Abstract
Shiga toxin-producing Escherichia coli (STEC)-associated enteric illness is attributed to O157 and non-O157 serotypes; however, traditional culture-based methods underdetect non-O157 STEC. Labor and cost of consumables are major barriers to implementation of the CDC recommendation to test all stools for both O157 and non-O157 serotypes. We evaluated the feasibility of a pooled nucleic acid amplification test (NAAT) as an approach for screening stool specimens for STEC. For retrospective evaluation, 300 stool specimens were used to create pools of 10 samples each. The sensitivity was 83% for the preenrichment pooling strategy and 100% for the postenrichment pooling strategy compared with those for individual NAAT results. The difference in cycle threshold (CT) between individual and pooled NAAT results for specimens was significantly lower and more consistent for postenrichment pooling (stx1 mean = 3.90, stx2 mean = 4.28) than those for preenrichment pooling (excluding undetected specimens; stx1 mean = 9.34, stx2 mean = 8.96) (P ≤ 0.0013). Cost of consumables and labor time savings of 48 to 81% and 6 to 66%, respectively, were estimated for the testing of 90 specimens by the postenrichment pooled NAAT strategy on the basis of an expected 1 to 2% positivity rate. A 30-day prospective head-to-head clinical trial involving 512 specimens confirmed the sensitivity and labor savings associated with the postenrichment pooled NAAT strategy. The postenrichment pooled NAAT strategy described here is suitable for efficient large-scale surveillance of all STEC serotypes. Comprehensive detection of STEC will result in accurate estimation of STEC burden and, consequently, appropriate public health interventions.
INTRODUCTION
Shiga toxin-producing Escherichia coli (STEC) is a major cause of sporadic and outbreak-associated enteric illness worldwide. While more than 100 STEC serotypes can cause illness in humans (1), traditional testing methods focus primarily on the most common serotype, O157:H7, owing to the ease of its detection using culture-based media. These methods underdetect non-O157 serogroups, and as such, their clinical burden is not well understood (2–4). Moreover, recent evidence suggests that infections attributed to non-O157 STEC may be more prevalent than those attributed to O157 (1, 5), with common serogroups being O26, O45, O103, O111, O121, and O145 in the United States (6, 7) and in other countries (1). Although non-O157 STEC serotypes are generally associated with milder disease than O157 STEC, infections caused by non-O157 STEC can also lead to hemolytic uremic syndrome (1) and have been associated with major outbreaks, most notably a 2011 outbreak in Germany that involved 3,816 cases and 54 deaths (8). Enhanced surveillance evidence is needed to determine the true burden of non-O157 STEC for public health investigations of potential exposure sources. Such efforts are ongoing in Canada (9) but require improved and comprehensive screening methods.
The Centers for Disease Control and Prevention (CDC) recommend that, in addition to O157-selective culture-based screening, all stools submitted for testing from patients with acute community-acquired diarrhea be assayed for non-O157 STEC with a test that detects the Shiga toxins or their genetic determinants (stx1 and stx2) (10). Given the low prevalence of STEC infections (1 to 2%) (11–13), the associated costs for clinical laboratories for personnel, equipment, and reagents are major barriers for implementation of the recommended universal screening of stool for STEC (4, 14). To reduce testing costs, we evaluated the feasibility of a pooled nucleic acid amplification test (NAAT) as an approach for low-cost high-throughput screening for stx1 and stx2 from stool. Similar approaches have been successfully applied for the detection of HIV (15, 16), hepatitis C (16, 17), and malaria (18, 19). Pooled NAAT strategies are best suited for scenarios where testing volumes are high but disease prevalence is low, as would be the case for universal stool screening.
MATERIALS AND METHODS
Retrospective clinical samples.
Stool specimens are routinely submitted to the British Columbia Centre for Disease Control (BCCDC) Public Health Laboratory (PHL) for stx detection by quantitative PCR (qPCR). Specimens that were positive by qPCR were subsequently cultured on sorbitol MacConkey agar (SMAC); suspect O157 (colorless) or non-O157 (pink) colonies were confirmed to be Shiga toxin producing by qPCR and serotyped using the Kaufmann classification scheme. Specimens were selected for the retrospective study on the basis of previous qPCR cycle threshold (CT) values and recovered isolate serogroups. Specifically, samples were chosen to represent a wide range of CT and serogroup findings. Previously stx-positive specimens (n = 30) were stored at −80°C prior to testing and evenly represented stx1, stx2, and stx1, and stx2 genotypes from recovered isolates (n = 10 each) (Table 1). Previously stx-negative specimens (n = 270) were randomly chosen from fresh samples kept at 4°C for up to 3 weeks.
TABLE 1.
Stool samples used in retrospective evaluation by STEC genotype, serotype, and individual NAAT CT value
| Serotypea | qPCR CT for samplesb |
|
|---|---|---|
| stx1 positive | stx2 positive | |
| O26:H11 | 20.00 | |
| O103:H2 | 17.68 | |
| O126:H27 | 17.52 | |
| O1:NM | 14.65 | |
| O103:H2 | 18.09 | |
| O71:H11 | 23.95 | |
| O43:H19 | 20.02 | |
| O76:H19 | 20.75 | |
| O43:H11 | 24.80 | |
| O111:NM | 22.38 | |
| O44:NM | 28.34 | |
| O145:NM | 14.08 | |
| O121:UT | 15.99 | |
| O150:H8 | 28.18 | |
| O43:H2 | 21.33 | |
| O174:UK | 15.19 | |
| O26:H11 | 15.64 | |
| O26:H11 | 21.56 | |
| O165:NM | 17.01 | |
| O121:H19 | 25.79 | |
| O Rough:H2 | 32.46 | 33.75 |
| O157:H7 | 28.96 | 25.25 |
| O157:H7 | 19.85 | 16.35 |
| O157:NM | 22.40 | 18.84 |
| O111:NM | 17.49 | 19.39 |
| O111:NM | 17.17 | 19.38 |
| O121:H19 | 28.47 | 25.36 |
| O157:H7 | 19.14 | 21.45 |
| O157:H7 | 17.75 | 20.13 |
| O157:H7 | 33.87 | 30.23 |
NM, nonmotile; UT, untypeable; UK, unknown.
Value from postenrichment sample.
Bacterial enrichment, DNA extraction, and qPCR.
Overnight enrichments (200 μl or “pea” size) in 5 ml brain heart infusion (BHI) broth (Bacto, Mississauga, ON, Canada), DNA extraction, and duplex quantitative PCR (qPCR) methods for stx detection from stool were performed as previously described (20) with the following modifications: (i) PerfeCTa qPCR ToughMix (UNG, Low ROX; Quanta Biosciences, Gaithersburg, MD, USA) was used for qPCR with an ABI 7500 Fast System (Life Technologies, Grand Island, NY, USA) and an initial 5-min incubation at 45°C, and (ii) FAM/ZEN/IABkFQ-labeled stx1 and MAX/IABkFQ-labeled stx2 TaqMan probes (Integrated DNA Technologies, Coralville, IA, USA) were used. The limit of detection (LoD) was determined from serial dilutions of cultures of three stx1 and three stx2 clinical isolates spiked into an enriched stx-negative specimen performed in triplicate. Briefly, isolates grown overnight in BHI broth were serially diluted 10-fold to achieve dilutions of approximately 109 to 101 CFU/ml. Overnight-enriched STEC-negative stool was spiked with each dilution at a 10:1 ratio, and subsequent DNA extraction and qPCR was performed as described above. In addition, 100-μl aliquots of 104, 103, and 102 dilutions were plated in duplicates for colony counts taken after 24 h at 37°C. These counts were used to calculate the average actual numbers of CFU/ml of the dilutions used. The LoD was defined as the CFU/ml concentration that was consistently detected by three technical repeats of qPCR.
Retrospective evaluation of preenrichment and postenrichment pooling strategies.
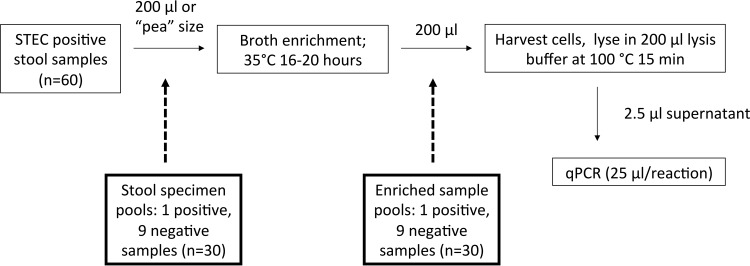
Pooling of stool specimens was performed using two approaches, where pooling occurred either prior to enrichment in BHI broth or after enrichment (Fig. 1). Pea-sized samples of solid stool or 200 μl of liquid stool were used. For preenrichment pooling, nine stx-negative stools were mixed with one stx-positive stool in 5 ml BHI broth prior to overnight enrichment. Enrichment, DNA extraction, and qPCR were then performed as described above. For postenrichment pooling, 10 stool samples were enriched separately overnight in 5 ml BHI broth before 200-μl samples from each specimen were combined. Accuracy and sensitivity, as determined by the change in CT (ΔCT), were evaluated for the two pooling strategies relative to individual specimen testing that was performed in parallel on postenrichment specimens.
FIG 1.
Methods for the detection of STEC by enriched broth extraction and stx gene qPCR. Shown is the testing protocol with introduction of two pooled NAAT strategies, where stool specimens are pooled either preenrichment or postenrichment.
Calculation of cost and time requirements.
The cost of consumables was calculated on the basis of U.S. catalog pricing at the time of the study and is shown in dollars. Consumables included plastics required for extraction and qPCR procedures, primers, probes, buffers, and disinfecting agents. Cost calculations incorporated amounts of consumables required to run two positive controls and one negative (no-template) control. Labor costs were not included in these calculations because they vary greatly among institutions. Labor time for calculations included hands-on time for extraction and qPCR procedures, room preparation and sample retrieval to room cleanup, sample storage, and result analysis and reporting. Labor time excluded events with set times, such as incubations and instrument run times; these, combined with hands-on time, were captured in the reported turn-around-times.
Prospective clinical evaluation of postenrichment pooling.
Over 30 days during June, August, September, and October 2014, postenrichment pooling for stx detection by qPCR was performed in parallel with routine individual specimen stx qPCR at the BCCDC PHL using the same enrichment broth. Historical data indicate that peak STEC detection in British Columbia occurs from June to October. A total of 512 specimens were tested. Resolution of the positive pools was performed the following day because pooled NAATs were started in the afternoon. To accurately compare hands-on time between the individual and pooled NAAT approaches, each day, the technical staff recorded the start and end times associated with procedures that varied depending on the number of samples and/or the testing strategy used. Start time was defined as the time of room preparation for DNA extraction of overnight stool cultures. End time was defined as the time following room cleanup after the start of stx qPCR.
Statistical analysis.
Differences between pooling strategies were determined by paired Student's t tests using GraphPad QuickCalcs Software (La Jolla, CA). The performances of the pooled qPCR strategies (sensitivity) were calculated in comparison to individual qPCR results.
RESULTS
Retrospective evaluation of pre- and postenrichment pooling strategies for STEC detection by NAAT.
The qPCR limits of detection for specimens tested individually were 7.23 × 102 for stx1 and 1.03 × 103 CFU/ml for stx2. A total of 300 patient stool specimens (30 stx positive, 270 stx negative) were used to evaluate two different pooling strategies simultaneously as approaches for high-throughput stx detection. Pool resolution, which would normally follow positive pool test results, was not performed at this time. The overall sensitivity for positive samples was 83% for the preenrichment pooling strategy and 100% for the postenrichment pooling strategy relative to individual qPCR results (Table 2). Two stx1, one stx2, and two stx1 and stx2 samples positive by individual NAAT (10 of each were tested in total) were not detected by the preenrichment pooled NAAT. Prepooling CT values of samples undetected in preenrichment pools ranged from 17.52 to 33.75, suggesting that factors other than the concentration of STEC present in the specimen (as reflected by the relative CT values) were at play. Therefore, the loss of detection in the preenrichment pooling evaluation were not predicted using the stx genotype or CT value of individually tested specimens. Furthermore, for both stx genes, the ΔCT between individual and pooled NAAT results for specimens was significantly lower and more consistent for postenrichment pooling (overall means ± standard deviation [SD]: stx1 = 3.90 ± 1.08, stx2 = 4.28 ± 1.36) than those from preenrichment pooling (overall mean ± SD excluding specimens that were undetected in pools: stx1 = 9.34 ± 5.18, stx2 = 8.96 ± 5.83) (P ≤ 0.0013) (Table 2). On the basis of average pooling ΔCT results, the theoretical qPCR LoDs for preenrichment pooling were 6.96 × 105 for stx1 and 8.67 × 105 CFU/ml for stx2, while the qPCR LoDs for postenrichment pooling were 1.27 × 104 for stx1 and 2.57 × 104 CFU/ml for stx2.
TABLE 2.
Results of a retrospective evaluation of two pooled NAAT techniques for STEC detection
| Category | Preenrichment poolinga |
Postenrichment pooling |
||||
|---|---|---|---|---|---|---|
| stx1 | stx2 | stx1, stx2 | stx1 | stx2 | stx1, stx2 | |
| No. of positive samples detected/total No. of samples (%) | 8/10 (80) | 9/10 (90) | 8/10 (80) | 10/10 (100) | 10/10 (100) | 10/10 (100) |
| qPCR CT by pooled testingb | ||||||
| Mean (SD) | 32.73 (3.76) | 31.75 (3.36) | 29.48 (5.73), 27.35 (4.32) | 24.17 (2.80) | 25.37 (4.98) | 27.36 (6.61), 26.51 (5.70) |
| Range | 27.52–36.97 | 27.08–36.57 | 20.48–37.05, 20.95–31.8 | 20.05–28.71 | 20.12–33.62 | 21.09–37.12, 18.48–37.08 |
| ΔCT (pooled vs individual qPCR) | ||||||
| Mean (SD) | 12.44 (1.81) | 12.04 (1.87) | 6.24 (1.09), 5.49 (1.40) | 4.18 (0.34) | 5.06 (0.38) | 3.61 (0.34), 3.50 (0.33) |
| Range | 5.14–19.29 | 1.28–19.76 | 2.99–11.37, 1.56–12.12 | 2.22–5.71 | 2.58–6.58 | 2.14–5.40, 1.97–5.62 |
Mean (SD) values exclude specimens that were undetected in pools.
P < 0.01 compared with prepooled CT by paired Student's t test.
Cost and workflow analyses of postenrichment pooling for STEC detection by NAAT.
To assess the operational requirements of the promising postenrichment pooled NAAT, cost and workflow analyses were performed to evaluate the costs of consumables and technician hands-on time (labor). Cost of consumables and labor time for the testing of 90 specimens simultaneously were estimated to be $223.47 ($76.50 for extraction procedures and $146.97 for qPCR procedures) and 4.75 h (3.47 h for extraction procedures and 1.28 h for qPCR procedures), respectively. Major consumables cost and labor savings (48 to 81% and 6 to 66%, respectively, based on an expected 1 to 2% positivity rate) were estimated with the postenrichment pooled NAAT for the same number of specimens, which corresponds to the maximum of 9 pools of 10 samples that would fit on a 96-well plate, although savings were impacted by the expected frequency of pool resolution (Table 3). Consumables cost savings were equally associated with extraction and qPCR procedures (80 and 81%, respectively, with no pool resolution), while extraction procedures gave greater labor savings over those of qPCR procedures (71 versus 53%, respectively, with no pool resolution). The estimated cost of consumables and labor time savings were highest with the greatest number of pools tested (9) and were decreased when fewer pools were tested given the incremental loss in efficiency (examples shown in Table 3). Moreover, while resolving positive pools necessitates additional testing, the turnaround time of the pooled postenrichment NAAT remains the same (within 1 day) as that of individual testing, given the time savings associated with the pooled strategy.
TABLE 3.
Estimated cost of consumables and labor time that result from postenrichment pooled NAAT
| No. of specimens tested (no. of pools tested) | Cost ($a [% savings per pool resolvedb]) |
Time (h [% savings per pool resolvedb]) |
||||
|---|---|---|---|---|---|---|
| 0 resolved | 1 resolved | 2 resolved | 0 resolved | 1 resolved | 2 resolved | |
| 90 (9) | 42.63 (81) | 79.43 (65) | 105.20 (53) | 1.62 (66) | 3.17 (33) | 3.57 (25) |
| 70 (7) | 36.55 (80) | 73.36 (59) | 99.12 (44) | 1.51 (62) | 3.06 (22) | 3.46 (12) |
| 50 (5) | 31.06 (76) | 67.87 (48) | 93.63 (29) | 1.41 (55) | 2.96 (6) | 3.36 (−7) |
| 30 (3) | 25.27 (71) | 62.08 (27) | 87.84 (−3) | 1.30 (44) | 2.85 (−22) | 3.25 (−39) |
| 10 (1) | 19.49 (51) | 56.30 (−42) | 82.06 (−107) | 1.20 (23) | 2.75 (−78) | 3.15 (−103) |
Supply costs were based on online listings excluding shipping as of March 2014 and included extraction and qPCR controls.
A positive percent savings (i.e., 82%) indicates that the pooled method provides benefits over the unpooled method (i.e., 82% savings). A negative percent savings (i.e., −42%) indicates that the pooled method is more expensive or timely than the unpooled methods (i.e., costs 42% more).
Prospective clinical evaluation of a postenrichment pooling strategy for STEC detection by NAAT.
Evaluation of the postenrichment pooled NAAT involved a 30-day direct comparison between the pooling strategy and the routine individual specimen testing currently performed at the BCCDC PHL. A total of 512 specimens were tested, with 9 to 27 specimens tested per day and consequently 1 (n = 1), 2 (n = 24), or 3 (n = 5) pools of 10 specimens or less per day (Table 4). An overall sensitivity of 100% was achieved once again for the postenrichment pooling strategy. Specimens positive for stx genes were identified on 7/30 days (23.3%), during which only 1 pool was positive on each day. Resolved pools contained 1 (n = 5), 2 (n = 1), or 4 (n = 1) positive specimens, for a total of 11 specimens (7 positive for stx1, 4 positive for stx1 and stx2)from 8 unique patients and, therefore, a positivity rate of 3% (2% excluding repeat patient samples). Isolates were recovered from 10 of the 11 specimens and included stx1- and stx2-positive O157:H7 (n = 4, two from one patient) and stx1-positive O103:H2 (n = 1), O118:H untypeable (n = 1), O126:H8 (n = 3, two from one patient), and O186:H2 (n = 1) serotypes. Consistent with retrospective evaluation findings, for both stx genes, the mean and range ΔCT values between the routine (individual) and pooled NAAT results for specimens in pools containing one positive specimen were 4.99 (2.00 to 7.66) for stx1 (n = 5) and 4.65 (4.41 to 4.88) for stx2 (n = 2). For the pool with two positive specimens, the pooled NAAT stx1 CT was 26.96, while individual NAAT stx1 CT values were 19.85 and 33.36. For the pool with four positive specimens, the pooled NAAT stx1 CT was 23.29, and the stx2 CT was 21.93, while individual NAAT CT values ranged from 19.78 to 32.05 (n = 4) and from 18.39 to 18.72 (n = 2), respectively. Interestingly, the CT values of the individual postresolution tests were generally higher than those of the individual routine tests that were conducted prior to pooling in this study (overall means: stx1, 27.02 versus 24.66, respectively; stx2, 19.63 versus 18.24 , respectively), resulting in ΔCT values of only 2.35 and 4.21 between postresolution and pooled NAAT results for specimens in pools containing one positive specimen. As expected, overall labor time savings were associated with routine performance of the postenrichment pooled NAAT compared with those of the individual NAAT, even in a scenario with only 2 pools per test (Table 4).
TABLE 4.
Prospective direct comparison of individual and postenrichment pooled NAAT techniques for STEC detection
| Category (n) | No. of samples (mean/day) | No. of pools (no. resolved) | Mean individual NAAT labor time (h) | Mean pooled NAAT labor time (h) | Pooled − individual NAAT Δtime (h) |
|---|---|---|---|---|---|
| No. of positive sample days (23) | 398 (17.3) | 50 (0) | 1.53 | 0.93 | 0.6 |
| Positive sample days (7) | 114 (16.3) | 14 (7) | 1.70 | 1.86 | −0.16 |
| Total days (30) | 512 (17.1) | 64 (7) | 1.57 | 1.15 | 0.42 |
DISCUSSION
As an approach for stx1 and stx2 detection from stool specimens, the postenrichment pooling strategy outperformed the preenrichment pooling strategy for all STEC genotypes. The observed loss of detection for preenrichment pooling was not investigated beyond the correlation with the CT value of individually tested specimens, but it might be attributable to a number of factors that result from the pooling of multiple samples, such as sample dilution, nutrient limitation within growth media, and competition for growth among organisms that are present. Notably, the performance of the postenrichment pooled strategy during the prospective clinical evaluation is better than that initially described in the retrospective evaluation. This might be attributable to technical variations (during the prospective evaluation, routine individual NAATs were performed by technologists who were not part of the study). Further, with an expected positivity rate of 1 to 2%, pooling was estimated to be a cost- and time-efficient strategy regardless of the number of pools included in the testing. Although labor costs were not included in these calculations owing to differences among institutions, they would increase the savings associated with pooled NAAT implementation.
A limitation of the two pooled NAAT strategies was that the detection threshold was greater with pooling than with individual specimen testing. However, the LoD of the postenrichment pooled NAAT is comparable to the LoD of other described in-house and commercial PCRs for STEC detection (21–23). To overcome any potential loss of detection, it may be possible to make method improvements, such as by concentrating enriched pools with centrifugation prior to lysis.
In high-volume testing scenarios, the postenrichment pooled NAAT strategy described here has considerable cost and workflow benefits. This high-throughput pooled NAAT is suited for comprehensive STEC surveillance to enhance the detection of non-O157 STEC serotypes at the community level and thereby inform burden estimates for all STEC serotypes. These burden estimates can be further supplemented by recovery of the isolate following the qPCR screening for serotype determination and subsequent characterization, such as with pulsed-field gel electrophoresis (PFGE) and whole-genome sequencing (WGS). This qPCR screening approach may also be suitable for STEC detection in a diagnostic laboratory setting and surge capacity during a large-scale STEC outbreak, but additional quality requirements, such as inclusion of an internal control, must be explored. The potential benefits of the proposed pooled NAAT outweigh the observed analytical limitations, given that the method is a solution for overcoming current cost-based barriers associated with adoption of CDC recommendations for universal stool screening for non-O157 STEC. Public health interventions based on accurate estimates of STEC prevalence have the potential to reduce the burden of STEC infections.
ACKNOWLEDGMENTS
This work was supported by the Public Health Agency of Canada's FoodNet Canada program and could not have been accomplished without the assistance of the Advanced Public Health Bacteriology and Mycology Section at the BCCDC PHL and, in particular, Loretta Janz. We also thank Justin Sorge at the BCCDC for providing positivity rate data.
Funding Statement
This project was funded through the Public Health Agency of Canada's FoodNet program.
REFERENCES
- 1.Johnson KE, Thorpe CM, Sears CL. 2006. The emerging clinical importance of non-O157 Shiga toxin-producing Escherichia coli. Clin Infect Dis 43:1587–1595. doi: 10.1086/509573. [DOI] [PubMed] [Google Scholar]
- 2.Hoefer D, Hurd S, Medus C, Cronquist A, Hanna S, Hatch J, Hayes T, Larson K, Nicholson C, Wymore K, Tobin-D'Angelo M, Strockbine N, Snippes P, Atkinson R, Griffin PM, Gould LH. 2011. Laboratory practices for the identification of Shiga toxin-producing Escherichia coli in the United States, FoodNet sites, 2007. Foodborne Pathog Dis 8:555–560. doi: 10.1089/fpd.2010.0764. [DOI] [PubMed] [Google Scholar]
- 3.Rosin P, Niskanen T, Palm D, Struelens M, Takkinen J; Shiga Toxin-Producing Escherichia coli Experts of European Union Food- and Waterborne Diseases and Zoonoses Network.2013. Laboratory preparedness for detection and monitoring of Shiga toxin 2-producing Escherichia coli O104:H4 in Europe and response to the 2011 outbreak. Euro Surveill 18:pii=20508 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20508. [DOI] [PubMed] [Google Scholar]
- 4.Stigi KA, Macdonald JK, Tellez-Marfin AA, Lofy KH. 2012. Laboratory practices and incidence of non-O157 Shiga toxin-producing Escherichia coli infections. Emerg Infect Dis 18:477–479. doi: 10.3201/eid1803.111358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. 2013. Incidence and trends of infection with pathogens transmitted commonly through food—Foodborne Diseases Active Surveillance Network, 10 U.S. sites, 1996–2012. MMWR Morb Mortal Wkly Rep 62:283–287. [PMC free article] [PubMed] [Google Scholar]
- 6.Brooks JT, Sowers EG, Wells JG, Greene KD, Griffin PM, Hoekstra RM, Strockbine NA. 2005. Non-O157 Shiga toxin-producing Escherichia coli infections in the United States, 1983–2002. J Infect Dis 192:1422–1429. doi: 10.1086/466536. [DOI] [PubMed] [Google Scholar]
- 7.Gould LH, Mody RK, Ong KL, Clogher P, Cronquist AB, Garman KN, Lathrop S, Medus C, Spina NL, Webb TH, White PL, Wymore K, Gierke RE, Mahon BE, Griffin PM. 2013. Increased recognition of non-O157 Shiga toxin-producing Escherichia coli infections in the United States during 2000–2010: epidemiologic features and comparison with E. coli O157 infections. Foodborne Pathog Dis 10:453–460. doi: 10.1089/fpd.2012.1401. [DOI] [PubMed] [Google Scholar]
- 8.Frank C, Werber D, Cramer JP, Askar M, Faber M, an der Heiden M, Bernard H, Fruth A, Prager R, Spode A, Wadl M, Zoufaly A, Jordan S, Kemper MJ, Follin P, Müller L, King LA, Rosner B, Buchholz U, Stark K, Krause G; Investigation Team HUS. 2011. Epidemic profile of Shiga-toxin-producing Escherichia coli O104:H4 outbreak in Germany. N Engl J Med 365:1771–1780. doi: 10.1056/NEJMoa1106483. [DOI] [PubMed] [Google Scholar]
- 9.Public Health Agency of Canada. 2014. FoodNet Canada 2012 short report. http://www.phac-aspc.gc.ca/foodnetcanada/report-rapport-2012-eng.php Accessed 13 May 2014.
- 10.Gould LH, Bopp C, Strockbine N, Atkinson R, Baselski V, Body B, Carey R, Crandall C, Hurd S, Kaplan R, Neill M, Shea S, Somsel P, Tobin-D'Angelo M, Griffin PM, Gerner-Smidt P; Centers for Disease Control and Prevention (CDC). 2009. Recommendations for diagnosis of Shiga toxin-producing Escherichia coli infections by clinical laboratories. MMWR Recomm Rep 58:1–14. [PubMed] [Google Scholar]
- 11.Buchan BW, Olson WJ, Pezewski M, Marcon MJ, Novicki T, Uphoff TS, Chandramohan L, Revell P, Ledeboer NA. 2013. Clinical evaluation of a real-time PCR assay for identification of Salmonella, Shigella, Campylobacter (Campylobacter jejuni and C. coli), and Shiga toxin-producing Escherichia coli isolates in stool specimens. J Clin Microbiol 51:4001–4007. doi: 10.1128/JCM.02056-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chui L, Lee M-C, Malejczyk K, Lim L, Fok D, Kwong P. 2011. Prevalence of Shiga toxin-producing Escherichia coli as detected by enzyme-linked immunoassays and real-time PCR during the summer months in northern Alberta, Canada. J Clin Microbiol 49:4307–4310. doi: 10.1128/JCM.05211-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lefterova MI, Slater KA, Budvytiene I, Dadone PA, Banaei N. 2013. A sensitive multiplex, real-time PCR assay for prospective detection of Shiga toxin-producing Escherichia coli from stool samples reveals similar incidences but variable severities of non-O157 and O157 infections in northern California. J Clin Microbiol 51:3000–3005. doi: 10.1128/JCM.00991-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kiska DL, Riddell SW. 2011. Point-counterpoint: should all stools be screened for Shiga toxin-producing Escherichia coli? J Clin Microbiol 49:2394–2397. doi: 10.1128/JCM.00817-11. [DOI] [PubMed] [Google Scholar]
- 15.Pilcher CD, McPherson JT, Leone PA, Smurzynski M, Owen-O'Dowd J, Peace-Brewer AL, Harris J, Hicks CB, Eron JJ, Fiscus SA. 2002. Real-time, universal screening for acute HIV infection in a routine HIV counseling and testing population. JAMA 288:216–221. doi: 10.1001/jama.288.2.216. [DOI] [PubMed] [Google Scholar]
- 16.Stramer SL, Glynn SA, Kleinman SH, Strong DM, Caglioti S, Wright DJ, Dodd RY, Busch MP. 2004. Detection of HIV-1 and HCV infections among antibody-negative blood donors by nucleic acid-amplification testing. N Engl J Med 351:760–768. doi: 10.1056/NEJMoa040085. [DOI] [PubMed] [Google Scholar]
- 17.Brant LJ, Ramsay ME, Balogun MA, Boxall E, Hale A, Hurrelle M, Kaluba L, Klapper P, Lewis D, Patel BC, Parry J, Irving WL. 2008. Diagnosis of acute hepatitis C virus infection and estimated incidence in low- and high-risk English populations. J Viral Hepat 15:871–877. doi: 10.1111/j.1365-2893.2008.01009.x. [DOI] [PubMed] [Google Scholar]
- 18.Hsiang MS, Lin M, Dokomajilar C, Kemere J, Pilcher CD, Dorsey G, Greenhouse B. 2010. PCR-based pooling of dried blood spots for detection of malaria parasites: optimization and application to a cohort of Ugandan children. J Clin Microbiol 48:3539–3543. doi: 10.1128/JCM.00522-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsiang MS, Hwang J, Kunene S, Drakeley C, Kandula D, Novotny J, Parizo J, Jensen T, Tong M, Kemere J, Dlamini S, Moonen B, Angov E, Dutta S, Ockenhouse C, Dorsey G, Greenhouse B. 2012. Surveillance for malaria elimination in Swaziland: a national cross-sectional study using pooled PCR and serology. PLoS One 7:e29550. doi: 10.1371/journal.pone.0029550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chui L, Couturier MR, Chiu T, Wang G, Olson AB, McDonald RR, Antonishyn NA, Horsman G, Gilmour MW. 2010. Comparison of Shiga toxin-producing Escherichia coli detection methods using clinical stool samples. J Mol Diagn 12:469–475. doi: 10.2353/jmoldx.2010.090221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang W, Bielaszewska M, Bauwens A, Fruth A, Mellmann A, Karch H. 2012. Real-time multiplex PCR for detecting Shiga toxin 2-producing Escherichia coli O104:H4 in human stools. J Clin Microbiol 50:1752–1754. doi: 10.1128/JCM.06817-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.US Food and Drug Administration. 2012. Evaluation of automatic class III designation (de novo) for xTAG gastrointestinal pathogen panel (GPP) decision summary. http://www.accessdata.fda.gov/cdrh_docs/reviews/K121454.pdf Accessed 15 March 2016.
- 23.US Food and Drug Administration. 2014. 510(k) substantial equivalence determination decision summary. http://www.accessdata.fda.gov/cdrh_docs/reviews/k140407.pdf Accessed 15 March 2016.