Abstract
Although cancer-associated fibroblasts (CAFs) are viewed as a promising therapeutic target, the design of rational therapy has been hampered by two key obstacles. First, attempts to ablate CAFs have resulted in significant toxicity because currently used biomarkers cannot effectively distinguish activated CAFs from non-cancer associated fibroblasts and mesenchymal progenitor cells. Second, it is unclear whether CAFs in different organs have different molecular and functional properties that necessitate organ-specific therapeutic designs. Our analyses uncovered COL11A1 as a highly specific biomarker of activated CAFs. Using COL11A1 as a ‘seed’, we identified co-expressed genes in 13 types of primary carcinoma in The Cancer Genome Atlas. We demonstrated that a molecular signature of activated CAFs is conserved in epithelial cancers regardless of organ site and transforming events within cancer cells, suggesting that targeting fibroblast activation should be effective in multiple cancers. We prioritized several potential pan-cancer therapeutic targets that are likely to have high specificity for activated CAFs and minimal toxicity in normal tissues.
Keywords: cancer-associated fibroblasts, myofibroblasts, pan-cancer, therapeutic targets, tumor microenvironment
1. INTRODUCTION
Under normal physiological conditions, collagen-rich fibroblasts maintain tissue architecture and serve as a barrier to epithelial cell migration. However, cancer cells have the ability to convert the surrounding fibroblasts into activated CAFs, which secrete specific collagens, growth factors, and enzymes that promote cancer growth, angiogenesis, invasion, and metastasis [1-3]. At the same time, these CAFs suppress anticancer immunity, confer drug resistance and/or limit the access of chemotherapies, anti-angiogenic therapies, and immunotherapies [1-3]. Although the exact mechanisms by which activated CAFs contribute to such diverse aspects of cancer progression are unclear, it is thought that fibroblasts together with increased collagen deposition and altered extracellular matrix (ECM) remodeling serve as a rich depot of cancer-promoting growth factors, cytokines, and chemokines [1, 2]. Additionally, altered levels of enzymes responsible for collagen cross-link formation, such as lysyl oxidase (LOX) [4], increase tissue stiffness and modify mechanotransduction resulting in the reorganization of loose connective tissue into tense linear tracks of fibers that serve as highways to promote chemotaxis of cancer cells [5, 6].
Recognizing the crucial role of CAFs in most aspects of cancer progression, it has been proposed that rational anticancer therapy design should not only target the cancer cells but also the CAFs [7, 8]. Unlike cancer cells, CAFs are genetically stable [9], which reduces the risk of therapy-induced clonal selection, resistance, and cancer recurrence. Furthermore, targeting CAFs could potentially affect multiple biochemical pathways to prevent cancer progression and recurrence. CAF-targeting therapeutic approaches in different experimental mouse cancer models have been shown to improve tumoral immune response, intratumoral drug delivery, and therapeutic efficacy [10-15]. These studies confirm the key role of CAFs in cancer progression and demonstrate their effectiveness as a therapeutic target. However, targeting CAFs in some cancer models actually promoted cancer progression. For example, depletion of αSMA+ stroma in a mouse pancreatic cancer model resulted in increased cancer aggressiveness, enhanced hypoxia and epithelial-mesenchymal transition (EMT), suppressed anticancer immunity, and reduced survival [16].
The contradictory results in different cancer models could be explained by different roles of CAFs in different cancer types, i.e. CAFs could be promoting breast cancer and inhibiting pancreatic cancer. Alternatively, in all cancer types CAFs prevent cancer progression until they receive activating signals from cancer cells and convert into ‘activated CAFs’, which in turn confer invasive and metastatic abilities upon cancer cells [7]. Therapies that target all CAFs are counterproductive and likely to result in the death of normal fibroblasts and significant toxicity. Preferential targeting of activated CAFs has been challenging because activated CAFs are poorly understood at the molecular level. During activation, CAFs exhibit phenotypic changes that partially overlap with myofibroblastic changes during wound healing, inflammation, and fibrosis, including secretion of specific ECM components, cytokines and growth factors [1, 17]. Several markers have been used to distinguish activated from non-activated CAFs: α-smooth muscle actin (αSMA, encoded by gene ACTA2) [3], fibroblast activation protein (FAP) [12], podoplanin (PDPN) [18], palladin (PALLD) [19, 20], tenascin-C (TNC) [21], platelet-derived growth factor receptor α (PDGFRα) [22, 23], and chondroitin sulfate proteoglycan 4 (CSPG4) [24]. However, these markers are frequently expressed in other cells within the cancer stroma, such as vascular smooth muscle cells, pericytes, and mesenchymal stem cells. This lack of specificity could pose problems in therapeutic targeting and underscores the need to better understand the molecular characteristics of activated CAFs in order to develop more precise and less toxic targeted therapies.
COL11A1 encodes the α1 chain of collagen XI, a minor fibrillar collagen expressed by chondrocytes and osteoblasts but not quiescent fibroblasts [25, 26]. The absence of functional collagen XI leads to abnormally thickened cartilage and tendon fibrils, suggesting the role of collagen XI in maintaining proper fibril diameter [27, 28]. Studies have demonstrated that COL11A1 mRNA is markedly elevated in cancers of the oral cavity/pharynx, head and neck, breast, lung, esophagus, stomach, pancreas, colon, and ovary, but not in matched normal tissues (reviewed in [25, 26]). COL11A1 has been identified as part of gene signatures associated with adverse clinical outcomes including resistance to neoadjuvant therapy in breast cancer [29], time to recurrence in glioblastoma [30], poor survival in kidney cancer [31], and time to recurrence and overall survival in ovarian cancer [32, 33]. In situ hybridization in ovarian cancer and immunohistochemistry in pancreatic cancer revealed that COL11A1 mRNA and pro-protein are primarily expressed in CAFs [25, 32]. The restricted expression of COL11A1 in normal tissues and its enrichment in CAFs during cancer progression combined with its association with adverse clinical outcomes in multiple types of cancer support its candidacy as a specific marker of fibroblast activation in diverse cancers. Here, we explore the suitability of COL11A1 as a pan-cancer marker of activated CAFs and use it as a ‘seed’ to identify the transcription signature of activated CAFs in 13 epithelial cancer types. We show that the COL11A1-coexpressed gene set is highly conserved in these 13 cancer types, indicating that the fibroblast reaction to cancer cells is independent of the organ site-of-origin and of the transforming events within cancer cells. Finally, by combining drug target databases with cancer vs. normal tissue expression databases, we identify several potential therapeutic targets that should have high specificity for activated CAFs and minimal toxicity in normal tissues.
2. MATERIALS AND METHODS
2.1 Human tissues
Studies involving human tissue samples were approved by the Cedars-Sinai Institutional Review Board (IRB 15425). The samples included a tissue microarray from 42 patients with matched primary, metastatic, and recurrent ovarian cancer.
2.2 In situ hybridization
The RNA hybridization kit (RNAscope 2.0 FFPE Assay) and probes for COL11A1, the bacterial gene dapB (negative control), and the housekeeping gene HPRT (positive control), were from Advanced Cell Diagnostics, Inc. Formalin-fixed, paraffin-embedded tissue section slides were processed by the Cedars-Sinai Biobank and Translational Research Core following the protocol provided with the RNAscope In Situ Hybridization kit from Advanced Cell Diagnostics, Inc. The slides were counterstained with Mayer’s hematoxylin.
2.3 Immunohistochemistry
Immunohistochemical detection of αSMA was performed on formalin-fixed, paraffin-embedded tissue sections using the protocol provided with the pre-diluted asm-1 clone antibody from Leica Microsystems. Staining was done by the Cedars-Sinai Pathology Service on the Ventana Benchmark Ultra automated slide stainer. The staining was visualized using the Ventana OptiView DAB Detection System. The slides were counterstained with Mayer’s hematoxylin.
2.4 In vitro co-culture experiments
The ovarian cancer cell lines OVCAR3-GFP, KURAMOCHI-GFP and OVSAHO-GFP were maintained in RPMI-1640 (Corning) supplemented with 10% fetal bovine serum (FBS) and 1x penicillin/streptomycin (Corning). Cell line authenticity was confirmed by Laragen using the short tandem repeat (STR) method. The immortalized normal ovarian fibroblasts INOF-tdTomato cell lines [34] were maintained in a 1:1 ratio of MCDB 105 (Sigma-Aldrich) and Medium 199 (GIBCO) with 10% FBS, 50 U/ml penicillin and 50 μg/ml streptomycin. Immortalized normal ovarian fibroblasts and ovarian cancer cells were co-cultured in 1% FBS supplemented media (1:1:2 ratio of MCDB 105, Medium 199 and RPMI-1640) using 6-well plates, either by directly plating ovarian cancer cells (105 cells/well) onto a 70% confluent layer of normal ovarian fibroblasts or onto a 0.4 μm Transwell membrane. Media were replaced every 2 days. After 4 days of co-culture, GFP-labeled ovarian cancer cells and tdTomato-labeled fibroblasts were separated by FACS in PBS with 0.5% FBS. RNA extraction from fibroblasts and ovarian cancer cell lines was performed using the RNeasy Mini Kit (Qiagen) and reverse transcribed to cDNA using the Quantitect Reverse Transcription Kit (Qiagen). For qRT-PCR, 50 ng of cDNA was mixed with COL11A1 primers (Forward: 5′-GACTATCCCCTCTTCAGAACTGTTAAC-3′; Reverse: 5′- CTTCTATCAAGTGGTTTCGTGGTTT-3′) and the iQ SYBR-Green Supermix (BioRad) and run on the CFX96 Real-Time System (BioRad). Data were analyzed using the 2−ΔCT method and normalized to INOF-tdTomato control to present the fold change ratios. All mRNA data were normalized to RPL32 expression (Forward: 5′-ACAAAGCACATGCTGCCCAGTG-3′; Reverse: 5′-TTCCACGATGGCTTTGCGGTTC-3′). The statistical analyses were performed using GraphPad Prism (version 6.0; GraphPad Software). The unpaired t test was used for data analyses.
2.5 Public database portals and dataset analyses
Data from public portals were used as provided by individual portals without additional processing or normalization, unless otherwise indicated. Box plots of COL11A1 expression in normal tissues and cancers were generated using the Gene Expression across Normal and Tumor tissue (GENT) portal (medical-genome.kribb.re.kr/GENT) in which data from multiple datasets were processed and normalized as previously described [35]. COL11A1 expression level diagrams for inflammatory bowel disease, lung fibrosis, and cancers of the colon and lung were generated using the R2 MegaSampler public portal (hgserver1.amc.nl/cgi-bin/r2/main.cgi). A description of the methods used for data processing and normalization is available through the portal. Survival z-scores for individual genes and cancer types were obtained from the PREdiction of Clinical Outcomes from Genomic Profiles (PRECOG) portal (precog.stanford.edu). Methods for calculating PRECOG z-scores have been published [36]. Ranking of the COL11A1-correlated genes in 13 TCGA carcinoma types was determined using data from individual cancer datasets that were processed by cBio Portal (cbioportal.org) as previously described [37]. Kaplan-Meier survival plots and plots of COL11A1 expression in individual molecular subtypes of ovarian carcinoma were generated using the ovarian cancer microarray gene expression database CSIOVDB (csibio.nus.edu.sg/CSIOVDB/CSIOVDB.html), which has been previously described [38]. The dataset for fibroblast and ovarian epithelial cell co-culture was imported from the Gene Expression Omnibus (ncbi.nlm.nih.gov/geo). The Euclidean distance clustering analysis heatmap for the e-mtab-991 [39] and GSE40595 [40] datasets was generated using the public R2 GeneSet Clustering Analysis portal (hgserver1.amc.nl/cgi-bin/r2/main.cgi), which also describes methods that were used to process and normalize data from datasets included in the portal.
3. RESULTS
3.1 COL11A1 is expressed in a subset of αSMA-positive CAFs and can be induced in normal fibroblasts by the presence of cancer cells
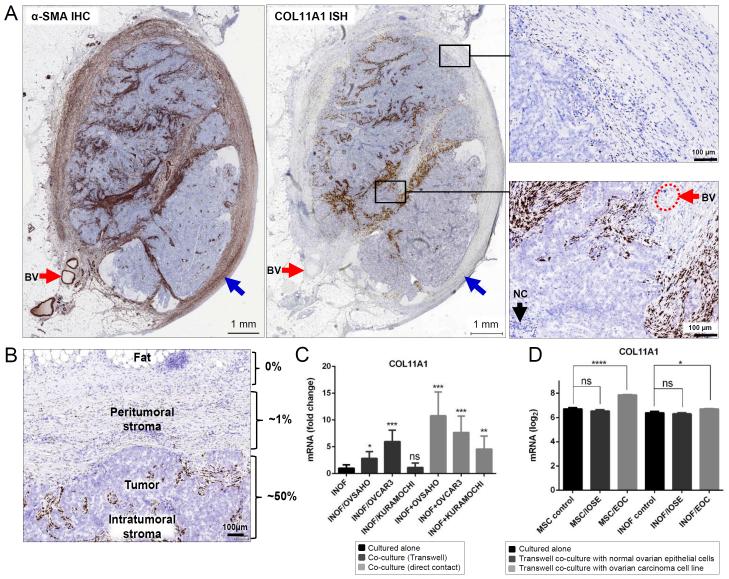
To determine if COL11A1 expression is associated with fibroblast activation, we used αSMA as a marker of activated CAFs [3]. Comparison of αSMA immunohistochemistry and COL11A1 in situ hybridization in a tissue microarray consisting of primary, metastatic and recurrent ovarian cancers from 42 patients showed that COL11A1 is expressed in a subset of αSMA+ CAFs (Fig. 1A). Unlike αSMA, COL11A1 was not expressed in blood vessels (red arrows) or in fibroblasts surrounding the cancer (blue arrows) (Fig. 1A).
Fig. 1. COL11A1 is expressed in CAFs.
(A) Comparison of αSMA immunohistochemistry and COL11A1 in situ hybridization in a metastatic ovarian cancer sample. Red arrows indicate blood vessels. Blue arrows indicate fibroblasts surrounding the tumor nodule. A high magnification of peritumoral and intratumoral regions in the black rectangles is shown in panels on the left. IHC, immunohistochemistry; ISH, in situ hybridization; BV, blood vessel; NC, necrosis. (B) Distribution of in situ hybridization COL11A1-positive CAFs in relation to cancer cells. The estimated percent of COL11A1-positive CAFs is shown on the right. The image is representative of metastatic and recurrent ovarian cancer samples, which typically express higher levels of COL11A1 than primary ovarian cancers. (C) Quantitative RT-PCR levels of COL11A1 in sorted (FACS) immortalized normal ovarian fibroblasts (INOFs) grown alone or co-cultured with ovarian cancer cell lines (OVSAHO, OVCAR3, KURAMOCHI) that were either separated from INOFs by a Transwell membrane or directly mixed with INOFs. Statistical analyses were performed between INOFs grown alone and INOFs co-cultured with ovarian cancer cells (*p<0.05; **p<0.01; ***p<0.001; ns, not significant). Error bars indicate standard deviation. (D) Levels of COL11A1 in the GSE52104 expression dataset in which mesenchymal stem cells (MSCs) or immortalized normal ovarian fibroblasts (INOFs) were either cultured alone or co-cultured with IOSE4 normal epithelial cells (IOSE) or HEYA8 epithelial ovarian cancer cells (EOC) using a Transwell membrane. Inverse-log2 values of the Robust Multi-array Average (RMA) scores from different COL11A1 probes were averaged, then log2-transformed. The data were extracted for statistical analyses using GraphPad Prism 6. Data are represented as the mean ± SEM. Intergroup differences were assessed by the Student’s t-test. *p<0.05; ****p<0.0001.
In sections of metastatic ovarian cancer, we observed that COL11A1-positive cells are confined to the intratumoral and immediate peritumoral CAFs (Fig. 1B), suggesting that COL11A1 expression may be induced by cues received from epithelial cancer cells. To test if cancer cells can induce COL11A1 expression in fibroblasts, we co-cultured immortalized normal ovarian fibroblasts (INOFs) with three different ovarian cancer cell lines (OVSAHO, OVCAR3, and KURAMOCHI). COL11A1 expression in INOFs was most strongly induced by direct co-culture with ovarian cancer cell lines although weak induction occurred by indirect co-culture on a Transwell membrane (Fig. 1C). The induction of COL11A1 in fibroblasts in the presence of cancer cells was confirmed by analysis of the public expression dataset GSE52104 in which two types of presumptive cancer-associated fibroblast precursor cells, mesenchymal stem cells (MSCs) and immortalized normal ovarian fibroblasts (INOFs), were either cultured alone or co-cultured with normal ovarian surface epithelial cells (IOSE) or epithelial ovarian cancer cells (EOC) using a Transwell membrane [41]. COL11A1 mRNA was statistically significantly upregulated when MSCs and INOFs were co-cultured with EOC but not IOSE (Fig. 1D), indicating that cancer cells have a greater capacity than normal cells to induce COL11A1 expression in fibroblasts.
3.2 COL11A1 is associated with cancer progression and poor survival
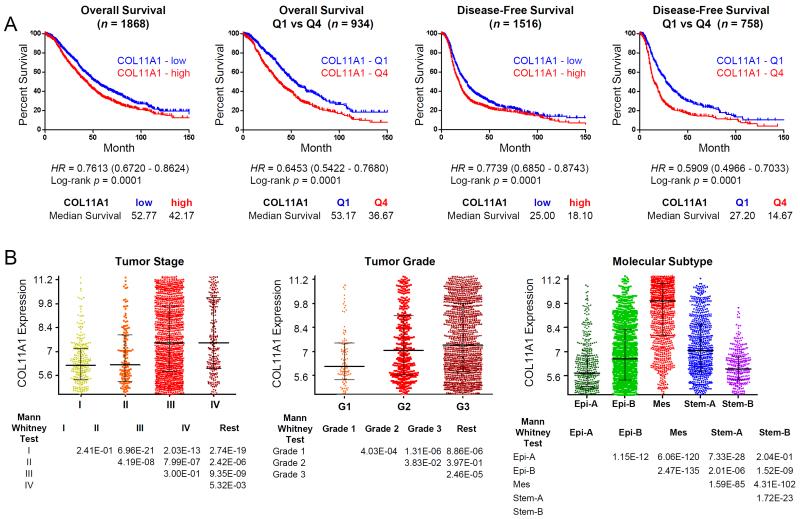
COL11A1 mRNA expression has been associated with poor survival in ovarian cancer [32, 33] and kidney cancer [31]. To elucidate the underlying biology that could result in poor survival, we investigated its expression in ovarian and colon cancers. Using a comprehensively annotated microarray database for 3431 human ovarian cancers [38], we show that increased expression of COL11A1 mRNA is associated with overall survival and disease-free survival (Fig. 2A) as well as with clinical and molecular parameters such as increased cancer stage and grade and mesenchymal molecular subtype (Fig. 2B). The association of COL11A1 expression with poor survival is unlikely to be a manifestation of the total amount of stromal fibroblasts because a general marker of fibroblasts, vimentin (VIM), is not associated with poor survival in the same cohort of ovarian cancer patients (Table S1). The association of COL11A1 with adverse outcomes is also not restricted to ovarian cancer. We show that in 1820 colon cancers [42], increased expression of COL11A1 mRNA is associated with poor disease-specific and disease-free survival as well as with clinical and molecular parameters, such as increased cancer stage and microsatellite instability and CMS4 (mesenchymal) molecular subtype (Fig. S1A,B).
Fig. 2. COL11A1 expression is associated with adverse clinical parameters.
(A) Kaplan-Meier analyses of overall survival (left two panels) and disease-free survival (right two panels) based on COL11A1 expression in ovarian carcinoma. Disease-free survival includes progression- and recurrence-free survival. Patients were stratified to COL11A1-high (red) or COL11A1-low (blue) based on the median expression of COL11A1, and to COL11A1-Q4 (highest 25% expression; red) or COL11A1-Q1 (lowest 25% expression; blue). (B) COL11A1 expression profiles were stratified by FIGO stage (left), FIGO grade (middle), and molecular subtype (right). Data were obtained from the ovarian microarray gene expression database CSIOVDB (csibio.nus.edu.sg/CSIOVDB/CSIOVDB.html). HR, hazard ratio.
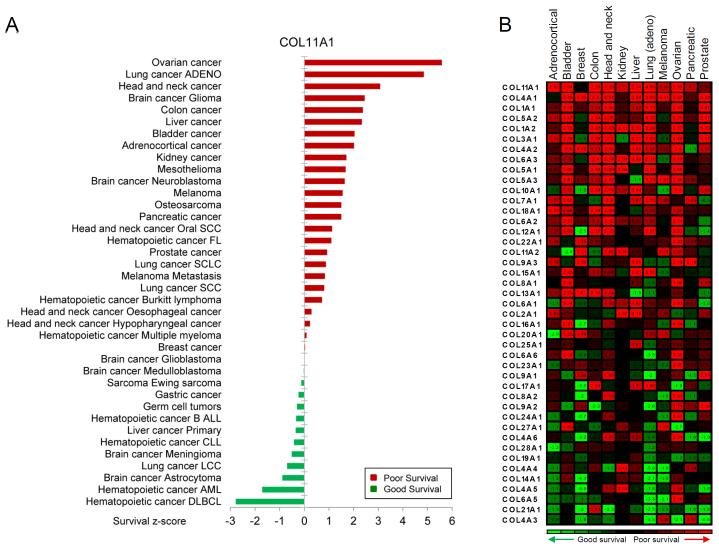
To systematically investigate an association between COL11A1 mRNA expression and survival in various solid and liquid cancers, we plotted COL11A1 z-score values as determined by the pan-cancer PREdiction of Clinical Outcomes from Genomic Profiles (PRECOG) analysis of ~18000 cases in 166 cancer datasets [36]. In most epithelial cancers, COL11A1 expression was associated with poor survival (Fig. 3A). Associations between expression of 43 collagen genes and survival z-scores in 12 common epithelial cancer types revealed that for the majority of collagens, increased expressions of mRNA were associated with poor survival, with COL11A1 having the strongest association (Fig. 3B).
Fig. 3. COL11A1 expression in cancer is associated with poor survival in multiple cancer types.
(A) Survival z-scores in different cancer types associated with expression of COL11A1 mRNA. (B) Survival z-scores associated with mRNA expression of different collagen genes. The data were obtained from the PREdiction of Clinical Outcomes from Genomic Profiles (PRECOG) database (precog.stanford.edu).
3.3 COL11A1 is among the most differentially expressed genes between cancers and corresponding benign tissues
In the Genotype-Tissue Expression (GTEx) project database [43], COL11A1 mRNA is expressed at appreciable levels in transformed skin fibroblasts but not in non-transformed skin fibroblasts or other normal tissues (Fig. S2). Additional analyses of various expression datasets containing normal adult mouse and human tissues revealed that COL11A1 is expressed in cartilage and collagen-producing cells in the eye and brain, with negligible levels in most other tissues that have been profiled including mesenchymal stem cells in the bone marrow, muscle, and fat (data not shown).
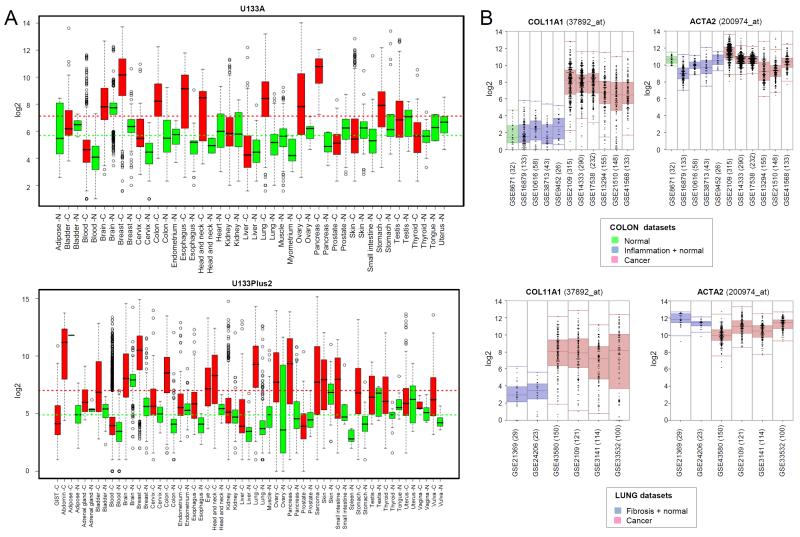
Comparison of COL11A1 expression in 17931 cancers and 3503 normal tissues (U133Plus2 platform) and 9258 cancers and 4087 normal tissues (U133A platform) using the Gene Expression across Normal and Tumor tissue (GENT) portal [35] revealed that COL11A1 mRNA is elevated in most cancers in comparison to their corresponding normal tissues (Fig. 4A). In some cancers, COL11A1 was ranked among the most statistically significant differentially expressed genes when cancer and its corresponding normal tissue were compared. For example, comparison of cancer and normal tissues in The Cancer Genome Atlas (TCGA) datasets for colon cancer and invasive breast cancer ranked COL11A1 as the first and third most differentially expressed gene, respectively (Fig. S3).
Fig. 4. Increased expression of COL11A1 in cancer and low expression in normal tissues, inflammation and fibrosis.
(A) Comparison of COL11A1 mRNA expression in normal tissues and corresponding cancers. Box plots for two different platforms (U133Plus2 and U133A) were generated using datasets and software available through the Gene Expression across Normal and Tumor tissue (GENT) portal (medical-genome.kribb.re.kr/GENT). The y axis shows log2 mRNA levels. Average expression levels in normal tissues and cancer tissues are indicated by vertical dotted green and red lines, respectively. (B) COL11A1 and ACTA2 mRNA expression in normal, inflammatory and fibrotic conditions in comparison to cancer. The graphs were generated using the public R2 MegaSampler software (hgserver1.amc.nl/cgi-bin/r2/main.cgi) for the processing and normalization of individual datasets imported from the Gene Expression Omnibus (u133p2, MAS5.0 platform). The number of samples in each GSE dataset are indicated in parentheses.
As many collagens and collagen-remodeling genes are frequently upregulated in fibroblast activation associated with inflammation and fibrosis in the absence of cancer, use of these genes as therapeutic targets in cancer could be problematic. Analysis of expression profile datasets show that levels of COL11A1 mRNA in inflamed colonic tissue from inflammatory bowel disease and fibrotic lung tissues are not significantly different from those in corresponding unaffected colon and lung and that COL11A1 levels associated with colon inflammation and lung fibrosis are minimal and markedly different from those associated with colon and lung cancers (Fig. 4B). In contrast, levels of ACTA2, the gene encoding the prototypical marker of myofibroblast differentiation, αSMA, is expressed at similar levels in cancers and inflamed or fibrotic tissues (Fig. 4B).
3.4 A consistent set of genes is co-expressed with COL11A1 across different cancers
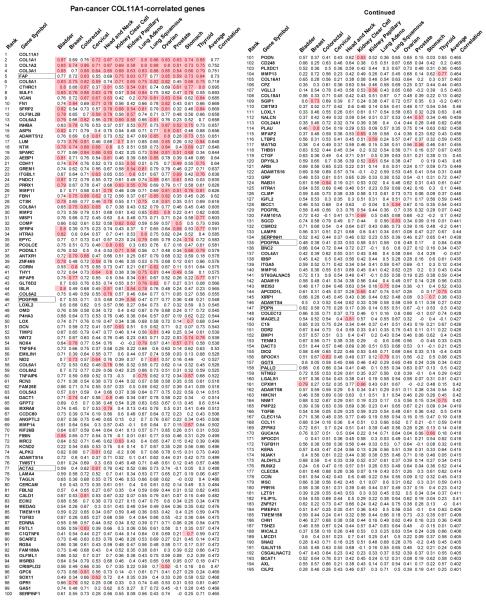
To better understand the biology of cancers with high levels of COL11A1, we identified genes that most closely correlate with COL11A1 mRNA expression in 13 TCGA datasets representing different cancer types. Spearman’s rank correlations between COL11A1 and its co-expressed genes for each cancer type were calculated. The genes were then ranked based on the average correlation of each gene across the 13 cancer types. The top 195 correlated genes were selected based on an average correlation of > 0.4 COL11A1-correlated genes were then ranked based on the average of the absolute correlation values (Table 1 and Table S2). The top 10% most highly correlated genes in each cancer type are highlighted in pink (Table 1). Notably, COL11A1-correlated genes with high average correlation scores also tended to be among the top 10% highest scored genes in each cancer type (indicated in pink in Table 1). In contrast, the top 10% COL11A1-anticorrelated genes were not conserved across these cancer types (Table S3). Some of the top ranked COL11A1-anticorrelated genes in individual cancer types were associated with normal functions of these organs suggesting that they may represent normal tissue or a noninvasive tumor component. For example, the ovarian cancer top 100 COL11A1-anticorrelated genes (Table S3) present in the GSE12172 ovarian cancer dataset were primarily expressed in ovarian tumors of low malignant potential (Fig. S4).
Table 1.
COL11A1-correlated genes (Spearman’s rank correlation) across 13 different TCGA carcinoma types, each represented by >100 primary tumors from therapy-naive patients. Pink denotes the top 10% COL11A1-correlated genes in each individual carcinoma type. Rectangles denote genes frequently used as markers of activated CAFs.
3.5 Pan-cancer COL11A1-correlated genes are induced in CAFs
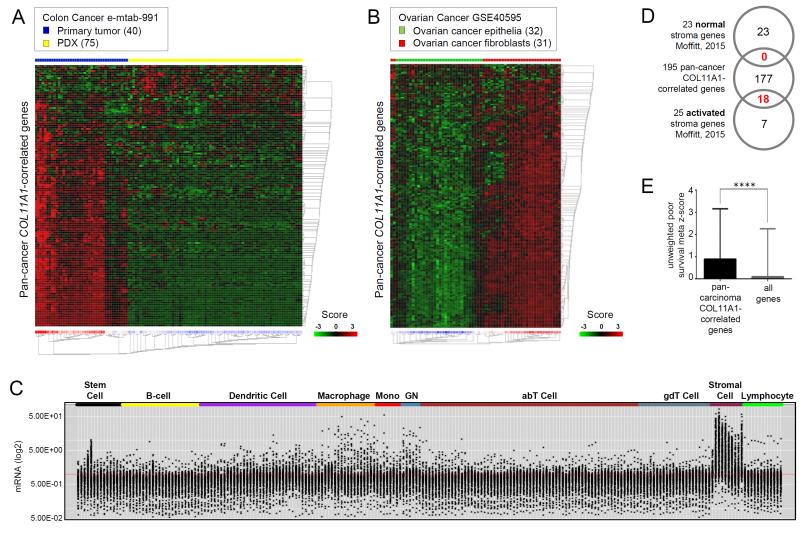
Consistent with the induced expression of COL11A1 in the in vitro co-culture model (Fig. 1D), the average expression of the pan-cancer COL11A1-correlated gene set was significantly induced in mesenchymal stem cells and normal ovarian fibroblasts co-cultured with ovarian cancer cells but not with normal ovarian epithelial cells (Fig. S5). Since epithelial cells were not profiled in this experiment, it is unknown if fibroblasts also induce expression of the pan-cancer COL11A1-correlated genes in epithelial cells. This is relevant because several of the 195 pan-cancer COL11A1-correlated genes have been shown to play a role in EMT [42] and malignant cells undergoing EMT have been proposed as one possible source of CAFs [44]. To determine if the pan-cancer COL11A1-correlated gene set is preferentially expressed in cancer cells undergoing EMT or in host-derived fibroblasts, we used the e-mtab-991 public transcription profile dataset of primary patient-derived colon cancers and their patient-derived xenografts (PDX) in nude mice [39]. Presumably, in PDX samples, fast-proliferating human cancer cells continued to grow in mice while slow-proliferating human CAFs were lost and eventually replaced by mouse fibroblasts, which can be distinguished from human cells by species-specific gene probes [39]. GeneSet clustering analysis showed that most of the pan-cancer COL11A1-correlated genes had diminished levels in PDX samples in comparison to primary cancers (Fig. 5A), suggesting that the genes are enriched in the CAFs rather than in the cancer cells. However, it is also possible that the pan-cancer COL11A1-correlated genes are expressed in epithelial cells in primary colon tumors but become silenced upon adaptation of human cancer cells to the mouse microenvironment. Thus, we conducted GeneSet clustering analysis of the primary ovarian cancer dataset GSE40595 in which ovarian CAFs and epithelial cancer cells were isolated by laser-capture microdissection [40]. The pan-cancer COL11A1-correlated genes were preferentially expressed in CAFs in this dataset (Fig. 5B).
Fig. 5. The pan-cancer COL11A1-correlated gene set is expressed in CAFs and associated with poor patient survival in multiple cancer types.
(A) GeneSet expression clustering analysis of 40 primary colon cancer samples and 75 patient-derived xenograft (PDX) samples in the e-mtab-991 dataset using the pan-cancer COL11A1-correlated genes. (B) GeneSet expression clustering analysis of laser-microdissected ovarian cancer epithelial cells (32 samples) and CAFs (31 samples) in high grade serous ovarian cancer in the GSE40595 dataset using the pan-cancer COL11A1-correlated genes. The Euclidean distance clustering analysis heatmaps in (A) and (B) were generated using the public R2 GeneSet Clustering Analysis tool (hgserver1.amc.nl/cgi-bin/r2/main.cgi). (C) Expression of the pan-cancer COL11A1-correlated genes mapped on the transcriptome of individual murine hematopoietic and stromal cell types in the ImmGene project (immgen.com). The plot was generated using MyGeneset tool (rstats.immgen.org/MyGeneSet). Black dots represent mRNA levels (y axis) of 186 pan-cancer COL11A1-correlated genes (9 genes were not present in the database) across 230 individual cell types (X axis) grouped into 10 main groups. (D) Overlap of the 195 pan-cancer COL11A1-correlated genes with 23 ‘normal stroma’ and 25 ‘activated stroma’ genes defined by Moffitt et al. (E) Unweighted meta z-scores of 191 COL11A1-correlated genes (4 genes were not available in the PRECOG database) were compared with those of all genes (total 23287 genes) in the PRECOG database using unpaired t test. The plot was generated using GraphPad Prism software version 6.0. Intergroup differences were assessed by the Student’s t-test. Mean ± SEM of pan-cancer COL11A1-correlated genes (0.8916 ± 0.1641 N=191); Mean ± SEM of all genes (0.09918 ± 0.01416 N=23287); ****p<0.0001.
In addition to CAFs, immune cells are a major component of the tissue microenvironment. To exclude the possibility that the pan-cancer COL11A1-correlated gene set represents immune cells in the tumor microenvironment, we used the expression profile of 230 mouse hematopoietic cell types generated by the Immunological Genome Project (ImmGen) compendium [45]. In addition to hematopoietic cell lineages, the dataset contains expression profiles of skin fibroblasts and fibroblasts residing in the thymus, lymph nodes, and spleen. The pan-cancer COL11A1-correlated gene set was highly represented in fibroblasts but not in hematopoietic cell lineages (Fig. 5C).
CAFs have a different expression profile than normal fibroblasts. Moffitt and colleagues defined a 23-gene signature of ‘normal stroma’ and a 25-gene signature of ‘activated stroma’ [46] using non-negative matrix factorization for virtual microdissection of primary and metastatic pancreatic ductal cancer samples into cell subsets with prognostic and biologic relevance. None of the 23 (0%) ‘normal stroma’ genes in contrast to 18 of 25 (72%) ‘activated stroma’ genes were present in the COL11A1-correlated gene set, respectively (Fig. 5D), suggesting that the COL11A1-correlated gene set represents CAFs.
3.6 Biological processes associated with fibroblast activation in cancer
The remarkable uniformity of COL11A1-correlated genes across 13 different cancer types suggests involvement of these genes in common biological processes that are independent of the organ site and of the phenotypic and genetic diversity observed in individual cancer types. To gain insight into the biology of this conserved gene set, we conducted several analyses that identified overlap between the 195 pan-cancer COL11A1-correlated genes and genes in various datasets with characterized biological features. The Gene Ontology (GO) Biological Process (BP) analysis revealed that the pan-cancer COL11A1-correlated genes are primarily involved in extracellular matrix modification and collagen remodeling (Table S4). Additionally, we used SABiosciences array gene tables, which consist of literature-based curated molecular pathways where each pathway was represented by 84 genes. Analysis of gene overlap between the pan-cancer COL11A1-correlated gene set and genes representative of 67 different pathways in SABiosciences arrays showed the largest overlap for pathways associated with extracellular matrix, fibrosis, osteogenesis, wound healing, EMT, cardiovascular disease, and transforming growth factor β (TGFβ) signaling (Table S5). COL11A1-correlated gene set enrichment analysis of chemical and genetic perturbations (CGP) showed the most significant overlap with genes up-regulated in association with cancer invasiveness, advanced stage, stromal cell stemness, and epithelial-mesenchymal transition (EMT) (Table S6). The uniformity of the COL11A1-correlated genes across different cancers might also indicate that these genes are regulated by a common mechanism. Ingenuity Pathway Analysis showed that transforming growth factor beta 1 (TGFB1) is the most strongly associated upstream regulator of the pan-cancer COL11A1-correlated genes (Table S7).
3.7 COL11A1-correlated genes are associated with poor patient survival and represent potential therapeutic targets
To determine whether the pan-cancer COL11A1-correlated gene set is associated with patient survival in the ~18,000 cases of liquid and solid malignancies in the PRECOG dataset [36], we compared survival z-scores for the 195 pan-cancer COL11A1-correlated genes with the survival z-scores for all genes in the dataset. This analysis showed that expression of the pan-cancer COL11A1-correlated gene set is significantly associated with poor survival (Fig. 5E).
Expression profile analyses have identified a mesenchymal molecular subtype of cancer associated with poor survival in multiple cancers including ovarian [47], pancreatic [46], gastric [48] and colon [49]. In colon cancer, it has been shown that the mesenchymal molecular subtype, which constitutes approximately 23% of colon cancers, has no significant enrichment for targetable mutations or copy number changes in candidate driver genes [49]. Even if future research identifies targetable events in cancer cells of the mesenchymal subtype, it is predicted that enrichment in CAFs and excessive ECM deposition will reduce therapeutic efficacy by creating a physical barrier for drug transport. Thus, simultaneous targeting of CAFs and cancer cells may be necessary for chemotherapeutic accessibility.
To identify therapies that preferentially target activated CAFs and spare normal tissues, we combined drug target searches with expression profile datasets in cancers and normal tissues. Ingenuity Pathway Analysis and searches of the Clinicaltrials.gov (clinicaltrials.gov) and ChEMBL [50] (ebi.ac.uk/chembl) databases revealed that, of the 195 pan-cancer COL11A1-correlated genes, 16 are targets of drugs used in clinical trials (Table S8) and 30 are targets of bioactive compounds (Table S9).
To test whether any of the drug/bioactive compound target genes in Tables S8 and S9 are exclusively expressed in activated CAFs, we determined the expression levels of each gene in normal tissues in the GTEx database [43] and in normal vs. cancer tissues in the GENT database [35]. Additionally, to test whether selected genes were exclusively overexpressed in cancer tissues and not in non-cancer associated pathologies such as inflammation and fibrosis, we compared expression of these genes in normal tissues, inflamed/fibrotic tissues and cancer tissues of the colon and lung. Unlike COL11A1, which has restricted expression in normal tissues (Fig. S2) and is highly elevated in cancer vs. normal tissues (Fig. 4A) and in cancer vs. inflamed/fibrotic tissues (Fig. 4B) most of the target genes were expressed at high levels in at least one normal tissue and/or exhibited equivalent expression levels in cancers vs. normal tissues, and cancers vs. inflamed/fibrotic tissues. One example of this pattern of expression is CTGF (Fig. S6 and S7). However, FN1, MMP13, MMP14, FAP, LOX and COL1A2 exhibited restricted expression in normal tissues and elevated expression in cancer vs. normal tissues. One example of this pattern of expression is FAP (Fig. S8 and S9). Among FN1, MMP13, MMP14, FAP, LOX and COL1A2, FAP was also differentially expressed between inflamed/fibrotic tissues and cancer tissues although this difference in expression was not as prominent as for COL11A1 (compare Fig. S9B and Fig. 4B).
4. DISCUSSION
Whereas in the past most therapeutic approaches have focused on the cancer cell and its genetic alterations, it is becoming apparent that the microenvironment plays an equally important role in cancer evolution. We now recognize that the cancer stroma not only serves as a scaffold for tissue organization and integrity but also provides key biomechanical and molecular signals that can affect various aspects of cancer growth and biology, including proliferation, survival, metabolism, stem cell fate, and response to chemotherapy [51, 52]. As the genetically stable subpopulations of the cancer microenvironment are increasingly recognized as potentially effective therapeutic targets, a comprehensive definition of their molecular characteristics will be a prerequisite for the development of more precise and less toxic therapies. Currently, there are no reliable methods to distinguish activated CAFs from non-activated CAFs, which although frequently abundant within cancers do not necessarily contribute to adverse outcome. We identified COL11A1 among the top differentially expressed genes in multiple cancer types when cancer tissues and their corresponding normal tissues were compared. We showed that an increase in COL11A1 expression is associated with progression and poor survival in most cancer types. COL11A1 is a particularly attractive therapeutic target because of its restricted expression in normal tissues and non-cancer conditions, such as inflammation and fibrosis.
The identification of a highly conserved set of genes associated with COL11A1 expression in breast, lung, pancreas, stomach, urinary bladder, colon, thyroid, cervix, head and neck, thyroid, ovary, and prostate cancers was somewhat surprising in light of the genetic and phenotypic diversity among these cancer types. The conserved expression signature indicates that the reaction of stromal tissues to invading epithelial cancer cells may be similar regardless of the organ of origin or genetic alterations. This has significant implications for the development of pan-cancer therapeutic strategies. Our analysis of potential upstream regulators of the pan-cancer COL11A1-correlated genes revealed TGFB1 as the most likely candidate. Dysregulation of TGFβ signaling is recognized as the main driver of fibroblast activation and represents the most logical therapeutic target [53]. In immortalized normal ovary fibroblast cell culture, recombinant TGFB1 has been shown to upregulate expression of COL11A1 and several other COL11A1-correlated genes; this effect was abrogated by the TGFβ receptor inhibitor A83-01 [32]. However, the pleiotropic nature of TGFβ signaling carries the risk of adverse effects in patients [54]. In order to abrogate fibroblast activation without the negative effects of pan-TGFβ therapy, it will be necessary to design therapies for more specific targets.
Sixteen of the 195 pan-cancer COL11A1-correlated genes are targets of drugs in clinical trials. These targets include CTGF, a matricellular protein involved in myofibroblast formation in cancer as a binding factor of fibronectin and a downstream mediator of TGFβ. A clinical trial (clinicaltrials.gov; NCT02210559) is currently enrolling patients with unresectable pancreatic cancer to test a combination of conventional chemotherapy and FG-3019, the human monoclonal antibody that interferes with the action of CTGF. Our expression analyses, consistent with the published literature (reviewed in [55]), shows that CTGF is expressed at similar levels in normal tissues and cancers and therefore unlikely to be a safe therapeutic target for cancer treatment. In contrast, targets such as FN1, FAP, MMP13, LOX and COL2A1 are markedly increased in cancer/inflammation/fibrosis compared to normal tissues and are thus predicted to have a better safety profile than agents targeting CTGF. Our assessment is consistent with the published moderate and reversible toxicity of the FN1-targeting monoclonal antibody-cytokine fusion protein L19-IL2, which is in a phase I/II study for patients with solid cancers (clinicaltrials.gov; NCT01058538) [56, 57]. Our expression analyses show that one of the targets of bioactive compounds, FAP, has low expression in normal tissues and also lower expression in inflamed/fibrotic tissues than in cancer. Yet, this difference in expression may not be sufficient for specific targeting of activated CAFs as studies in mouse models have shown that depletion of the FAP+ stroma can induce toxicity due to expression of FAP in the mesenchymal cells of bone marrow, muscle and adipose tissue [58, 59]. Future efforts to specifically target activated CAFs can be improved by designing novel therapies to target genes that exhibit restricted expression in nonmalignant tissues. When considering COL11A1 as a cancer-specific biomarker and therapeutic target, it is important to note that several normal tissues express COL11A1. The potential side effects of COL11A1 targeting can be predicted based on the phenotypes of mice and humans expressing mutant nonfunctional forms of COL11A1. A homozygous truncating mutation of COL11A1 in mice results in poorly formed cartilage [60], while human COL11A1 mutations are associated with articular hypermobility, dermal hyperelasticity and widespread tissue fragility [61]. Of note, these collagenopathies are associated with the absence of COL11A1 function throughout development and are unlikely to manifest upon transient targeting of COL11A1 in adults. Additionally, since COL11A1 and many of the pan-cancer COL11A1-coexpressed genes have multiple tissue-specific mRNA splicing isoforms, it will be valuable for future targeting purposes to determine if any mRNA isoforms are specifically expressed in activated CAFs [62].
Supplementary Material
Supplementary Figure Legend
Fig. S1. Colon cancer survival (A), clinical and molecular (B) parameters associated with COL11A1 expression. Kaplan-Meier survival plots and plots of COL11A1 expression in individual subsets of colon cancer were generated using the data and software described previously by Tan and colleagues (Tan et al., EMBO Mol Med, 2014).
Fig. S2. Expression of COL11A1 in normal tissues and transformed cell lines. The data were obtained from the Genotype-Tissue Expression (GTEx) portal (gtexportal.com; date of analysis: 05/28/16).
Fig. S3. COL11A1 expression in invasive ductal breast cancer and colon cancer and their corresponding normal tissues. The data were obtained using TCGA datasets in the Oncomine portal (oncomine.com; date of analysis: 12/14/15.
Fig. S4. Ovarian cancer COL11A1-anticorrelated genes are enriched in ovarian tumors of low malignant potential. GeneSet clustering analysis of low malignant potential serous ovarian tumors (n=32) and invasive serous ovarian tumors (n=58) using the top 100 COL11A1-anticorrelated genes (Spearman’s correlation, Table S3) from the ovarian cancer TCGA dataset. The clustering analysis was performed using R2 (http://hgserver1.amc.nl/cgi-bin/r2/main.cgi; date of analysis: 03/20/16).
Fig. S5. Levels of the pan-cancer COL11A1-correlated genes in the GSE52104 expression dataset in which mesenchymal stem cells (MSCs) or immortalized normal ovarian fibroblasts (INOFs) were either cultured alone or co-cultured with IOSE4 normal epithelial cells (IOSE) or HEYA8 epithelial ovarian cancer cells (EOC) using a Transwell membrane. Inverse-log2 data of different probes of the same gene were averaged, then log2-transformed. The average of log2 values for COL11A1 and the 194 COL11A1-correlated genes (one gene was not represented in the dataset) were extracted for statistical analyses using GraphPad Prism 6. Data were expressed as the mean ± SEM. Intergroup differences were assessed by the Student’s t-test. *p<0.05.
Fig. S6. Expression of CTGF in normal tissues and transformed cell lines. The data were obtained from the Genotype-Tissue Expression (GTEx) portal (gtexportal.com; date of analysis: 05/28/16).
Fig. S7. CTGF may not be an adequate therapeutic target in cancer because of the similar expression levels in cancer and normal tissues. (A) Comparison of CTGF mRNA expression in normal tissues and corresponding cancers. Box plots for two different platforms (U133Plus2 and U133A) were generated using datasets and software available through the Gene Expression across Normal and Tumor tissue (GENT) portal (medical-genome.kribb.re.kr/GENT). The y axis shows log2 mRNA levels. Average expression levels in normal tissues and cancer tissues are indicated by vertical dotted green and red lines, respectively. (B) CTGF mRNA expression in normal, inflammatory and fibrotic conditions in comparison to cancer. The graphs were generated using the public R2 MegaSampler software (hgserver1.amc.nl/cgi-bin/r2/main.cgi) for the processing and normalization of individual datasets imported from the Gene Expression Omnibus (u133p2, MAS5.0 platform). The number of samples in each GSE dataset are indicated in parentheses.
Fig. S8. Expression of FAP in normal tissues and transformed cell lines. The data were obtained from the Genotype-Tissue Expression (GTEx) portal (gtexportal.com; date of analysis: 05/28/16).
Fig. S9. Increased expression of FAP in cancer. (A) Comparison of FAP mRNA expression in normal tissues and corresponding cancers. Box plots for two different platforms (U133Plus2 and U133A) were generated using datasets and software available through the Gene Expression across Normal and Tumor tissue (GENT) portal (medical-genome.kribb.re.kr/GENT). The y axis shows log2 mRNA levels. Average expression levels in normal tissues and cancer tissues are indicated by vertical dotted green and red lines, respectively. (B) FAP mRNA expression in normal, inflammatory and fibrotic conditions in comparison to cancer. The graphs were generated using the public R2 MegaSampler software (hgserver1.amc.nl/cgi-bin/r2/main.cgi) for the processing and normalization of individual datasets imported from the Gene Expression Omnibus (u133p2, MAS5.0 platform). The number of samples in each GSE dataset are indicated in parentheses.
Supplementary Table Legend
Table S1. Cox multivariate analysis of COL11A1 (marker of activated CAFs) and VIM (general marker of fibroblasts) related to survival in ovarian cancer patients. Data were obtained from the ovarian microarray gene expression database CSIOVDB (csibio.nus.edu.sg/CSIOVDB/CSIOVDB.html).
Table S2. Pan-cancer COL11A1-correlated genes ranked by average correlation to COL11A1 in 13 TCGA cancer types and annotated by gene name, cellular localization and molecule type.
Table S3. COL11A1-anticorrelated genes (Spearman’s rank correlation) across 13 different TCGA carcinoma types, each represented by >100 primary tumors from therapy-naive patients. Pink denotes the top 10% COL11A1-anticorrelated genes in each individual carcinoma type. Data were analyzed using cBio Portal (cbioportal.org; date of analysis: 01/16/16).
Table S4. Gene Ontology (GO) enrichment analysis of biological processes associated with the pan-cancer COL11A1-correlated gene set. Data were analyzed using the Gene Ontology Consortium portal (geneontology.org/; date of analysis: 02/01/16).
Table S5. Molecular pathways associated with the pan-cancer COL11A1-correlated gene set. Overlap between the 195 pan-cancer COL11A1-correlated genes and common molecular pathways defined by 84 representative genes in the SABiosciences Pathway Arrays.
Table S6. Pan-cancer COL11A1-correlated gene set analysis of chemical and genetic perturbations (CGP) using the GSEA portal (software.broadinstitute.org/gsea/index.jsp; date of analysis: 02/08/16).
Table S7. Upstream regulators of pan-cancer COL11A1-correlated genes. Ingenuity Pathway Analysis (ingenuity.com; date of analysis: 02/08/16).
Table S8. Pan-cancer COL11A1-correlated genes that are targets of drugs in clinical trials. Ingenuity Pathway Analysis (ingenuity.com; date of analysis: 05/28/16).
Table S9. Pan-cancer COL11A1-correlated genes that are targets of bioactive compounds. ChEMBL database analysis (ebi.ac.uk/chembl; date of analysis: 05/28/16).
Highlights.
COL11A1 is a highly specific biomarker of activated CAFs in multiple epithelial cancer types.
COL11A1 is not expressed in mesenchymal precursors or fibroblasts associated with non-cancerous conditions, such as inflammation and organ fibrosis.
Activated CAFs across genetically different epithelial cancer types express a highly conserved gene signature.
ACKNOWLEDGMENTS
We thank S. Swartwood (Biobank and Translational Research Core) for in situ hybridization; F. Chung (Pathology Service) for immunohistochemistry; G. Martins (Flow Cytometry Core) for assistance with FACS analysis; and K. Daniels for assistance with manuscript preparation. SO is supported by the Office of the Assistant Secretary of Defense for Health Affairs through the Ovarian Cancer Research Program Award No. W81XWH-14-1-0107 and W81XWH-16-1-0190, the National Cancer Institute grants R21CA194626-01 and 1R01CA208753-01, and the Cantor-UCLA Women’s Health Center Executive Advisory Board & NCATS UCLA Clinical and Translational Science Institute grant UL1TR000124. BYK is supported by the American Cancer Society Early Detection Professorship Grant (SIOP-06-258-01-COUN). BYK, SO, KL and WRW are supported by the Ovarian Cancer Research Fund Program Project Development Award and DJC is supported by the Ovarian Cancer Research Fund Ann Shreiber Mentored Investigator Award. Opinions, interpretations, conclusions and recommendations are those of the authors and are not necessarily endorsed by the funding agencies.
Abbreviations
- αSMA
α-smooth muscle actin
- CAFs
cancer-associated fibroblasts
- CSPG4
chondroitin sulfate proteoglycan 4
- ECM
extracellular matrix
- LOX
lysyl oxidase
- EMT
epithelial-mesenchymal transition
- FAP
fibroblast activation protein
- PALLD
palladin
- PDGFRα
platelet-derived growth factor receptor α
- PDPN
podoplanin
- TGFβ
transforming growth factor β
- TNC
tenascin-C
- PRECOG
PREdiction of Clinical Outcomes from Genomic Profiles
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONFLICT OF INTEREST
Authors declare no conflict of interest.
REFERENCES
- [1].Kalluri R, Zeisberg M. Fibroblasts in cancer. Nature reviews. Cancer. 2006;6:392–401. doi: 10.1038/nrc1877. [DOI] [PubMed] [Google Scholar]
- [2].Hinz B, Phan SH, Thannickal VJ, Prunotto M, Desmouliere A, Varga J, De Wever O, Mareel M, Gabbiani G. Recent developments in myofibroblast biology: paradigms for connective tissue remodeling. The American journal of pathology. 2012;180:1340–1355. doi: 10.1016/j.ajpath.2012.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Desmouliere A, Guyot C, Gabbiani G. The stroma reaction myofibroblast: a key player in the control of tumor cell behavior. Int J Dev Biol. 2004;48:509–517. doi: 10.1387/ijdb.041802ad. [DOI] [PubMed] [Google Scholar]
- [4].Barker HE, Cox TR, Erler JT. The rationale for targeting the LOX family in cancer. Nature reviews. Cancer. 2012;12:540–552. doi: 10.1038/nrc3319. [DOI] [PubMed] [Google Scholar]
- [5].Paszek MJ, Zahir N, Johnson KR, Lakins JN, Rozenberg GI, Gefen A, Reinhart-King CA, Margulies SS, Dembo M, Boettiger D, Hammer DA, Weaver VM. Tensional homeostasis and the malignant phenotype. Cancer Cell. 2005;8:241–254. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- [6].Provenzano PP, Inman DR, Eliceiri KW, Knittel JG, Yan L, Rueden CT, White JG, Keely PJ. Collagen density promotes mammary tumor initiation and progression. BMC Med. 2008;6:11. doi: 10.1186/1741-7015-6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med. 2013;19:1423–1437. doi: 10.1038/nm.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Jain RK. Normalizing tumor microenvironment to treat cancer: bench to bedside to biomarkers. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2013;31:2205–2218. doi: 10.1200/JCO.2012.46.3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Qiu W, Hu M, Sridhar A, Opeskin K, Fox S, Shipitsin M, Trivett M, Thompson ER, Ramakrishna M, Gorringe KL, Polyak K, Haviv I, Campbell IG. No evidence of clonal somatic genetic alterations in cancer-associated fibroblasts from human breast and ovarian carcinomas. Nat Genet. 2008;40:650–655. doi: 10.1038/ng.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Arnold JN, Magiera L, Kraman M, Fearon DT. Tumoral immune suppression by macrophages expressing fibroblast activation protein-alpha and heme oxygenase-1. Cancer immunology research. 2014;2:121–126. doi: 10.1158/2326-6066.CIR-13-0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Feig C, Jones JO, Kraman M, Wells RJ, Deonarine A, Chan DS, Connell CM, Roberts EW, Zhao Q, Caballero OL, Teichmann SA, Janowitz T, Jodrell DI, Tuveson DA, Fearon DT. Targeting CXCL12 from FAP-expressing carcinoma-associated fibroblasts synergizes with anti-PD-L1 immunotherapy in pancreatic cancer. Proc Natl Acad Sci U S A. 2013;110:20212–20217. doi: 10.1073/pnas.1320318110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Kraman M, Bambrough PJ, Arnold JN, Roberts EW, Magiera L, Jones JO, Gopinathan A, Tuveson DA, Fearon DT. Suppression of antitumor immunity by stromal cells expressing fibroblast activation protein-alpha. Science. 2010;330:827–830. doi: 10.1126/science.1195300. [DOI] [PubMed] [Google Scholar]
- [13].Lo A, Wang LC, Scholler J, Monslow J, Avery D, Newick K, O'Brien S, Evans RA, Bajor DJ, Clendenin C, Durham AC, Buza EL, Vonderheide RH, June CH, Albelda SM, Pure E. Tumor-Promoting Desmoplasia Is Disrupted by Depleting FAP-Expressing Stromal Cells. Cancer research. 2015;75:2800–2810. doi: 10.1158/0008-5472.CAN-14-3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Loeffler M, Kruger JA, Niethammer AG, Reisfeld RA. Targeting tumor-associated fibroblasts improves cancer chemotherapy by increasing intratumoral drug uptake. J Clin Invest. 2006;116:1955–1962. doi: 10.1172/JCI26532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Provenzano PP, Cuevas C, Chang AE, Goel VK, Von Hoff DD, Hingorani SR. Enzymatic targeting of the stroma ablates physical barriers to treatment of pancreatic ductal adenocarcinoma. Cancer Cell. 2012;21:418–429. doi: 10.1016/j.ccr.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Ozdemir BC, Pentcheva-Hoang T, Carstens JL, Zheng X, Wu CC, Simpson TR, Laklai H, Sugimoto H, Kahlert C, Novitskiy SV, De Jesus-Acosta A, Sharma P, Heidari P, Mahmood U, Chin L, Moses HL, Weaver VM, Maitra A, Allison JP, LeBleu VS, Kalluri R. Depletion of carcinoma-associated fibroblasts and fibrosis induces immunosuppression and accelerates pancreas cancer with reduced survival. Cancer Cell. 2014;25:719–734. doi: 10.1016/j.ccr.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Junttila MR, de Sauvage FJ. Influence of tumour micro-environment heterogeneity on therapeutic response. Nature. 2013;501:346–354. doi: 10.1038/nature12626. [DOI] [PubMed] [Google Scholar]
- [18].Kawase A, Ishii G, Nagai K, Ito T, Nagano T, Murata Y, Hishida T, Nishimura M, Yoshida J, Suzuki K, Ochiai A. Podoplanin expression by cancer associated fibroblasts predicts poor prognosis of lung adenocarcinoma. Int J Cancer. 2008;123:1053–1059. doi: 10.1002/ijc.23611. [DOI] [PubMed] [Google Scholar]
- [19].Ronty MJ, Leivonen SK, Hinz B, Rachlin A, Otey CA, Kahari VM, Carpen OM. Isoform-specific regulation of the actin-organizing protein palladin during TGF-beta1-induced myofibroblast differentiation. The Journal of investigative dermatology. 2006;126:2387–2396. doi: 10.1038/sj.jid.5700427. [DOI] [PubMed] [Google Scholar]
- [20].Brentnall TA, Lai LA, Coleman J, Bronner MP, Pan S, Chen R. Arousal of cancer-associated stroma: overexpression of palladin activates fibroblasts to promote tumor invasion. PLoS One. 2012;7:e30219. doi: 10.1371/journal.pone.0030219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].De Wever O, Nguyen QD, Van Hoorde L, Bracke M, Bruyneel E, Gespach C, Mareel M. Tenascin-C and SF/HGF produced by myofibroblasts in vitro provide convergent pro-invasive signals to human colon cancer cells through RhoA and Rac. FASEB J. 2004;18:1016–1018. doi: 10.1096/fj.03-1110fje. [DOI] [PubMed] [Google Scholar]
- [22].Pietras K, Pahler J, Bergers G, Hanahan D. Functions of paracrine PDGF signaling in the proangiogenic tumor stroma revealed by pharmacological targeting. PLoS Med. 2008;5:e19. doi: 10.1371/journal.pmed.0050019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Erez N, Truitt M, Olson P, Arron ST, Hanahan D. Cancer-Associated Fibroblasts Are Activated in Incipient Neoplasia to Orchestrate Tumor-Promoting Inflammation in an NF-kappaB-Dependent Manner. Cancer Cell. 2010;17:135–147. doi: 10.1016/j.ccr.2009.12.041. [DOI] [PubMed] [Google Scholar]
- [24].Sugimoto H, Mundel TM, Kieran MW, Kalluri R. Identification of fibroblast heterogeneity in the tumor microenvironment. Cancer Biol Ther. 2006;5:1640–1646. doi: 10.4161/cbt.5.12.3354. [DOI] [PubMed] [Google Scholar]
- [25].Vazquez-Villa F, Garcia-Ocana M, Galvan JA, Garcia-Martinez J, Garcia-Pravia C, Menendez-Rodriguez P, Gonzalez-del Rey C, Barneo-Serra L, de Los Toyos JR. COL11A1/(pro)collagen 11A1 expression is a remarkable biomarker of human invasive carcinoma-associated stromal cells and carcinoma progression. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine. 2015;36:2213–2222. doi: 10.1007/s13277-015-3295-4. [DOI] [PubMed] [Google Scholar]
- [26].Raglow Z, Thomas SM. Tumor matrix protein collagen XIalpha1 in cancer. Cancer letters. 2015;357:448–453. doi: 10.1016/j.canlet.2014.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Wenstrup RJ, Smith SM, Florer JB, Zhang G, Beason DP, Seegmiller RE, Soslowsky LJ, Birk DE. Regulation of collagen fibril nucleation and initial fibril assembly involves coordinate interactions with collagens V and XI in developing tendon. The Journal of biological chemistry. 2011;286:20455–20465. doi: 10.1074/jbc.M111.223693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Smith SM, Birk DE. Focus on molecules: collagens V and XI. Experimental eye research. 2012;98:105–106. doi: 10.1016/j.exer.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Farmer P, Bonnefoi H, Anderle P, Cameron D, Wirapati P, Becette V, Andre S, Piccart M, Campone M, Brain E, Macgrogan G, Petit T, Jassem J, Bibeau F, Blot E, Bogaerts J, Aguet M, Bergh J, Iggo R, Delorenzi M. A stroma-related gene signature predicts resistance to neoadjuvant chemotherapy in breast cancer. Nat Med. 2009;15:68–74. doi: 10.1038/nm.1908. [DOI] [PubMed] [Google Scholar]
- [30].Cheng WY, Kandel JJ, Yamashiro DJ, Canoll P, Anastassiou D. A multi-cancer mesenchymal transition gene expression signature is associated with prolonged time to recurrence in glioblastoma. PLoS One. 2012;7:e34705. doi: 10.1371/journal.pone.0034705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Boguslawska J, Kedzierska H, Poplawski P, Rybicka B, Tanski Z, Piekielko-Witkowska A. Expression of genes involved in cellular adhesion and ECM-remodelling correlates with poor survival of renal cancer patients. The Journal of urology. 2015 doi: 10.1016/j.juro.2015.11.050. [DOI] [PubMed] [Google Scholar]
- [32].Cheon DJ, Tong Y, Sim MS, Dering J, Berel D, Cui X, Lester J, Beach JA, Tighiouart M, Walts AE, Karlan BY, Orsulic S. A collagen-remodeling gene signature regulated by TGF-beta signaling is associated with metastasis and poor survival in serous ovarian cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2014;20:711–723. doi: 10.1158/1078-0432.CCR-13-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Wu YH, Chang TH, Huang YF, Huang HD, Chou CY. COL11A1 promotes tumor progression and predicts poor clinical outcome in ovarian cancer. Oncogene. 2014;33:3432–3440. doi: 10.1038/onc.2013.307. [DOI] [PubMed] [Google Scholar]
- [34].Beach JA, Aspuria PJ, Cheon DJ, Lawrenson K, Agadjanian H, Walsh CS, Karlan BY, Orsulic S. Sphingosine kinase 1 is required for TGF-beta mediated fibroblast-to-myofibroblast differentiation in ovarian cancer. Oncotarget. 2016;7:4167–4182. doi: 10.18632/oncotarget.6703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Shin G, Kang TW, Yang S, Baek SJ, Jeong YS, Kim SY. GENT: gene expression database of normal and tumor tissues. Cancer Inform. 2011;10:149–157. doi: 10.4137/CIN.S7226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Gentles AJ, Bratman SV, Lee LJ, Harris JP, Feng W, Nair RV, Shultz DB, Nair VS, Hoang CD, West RB, Plevritis SK, Alizadeh AA, Diehn M. Integrating Tumor and Stromal Gene Expression Signatures With Clinical Indices for Survival Stratification of Early-Stage Non-Small Cell Lung Cancer. Journal of the National Cancer Institute. 2015;107 doi: 10.1093/jnci/djv211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer discovery. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Tan TZ, Yang H, Ye JR, Low J, Choolani M, Tan DSP, Thiery JP, Huang RYJ. CSIOVDB: a microarray gene expression database of epithelial ovarian cancer subtype. Oncotarget. 2015;6:43843–43852. doi: 10.18632/oncotarget.5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Julien S, Merino-Trigo A, Lacroix L, Pocard M, Goere D, Mariani P, Landron S, Bigot L, Nemati F, Dartigues P, Weiswald LB, Lantuas D, Morgand L, Pham E, Gonin P, Dangles-Marie V, Job B, Dessen P, Bruno A, Pierre A, De The H, Soliman H, Nunes M, Lardier G, Calvet L, Demers B, Prevost G, Vrignaud P, Roman-Roman S, Duchamp O, Berthet C. Characterization of a large panel of patient-derived tumor xenografts representing the clinical heterogeneity of human colorectal cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2012;18:5314–5328. doi: 10.1158/1078-0432.CCR-12-0372. [DOI] [PubMed] [Google Scholar]
- [40].Yeung TL, Leung CS, Wong KK, Samimi G, Thompson MS, Liu J, Zaid TM, Ghosh S, Birrer MJ, Mok SC. TGF-beta modulates ovarian cancer invasion by upregulating CAF-derived versican in the tumor microenvironment. Cancer research. 2013;73:5016–5028. doi: 10.1158/0008-5472.CAN-13-0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Lawrenson K, Grun B, Lee N, Mhawech-Fauceglia P, Kan J, Swenson S, Lin YG, Pejovic T, Millstein J, Gayther SA. NPPB is a novel candidate biomarker expressed by cancer-associated fibroblasts in epithelial ovarian cancer. Int J Cancer. 2015;136:1390–1401. doi: 10.1002/ijc.29092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Tan TZ, Miow QH, Miki Y, Noda T, Mori S, Huang RY, Thiery JP. Epithelial-mesenchymal transition spectrum quantification and its efficacy in deciphering survival and drug responses of cancer patients. EMBO Mol Med. 2014;6:1279–1293. doi: 10.15252/emmm.201404208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Consortium G. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. Journal of Clinical Investigation. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Shay T, Kang J. Immunological Genome Project and systems immunology. Trends in immunology. 2013;34:602–609. doi: 10.1016/j.it.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Moffitt RA, Marayati R, Flate EL, Volmar KE, Loeza SG, Hoadley KA, Rashid NU, Williams LA, Eaton SC, Chung AH, Smyla JK, Anderson JM, Kim HJ, Bentrem DJ, Talamonti MS, Iacobuzio-Donahue CA, Hollingsworth MA, Yeh JJ. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat Genet. 2015;47:1168–1178. doi: 10.1038/ng.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Verhaak RG, Tamayo P, Yang JY, Hubbard D, Zhang H, Creighton CJ, Fereday S, Lawrence M, Carter SL, Mermel CH, Kostic AD, Etemadmoghadam D, Saksena G, Cibulskis K, Duraisamy S, Levanon K, Sougnez C, Tsherniak A, Gomez S, Onofrio R, Gabriel S, Chin L, Zhang N, Spellman PT, Zhang Y, Akbani R, Hoadley KA, Kahn A, Kobel M, Huntsman D, Soslow RA, Defazio A, Birrer MJ, Gray JW, Weinstein JN, Bowtell DD, Drapkin R, Mesirov JP, Getz G, Levine DA, Meyerson M, N. Cancer Genome Atlas Research Prognostically relevant gene signatures of high-grade serous ovarian carcinoma. J Clin Invest. 2013;123:517–525. doi: 10.1172/JCI65833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Cristescu R, Lee J, Nebozhyn M, Kim KM, Ting JC, Wong SS, Liu J, Yue YG, Wang J, Yu K, Ye XS, Do IG, Liu S, Gong L, Fu J, Jin JG, Choi MG, Sohn TS, Lee JH, Bae JM, Kim ST, Park SH, Sohn I, Jung SH, Tan P, Chen R, Hardwick J, Kang WK, Ayers M, Hongyue D, Reinhard C, Loboda A, Kim S, Aggarwal A. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat Med. 2015;21:449–456. doi: 10.1038/nm.3850. [DOI] [PubMed] [Google Scholar]
- [49].Guinney J, Dienstmann R, Wang X, de Reynies A, Schlicker A, Soneson C, Marisa L, Roepman P, Nyamundanda G, Angelino P, Bot BM, Morris JS, Simon IM, Gerster S, Fessler E, De Sousa EMF, Missiaglia E, Ramay H, Barras D, Homicsko K, Maru D, Manyam GC, Broom B, Boige V, Perez-Villamil B, Laderas T, Salazar R, Gray JW, Hanahan D, Tabernero J, Bernards R, Friend SH, Laurent-Puig P, Medema JP, Sadanandam A, Wessels L, Delorenzi M, Kopetz S, Vermeulen L, Tejpar S. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015;21:1350–1356. doi: 10.1038/nm.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Bento AP, Gaulton A, Hersey A, Bellis LJ, Chambers J, Davies M, Kruger FA, Light Y, Mak L, McGlinchey S, Nowotka M, Papadatos G, Santos R, Overington JP. The ChEMBL bioactivity database: an update. Nucleic acids research. 2014;42:D1083–1090. doi: 10.1093/nar/gkt1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Egeblad M, Rasch MG, Weaver VM. Dynamic interplay between the collagen scaffold and tumor evolution. Curr Opin Cell Biol. 2010;22:697–706. doi: 10.1016/j.ceb.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Malanchi I, Santamaria-Martinez A, Susanto E, Peng H, Lehr HA, Delaloye JF, Huelsken J. Interactions between cancer stem cells and their niche govern metastatic colonization. Nature. 2012;481:85–89. doi: 10.1038/nature10694. [DOI] [PubMed] [Google Scholar]
- [53].Massague J. TGFbeta signalling in context. Nature reviews. Molecular cell biology. 2012;13:616–630. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Connolly EC, Freimuth J, Akhurst RJ. Complexities of TGF-beta targeted cancer therapy. Int J Biol Sci. 2012;8:964–978. doi: 10.7150/ijbs.4564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Kubota S, Takigawa M. Cellular and molecular actions of CCN2/CTGF and its role under physiological and pathological conditions. Clin Sci (Lond) 2015;128:181–196. doi: 10.1042/CS20140264. [DOI] [PubMed] [Google Scholar]
- [56].Johannsen M, Spitaleri G, Curigliano G, Roigas J, Weikert S, Kempkensteffen C, Roemer A, Kloeters C, Rogalla P, Pecher G, Miller K, Berndt A, Kosmehl H, Trachsel E, Kaspar M, Lovato V, Gonzalez-Iglesias R, Giovannoni L, Menssen HD, Neri D, de Braud F. The tumour-targeting human L19-IL2 immunocytokine: preclinical safety studies, phase I clinical trial in patients with solid tumours and expansion into patients with advanced renal cell carcinoma. Eur J Cancer. 2010;46:2926–2935. doi: 10.1016/j.ejca.2010.07.033. [DOI] [PubMed] [Google Scholar]
- [57].Eigentler TK, Weide B, de Braud F, Spitaleri G, Romanini A, Pflugfelder A, Gonzalez-Iglesias R, Tasciotti A, Giovannoni L, Schwager K, Lovato V, Kaspar M, Trachsel E, Menssen HD, Neri D, Garbe C. A dose-escalation and signal-generating study of the immunocytokine L19-IL2 in combination with dacarbazine for the therapy of patients with metastatic melanoma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2011;17:7732–7742. doi: 10.1158/1078-0432.CCR-11-1203. [DOI] [PubMed] [Google Scholar]
- [58].Tran E, Chinnasamy D, Yu Z, Morgan RA, Lee CC, Restifo NP, Rosenberg SA. Immune targeting of fibroblast activation protein triggers recognition of multipotent bone marrow stromal cells and cachexia. The Journal of experimental medicine. 2013;210:1125–1135. doi: 10.1084/jem.20130110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Roberts EW, Deonarine A, Jones JO, Denton AE, Feig C, Lyons SK, Espeli M, Kraman M, McKenna B, Wells RJ, Zhao Q, Caballero OL, Larder R, Coll AP, O'Rahilly S, Brindle KM, Teichmann SA, Tuveson DA, Fearon DT. Depletion of stromal cells expressing fibroblast activation protein-alpha from skeletal muscle and bone marrow results in cachexia and anemia. The Journal of experimental medicine. 2013;210:1137–1151. doi: 10.1084/jem.20122344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Li Y, Lacerda DA, Warman ML, Beier DR, Yoshioka H, Ninomiya Y, Oxford JT, Morris NP, Andrikopoulos K, Ramirez F, et al. A fibrillar collagen gene, Col11a1, is essential for skeletal morphogenesis. Cell. 1995;80:423–430. doi: 10.1016/0092-8674(95)90492-1. [DOI] [PubMed] [Google Scholar]
- [61].Spranger J. The type XI collagenopathies. Pediatric radiology. 1998;28:745–750. doi: 10.1007/s002470050459. [DOI] [PubMed] [Google Scholar]
- [62].Weidle UH, Maisel D, Klostermann S, Weiss EH, Schmitt M. Differential splicing generates new transmembrane receptor and extracellular matrix-related targets for antibody-based therapy of cancer. Cancer Genomics Proteomics. 2011;8:211–226. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure Legend
Fig. S1. Colon cancer survival (A), clinical and molecular (B) parameters associated with COL11A1 expression. Kaplan-Meier survival plots and plots of COL11A1 expression in individual subsets of colon cancer were generated using the data and software described previously by Tan and colleagues (Tan et al., EMBO Mol Med, 2014).
Fig. S2. Expression of COL11A1 in normal tissues and transformed cell lines. The data were obtained from the Genotype-Tissue Expression (GTEx) portal (gtexportal.com; date of analysis: 05/28/16).
Fig. S3. COL11A1 expression in invasive ductal breast cancer and colon cancer and their corresponding normal tissues. The data were obtained using TCGA datasets in the Oncomine portal (oncomine.com; date of analysis: 12/14/15.
Fig. S4. Ovarian cancer COL11A1-anticorrelated genes are enriched in ovarian tumors of low malignant potential. GeneSet clustering analysis of low malignant potential serous ovarian tumors (n=32) and invasive serous ovarian tumors (n=58) using the top 100 COL11A1-anticorrelated genes (Spearman’s correlation, Table S3) from the ovarian cancer TCGA dataset. The clustering analysis was performed using R2 (http://hgserver1.amc.nl/cgi-bin/r2/main.cgi; date of analysis: 03/20/16).
Fig. S5. Levels of the pan-cancer COL11A1-correlated genes in the GSE52104 expression dataset in which mesenchymal stem cells (MSCs) or immortalized normal ovarian fibroblasts (INOFs) were either cultured alone or co-cultured with IOSE4 normal epithelial cells (IOSE) or HEYA8 epithelial ovarian cancer cells (EOC) using a Transwell membrane. Inverse-log2 data of different probes of the same gene were averaged, then log2-transformed. The average of log2 values for COL11A1 and the 194 COL11A1-correlated genes (one gene was not represented in the dataset) were extracted for statistical analyses using GraphPad Prism 6. Data were expressed as the mean ± SEM. Intergroup differences were assessed by the Student’s t-test. *p<0.05.
Fig. S6. Expression of CTGF in normal tissues and transformed cell lines. The data were obtained from the Genotype-Tissue Expression (GTEx) portal (gtexportal.com; date of analysis: 05/28/16).
Fig. S7. CTGF may not be an adequate therapeutic target in cancer because of the similar expression levels in cancer and normal tissues. (A) Comparison of CTGF mRNA expression in normal tissues and corresponding cancers. Box plots for two different platforms (U133Plus2 and U133A) were generated using datasets and software available through the Gene Expression across Normal and Tumor tissue (GENT) portal (medical-genome.kribb.re.kr/GENT). The y axis shows log2 mRNA levels. Average expression levels in normal tissues and cancer tissues are indicated by vertical dotted green and red lines, respectively. (B) CTGF mRNA expression in normal, inflammatory and fibrotic conditions in comparison to cancer. The graphs were generated using the public R2 MegaSampler software (hgserver1.amc.nl/cgi-bin/r2/main.cgi) for the processing and normalization of individual datasets imported from the Gene Expression Omnibus (u133p2, MAS5.0 platform). The number of samples in each GSE dataset are indicated in parentheses.
Fig. S8. Expression of FAP in normal tissues and transformed cell lines. The data were obtained from the Genotype-Tissue Expression (GTEx) portal (gtexportal.com; date of analysis: 05/28/16).
Fig. S9. Increased expression of FAP in cancer. (A) Comparison of FAP mRNA expression in normal tissues and corresponding cancers. Box plots for two different platforms (U133Plus2 and U133A) were generated using datasets and software available through the Gene Expression across Normal and Tumor tissue (GENT) portal (medical-genome.kribb.re.kr/GENT). The y axis shows log2 mRNA levels. Average expression levels in normal tissues and cancer tissues are indicated by vertical dotted green and red lines, respectively. (B) FAP mRNA expression in normal, inflammatory and fibrotic conditions in comparison to cancer. The graphs were generated using the public R2 MegaSampler software (hgserver1.amc.nl/cgi-bin/r2/main.cgi) for the processing and normalization of individual datasets imported from the Gene Expression Omnibus (u133p2, MAS5.0 platform). The number of samples in each GSE dataset are indicated in parentheses.
Supplementary Table Legend
Table S1. Cox multivariate analysis of COL11A1 (marker of activated CAFs) and VIM (general marker of fibroblasts) related to survival in ovarian cancer patients. Data were obtained from the ovarian microarray gene expression database CSIOVDB (csibio.nus.edu.sg/CSIOVDB/CSIOVDB.html).
Table S2. Pan-cancer COL11A1-correlated genes ranked by average correlation to COL11A1 in 13 TCGA cancer types and annotated by gene name, cellular localization and molecule type.
Table S3. COL11A1-anticorrelated genes (Spearman’s rank correlation) across 13 different TCGA carcinoma types, each represented by >100 primary tumors from therapy-naive patients. Pink denotes the top 10% COL11A1-anticorrelated genes in each individual carcinoma type. Data were analyzed using cBio Portal (cbioportal.org; date of analysis: 01/16/16).
Table S4. Gene Ontology (GO) enrichment analysis of biological processes associated with the pan-cancer COL11A1-correlated gene set. Data were analyzed using the Gene Ontology Consortium portal (geneontology.org/; date of analysis: 02/01/16).
Table S5. Molecular pathways associated with the pan-cancer COL11A1-correlated gene set. Overlap between the 195 pan-cancer COL11A1-correlated genes and common molecular pathways defined by 84 representative genes in the SABiosciences Pathway Arrays.
Table S6. Pan-cancer COL11A1-correlated gene set analysis of chemical and genetic perturbations (CGP) using the GSEA portal (software.broadinstitute.org/gsea/index.jsp; date of analysis: 02/08/16).
Table S7. Upstream regulators of pan-cancer COL11A1-correlated genes. Ingenuity Pathway Analysis (ingenuity.com; date of analysis: 02/08/16).
Table S8. Pan-cancer COL11A1-correlated genes that are targets of drugs in clinical trials. Ingenuity Pathway Analysis (ingenuity.com; date of analysis: 05/28/16).
Table S9. Pan-cancer COL11A1-correlated genes that are targets of bioactive compounds. ChEMBL database analysis (ebi.ac.uk/chembl; date of analysis: 05/28/16).