Abstract
There are inherent limitations with traditional methods to study protein behavior or to determine the constituency of proteins in discrete subcellular compartments. In response to these limitations, several methods have recently been developed that use proximity-dependent labeling. By fusing proteins to enzymes that generate reactive molecules, most commonly biotin, proximate proteins are covalently labeled to enable their isolation and identification. In this review, we describe current methods for proximity-dependent labeling in living cells, and discuss their applications and future use in the study of protein behavior.
Keywords: proximity-dependent labeling, APEX, BioID, protein-protein interactions, subcellular proteome, proteomics
Unmet needs: The rationale for proximity-dependent labeling
Protein-protein interactions are involved in all cellular processes. Mapping of these interaction networks to reveal the organization of the proteome into functional units is of prime importance to understand complex biological processes. While a large number of methods have been developed to screen for protein-protein interactions (PPIs), the utility of conventional methods, such as affinity complex purification or yeast-2-hybrid (Y2H) is limited to the isolation of high affinity PPIs in vitro or under nonphysiological conditions. Similarly, there are inherent biochemical limitations in the isolation and proteome mapping of organelles and loosely associated factors, which is crucial for discerning the protein ‘interactome’.
The recent development of several proximity-dependent labeling methods in living cells partially addresses these deficiencies. PPIs or subcellular proteomes can be screened under more physiologically relevant conditions and be applied to insoluble proteins. These methods can also detect low affinity and/or transient PPIs as well as protein constituents of subcellular domains. Proximity-dependent labeling utilizes enzymes that produce reactive molecules that covalently interact with neighboring proteins. These labeled proteins can be isolated via conventional affinity purification and identified, most often by immunoblot analysis or protein mass spectrometry (MS). Fusion of the enzyme to a specific protein enables labeling of proximate proteins as a means to identify candidate PPIs, whereas fusion to a minimal targeting motif that restricts the enzyme to a specific subcellular compartment or structure can be used to map the protein population therein (Figure 1, Key Figure).
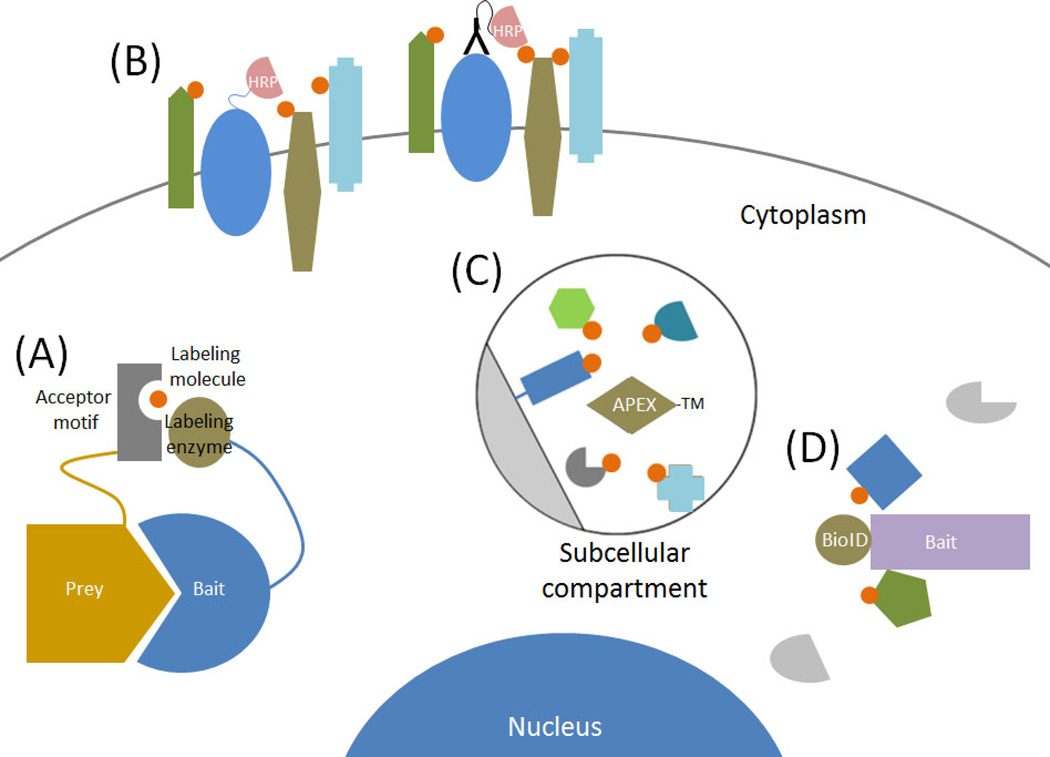
Figure 1. Key Figure: Proximity-dependent protein biotinylation to study protein behavior and subcellular proteomics in live cells.
There are a multiple methods that use proximity-dependent protein labeling to study protein behavior and constituency in living cells. (A) Binary candidate methods utilize fusion of a labeling enzyme or an acceptor motif to bait and prey proteins to assess protein interactions. (B) HRP-based methods are used at the cell surface to label proteins associated with a bait protein. (C) With APEX-methods a peroxidase is fused to a targeting motif (TM) or protein and used to label proximate to map the proteome of discrete subcellular compartments. (D) Using a promiscuous biotin ligase fused to a bait protein BioID-methods generate a history of protein association over time to screen for candidate protein-protein interactions or the constituency of subcellular structures.
Depending on the enzyme utilized, these proximity-dependent labeling approaches can generate a history of protein associations over time in living cells or capture a snapshot of associated proteins. This review focuses on proximity-labeling based methods that have proven applicable to the study of proteins in living cells for the identification of either candidate PPIs or the constituency of subcellular domains and compartments.
Biotin ligase-based methods for proximity labeling
Fundamental to cellular life, biotin ligases are enzymes that naturally biotinylate proteins, predominantly modifying a subset of carboxylases. All cells appear to express a biotin ligase with a moderate degree of conservation within the active site of the enzyme. The most comprehensively studied biotin ligase is BirA, a bifunctional protein expressed in E. coli that mediates biotinylation on a specific lysine residue of a subunit of the acetyl-CoA carboxylase and also transcriptionally regulates the biotin synthetic operon [1]. Biotinylation takes places when protein is incubated with BirA in the presence of biotin and ATP, which allows for the generation of reactive biotinoyl-5’-AMP (bioAMP). BirA retains bioAMP at the active site and facilitates transfer to a specific lysine on a target protein [2].
Proximity-dependent labeling techniques have exploited biotinylation for the rapid and specific isolation of tagged proteins with avidin or streptavidin, which have extremely high affinity to biotin. Because biotinylation is a covalent protein modification, biotinylated proteins can also withstand stringent lysis and wash conditions, which maximize the purity of the isolated proteins.
Proximity-dependent labeling of candidate binary interactions
BirA has high sequence specificity that can be exploited to generate biotinylated proteins of interest. To test for candidate PPIs in living cells, BirA that is fused to a bait protein is coexpressed in the same vector with the protein of interest (prey protein) that is fused to a short biotin acceptor peptide (BAP). The high sequence specificity of BirA allows the BAP to be uniquely labeled. Thus, if the BAP and BirA are brought together by interaction or proximity of the bait and prey, the prey will be biotinylated, enabling detection of the PPI by streptavidin staining (Figure 2).
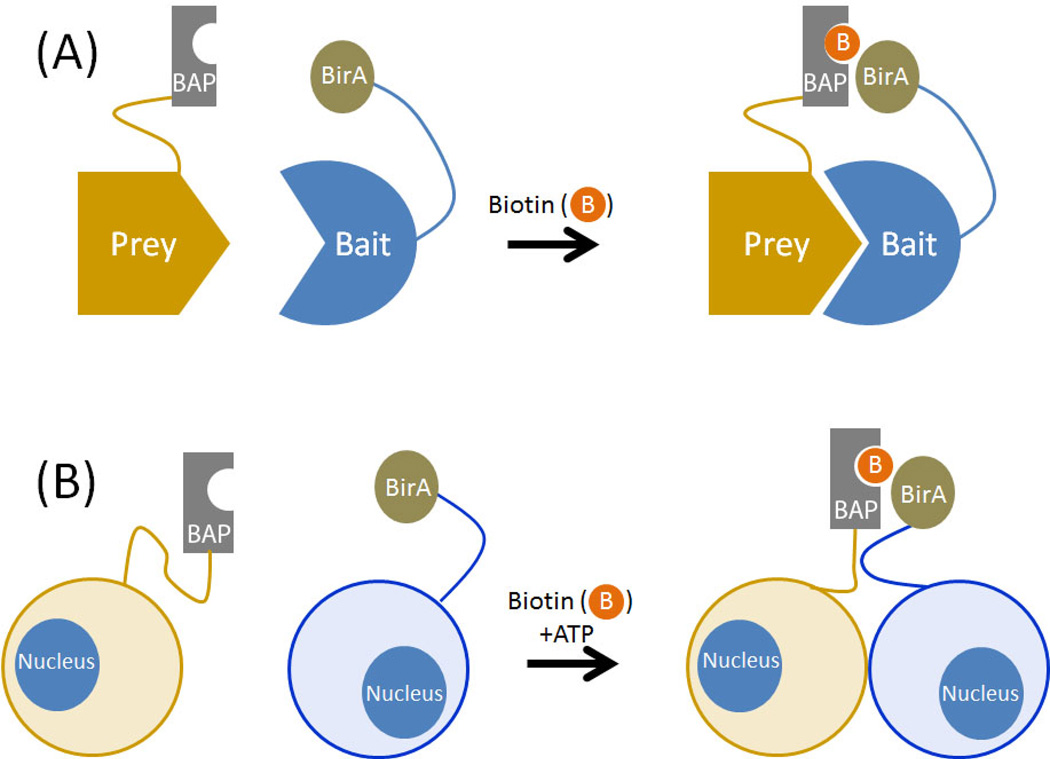
Figure 2. Proximity-dependent protein labeling can be used to detect candidate binary interactions.
The biotin ligase BirA is fused to a bait protein and a biotin acceptor peptide (BAP) is fused to a candidate prey protein. Upon cellular expression of both proteins, biotinylation (denoted by orange circles with B) of the BAP-fused prey protein indicates their proximity and suggests that an interaction between bait and prey has occurred. This approach can be used (A) within a cell or (B) between two different cells.
Despite the utility of this method for detecting PPIs in vitro and in cells, the high sequence affinity of BAP to BirA can artificially generate interactions. Therefore, a modified BAP with reduced affinity to BirA was developed and validated with known PPIs FKBP and FRB in vitro and in cells as well as CDC25C and 14-3-3 epsilon [3]. This approach was further modified by altering the amino acid sequence of the BAP to facilitate its detection by protein MS [4]. The advantages of PPI detection by these modified proximity-dependent biotinylation methods include low background and high sensitivity and spatial resolution in cells.
A variation on proximity biotinylation with BirA/BAP named BLINC (biotin labeling of intercellular contacts) monitors PPIs at the cell surface using microscopy. Cells that express a BirA-fused cell surface protein are co-cultured with cells that express a BAP-conjugated cell surface protein, in the presence of exogenous ATP and biotin (Figure 2).
BLINC was used to visualize the interaction between neuronal surface receptors neurogligin and neurexin in live cells using fluorescence-conjugated streptavidin [5]. In yeast and human cells, BirA/BAP was utilized to study protein synthesis at specific cellular sites by fusing BAP to ribosomes and targeting BirA to the cytoplasm, ER or mitochondrial membranes [6]. The BirA/BAP has also been applied to indirectly screen for DNA-protein interactions by fusing BAP to histones and BirA to a nuclear protein. Named PUB-NChIP, this method uses native chromatin-immunopurification techniques, but with biotin affinity purification in lieu of antibody-based purification [7].
A caveat of these binary-candidate methods is the potential for false-negatives. Given that proximity-dependent labeling is based on the direct interaction of the BAP with BirA, the point of contact or surface accessibility of the protein interaction needs to be carefully considered for successful screening. Even if the labeling enzyme and its acceptor peptide are placed on the point of contact, accessibility between the enzyme and the acceptor peptide might be affected by protein structure, orientation, and size. It is also possible that the need for a direct BirA/BAP interaction could artificially drive associations between proximate but not interacting proteins, creating the possibility of false positives. However, considering the methodological simplicity compared to protein-fragment complementation (e.g. split-GFP) [8, 9] these candidate based proximity-labeling methods are powerful tools to identify PPIs between known proteins.
Proximity-dependent biotin identification
A proximity-dependent biotinylation method called BioID (proximity-dependent dependent biotin identification) does not require a binary-candidate approach but instead labels any proximate proteins [10, 11]. BioID is based on promiscuous biotinylation generated by a mutated BirA [2]. The enzyme ligase contains a mutation within the conserved biotin- and bioAMP-binding domain (hereafter this enzyme is called BioID). The mutation substantially reduces its affinity to bioAMP [2], presumably releasing reactive biotin to covalently interact with primary amines (e.g. lysine side chains) on proximate proteins within the estimated range of approximately 10 nm [12]. Therefore, it is important to note that a lack of amines at the surface of a protein proximate to the bait may lead to false-negatives. Cellular expression of a BioID-fusion protein with a 15–18 hour optimal labeling period, which is induced by biotin supplementation, leads to biotinylation of endogenous proteins that are proximate to the bait (Figure 3) [10, 11]. In this way, the BioID method generates a “history” of candidate PPIs under relatively physiological conditions in living cells.
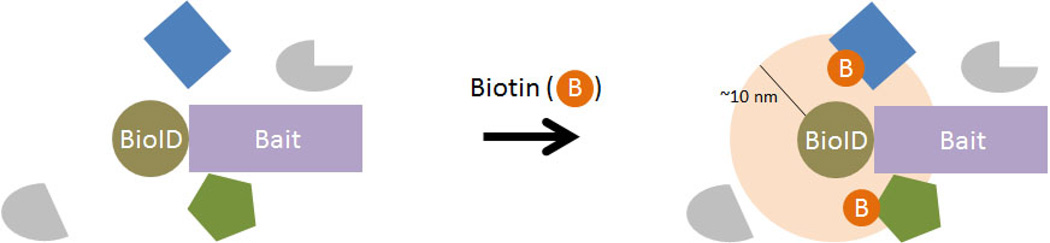
Figure 3. Using BioID to generate a history of PPIs or to identify constituents of subcellular domains.
Cellular expression of a protein of interest (bait) fused to a promiscuous biotin ligase called BioID enables biotinylation (denoted by orange circles with B) of proximate proteins within approximately 10nm (light orange disc). This protein labeling is initiated by the addition of a supraphysiological concentration of biotin and requires many hours to generate substantial labeling. Following cell lysis and protein denaturation, biotinylated proteins are captured by affinity purification for identification by mass spectrometry to reveal a history of protein associations that occurred during the labeling period.
BioID has been used both in vitro and in vivo, to reveal associations for proteins that exhibit a wide variety of subcellular localizations and dynamics (Table 1). Indeed, BioID was originally applied to the study of PPIs of insoluble nuclear lamins [10]. The method revealed abundant interactions of lamin A with integral proteins of the inner nuclear membrane and nuclear pore complex. Additional, but less abundant candidates were also identified that had functions in transcription, chromatin regulation, RNA processing, and DNA repair, suggesting potentially unknown functional roles for lamin A in these processes. More importantly, BioID identified a novel protein interacting partner with lamin A, SLAP75, which was found to associate with the nuclear envelope. BioID has been similarly used to identify uncharacterized PPIs and constituents within largely insoluble cellular structures such as the mammalian cell-cell junctions [13–18] and trypanosome bilobe and flagella [19–22], structures that are often refractory to traditional study with methods such as affinity complex purification. As the labeling radius of BioID is limited, it can be used to map the population and spatial distribution of proteins within large structures by application to proteins throughout the structure, as was demonstrated at the nuclear pore complex [12]. If a larger labeling radius is desired, for example to capture more constituents within a large protein complex, it can be increased by insertion of flexible linkers between the bait and the ligase [23]. BioID also captures protein associations over a period of time and independent of stable interactions, making it an effective approach in identifying novel substrates of proteins with dynamic and transient protein associations such as the E3 ubiquitin ligase, β-TrCP1 [24].
Table 1.
Applications of BioID to proximity-dependent protein labeling
| Applications | Organisms | Subcellular locations or cellular mechanisms |
Fusion proteins | References |
|---|---|---|---|---|
| Structural proteins |
Mammalian cells |
Nuclear envelope | Lamin A | [10] |
| Lamin A (WT and progerin) | [49] | |||
| Lamin B1 | [50] | |||
| Nuclear pore complex |
Y-complex, Nup53 | [12] | ||
| Centrosome | CEP120 | [51] | ||
| 8 members of the centrosome (PLK4, CEP192, CEP63, CEP152, CPAP, CCDC67, CCDC14, and KIAA0753) |
[52] | |||
| 58 members of the centrosome | [26] | |||
| Cell junction | ZO-1 | [17] | ||
| E-Cadherin | [14, 18] | |||
| Occludin, Claudin-4 | [13] | |||
| MarvelD3 | [15] | |||
| α-catenin | [16] | |||
| Quantitative analysis (SILAC) of focal adhesion |
Paxillin | [53] | ||
|
Trypanosoma brucei |
Cytoskeleton (bilobe) complex |
TbMORN1 | [21] | |
| Flagellum attachment zone |
TbSAS-4 | [19] | ||
| Bilobe, flagellum attachment zone |
TbPLK | [20] | ||
| Flagellum attachment zone |
CIF1 | [22] | ||
|
Toxoplasma gondii |
Inner membrane complex |
ISP3, AC2 | [54] | |
|
Dictyostelium discoidem |
Nuclear envelope | Lamin NE81 | [55] | |
| Dynamic proteins |
Mammalian cells |
Mitochondrial roles in cancer cell development |
CHCHD2 | [56] |
| Mitochondrial roles in epidermal differentiation |
MPZL3 | [57] | ||
| Mitochondrial proteolysis |
ClpP | [58] | ||
| Autophagy | TBC1D14 | [59] | ||
| Hippo signaling | 19 members of the Hippo pathway (AMOT, ANKRD28, LATS2, MOB1A, MOB1N, PPP6R1, PPP6R2, PPP6R3, RASSF2, SAV1, MST2, MST1, YAP1, CTTNBP2NL, LATS1, SLMAP, STRN, STRN3, TJP2) |
[60] | ||
| MAP signaling | MEKK3 | [61] | ||
| Lipid converting metabolism |
PPP2R5C | [62] | ||
| Keratinocyte differentiation |
CALML5, SFN | [63] | ||
| Oncogenic transcription factors |
EWS-Fli-1 | [64] | ||
| Xenografted oncogenic transcriptional factors |
C-Myc | [25] | ||
| Chromatin binding proteins |
Med4, Med20, Med23, Histone H2B, Histone H3 |
[65] | ||
| DNA modifying enzymes in mESC |
Tet1 | [66] | ||
| Mitosis | ULK3 | [67] | ||
| Mitosis | USP37 | [68] | ||
| Endocytosis | PLLP | [69] | ||
| Structural maintenance of the myelin sheath |
SH3TC2 | [70] | ||
| mRNA turnover | UPF1, UFP2, SMG5 | [71] | ||
| mRNA turnover | 4E–T | [72] | ||
| ER-associated degradation |
HMG-CoA | [73] | ||
| Ubiquitin-mediated degradation |
β-TrCP1/2 | [24] | ||
| Ubiquitin-mediated degradation |
USP12 | [74] | ||
| Protein interaction in endosomes |
RhoB | [75] | ||
| Mitochondria-ER interaction |
SLC25A46 | [76] | ||
| Protein interactions in redox and glucose- dependent conditions |
Txnip | [77] | ||
| Cellular response to Chlamydia trachomatis |
Syntaxin 6 (WT and ΔYGRL) | [78] | ||
| Cellular response to Chlamydia psittaci |
SINC | [79] | ||
| Cellular response to HIV protein |
Gag | [80] | ||
| Cellular response to HIV protein |
Gag (WT and ΔMA Gag) | [81] | ||
| Cellular response to HIV protein |
Vpu | [82] | ||
| Cellular response to Epstein-Barr virus |
LMP1 (WT and mutants: LMP1-A5, LMP1-GG, and LMP1-A5-GG) |
[83] | ||
|
Toxoplasma gondii |
Kinase dynamics | TgCDPK3 | [84] | |
|
Plasmodium berghei |
Secretory vesicular proteome of malaria |
MDV1/PEG3 | [27] |
Methods such as BioID can also be used in complex model systems, which can provide more insight into protein behaviors as they occur in their native biological settings in vivo. For example, the BioID method has been applied to xenografted tumors in mice [25]. Tumor cells expressing the oncoprotein Myc-BioID were injected subcutaneously into NOD/SCID mice and later treated with biotin. Biotinylated proteins were isolated and candidate Myc interactors were identified by mass spectrometry [25]. This study sets the stage for more advanced applications of BioID in mice genetically modified to express BioID fusion proteins in specific cell types, thus overcoming many of the limitations of cell culture. However, it is important to note that BioID methods exhibit substantially reduced efficacy below 37°C [23] and may not be optimal for use in some model organisms.
In addition to screening for PPIs, the BioID method can be used to map the proteome of discrete subcellular domains including those that are not membrane bound and/or are refractory to isolation. An in depth BioID analysis of 58 components of the centrosome-cilium interface and centriolar satellites generated over 7,000 candidate PPIs [26]. Using this data, a map was generated for higher-order organization of the centrosome-cilium interface [26]. BioID was also used to help create a proteome of the membrane bound secretory vesicles from gametocytes of Plasmodium berghei that were harvested from the blood of infected mice, highlighting the application of BioID both for compartmental proteomics and for complex in vivo systems [27].
Like any approach, BioID has limitations. One limitation, not unique to BioID, is that the fusion of a 35 kDa biotin ligase to a protein of interest may impair its localization and/or function. To improve targeting of BioID fusion proteins, a smaller ligase from Aquifex aeolicus was made promiscuous by mutation of the reactive site [23]. Named BioID2, this variant (233 a.a) is substantially smaller than BioID (321 a.a.), thus enhancing selective targeting of BioID2 fusion proteins [23]. BioID2 requires less biotin than other methods, which is potentially useful in model systems where sufficient biotin supplementation may be difficult [23].
While some changes in the sequence of biotin ligases can improve tagging, biotin based methods have inherent limitations. Biotin is actively imported into the cytoplasm of mammalian cells [28] and free to diffuse to the nucleus, but it may not be as accessible in the secretory pathway, thus reducing labeling efficacy in that compartment. In addition, the long labeling time required for BioID methods prevents isolated analysis of events that occur over shorter time periods. Since the protein labeling is based on the release of reactive biotin from the enzyme, proteins identified by BioID may not be direct binding partners, but merely proximate proteins. Although biotin itself is not toxic, the biotinylation of proteins over the long BioID labeling periods may impact protein function. The use of a BioID-only control is required to confidently exclude those candidates identified by association with the BioID ligase itself and not the bait. Of particular importance is the realization that detection of candidate protein associations does not inform on their biological relevance, with low abundance candidates potentially being more relevant than abundant ones.
Peroxidase-based methods for proximity labeling
A more rapid labeling of proximate proteins can be obtained by using the enzymatic activity of peroxidases instead of biotin ligases. Peroxidases can generate short-lived free radicals from molecules such as phenolic aryl azide derivatives [29] or tyramide derivatives and H2O2 [30]. These free radicals can be covalently attached to electron rich side chains of amino acids (e.g. aromatic ring side chain of tryptophan or tyrosine) [30, 31]. In this way, peroxidases mediate biotin or fluorescein labeling of proteins using biotin-tyramide or biotin-phenolic aryl azide derivatives. Peroxidase-dependent labeling harnesses short-lived reactive labels that have a limited range, estimated to fall within <20–300 nm, depending on the method [32–34]. This approach has been used to label proteins for the identification of candidate PPIs and to map the protein constituency of subcellular compartments (Table 2).
Table 2.
Application of peroxidase-based approaches to proximity-dependent protein labeling
| Applications | Methods | Targeting locations |
Conjugated molecules | References |
|---|---|---|---|---|
| Horse radish peroxidase |
EMARS | Lipid raft | Glycosylphosphatidylinositol (GPI) targeting motifs |
[36] |
| anti-β1 integrin monoclonal antibody, Cholera toxin subunit B (CTxB) |
[35] | |||
| anti-ganglioside GD3 monoclonal antibody |
[85] | |||
| anti-β1 integrin monoclonal antibody, anti-EGFR monoclonal antibody, anti-IGF1R monoclonal antibody, anti-EphA2 monoclonal antibody, CTxB anti-CD3ε antibody |
[32] | |||
| Cell surface molecules associated with Thy1 |
anti- Thy1 monoclonal antibody | [86] | ||
| Cell surface molecules associated with β1 integrin |
anti-β1 integrin monoclonal antibody |
[87] | ||
| Cell surface molecules associated with CD20 |
Rituximab, anti-CD20 monoclonal antibody |
[88] | ||
| Proteins associated with PrPC |
Anti-PrPC antibody | [89] | ||
| SPLAAT | BRC cluster at the membrane |
anti-chicken IgM (target receptor BCR is a IgM) |
[37] | |
| Engineered- ascorbate peroxidase |
APEX | Proteomics of mammalian cell mitochondria matrix |
Mitochondria matrix targeting motif | [34] |
| Proteomics in D. melanogaster mitochondria |
Mitochondria matrix targeting motif | [44] | ||
| Proteomics of mammalian mitochondrial intermembrane space |
Mitochondria intermembrane targeting sequence |
[46] | ||
| Proteomics of mammalian cilia |
N-terminal cilia targeting sequence of NPHP3 |
[42] | ||
| APEX2 | Ultrastructural localization of ER and mitochondria proteins |
NES (6–17 residues of HIV1 Rev protein), MICU1, mitochondria outer membrane targeting motif (C-terminal 31 residues of MAVS), ER targeting motif (N-terminal 27 residues of rabbit p450) |
[41] | |
| Ultrastructural localization of ER and mitochondria proteins |
ER targeting motif, Tom20, H2B, HMOX1, TRMT61B, MGST3, Sco1, Sec61B, AURKAIP1, BIT1/PTRH2, COX14, MRPL12, MRM1, MRPS15, PLGRKT, QIL1/MIC13, SFXN1, SVH/ARMC10, TRMT2B, Pdk1 |
[90] | ||
| Proteomics of mammalian mitochondrial matrix and intermembrane space |
Mitochondria matrix and intermembrane targeting motifs |
[45] | ||
| Yeast proteomics | Golgi | [91] | ||
| Mammalian ER-PM junctions | STIM1 | [43] |
Horseradish Peroxidase-Dependent Labeling approaches
Enzyme mediated activation of radical source (EMARS) utilizes horseradish peroxidase (HRP) conjugated antibodies or expression of HRP-fusion proteins to rapidly label proximate proteins (Figure 4). In the presence of biotin- or fluorescein-labeled aryl azide and H2O2, the HRP generates an active radical species that labels proteins with biotin or fluorescein within a 200–300 nm radius from the enzyme [32]. Fluorescein-aryl azide is used to overcome cytoplasmic background generated by membrane permeable biotin-aryl azide [35]. Although most applications of EMARS use an HRP-conjugated antibody, cellular expression of HRP-fusion proteins has been used to study associations between glycosylphosphatidylinositol-anchored proteins and receptor tyrosine kinases (RTKs) in lipid rafts on the cell surface of living cells [36]. Using an RTK-antibody array, RTKs proximate to decay accelerating factor and Thy-1 were detected following EMARS reactions using fluorescein-aryl azide. However, this approach appears to be limited by the need to identify proteins with specific antibodies [35].
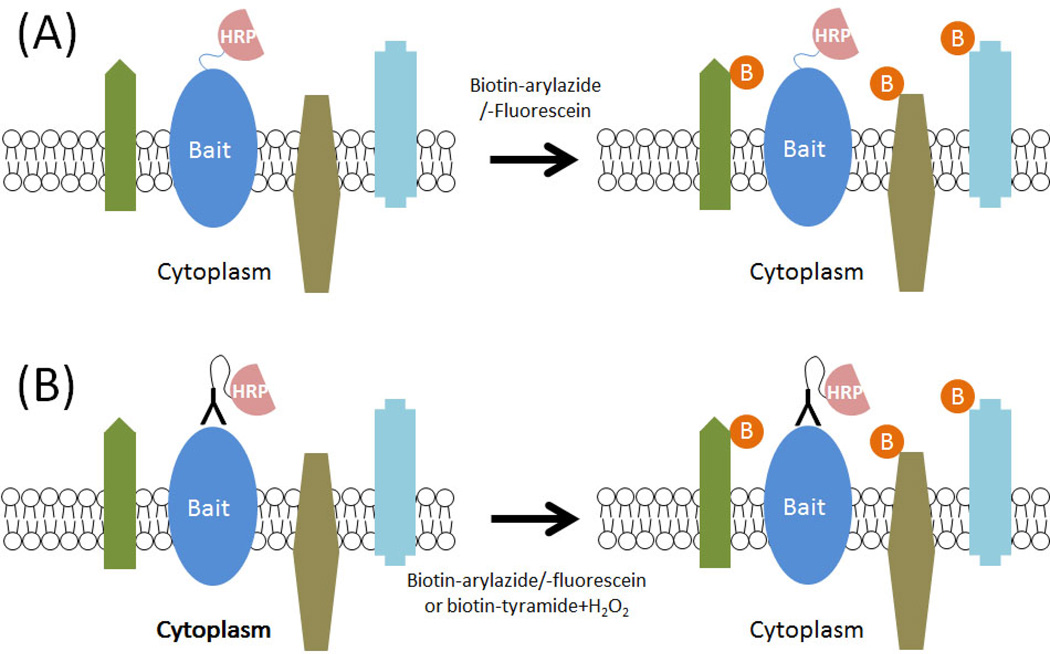
Figure 4. Proximity dependent labeling on the cell surface with HRP.
Horseradish peroxidase is not active with cells but when it is either (A) expressed on the cell surface as a fusion protein or (B) targeted to the cell surface by fusion with an antibody it can be used to label proximate proteins. By providing reactive molecules, such as arylazides or tyramines and H2O2, conjugated to biotin or fluorescein, the HRP generates a reactive molecule that covalently labels with proximate proteins within 300 nm. The biotin or fluorescein label enables protein identification. Methods that use this approach, including EMARS and SPPLAT, can be used to identify proximate proteins on the cell surface.
Another HRP-based approach called selective proteomic proximity labeling assay using tyramide (SPPLAT) [37, 38] utilizes HRP conjugated antibodies or ligands with biotin tyramide compounds and H2O2 to biotinylate proximate proteins on the cell surface. The biotin-tyramide derivatives have a cleavable linker that facilitates access of streptavidin to biotinylated proteins, recovery of biotinylated proteins from streptavidin, and reduced membrane permeability [37, 38]. Used in combination with stable isotope labeling for quantitative proteomics in a lymphocyte cell line, SPPLAT was applied to study the composition of B-cell receptor (BCR) cluster with an HRP-conjugated anti-IgM antibody. Six novel proteins and six known constituents of the BRC were identified with SPPLAT, including three proteins on the cytosolic face of the membrane, thereby demonstrating membrane diffusion of the biotin-tyramide.
Since HRP is not functional inside most cellular compartments, these methods are best suited for use on the cell surface. One limitation observed with HRP-based methods has been membrane permeability of some labeling reagents that may potentially lead to false positives [29, 37]. It is difficult to distinguish a clear advantage between EMARS or SPPLAT, except for the difference in labeling reagents. In the future, these HRP-based proximity-dependent protein-labeling methods should be used to address questions at the cell surface, such as the characterization of orphan receptors and/or ligands.
Engineered ascorbate peroxidase approaches
Proteomic mapping of subcellular compartments and structures relies on the quality of the purified sample. However, many such compartments and structures of interest cannot be purified, such as the mitochondrial intermembrane space, limiting their proteomic analysis. A recent approach known as APEX (engineered ascorbate peroxidase), which is based on mutations of a plant ascorbate peroxidase and initially applied to electron microscopy (EM) studies [39], was further developed for proximity-dependent protein biotinylation [34]. Unlike HRP [40], APEX is active within subcellular compartments of live cells, and maintains protein complex and membrane integrity; therefore, bypassing the requirement for organelle and protein complex purification. A monomeric APEX with enhanced catalytic activity oxidizes biotin-phenol in the presence of H2O2, generating short-lived radicals that covalently react with tyrosine, and potentially other electron-rich amino acids tryptophan, histidine and cysteine (Figure 5) [34, 39]. Following incubation with biotin-phenol, cells expressing APEX fusion proteins are exposed to H2O2 for approximately a minute before quenching the reaction [2]. An improved variant of APEX, called APEX2, exhibits enhanced catalytic efficiency enabling lower cellular expression [41].
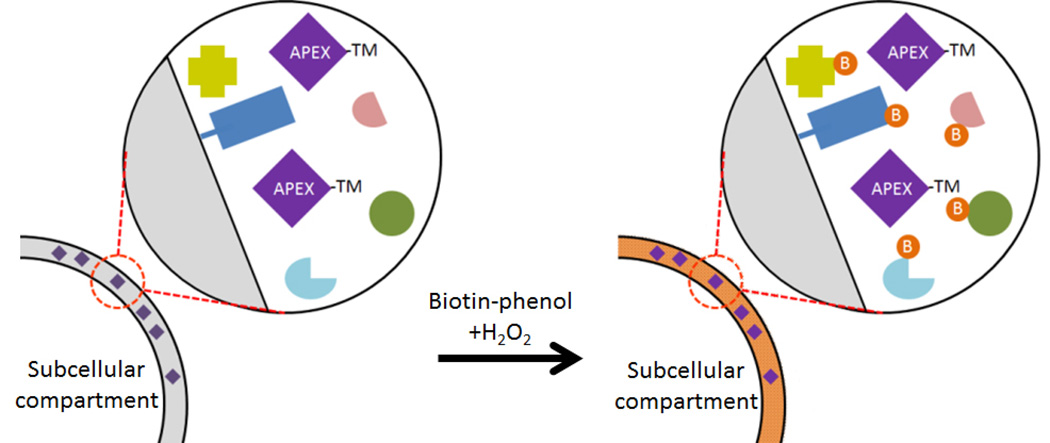
Figure 5. Mapping the proteome of subcellular compartments with APEX.
An enzyme that promiscuously labels proteins, such as the mutant peroxidase used for APEX, is specifically targeted to a distinct subcellular compartment by fusion of a strong targeting motif (TM). The motif could be a minimal sequence needed for targeting or a full-length protein that is mobile within the compartment. Following incubation with biotin-phenol and a pulse of H2O2, the APEX enzyme rapidly produces a reactive biotin that covalently biotinylates (denoted by orange circles with B) proteins constituents within the compartment enabling their selective isolation and identification by mass spectrometry.
With their robust enzyme kinetics and ability to function intracellularly, APEX-based methods are uniquely capable of generating a snapshot of proximate proteins with a rapid labeling time (~1 min) and in a variety of subcellular compartments. This is in stark contrast to BioID that requires 15–18 hrs for maximum protein labeling. The rapid kinetics of APEX also enable substantial labeling of proteins sufficient to identity the vast majority of constituents within discrete subcellular compartments and proximate proteins. Indeed, APEX fused to a mitochondrial matrix-targeting motif was used to map the proteome of the mitochondrial matrix in mammalian cells, identifying almost 500 proteins including over 30 novel candidate constituents [34]. APEX was also used to map the proteome of primary cilia, a non-membrane enclosed compartment, identifying not just known stable ciliary constituents but several more mobile signaling components [42]. In addition, APEX could detect the defective accumulation of factors in dysfunctional cilia [42]. Mapping of the proteome of ER-plasma membrane junctions in living cells with APEX coupled to mass spectrometry identified an ER-resident multi-transmembrane protein, STIMATE (STIM-activating enhancer, encoded by TMEM110) as an ER-resident Ca2+ sensor [43].
Similar to BioID, APEX can be performed in in vivo model systems as recently observed in Drosophila melanogaster [44]. APEX labeling functioned in multiple tissues and in various subcellular compartments, and provided an inventory of Drosophila mitochondrial proteins that could be annotated by subcompartments. In addition, APEX methods have been applied to ultrastructural imaging since the H2O2-generated free radicals induce polymerization of 3,3′-diaminobenzidine that generates a precipitate visible by electron microscopy after OsO4 treatment (Figure 5) [34, 45].
Studies with APEX often utilize quantitative SILAC-based radiometric methods for proteomic analysis. This was perhaps most useful when APEX methods were applied to map the proteome of the mitochondrial intermembrane space (IMS) but were also found to label cytoplasmic components, presumably via diffusion of the reactive biotin through pores in the outer mitochondrial membrane [45, 46]. A cytoplasmic-restricted APEX was used in combination with quantitative SILAC-based radiometric proteomic to exclude proteins labeled in the cytoplasm and enable the identification of specific IMS proteins including novel candidate constituents. Although this radiometric approach can improve the specificity of identified candidates it removes proteins that reside both within and outside the region of interest.
It is clear that APEX methods excel at compartmental proteomics, providing a substantial improvement over conventional methods that require isolation of purified organelles, most of which do not provide the spatial resolution of APEX. And similar to BioID, APEX is also applicable to the study of PPIs. Unlike BioID, APEX does not provide a history of protein associations over time, which may prevent detection of some transient protein associations, such as those that occur during various stages of the cell cycle. Instead, its rapid kinetics allows APEX to label proteins during a very specific event for comparative experiments, such as specific times before or after a pharmacological treatment. However, even with a short labeling period, there may be concerns over the cellular impact of biotin-phenol due to hydrophobic effects and/or oxidative damage from H2O2 on some signal transduction pathways, organelle dynamics, and protein structure and interactions. It is important to note that because peroxidase-based labeling is often specific to low abundance amino acids such as amino acids tyrosine [47, 48], it is possible that labeling will not occur if surface exposed tyrosine is unavailable.
Concluding remarks
Proximity-dependent labeling in living cells has emerged as a powerful approach to study protein behavior and the constituency of subcellular compartments and protein structures. While not likely to completely replace conventional approaches such as Y2H or complex purification, proximity-dependent biotinylation provides a complementary approach that brings unique potential, such as the ability to generate a history of protein-protein associations in live cells or to efficiently determine the proteome of discrete subcellular compartments. Despite these fundamental capabilities, methods for proximity-dependent labeling in living cells have a variety of limitations, some of which are likely to be overcome through additional method development (see Outstanding Questions).
Predominantly based on cellular expression of a promiscuous labeling enzyme fused to a protein of interest or subcellular compartment targeting motif, current proximity-dependent methods are most easily utilized in tractable cell culture models. However, PPIs and compartmental proteomics will have far reaching implications if physiologically relevant conditions, such as those found in vivo, are used. In support of this concept, APEX has been applied to subcellular proteomes in D. melanogaster [44] and BioID has been applied to mouse xenografted tumors [25]; however, it remains to be seen if BioID is effective in a wide variety of common model organisms.
By using APEX methods [34] and the BioID method (Kim and Roux, unpublished data) a limited number of biotinylated peptides can be detected by MS. If consistency and efficiency in detection of these biotinylated peptides can be improved, it is possible that these techniques may inform on not just the identities of the proteins but also the protein domains involved in PPIs. We have already seen the creation of second-generation tools, namely APEX2 and BioID2, each with their own unique enhancement of the original enzyme. It is predictable that more modifications may be expected, for example enhancing BioID kinetics to reduce the labeling time, harnessing biotin ligases from organisms that have evolved to function at lower temperatures, or enabling a slower labeling period for APEX to capture a longer history of protein associations. The future seems bright for expansion of proximity-dependent protein labeling techniques, whether it be novel applications, enhancements to existing approaches to provide a ‘toolbox’ of variants each optimized for a specific task, or entirely new approaches based on different technologies.
Outstanding Box.
Can proximity-dependent labeling methods produce the resolution necessary to determine sites of protein-protein interaction?
Can BioID approaches to identify PPIs or protein constituents of subcellular structures be applied to a wider variety of physiologically relevant model systems such as Caenorhabditis elegans and Arabidopsis thaliana that grow at temperatures below 37°C?
Will proximity-dependent methods such as APEX or BioID be effective for the study of complex higher organisms such as genetically modified mice?
Can BioID methods be applied in acidified compartments, such as lysosomes, or in the extracellular space that contains abundant ATP hydrolases?
Trends Box.
Proximity-dependent protein labeling in living cells is based on enzymes that generate a reactive protein label that has useful properties, most commonly biotin to enable their isolation and identification by MS.
Peroxidase-based APEX methods are predominantly being used to map the protein constituency of distinct subcellular compartments that are unable to be purified for conventional analysis.
BioID harnesses promiscuous biotin ligases to label proximate proteins when fused to a bait protein. This approach is most commonly used to identify candidate PPIs and constituents of subcellular structures refractory to isolation.
Acknowledgments
This manuscript was supported by grants RO1GM102203, RO1GM102486, and RO1EB014869 (to K.J.R.) from the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chakravartty V, Cronan JE. Altered regulation of Escherichia coli biotin biosynthesis in BirA superrepressor mutant strains. J Bacteriol. 2012;194(5):1113–1126. doi: 10.1128/JB.06549-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Choi-Rhee E, et al. Promiscuous protein biotinylation by Escherichia coli biotin protein ligase. Protein Sci. 2004;13(11):3043–3050. doi: 10.1110/ps.04911804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fernandez-Suarez M, et al. Protein-protein interaction detection in vitro and in cells by proximity biotinylation. J Am Chem Soc. 2008;130(29):9251–9253. doi: 10.1021/ja801445p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kulyyassov A, et al. PUB-MS: a mass spectrometry-based method to monitor protein-protein proximity in vivo. J Proteome Res. 2011;10(10):4416–4427. doi: 10.1021/pr200189p. [DOI] [PubMed] [Google Scholar]
- 5.Liu DS, et al. Imaging trans-cellular neurexin-neuroligin interactions by enzymatic probe ligation. PLoS One. 2013;8(2):e52823. doi: 10.1371/journal.pone.0052823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jan CH, et al. Principles of ER cotranslational translocation revealed by proximity-specific ribosome profiling. Science. 2014;346(6210):1257521. doi: 10.1126/science.1257521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shoaib M, et al. PUB-NChIP--"in vivo biotinylation" approach to study chromatin in proximity to a protein of interest. Genome Res. 2013;23(2):331–340. doi: 10.1101/gr.134874.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kodama Y, Hu CD. An improved bimolecular fluorescence complementation assay with a high signal-to-noise ratio. Biotechniques. 2010;49(5):793–805. doi: 10.2144/000113519. [DOI] [PubMed] [Google Scholar]
- 9.Cabantous S, et al. A new protein-protein interaction sensor based on tripartite split-GFP association. Sci Rep. 2013;3:2854. doi: 10.1038/srep02854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roux KJ, et al. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J Cell Biol. 2012;196(6):801–810. doi: 10.1083/jcb.201112098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roux KJ, et al. BioID: a screen for protein-protein interactions. Curr Protoc Protein Sci. 2013;74 doi: 10.1002/0471140864.ps1923s74. Unit 19 23. [DOI] [PubMed] [Google Scholar]
- 12.Kim DI, et al. Probing nuclear pore complex architecture with proximity-dependent biotinylation. Proc Natl Acad Sci U S A. 2014;111(24):E2453–e2461. doi: 10.1073/pnas.1406459111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fredriksson K, et al. Proteomic analysis of proteins surrounding occludin and claudin-4 reveals their proximity to signaling and trafficking networks. PLoS One. 2015;10(3):e0117074. doi: 10.1371/journal.pone.0117074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo Z, et al. E-cadherin interactome complexity and robustness resolved by quantitative proteomics. Sci Signal. 2014;7(354):rs7. doi: 10.1126/scisignal.2005473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Steed E, et al. MarvelD3 couples tight junctions to the MEKK1-JNK pathway to regulate cell behavior and survival. J Cell Biol. 2014;204(5):821–838. doi: 10.1083/jcb.201304115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ueda S, et al. Force dependent biotinylation of myosin IIA by alpha-catenin tagged with a promiscuous biotin ligase. PLoS One. 2015;10(3):e0122886. doi: 10.1371/journal.pone.0122886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Itallie CM, et al. The N and C termini of ZO-1 are surrounded by distinct proteins and functional protein networks. J Biol Chem. 2013;288(19):13775–13788. doi: 10.1074/jbc.M113.466193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Itallie CM, et al. Biotin ligase tagging identifies proteins proximal to E-cadherin, including lipoma preferred partner, a regulator of epithelial cell-cell and cell-substrate adhesion. J Cell Sci. 2014;127(Pt 4):885–895. doi: 10.1242/jcs.140475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu H, et al. SAS-4 Protein in Trypanosoma brucei Controls Life Cycle Transitions by Modulating the Length of the Flagellum Attachment Zone Filament. J Biol Chem. 2015;290(51):30453–30463. doi: 10.1074/jbc.M115.694109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McAllaster MR, et al. Proteomic identification of novel cytoskeletal proteins associated with TbPLK, an essential regulator of cell morphogenesis in Trypanosoma brucei. Mol Biol Cell. 2015;26(17):3013–3029. doi: 10.1091/mbc.E15-04-0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morriswood B, et al. Novel bilobe components in Trypanosoma brucei identified using proximity-dependent biotinylation. Eukaryot Cell. 2013;12(2):356–367. doi: 10.1128/EC.00326-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou Q, et al. An EF-hand-containing Protein in Trypanosoma brucei Regulates Cytokinesis Initiation by Maintaining the Stability of the Cytokinesis Initiation Factor CIF1. The Journal of biological chemistry. 2016;291(28):14395–14409. doi: 10.1074/jbc.M116.726133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim DI, et al. An improved smaller biotin ligase for BioID proximity labeling. Mol Biol Cell. 2016 doi: 10.1091/mbc.E15-12-0844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coyaud E, et al. BioID-based Identification of Skp Cullin F-box (SCF)beta-TrCP1/2 E3 Ligase Substrates. Mol Cell Proteomics. 2015;14(7):1781–1795. doi: 10.1074/mcp.M114.045658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dingar D, et al. BioID identifies novel c-MYC interacting partners in cultured cells and xenograft tumors. J Proteomics. 2015;118:95–111. doi: 10.1016/j.jprot.2014.09.029. [DOI] [PubMed] [Google Scholar]
- 26.Gupta GD, et al. A Dynamic Protein Interaction Landscape of the Human Centrosome-Cilium Interface. Cell. 2015;163(6):1484–1499. doi: 10.1016/j.cell.2015.10.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kehrer J, et al. Proteomic analysis of the Plasmodium berghei gametocyte egressome and vesicular bioID of osmiophilic body proteins identifies MTRAP as an essential factor for parasite transmission. Molecular & cellular proteomics : MCP. 2016 doi: 10.1074/mcp.M116.058263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zempleni J. Uptake, localization, and noncarboxylase roles of biotin. Annu Rev Nutr. 2005;25:175–196. doi: 10.1146/annurev.nutr.25.121304.131724. [DOI] [PubMed] [Google Scholar]
- 29.Honke K, Kotani N. The enzyme-mediated activation of radical source reaction: a new approach to identify partners of a given molecule in membrane microdomains. J Neurochem. 2011;116(5):690–695. doi: 10.1111/j.1471-4159.2010.07027.x. [DOI] [PubMed] [Google Scholar]
- 30.Gross AJ, Sizer IW. The oxidation of tyramine, tyrosine, and related compounds by peroxidase. J Biol Chem. 1959;234(6):1611–1614. [PubMed] [Google Scholar]
- 31.Schonhuber W, et al. Improved sensitivity of whole-cell hybridization by the combination of horseradish peroxidase-labeled oligonucleotides and tyramide signal amplification. Appl Environ Microbiol. 1997;63(8):3268–3273. doi: 10.1128/aem.63.8.3268-3273.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kotani N, et al. Biochemical visualization of cell surface molecular clustering in living cells. Proc Natl Acad Sci U S A. 2008;105(21):7405–7409. doi: 10.1073/pnas.0710346105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schofer C, et al. Signal amplification at the ultrastructural level using biotinylated tyramides and immunogold detection. Histochem Cell Biol. 1997;108(4–5):313–319. doi: 10.1007/s004180050171. [DOI] [PubMed] [Google Scholar]
- 34.Rhee HW, et al. Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science. 2013;339(6125):1328–1331. doi: 10.1126/science.1230593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang S, et al. A proteomics approach to the cell-surface interactome using the enzyme-mediated activation of radical sources reaction. Proteomics. 2012;12(1):54–62. doi: 10.1002/pmic.201100551. [DOI] [PubMed] [Google Scholar]
- 36.Miyagawa-Yamaguchi A, et al. Expressed glycosylphosphatidylinositol-anchored horseradish peroxidase identifies co-clustering molecules in individual lipid raft domains. PLoS One. 2014;9(3):e93054. doi: 10.1371/journal.pone.0093054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li XW, et al. New insights into the DT40 B cell receptor cluster using a proteomic proximity labeling assay. J Biol Chem. 2014;289(21):14434–14447. doi: 10.1074/jbc.M113.529578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rees JS, et al. Selective Proteomic Proximity Labeling Assay Using Tyramide (SPPLAT): A Quantitative Method for the Proteomic Analysis of Localized Membrane-Bound Protein Clusters. Curr Protoc Protein Sci. 2015;80 doi: 10.1002/0471140864.ps1927s80. 19 27 1–18. [DOI] [PubMed] [Google Scholar]
- 39.Martell JD, et al. Engineered ascorbate peroxidase as a genetically encoded reporter for electron microscopy. Nat Biotechnol. 2012;30(11):1143–1148. doi: 10.1038/nbt.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hopkins C, et al. Chimeric molecules employing horseradish peroxidase as reporter enzyme for protein localization in the electron microscope. Methods Enzymol. 2000;327:35–45. doi: 10.1016/s0076-6879(00)27265-0. [DOI] [PubMed] [Google Scholar]
- 41.Lam SS, et al. Directed evolution of APEX2 for electron microscopy and proximity labeling. Nat Methods. 2015;12(1):51–54. doi: 10.1038/nmeth.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mick DU, et al. Proteomics of Primary Cilia by Proximity Labeling. Dev Cell. 2015;35(4):497–512. doi: 10.1016/j.devcel.2015.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jing J, et al. Proteomic mapping of ER-PM junctions identifies STIMATE as a regulator of Ca(2)(+) influx. Nature cell biology. 2015;17(10):1339–1347. doi: 10.1038/ncb3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen CL, et al. Proteomic mapping in live Drosophila tissues using an engineered ascorbate peroxidase. Proc Natl Acad Sci U S A. 2015;112(39):12093–12098. doi: 10.1073/pnas.1515623112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hung V, et al. Spatially resolved proteomic mapping in living cells with the engineered peroxidase APEX2. Nat Protoc. 2016;11(3):456–475. doi: 10.1038/nprot.2016.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hung V, et al. Proteomic mapping of the human mitochondrial intermembrane space in live cells via ratiometric APEX tagging. Mol Cell. 2014;55(2):332–341. doi: 10.1016/j.molcel.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Echols N, et al. Comprehensive analysis of amino acid and nucleotide composition in eukaryotic genomes, comparing genes and pseudogenes. Nucleic Acids Res. 2002;30(11):2515–2523. doi: 10.1093/nar/30.11.2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tourasse NJ, Li WH. Selective constraints, amino acid composition, and the rate of protein evolution. Mol Biol Evol. 2000;17(4):656–664. doi: 10.1093/oxfordjournals.molbev.a026344. [DOI] [PubMed] [Google Scholar]
- 49.Chojnowski A, et al. Progerin reduces LAP2alpha-telomere association in Hutchinson-Gilford progeria. Elife. 2015:4. doi: 10.7554/eLife.07759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fu Y, et al. MacroH2A1 associates with nuclear lamina and maintains chromatin architecture in mouse liver cells. Sci Rep. 2015;5:17186. doi: 10.1038/srep17186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Comartin D, et al. CEP120 and SPICE1 cooperate with CPAP in centriole elongation. Curr Biol. 2013;23(14):1360–1366. doi: 10.1016/j.cub.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 52.Firat-Karalar EN, et al. Proximity interactions among centrosome components identify regulators of centriole duplication. Curr Biol. 2014;24(6):664–670. doi: 10.1016/j.cub.2014.01.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dong JM, et al. Proximity biotinylation provides insight into the molecular composition of focal adhesions at the nanometer scale. Science signaling. 2016;9(432):rs4. doi: 10.1126/scisignal.aaf3572. [DOI] [PubMed] [Google Scholar]
- 54.Chen AL, et al. Novel components of the Toxoplasma inner membrane complex revealed by BioID. MBio. 2015;6(1):e02357–e02314. doi: 10.1128/mBio.02357-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Batsios P, et al. Src1 is a Protein of the Inner Nuclear Membrane Interacting with the Dictyostelium Lamin NE81. Cells. 2016;5(1) doi: 10.3390/cells5010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wei Y, et al. CHCHD2 Is Coamplified with EGFR in NSCLC and Regulates Mitochondrial Function and Cell Migration. Mol Cancer Res. 2015;13(7):1119–1129. doi: 10.1158/1541-7786.MCR-14-0165-T. [DOI] [PubMed] [Google Scholar]
- 57.Bhaduri A, et al. Network Analysis Identifies Mitochondrial Regulation of Epidermal Differentiation by MPZL3 and FDXR. Dev Cell. 2015;35(4):444–457. doi: 10.1016/j.devcel.2015.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cole A, et al. Inhibition of the Mitochondrial Protease ClpP as a Therapeutic Strategy for Human Acute Myeloid Leukemia. Cancer Cell. 2015;27(6):864–876. doi: 10.1016/j.ccell.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lamb CA, et al. TBC1D14 regulates autophagy via the TRAPP complex and ATG9 traffic. EMBO J. 2016;35(3):281–301. doi: 10.15252/embj.201592695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Couzens AL, et al. Protein interaction network of the mammalian Hippo pathway reveals mechanisms of kinase-phosphatase interactions. Sci Signal. 2013;6(302):rs15. doi: 10.1126/scisignal.2004712. [DOI] [PubMed] [Google Scholar]
- 61.Zhou Z, et al. The cerebral cavernous malformation pathway controls cardiac development via regulation of endocardial MEKK3 signaling and KLF expression. Developmental cell. 2015;32(2):168–180. doi: 10.1016/j.devcel.2014.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cheng YS, et al. PPP2R5C Couples Hepatic Glucose and Lipid Homeostasis. PLoS Genet. 2015;11(10):e1005561. doi: 10.1371/journal.pgen.1005561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sun BK, et al. CALML5 is a ZNF750- and TINCR-induced protein that binds stratifin to regulate epidermal differentiation. Genes & development. 2015;29(21):2225–2230. doi: 10.1101/gad.267708.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Elzi DJ, et al. Proteomic Analysis of the EWS-Fli-1 Interactome Reveals the Role of the Lysosome in EWS-Fli-1 Turnover. J Proteome Res. 2014;13(8):3783–3791. doi: 10.1021/pr500387m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lambert JP, et al. Proximity biotinylation and affinity purification are complementary approaches for the interactome mapping of chromatin-associated protein complexes. J Proteomics. 2015;118:81–94. doi: 10.1016/j.jprot.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mulholland CB, et al. A modular open platform for systematic functional studies under physiological conditions. Nucleic Acids Res. 2015;43(17):e112. doi: 10.1093/nar/gkv550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Caballe A, et al. ULK3 regulates cytokinetic abscission by phosphorylating ESCRT-III proteins. Elife. 2015;4:e06547. doi: 10.7554/eLife.06547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yeh C, et al. The Deubiquitinase USP37 Regulates Chromosome Cohesion and Mitotic Progression. Curr Biol. 2015;25(17):2290–2299. doi: 10.1016/j.cub.2015.07.025. [DOI] [PubMed] [Google Scholar]
- 69.Rodriguez-Fraticelli AE, et al. Developmental regulation of apical endocytosis controls epithelial patterning in vertebrate tubular organs. Nat Cell Biol. 2015;17(3):241–250. doi: 10.1038/ncb3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vijay S, et al. Exclusive expression of the Rab11 effector SH3TC2 in Schwann cells links integrin-alpha and myelin maintenance to Charcot-Marie-Tooth disease type 4C. Biochim Biophys Acta. 2016 doi: 10.1016/j.bbadis.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schweingruber C, et al. Identification of Interactions in the NMD Complex Using Proximity-Dependent Biotinylation (BioID) PLoS One. 2016;11(3):e0150239. doi: 10.1371/journal.pone.0150239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nishimura T, et al. The eIF4E–Binding Protein 4E–T Is a Component of the mRNA Decay Machinery that Bridges the 5' and 3' Termini of Target mRNAs. Cell Rep. 2015;11(9):1425–1436. doi: 10.1016/j.celrep.2015.04.065. [DOI] [PubMed] [Google Scholar]
- 73.Schumacher MM, et al. The prenyltransferase UBIAD1 is the target of geranylgeraniol in degradation of HMG CoA reductase. Elife. 2015:4. doi: 10.7554/eLife.05560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jahan AS, et al. Usp12 stabilizes the T-cell receptor complex at the cell surface during signaling. Proc Natl Acad Sci U S A. 2016;113(6):E705–E714. doi: 10.1073/pnas.1521763113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Marcos-Ramiro B, et al. RhoB controls endothelial barrier recovery by inhibiting Rac1 trafficking to the cell border. The Journal of cell biology. 2016;213(3):385–402. doi: 10.1083/jcb.201504038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Janer A, et al. SLC25A46 is required for mitochondrial lipid homeostasis and cristae maintenance and is responsible for Leigh syndrome. EMBO molecular medicine. 2016 doi: 10.15252/emmm.201506159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Forred BJ, et al. Identification of Redox and Glucose-Dependent Txnip Protein Interactions. Oxidative medicine and cellular longevity. 2016;2016:5829063. doi: 10.1155/2016/5829063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kabeiseman EJ, et al. The eukaryotic signal sequence, YGRL, targets the chlamydial inclusion. Front Cell Infect Microbiol. 2014;4:129. doi: 10.3389/fcimb.2014.00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mojica SA, et al. SINC, a type III secreted protein of Chlamydia psittaci, targets the inner nuclear membrane of infected cells and uninfected neighbors. Mol Biol Cell. 2015;26(10):1918–1934. doi: 10.1091/mbc.E14-11-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Le Sage V, et al. Proteomic analysis of HIV-1 Gag interacting partners using proximity-dependent biotinylation. Virol J. 2015;12:138. doi: 10.1186/s12985-015-0365-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ritchie C, et al. Analysis of HIV-1 Gag protein interactions via biotin ligase tagging. J Virol. 2015;89(7):3988–4001. doi: 10.1128/JVI.03584-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kueck T, et al. Serine Phosphorylation of HIV-1 Vpu and Its Binding to Tetherin Regulates Interaction with Clathrin Adaptors. PLoS Pathog. 2015;11(8):e1005141. doi: 10.1371/journal.ppat.1005141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Holthusen K, et al. Regulation of Latent Membrane Protein 1 Signaling through Interaction with Cytoskeletal Proteins. J Virol. 2015;89(14):7277–7290. doi: 10.1128/JVI.00321-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gaji RY, et al. Phosphorylation of a Myosin Motor by TgCDPK3 Facilitates Rapid Initiation of Motility during Toxoplasma gondii egress. PLoS Pathog. 2015;11(11):e1005268. doi: 10.1371/journal.ppat.1005268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hashimoto N, et al. Proteomic analysis of ganglioside-associated membrane molecules: substantial basis for molecular clustering. Proteomics. 2012;12(21):3154–3163. doi: 10.1002/pmic.201200279. [DOI] [PubMed] [Google Scholar]
- 86.Ishiura Y, et al. Anomalous expression of Thy1 (CD90) in B-cell lymphoma cells and proliferation inhibition by anti-Thy1 antibody treatment. Biochemical and biophysical research communications. 2010;396(2):329–334. doi: 10.1016/j.bbrc.2010.04.092. [DOI] [PubMed] [Google Scholar]
- 87.Yamashita R, et al. Spatiotemporally-regulated interaction between beta1 integrin and ErbB4 that is involved in fibronectin-dependent cell migration. Journal of biochemistry. 2011;149(3):347–355. doi: 10.1093/jb/mvq148. [DOI] [PubMed] [Google Scholar]
- 88.Kotani N, et al. Fibroblast growth factor receptor 3 (FGFR3) associated with the CD20 antigen regulates the rituximab-induced proliferation inhibition in B-cell lymphoma cells. J Biol Chem. 2012;287(44):37109–37118. doi: 10.1074/jbc.M112.404178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Iwamaru Y, et al. Proximity of SCG10 and prion protein in membrane rafts. J Neurochem. 2015 doi: 10.1111/jnc.13488. [DOI] [PubMed] [Google Scholar]
- 90.Lee SY, et al. APEX Fingerprinting Reveals the Subcellular Localization of Proteins of Interest. Cell reports. 2016;15(8):1837–1847. doi: 10.1016/j.celrep.2016.04.064. [DOI] [PubMed] [Google Scholar]
- 91.Hwang J, Espenshade PJ. Proximity-dependent biotin labeling in yeast using the engineered ascorbate peroxidase APEX2. The Biochemical journal. 2016 doi: 10.1042/BCJ20160106. [DOI] [PMC free article] [PubMed] [Google Scholar]