitions generated with our approach (the rest did contain clusters with more than 2 concepts found in NDF-RT). The number of the concepts in different partitions varies from 156 concepts for the partition “Pharmacologic Substance” to 11 concepts for partition “Hormone”. For one partition (p5, semantic type: “125-Hormone”), we notice low values of cluster purity and precision. On the other side, for the other partitions we observe higher values of the measured metrics, an average of 0.76 for purity and 0.72 for precision, even reaching 1.0 for partition p6 (semantic type: “118-Carbohydrates”). This partition is characterized of a small number of concepts (6 concepts). The method yields very promising average purity and precision for all the partitions (0.71 and 0.7, accordingly).
Table 5.
Quality of the taxonomy learned by Ontofier for each partition in dataset D4
| Partition | No. concepts | Purity | Precision |
|---|---|---|---|
| p1-Organic Chemical | 146 | 0.72 | 0.72 |
| p2-Pharmacologic Substance | 156 | 0.60 | 0.59 |
| p3-Amino Acid, Peptide, or Protein | 11 | 0.83 | 0.67 |
| p4-Antibiotic | 23 | 0.69 | 0.65 |
| p5-Hormone | 11 | 0.39 | 0.44 |
| p6-Carbohydrate | 6 | 1.0 | 1.0 |
| p7-Steroid | 11 | 0.70 | 0.68 |
| Total: 364 | Avg: 0.71 | Avg: 0.70 |
In Table 6, we illustrate some examples of good clusters that are cohesive, correctly containing sibling concepts that share same parent in NDF-RT as well. We also show examples of mixed clusters, e.g. cluster C11 in partition P1 “Organic Chemical” contains two concepts (“cerivastatin”, “simvastatin”) that are not siblings with the rest.
Table 6.
Examples of good clusters and partly correct clusters (UMLS CUI accompanies each concept name). In red are the concepts incorrectly assigned as siblings, e.g. in cluster C11: cerivastatin, atorvastatin are not assessed as siblings i.e. in NDF-RT they share as common ancestor only root nodes such as “Pharmaceutical Preparations”, which we have excluded from the valid ancestors list because they are very general (same for cerivastatin and other concepts in C11); cluster C3: rofecoxib is incorrectly assigned as sibling with the other 8 concepts.
| Examples of good clusters Ci in two partitions P1-Organic Chemical and P6-Carbohydrate: | ||
|
| ||
| P6:C2 | P1:C8 | P1:C22 |
|
| ||
| digoxin:c0012265 | ampicillin:c0002680 | ciprofloxacin:c0008809 |
|
| ||
| neomycin:c0027603 | cefazolin:c0007546 | enoxacin:c0014310 |
|
| ||
| ouabain:c0029904 | chloramphenicol:c0008168 | lomefloxacin:c0065162 |
|
| ||
| teniposide:c0039512 | oxytetracycline:c0030092 | norfloxacin:c0028365 |
|
| ||
| trovafloxacin:c0379881 | ||
|
| ||
| Examples of partly correct clusters: | ||
|
| ||
| P1:C11 | P1:C3 | P1:C4 |
|
| ||
| atorvastatin:c0286651 | allopurinol:c0002144 | aminopyrine:c0002586 |
| lovastatin:c0024027 | ascorbic-acid:c0003968 | amiodarone:c0002598 |
| pravastatin:c0085542 | carbamazepine:c0006949 | clarithromycin:c0055856 |
| cerivastatin:c0528023 | diazepam:c0012010 | clozapine:c0009079 |
| simvastatin:c0074554 | etodolac:c0059865 | dicumarol:c0005640 |
| loperamide:c0023992 | mazindol:c0024977 | |
| midazolam:c0026056 | phenobarbital-sodium:c0282303 | |
| nelfinavir:c0525005 | temazepam:c0039468 | |
| nimodipine:c0028094 | ||
| rofecoxib:c0762662 | ||
| tannic-acid:c0039294 | ||
We compared our approach to existing taxonomy extraction methods (HAC, Subsumption, Ontolearn) described in the Experimental Protocols section. The results in terms of purity and precision metrics are shown in Table 7.
Table 7.
Comparative results of taxonomy quality between our approach Ontofier and other methods in dataset D4
| Methods | Subsumption | ADTCT-HAC | Ontolearn | Ontofier | ||
|---|---|---|---|---|---|---|
| Window-based | Document-based | Window-based | Document-based | |||
| Purity | 0.25 | 0.25 | 0 | 0.29 | 0 | 0.71 |
| Precision | 0.26 | 0.26 | 0 | 0.29 | 0 | 0.70 |
We observed that the subsumption method generates a more shallow taxonomy, whereas HAC-method a very deep, nested one where the concept labeled drugs is repetitively created as parent of other concepts. These tests show that our method is superior to the others both in concept coverage (reported in the Supplementary Material) and quality of relations (i.e. concepts under the same parent are correctly assigned as siblings when compared to the NDF-RT taxonomy taken as gold standard).
For ADTCT-HAC, from the 28 concepts correctly extracted as drugs (as in MEDLINE annotations), there are only 5 relations among them in the taxonomy that could be checked (the other terms stood alone without siblings under a parent concept). Yet, for the window-based version none of these relation resulted to be correct, e.g. there are siblings such as “drugs”-“gentamicin”, “drugs”-“estradiol”, etc. Whereas the document-based version has a few correct relations, resulting in 0.29% purity and precision. For Sub-method, the results show 0.25% purity and 0.26% precision for both versions of the approach.
Regarding the method Ontolearn, from the 14 concepts that could match MEDLINE annotations, there were no pairs having a correct taxonomic or sibling relation. Table 8 shows the list of relations of the taxonomy, containing as a child a concept that could be mapped to NDF-RT.
Table 8.
List of parent-child relations of Ontolearn taxonomy in which the child concept can be mapped to MEDLINE annotations for evaluation purpose.
| Child-parent relation | Child-parent relation |
|---|---|
|
| |
| sodium bicarbonate - salt | valproic acid - compound |
| sodium thiosulfate - compound | sildenafil citrate - virility drug |
| tannic acid various - complex phenolic substance | ethanol - antagonis |
| castor oil - vegetable oil | ethyl alcohol - agent |
| magnesium salt - salt | chloral hydrate - sedative |
| lithium carbonate - compound | ascorbic acid - water soluble |
We observe that Ontolearn infers rather generic relations (e.g. “ethyl alcohol”-“agent”, or “lithium carbonate”-“compound”), yet there are no groupings of closely related concepts under a domain-specific parent concept (e.g. placing ‘ethyl alcohol” and ‘ethanol” under the same parent). The extracted relations are rather inferred from sentences of a definition form, resulting in highly generic taxonomic relations.
5. Discussion
The contribution of our approach lies in the formation of new concepts and induction of taxonomic relations that are not present in other KBs (UMLS, SNOMED CT). E.g. only a part (70%) of the initially extracted concepts are in SNOMED CT, let alone hierarchical relations. Using this approach as a basis for other semantic relations, also our future goal, we discover richer relations not existing in such KBs. Let’s take the examples illustrated in the taxonomy of Figure 1. Using the clustering/grouping and subsumption, we create new concepts as parents of the siblings in the cluster, e.g. “PG jew”, “PG american”, “MR activity”. Furthermore, these parent concepts are not in UMLS, and no parent (PAR) or child (CHD) relation can even be found in UMLS for the sibling concepts (e.g. “jew follow of religion” and “ashkenazi jew”). Further examples, when learning the taxonomy from the text of Alzheimers protocols, we infer e.g. that “retinal-diseases” and “myopia” and “age-related-macular-degeneration” are three siblings subsumed by the same new concepts and hence separated them from other diseases, e.g. brain diseases). This method can potentially improve the granularity of an existing ontology and enrich its taxonomic relations.
Secondly, this approach helps infer further synonymies: people express different formulations of terms in text, hence we can learn richer lexical variations of concepts (i.e. atoms). With respect to the exploitation of external knowledge bases, such as UMLS, they are used in our approach for term extraction and initial partitioning based on semantic types. To support disambiguation, we form distinct concepts for each partition (semantic type) (Section 3.1).
It is also important to note that the methodology is not specific to any domain. The approach is agnostic to the text collection and domain concepts it contains. It is necessary to note though that our method relies on the presence of external knowledge bases for initial term detection, which can be of general nature like WordNet and DBPedia, or domain-specific.
With respect to the methodology, hierarchical clustering is used in other previous works to organize sets of terms in a hierarchy, which can then be transformed into an ontology. While one class of taxonomy learning methods is based on extraction patterns [44, 45, 40] using explicit clues like Hearst-patterns [46], another class of approaches, similar to ours, are based on distributional similarity [47, 48, 49, 18]. Works such as [50, 51] combine both pattern-based and distributional approaches to cluster nouns based on distributional similarity, additionally using Hearst-patterns and WordNet as background knowledge for constructing a hypernyms hierarchy. Yang and Callan [52] also integrate lexico-syntactic patterns and concept clustering with contextual, co-occurrence features in a semi-supervised taxonomy induction framework.
Distributional methods generally apply syntactic approaches to find out the similarity regarding predicate-argument relations (i.e. verb-subject and verb-object relations). A set of syntactic approaches is incorporated in the MoK workbench [49], which provides a framework to define hierarchical term clustering methods based on similarity of contexts that are limited to particular syntactic constructions. In another work, Neshati et al. [53] propose a method for taxonomy learning that exploits the similarity of words in knowledge bases such as WordNet and Web pages. Their similarity measure is a combination of several statistics- and syntactic-based methods trained on a Neural Network with supervised knowledge.
In a work related to ours, Knijff et al. [41] propose the ADTCT framework to automatically construct taxonomies from text documents, where concepts are arranged hierarchically by applying a statistics-based subsumption method [42] or hierarchical agglomerative clustering. The clustering algorithm applies two similarity measures based on the co-occurrence frequency of words in text documents or in user-specified word windows. Our experiments showed the superiority of our approach to these methods.
There are three main aspects that distinguish our work from the existing approaches. First, our method does not rely on the design of pre-defined patterns or specific syntactic constructions. Second, we leverage syntactic similarity functions embedded in the distance measure used for clustering with additional semantic-based features. Last, we introduce a novel way to perform the hierarchy cut-off automatically based on the concept similarity distribution in the domain at hand, without requiring manually-defined parameters.
Limitations
This work has its own limitations that we need to point out. First, the approach applies partitioning of extracted terms by relying directly on the categories of the external knowledge base used (e.g. semantic types in the case of UMLS). One shortcoming is that we might carry along modeling errors that could exist in the original assignment of terms in those knowledge bases. Another issue is related to the depth of the generated taxonomies. The presented clustering method is restricted to the discovery of one level of parent-children relation. However, in several cases it is interesting to obtain a deeper taxonomy with more granularity in the hierarchical organization of concepts.
This work has a few limitations with respect to the experimental evaluation. In Experiment 2, we evaluate the performance of the relation learning task in terms of different aspects of the quality of clusters from which we infer the subsumption relations. The evaluation in this experiment focuses on the quality of taxons (i.e., groups of siblings). The evaluation of the quality of the parent concept with regard to its naming and appropriateness is beyond the scope of this paper.
We also compare our method to two other taxonomy learning methods for which we have applied their default parameters. An extension of Experiment 2 with different variations of these parameters will be addressed in our future work.
Additionally, the use of the DBPedia knowledge base for the computation of the semantic similarity function (f5) is also a feature whose impact is not covered in the presented evaluation.
Finally, even though our evaluation involves two completely different types of data (clinical trial descriptions and MEDLINE publication abstracts), they are restricted to the biomedical domain. We need to test our approach with corpora from other domains and diverse knowledge bases (e.g. DBPedia) to assess its performance and generalizability. We will address these limitations in our future work.
6. Conclusions
We present an unsupervised approach to concept formation and taxonomic relation extraction from text. We exploit structured Web data and syntactic matching to measure relatedness of concepts. They are grouped using a novel approach of dynamic dendrogram pruning for cluster detection, which is shown to outperform other methods. Clusters are used to infer subsumption relations between concepts. Several experiments demonstrate the superiority of our approach with respect to the high coverage of the domain concepts extracted from biomedical text, and the quality of taxonomic relation learning.
In our future work, we will investigate how to obtain more fine-granular clusters that provide deeper hierarchy levels. We will also advance this work for automated extraction of other types of semantic relations.
Supplementary Material
Acknowledgments
This work was supported by R01 LM009886 (Bridging the semantic gap between research eligibility criteria and clinical data: PI: Weng), and U01 CA180940 (PI: Jiang).
Footnotes
Text is extracted from the public portal http://www.clinicaltrials.gov
UMLS 2015AA Release
SNOMED CT 2014AA Release
The original lemma contains definition for two knee points covering both convex/concave curve. We have strictly concave curve, hence we use only point XF1 (reusing notation), proof remaining as the original.
The differentiability class C(n) shows that the derivatives f′, f″, …,fk exist and are continuous.
We set the values of the minimum cluster size to 2 and 3 (instead of the default value 20) to avoid large number of clusters in the case of small partitions with few concepts.
References
- 1.Manning CD, Schutze H. Foundations of Statistical Natural Language Processing. MIT Press; 1999. [Google Scholar]
- 2.Manning CD, Raghavan P, Schtze H. Introduction to Information Retrieval. Cambridge University Press; 2008. [Google Scholar]
- 3.Etzioni O, Banko M, Cafarella MJ. Machine reading. 2007 AAAI Spring Symposium, Technical Report SS-07-06; 2007. [Google Scholar]
- 4.Poon H, Domingos P. Machine reading: A ”killer app” for statistical relational ai. Workshop on Statistical Relational AI; 2010. [Google Scholar]
- 5.Lehmann J, Vlker J. Perspectives on Ontology Learning. Amsterdam: IOS Press; 2014. [Google Scholar]
- 6.Uschold M, Gruninger M. Ontologies and semantics for seamless connectivity. SIGMOD Rec. 2004;33(4):58–64. doi: 10.1145/1041410.1041420. [DOI] [Google Scholar]
- 7.Yu S, Liao KP, Shaw SY, Gainer VS, Churchill SE, Szolovits P, Murphy SN, Kohane IS, Cai T. Toward high-throughput phenotyping: unbiased automated feature extraction and selection from knowledge sources. Journal of the American Medical Informatics Association. 2015;22(5):993–1000. doi: 10.1093/jamia/ocv034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wong W, Liu W, Bennamoun M. Ontology learning from text: A look back and into the future. ACM Computing Surveys. 2012;44(4):1–36. doi: 10.1145/2333112.2333115. [DOI] [Google Scholar]
- 9.Liu K, Hogan WR, Crowley RS. Natural language processing methods and systems for biomedical ontology learning. Journal of Biomedical Informatics. 2011;44:163179. doi: 10.1016/j.jbi.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hazman M, El-Beltagy SR, Rafea A. A survey of ontology learning approaches. International Journal of Computer Applications. 2011;22(8):3643. [Google Scholar]
- 11.Biemann C. Ontology learning from text: A survey of methods. LDV Forum. 2005;20(2):75–93. http://www.jlcl.org/2005_Heft2/Chris_Biemann.pdf. [Google Scholar]
- 12.Buitelaar P, Cimiano P, Magnini B. Ontology Learning from Text: Methods, Applications and Evaluation. IOS Press; 2005. Ontology learning from text: An overview; pp. 3–12. [Google Scholar]
- 13.Geffet M, Dagan I. The distributional inclusion hypotheses and lexical entailment. Proceedings of the 43rd Annual Meeting on Association for Computational Linguistics, ACL ’05, Association for Computational Linguistics; Stroudsburg, PA, USA. 2005; pp. 107–114. [DOI] [Google Scholar]
- 14.Faatz A, Steinmetz R. Ontology enrichment with texts from the www. Proceedings of the ECML-Semantic Web Mining Workshop; 2002; ftp://www.kom.tu-darmstadt.de/papers/FS02-1.pdf. [Google Scholar]
- 15.Agirre E, Ansa O, Hovy E, Martinez D. Enriching very large ontologies using the www. Proceedings of the ECAI Workshop on Ontology Learning; 2000; http://arxiv.org/abs/cs/0010026. [Google Scholar]
- 16.Linden K, Piitulainen J. Discovering synonyms and other related words. Proceedings of the 3rd International Workshop on Computational Terminology (CompuTerm 2004); 2004; pp. 63–70. http://www.aclweb.org/anthology/W04-1808.pdf. [Google Scholar]
- 17.Fotzo HN, Gallinari P. Learning generalization/specialization relations between concepts: Application for automatically building thematic document hierarchies. Coupling Approaches, Coupling Media and Coupling Languages for Information Retrieval, RIAO ’04; Vaucluse, France. 2004; pp. 143–155. [Google Scholar]
- 18.Cimiano P, Hotho A, Staab S. Learning concept hierarchies from text corpora using formal concept analysis. Journal of Artificial Intelligence Research. 2005;24(1):305–339. http://dl.acm.org/citation.cfm?id=1622519.1622528. [Google Scholar]
- 19.Drymonas E, Zervanou K, Petrakis EGM. Unsupervised ontology acquisition from plain texts: The ontogain system. Proceedings of the Natural Language Processing and Information Systems, and 15th International Conference on Applications of Natural Language to Information Systems, NLDB’10; Berlin, Heidelberg: Springer-Verlag; 2010. pp. 277–287. http://dl.acm.org/citation.cfm?id=1894525.1894563. [Google Scholar]
- 20.Weber N, Buitelaar P. Web-based ontology learning with ISOLDE. Proceedings of the Workshop on Web Content Mining with Human Language at the International Semantic Web Conference; 2006; http://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.73.1845. [Google Scholar]
- 21.Pei M, Nakayama K, Hara T, Nishio S. Constructing a global ontology by concept mapping using wikipedia thesaurus. International Conference on Advanced Information Networking and Applications, IEEE; 2008; pp. 1205–1210. [Google Scholar]
- 22.Liu Q, Zhu K, Zhang L, Wang H, Yu Y, Pan Y. Catriple: Extracting triples from wikipedia categories. 3rd Asian Semantic Web Conference (ASWC); Springer; 2008. pp. 330–344. [Google Scholar]
- 23.Mintz M, Bills S, Snow R, Jurafsky D. Distant supervision for relation extraction without labeled data. Proceedings of the Joint Conference of the 47th Annual Meeting of the ACL and the 4th International Joint Conference on Natural Language Processing of the AFNLP: Volume 2, ACL ’09; Stroudsburg, PA, USA: Association for Computational Linguistics; 2009. pp. 1003–1011. http://dl.acm.org/citation.cfm?id=1690219.1690287. [Google Scholar]
- 24.Wong W, Liu W, Bennamoun M. Acquiring semantic relations using the web for constructing lightweight ontologies. Proceedings of the 13th Pacific-Asia Conference on Advances in Knowledge Discovery and Data Mining, PAKDD ’09; Berlin, Heidelberg: Springer-Verlag; 2009. pp. 266–277. http://dx.doi.org/10.1007/978-3-642-01307-2_26. [DOI] [Google Scholar]
- 25.Fan JW, Friedman C. Semantic classification of biomedical concepts using distributional similarity. Journal of the American Medical Informatics Association. 2007;14(4):467–477. doi: 10.1197/jamia.M2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Booshehri M, Luksch P. An ontology enrichment approach by using dbpedia. Proceedings of the 5th International Conference on Web Intelligence, Mining and Semantics, WIMS ’15; New York, NY, USA: ACM; 2015. pp. 5:1–5:11. [DOI] [Google Scholar]
- 27.Baader F, Nutt W. The description logic handbook. Cambridge University Press; New York, NY, USA: 2003. pp. 43–95. Ch. Basic Description Logics. http://dl.acm.org/citation.cfm?id=885746.885749. [Google Scholar]
- 28.Ausubel D. The Acquisition and Retention of Knowledge: A Cognitive View. Springer; 2000. [Google Scholar]
- 29.McCray AT. The umls semantic network. Proceedings of the 13th Annual Symposium on Computer Applications in Medical Care; IEEE Computer Society Press; 1989. pp. 475–480. [Google Scholar]
- 30.Weng C, Wu X, Luo Z, Boland MR, Theodoratos D, Johnson SB. Elixr:an approach to eligibility criteria extraction and representation. Journal of the American Medical Informatics Association. 2011;18:116–124. doi: 10.1136/amiajnl-2011-000321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.HXH, Stenner S, Doan S, Johnson K, Waitman L, Denny J. Medex: a medication information extraction system for clinical narratives. Journal of the American Medical Informatics Association. 2010;17:19–24. doi: 10.1197/jamia.M3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luo Z, Duffy R, Johnson S, Weng C. Corpus-based approach to creating a semantic lexicon for clinical research eligibility criteria from umls. AMIA Summit on Translational Informatics. 2010:26–30. [PMC free article] [PubMed] [Google Scholar]
- 33.National library of medicine, rxnorm. [accessed: 2015-12-14]; http://www.nlm.nih.gov/research/umls/rxnorm/
- 34.Johnson SB. A semantic lexicon for medical language processing. Journal of the American Medical Informatics Association. 1999;6(3):205–218. doi: 10.1136/jamia.1999.0060205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Black PE. [accessed: 2015-12-14];Levenshtein distance: Dictionary of algorithms and data structures. 2008 https://xlinux.nist.gov/dads//HTML/Levenshtein.html.
- 36.Everitt S, Landau S, Leese M. Cluster Analysis. 4. Oxford University Press; Oxford: 2001. [Google Scholar]
- 37.Christopoulos DT. Developing methods for identifying the inflection point of a convex/concave curve. arXiv:1206.5478 [math.NA] 2012 [Google Scholar]
- 38.Langfelder P, Zhang B, Horvath S. Defining clusters from a hierarchical cluster tree: the dynamic tree cut package for r. Bioinformatics. 2008;24(5):719–720. doi: 10.1093/bioinformatics/btm563. [DOI] [PubMed] [Google Scholar]
- 39.Rousseeuw PJ. Silhouettes: a graphical aid to the interpretation and validation of cluster analysis. Computational and Applied Mathematics. 1987;20:53–65. doi: 10.1016/0377-0427(87)90125-7. [DOI] [Google Scholar]
- 40.Velardi P, Faralli S, Navigli R. Ontolearn reloaded: A graph-based algorithm for taxonomy induction. Computational Linguistics. 2013;39(3):665–707. [Google Scholar]
- 41.de Knijff J, Frasincar F, Hogenboom F. Domain taxonomy learning from text: The subsumption method versus hierarchical clustering. Data Knowl Eng. 2013;83:54–69. doi: 10.1016/j.datak.2012.10.002. [DOI] [Google Scholar]
- 42.Sanderson M, Croft B. Deriving concept hierarchies from text. Proceedings of the 22Nd Annual International ACM SIGIR Conference on Research and Development in Information Retrieval, SIGIR ’99, ACM; 1999; pp. 206–213. [DOI] [Google Scholar]
- 43.Moutari S. [accessed: 2015-12-10];Unsupervised learning: Clustering. 2015 http://www.bio-complexity.com/QUBsscb13/SSCB2013_SM.pdf.
- 44.Kozareva Z, Hovy E. A semi-supervised method to learn and construct taxonomies using the web. Proceedings of the 2010 Conference on Empirical Methods in Natural Language Processing, EMNLP ’10; Cambridge, Massachusetts. 2010; pp. 1110–1118. http://dl.acm.org/citation.cfm?id=1870658.1870766. [Google Scholar]
- 45.Bennacer N, Karoui L. A framework for retrieving conceptual knowledge from web pages. 2nd Italian Semantic Web Workshop, CEUR Proceedings; 2005; http://ceur-ws.org/Vol-166/43.pdf. [Google Scholar]
- 46.Hearst MA. Automatic acquisition of hyponyms from large text corpora. Proceedings of the 14th Conference on Computational Linguistics - Volume 2, COLING ’92; 1992; pp. 539–545. [DOI] [Google Scholar]
- 47.Grefenstette G. Explorations in Automatic Thesaurus Discovery. Kluwer Academic Publishers; Norwell, MA, USA: 1994. [Google Scholar]
- 48.Caraballo SA. Automatic construction of a hypernym-labeled noun hierarchy from text. Proceedings of the 37th Annual Meeting of the Association for Computational Linguistics on Computational Linguistics, ACL ’99; 1999; pp. 120–126. [DOI] [Google Scholar]
- 49.Bisson G, Nedellec C, Canamero L. Designing clustering methods for ontology building - the mok workbench. ECAI Ontology Learning Workshop; 2000; pp. 13–19. [Google Scholar]
- 50.El Sayed A, Hacid H, Zighed D. A new framework for taxonomy discovery from text. Proceedings of the 12th Pacific-Asia Conference on Advances in Knowledge Discovery and Data Mining, PAKDD’08; Berlin, Heidelberg: Springer-Verlag; 2008. pp. 985–991. http://dl.acm.org/citation.cfm?id=1786574.1786682. [Google Scholar]
- 51.Cimiano P, Staab S. Learning concept hierarchies from text with a guided agglomerative clustering algorithm. Proceedings of the Workshop on Learning and Extending Lexical Ontologies with Machine Learning Methods, OntoML’05; Bonn, Germany. 2005; http://citeseerx.ist.psu.edu/viewdoc/summary?doi=10.1.1.140.7472. [Google Scholar]
- 52.Yang H, Callan J. A metric-based framework for automatic taxonomy induction. Proceedings of the Joint Conference of the 47th Annual Meeting of the ACL and the 4th International Joint Conference on Natural Language Processing of the AFNLP: Volume 1 - Volume 1, ACL ’09; Suntec, Singapore. 2009; pp. 271–279. http://dl.acm.org/citation.cfm?id=1687878.1687918. [Google Scholar]
- 53.Neshati M, Hassanabadi LS. Taxonomy construction using compound similarity measure. Proceedings of the 2007 OTM Confederated International Conference on On the Move to Meaningful Internet Systems: CoopIS, DOA, ODBASE, GADA, and IS - Volume Part I, OTM’07; 2007; pp. 915–932. http://dl.acm.org/citation.cfm?id=1784607.1784688. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.