Abstract
Introduction
Altered DNA methylation (DNAm) levels of hypothalamic-pituitary-adrenal (HPA) axis genes has been associated with exposure to childhood maltreatment (CM) and depression; however, it is unknown whether CM and depression have joint and potentially interacting effects on the glucocorticoid receptor (NR3C1) DNAm. We investigated the impact of CM and lifetime major depressive disorder (MDD) on NR3C1 DNAm and gene expression (GE) in 147 adult participants from the Detroit Neighborhood Health Study.
Methods
NR3C1 promoter region DNAm was assessed via pyrosequencing using whole blood-derived DNA. Quantitative RT-PCR assays measured GE from leukocyte-derived RNA. Linear regression models were used to examine the relationship among CM, MDD, and DNAm.
Results
Both CM and MDD were significant predictors of NR3C1 DNAm: CM was associated with an increase in DNAm in an EGR1 transcription factor binding site (TFBS), whereas MDD was associated with a decrease in DNAm downstream of the TFBS. No significant CM-MDD interactions were observed. CM alone was associated with significantly lower NR3C1 GE.
Limitations
Our report of CM is a retrospective self-report of abuse, which may introduce recall bias. DNAm was measured in whole blood and may not reflect brain-derived DNAm levels.
Conclusions
CM and MDD are both associated with altered DNAm levels in the NR3C1 promoter region, however the location and direction of effects differ between the two exposures, and the functional effects, as measured by GE, appear to be limited to CM exposure alone. CM exposure may be biologically embedded in this key HPA axis gene.
Keywords: Development, Epigenetics, Gene expression, Childhood maltreatment, Depression
1. Introduction
There is a well-established link between early life adversity and poor mental health later in life (Afifi et al., 2008; McEwen, 2003). Specifically, childhood maltreatment has been strongly associated with the onset of major depressive disorder (MDD) (Kendler et al., 2004; Nemeroff, 2004) and other mental illnesses during adulthood, including posttraumatic stress disorder (PTSD) (Green et al., 2010), bipolar disorder (Afifi et al., 2008), and anxiety disorders (Kessler et al., 1997). Several decades of work in rodents, humans, and non-human primates has demonstrated the importance of early environment on the molecular pathways regulating the stress response (reviewed in (Klengel et al., 2014)). These studies have largely focused on examining the epigenetics of hypothalamic-pituitary-adrenal (HPA) axis genes due to the primary role of the HPA axis in regulating the body's stress response. Dysregulation of the HPA axis results in an altered stress response (Nemeroff, 2004), producing an increased risk for mood and anxiety disorders, as well as physical disorders such as diabetes and cardiovascular disease (Irwin and Cole, 2011; Nemeroff, 2004; Radtke et al., 2011). However, the molecular mechanisms that underlie HPA axis dysregulation, and their possible associations with commonly occurring mental disorders such as depression remain unclear. DNA methylation is a stable, but modifiable, epigenetic mark that is characterized by a chemical alteration to the nucleotides that comprise DNA (Whitelaw and Whitelaw, 2006). This chemical modification does not alter the underlying DNA sequence but rather serves to regulate chromatin structure and DNA accessibility, often resulting in altered transcription. DNA methylation is characterized by the addition of a methyl group, –CH3, to the 5′ position of cytosine – typically when cytosine is coupled to guanine on the same strand of DNA – and is stable over time (Heijmans et al., 2008; Tyrka et al., 2008), tissue specific (Gama-Sosa et al., 1983), and responsive to environmental exposures (Fraga et al., 2005). From a functional perspective, DNA methylation of transcription factor binding sites (TFBS) within gene promoter regions typically results in reduced gene expression (Brenet et al., 2011), and, in particular, has been associated with early life changes to stress-relevant phenotypes that persist into adulthood (Weaver et al., 2004). Thus, as a stable, but modifiable, molecular mechanism with functional effects, epigenetic regulation is a potential contributor to the etiology of mental disorders resulting from adverse early life experiences.
A substantial body of evidence has shown that early life adversity, in particular childhood maltreatment, is associated with DNA methylation differences in HPA axis gene nuclear receptor subfamily 3, group member 1 (NR3C1), whose product is commonly known as the glucocorticoid receptor (McGowan et al., 2009; Tyrka et al., 2012). The glucocorticoid receptor plays an important role in the body's stress response as it not only binds to the stress hormone cortisol, but also modulates the negative feedback of the HPA axis. High levels of cortisol tamp down the stress response by reducing corticotropin releasing hormone (CRH), the hormone released by the hypothalamus that triggers the stress response cascade (Binder, 2009); this negative feedback is facilitated by the binding of cortisol to the glucocorticoid receptor (Binder, 2009). Initial rodent studies in NR3C1 demonstrated that early environment, specifically level of maternal care, was associated with alterations to NR3C1 DNA methylation, resulting in changes in stress sensitivity that last into adulthood (Weaver et al., 2004). Similarly, in humans, maternal depressed/anxious mood during the third trimester was associated with increased NR3C1 DNA methylation in a TFBS in cord derived-blood, and with increased infant cortisol responses 3 months postnatally (Oberlander et al., 2008). Subsequent human studies have confirmed exposure to childhood maltreatment and other early life adversities are associated with increased NR3C1 DNA methylation measured in both blood and post mortem brain tissue (Labonte et al., 2012; Martin-Blanco et al., 2014; McGowan et al., 2009; Tyrka et al., 2012). Maternal experiences during pregnancy have also been associated with DNA methylation differences in NR3C1 in their offspring, reviewed and summarized in a recent meta-analysis (Palma-Gudiel et al., 2015). These studies underscore the importance of early life environments and, in particular, the long-term impact of early life adversity (ELA)-induced changes to DNA methylation on mental health.
Despite the extensive research supporting the link between ELA and altered DNA methylation, few studies have examined the link between depression and NR3C1 DNA methylation. To date, only two studies have directly examined NR3C1 promoter region DNA methylation and MDD. A recent study examined NR3C1 DNA methylation and hippocampal volume in a group of participants with MDD compared to healthy controls, finding MDD patients had significantly lower levels of DNA methylation within the promoter region of NR3C1 (Na et al., 2014). Additionally, a second paper examined NR3C1 DNA methylation in a group of participants with and without MDD, reporting a significant increase in NR3C1 DNA methylation at a single CpG site associated with the disorder (Nantharat et al., 2015). These studies report opposite associations between MDD and NR3C1 DNA methylation levels measured in blood; however, the relationship among childhood maltreatment, MDD, and NR3C1 DNA methylation was not addressed in either work. To address this gap in knowledge, we sought to examine the impact of childhood maltreatment and MDD on NR3C1 DNA methylation and gene expression levels among adults. We hypothesized that (1) childhood maltreatment and MDD affect NR3C1 DNA methylation in a joint and potentially interacting manner, and, secondarily, that (2) DNA methylation differences resulting from childhood maltreatment and/or MDD would contribute to functional consequences in NR3C1 as measured by gene expression levels.
2. Methods
2.1. Participant selection
The Detroit Neighborhood Health Study (DNHS) was approved by the institutional review boards at the University of Michigan and the University of North Carolina at Chapel Hill. Participants (N=152) were selected from the DNHS, a longitudinal, population-based representative sample of adult residents from Detroit, MI (Uddin et al., 2010). All participants provided informed consent prior to participation in the DNHS. Selection for inclusion within this study was based on the availability of whole-blood derived DNA, leukocyte-derived RNA, and complete survey data regarding childhood maltreatment and depression histories. In our study population of 152 DNHS participants, 94 were female and 58 were male; 26 self-identified as European-American, 116 as African-American, and 10 as “other”. The average age was 49.6 years.
2.2. Childhood maltreatment
Participant survey data regarding childhood maltreatment history were collected via structured telephone interviews on the severity, duration, and frequency of each event type. Assessment of childhood maltreatment was based on the Conflict Tactics Scale (CTS) (Straus, 1979) and the Childhood Trauma Questionnaire (CTQ) (Bernstein et al., 1997), as previously described (Keyes et al., 2012; Uddin et al., 2013). CTS items assessed physical and emotional abuse before age 11, with responses rated on a 5-point scale. CTQ assessed physical and sexual abuse before age 18, rating responses on a 3-point scale. The childhood maltreatment score variable is a continuous measure ranging from 0 to 22, as previously described (Keyes et al., 2012; Uddin et al., 2013). In this study, participants with childhood maltreatment exposure (N=76) were defined as any individual belonging to the upper quartile for childhood maltreatment score within the full DNHS survey sample (N=1547). Participants without childhood maltreatment exposure (N=76) belonged to the bottom quartile of childhood maltreatment score within the full DNHS survey sample.
2.3. Depression measure
MDD was assessed using the Patient Health Questionnaire (PHQ-9) (Kroenke et al., 2001) with additional questions that assessed timing and duration of symptoms, consistent with DSM-IV criteria (American Psychiatric, 1994). The PHQ-9 is a 9-item instrument rating responses on a 4-point scale ranging from 0 (not at all) to 3 (nearly every day), with total scores ranging from 0 to 27. The measure has been previously validated (Uddin et al., 2011). MDD was defined as the presence of lifetime MDD (cases N=76, controls N=76).
2.4. Antidepressant medication
Participant medication information was taken during the in-home visit at the time of biologic sample collection (see Section 2.5). Participants were instructed to provide all current prescribed and over the counter medication to the phlebotomist, who recorded the medication name, dosage, and frequency each medication was taken. Antidepressant medication use for this study was determined based on participant medication information from the appropriate wave. SAS Enterprise Guide 7.1 (SAS Institute Incorporated, NC) was used to code medications as antidepressants.
2.5. Sample preparation
2.5.1. DNA
Whole blood was collected via venipuncture from study participants during scheduled in home visits by a trained phlebotomist. DNA was isolated from whole blood using Qiagen's QIAamp DNA Blood Mini Kit (Qiagen, Valencia, CA) and LifeSciences's Quickgene DNA Whole Blood Kit (St. Petersburg, FL) following the manufacturer's recommended protocols. DNA concentrations were verified using the NanoDrop 1000 (ThermoScientific, Waltham, MA) following the manufacturer's recommended protocol.
2.5.2. RNA
RNA was obtained from leukocytes using Leukolock kits following the manufacturer's alternative protocol to preserve total RNA (Ambion, Austin, TX). Quality control criteria was used to ensure high quality RNA was obtained, including a RNA integrity number (RIN) ≥ 5, 28s/18s ≥ 1.0, and 260/280 ≥ 1.7 (Fleige and Pfaffl., 2006; Fleige et al., 2006). RNA sample RIN values and 28s/18s ratios were calculated using the 2100 Bioanalyzer (Agilent, Wilmington, DE) to determine RNA quality. RNA concentration for each sample was determined using the NanoDrop 1000 (ThermoScientific).
2.5.3. Peripheral blood mononuclear cell counts
At the time of blood draw, two FICOLL gradient containing 8 ml BD Vacutainers CPT™ with sodium citrate (Franklin Lakes, NJ) were used for the collection of peripheral blood mononuclear cells (PBMC) samples. Participant tubes were spun in a centrifuge within two hours of collection and processed immediately. During processing, mononuclear cells were isolated, assessed for viability, and counted using Invitrogen's Countess automated cell counter (Carlsbad, CA). A small number of PBMC samples used in this study were measured using TPP PCV Packed CellVolume tubes (Trasadingen, Switzerland) and assessed for viability using a hemacytometer (Fisher Scientific, Pittsburgh, PA).
2.5.4. Bisulfite conversion
750 ng of DNA from each participant was bisulfite converted using Qiagen's Epitect Bisulfite Kit following the manufacturer's recommended protocol. Negative controls containing RNA/DNA free water in place of DNA were included with each bisulfite conversion. High and low methylation control DNA purchased from Zymo Research (Irvine, CA) were bisulfite converted along with the participant samples in order to assess assay performance.
2.6. PCR amplification and pyrosequencing
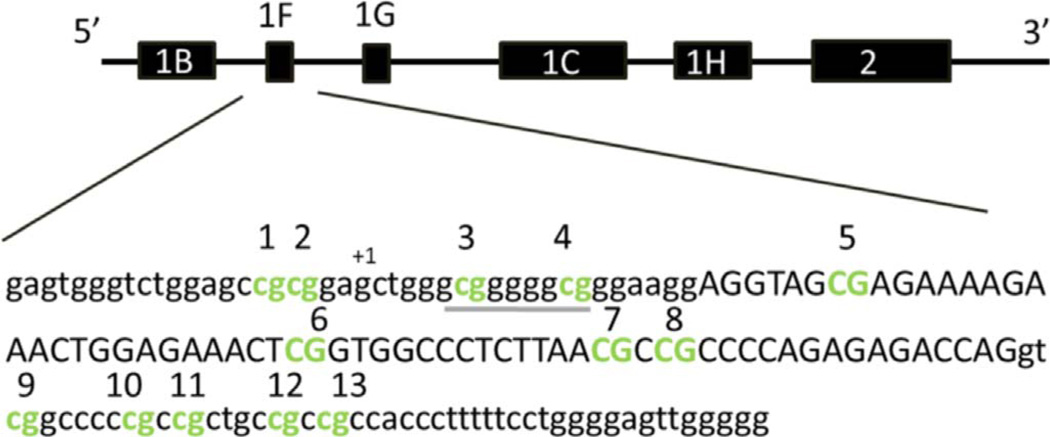
DNA methylation for 13 CpG sites within the promoter region of NR3C1 were assessed via pyrosequencing (CHR5: 142,783,655–142,783,501). The 13 CpG sites targeted in our analyses encompass a 155 bp region and contain an EGR1 TFBS (also known as NGF1-A) (Fig. 1). This locus has been the focus of several other previous studies (McGowan et al., 2009; Oberlander et al., 2008; Radtke et al., 2011; Tyrka et al., 2012). Primers were newly developed for this study (see below) using the PyroMark Q24 Assay Design Software 2.0 (Qiagen). Validation experiments were carried out according to recommendations in the PyroMark manual on all custom assays to ensure high quality primers were used. PCRs were run in duplicate and contained 20 ng of bisulfite converted DNA as starting template. No template controls were also run in duplicate with each set of PCRs as a negative control. Each primer was also tested using bisulfite converted DNA from high and low methylation controls (Zymo). Qiagen's PyroMark Q24 Pyrosequencer was used to detect DNA methylation levels following manufacturer's protocols and default settings.
1 to 4CpG.
Forward PCR primer: 5′-AGTTTTAGAGTGGGTTTGGAG-3′.
Reverse PCR primer (biotinylated): 5′-ACCACCCAATTTCTCCAATTTCTTTTCTC-3′.
Sequencing primer: 5′-GAGTGGGTTTGGAGT-3′.
5 to 13CpG.
Forward PCR primer: 5′-GGGGGAGGGAAGGAGGTA-3′.
Reverse PCR primer (biotinylated): 5′-CCCCCAACTCCCCAAAAA-3′.
Sequencing primer: 5′-GGGAGGGAAGGAGGTAG-3′.
9 to 13CpG.
Forward PCR primer: 5′-GGAAGGAGGTAGAGAGAAAAGAAATTGG-3′.
Reverse PCR primer (biotinylated): 5′-CCCCCAACTCCCCAAAAA-3′.
Sequencing primer: 5′-GGAGAAATTAGGTTTTTTTAA-3′.
- PCR Program: (same for all primer sets).
Initial 15 minutes 95°C Denaturation 30 seconds 94°C 
Annealing 30 seconds 56°C 50 cycles Extension 30 seconds 72°C Final 10 minutes 72°C Hold 4°C indefinitely
Fig. 1.
Promoter region of NR3C1 examined within this study (CHR5: 142,783,501–142,783,655 UCSC Genome Browser Build 2009/hg19). CpG sites tested in this study are depicted in bold green font (online version) and are also numbered. The EGR1 transcription factor binding site is located at CpG sites 3 and 4 and is denoted by underline. The transcription start site is indicated by “+1”. Lowercase nucleotides represent intronic regions, while uppercase nucleotides represent exon 1F. Figure was adapted from “Prenatal exposure to maternal depression, neonatal methylation of human glucocorticoid receptor gene (NR3C1) and infant cortisol stress responses”, Oberlander et al. Epigenetics April 2008 with permission of the publisher (Taylor & Francis Ltd: http://www.tandfonline.com).
2.7. Reverse transcription and Real Time PCR
To analyze gene expression levels, RNA was reverse transcribed into cDNA following the manufacturer's protocol using the High Capacity Reverse Transcription Kit purchased from Applied Bio-Systems (Foster City, CA). Ready-made Taqman gene expression assays (Applied Biosystems) were used to measure relative transcript levels of the target gene NR3C1 (Hs00353740_m1) and the control gene PGK1 (Hs00943178_g1) run in separate wells. The NR3C1 Taqman assay specifically targeted the GRα isoform. Reactions were performed in triplicate for each locus, with each replicate tested in a 20 µl reaction containing 10 ng of participant cDNA. Reactions were run on a HT7500 Fast Real Time PCR machine (Applied BioSystems, Foster City, CA) following the manufacturer's recommended protocol for standard reactions.
2.8. Primary analyses
Previous studies have reported significant associations between DNA methylation at CpG sites 1–4 and early life experiences, while CpG sites 5–13 have been grouped together for exploratory analyses (McGowan et al., 2009; Oberlander et al., 2008; Tyrka et al., 2012). Additionally, the ERG1 TFBS, previously implicated in multiple studies of early-life adversity and NR3C1 DNA methylation (Romens et al., 2015; Tyrka et al., 2012; van der Knaap et al., 2014) encompasses CpG site 3 and 4, while other TFBS are located within bin 5–13 as identified using the UCSC genome browser track “Encode Regulation” ChIP-seq data. The TFBSs within the 5–13CpG region have been confirmed in a variety of cell lines including those derived from blood and brain tissues. Similar binning approaches have been applied to analyses of stress-related effects on other HPA axis genes, in which functionally distinct regions are grouped together in bins (e.g. FKBP5; (Klengel et al., 2013; Yehuda et al., 2015). Therefore, in our study, DNA methylation levels were analyzed in two bins, 1–4 and 5–13. Bin 1–4 was created by averaging DNA methylation levels within participants across all 4 sites. To create bin 5–13, we pooled data generated from both the 5to13CpG and 9to13CpG primer sets, and then averaged DNA methylation levels within participants across all 9 sites. Once data collection was complete, the full DNA methylation dataset N=152 was examined for normality according to bin (1–4 or 5–13) using boxplots, histograms, and the Shapiro-Wilk test of normality in IBM SPSS Statistics for Windows, version 22.0 (IBM Corp., Armonk, NY). Extreme outliers (more than 3 interquartile ranges from the nearest edge of the boxplot) were removed from the dataset to facilitate normality, resulting in a final dataset comprised of N=147.
2.8.1. T-tests
Independent samples t-tests were used to test for bivariate associations between childhood maltreatment exposure and demographic covariates, as well as MDD and demographic covariates, using IBM SPSS Statistics for Windows version 22.0 (IBM Corp., Armonk, NY). Chi-square tests were used to test for associations between childhood maltreatment exposure and the demographic variables of sex and race. Similarly, Chi-square tests were used to test for the association between MDD history and sex and race. All statistical tests were two-tailed and results were considered significant with an uncorrected p < 0.05.
2.8.2. Regression
Linear regressions were performed separately on each bin to test whether childhood maltreatment and MDD have joint and potentially interacting relationships on DNA methylation in these regions. In addition to our main variables of interest, all regression models included age, sex, race, PBMC viability count and anti-depressant medication information as covariates. Main effect models were run first, followed by interaction models. We addressed the concern of multiple hypothesis testing by calculating the false discovery rate (FDR) using the Benjamini Liu method (Benjamini et al., 2001) for our primary study hypotheses, such that results were accepted as significant when pcorrected ≤ 0.012.
2.8.3. Secondary analyses
2.8.3.1. Gene expression
To examine the potential functional consequences of observed DNA methylation differences, we analyzed NR3C1 gene expression values using real-time PCR data. Cycle threshold (CT) values for each replicate were averaged to obtain a mean CT value for each participant used in our analysis. All data was examined for outliers, and any replicates with a standard deviation greater than 0.3 were removed (n=6), and the mean CT was re-calculated from remaining data points. Gene expression data were analyzed using the comparative CT method (Schmittgen and Livak, 2008), normalizing NR3C1 gene expression against the control gene PGK1. Resulting data were analyzed by student's t-test to compare expression levels according to the main study variables, as warranted by the DNA methylation data, and results were accepted as significant if p < 0.05.
3. Results
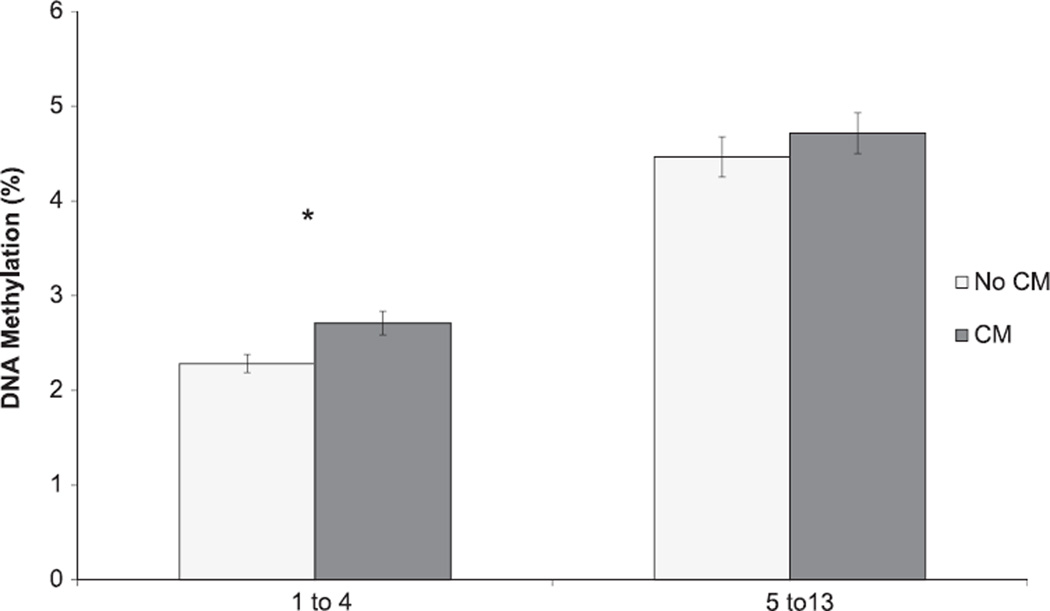
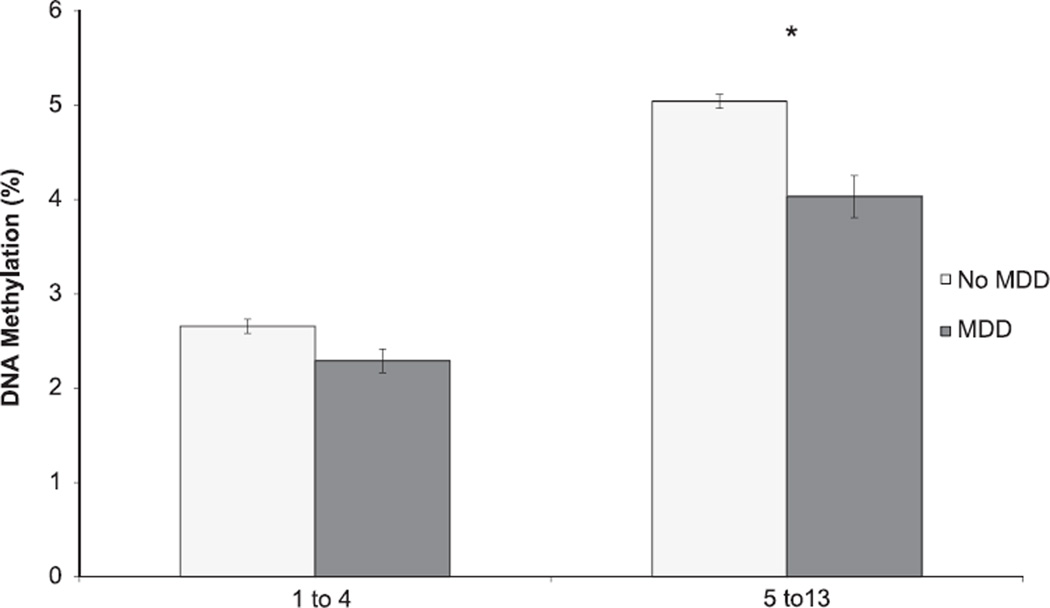
Participants with exposure to childhood maltreatment did not differ significantly from those without childhood maltreatment exposure in terms of age, sex, or race (Table 1). As expected, participants with childhood maltreatment exposure differed from those without such exposure for childhood maltreatment score (Table 1); in addition, DNA methylation over CpG sites 1–4 was significantly higher in those with vs. without childhood maltreatment exposure (Fig. 2). Similarly, MDD cases and controls were significantly different for childhood maltreatment score (Table 1); in addition, DNA methylation was significantly lower over CpG sites 5–13 in those with vs. without MDD (Fig. 3).
Table 1.
Demographic and survey data summary of the study participants. Values indicate the counts or mean ± standard deviation.
| Total N = 147 | ||||||||
|---|---|---|---|---|---|---|---|---|
| Measures | CM− | CM+ | t-test p-value | X2 p-value | MDD− | MDD+ | t-test p-value | X2 p-value |
| Age | 49.95 ± 12.68 | 50.07 ± 12.33 | 0.953 | 50.30 ± 13.80 | 49.63 ± 10.64 | 0.739 | ||
| Sex | 0.948 | 0.672 | ||||||
| Male | 28 | 28 | 30 | 26 | ||||
| Female | 46 | 45 | 52 | 39 | ||||
| Race | 0.553 | 0.915 | ||||||
| European American | 15 | 10 | 13 | 12 | ||||
| African American | 55 | 58 | 64 | 49 | ||||
| Other | 4 | 5 | 5 | 4 | ||||
| PBMC count (million cells) | 4.50 ± 1.28 | 4.55 ± 1.73 | 0.858 | 4.47 ± 1.46 | 4.61 ± 1.58 | 0.598 | ||
| Percent DNA Methylation | ||||||||
| 1–4 CpG | 2.28 ± 0.82 | 2.81 ± 1.35 | 0.005 | 2.64 ± 0.92 | 2.43 ± 1.38 | 0.262 | ||
| 5– 13 CpG | 4.46 ± 1.82 | 4.72 ± 1.84 | 0.411 | 5.01 ± 1.75 | 4.06 ± 1.80 | 0.002 | ||
| CM score | 1.54 ± 1.39 | 11.01 ± 3.88 | < 0.001 | 5.20 ± 4.82 | 7.57 ± 6.17 | 0.012 | ||
Fig. 2.
Average DNA methylation among participants with vs. without childhood maltreatment (CM) exposure at CpG sites 1–4 and 5–13 within NR3C1, N=147. * denotes statistical significance (pcorrected <0.012). The error bars represent standard error of the mean within each binned CpG region.
Fig. 3.
Average DNA methylation among participants with vs. without MDD history at CpG sites 1–4 and 5–13 in NR3C1, N=147. * denotes statistical significance (pcorrected < 0.012). The error bars represent standard error of the mean within each binned CpG region.
3.1. Main effect and interaction of childhood maltreatment and MDD to predict NR3C1 DNA methylation levels
3.1.1. Average DNA methylation CpG sites 1–4
We first examined whether childhood maltreatment and MDD predict DNA methylation levels in our study participants, with DNA methylation averaged at CpGs 1–4 and 5–13, respectively, as the outcome (Table 2). In the main effects model for the average DNA methylation of CpG sites 1–4, childhood maltreatment significantly predicted DNA methylation levels at CpG sites 1–4, such that participants with childhood maltreatment exposure showed increased DNA methylation (β=0.038 SE 0.015, pcorrected=0.001); MDD was not associated with DNA methylation in this region (pcorrected=0.019) following FDR adjustment, which attenuated the unadjusted significant p-value (p=0.026). Age, sex, race, PBMC, and antidepressant medication were also not significant in this model (Table 2). In addition, the interaction model showed no significant synergistic effect of childhood maltreatment and MDD on NR3C1 DNA methylation (Table 2).
Table 2.
Regression results. Linear regression results showing the main effect and interaction models for CpG sites 1–4 and 5–13. The β value, 95% confidence interval (CI), uncorrected p-values, and FDR corrected p-values are shown for each variable within the model. Age, sex, race, peripheral blood mononuclear cell (PBMC) counts, and antidepressant medication use were included as covariates. Bold values indicate the variable is a significant predictor of DNA methylation following adjustment for multiple hypothesis testing.
| Main effect | Interaction | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| β | 95% CI | p-Value | FDR | β | 95% CI | p-Value | FDR | |||
| CpG sites 1–4 | ||||||||||
| CM | 0.038 | 0.009 | 0.068 | 0.01 | 0.012 | 0.049 | 0.012 | 0.087 | 0.01 | |
| MDD | −0.385 | −0.722 | −0.048 | 0.026 | 0.019 | −0.267 | −0.688 | 0.153 | 0.211 | |
| Age | −0.005 | −0.018 | 0.008 | 0.431 | −0.005 | −0.018 | 0.008 | 0.445 | ||
| Sex | −0.071 | −0.392 | 0.251 | 0.665 | −0.066 | −0.388 | 0.256 | 0.687 | ||
| Race | 0.348 | 0.001 | 0.695 | 0.049 | 0.366 | 0.016 | 0.715 | 0.04 | ||
| PBMC (counts) | 0.04 | −0.064 | 0.144 | 0.45 | 0.043 | −0.062 | 0.147 | 0.419 | ||
| Antidepressant (Y/N) | −0.09 | −0.604 | 0.425 | 0.731 | −0.121 | −0.64 | 0.399 | 0.647 | ||
| CM*MDD | – | – | – | – | −0.289 | −0.903 | 0.325 | 0.354 | 0.05 | |
| Main effect | Interaction | |||||||||
| β | 95% CI | p-Value | FDR | β | 95% CI | p-Value | FDR | |||
| CpG sites 5–13 | ||||||||||
| CM | 0.023 | −0.031 | 0.078 | 0.4 | 0.033 | 0.041 | −0.029 | 0.11 | 0.251 | |
| MDD | −0.983 | −1.611 | −0.356 | 0.002 | 0.008 | −0.794 | −1.581 | −0.007 | 0.048 | |
| Age | −0.022 | −0.046 | 0.002 | 0.067 | −0.022 | −0.046 | 0.002 | 0.07 | ||
| Sex | −0.223 | −0.823 | 0.378 | 0.465 | −0.217 | −0.818 | 0.385 | 0.477 | ||
| Race | 0.43 | −0.219 | 1.079 | 0.193 | 0.458 | −0.196 | 1.112 | 0.168 | ||
| PBMC (counts) | 0.104 | −0.091 | 0.299 | 0.293 | 0.108 | −0.087 | 0.304 | 0.275 | ||
| Antidepressant (Y/N) | 0.206 | −0.754 | 1.167 | 0.671 | 0.155 | −0.816 | 1.126 | 0.753 | ||
| CM*MDD | – | – | – | – | −0.456 | −1.597 | 0.685 | 0.43 | 0.05 | |
3.1.2. Average DNA methylation CpG sites 5–13
For DNA methylation averaged across CpG sites 5–13, the main effects models revealed that MDD significantly predicted lower DNA methylation (β=−1.038 SE 0.315, pcorrected=0.008). childhood maltreatment was not associated with DNA methylation in this region (pcorrected=0.05). In addition, age, sex, PBMCs, and antidepressant medication were not significant in this model (Table 2). As with sites 1–4, analyses of sites 5–13 showed no significant synergistic effect of childhood maltreatment and MDD on NR3C1 DNA methylation (pcorrected=0.05; Table 2).
3.2. Gene expression of peripheral leukocytes
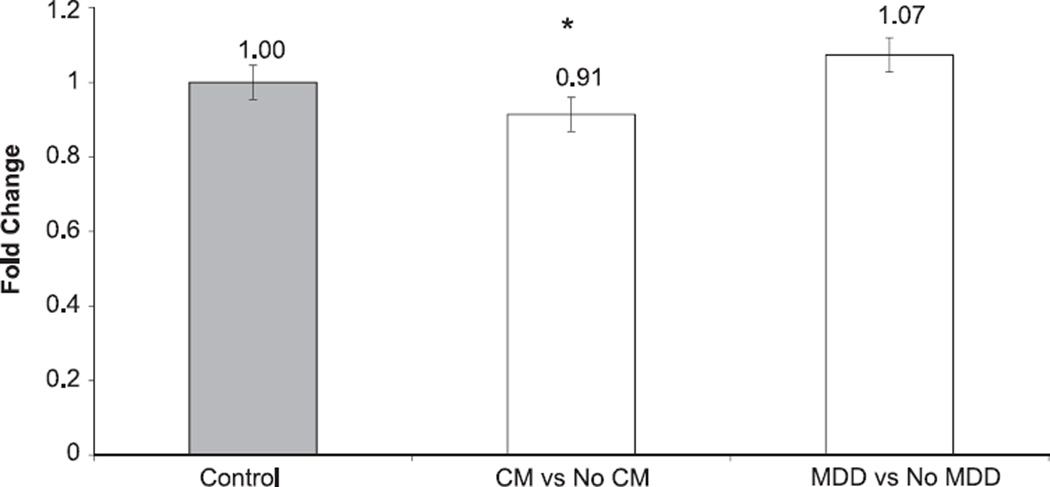
Results from the regression models suggested independent effects of childhood maltreatment and MDD on DNA methylation, thus we chose to separately examine the potential effect of childhood maltreatment and MDD on gene expression. T-tests were used to determine significant differences in gene expression levels within groups (childhood maltreatment vs No childhood maltreatment and MDD vs. No MDD). Results indicated a significant (p=0.037) decrease in fold change in the childhood maltreatment group compared to the No childhood maltreatment group; no significant expression differences were observed for MDD (p=0.27) (Fig. 4).
Fig. 4.
Fold change of NR3C1 gene expression levels between individuals with vs. without childhood maltreatment (CM) exposure and for participants with and without MDD history. Paired T-tests show a significant (p = 0.037) decrease in expression among CM cases and controls for NR3C1. No significant difference was observed for MDD cases compared to controls. Error bars represent standard error of the mean.
4. Discussion
The overall goal of this study was to investigate the association of childhood maltreatment and MDD with DNA methylation and gene expression of the glucocorticoid receptor in an adult population. Our primary analyses sought to test whether childhood maltreatment and MDD have a joint and potentially interacting association with NR3C1 DNA methylation. Our secondary analyses assessed whether childhood maltreatment and/or MDD-associated differences in NR3C1 DNA methylation levels were also associated with differences in gene expression. Results from our primary analyses showed that childhood maltreatment exposure was associated with an increase in DNA methylation in the upstream region of NR3C1 tested in our assays, i.e. across the four CpG sites spanning the EGR1 TFBS. Conversely, MDD was associated with significant decreases in DNA methylation in the latter half of the CpG sites examined (i.e. sites 5–13). No significant childhood maltreatment × MDD interactions were observed in either region (1–4 or 5–13), suggesting that there is no effect heterogeneity for childhood maltreatment and MDD on DNA methylation within these sites of the NR3C1 promoter region. Taken together, results indicate that while childhood maltreatment and MDD are both associated with DNA methylation differences the NR3C1 promoter region, the location and direction of effects differ between the two exposures.
Our finding of increased NR3C1 DNA methylation associated with exposure to childhood maltreatment is consistent with previous literature focusing on either childhood maltreatment (Martin-Blanco et al., 2014; McGowan et al., 2009; Tyrka et al., 2012) or early life experiences (Oberlander et al., 2008; Radtke et al., 2011). Similarly, our MDD results are also consistent with a recent study examining NR3C1 DNA methylation levels which reported decreased DNA methylation (Na et al., 2014). It should be noted that one other study that examined NR3C1 promoter region DNA methylation levels reported a significant increase at their CpG site 7 (corresponding to CpG site 5 in this study) in MDD cases compared to healthy controls (Nantharat et al., 2015). Additionally, only one of their CpG sites overlaps with the region we found to be significantly associated with MDD in our study, which is a point of consideration when comparing results. However, neither of these studies assessed childhood maltreatment or other adverse life events in their population. Despite these findings, our study is distinct from this more recent work as we examined the joint and potentially interacting effects of both childhood maltreatment and MDD on NR3C1 DNA methylation. Interestingly, despite this difference in focus from previous studies, we did detect significant differences in DNA methylation associated with childhood maltreatment and MDD, which are consistent with previous studies that examined these outcomes separately.
To date, research on NR3C1 DNA methylation has largely focused on the importance of early life environment (Oberlander et al., 2008; Radtke et al., 2011; Tyrka et al., 2012). Our results, however, identify a significant association between MDD and DNA methylation in adults, suggesting that NR3C1 DNA methylation levels are potentially more plastic than previously thought; early life environment may not be the only critical window where alterations of DNA methylation levels can occur within HPA axis genes. Additionally, we did not observe a significant interaction between childhood maltreatment and MDD for CpG sites in either of the two bins (1–4 or 5–13). This finding is somewhat surprising given the strong body of evidence linking childhood maltreatment exposure with MDD onset (Kendler et al., 2004; Nemeroff, 2004) and our significant association of childhood maltreatment and MDD with DNA methylation levels. The lack of significant interaction suggests childhood maltreatment and MDD influence NR3C1 promoter region DNA methylation levels independently of one another.
Our secondary analyses tested whether the observed significant DNA methylation differences were reflective of gene expression levels in NR3C1. We found a significant decrease in gene expression between individuals with vs. without childhood maltreatment exposure. This finding is consistent with previous reports in the literature linking increased DNA methylation within the promoter to decreased gene expression (Brenet et al., 2011; McGowan et al., 2009), in particular at CpG sites spanning the EGR1 TFBS (Weaver et al., 2007). Despite finding significantly lower DNA methylation in the downstream region of NR3C1 CpG sites associated with MDD, we did not observe significant gene expression differences associated with MDD status; however, MDD does not significantly influence DNA methylation at the EGR1-associated CpG sites, suggesting that its functional consequences (as measured by NR3C1 expression) may be limited compared to the effects of childhood maltreatment exposure. This null gene expression finding associated with MDD is consistent with a previous study examining DNA methylation and gene expression in a group of MDD patients and healthy controls (Nantharat et al., 2015).
It should be noted that several transcripts of NR3C1 exist as a result of alternative splicing. The presence of these different transcripts can result in altered glucocorticoid sensitivity (Lewis-Tuffin and Cidlowski, 2006; Shahidi et al., 1999). Two of the more prominent products of alternative splicing include: glucocorticoid receptor α (GRα) and β (GRβ). Multiple studies have reported GRα as the main active isoform (Labonte et al., 2014, 2012; Pujols et al., 2002) which works to bind glucocorticoids, while GRβ negatively regulates GRα (Hagendorf et al., 2005; Oakley et al., 1996; Pujols et al., 2002). GRα is more prevalent in peripheral leukocytes and monocytes compared to GRβ (Hagendorf et al., 2005; Pujols et al., 2002), the same tissues used in this study to measure relative gene expression and DNA methylation levels of NR3C1, respectively. Our Taqman gene expression assay specifically targeted the GRα isoform. Taking into account our finding of decreased mRNA expression in those with a history of childhood maltreatment, is it likely that GR-α is down regulated among those with this exposure, possibly leading to decreased cortisol sensitivity. This notion is supported by a study reporting significantly decreased GRα mRNA expression in participants with PTSD compared to healthy controls. Of particular relevance to the current work is the finding that decreased GRα expression was modulated by a dose-response effect of trauma irrespective of PTSD status, such that those with higher trauma loads showed more marked decrease in GRα expression (Gola et al., 2014).
Of particular importance to this study is the issue of tissue specificity in DNA methylation. In this study, we report DNA methylation levels derived from a peripheral tissue, blood, and do not assess DNA methylation in brain, as our population is composed of living individuals. Nevertheless, several studies have examined post-mortem brain tissue samples and report similar patterns of DNA methylation as those observed in peripheral tissue (Farre et al., 2015; McGowan et al., 2009). To assess whether DNA methylation levels within NR3C1 were consistent between blood and brain tissues, we visualized our regions of interest (see methods for coordinates) using the UCSC genome browser (genome.ucsc.edu) and MARMAL-AID (marmal-aid.org). NR3C1 DNA methylation levels were similar in both tissues, providing further support that the observed changes in blood are potentially a useful biomarker of changes occurring in the brain. Additional work in rodents suggests that GR-responsive genes, including NR3C1, show concordant gene expression changes in brain and blood in relation to stress-related phenotypes (Daskalakis et al., 2014), further supporting the use of select blood-based measures as biomarkers of stress-related conditions.
4.1. Limitations
It is important to note limitations one must consider when interpreting our results. First, the childhood maltreatment variable used in our study is a retrospective self-reported measure from each individual of abuses occurring before the age of 18. Even though early life experiences have been shown to be long lasting and detected into adulthood, self-reported measures may introduce recall bias. Previously, retrospective self-reports of childhood maltreatment from adults with documented cases were associated with underreporting of physical and sexual abuse (Widom and Morris, 1997; Widom and Shepard, 1996). Therefore, our measure of childhood maltreatment may well be an underestimate of previous abuse. Second, we report small, yet significant DNA methylation changes and observed effect sizes within our data; however, these results are consistent with previous childhood maltreatment and depression-related reports of NR3C1 DNA methylation (Labonte et al., 2014; Oberlander et al., 2008; Tyrka et al., 2012). In addition, the functional effect of childhood maltreatment-associated DNA methylation differences on gene expression lends support to the relevance of these small effects. Third, although we controlled for PBMC viability counts, which did not differ among participants with vs. without childhood maltreatment or MDD, we were not able to control for differences in white blood cell subsets. Thus it is possible that the childhood maltreatment and/or MDD-associated DNA methylation differences might be localized to a particular cell type. Fourth, studies have shown that atypical and melancholic subtypes of MDD manifest in different biologic pathways (Charmandari et al., 2005; Lamers et al., 2013); however, in our work we were unable to subdivide our participants into atypical or melancholic depression based on the instrument used within the parent study preventing a more detailed interpretation of our findings with regard to MDD subtype. Finally, we were unable to assess whether our observed DNA methylation and expression differences in NR3C1 were also associated with differences in cortisol levels, as such samples were not collected in the original parent study. However, related studies suggest that DNA methylation differences are indeed associated with differences in cortisol levels (Tyrka et al., 2012), with increased NR3C1 DNA methylation associated with reductions in plasma cortisol.
This study also has many strengths. Our study is unique in that we are testing samples from a non-clinical, community-based cohort of adult residents in Detroit, providing the opportunity to generalize our results to the larger Detroit population. Additionally, our analyses included participant antidepressant medication information: controlling for medication use in analyses is imperative to accurately interpret data and results, due to multiple reports of altered epigenetic signatures following antidepressant medication use (Melas et al., 2012; Perisic et al., 2010). Furthermore, our study extends the current literature, as we simultaneously examined the association of childhood maltreatment and MDD on DNA methylation and gene expression levels within the same study participants. Finally, we tested study participants for both gene expression and DNA methylation levels, allowing us to infer the potential downstream impact of differences in DNA methylation levels.
5. Conclusion
In conclusion, we report significant DNA methylation differences within the promoter region of NR3C1 associated with both childhood maltreatment and MDD. We also found a significant decrease in NR3C1 gene expression among those exposed to childhood maltreatment, however no significant difference in relative gene expression levels was observed in MDD. Future work on other HPA axis genes should provide additional insight into the joint and potentially interacting effects of childhood maltreatment and MDD on stress-relevant DNA methylation.
Acknowledgments
The authors would like to thank Sarah Lee for assisting with medication data entry.
Funding
This work was funded by NIH grants DA022720 and MH088283.
Footnotes
Conflict of interest
None.
References
- Afifi TO, Enns MW, Cox BJ, Asmundson GJ, Stein MB, Sareen J. Population attributable fractions of psychiatric disorders and suicide ideation and attempts associated with adverse childhood experiences. Am. J. Public Health. 2008;98:946–952. doi: 10.2105/AJPH.2007.120253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Psychiatric, A. Diagnostic and Statistical Manual of Mental Disorders, Fourth edition. Washintgon, DC: American Psychiatric Association; 1994. [Google Scholar]
- Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav. Brain Res. 2001;125:279–284. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- Bernstein DP, Ahluvalia T, Pogge D, Handelsman L. Validity of the Childhood Trauma Questionnaire in an adolescent psychiatric population. J. Am. Acad. Child and Adolesc. Psychiatry. 1997;36(3):340–348. doi: 10.1097/00004583-199703000-00012. [DOI] [PubMed] [Google Scholar]
- Binder EB. The role of FKBP5, a co-chaperone of the glucocorticoid receptor in the pathogenesis and therapy of affective and anxiety disorders. Psycho-neuroendocrinology. 2009;34(Suppl. 1):S186–S195. doi: 10.1016/j.psyneuen.2009.05.021. [DOI] [PubMed] [Google Scholar]
- Brenet F, Moh M, Funk P, Feierstein E, Viale AJ, Socci ND, Scandura JM. DNA methylation of the first exon is tightly linked to transcriptional silencing. PLoS One. 2011;6:e14524. doi: 10.1371/journal.pone.0014524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charmandari E, Tsigos C, Chrousos G. Endocrinology of the stress response. Annu. Rev. Physiol. 2005;67:259–284. doi: 10.1146/annurev.physiol.67.040403.120816. [DOI] [PubMed] [Google Scholar]
- Daskalakis NP, Cohen H, Cai G, Buxbaum JD, Yehuda R. Expression profiling associates blood and brain glucocorticoid receptor signaling with trauma-related individual differences in both sexes. Proc. Natl. Acad. Sci. USA. 2014;111:13529–13534. doi: 10.1073/pnas.1401660111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farre P, Jones MJ, Meaney MJ, Emberly E, Turecki G, Kobor MS. Concordant and discordant DNA methylation signatures of aging in human blood and brain. Epigenetics Chromatin. 2015;8:19. doi: 10.1186/s13072-015-0011-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleige S, Pfaffl MW. RNA integrity and the effect on the real-time qRT-PCR performance. Mol. Asp. Med. 2006;27:126–139. doi: 10.1016/j.mam.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Fleige S, Walf V, Huch S, Prgomet C, Sehm J, Pfaffl MW. Comparison of relative mRNA quantification models and the impact of RNA integrity in quantitative real-time RT-PCR. Biotechnol. Lett. 2006;28:1601–1613. doi: 10.1007/s10529-006-9127-2. [DOI] [PubMed] [Google Scholar]
- Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar ML, Heine-Suner D, Cigudosa JC, Urioste M, Benitez J, Boix-Chornet M, Sanchez-Aguilera A, Ling C, Carlsson E, Poulsen P, Vaag A, Stephan Z, Spector TD, Wu YZ, Plass C, Esteller M. Epigenetic differences arise during the lifetime of monozygotic twins. Proc. Natl. Acad. Sci. USA. 2005;102:10604–10609. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gama-Sosa MA, Midgett RM, Slagel VA, Githens S, Kuo KC, Gehrke CW, Ehrlich M. Tissue-specific differences in DNA methylation in various mammals. Biochim. Biophys. Acta. 1983;740:212–219. doi: 10.1016/0167-4781(83)90079-9. [DOI] [PubMed] [Google Scholar]
- Gola H, Engler A, Morath J, Adenauer H, Elbert T, Kolassa IT, Engler H. Reduced peripheral expression of the glucocorticoid receptor alpha isoform in individuals with posttraumatic stress disorder: a cumulative effect of trauma burden. PLoS One. 2014;9:e86333. doi: 10.1371/journal.pone.0086333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green JG, McLaughlin KA, Berglund PA, Gruber MJ, Sampson NA, Zaslavsky AM, Kessler RC. Childhood adversities and adult psychiatric disorders in the national comorbidity survey replication I: associations with first onset of DSM-IV disorders. Arch. Gen. psychiatry. 2010;67:113–123. doi: 10.1001/archgenpsychiatry.2009.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagendorf A, Koper JW, de Jong FH, Brinkmann AO, Lamberts SW, Feelders RA. Expression of the human glucocorticoid receptor splice variants alpha, beta, and P in peripheral blood mononuclear leukocytes in healthy controls and in patients with hyper- and hypocortisolism. J. Clin. Endocrinol. Metab. 2005;90:6237–6243. doi: 10.1210/jc.2005-1042. [DOI] [PubMed] [Google Scholar]
- Heijmans BT, Tobi EW, Stein AD, Putter H, Blauw GJ, Susser ES, Slagboom PE, Lumey LH. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc. Natl. Acad. Sci. USA. 2008;105:17046–17049. doi: 10.1073/pnas.0806560105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin MR, Cole SW. Reciprocal regulation of the neural and innate immune systems. Nat. Rev. Immunol. 2011;11:625–632. doi: 10.1038/nri3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS, Kuhn JW, Prescott CA. Childhood sexual abuse, stressful life events and risk for major depression in women. Psychol. Med. 2004;34:1475–1482. doi: 10.1017/s003329170400265x. [DOI] [PubMed] [Google Scholar]
- Kessler RC, Davis CG, Kendler KS. Childhood adversity and adult psychiatric disorder in the US National Comorbidity Survey. Psychol. Med. 1997;27:1101–1119. doi: 10.1017/s0033291797005588. [DOI] [PubMed] [Google Scholar]
- Keyes KM, McLaughlin KA, Koenen KC, Goldmann E, Uddin M, Galea S. Child maltreatment increases sensitivity to adverse social contexts: Neighborhood physical disorder and incident binge drinking in Detroit. Drug Alcohol Depend. 2012;122:77–85. doi: 10.1016/j.drugalcdep.2011.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klengel T, Mehta D, Anacker C, Rex-Haffner M, Pruessner JC, Pariante CM, Pace TWW, Mercer KB, Mayberg HS, Bradley B, Nemeroff CB, Holsboer F, Heim CM, Ressler KJ, Rein T, Binder EB. Allele-specific FKBP5 DNA demethylation mediates gene-childhood trauma interactions. Nat. Neurosci. 2013;16:33–41. doi: 10.1038/nn.3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klengel T, Pape J, Binder EB, Mehta D. The role of DNA methylation in stress-related psychiatric disorders. Neuropharmacology. 2014;80:115–132. doi: 10.1016/j.neuropharm.2014.01.013. [DOI] [PubMed] [Google Scholar]
- Kroenke K, Spitzer RL, Williams JB. The PHQ-9: validity of a brief depression severity measure. J. Gen. Intern. Med.: Off. J. Soc. Res. Educ. Prim. Care Intern. Med. 2001;16:606–613. doi: 10.1046/j.1525-1497.2001.016009606.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labonte B, Azoulay N, Yerko V, Turecki G, Brunet A. Epigenetic modulation of glucocorticoid receptors in posttraumatic stress disorder. Transl. Psychiatry. 2014;4:e368. doi: 10.1038/tp.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labonte B, Yerko V, Gross J, Mechawar N, Meaney MJ, Szyf M, Turecki G. Differential glucocorticoid receptor exon 1(B), 1(C), and 1(H) expression and methylation in suicide completers with a history of childhood abuse. Biol. Psychiatry. 2012;72:41–48. doi: 10.1016/j.biopsych.2012.01.034. [DOI] [PubMed] [Google Scholar]
- Lamers F, Vogelzangs N, Merikangas KR, de Jonge P, Beekman AT, Penninx BW. Evidence for a differential role of HPA-axis function, inflammation and metabolic syndrome in melancholic versus atypical depression. Mol. Psychiatry. 2013;18:692–699. doi: 10.1038/mp.2012.144. [DOI] [PubMed] [Google Scholar]
- Lewis-Tuffin LJ, Cidlowski JA. The physiology of human glucocorticoid receptor beta (hGRbeta) and glucocorticoid resistance. Ann. N.Y. Acad. Sci. 2006;1069:1–9. doi: 10.1196/annals.1351.001. [DOI] [PubMed] [Google Scholar]
- Martin-Blanco A, Ferrer M, Soler J, Salazar J, Vega D, Andion O, Sanchez-Mora C, Arranz MJ, Ribases M, Feliu-Soler A, Perez V, Pascual JC. Association between methylation of the glucocorticoid receptor gene, childhood maltreatment, and clinical severity in borderline personality disorder. J. Psychiatr. Res. 2014;57:34–40. doi: 10.1016/j.jpsychires.2014.06.011. [DOI] [PubMed] [Google Scholar]
- McEwen BS. Early life influences on life-long patterns of behavior and health. Ment. Retard. Dev. Disabil. Res. Rev. 2003;9:149–154. doi: 10.1002/mrdd.10074. [DOI] [PubMed] [Google Scholar]
- McGowan PO, Sasaki A, D’Alessio AC, Dymov S, Labonte B, Szyf M, Turecki G, Meaney MJ. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat. Neurosci. 2009;12:342–348. doi: 10.1038/nn.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melas PA, Rogdaki M, Lennartsson A, Bjork K, Qi H, Witasp A, Werme M, Wegener G, Mathe AA, Svenningsson P, Lavebratt C. Antidepressant treatment is associated with epigenetic alterations in the promoter of P11 in a genetic model of depression. Int. J. Neuropsychopharmacol./Off. Sci. J. Coll. Int. Neuropsychopharmacol. 2012;15:669–679. doi: 10.1017/S1461145711000940. [DOI] [PubMed] [Google Scholar]
- Na KS, Chang HS, Won E, Han KM, Choi S, Tae WS, Yoon HK, Kim YK, Joe SH, Jung IK, Lee MS, Ham BJ. Association between glucocorticoid receptor methylation and hippocampal subfields in major depressive disorder. PLoS One. 2014;9:e85425. doi: 10.1371/journal.pone.0085425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nantharat M, Wanitchanon T, Amesbutr M, Tammachote R, Praphanphoj V. Glucocorticoid receptor gene (NR3C1) promoter is hypermethylated in Thai females with major depressive disorder. Genet. Mol. Res.: GMR. 2015;14:19071–19079. doi: 10.4238/2015.December.29.15. [DOI] [PubMed] [Google Scholar]
- Nemeroff CB. Neurobiological consequences of childhood trauma. J. Clin. Psychiatry. 2004;65(Suppl. 1):18–28. [PubMed] [Google Scholar]
- Oakley RH, Sar M, Cidlowski JA. The human glucocorticoid receptor beta isoform. Expression, biochemical properties, and putative function. J. Biol. Chem. 1996;271:9550–9559. doi: 10.1074/jbc.271.16.9550. [DOI] [PubMed] [Google Scholar]
- Oberlander TF, Weinberg J, Papsdorf M, Grunau R, Misri S, Devlin AM. Prenatal exposure to maternal depression, neonatal methylation of human glucocorticoid receptor gene (NR3C1) and infant cortisol stress responses. Epigenetics: Off. J. DNA Methylation Soc. 2008;3:97–106. doi: 10.4161/epi.3.2.6034. [DOI] [PubMed] [Google Scholar]
- Palma-Gudiel H, Cordova-Palomera A, Eixarch E, Deuschle M, Fananas L. Maternal psychosocial stress during pregnancy alters the epigenetic signature of the glucocorticoid receptor gene promoter in their offspring: a meta-analysis. Epigenetics: Off. J. DNA Methylation Soc. 2015;10:893–902. doi: 10.1080/15592294.2015.1088630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perisic T, Zimmermann N, Kirmeier T, Asmus M, Tuorto F, Uhr M, Holsboer F, Rein T, Zschocke J. Valproate and amitriptyline exert common and divergent influences on global and gene promoter-specific chromatin modifications in rat primary astrocytes. Neuropsychopharmacol.: Off. Publ. Am. Coll. Neuropsychopharmacol. 2010;35:792–805. doi: 10.1038/npp.2009.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujols L, Mullol J, Roca-Ferrer J, Torrego A, Xaubet A, Cidlowski JA, Picado C. Expression of glucocorticoid receptor alpha- and beta-isoforms in human cells and tissues. Am. J. Physiol. Cell Physiol. 2002;283:C1324–C1331. doi: 10.1152/ajpcell.00363.2001. [DOI] [PubMed] [Google Scholar]
- Radtke KM, Ruf M, Gunter HM, Dohrmann K, Schauer M, Meyer A, Elbert T. Transgenerational impact of intimate partner violence on methylation in the promoter of the glucocorticoid receptor. Transl. Psychiatry. 2011;1:e21. doi: 10.1038/tp.2011.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romens SE, McDonald J, Svaren J, Pollak SD. Associations between early life stress and gene methylation in children. Child. Dev. 2015;86:303–309. doi: 10.1111/cdev.12270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Shahidi H, Vottero A, Stratakis CA, Taymans SE, Karl M, Longui CA, Chrousos GP, Daughaday WH, Gregory SA, Plate JM. Imbalanced expression of the glucocorticoid receptor isoforms in cultured lymphocytes from a patient with systemic glucocorticoid resistance and chronic lymphocytic leukemia. Biochem. Biophys. Res. Commun. 1999;254:559–565. doi: 10.1006/bbrc.1998.9980. [DOI] [PubMed] [Google Scholar]
- Straus MA. Measuring intrafamily Conflict and violence: the conflict tactics (CT) (scales) J. Marriage Fam. 1979;41:75–78. [Google Scholar]
- Tyrka AR, Price LH, Marsit C, Walters OC, Carpenter LL. Childhood adversity and epigenetic modulation of the leukocyte glucocorticoid receptor: preliminary findings in healthy adults. PLoS One. 2012;7:e30148. doi: 10.1371/journal.pone.0030148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyrka AR, Wier L, Price LH, Ross N, Anderson GM, Wilkinson CW, Carpenter LL. Childhood parental loss and adult hypothalamic-pituitary-adrenal function. Biol. Psychiatry. 2008;63:1147–1154. doi: 10.1016/j.biopsych.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin M, Aiello AE, Wildman DE, Koenen KC, Pawelec G, de Los Santos R, Goldmann E, Galea S. Epigenetic and immune function profiles associated with posttraumatic stress disorder. Proc. Natl. Acad. Sci. USA. 2010;107:9470–9475. doi: 10.1073/pnas.0910794107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin M, Chang SC, Zhang C, Ressler K, Mercer KB, Galea S, Keyes KM, McLaughlin KA, Wildman DE, Aiello AE, Koenen KC. Adcyap1r1 genotype, posttraumatic stress disorder, and depression among women exposed to childhood maltreatment. Depress. Anxiety. 2013;30:251–258. doi: 10.1002/da.22037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin M, Koenen KC, Aiello AE, Wildman DE, de los Santos R, Galea S. Epigenetic and inflammatory marker profiles associated with depression in a community-based epidemiologic sample. Psychol. Med. 2011;41:997–1007. doi: 10.1017/S0033291710001674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Knaap LJ, Riese H, Hudziak JJ, Verbiest MM, Verhulst FC, Oldehinkel AJ, van Oort FV. Glucocorticoid receptor gene (NR3C1) methylation following stressful events between birth and adolescence. The TRAILS study. Transl. Psychiatry. 2014;4:e381. doi: 10.1038/tp.2014.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver IC, Cervoni N, Champagne FA, D’Alessio AC, Sharma S, Seckl JR, Dymov S, Szyf M, Meaney MJ. Epigenetic programming by maternal behavior. Nat. Neurosci. 2004;7:847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- Weaver IC, D’Alessio AC, Brown SE, Hellstrom IC, Dymov S, Sharma S, Szyf M, Meaney MJ. The transcription factor nerve growth factor-inducible protein a mediates epigenetic programming: altering epigenetic marks by immediate-early genes. J. Neurosci.: Off. J. Soc. Neurosci. 2007;27:1756–1768. doi: 10.1523/JNEUROSCI.4164-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelaw NC, Whitelaw E. How lifetimes shape epigenotype within and across generations. Hum. Mol. Genet. 2006;15(2):R131–R137. doi: 10.1093/hmg/ddl200. [DOI] [PubMed] [Google Scholar]
- Widom CS, Morris S. Accuracy of adult recollections of childhood victimization: Part 2. Childhood sexual abuse. Psychol. Assess. 1997;9:34–46. [Google Scholar]
- Widom CS, Shepard RL. Accuracy of adult recollections of childhood victimization: Part 1. Childhood physical abuse. Psychol. Assess. 1996;8:412–421. [Google Scholar]
- Yehuda R, Daskalakis NP, Bierer LM, Bader HN, Klengel T, Holsboer F, Binder EB. Holocaust exposure induced intergenerational effects on FKBP5 methylation. Biol. Psychiatry. 2015 doi: 10.1016/j.biopsych.2015.08.005. [DOI] [PubMed] [Google Scholar]