Abstract
Multilocus DNA sequence data were used to assess the genetic diversity and evolutionary relationships of 67 Fusarium strains from veterinary sources, most of which were from the United States. Molecular phylogenetic analyses revealed that the strains comprised 23 phylogenetically distinct species, all but two of which were previously known to infect humans, distributed among eight species complexes. The majority of the veterinary isolates (47/67 = 70.1%) were nested within the Fusarium solani species complex (FSSC), and these included 8 phylospecies and 33 unique 3-locus sequence types (STs). Three of the FSSC species (Fusarium falciforme, Fusarium keratoplasticum, and Fusarium sp. FSSC 12) accounted for four-fifths of the veterinary strains (38/47) and STs (27/33) within this clade. Most of the F. falciforme strains (12/15) were recovered from equine keratitis infections; however, strains of F. keratoplasticum and Fusarium sp. FSSC 12 were mostly (25/27) isolated from marine vertebrates and invertebrates. Our sampling suggests that the Fusarium incarnatum-equiseti species complex (FIESC), with eight mycoses-associated species, may represent the second most important clade of veterinary relevance within Fusarium. Six of the multilocus STs within the FSSC (3+4-eee, 1-b, 12-a, 12-b, 12-f, and 12-h) and one each within the FIESC (1-a) and the Fusarium oxysporum species complex (ST-33) were widespread geographically, including three STs with transoceanic disjunctions. In conclusion, fusaria associated with veterinary mycoses are phylogenetically diverse and typically can only be identified to the species level using DNA sequence data from portions of one or more informative genes.
INTRODUCTION
Best known as one of the most important genera of mycotoxigenic plant pathogens (1), Fusarium species (Hypocreales, Ascomycota) and their toxins are responsible for multibillion U.S. dollar losses annually to the world's agricultural economy. Due to the increase in immunocompromised and artificially immunosuppressed patient populations over the past 3 decades, Fusarium has also emerged as an important mycotic agent, causing life-threatening infections in humans (2, 3). Because fusaria are broadly resistant to most of the currently available antifungals, particularly high levels of mortality occur in patients who are persistently neutropenic (4). The greatest impact of Fusarium species on veterinary science is due to the diverse mycotoxicoses that they cause in animals that consume feed contaminated with fusarial toxins, such as trichothecenes, fumonisins, and estrogenic compounds (5, 6).
Judging from isolated reports in the literature, veterinary fusarioses appear to be relatively rare, and those reported typically involve economically important domesticated animals or animals housed in zoological parks where a veterinarian is on staff. Fusaria are responsible for a broad spectrum of mycotic infections in animals, with keratitis and dermatitis being the most common (7, 8). Less common fusarial infections in animals include melanized lesions that affect the abdominal segments and cephalothorax of crustaceans (9), dysecdysis in snakes (10), embryonic death in sea turtle eggs (11), and invasive sinusitis and facial mycetoma in dogs (12). In the past, morphology alone was typically used to identify the etiological agent (1, 8); however, reports of mycotic infections caused by Fusarium solani, Fusarium moniliforme, and Fusarium incarnatum that were identified using morphological data are largely unreliable because these morphospecies have been resolved as diverse complexes, comprising >100 phylogenetically recognizable species (12). Multilocus phylogenetic species recognition of medically important fusaria and important veterinary fusaria based on genealogical exclusivity (13) has revealed that approximately three-fourths of clinically relevant Fusarium species cannot be identified using phenotypic data (see reference 12 and references therein). To date, only a relatively small number of Fusarium strains from veterinary sources have been included in multilocus molecular phylogenetic studies, and they comprised 17 phylogenetically distinct species distributed among six species complexes, i.e., Fusarium incarnatum-equiseti, Fusarium chlamydosporum, Fusarium sambucinum, Fusarium oxysporum, Fusarium fujikuroi, and F. solani (12, 14–19).
The above summary highlights the need for a molecular phylogenetic study that is focused on elucidating the genetic diversity of fusaria of veterinary importance. Toward this end, the present study of 67 fusaria from veterinary sources was initiated with the following three objectives: (i) elucidate the evolutionary relationships and the species haplotype diversity of fusaria associated with veterinary fusarioses within the United States using genealogical concordance phylogenetic species recognition (GCPSR) (13), (ii) deposit the sequence data in the web-accessible Fusarium MLST (http://www.cbs.knaw.nl/Fusarium) and Fusarium-ID (http://fusariumdb.org) databases to facilitate the identification of veterinary fusaria via the Internet (12, 20, 21), and (iii) make the genetically characterized germplasm available to the scientific community via the ARS Culture Collection (NRRL) to promote further study.
MATERIALS AND METHODS
Fusarium isolates analyzed.
Of the 76 isolates analyzed in the present study, 67 were cultured from veterinary sources (Table 1) and 62 of these were from the United States. Strains cultured from veterinary sources were nested within 8 of the 10 species complexes (Fig. 1). The nine strains from nonveterinary sources were chosen primarily to provide representatives of five clades that contain human-pathogenic strains, including three members of the Fusarium dimerum species complex for rooting the phylogeny (Fig. 1). All of the strains that were included in this study are available upon request from the Agricultural Research Service Culture Collection, National Center for Agricultural Utilization Research, Peoria, IL (NRRL; http://nrrl.ncaur.usda.gov/TheCollection/index.html).
TABLE 1.
Isolates identified using multilocus DNA sequence data
| Fusarium species complexa | Species⃥MLSTb | NRRL no.c | Equivalent no.d | Host | Geographic origin |
|---|---|---|---|---|---|
| F. chlamydosporum | FCSC 2-b | 34015 | DI16-453 | Equine eye | Alabama |
| F. concolor | F. concolor | 25728 | CBS 463.91 | Human nail | Germany |
| F. dimerum | F. delphinoides | 36160 | CBS 110140 | Human eye | Florida |
| F. dimerum | F. dimerum | 36140 | CBS 108944 | Human blood | The Netherlands |
| F. dimerum | F. penzigii | 20711 | ATCC 15621 | Human eye | Sri Lanka |
| F. fujikuroi | F. proliferatum | 54994 | DI16-447 | Rhinoceros horn | California |
| F. fujikuroi | F. proliferatum | 62546 | DI16-448 | Equine eye | Florida |
| F. fujikuroi | F. verticillioides | 54997 | DI16-452 | Feline sinus exudate | Illinois |
| F. incarnatum-equiseti | FIESC 1-a | 43637 | DI16-456 | Canine skin lesion | Pennsylvania |
| F. incarnatum-equiseti | FIESC 1-a | 43640 | DI16-457 | Canine nose | Texas |
| F. incarnatum-equiseti | FIESC 1-a | 45996 | DI16-458 | Canine nasal cavity | New York |
| F. incarnatum-equiseti | FIESC 4-a | 20423 | IMI 300797 | Lizard skin | India |
| F. incarnatum-equiseti | FIESC 6-a | 43638 | FRC R-3500 | Manatee skin | Florida |
| F. incarnatum-equiseti | FIESC 13-a | 43635 | DI16-454 | Equine placenta | Nebraska |
| F. incarnatum-equiseti | F. equiseti (FIESC 14-c) | 43636 | DI16-417 | Canine | Texas |
| F. incarnatum-equiseti | FIESC 17-c | 34070 | Loyola W37591 | Tortoise | Illinois |
| F. incarnatum-equiseti | FIESC 19-a | 43639 | DI16-455 | Manatee skin | Florida |
| F. incarnatum-equiseti | FIESC 20-c | 54973 | DI16-459 | Rhinoceros eye | Ohio |
| F. oxysporum | F. oxysporum ST-33 | 54984 | DI16-437 | Mouse mucosa | Massachusetts |
| F. oxysporum | F. oxysporum ST-33 | 54996 | DI16-438 | Little blue penguin foot | Massachusetts |
| F. oxysporum | F. oxysporum ST-33 | 62542 | DI16-439 | Unknown feces | Texas |
| F. oxysporum | F. oxysporum ST-33 | 62545 | DI16-440 | Endoscope from veterinary clinic | California |
| F. oxysporum | F. oxysporum ST-33 | 62547 | DI16-441 | Canine stomach | California |
| F. redolens | F. redolens | 54967 | DI16-449 | Feline granuloma | California |
| F. sambucinum | F. armeniacum | 43641 | DI16-416 | Equine eye | Missouri |
| F. solani | F. falciforme (FSSC 3+4-d) | 54989 | DI16-418 | Equine eye | Texas |
| F. solani | F. falciforme (FSSC 3+4-dddd) | 54976 | DI16-419 | Equine corneal ulcer | Florida |
| F. solani | F. falciforme (FSSC 3+4-eee) | 32872 | JCM 11488 | Canine facial mycetoma | Japan |
| F. solani | F. falciforme (FSSC 3+4-eee) | 54964 | DI16-420 | Equine eye | Tennessee |
| F. solani | F. falciforme (FSSC 3+4-eeee) | 54978 | DI16-421 | Equine corneal ulcer | Florida |
| F. solani | F. falciforme (FSSC 3+4-eeee) | 54987 | DI16-422 | Equine eye | Florida |
| F. solani | F. falciforme (FSSC 3+4-eeee) | 54991 | DI16-423 | Aldabra tortoise carapace lesion | Florida |
| F. solani | F. falciforme (FSSC 3+4-eeee) | 62543 | DI16-424 | Equine eye | Florida |
| F. solani | F. falciforme (FSSC 3+4-ffff) | 54966 | DI16-425 | Equine eye | Georgia |
| F. solani | F. falciforme (FSSC 3+4-gggg) | 54983 | DI16-426 | Equine eye | Florida |
| F. solani | F. falciforme (FSSC 3+4-hhhh) | 54977 | DI16-427 | Equine corneal ulcer | Florida |
| F. solani | F. falciforme (FSSC 3+4-uu) | 62548 | DI16-428 | Equine eye | Florida |
| F. solani | F. falciforme (FSSC 3+4-uuu) | 54980 | DI16-429 | Equine eye | Florida |
| F. solani | F. falciforme (FSSC 3+4-ww) | 32754 | FRC S-0449 | Turtle head lesion | Florida |
| F. solani | F. falciforme (FSSC 3+4-yy) | 32778 | FRC S-0802 | Equine eye | Kentucky |
| F. solani | F. keratoplasticum (FSSC 2-dd) | 54162 | G8 lesion | Indigo snake skin | Alabama |
| F. solani | F. keratoplasticum (FSSC 2-ff) | 54975 | DI16-431 | Equine corneal ulcer | Florida |
| F. solani | F. keratoplasticum (FSSC 2-g) | 54998 | DI16-435 | Gray seal nail | California |
| F. solani | F. keratoplasticum (FSSC 2-g) | 54999 | DI16-436 | Gray seal nail | California |
| F. solani | F. keratoplasticum (FSSC 2-g) | 62544 | DI16-432 | Gray seal nail | California |
| F. solani | F. keratoplasticum (FSSC 2-h) | 22791 | IMI 095994 | Iguana tail | England |
| F. solani | F. keratoplasticum (FSSC 2-i) | 54965 | DI16-433 | Lung fish skin | Massachusetts |
| F. solani | F. keratoplasticum (FSSC 2-m) | 22645 | ATCC 46940 | Kuruma shrimp gill | Hawaii |
| F. solani | F. keratoplasticum (FSSC 2-p) | 25391 | ATCC 32751 | California brown shrimp gill | California |
| F. solani | F. keratoplasticum (FSSC 2-u) | 32780 | FRC S-0906 | Sea turtle head | Texas |
| F. solani | F. keratoplasticum (FSSC 2-ww) | 54963 | DI16-434 | Bonnet head shark eye | Florida |
| F. solani | F. petroliphilum (FSSC 1-a) | 54985 | DI16-442 | Dolphin digit bone | Georgia |
| F. solani | F. petroliphilum (FSSC 1-a) | 54988 | DI16-443 | Whale skin | Georgia |
| F. solani | F. petroliphilum (FSSC 1-b) | 54986 | DI16-444 | Pygmy sweeper skin | Georgia |
| F. solani | F. petroliphilum (FSSC 1-b) | 54995 | DI16-445 | Dolphin stomach | Bahamas |
| F. solani | F. petroliphilum (FSSC 1-c) | 32317 | DI16-446 | Treefish eye | Texas |
| F. solani | F. solani (FSSC 5-gg) | 54969 | DI16-450 | Canine foot mass biopsy | California |
| F. solani | F. solani (FSSC 5-v) | 54981 | DI16-451 | Unknown | California |
| F. solani | FSSC 9-a | 32755 | FRC S-0452 | Turtle head lesion | Florida |
| F. solani | FSSC 12-a | 22642 | ATCC 38341 | Kuruma shrimp gill | Japan |
| F. solani | FSSC 12-a | 54720 | DI16-460 | Lined sea horse | Georgia |
| F. solani | FSSC 12-a | 54974 | DI16-461 | Honeycomb cowfish facial lesion | Massachusetts |
| F. solani | FSSC 12-b | 25392 | ATCC 32752 | American lobster gill | New York |
| F. solani | FSSC 12-b | 54970 | DI16-462 | Antler crab tissue | Connecticut |
| F. solani | FSSC 12-e | 32821 | FRC S-1230 | Turtle egg | Texas |
| F. solani | FSSC 12-f | 54979 | DI16-463 | Kemps Ridley turtle flipper | Massachusetts |
| F. solani | FSSC 12-f | 62549 | DI16-464 | Horseshoe crab gill | Maryland |
| F. solani | FSSC 12-g | 54968 | DI16-465 | Bonnet head shark | New York |
| F. solani | FSSC 12-h | 54971 | DI16-466 | Reptile bronchus | Florida |
| F. solani | FSSC 12-h | 54982 | DI16-467 | Kemps Ridley turtle carapace lesion | Massachusetts |
| F. solani | FSSC 20-g | 54972 | DI16-468 | Equine eye | Florida |
| F. solani | FSSC 43-a | 54992 | DI16-469 | Zebra shark multiple tissues | Georgia |
| F. solani | FSSC 43-a | 54993 | DI16-470 | Zebra shark multiple tissues | Georgia |
| F. tricinctum | F. acuminatum | 36147 | CBS 109232 | Human bronchial secretion | Unknown |
| F. tricinctum | F. acuminatum | 45994 | DI16-415 | Cloaca | Texas |
| F. tricinctum | F. flocciferum | 45999 | DI16-430 | Human scalp | California |
Species complexes are as previously defined based on phylogenetic analysis of partial RPB1 plus RPB2 sequence data (22).
Multilocus sequence typing (MLST) followed published protocols (12, 16–18). Arabic numerals identify species, and lowercase roman letters identify unique haplotypes within each species. FCSC, Fusarium chlamydosporum species complex; FIESC, F. incarnatum-equiseti species complex; FSSC, F. solani species complex. F. oxysporum ST33 was identified using a portion of TEF1 and the entire IGS rDNA (17).
NRRL, ARS Culture Collection, NCAUR-ARS-USDA, Peoria, IL.
Abbreviations: ATCC, American Type Culture Collection, Manassas, VA; CBS, Centraalbureau voor Schimmelcultures-KNAW—Fungal Biodiversity Center, Utrecht, the Netherlands; DI16, University of Texas Health Sciences Center, San Antonio, TX; FRC, Fusarium Research Center, The Pennsylvania State University, State College, PA; JCM, Japanese Collection of Microorganisms, Tokyo, Japan; Loyola, Loyola University, Chicago, IL; IMI, CABI Biosciences, Egham, Surrey, England.
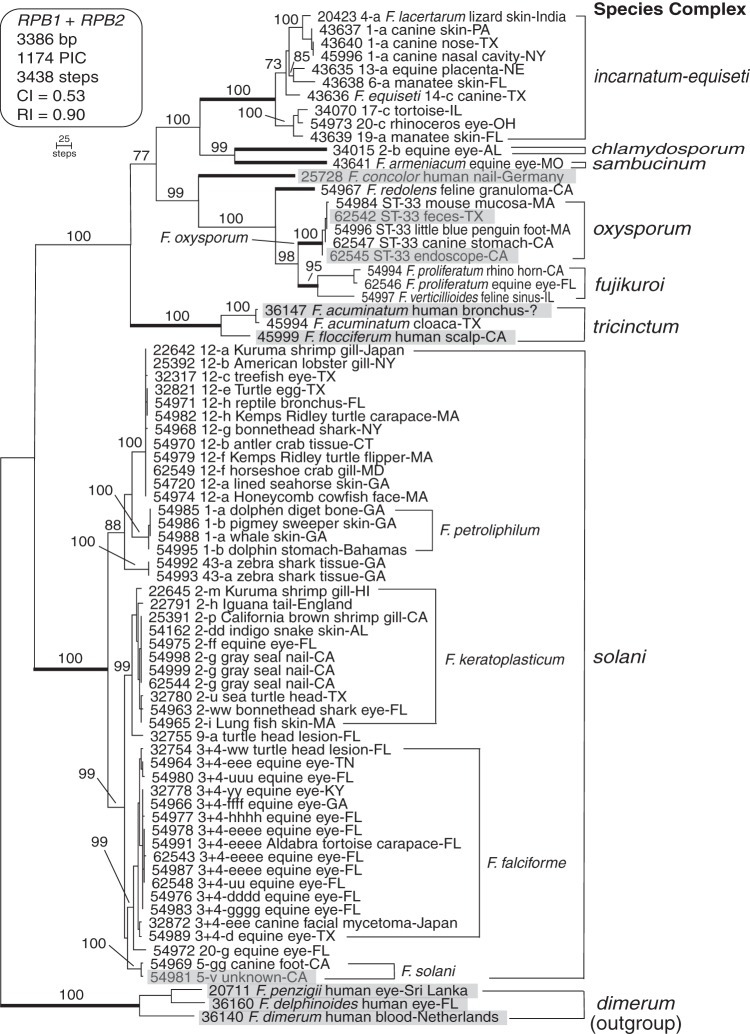
FIG 1.
Maximum parsimony phylogram inferred from a 2-locus data set comprising portions of the DNA-directed RNA polymerase II largest (RPB1, 1,607 bp) and second largest (RPB2, 1,779 bp) subunits for 67 veterinary strains and nine fusaria from other sources. Gray highlight is used to identify the fusaria from other sources. Sequences of three medically important species within the F. dimerum species complex (32) were used to root the phylogram based on more inclusive analyses (22). Isolates of veterinary importance are nested within eight species complexes (identified using bold internodes) and comprise 23 phylogenetically distinct species. Each isolate is identified by a 5-digit ARS Culture Collection accession number (http://nrrl.ncaur.usda.gov/). Because Latin binomials cannot be applied with confidence, Arabic numbers and lowercase Roman letters, respectively, were used to identify species and 3-locus haplotypes within the F. solani, F. incarnatum-equiseti, and F. chlamydosporum species complexes as previously reported (16, 18). In addition, a 2-locus typing scheme (17) was used to identify five isolates within the F. oxysporum species complex as ST33. Numbers above internodes represent maximum parsimony bootstrap support based on 1,000 pseudoreplicates of the data. CI, consistency index; PIC, parsimony informative character; RI, retention index.
Molecular biology.
A cetyltrimethylammonium bromide (CTAB) (Sigma-Aldrich, St. Louis, MO) protocol was used to extract total genomic DNA from freeze-dried mycelium prepared from yeast malt broth cultures as previously described (12). Portions of the DNA-directed RNA polymerase subunit 1 (RPB1) and the second largest subunit (RPB2) were PCR amplified and sequenced using published primers and protocols (see Table 2 in reference 12). The 2-locus sequence types (STs) of strains nested within the Fusarium oxysporum species complex (FOSC) were determined by PCR amplifying and sequencing a portion of the translation elongation factor (TEF1) gene and the entire nuclear ribosomal intergenic spacer (IGS) region (IGS rDNA) using published primers and protocols (12, 17). We employed a published 3-locus typing scheme that included a portion of TEF1, RPB2, and the nuclear ribosomal internal transcribed spacer (ITS) region plus domains D1 and D2 of the nuclear large-subunit (LSU) rRNA to identify species haplotypes within the F. solani, F. incarnatum-equiseti, and F. chlamydosporum species complexes (12, 16, 18). All PCR amplifications were conducted in an Applied Biosystems (ABI) 9700 thermocycler (Emeryville, CA) using Platinum Taq DNA polymerase (Invitrogen, Carlsbad, CA) and the following program: 1 cycle of 90 s at 94°C and 40 cycles of 30 s at 94°C, 90 s at 55°C, and 2 min at 68°C, followed by 1 cycle of 5 min at 68°C and a 4°C soak. After amplicons were electrophoretically size fractionated in 1.5% agarose gels (Invitrogen), they were stained with ethidium bromide and then photographed over a UV transilluminator (Fotodyne Inc., Hartland, WI). Montage96 filter plates (Millipore Corp., Billerica, MA) were used to purify amplicons prior to cycle sequencing using an ABI BigDye Terminator version 3.1 reaction mix. Prior to running sequencing reaction mixes on an ABI 3730 48-capillary automated sequencer, they were purified using an ABI XTerminator purification kit.
Phylogenetic analysis.
Raw ABI chromatograms were edited and aligned using Sequencher version 5.2.4 (Gene Codes, Ann Arbor, MI), exported as Nexus files, and then edited manually using TextPad version 5.1.0 for Windows (Helios Software Solutions, Longridge, England). The comprehensive partial RPB1 and RPB2 data set that was published previously (22) was used as a reference for placing indels in the alignment. Similarly, the TEF1 plus IGS rDNA sequences of five strains within the FOSC were aligned with a published data set for this complex (17), and then Collapse version 1.1 (http://www.softpedia.com/get/Science-CAD/Collapse.shtml) was used to identify the 2-locus ST of each strain. Paup version 4.0b10 (23) was used to conduct unweighted maximum parsimony (MP) analyses on the RPB1 plus RPB2 and TEF1 plus RPB2 plus rRNA data sets as previously described (16, 17). Nonparametric MP bootstrapping, based on 1,000 pseudoreplicates of the data, was used to assess clade stability.
Accession number(s).
DNA sequences generated in this study were deposited in GenBank under accession numbers KC808189 through KC808371 and KX768759 through KX768764, and alignments and phylogenetic trees were deposited in TreeBASE under accession numbers S19744 and Tr98123 through Tr98125. In addition, all of the sequences have been deposited in the web-accessible Fusarium MLST and Fusarium-ID databases.
RESULTS AND DISCUSSION
Multilocus DNA sequence-based genotyping of strains.
This study was conducted to characterize the phylogenetic diversity of the species and multilocus sequence types (STs) associated with veterinary fusarioses within the United States. Sixty-seven of the 76 analyzed isolates were cultured from veterinary sources (Table 1), and 62 of these were from the United States. To obtain an initial estimate of their diversity, portions of the DNA-directed RNA polymerase II largest (RPB1, 1,606-bp alignment) and second largest subunit (RPB2, 1,780-bp alignment) were sequenced and analyzed phylogenetically using MP (23). Sequences of three human-pathogenic isolates within the Fusarium dimerum species complex were used to root the phylogenies based on a more inclusive analysis (22). MP bootstrapping (BS) of the combined data set (3,386 bp) strongly supported the monophyly (MP-BS = 95% to 100%) of the six species complexes that contain two or more strains (Fig. 1). Evolutionary relationships among the species complexes received moderate to strong bootstrap support (MP-BS = 77% to 100%). The molecular phylogeny revealed that 23 Fusarium species distributed among eight species complexes were associated with veterinary mycoses (Fig. 1).
To identify 3-locus haplotypes of strains within the F. solani, F. incarnatum-equiseti, and F. chlamydosporum species complexes, portions of the following three loci were sequenced and analyzed phylogenetically as described previously (16, 18): TEF1 (692- to 710-bp alignments), RPB2 (1,766- to 1,866-bp alignments), and the internal transcribed spacer (ITS) region plus domains D1 and D2 of the nuclear large-subunit (LSU) rRNA (1,009- to 1,153-bp alignments). Because Latin binomials could be applied with confidence to only 11 of the 23 fusaria of veterinary importance, the unnamed species were identified using an informal ad hoc alphanumeric typing scheme for species haplotypes developed for undescribed species in these groups (12, 16, 18). STs within the F. oxysporum species complex (FOSC) were identified using a 2-locus typing scheme (18) that included a portion of TEF1 (634-bp alignment) and the entire nuclear ribosomal intergenic spacer region (IGS rDNA, 2,220-bp alignment).
The most common animal infections were ocular (21/67 = 31.3%), followed by dermatomycoses and onychomycoses (Fig. 1; Table 1). Eight phylogenetically diverse fusaria distributed among five species complexes were associated with keratitis infections of veterinary animals. Consistent with previous findings on medically important fusaria (12, 16, 24), most of the veterinary isolates (47/67 = 70.1%) were nested within the F. solani species complex (FSSC), and these comprised 8 phylospecies and 33 unique 3-locus STs (Fig. 1; see also Fig. S1 in the supplemental material). With the exception of phylospecies FSSC 43-a from a zebra shark (Stegostoma fasciatum), the seven other FSSC species have been reported from human mycoses (16). Three of the FSSC species (Fusarium falciforme, Fusarium keratoplasticum, and Fusarium sp. FSSC 12) accounted for 38/47 of the veterinary isolates and 27/33 of the 3-locus STs within this complex. It is noteworthy that all but three of the 15 genetically diverse F. falciforme strains were recovered from equines in five states (Florida, Georgia, Kentucky, Tennessee, and Texas; Table 1) with keratitis infections. Nine of the 11 unique STs within F. falciforme were singletons. However, F. falciforme 3+4-eee was isolated from a canine facial mycetoma in Japan and an equine eye in Tennessee. In addition, F. falciforme 3+4-eeee was associated with ocular mycoses of three equines and a lesion on the carapace of an Aldabra tortoise (Geochelone gigantea) in Florida. Each of these strains was collected in a different year between 2006 and 2011. Two of the equines were in Ocala and the third in Gainesville, which is only 38 miles away.
By way of contrast, 19 of the 22 strains of F. keratoplasticum and Fusarium sp. FSSC 12 were isolated from marine vertebrates and invertebrates, which suggests they may be adapted to the habitats of these hosts (Fig. 1; Table 1). Nine of the 11 STs within F. keratoplasticum were singletons compared with only two of the six STs within Fusarium sp. FSSC 12. Four of the six STs in the latter species were represented by geographically distant STs (i.e., 12-a, 12-b, 12-f, and 12-h). Fusarium sp. FSSC 12 has been reported to cause mycotic infections in humans (16) and in a number of captive marine animals, including a recent epizootic at a public aquarium in the United States that resulted in 100% mortality of lined seahorses (Hippocampus erectus) (19), cutaneous lesions in scalloped hammerhead sharks from Japan in a public aquarium in Hong Kong (Sphyrna lewini) (14), black gill disease of kuruma prawn (Penaeus japonicas) in Japan (25), and fatal mycotic infections of lobsters in Australia and New York (26). The etiological agents in the latter two studies were reported as F. solani and Fusarium sp., respectively, based on morphological species recognition. However, it should be noted that F. solani was recently epitypified and corresponds to phylospecies FSSC 5 (27). Prior to the application of GCPSR within the F. solani species complex (16), most of the >60 species within this clade were reported as F. solani in the medical and plant pathological literature (1).
Our sampling suggests that the F. incarnatum-equiseti species complex (FIESC), with 10/67 (14.9%) of the isolates, 8/23 species, and 8 unique STs, may represent the second most important clade of veterinary relevance within Fusarium (Fig. 1; see also Fig. S2 in the supplementary material). Members of this clade have only rarely been reported to cause fusarioses (18, 28). Although this complex contains at least 28 phylospecies (18), Latin binomials cannot not be applied to 25 of them. Therefore, six of the FIESC species included in this study were reported here using an informal alphanumeric nomenclature (Table 1), and they include FIESC 1-a from canines in three different states (Table 1), FIESC 6-a and 19-a from manatee skin in Florida (Trichechus manatus), FIESC 17-c from a tortoise in Illinois, FIESC 19-a from an equine placenta in Nebraska, and FIESC 20-c from the eye of a rhinoceros in Ohio. The two named animal-associated species that we analyzed within this complex included Fusarium lacertarum (FIESC 4-a) from lizard skin in India and Fusarium equiseti (FIESC 14-c) from the nose of a dog in Texas. The fact that many members of this complex are found in soil may help explain a possible epidemiological link between FIESC 1-a and nasal infections of canines and humans (18).
We determined that the five isolates within the F. oxysporum species complex (FOSC) were all members of the widespread clonal lineage ST33 using the TEF1 plus IGS rDNA 2-locus typing scheme (17). Similar to other medically important fusaria (28), F. oxysporum clonal lineage ST33 is common in plumbing systems and appears to be responsible for the majority of mycoses in human and other animals caused by members of the FOSC (17, 29), as well as a pseudoepidemic associated with bronchoscopy specimens at a San Antonio, TX hospital in 1997 and 1998 (30). Phylogenetically diverse fusaria are known to colonize plumbing systems via biofilms (16, 31–33), and this may explain their near exclusivity in the contact lens solution-associated keratitis outbreaks in the United States, Singapore, and Hong Kong (34, 35). Our finding that Fusarium redolens was associated with the granuloma of a cat in California to our knowledge represents only the second report of this soilborne fungus associated with a mycotic infection. This species was originally reported as causing human onychomycosis in Malaysia as F. oxysporum var. redolens (36); however, the F. redolens and F. oxysporum clades are phylogenetically distinct (22, 37).
All of the DNA sequence data generated in this study have been incorporated into the following web-accessible sites dedicated to facilitating accurate identification of fusaria via the Internet: Fusarium-ID at the Fusarium Research Center (FRC), Pennsylvania State University (20, 21), and Fusarium MLST at the Centraalbureau voor Schimmelcultures (CBS-KNAW) Fungal Biodiversity Center, Utrecht, the Netherlands (12). For these databases to reach their full potential, veterinarians and clinical microbiologists are encouraged to deposit strains in a publically accessible microbial culture collection, such as the CBS-KNAW, FRC, or the ARS Culture Collection (NRRL), to facilitate future studies.
We recommend that veterinarian researchers and diagnosticians who are interested in identifying isolates molecularly to the species level first generate a partial TEF1 or RPB2 sequence and use it to conduct a blastn query of Fusarium-ID and/or Fusarium MLST and carefully examine the accessions with the top maximum identity scores (38). If similar BLAST queries are made of GenBank (http://www.ncbi.nlm.nih.gov/), researchers may find instances where different taxon names are applied to records with similar maximum identity scores. When encountered, this should alert individuals that one or more accessions is probably misidentified. Researchers need to be aware that if a precise species identification is required, additional loci may need to be sequenced if the result of a blastn query using a partial TEF1 or RPB2 is equivocal. It is also worth noting that loci, such as the nuclear ribosomal ITS rRNA and mitochondrial cytochrome b gene, which were used, respectively, to identify the etiological agent associated with the ulcerative dermatitis of a canine in Japan as Fusarium sporotrichioides (38) and the granulomatous dermatitis in a cat as Fusarium proliferatum (39), are too conserved to obtain a reliable species-level identification in Fusarium.
Looking to the future, Fusarium comparative genomics (40) offers enormous potential for accelerating the discovery of additional phylogenetically informative loci, thereby greatly advancing our understanding of the population structure, reproductive mode, and species limits of fusaria of medical and veterinary importance. This information will become increasingly more valuable given that climate change has been predicted to contribute to the emergence of novel fungal diseases of domestic animals and wildlife worldwide (41).
Supplementary Material
ACKNOWLEDGMENTS
Special thanks are due to Stacy Sink for generating all of the DNA sequence data reported in this study, Nathane Orwig for running the sequences in NCAUR's DNA core facility, and the culture collections and individuals who supplied isolates used in this study. We also thank Marcelo Sandoval (CBS-KNAW) for helpful comments for improving the manuscript.
Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.01607-16.
REFERENCES
- 1.Leslie JF, Summerell BA. 2006. The Fusarium laboratory manual. Blackwell Publishing, Ames, IA. [Google Scholar]
- 2.Dignani MC, Anaissie E. 2004. Human fusariosis. Clin Microbiol Infect 10(Suppl):S67–S75. [DOI] [PubMed] [Google Scholar]
- 3.Zhang SX, O'Donnell K, Sutton DA. 2015. Fusarium and other opportunistic hyaline fungi, p 2057–2086. In Jorgensen J, Pfaller M, Carroll K, Funke G, Landry M, Richter S, Warnock D (ed), Manual of clinical microbiology, 11th ed ASM Press, Washington, DC. [Google Scholar]
- 4.Nucci M, Anaissie E. 2007. Fusarium infections in immunocompromised patients. Clin Microbiol Rev 20:695–704. doi: 10.1128/CMR.00014-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alexander NJ, Proctor RH, McCormick SP. 2009. Genes, gene clusters, and biosynthesis of trichothecenes and fumonisins in Fusarium. Toxin Rev 24:198–215. [Google Scholar]
- 6.Marasas WFO, Nelson PE, Toussoun TA. 1984. Toxigenic Fusarium species: identity and mycotoxicology. Pennsylvania State University Press, University Park, PA. [Google Scholar]
- 7.Cabañes FJ, Alonso JM, Castellá G, Alegre F. 1997. Cutaneous hyalophyphomycosis caused by Fusarium solani in a loggerhead sea turtle (Caretta caretta L.). J Clin Microbiol 35:3343–3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rebell G. 1981. Fusarium infections in human and veterinary medicine, p 210–220. In Nelson PE, Toussoun TA, Cook RJ (ed), Fusarium: diseases, biology, and taxonomy. Pennsylvania State University Press, University Park, PA. [Google Scholar]
- 9.Hose JE, Lightner DV, Redman RM, Danald DA. 1984. Observations on the pathogenesis of the imperfect fungus, Fusarium solani, in the California brown shrimp Penaeus californienis. J Invertebr Pathol 44:292–303. doi: 10.1016/0022-2011(84)90027-2. [DOI] [Google Scholar]
- 10.Dadone LI, Klaphake E, Garner MM, Schwahn D, Sigler L, Trupkiewicz JG, Myers G, Barrie MT. 2010. Pituitary cystadenoma, enterolipidosis, and cutaneous mycosis in an Everglades ratsnake (Elaphe obsoleta Rossalleni). J Zoo Wildl Med 41:538–541. doi: 10.1638/2009-0124.1. [DOI] [PubMed] [Google Scholar]
- 11.Sarmiento-Ramírez JM, Abella-Pérez E, Phillott AD, Sim J, van West P, Martín MP, Marco A, Diéguez-Uribeondo JD. 2014. Global distribution of two fungal pathogens threatening endangered sea turtles. PLoS One 9(1):e85853. doi: 10.1371/journal.pone.0085853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O'Donnell K, Sutton DA, Rinaldi MG, Sarver BJA, Balajee SA, Schroers HJ, Summerbell RC, Robert VARG, Crous PW, Zhang N, Aoki T, Jung K, Park J, Lee YH, Kang S, Park B, Geiser DM. 2010. Internet-accessible DNA sequence database for identifying fusaria from human and animal infections. J Clin Microbiol 48:3708–3718. doi: 10.1128/JCM.00989-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taylor JW, Jacobson DJ, Kroken S, Kasuga T, Geiser DM, Hibbett DS, Fisher MC. 2000. Phylogenetic species recognition and species concepts in fungi. Fungal Genet Biol 31:21–32. doi: 10.1006/fgbi.2000.1228. [DOI] [PubMed] [Google Scholar]
- 14.Fernando N, Hui SW, Tsang CC, Leung SY, Ngan AHY, Leung RWW, Groff JM, Lau SKP, Woo PCY. 2015. Fatal Fusarium solani species complex infections in elasmobranchs: the first case report for black spotted stingray (Taeniura melanopsila) and a literature review. Mycoses 58:422–431. doi: 10.1111/myc.12342. [DOI] [PubMed] [Google Scholar]
- 15.O'Donnell K, Cigelnik E, Nirenberg H. 1998. Molecular systematics and phylogeography of the Gibberella fujikuroi species complex. Mycologia 90:465–493. doi: 10.2307/3761407. [DOI] [Google Scholar]
- 16.O'Donnell K, Sutton DA, Fothergill A, McCarthy D, Rinaldi MG, Brandt ME, Zhang N, Geiser DM. 2008. Molecular phylogenetic diversity, multilocus haplotype nomenclature, and in vitro antifungal resistance within the Fusarium solani species complex. J Clin Microbiol 46:2477–2490. doi: 10.1128/JCM.02371-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O'Donnell K, Gueidan C, Sink S, Johnston PR, Crous PW, Glenn A, Riley R, Zitomer NC, Colyer P, Waalwijk C, van der Lee T, Moretti A, Kang S, Kim HS, Geiser DM, Juba JH, Baayen PR, Cromey MG, Bithell S, Sutton DA, Skovgaard K, Ploetz R, Kistler HC, Elliott M, Davis M, Sarver BAJ. 2009. A two-locus DNA sequence database for typing plant and human pathogens within the Fusarium oxysporum species complex. Fungal Genet Biol 46:936–948. doi: 10.1016/j.fgb.2009.08.006. [DOI] [PubMed] [Google Scholar]
- 18.O'Donnell K, Sutton DA, Rinaldi MG, Gueidan C, Crous PW, Geiser DM. 2009. Novel multilocus sequence typing scheme reveals high genetic diversity of human pathogenic members of the Fusarium incarnatum-F. equiseti and F. chlamydosporum species complexes within the United States. J Clin Microbiol 47:3851–3861. doi: 10.1128/JCM.01616-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salter CE, O'Donnell K, Sutton DA, Marancik DP, Knowles S, Clauss TM, Berliner AL, Camus AC. 2012. Dermatitis and systemic mycosis in lined seahorses Hippocampus erectus associated with a marine-adapted Fusarium solani species complex pathogen. Dis Aquat Organ 101:23–31. doi: 10.3354/dao02506. [DOI] [PubMed] [Google Scholar]
- 20.Geiser DM, del Jiménez-Gasco M, Kang S, Makalowska I, Veeraraghavan N, Ward TJ, Zhang N, Kuldau GA, O'Donnell K. 2004. FUSARIUM-ID v. 1.0: a DNA sequence database for identifying Fusarium. Eur J Plant Pathol 110:473–479. doi: 10.1023/B:EJPP.0000032386.75915.a0. [DOI] [Google Scholar]
- 21.Park B, Park J, Cheong KC, Choi J, Jung K, Kim D, Lee YH, Ward TJ, O'Donnell K, Geiser DM, Kang S. 2011. Cyber infrastructure for Fusarium: three integrated platforms supporting strain identification, phylogenetics, comparative genomics, and knowledge sharing. Nucleic Acids Res 39:D640–D646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O'Donnell K, Rooney AP, Proctor RH, Brown DW, McCormick SP, Ward TJ, Frandsen RJN, Lysøe E, Rehner SA, Aoki T, Robert VARG, Crous PW, Groenewald JZ, Kang S, Geiser DM. 2013. Phylogenetic analyses of RPB1 and RPB2 support a middle Cretaceous origin for a clade comprising all agriculturally and medically important fusaria. Fungal Genet Biol 52:20–31. doi: 10.1016/j.fgb.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 23.Swofford DL. 2002. PAUP*. Phylogenetic analysis using parsimony (*and other methods). version 4. Sinauer Associates, Sunderland, MA. [Google Scholar]
- 24.Zhang N, O'Donnell K, Sutton DA, Nalim FA, Summerbell RC, Padhye AA, Geiser DM. 2006. Members of the Fusarium solani species complex that cause infections in both humans and plants are common in the environment. J Clin Microbiol 44:2186–2190. doi: 10.1128/JCM.00120-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hatai K, Furuya K, Egusa S. 1978. Studies on the pathogenic fungus associated with black gill disease of kuruma prawn, Penaeus japonicus. I. Isolation and identification of the BG-Fusarium. Fish Pathol 12:219–224. [Google Scholar]
- 26.Lightner DV, Fontaine CT. 1975. A mycosis of the American lobster, Homarus americanus, caused by Fusarium sp. J Invertebr Pathol 25:239–245. doi: 10.1016/0022-2011(75)90074-9. [DOI] [PubMed] [Google Scholar]
- 27.Schroers HJ, Samuels GJ, Zhang N, Short DPG, Juba JH, Geiser DM. 2016. Epitypification of Fusisporium (Fusarium) solani and its assignment to a common phylogenetics species in the Fusarium solani species complex. Mycologia 108:806–819. doi: 10.3852/15-255. [DOI] [PubMed] [Google Scholar]
- 28.Khoa LV, Hatai K, Aoki T. 2004. Fusarium incarnatum isolated from black tiger shrimp, Penaeus monodon Fabricius, with black gill disease cultured in Vietnam. J Fish Dis 27:507–515. doi: 10.1111/j.1365-2761.2004.00562.x. [DOI] [PubMed] [Google Scholar]
- 29.Migheli Q, Balmas V, Harak H, Sanna S, Scherm B, Aoki T, O'Donnell K. 2010. Molecular phylogenetic diversity of dermatologic and other human pathogenic fusarial isolates from hospitals in northern and central Italy. J Clin Microbiol 48:1076–1084. doi: 10.1128/JCM.01765-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O'Donnell K, Sutton DA, Rinaldi MG, Magnon KC, Cox PA, Revankar SG, Sanche S, Geiser DM, Juba JH, van Burik JAH, Padhye A, Anaissie EJ, Francesconi A, Walsh TJ, Robinson JS. 2004. Genetic diversity of human pathogenic members of the Fusarium oxysporum complex inferred from multilocus DNA sequence data and amplified fragment length polymorphism analyses: evidence for the recent dispersion of a geographically widespread clonal lineage and nosocomial origin. J Clin Microbiol 42:5109–5120. doi: 10.1128/JCM.42.11.5109-5120.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Imamura Y, Chandra J, Mukherjee PK, Lattif AA, Szczotka-Flynn LB, Pearlman E, Lass JH, O'Donnell K, Ghannoum MA. 2008. Fusarium and Candida albicans biofilms on soft contact lenses: model development, influence of lens type, and susceptibility to lens care solutions. Antimicrob Agents Chemother 52:171–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schroers HJ, O'Donnell K, Lamprecht SC, Kammeyer PL, Johnson S, Sutton DA, Rinaldi MG, Geiser DM, Summerbell RC. 2009. Taxonomy and phylogeny of the Fusarium dimerum species group. Mycologia 101:44–70. doi: 10.3852/08-002. [DOI] [PubMed] [Google Scholar]
- 33.Short DPG, O'Donnell K, Zhang N, Juba JH, Geiser DM. 2011. Widespread occurrence of diverse human pathogenic types of the fungus Fusarium detected in plumbing drains. J Clin Microbiol 49:4264–4272. doi: 10.1128/JCM.05468-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chang DC, Grant GB, O'Donnell K, Wannemuehler KA, Nobel-Wang J, Rao CY, Jacobson LM, Crowell CS, Sneed RS, Lewis FMT, Schaffzin JK, Kainer MA, Alfonso EC, Jones DB, Srinivasan A, Fridkin SK, Park BJ. 2006. Multistate outbreak of Fusarium keratitis associated with use of a contact lens solution. JAMA 296:953–963. doi: 10.1001/jama.296.8.953. [DOI] [PubMed] [Google Scholar]
- 35.O'Donnell K, Sarver BAJ, Brandt M, Chang DC, Nobel-Wang J, Park BJ, Sutton DA, Benjamin L, Lindsley M, Padhye A, Geiser DM, Ward TJ. 2007. Phylogenetic diversity and microsphere array-based genotyping of human pathogenic fusaria, including isolates from the multistate contact lens-associated U.S. keratitis outbreaks of 2005 and 2006. J Clin Microbiol 45:2235–2248. doi: 10.1128/JCM.00533-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salleh D, Strange RN. 1988. Toxigenicity of some fusaria associated with plant and human diseases in the Malaysian peninsula. J Gen Microbiol 134:841–847. [DOI] [PubMed] [Google Scholar]
- 37.Baayen RP, O'Donnell K, Breeuwsma S, Geiser DM, Waalwijk C. 2001. Molecular relationships of fungi within the Fusarium redolens-F. hostae clade. Phytopathology 91:1037–1044. doi: 10.1094/PHYTO.2001.91.11.1037. [DOI] [PubMed] [Google Scholar]
- 38.O'Donnell K, Ward TJ, Robert VARG, Crous PW, Geiser DM, Kang S. 2015. DNA sequence-based identification of Fusarium: current status and future directions. Phytoparasitica 43:583–595. doi: 10.1007/s12600-015-0484-z. [DOI] [Google Scholar]
- 39.Sugahara GA, Kiuchi A, Usui R, Usui R, Mineshige T, Kamiie J, Shirota S. 2014. Granulomatous pododermatitis in the digits caused by Fusarium proliferatum in a cat. J Vet Med Sci 76:435–438. doi: 10.1292/jvms.13-0449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma LJ, Geiser DM, Proctor RH, Rooney AP, O'Donnell K, Trail F, Gardiner DM, Manners JM, Kazan K. 2013. Fusarium pathogenomics. Annu Rev Microbiol 67:399–416. doi: 10.1146/annurev-micro-092412-155650. [DOI] [PubMed] [Google Scholar]
- 41.Fisher MC, Henk DA, Briggs CJ, Brownstein JS, Madoff LC, McCraw SL, Gurr SJ. 2012. Emerging fungal threats to animal, plant and ecosystem health. Nature 484:186–194. doi: 10.1038/nature10947. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.