Abstract
Recently, the Fusarium genus has been narrowed based upon phylogenetic analyses and a Fusarium-like clade was adopted. The few species of the Fusarium-like clade were moved to new, re-installed or existing genera or provisionally retained as "Fusarium." Only a limited number of reference strains and DNA marker sequences are available for this clade and not much is known about its actual species diversity. Here, we report six strains, preserved by the Belgian fungal culture collection BCCM/IHEM as a Fusarium species, that belong to the Fusarium-like clade. They showed a slow growth and produced pionnotes, typical morphological characteristics of many Fusarium-like species. Multilocus sequencing with comparative sequence analyses in GenBank and phylogenetic analyses, using reference sequences of type material, confirmed that they were indeed member of the Fusarium-like clade. One strain was identified as "Fusarium" ciliatum whereas another strain was identified as Fusicolla merismoides. The four remaining strains were shown to represent a unique phylogenetic lineage in the Fusarium-like clade and were also found morphologically distinct from other members of the Fusarium-like clade. Based upon phylogenetic considerations, a new genus, Pseudofusicolla gen. nov., and a new species, Pseudofusicolla belgica sp. nov., were installed for this lineage. A formal description is provided in this study. Additional sampling will be required to gather isolates other than the historical strains presented in the present study as well as to further reveal the actual species diversity in the Fusarium-like clade.
Keywords: Fusarium, Fusicolla, Microcera, Nectriaceae, Phylogeny, Pseudofusicolla
In 2011, a phylogenetic study from Gräfenhan et al. [1] showed that the Fusarium genus (Hypocreales, Nectriaceae) was not monophyletic. They found that the genus was divided into two groups in their phylogenic tree of Nectriaceae, separated by a large number of species classified in genera such as Neonectria and Volutella [1]. These groups were referred to as the "basal Fusarium clade" and the "terminal Fusarium clade." In order to retain the monophyly of Fusarium, Gräfenhan et al. [1] narrowed the generic concept of Fusarium and the basal Fusarium clade was no longer considered as Fusarium sensu stricto. The few species and lineages represented in this Fusarium-like clade were moved to new, re-installed or existing genera (i.e., Atractium, Microcera, Macroconia, Fusicolla, Dialonectria, and Stylonectria), or provisionally retained as "Fusarium" or as one of its teleomorphs (i.e., Nectria and Cosmospora), based upon phylogenetic and morphological analyses [1,2].
Recently, Lombard et al. [3] phylogenetically re-evaluated the generic concepts in the Nectriaceae and confirmed the monophyly of each of these Fusarium-like genera. Lombard et al. [3] also further narrowed Fusarium sensu Gräfenhan et al. [1] according to its internal phylogenetic structure and thereby applied the "one fungus, one name" concept adopted in the International Code of Nomenclature for Algae, Fungi and Plants as of January 2013 in order to abolish the dual naming system (cfr. teleomorphic name is preferred). They installed a new genus Bisifusarium and applied several teleomorphic names for some important groups of Fusarium anamorphs. In doing so, Lombard et al. [3] rejected the "Fusarium-first" proposal made by the Fusarium working community [2] to conserve the name Fusarium above all linked teleomorphic names for the sake of nomenclatural stability as well as rejected their broad phylogenetic definition of Fusarium, which was based upon Fusarium sensu Gräfenhan et al. [1] and the refinements made by O'Donnell et al. [4].
Species of the Fusarium-like clade are mostly slow growing and produce a characteristic orange, conidial slime, known as pionnotes, rather than an aerial mycelium [1]. Species identification based upon morphology is difficult in the Fusarium-like clade. They often occur as saprophytes in the soil, on trees or other fungi, and in aquatic environments [1,2]. In contrast to the well-studied species diversity of Fusarium sensu Gräfenhan et al. [1], they have not been reported as plant pathogens or opportunistic human pathogens and only a few have been described to produce mycotoxins [2,5]. Members of the Fusarium-like clade have therefore been largely neglected for a long period of time. As a consequence, not much is known about the actual phylogenetic species diversity in this clade. It was only after the taxonomical revision of Gräfenhan et al. [1] that some DNA marker sequences became available.
Among the Fusarium strains preserved by the Belgian culture collection of biomedical fungi BCCM/IHEM, six appeared to belong to the Fusarium-like clade based upon their morphological appearance during re-identification [6]. All six strains were slow growing and produced pionnotes, i.e., typical morphological characteristics of only some Fusarium, but many Fusarium-like species. The objective of the present study was to determine whether these strains indeed belong to the Fusarium-like clade in the phylogeny of the Nectriaceae as well as to confirm their identity up to the genus and species level. This was achieved by performing multilocus sequencing and phylogenetic analyses using reference sequences of type material. Also morphological analyses and mating experiments were performed.
MATERIALS AND METHODS
Strains
The six strains, used in this study, were collected and preserved by the BCCM/IHEM fungal culture collection after their isolation from different substrates (Table 1). All were identified based upon their morphology and identified as a Fusarium species at the time of their deposit in the collection, more than 20 years ago. They were recently subjected to re-identification, together with the other Fusarium strains in the BCCM/IHEM collection [6].
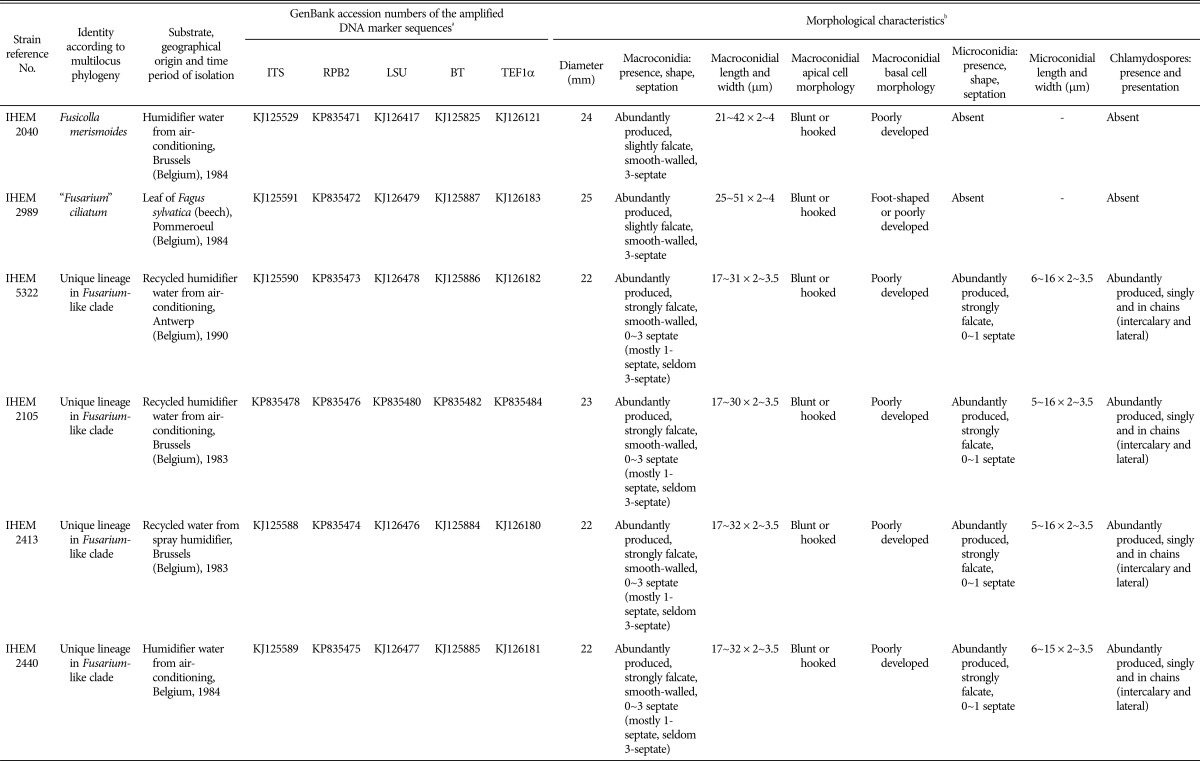
Table 1. Strains of the Belgian fungal culture collection BCCM/IHEM used in this study.
aDNA markers that were sequenced include the internal transcribed spacer (ITS), the ribosomal large subunit (LSU), the translation elongation factor 1-alpha (TEF1α), the beta-tubulin (BT) and the second largest subunit of RNA polymerase II (RPB2).
bDiameter after 14 days of growth on potato dextrose agar at 23℃ under alternating cycles of light (i.e., day) and dark (i.e., night) conditions. Other morphological characteristics were assessed from the synthetic nutrient agar plates after 30 days of growth at 23℃ under alternating cycles of light (i.e., day) and dark (i.e., night) conditions.
Morphological re-identification
The six strains were re-analyzed morphologically, according to the procedure previously applied by Hosoya and Tubaki [7] for their description of Fusicolla matuoi, a member of the Fusarium-like clade. This involved cultivation on nutrient-rich agar, i.e., potato dextrose agar (PDA), as well as nutrient-poor agar, i.e., synthetic nutrient agar (SNA) with or without fragments of carnation leaf, at 23℃ for 30 days under alternating cycles of light (i.e., day) and dark (i.e., night) conditions. Growth rates were measured from the PDA plates. Colony morphology, pigmentation, conidiogenesis, conidial characteristics, and presence of chlamydospores or other features were described from both the PDA and SNA plates. Conidial measurements were taken from 20 randomly selected conidia. A Nikon eclipse E600 microscope (Nikon, Tokyo, Japan) was used and pictures were recorded with the NIS-Elements BR 4.0 imaging software. The strains were described according to the terminology applied in "The Fusarium laboratory manual" of Leslie et al. [8].
Re-identification by multilocus sequencing
Multilocus sequencing was applied on the six strains and comparative sequence analyses were performed in GenBank. The internal transcribed spacer (ITS) region and part of the ribosomal large subunit (LSU) were amplified as well as partial fragments of the commonly used Fusarium DNA marker genes: translation elongation factor 1-alpha (TEF1α), beta-tubulin (BT), and the second largest subunit of RNA polymerase II (RPB2). DNA extraction, PCR amplification, and sequencing were achieved according to the protocol applied by Beguin et al. [9]. Primers were described previously: for ITS by White et al. [10], for LSU by Hopple and Vilgalys [11], for TEF1α by Carbone and Kohn [12], for BT by Glass and Donaldson [13], and for RPB2 by Van Hove et al. [14].
Phylogenetic analyses
In order to determine whether our six strains belong to the Fusarium-like clade in the phylogeny of the Nectriaceae, a Bayesian phylogenetic analysis was conducted. Therefore, we used the ITS and RPB2 reference sequences of type material published by Gräfenhan et al. [1] and included our strains. The combined sequence dataset was first aligned by the ClustalW algorithm in MEGA4 [15] and edited manually. Four ambiguously aligned regions (two in the ITS and two in the RPB2 gene marker) were excluded from the analysis. Subsequently, Bayesian inference analysis was executed with MrBayes3.2 [16], applying the GTR + I + Γ model of evolution for both loci and estimating parameters separately for each locus. The Monte Carlo Markov chain method was used with runs of one million generations and sampling a tree every 100 generations. The first 25% of sampled trees were discarded (i.e., burn-in) and the consensus tree, with posterior probabilities, was assessed from the remaining trees. The Acremonium sp. A104 strain from Grum-Grzhimaylo et al. [17] was chosen as outgroup. Tracer v1.5 [18] was used to check the convergence of the likelihood scores and the effective sample sizes for the different parameters.
In order to determine the phylogenetic position of our six strains, taking into account all currently known phylogenetic species diversity of the Fusarium-like clade, three other Bayesian phylogenetic trees were constructed according to the same methodology. A first by using the RPB2 reference sequences of type material from the Fusarium-like clade published by Gräfenhan et al. [1]. In doing so, more type material from the Fusarium-like clade could be included compared to the ITS-RPB2 combined tree, since all reference strains from Gräfenhan et al. [1] had been sequenced for RPB2, but not all of them had been sequenced for ITS. A second tree was constructed by using type material published by Bills et al. [5] from the "Fusarium" larvarum complex and the "Fusarium" merismoides complex, of which the members were respectively moved to Microcera and Fusicolla by Gräfenhan et al. [1]. The ITS, LSU, and BT reference sequences were used, excluding one ambiguously aligned region for the BT gene marker and using the Viridispora alata CBS 421.88 strain from Bills et al. [5] as outgroup. A third Bayesian phylogenetic tree was constructed by using the ITS reference sequences of the Fusarium-like type material from both the studies of Gräfenhan et al. [1] and Bills et al. [5]. Also for this tree, we used the Viridispora alata CBS 421.88 strain from Bills et al. [5] as outgroup.
Mating experiments
Sexual crossing experiments were carried out between IHEM 2105, IHEM 2413, IHEM 2440, and IHEM 5322 by co-incubating each time two strains on V8 Juice Agar (VJA) at 23℃ for 30 days under alternating cycles of light (i.e., day) and dark (i.e., night) conditions.
RESULTS
Morphological re-identification
The six strains showed a colony diameter between 22~25 mm after 14 days of growth on PDA. All six strains produced a dense sterile, white to (pale) orange, aerial mycelium on PDA (reverse cream colored) and pionnotes on SNA with or without fragments of carnation leaf. Orange pigmentation was more pronounced for IHEM 2040 than for the other strains. No aerial mycelium was observed on SNA for any of the strains and conidiogenesis started after a few days from phialides, consisting of branched conidiogenous cells or conidiogenous cells scattered along branched hyphae. Growth rates and conidial measurements as well as information about other morphological characteristics of the strains are enlisted in Table 1 and pictures from the macroscopic/microscopic observations are shown in Fig. 1A~1I and Fig. 2A~2C.
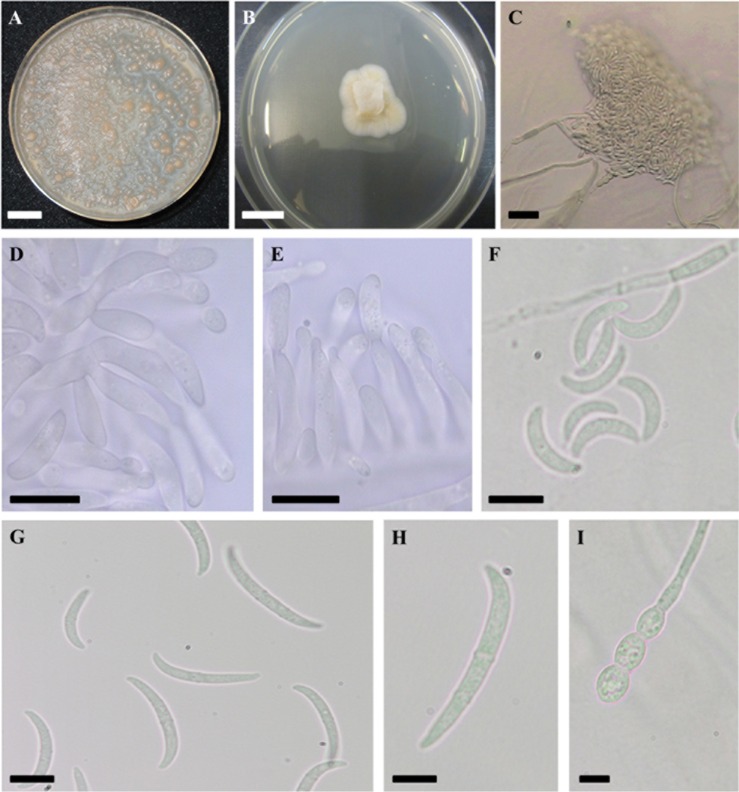
Fig. 1. Morphological characterization of Pseudofusicolla gen. nov. and Pseudofusicolla belgica sp. nov. (type strain IHEM 2413). A, Pionnotal growth after 30 days on synthetic nutrient agar (SNA); B, Sterile hyphal growth after 7 days on potato dextrose agar; C~I, Characteristics observed on SNA; C, Mass of conidia; D, E, Conidiogenesis; F, Microconidia; G, Macroconidia; H, Characteristic one-septate macroconidium; I, Chlamydospores (scale bars: A, B = 1 cm, C = 20 µm, D, E, H, I=5 µm, F, G = 10 µm).
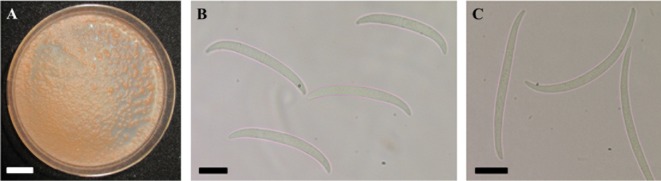
Fig. 2. Macroscopic and microscopic observations made for Fusicolla merismoides IHEM 2040 (A, B) and "Fusarium" ciliatum IHEM 2989 (C). A, Pionnotal growth after 30 days on synthetic nutrient agar (SNA); B, C, Macroconidia on SNA (scale bars: A = 1 cm, B, C = 10 µm).
Re-identification by multilocus sequencing
All sequences were deposited in GenBank with the accession numbers given in Table 1. By performing comparative sequence analyses in GenBank, using the amplified sequences for the different loci, we could provisionally identify two of our strains according to their RPB2 sequence. IHEM 2040 and IHEM 2989 were identified as Fusicolla epistroma (with 99% sequence similarity) and "Fusarium" ciliatum (with 100% sequence similarity), respectively, both members of the Fusarium-like clade. The identification of IHEM 2989 based upon its RPB2 sequence was confirmed by a BLAST query with its ITS sequence (99% sequence similarity), but not by a BLAST query with its BT sequence (only 94% similarity with a "Fusarium" merismoides strain). Also the RPB2 identification of IHEM 2040 could not be confirmed by a BLAST query with its ITS sequence (99% similarity with uncultured fungal strains) nor by a BLAST query with its BT sequence, which lead to a Fusicolla merismoides identification (99% sequence similarity).
No identification could be obtained for IHEM 2105, IHEM 2413, IHEM 2440, and IHEM 5322, of which the sequences were 100% identical for all five amplified DNA markers. Nevertheless, the highest similarity scores, using the RPB2 and BT sequences of these strains, were, although low (i.e., 85% and 92%, respectively), associated with members of the Fusarium-like clade (i.e., Fusicolla aquaeductuum and Microcera larvarum, respectively). Moreover, comparative sequence analyses performed with the ITS sequences resulted in highest similarity scores (i.e., 99%) associated with sequences of uncultured fungal strains from aquatic habitats or from soils. These strains could, as such, not be allocated to any described taxon.
For all six strains, screening of the LSU sequences resulted in highest similarities (i.e., 99%) with those of unidentified members of the Hypocreales, whereas with the TEF1α sequences, no matching sequences were found (query cover of < 20% for five of the six strains).
Phylogenetic analyses
The four Bayesian phylogenetic analyses had effective sample sizes higher than 100 for all parameters, showing sufficient sampling and acceptable mixing of the runs. The obtained consensus trees are illustrated (Figs. 3, 4, 5, 6) and were each generated from the 15002 remaining trees after burn-in.
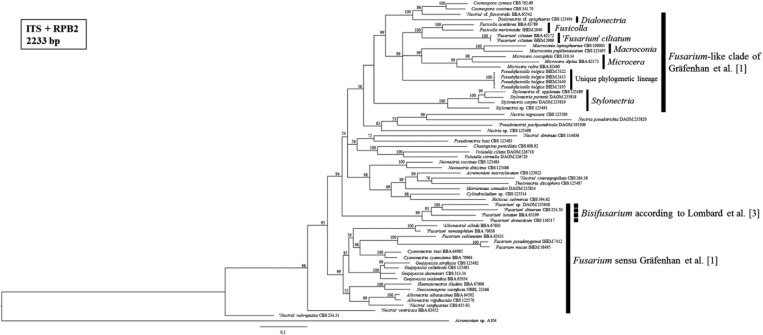
Fig. 3. Consensus tree from the Bayesian phylogenetic analysis, using the internal transcribed spacer (ITS) and second largest subunit of RNA polymerase II (RPB2) reference sequences of nectriaceous fungi published by Gräfenhan et al. [1] and including the strains of the Belgian fungal culture collection BCCM/IHEM used in this study (i.e., IHEM 2040, IHEM 2989, IHEM 5322, IHEM 2105, IHEM 2413, and IHEM 2440). Posterior probabilities (%) are represented at the nodes of the tree. An Acremonium sp. strain was chosen as outgroup. We can distinguish the Fusarium and Fusarium-like clade as defined by Gräfenhan et al. [1], being separated from each other by a large number of species from different genera. The Fusarium sensu Gräfenhan et al. [1] clade in our tree is not monophyletic and the aberrant taxa, for which the genus Bisifusarium was installed by Lombard et al. [3], are indicated by a dashed line. Our six strains with presumed Fusarium-like identities are all embedded in the Fusarium-like clade, for which the different genera, as defined by Gräfenhan et al. [1], are shown.
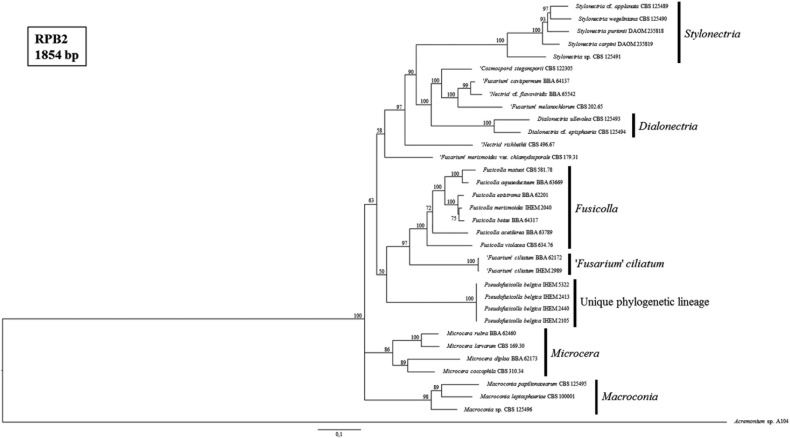
Fig. 4. Consensus tree from the Bayesian phylogenetic analysis, using the second largest subunit of RNA polymerase II (RPB2) reference sequences of species from the Fusarium-like clade published by Gräfenhan et al. [1] and including the strains of the Belgian fungal culture collection BCCM/IHEM used in this study (i.e., IHEM 2040, IHEM 2989, IHEM 5322, IHEM 2105, IHEM 2413, and IHEM 2440). Posterior probabilities (%) are represented at the nodes of the tree. An Acremonium sp. strain was chosen as outgroup. The different genera of the Fusarium-like clade, as defined by Gräfenhan et al. [1], are indicated.
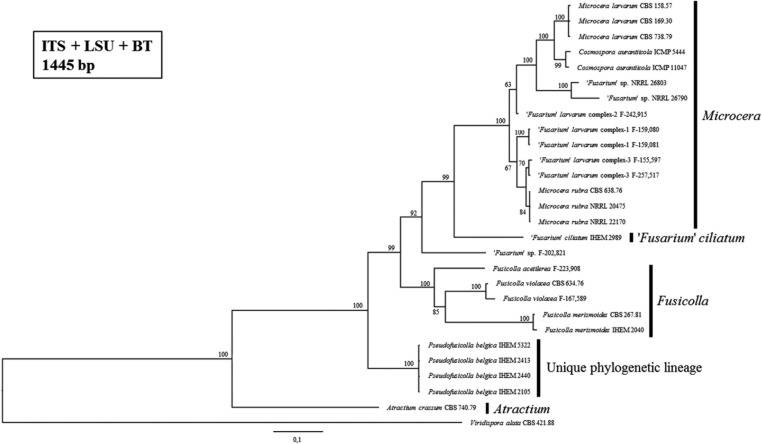
Fig. 5. Consensus tree from the Bayesian phylogenetic analysis, using the internal transcribed spacer (ITS), ribosomal large subunit (LSU), and beta-tubulin (BT) reference sequences of species from the Fusarium-like clade published by Bills et al. [5] and including the strains of the Belgian fungal culture collection BCCM/IHEM used in this study (i.e., IHEM 2040, IHEM 2989, IHEM 5322, IHEM 2105, IHEM 2413, and IHEM 2440). Posterior probabilities (%) are represented at the nodes of the tree. A Viridispora alata strain was chosen as outgroup. The different genera of the Fusarium-like clade, as defined by Gräfenhan et al. [1], are indicated.
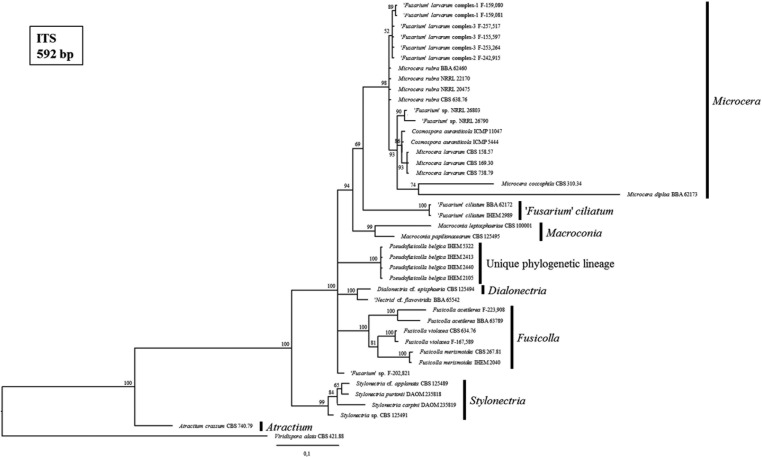
Fig. 6. Consensus tree from the Bayesian phylogenetic analysis, using the internal transcribed spacer (ITS) reference sequences of species from the Fusarium-like clade published by Gräfenhan et al. [1] as well as Bills et al. [5] and including the strains of the Belgian fungal culture collection BCCM/IHEM used in this study (i.e., IHEM 2040, IHEM 2989, IHEM 5322, IHEM 2105, IHEM 2413, and IHEM 2440). Posterior probabilities (%) are represented at the nodes of the tree. A Viridispora alata strain was chosen as outgroup. The different genera of the Fusarium-like clade, as defined by Gräfenhan et al. [1], are indicated.
In the phylogenetic tree of nectriaceous fungi (Fig. 3), we could distinguish the Fusarium and Fusarium-like clade as defined by Gräfenhan et al. [1]. Similar to their study, these groups were separated from each other by a large number of species classified in different genera. Though, in contrary to what is seen in the tree of Gräfenhan et al. [1], which was constructed using sequences of RPB2 and the larger subunit of ATP citrate lyase, Fusarium sensu Gräfenhan et al. [1] formed no monophyletic clade in our ITS-RPB2 tree of nectriaceous fungi. The aberrant taxa were the same as those who received a provisional status as "Fusarium" in the reference phylogeny of O'Donnell et al. [4] (used by Geiser et al. [2] for their definition of the Fusarium genus) and are now members of Bisifusarium, the new genus installed by Lombard et al. [3].
The BCCM/IHEM strains with presumed Fusarium-like identities were all embedded in the Fusarium-like clade (Fig. 3). IHEM 2040 and IHEM 2989 were respectively most closely related to the reference strain of the Fusicolla genus and the "Fusarium" ciliatum reference strain. The strains IHEM 2105, IHEM 2413, IHEM 2440, and IHEM 5322 formed a well-supported (i.e., 100% posterior probability) phylogenetic lineage in the Fusarium-like clade, distinct from all the monophyletic Fusarium-like genera as defined by Gräfenhan et al. [1]. This unique, distinct lineage was also observed and supported by a maximum posterior probability in all our phylogenetic trees of Fusarium-like species only (Figs. 4, 5, 6).
Similar as in our tree of nectriaceous fungi (Fig. 3), IHEM 2989 clustered together with the "Fusarium" ciliatum reference strain in a separate and well-supported clade in both the ITS and RPB2 tree of Fusarium-like species only (Figs. 4 and 6). This was not the case in the ITS-LSU-BT tree (Fig. 5), for which no "Fusarium" ciliatum reference strain could be included due to the absence of a BT reference sequence. These observations are in agreement with the identification obtained for IHEM 2989 by performing sequence similarity searches in GenBank. Consequently, IHEM 2989 is identified as "Fusarium" ciliatum.
IHEM 2040, on the other hand, appeared to be most closely related to Fusicolla merismoides in the ITS-LSU-BT tree as well as the ITS tree of Fusarium-like species only (Figs. 5 and 6). This was not the case in the ITS-RPB2 tree (Fig. 3) nor in the RPB2 tree (Fig. 4), for which no Fusicolla merismoides reference strain could be included due to the absence of a RPB2 reference sequence. Again, these observations are in agreement with the provisional identifications obtained for IHEM 2040 by performing sequence similarity searches in GenBank. Consequently, IHEM 2040 is identified as Fusicolla merismoides.
Mating experiments
All sexual crosses were negative and pionnotal growth was observed after a few days on VJA, together with a sparse, white aerial mycelium.
Taxonomic description
Pseudofusicolla
Triest, gen. nov. (Fig. 1A~1I). MycoBank No. MB 811910.
Etymology: Latin "Pseudofusicolla" = like Fusicolla; Pseudofusicolla is a genus in the Fusarium-like clade as defined by Gräfenhan et al. [1] and shows morphological resemblances with Fusicolla.
Description: Dense sterile, white to pale orange, aerial mycelium on PDA and pionnotal growth on SNA with or without fragments of carnation leaf. On SNA: conidiogenesis starting after a few days from phialides, consisting of branched conidiogenous cells or conidiogenous cells scattered along branched hyphae; microconidia strongly falcate, aseptate or one-septate; macroconidia strongly falcate, smooth-walled, aseptate to three-septate; chlamydospores present. Teleomorph unknown.
Type species: Pseudofusicolla belgica Triest, sp. nov. (Fig. 1A~1I). MycoBank No. MB 812587.
Description: On PDA: colony diameter 22~23 mm after 14 days at 23℃; sterile hyphal growth and dense, white to pale orange, aerial mycelium, reverse cream colored. On SNA with or without fragments of carnation leaf: slow pionnotal growth, no aerial mycelium; conidiogenesis starting after a few days from phialides, consisting of branched conidiogenous cells or conidiogenous cells scattered along branched hyphae; microconidia abundantly produced, strongly falcate, aseptate or one-septate, 5~16 × 2~3.5 µm; macroconidia abundantly produced, strongly falcate, smooth-walled, aseptate to three-septate (mostly one-septate, seldom three-septate), 17~32 × 2~3.5 µm, apical cell blunt or hooked, basal cell poorly developed; chlamydospores abundantly produced, singly and in chains (intercalary and lateral). Teleomorph not observed.
Type strain: Belgium, Brussels, isolated from recycled water from spray humidifier, collected by the BCCM/IHEM collection in 1983, permanently inactivated but living strain preserved by the BCCM/IHEM collection (holotype, IHEM 2413).
Additional specimens examined: Belgium, Antwerp, isolated from recycled humidifier water from air-conditioning, collected by the BCCM/IHEM collection in 1990, IHEM 5322; Belgium, Brussels, isolated from recycled humidifier water from air-conditioning, collected by the BCCM/IHEM collection in 1983, IHEM 2105; Belgium, isolated from humidifier water from air-conditioning, collected by the BCCM/IHEM collection in 1984, IHEM 2440; permanently inactivated but living strains preserved by the BCCM/IHEM collection.
Habitat: Recycled water from spray humidifier and airconditioners.
Known distribution: Belgium.
Etymology: "belgica" refers to Belgium, i.e., the country in which all Pseudofusicolla belgica sp. nov. strains, presented in this research, were isolated.
Notes: Pseudofusicolla belgica sp. nov. forms a distinct and well-supported phylogenetic lineage in multilocus phylogenies of the Fusarium-like clade. Pseudofusicolla gen. nov. is installed in order to retain the monophyly of the different genera as defined in the Fusarium-like clade according to Gräfenhan et al. [1] and Lombard et al. [3].
DISCUSSION
The Fusarium-like clade, of which most members were formerly classified in the Fusarium sections Arachnites, Eupionnotes, Macroconia, Pseudomicrocera, and Submicrocera [2], is clearly phylogenetically distinct from Fusarium sensu stricto. This was shown by Gräfenhan et al. [1] as well as Lombard et al. [3] and is also confirmed in the present study (Fig. 3).
Re-identification of six "Fusarium" strains from the BCCM/IHEM fungal culture collection revealed a Fusarium-like identity for each of them, supported both morphologically and phylogenetically. All strains were slow growing and produced pionnotes, i.e., typical morphological characteristics of many Fusarium-like species (Table 1, Figs. 1A and 2A). Since the majority of Fusarium-like species have descriptions which are often not well-documented or confirmed [1], reliable morphological identification up to the species level or even genus level was not possible. Because sequence data and phylogenetic analysis seems indispensable for species identification in the Fusarium-like clade, a multilocus phylogenetic approach, using reference sequences of type material, was applied.
We identified one strain as Fusicolla merismoides (i.e., IHEM 2040). Fusicolla, which has for a long time been considered a synonym of Fusarium, was installed by Gräfenhan et al. [1] as a separate genus containing most of the former "Fusarium" merismoides varieties of which some were raised to species rank. Another strain was identified as "Fusarium" ciliatum (i.e., IHEM 2989), a known member of the Fusarium-like clade provisionally retained as "Fusarium" by Gräfenhan et al. [1].
The remaining four strains (i.e., IHEM 2105, IHEM 2413, IHEM 2440, and IHEM 5322) were also embedded in the Fusarium-like clade of our phylogenetic tree of nectriaceous fungi (Fig. 3), but their sequences, which were identical for all tested DNA markers, showed limited similarity with those published by Gräfenhan et al. [1] or in public databases in order to perform species/genus identification. Moreover, they formed a distinct and maximum posterior probability supported lineage in this phylogenetic tree as well as in the ones of Fusarium-like species only (Figs. 4, 5, 6). Based upon these phylogenetic observations and in order to retain the monophyly of the different genera as defined in the Fusarium-like clade according to Gräfenhan et al. [1] as well as Lombard et al. [3], we installed a new genus, Pseudofusicolla gen. nov., and a new species, Pseudofusicolla belgica sp. nov., to describe this unique phylogenetic lineage. Morphologically, Pseudofusicolla belgica strains resemble somewhat to immature strains of Microcera larvarum and Dialonectria spp., which also form relatively small and strongly falcate aseptate to two-septate macroconidia [1,19]. Though, when mature, Microcera larvarum and Dialonectria spp. predominantly produce three-septate macroconidia, whereas this type of macroconidia is only rarely observed in the cultures of our Pseudofusicolla belgica strains (Fig. 1G). The predominant occurrence of one-septate macroconidia (Fig. 1H) appears to be discriminatory for the species as opposed to the other members of the Fusarium-like clade, which generally form macroconidia that are three-septate or more, as was seen in our cultures of IHEM 2989 and IHEM 2040 (Table 1, Fig. 2B and 2C). Microconidia were also detected in all SNA cultures of Pseudofusicolla belgica (Fig. 1F), though similar as in Fusicolla matuoi and other Fusicolla spp., these form a continuum in conidial shape and length with the macroconidia [1,7]. Moreover, our Pseudofusicolla belgica strains produced chlamydospores (Fig. 1I). But, no teleomorph was found after performing crossing experiments.
Five of the six IHEM Fusarium-like strains discussed in this study had been isolated from humidifier water of either a spray or air-conditioners from different localities in Belgium (Table 1). Only the "Fusarium" ciliatum strain (i.e., IHEM 2989) had been isolated from the leaf of a beech tree. Additional sampling is required to gather strains other than the historical strains presented in this study (cfr. collected over 20 years ago) as well as to tell us something more about the ecological niches of Fusarium-like species. For example: do humidifiers act as reservoirs for Fusarium-like species? Our generated and published DNA marker sequences will aid this search. However, sequence similarity searches in GenBank with the ITS sequences of our isolates, showed us that a substantial part of the Fusarium-like species diversity might be hidden due to the inability to culture them.
In conclusion, we found a unique phylogenetic lineage in the Fusarium-like clade after re-examining collection material from the Belgian fungal culture collection BCCM/IHEM and described this new taxon as Pseudofusicolla belgica, thereby expanding our knowledge about the Fusarium-like species diversity. More isolates will need to be gathered in order to further reveal the actual phylogenetic species diversity in the Fusarium-like clade.
ACKNOWLEDGEMENTS
This study was funded by the Belgian Science Policy (BELSPO). We would like to thank Dr. Dirk Stubbe (BCCM/IHEM Collection of Biomedical Fungi) and Hugues Beguin (BCCM/IHEM Collection of Biomedical Fungi) for the suggestions made to improve our manuscript.
References
- 1.Gräfenhan T, Schroers HJ, Nirenberg HI, Seifert KA. An overview of the taxonomy, phylogeny, and typification of nectriaceous fungi in Cosmospora, Acremonium, Fusarium, Stilbella, and Volutella. Stud Mycol. 2011;68:79–113. doi: 10.3114/sim.2011.68.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Geiser DM, Aoki T, Bacon CW, Baker SE, Bhattacharyya MK, Brandt ME, Brown DW, Burgess LW, Chulze S, Coleman JJ, et al. One fungus, one name: defining the genus Fusarium in a scientifically robust way that preserves the longstanding use. Phytopathology. 2013;103:400–408. doi: 10.1094/PHYTO-07-12-0150-LE. [DOI] [PubMed] [Google Scholar]
- 3.Lombard L, van der Merwe NA, Groenewald JZ, Crous PW. Generic concepts in Nectriaceae. Stud Mycol. 2015;80:189–245. doi: 10.1016/j.simyco.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O'Donnell K, Rooney AP, Proctor RH, Brown DW, McCormick SP, Ward TJ, Frandsen RJ, Lysøe E, Rehner SA, Aoki T, et al. Phylogenetic analyses of RPB1 and RPB2 support a middle Cretaceous origin for a clade comprising all agriculturally and medically important fusaria. Fungal Genet Biol. 2013;52:20–31. doi: 10.1016/j.fgb.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 5.Bills GF, Platas G, Overy DP, Collado J, Fillola A, Jiménez MR, Martín J, del Val AG, Vicente F, Tormo JR, et al. Discovery of the parnafungins, antifungal metabolites that inhibit mRNA polyadenylation, from the Fusarium larvarum complex and other Hypocrealean fungi. Mycologia. 2009;101:449–472. doi: 10.3852/08-163. [DOI] [PubMed] [Google Scholar]
- 6.Triest D, Stubbe D, De Cremer K, Piérard D, Normand AC, Piarroux R, Detandt M, Hendrickx M. Use of matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of molds of the Fusarium genus. J Clin Microbiol. 2015;53:465–476. doi: 10.1128/JCM.02213-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hosoya T, Tubaki K. Fusarium matuoi sp. nov. and its teleomorph Cosmospora matuoi sp. nov. Mycoscience. 2004;45:261–270. [Google Scholar]
- 8.Leslie JF, Summerell BA, Bullock S. The Fusarium laboratory manual. Hoboken (NJ): Wiley-Blackwell; 2006. [Google Scholar]
- 9.Beguin H, Pyck N, Hendrickx M, Planard C, Stubbe D, Detandt M. The taxonomic status of Trichophyton quinckeanum and T. interdigitale revisited: a multigene phylogenetic approach. Med Mycol. 2012;50:871–882. doi: 10.3109/13693786.2012.684153. [DOI] [PubMed] [Google Scholar]
- 10.White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. London: Academic Press; 1990. pp. 315–322. [Google Scholar]
- 11.Hopple JS, Jr, Vilgalys R. Phylogenetic relationships in the mushroom genus Coprinus and dark-spored allies based on sequence data from the nuclear gene coding for the large ribosomal subunit RNA: divergent domains, outgroups, and monophyly. Mol Phylogenet Evol. 1999;13:1–19. doi: 10.1006/mpev.1999.0634. [DOI] [PubMed] [Google Scholar]
- 12.Carbone I, Kohn LM. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia. 1999;91:553–556. [Google Scholar]
- 13.Glass NL, Donaldson GC. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl Environ Microbiol. 1995;61:1323–1330. doi: 10.1128/aem.61.4.1323-1330.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van Hove F, Waalwijk C, Logrieco A, Munaut F, Moretti A. Gibberella musae (Fusarium musae) sp. nov., a recently discovered species from banana is sister to F. verticillioides. Mycologia. 2011;103:570–585. doi: 10.3852/10-038. [DOI] [PubMed] [Google Scholar]
- 15.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 16.Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grum-Grzhimaylo AA, Georgieva ML, Debets AJ, Bilanenko EN. Are alkalitolerant fungi of the Emericellopsis lineage (Bionectriaceae) of marine origin? IMA Fungus. 2013;4:213–228. doi: 10.5598/imafungus.2013.04.02.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rambaut A, Suchard M, Drummond A. Tracer v1.5 software [Internet] Andrew Rambaut; 2007. [cited 2016 May 2]. Available from: http://tree.bio.ed.ac.uk/software/tracer/ [Google Scholar]
- 19.Booth C. The genus Fusarium. Kew: Commonwealth Mycological Institute; 1971. [Google Scholar]