Abstract
Haemonchus contortus is one of the most important parasitic nematodes of small ruminants around the world, particularly in tropical and subtropical regions. The control of haemonchosis relies mainly on anthelmintics, but the excessive and prolonged use of anthelmintics is causing serious drug resistance issues in many countries. As benzimidazole (BZ) anthelmintics have been broadly used in China, we hypothesized that resistance is widespread. Given the link between three known single nucleotide polymorphisms (SNPs, designated F167Y, E198A and F200Y) in the isotype-1 β-tubulin gene and BZ resistance, our goal here was to explore the presence of these mutations in H. contortus from small ruminants (sheep and goats) from eight provinces in China using PCR-coupled sequencing. In addition, the genetic diversity and genetic relationship of isotype-1 β-tubulin sequence haplotypes were also investigated. Among 192 H. contortus adult individuals representing the eight populations, we identified six distinct sequence types, five of which had SNP E198A (GCA) and/or F200Y (TAC). Sequence analysis showed that the frequencies of SNPs E198A and F200Y were 0–70% and 0–31%, respectively. SNP F167Y (TAC) was not detected in any population. In addition, high haplotype diversities (0.455–0.939) and nucleotide diversities (0.018–0.039) were calculated. A network analysis of the isotype-1 β-tubulin gene sequences showed that SNPs E198A and F200Y occurred in multiple distinct groupings, suggesting multiple independent origins of these SNPs. The findings of this first study of SNPs in the isotype-1 β-tubulin gene of H. contortus populations suggest that BZ resistance is prevalent in some regions of China, and that any control strategy might focus on monitoring BZ resistance in this country.
Keywords: Haemonchus contortus, Isotype-1 β-tubulin gene, Single nucleotide polymorphism (SNP), Benzimidazole resistance
Graphical abstract
Highlights
-
•
First genetic analysis of three β-tubulin gene SNPs in Chinese Haemonchus contortus.
-
•
E198A (GCA) occurred more frequently than did F200Y (TAC).
-
•
F167Y (TAC) was not detected in any H. contortus population in China.
-
•
F200Y (TAC) had multiple origins, and E198A (GCA) had two distinct origins.
-
•
High genetic diversity within population and high gene flow were confirmed.
1. Introduction
Haemonchus contortus is one of the most economically important trichostrongyloid nematodes affecting small ruminant livestock worldwide. This nematode has a complex three-week life cycle, with a parasitic and a free-living phase. Following the ingestion of infective third-stage larvae (L3s), hosts are affected mainly by the fourth-stage larvae and adults of H. contortus primarily due to their blood feeding activity in the abomasum, leading to anaemia and associated complications and often death in heavily infected animals (Waller et al., 2004, Nikolaou and Gasser, 2006, O'Connor et al., 2006).
The control of H. contortus and related gastrointestinal nematodes has relied on the treatment with anthelmintic drugs including benzimidazoles (BZs). As a consequence of their excessive use, resistance against BZ has developed and spread in H. contortus and related strongylid nematodes (Waller, 1997, Jackson and Coop, 2000, Kaplan, 2004). β-tubulin is the target for BZs (Kohler, 2001, Kotze et al., 2014), and, collectively, studies to date recognise three different single nucleotide polymorphisms (SNPs) in the isotype-1 β-tubulin gene at codons 167 (TTC to TAC; F167Y) (Silvestre and Cabaret, 2002), 198 (GAA to GCA; E198A) (Ghisi et al., 2007, Rufener et al., 2009) and 200 (TTC to TAC; F200Y) (Kwa et al., 1994, Kwa et al., 1995) to be associated with BZ resistance in H. contortus. F200Y has been detected at high prevalence in many countries and appears to be the commonest SNP linked to BZ resistance (Kotze et al., 2014). F167Y is less common than F200Y, with an apparently limited distribution in countries including Argentina, Brazil, Canada, France, the UK and the USA (Silvestre and Cabaret, 2002, Barrere et al., 2012, Barrere et al., 2013a, Barrere et al., 2013b, Brasil et al., 2012, Chaudhry et al., 2014, dos Santos et al., 2014, Redman et al., 2015). In addition, E198A has been found in three field-derived populations from South Africa and Australia and in another in vitro-selected population from Australia (Ghisi et al., 2007, Rufener et al., 2009, Kotze et al., 2012); recently, this mutation was detected in field populations of H. contortus in India (Chaudhry et al., 2015).
BZs and other anthelmintic drugs have been used heavily to control nematode infections in China. Based on some publications from this country (He et al., 1999a, He et al., 1999b, Cai et al., 2007a, Cai et al., 2007b, Zhao et al., 2010), we have reason to believe that BZ resistance in H. contortus is becoming a major issue. Although SNPs, F167Y, E198A and F200Y have been detected in H. contortus in many countries (Kotze et al., 2014), the three studies of isotype-1 β-tubulin gene in China have focused only on the F200Y (Bo and Li, 2005, Hao, 2007, Cai and Bai, 2009); thus, there is no information on the other two SNPs, in spite of the high prevalence of H. contortus and the excessive and uncontrolled use of BZs in this country (Shen, 2005).
Understanding the origin and spread of SNPs linked to BZ resistance is important (Skuce et al., 2010), which is why population genetic studies have been conducted recently. For example, Chaudhry et al. (2015) studied 13 H. contortus populations from sheep and goats in southern India, and indicated that SNP E198A spread from a single origin to multiple locations in this region, whereas F200Y likely had three independent origins. Similarly, Redman et al. (2015) provided evidence for multiple independent origins of both SNPs F200Y and F167Y in H. contortus in the UK. However, there is no such information for China.
Therefore, for the first time in China, we explored genetic variation and the three known BZ-associated SNPs (F167Y, E198A and F200Y) in the isotype-1 β-tubulin gene in populations of H. contortus from sheep and goats from eight provinces using PCR-coupled sequencing, to begin to understand the status of BZ resistance and the origin of these SNPs.
2. Materials and methods
2.1. Parasite populations and genomic DNA isolation
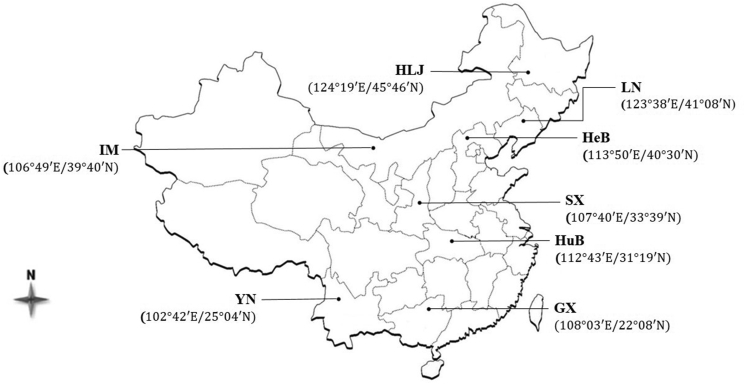
Animal ethics approval was granted (permit SYXK2015-0084) by the Science and Technology Department of Hubei province, China. Adult H. contortus populations were collected from eight different geographical regions from provinces in subtropical to tropical climatic zones of China (Fig. 1). Samples from Hubei, Yunnan, Shaanxi and Guangxi were from goats, whereas others were from sheep. The history of use of anthelmintics, including BZs or other drugs, was not clear in many cases. After collection from the abomasum, worms were washed in physiological saline, fixed in 75% ethanol and submitted to the College of Veterinary Medicine, Huazhong Agricultural University, Wuhan. Upon arrival, individual adult male worms were morphologically identified (Lichtenfels et al., 1994). Then, following the rehydration of worms, total genomic DNA was extracted from individual adults using sodium dodecyl-sulfate/proteinase K treatment, followed by spin-column purification (Wizard DNA Clean-Up, Promega) (Gasser et al., 2006). Genomic DNA samples were stored at −20 °C until use. For the identification of BZ-associated SNPs, genomic DNA from individual worms was used. For the analysis of genetic diversity within and among populations, ‘pooled’ DNA was subjected to PCR, cloned and then sequenced. To prepare DNA template representing individual populations, 2 μl aliquots of the genomic DNAs from each of the 24 individual worms from each population were pooled.
Fig. 1.
Geographical origins of Haemonchus contortus populations in China. Samples from Hebei (HeB), Inner Mongolia (IM), Liangning (LN) and Heilongjiang (HLJ) were isolated from sheep. Samples from Hubei (HuB), Yunnan (YN), Shaanxi (SX) and Guangxi (GX) were from goats.
2.2. Specific identification of individual H. contortus
The specific identity of each worm as H. contortus was confirmed by molecular means. A 265 bp region of the second internal transcribed spacer (ITS-2) of nuclear ribosomal DNA gene was specifically amplified by conventional PCR from individual genomic DNA samples using the primer pair HAE (5′-CAAATGGCATTTGTCTTTTAG-3′) + NC2 (5′-TTAGTTTCTTTTCCTCCGCT-3′) (Bott et al., 2009). In brief, PCR was performed in 25 μl of 10 mM Tris-HCl, pH 8.3, 50 mM KCl, 4 mM MgCl2, 250 μM each of dNTP, 100 pmol of each primer and 1 U Taq polymerase (TaKaRa) in thermal cycler (Eastwin Life Sciences, Inc., China) using the following protocol: 94 °C/5 min, followed by 35 cycles of 94 °C/30 s, 55 °C/30 s and 72 °C/30 s, with a final extension at 72 °C/7 min. Known positive and a negative (no-template) control samples were included in each PCR run. All amplicons were examined on 1.5% agarose gels, and detected upon ultraviolet transillumination using an electronic documentation system (Alphamager Mini) to confirm a single band of the appropriate size. Subsequently, the amplicons were column-purified (Wizard PCR-Preps, Promega) and then directly sequenced (in separate reactions) with both primer HAE and NC2 using the BigDye Terminator v.3.1 cycle sequencing kit (Applied Biosystems) in an automated sequencer (PRISM3730, ABI). The identity of each sequence was established based on the comparison with reference sequences of H. contortus ITS-2 available in the GenBank database (GenBank accession nos. X78803 and EU084691.1). Eventually, 24 individual adult male worms from each of the eight populations were identified as H. contortus (Table 1).
Table 1.
Number and frequency (%) of individual worm genotypes with “susceptible” and/or “resistant” single nucleotide polymorphisms (SNPs) at codons 198 and 200 associated with benzimidazole resistance in isotype-1 β-tubulin gene of eight Haemonchus contortus populations from goats and sheep in China.
| Populationa (number) | Number of genotypeb and frequency (%)c |
|||||
|---|---|---|---|---|---|---|
| Hs-198, Hs-200 | Het-198, Hs-200 | Hs-198, Het-200 | Het-198, Het-200 | Hs-198, HR-200 | HR-198, Hs-200 | |
| HuB (24) | 16 (66.7) | 4 (16.7) | 2 (8.3) | 0 (0) | 0 (0) | 2 (8.3) |
| HeB (24) | 19 (79.1) | 3 (12.5) | 1 (4.2) | 0 (0) | 0 (0) | 1 (4.2) |
| YN (24) | 0 (0) | 2 (8.3) | 9 (37.5) | 6 (25.0) | 0 (0) | 7 (29.2) |
| SX (24) | 24 (100.0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| GX (24) | 3 (12.5) | 7 (29.1) | 6 (25.0) | 4 (16.7) | 1 (4.2) | 3 (12.5) |
| IM (24) | 2 (8.3) | 1 (4.2) | 0 (0) | 7 (29.2) | 1 (4.2) | 13 (54.1) |
| LN (24) | 7 (29.2) | 5 (20.8) | 4 (16.7) | 2 (8.3) | 0 (0) | 6 (25.0) |
| HLJ (24) | 18 (75.0) | 2 (8.3) | 4 (16.7) | 0 (0) | 0 (0) | 0 (0) |
HuB-Hubei, HeB-Hebei, YN-Yunnan, SX-Shaanxi, GX-Guangxi, IM-Inner Mongolia, LN-Liaoning, HLJ-Heilongjiang.
Hs = homozygous susceptible; Het = heterozygote; HR = homozygous resistant.
Frequency (%) was obtained by dividing the number of genotype by the total number of worms in each population.
2.3. PCR amplification and direct sequencing of the isotype-1 β-tubulin gene fragment of H. contortus
A 385 bp region (including exons 4 and 5 and their intervening intron) of the isotype-1 β-tubulin gene was amplified from individual H. contortus genomic DNA samples using primer pair HcPy2PCR-F (5′-GACGCATTCACTTGGAGGAG-3′) and HcPy2PCR-R (5′-CATAGGTTGGATTTGTGAGTT-3′) (von Samson-Himmelstjerna et al., 2009) by PCR (as described in Subsection 2.2) using the following cycling protocol: 94 °C/3 min, followed by 40 cycles at 94 °C/40 s, 53 °C/40 s and 68 °C/40 s, with a final extension at 68 °C/7 min. Amplicons were subjected to agarose gel (1.5%) electrophoresis to confirm that each represented a single band, followed by individual PCR product purification and direct sequencing (as described in Subsection 2.2) with primers HcPy2PCR-F and HcPy2PCR-R (in both directions).
2.4. Determination of the frequencies of the isotype-1 β-tubulin F167Y, E198A and F200Y BZ resistance-associated SNPs in H. contortus populations
Sequence traces were examined using Chromas v.2.4.1 software, focusing on the SNPs F167Y, E198A and F200Y. As described by Kotze et al. (2012), in individual sequences, a secondary peak whose ‘height’ was at least 50% of the dominant peak at the same position was considered indicative of heterozygosity; the presence of a single peak was regarded as homozygous at that position. In the present study, the same threshold was used to identify the SNPs in the isotype-1 β-tubulin gene region for individual adult worms. Genotypic frequencies and allelic frequencies were calculated as described (Tiwari et al., 2006); a detailed description is given in Table 2.
Table 2.
Frequencies (%) of single nucleotide polymorphism (SNPs) that resulted in an amino acid change at codons E198A (GAA/GCA) and F200Y (TTC/TAC) associated with benzimidazole resistance at isotype-1 β-tubulin gene of Haemonchus contortus from goats and sheep in China.
| Populationa | P198 |
P200 |
||
|---|---|---|---|---|
| Resistantb | Susceptible | Resistantc | Susceptible | |
| HuB | 17 | 83 | 4 | 96 |
| HeB | 10 | 90 | 2 | 98 |
| YN | 46 | 54 | 31 | 69 |
| SX | 0 | 100 | 0 | 100 |
| GX | 35 | 65 | 25 | 75 |
| IM | 70 | 30 | 19 | 81 |
| LN | 40 | 60 | 12.5 | 87.5 |
| HLJ | 4 | 96 | 8 | 92 |
HuB-Hubei, HeB-Hebei, YN-Yunnan, SX-Shaanxi, GX-Guangxi, IM-Inner Mongolia, LN-Liaoning, HLJ-Heilongjiang.
Resistant = 198A; Susceptible = E198. The allele frequency for 198A was calculated using the data in Table 1 as below: ((Het-198, Hs-200) + (Het-198, Het-200) + 2(HR-198, Hs-200))/2 × 24; The allele frequency for E198 was calculated using the data in Table 1 as below: (2(Hs-198, Hs-200) + (Het-198, Hs-200) + 2(Hs-198, Het-200) + (Het-198, Het-200) + 2(Hs-198, HR-200))/2 × 24.
Resistant = 200Y; Susceptible = F200. The allele frequency for 200Y was calculated using the data in Table 1 as follow: ((Hs-198, Het-200) + (Het-198, Het-200) + 2(Hs-198, HR-200))/2 × 24; The allele frequency for F200 was calculated using the data in Table 1 as follow: (2(Hs-198, Hs-200) + 2(Het-198, Hs-200) + (Hs-198, Het-200) + (Het-198, Het-200) + 2(HR-198, Hs-200))/2 × 24.
2.5. Cloning and sequencing of the isotype-1 β-tubulin gene fragment and detection of SNPs F167Y, E198A and F200Y
The 385 bp of isotype-1 β-tubulin gene region was amplified from each pooled DNA sample (n = 8) as described in Subsection 2.3. The eight PCR products were then cloned into pGEM-T vector (Promega) according to manufacturer's instructions, and 15–17 clones were selected per population and fully sequenced in both directions. Using a published isotype-1 β-tubulin gene sequence (GenBank accession no. X67489; Kwa et al., 1994) as a reference, only SNPs occurring multiple times per population were recorded as ‘true’ SNPs, as recommended in recent publications (Chaudhry et al., 2015, Redman et al., 2015).
2.6. Sequence diversity, genetic recombination and phylogenetic network analysis
The raw sequences were aligned and edited using the program Clustal W within the suite MEGA 5.0 (Tamura et al., 2011), followed by the analysis of genetic recombination according to the methods of RDP, MaxChi, 3Seq, Chimera, Siscan, Bootscan and GenConv using software RDP4 (Heath et al., 2006). The program DnaSP, v.5.10 (see Subsection 2.5) was used to establish haplotypes and calculate genetic diversity (haplotype diversity: Hd; nucleotide diversity: π) in the isotype-1 β-tubulin gene within each population. Pairwise FST values were obtained using the same program (Librado and Rozas, 2009). In addition, a hierarchical analysis of molecular variance (AMOVA) was conducted to estimate the diversity within and among populations, and the possible factors that affect gene flow using the Arlequin 3.1 package (Excoffier et al., 1992). Then, circular (equal angle) split networks were generated from the haplotype data using the neighbor-net method within SplitsTrees4 software (Huson and Bryant, 2006). In addition, a median joining (MJ) network was drawn using the program Network v.4.6.1 to establish haplotype relationships (Bandelt et al., 1999). Furthermore, phylogenetic analysis of isotype-1 β-tubulin gene sequence data was conducted using the maximum likelihood (ML) method in MEGA 5.0 using 1000 bootstrap replicates (Tamura et al., 2011).
3. Results
3.1. The genotypes and SNP frequencies in the isotype-1 β-tubulin gene in eight H. contortus populations in China
For all 192 individual male adults from eight H. contortus populations, SNPs E198A (GAA to GCA) and F200Y (TTC to TAC) were detected, whereas F167Y (TTC to TAC) was not. The genotypes and SNP frequencies are given in Table 1, Table 2, respectively. In total, six genotypes were detected among the eight populations of H. contortus. The primary genotypes (Hs-198 and Hs-200) homozygous at both P198 and P200 had frequencies of 29.2–100% in five H. contortus populations (HuB, HeB, SX, LN and HLJ) and low frequencies (0–12.5%) in three others (YN, IM and GX). Interestingly, in population IM, the genotype (HR-198, Hs-200) was ‘homozygous resistant’ at P198 but ‘homozygous susceptible’ at P200, and was most frequent (54.1%). The genotype (Hs-198, Hs-200) was ‘homozygous resistant’ at P200 and ‘homozygous susceptible’ at P198 in populations GX and IM, and was infrequent (4.2%). Simultaneously, genotypes that were heterozygous at one position and homozygous at another position (Het-198, Hs-200; Hs-198, Het-200) were found in all populations except SX at frequencies ranging from 4.2 to 37.5%. A genotype that was heterozygous at two positions (Het-198, Het-200) was detected in populations YN, GX, IM and LN at frequencies ranging from 8.3 to 29.2% (Table 1). Genotypes that were ‘homozygous resistant’ at two positions or ‘homozygous resistant’ at one position and heterozygous at another position were not found in any population. In the eight H. contortus populations, the frequency of (resistant) SNP 198A ranged from 0 to 70% and that of (susceptible) SNP E198 ranged from 30% to 100%. The frequencies of (resistant) SNP 200Y ranged from 0 to 31%, and from 69 to 100% for (susceptible) SNP F200 (Table 2).
3.2. Genetic diversity and population structure of H. contortus populations
In order to study the genetic diversity in the isotype-1 β-tubulin gene of H. contortus representing eight populations, a 385 bp region (including partial exons 4 and 5 and their intervening intron) of isotype-1 β-tubulin gene was PCR-amplified, cloned and sequenced from pooled genomic DNA of each population, and 15–17 sequences were obtained per population (a total of 132). Following filtering, 4–9 haplotypes were identified per population, resulting in a total of 36 haplotypes from eight populations (GenBank accession nos. KX258881, KX258883–KX258889, KX258897–KX258899, KX258901–KX258903, KX258905, KX258907, KX258909–KX258910, KX258913, KX258915–KX258916, KX258919, KX258921, KX258923–KX258927, KX258930, KX258939–KX258944 and KX258946) (Supplementary Table S1). High degrees of haplotype diversity were observed within individual populations, ranging from 0.455 to 0.939, with nucleotide diversities ranging from 0.018 to 0.039 (Table 3).
Table 3.
Genetic diversity indices of isotype-1 β-tubulin gene of Haemonchus contortus from goats and sheep in China.
| Populationa | No. of sequences | No. of haplotypes | No. of haplotypes with F200Y | No. of haplotypes with E198A | Hd | π |
|---|---|---|---|---|---|---|
| HuB | 16 | 9 | 1 | 1 | 0.939 | 0.03910 |
| HeB | 17 | 5 | 0 | 1 | 0.788 | 0.03514 |
| YN | 15 | 9 | 3 | 1 | 0.912 | 0.02970 |
| SX | 17 | 8 | 0 | 0 | 0.808 | 0.02704 |
| GX | 17 | 9 | 2 | 1 | 0.936 | 0.02401 |
| IM | 17 | 4 | 0 | 2 | 0.455 | 0.01834 |
| LN | 17 | 9 | 4 | 2 | 0.912 | 0.03182 |
| HLJ | 16 | 8 | 1 | 0 | 0.894 | 0.01862 |
| Total | 132 | 36 | 7 | 2 | 0.918 | 0.03146 |
The number of sequences were obtained by cloning and sequencing from pooled DNA extracted from individual worms from each of the eight H. contortus populations.
The number of haplotypes and haplotype diversity (Hd) and nucleotide diversity (π) of partial β-tubulin isotype-1 gene sequences were defined using the program DnaSP5.1.
HuB-Hubei, HeB-Hebei, YN-Yunnan, SX-Shaanxi, GX-Guangxi, IM-Inner Mongolia, LN-Liaoning, HLJ-Heilongjiang.
To understand the population genetic structure, pairwise FST values were calculated from sequences of isotype-1 β-tubulin gene of H. contortus representing the eight populations; low pairwise values ranging from 0.00907 to 0.30732 between the eight populations were obtained (Table 4). To assess the possible factors that might affect gene flow among populations, a hierarchical analysis of molecular variance (AMOVA) was computed for the eight H. contortus populations. With the introduction of three levels in the analysis, it was shown that 93% of the genetic variance was partitioned within populations, with 7% among populations. Simultaneously, the different geographical origins and host species of H. contortus populations had little effect (<2%) on the genetic structure (Table 5).
Table 4.
Pairwise FST between the populations of Haemonchus contortus from eight different geographical locations in China, calculated from sequences of isotype-1 β-tubulin gene haplotypes.
| Populationsa | HuB | HeB | YN | SX | GX | IM | LN | HLJ |
|---|---|---|---|---|---|---|---|---|
| HuB | ||||||||
| HeB | −0.13345 | |||||||
| YN | 0.05049 | −0.02995 | ||||||
| SX | 0.01053 | −0.10238 | 0.00907 | |||||
| GX | 0.03682 | −0.04000 | 0.06851 | −0.03945 | ||||
| IM | −0.02288 | −0.09677 | −0.07324 | −0.05254 | 0.01355 | |||
| LN | −0.01507 | −0.07379 | −0.04376 | −0.04360 | −0.01188 | −0.10859 | ||
| HLJ | 0.11203 | 0.03163 | 0.29449 | 0.14961 | 0.11275 | 0.30732 | 0.21231 |
The value of FST were calculated using DnaSP5.1.
Negative values indicate that more nucleotide substitutions occur within than between populations.
HuB-Hubei, HeB-Hebei, YN-Yunnan, SX-Shaanxi, GX-Guangxi, IM-Inner Mongolia, LN-Liaoning, HLJ-Heilongjiang.
Table 5.
Analysis of Molecular Variance (AMOVA) for eight populations of Haemonchus contortus from China based on isotype-1 β-tubulin gene sequences.
| Variance component | Variance | % of total | P | F-statistic |
|---|---|---|---|---|
| Among populations | 0.03482 | 6.61 | – | – |
| Within populations | 0.43414 | 93.39 | 0c | FST = 0.06605 |
| Between two host speciesa | −0.00962 | −2.07 | 0.93 | FCT = −0.02069 |
| Among populations within host species | 0.04032 | 8.67 | 0c | FSC = 0.08498 |
| Within populations | 0.43414 | 92.70 | 0c | FST = 0.07300 |
| Among four groupsb | −0.00442 | −0.94 | 0.59 | FCT = −0.00943 |
| Among populations within groups | 0.03860 | 8.24 | 0c | FSC = 0.08166 |
The hierarchical analysis of molecular variance (AMOVA) was performed using the Arlequin 3.1 software.
Negative values might indicate more nucleotide differences between parasites within the same group than between different groups.
The eight populations were divided into two groups in the light of two different host species: Goat (Hubei, Yunnan, Shaanxi and Guangxi), Sheep (Hebei, Inner Mongolia, Liaoning and Heilongjiang).
The eight populations were divided into four groups according to their geographical origins: Southwest (Guangxi and Yunnan), Central (Hubei and Shaanxi), North (Hebei and Inner Mongolia), Northeast (Liaoning and Heilongjiang).
P < 0.05.
3.3. Genetic relationships among isotype-1 β-tubulin gene haplotypes of H. contortus
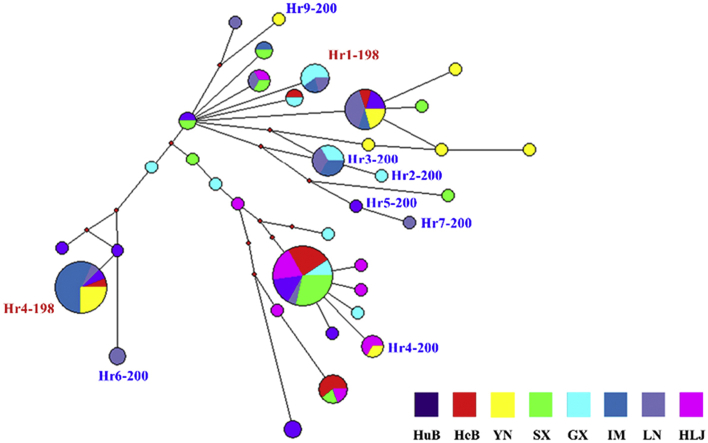
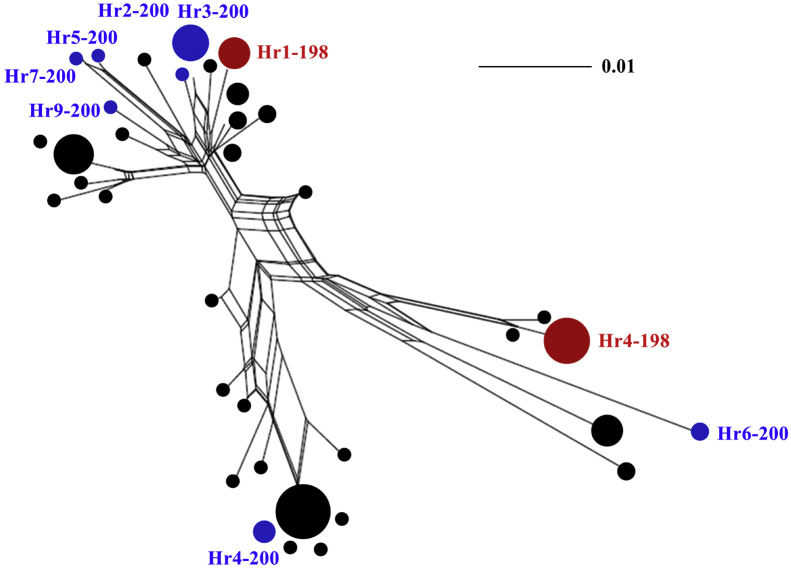
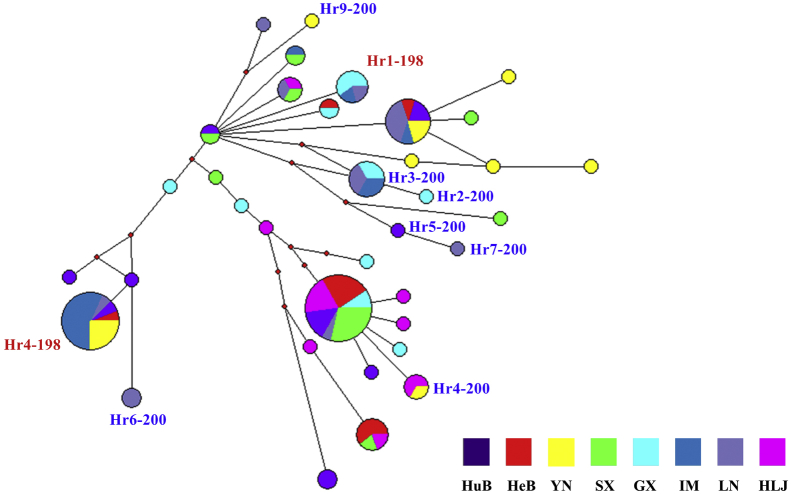
Thirty-six distinct isotype-1 β-tubulin gene haplotypes were identified among 132 sequences from eight H. contortus populations. Two of the 36 haplotypes had (resistant) SNP E198A and seven of them had (resistant) SNP F200Y; the remaining 27 haplotypes had BZ-susceptible SNPs at both P198 and P200. To test the hypothesis that BZ resistant SNPs have multiple origins, a network (SplitsTrees) was built with the 36 distinct isotype-1 β-tubulin gene haplotypes to examine their genetic relationships. The network revealed two haplotypes possessing resistant SNP E198A (Hr1-198 and Hr4-198) in two distinct groups in the network, with each containing at least one haplotype with a ‘susceptible’ SNP, indicating two independent origins of E198A (Fig. 2). The seven haplotypes possessing resistant SNP F200Y were found in three distinct groups in the network, and these haplotypes clustered with at least one haplotype with a ‘susceptible’ SNP, suggesting at least three different origins of this SNP (Fig. 2). The topology of a Median-Joining network built using data for all 36 haplotypes was consistent with that of SplitsTrees network, which supported the hypothesis of multiple origins of the resistant SNPs (Fig. 3). Two haplotypes (Hr3-200 and Hr4-200) of seven sequences containing resistant SNP F200Y were present in more than one population. The two haplotypes (Hr1-198 and Hr4-198) containing resistant SNP E198A were present in at least three populations (Fig. 3). A maximum likelihood (ML) tree built with MEGA5 also showed the same phylogenetic relationships (Supplementary Fig. S1).
Fig. 2.
Phylogenetic relationships among Haemonchus contortus isotype-1 β-tubulin gene sequences established using the neighbor-net method in SplitsTree 4.14 software (Huson and Bryant, 2006). This network was built with 36 different isotype-1 β-tubulin gene haplotypes from eight H. contortus populations. The distinct haplotypes represent three groups: black circles without corresponding text represent benzimidazole (BZ) ‘susceptible’ haplotypes containing F200Y (TTC)/F167Y (TTC)/E198A (GAA); the red and blue circles with corresponding text represent E198A-resistant haplotypes containing F200Y (TTC)/F167Y (TTC)/E198A (GCA) and F200Y-resistant haplotypes containing F200Y (TAC)/F167Y (TTC)/E198A (GAA), respectively. The size of individual circles is proportional to the frequency of individual haplotype in the overall dataset. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 3.
The median joining network of Haemonchus contortus isotype-1 β-tubulin gene sequences generated in Network v.4.6.1. This network was built using 36 different isotype-1 β-tubulin gene haplotypes from eight populations. The size of each circle representing each haplotype is proportional to its frequency within the sequence dataset. The different coloured squares at the bottom represent each population. Colours in each circle show the frequency in each population. Small red dots represent median vectors. The red and blue text indicate E198A - benzimidazole (BZ) resistant haplotypes containing F200Y (TTC)/F167Y (TTC)/E198A (GCA) and F200Y- BZ resistant haplotypes containing F200Y (TAC)/F167Y (TTC)/E198A (GAA), respectively. BZ susceptible haplotypes are not marked. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
In addition, we assessed whether the recombination might play a role in increasing the haplotype diversity in the isotype-1 β-tubulin gene. However, there was no significant signal of recombination among all haplotypes using distinct methods, showing that genetic recombination did not contribute to haplotypic diversity in the H. contortus populations studied.
4. Discussion
This is the first study to examine all three BZ resistance-associated SNPs at codons F167Y, E198A and F200Y in the isotype-1 β-tubulin gene in eight populations of H. contortus in China. The homozygous resistant genotypes linked to SNP E198A was most frequently encountered (Table 1, Table 2). Based on these and previous results for China (Bo and Li, 2005, Hao, 2007, Cai and Bai, 2009), we suggest that E198A plays a more significant role in BZ resistance than F200Y (which is the primary SNP associated with BZ resistance worldwide; Kotze et al., 2014).
It has been suggested that if ≥ 10% of the individuals in a population have a resistance genotype (designated as homozygous resistance at either P167 or P200 or heterozygote at both P167 and P200), the population is considered to be BZ resistant (Barrere et al., 2013a). However, in the present study, BZ resistance-associated SNP F167Y was not detected in any population, whereas E198A occurred in most populations. As SNPs F167Y, E198A and F200Y are known to be linked to BZ resistance (Kwa et al., 1994, Kwa et al., 1995, Silvestre and Cabaret, 2002, Ghisi et al., 2007, Rufener et al., 2009), the four populations (YN, GX, IM and LN) are predicted to be BZ resistant with frequencies of resistant, homozygous genotypes (Hs-198, HR-200 + HR-198, Hs-200) ranging from 16.7% to 58.3% (Table 1). In addition, both E198A and F200Y were detected in seven of the eight H. contortus populations, with frequencies ranging from 4% to 70% and 2% to 31%, respectively (Table 2). These results suggest that each population has a different BZ resistance status, and that the high frequencies of BZ resistant genotypes (homozygous resistant at P198 or P200) in YN, LN, GX and IM might indicate an excessive use of BZ drugs (over time) in these regions. This proposal is further supported by the evidence of heterozygosity at positions P198 and P200 in these four populations (Table 1). As no BZ resistant genotype was detected in worms from province SX, BZ drugs might still be applicable in this H. contortus population. For other populations (HuB, HeB and HLJ), heterozygosity at one position may suggest a risk of a gradual emergence of BZ resistance, which would need monitoring.
In the present study, no H. contortus genotypes that were ‘homozygous resistant’ at both codon 198 and 200 or ‘homozygous resistant’ at one codon (198 or 200) and heterozygous at another codon (200 or 198) were found in any population in China (Table 1). This finding might be explained by “mutual exclusion” in nature between the two homozygous alterations, as both mutations would result in worm lethality (Ghisi et al., 2007, Kotze et al., 2012).
Based on our findings, the occurrence of BZ resistance-associated SNPs in the isotype-1 β-tubulin gene in H. contortus populations from China is quite different from previous reports for other countries, which all detected F167Y and F200Y in field populations of H. contortus but not E198A (Silvestre and Cabaret, 2002, Barrere et al., 2012, Barrere et al., 2013a, Barrere et al., 2013b, Brasil et al., 2012, Chaudhry et al., 2014, dos Santos et al., 2014, Redman et al., 2015). In the present study, F167Y was not detected in any sequence, whereas E198A and F200Y had differing distributions, which is consistent with the findings from a recent study of H. contortus populations from southern India (Chaudhry et al., 2015). Thus, we propose that although E198A is the rarest of the three known SNPs (von Samson-Himmelstjerna et al., 2007, von Samson-Himmelstjerna et al., 2009), it appears to be more widespread in some regions of Asia than in Europe and the Americas, although one recent report from Pakistan did not detect E198A in any H. contortus population studied (Hussain et al., 2014).
The results of the present study are relatively consistent to those from India, the UK and Brazil, which indicate multiple independent origins of SNP F200Y (Brasil et al., 2012, Chaudhry et al., 2015, Redman et al., 2015). We found that seven haplotypes encoding F200Y (Hr2-200, Hr3-200, Hr4-200, Hr5-200, Hr6-200, Hr7-200 and Hr9-200) were represented in multiple distinct groupings in the network, suggesting multiple origins of this SNP and supporting its broad distribution throughout the world. In the case of mutation E198A in Chinese populations, it was found that two haplotypes (Hr1-198 and Hr4-198) encoding E198A were represented in two distinct groups of the network, coupling to at least one susceptible haplotype, which supports the hypothesis that this SNP has spread to multiple regions in China from two distinct origins (Fig. 2, Fig. 3). An interesting discovery was that haplotype Hr4-198 (385 bp in length) in our study contains 324 bp of nucleotide sequences identical to the sequences of the haplotype Hr24 (324 bp in length, GenBank accession no. KP792525) which is the only haplotype carrying E198A in H. contortus specimens from India (Chaudhry et al., 2015). Therefore, we speculate that SNP E198A in H. contortus from China and India might share a common origin. In addition, it was found that not only the susceptible haplotypes (n = 7) were identified in at least two populations, but that also resistant haplotypes (Hr3-200, Hr4-200, Hr1-198 and Hr4-198) were present in more than one population (Fig. 3). These results indicate a high gene flow among all populations, and that BZ drug resistance is likely a nation-wide problem in China. This problem is likely exacerbated by anthropogenic transportation of hosts across China, as discussed recently by Yin et al. (2013).
Consistent with previous findings using mitochondrial gene and microsatellites (Yin et al., 2013, Yin et al., 2016), results from the present study also revealed a high degree of genetic diversity within Chinese H. contortus populations. Also a study of the isotype-1 β-tubulin gene of H. contortus populations from Brazil revealed nucleotide diversities ranging from 0.025 to 0.038 (Brasil et al., 2012), similar to those calculated in the present study (ranging from 0.018 to 0.039). Nevertheless, a distinct genetic structure was absent for Chinese H. contortus populations, corroborating high gene flow among them. This proposal is supported by evidence of low pairwise FST values between Chinese populations (Table 4). Moreover, limited variance between H. contortus from sheep (HeB, IM, LN and HLJ) and from goats (HuB, YN, SX and GX) indicates cross infection between these host species, with little or no barrier (Table 5). Furthermore, different geographical origins appeared to have little influence on the genetic structure of H. contortus populations in China, suggesting a high gene flow among populations (Table 5). However, FST values between population HLJ and other H. contortus populations from China was higher than those between other populations (Table 4), which might be explained by infrequent host movement between HLJ and other populations (and vice versa), and possibly some other natural factors, including geographic and climatic variations, preventing dispersal and gene flow.
In order to evaluate the situation of BZ resistance in China, we compared studies that used conventional techniques with those that employed molecular methods of ‘resistance detection’. It was found that different degrees of BZ resistance were identified in H. contortus or other trichostrongyloid nematodes in some provinces using faecal egg count reduction testing (FECRT) or the egg hatch assay (EHA), indicating that BZ resistance is an increasing issue in China (He et al., 1999a, He et al., 1999b, Cai et al., 2007a, Cai et al., 2007b, Pu and Yue, 2009, Zhao et al., 2010), as FECRT and EHA were recommended by the WAAVP (World Association for the Advancement of Veterinary Parasitology) and the creditability of these two methods was demonstrated (Coles et al., 1992). Therefore, the findings from the present study emphasize the need, in the future, to screen for or monitor all three mutations (i.e. F167Y, E198A and F200Y) to assess BZ resistance in H. contortus populations in China.
In conclusion, this is the first genetic analysis of all three BZ resistance-associated SNPs F167Y, E198A and F200Y and of sequence diversity in the isotype-1 β-tubulin gene of H. contortus populations from sheep and goats in China. Different frequencies of E198A and F200Y were detected in all eight populations studied; E198A occurred more frequently than did F200Y in field populations, whereas F167Y was not detected. The results also confirmed a high genetic diversity within each population and a high gene flow among all H. contortus populations studied. Genetic relationship analysis suggested that F200Y had multiple origins in China, and E198A had two distinct origins. Considering that small ruminant production is one of the important livestock industries in China, the use of anthelmintics, including BZ, seems unavoidable at this point. Therefore, we conclude that it will be of considerable importance to develop guidelines for parasite control and management practices in China, aiming at reducing the selection pressures for anthelmintic resistance. It will also be important to develop parasite management strategies tailored to individual livestock farms, including quarantine treatments of imported livestock, employing suitable combinations of drugs for strategic treatment, identifying refugia in hosts with susceptible parasites and careful monitoring anthelmintic resistance using molecular tools. Finally, in our opinion, detailed studies of other resistances, particularly to macrocyclic lactones, in parasitic nematodes of livestock animals are urgently needed in China, in order to inform future integrated parasite control programs.
Conflicts of interest
The authors declare that they have no competing interests.
Acknowledgements
This study was supported by the “National Key Basic Research Program (973 Program) of China” (Grant No. 2015CB150300) and the “Special Fund for Agro-Scientific Research in the Public Interest” (Grant no. 201303037). Funding from Huazhong Agricultural University Scientific & Technological Self-innovation Foundation (Program No. 2015RC005), the Australian Research Council (ARC), the National Health and Medical Research Council (NHMRC) and Melbourne Water Corporation is gratefully acknowledged (RBG).
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.ijpddr.2016.10.001.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- Bandelt H.J., Forster P., Rohl A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- Barrere V., Alvarez L., Suarez G., Ceballos L., Moreno L., Lanusse C., Prichard R.K. Relationship between increased albendazole systemic exposure and changes in single nucleotide polymorphisms on the beta-tubulin isotype 1 encoding gene in Haemonchus contortus. Vet. Parasitol. 2012;186:344–349. doi: 10.1016/j.vetpar.2011.11.068. [DOI] [PubMed] [Google Scholar]
- Barrere V., Falzon L.C., Shakya K.P., Menzies P.I., Peregrine A.S., Prichard R.K. Assessment of benzimidazole resistance in Haemonchus contortus in sheep flocks in Ontario, Canada: comparison of detection methods for drug resistance. Vet. Parasitol. 2013;198:159–165. doi: 10.1016/j.vetpar.2013.07.040. [DOI] [PubMed] [Google Scholar]
- Barrere V., Keller K., von Samson-Himmelstjerna G., Prichard R.K. Efficiency of a genetic test to detect benzimidazole resistant Haemonchus contortus nematodes in sheep farms in Quebec, Canada. Parasitol. Int. 2013;62:464–470. doi: 10.1016/j.parint.2013.06.001. [DOI] [PubMed] [Google Scholar]
- Bo X.W., Li X.R. Multiplex PCR detection of allele on benzimidazole resistance or susceptibity in natural populations of Haemonchus contortus. Sci. Agric. Sin. 2005;38:826–830. (In Chinese) [Google Scholar]
- Bott N.J., Campbell B.E., Beveridge I., Chilton N.B., Rees D., Hunt P.W., Gasser R.B. A combined microscopic-molecular method for the diagnosis of strongylid infections in sheep. Int. J. Parasitol. 2009;39:1277–1287. doi: 10.1016/j.ijpara.2009.03.002. [DOI] [PubMed] [Google Scholar]
- Brasil B.S., Nunes R.L., Bastianetto E., Drummond M.G., Carvalho D.C., Leite R.C., Molento M.B., Oliveira D.A. Genetic diversity patterns of Haemonchus placei and Haemonchus contortus populations isolated from domestic ruminants in Brazil. Int. J. Parasitol. 2012;42:469–479. doi: 10.1016/j.ijpara.2012.03.003. [DOI] [PubMed] [Google Scholar]
- Cai K.Z., Yang X.Y., Yand C.S., Hao C., Wang X.L., Liang X.J., Wang R., Zhang X.D., Zhang Y.L., Li X., Han H.B. An egg hatch assay using albendazole for in vitro detection of resistance of sheep and goat nematodes to anthelmintics. Chin. Vet. Sci. 2007;37:721–725. (In Chinese) [Google Scholar]
- Cai K.Z., Yang X.Y., Wang X.L., Hao C., Yang A., Zhao Y.F. Investigation on resistance of gastrointestinal nematodes in sheep and goats to anthelmintics in Ningxia, China. Chin. Vet. Sci. 2007;37:491–495. (In Chinese) [Google Scholar]
- Cai K.Z., Bai J.L. Analysis of alleles resistant to benzimidazoles in Haemonchus contortus in sheep by PCR-RFLP. Chin. Vet. Sci. 2009;39:685–689. (In Chinese) [Google Scholar]
- Chaudhry U., Miller M., Yazwinski T., Kaplan R., Gilleard J. The presence of benzimidazole resistance mutations in Haemonchus placei from US cattle. Vet. Parasitol. 2014;204:411–415. doi: 10.1016/j.vetpar.2014.05.025. [DOI] [PubMed] [Google Scholar]
- Chaudhry U., Redman E.M., Raman M., Gilleard J.S. Genetic evidence for the spread of a benzimidazole resistance mutation across southern India from a single origin in the parasitic nematode Haemonchus contortus. Int. J. Parasitol. 2015;45:721–728. doi: 10.1016/j.ijpara.2015.04.007. [DOI] [PubMed] [Google Scholar]
- Coles G.C., Bauer C., Borgsteede F.H., Geerts S., Klei T.R., Taylor M.A., Waller P.J. World Association for the Advancement of Veterinary Parasitology (W.A.A.V.P.) methods for the detection of anthelmintic resistance in nematodes of veterinary importance. Vet. Parasitol. 1992;44:35–44. doi: 10.1016/0304-4017(92)90141-u. [DOI] [PubMed] [Google Scholar]
- dos Santos J.M., Monteiro J.P., Ribeiro W.L., Macedo I.T., Camurca-Vasconcelos A.L., Vieira Lda S., Bevilaqua C.M. Identification and quantification of benzimidazole resistance polymorphisms in Haemonchus contortus isolated in Northeastern Brazil. Vet. Parasitol. 2014;199:160–164. doi: 10.1016/j.vetpar.2013.11.006. [DOI] [PubMed] [Google Scholar]
- Excoffier L., Smouse P.E., Quattro J.M. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser R.B., Hu M., Chilton N.B., Campbell B.E., Jex A.J., Otranto D., Cafarchia C., Beveridge I., Zhu X. Single-strand conformation polymorphism (SSCP) for the analysis of genetic variation. Nat. Protoc. 2006;1:3121–3128. doi: 10.1038/nprot.2006.485. [DOI] [PubMed] [Google Scholar]
- Ghisi M., Kaminsky R., Maser P. Phenotyping and genotyping of Haemonchus contortus isolates reveals a new putative candidate mutation for benzimidazole resistance in nematodes. Vet. Parasitol. 2007;144:313–320. doi: 10.1016/j.vetpar.2006.10.003. [DOI] [PubMed] [Google Scholar]
- Hao C. Inner Mongolia Agricultural University; 2007. Study of Detection of Benzimidazoles Resistance and PCR-SSCP Analysis in Gastro-intestinal Nematodes of Small Domestic Ruminants. (Master's Degree Thesis) (In Chinese) [Google Scholar]
- He G.S., Gu Y.X., Cao J., Zhu S.H., Wang Q., Xu M.Q. Detection of anthelmintic resistance against Haemonchus contortus in goats using the egg hatch test. Chin. J. Vet. Parasitol. 1999;7:8–11. (In Chinese) [Google Scholar]
- He G.S., Wang C.R., Gu Y.X., Liu W.T., Cao G., Cao J. Detecting the anthelmintic resistance for nematodes of sheep in Da shan sheep ranch, Hei longjiang province. Chin. J. Vet. Parasitol. 1999;7:9–11. (In Chinese) [Google Scholar]
- Heath L., van der Walt E., Varsani A., Martin D.P. Recombination patterns in aphthoviruses mirror those found in other picornaviruses. J. Virol. 2006;80:11827–11832. doi: 10.1128/JVI.01100-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson D.H., Bryant D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- Hussain T., Periasamy K., Nadeem A., Babar M.E., Pichler R., Diallo A. Sympatric species distribution, genetic diversity and population structure of Haemonchus isolates from domestic ruminants in Pakistan. Vet. Parasitol. 2014;206:188–199. doi: 10.1016/j.vetpar.2014.10.026. [DOI] [PubMed] [Google Scholar]
- Jackson F., Coop R.L. The development of anthelmintic resistance in sheep nematodes. Parasitology. 2000;120(Suppl.):S95–S107. doi: 10.1017/s0031182099005740. [DOI] [PubMed] [Google Scholar]
- Kaplan R.M. Drug resistance in nematodes of veterinary importance: a status report. Trends Parasitol. 2004;20:477–481. doi: 10.1016/j.pt.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Kohler P. The biochemical basis of anthelmintic action and resistance. Int. J. Parasitol. 2001;31:336–345. doi: 10.1016/s0020-7519(01)00131-x. [DOI] [PubMed] [Google Scholar]
- Kotze A.C., Cowling K., Bagnall N.H., Hines B.M., Ruffell A.P., Hunt P.W., Coleman G.T. Relative level of thiabendazole resistance associated with the E198A and F200Y SNPs in larvae of a multi-drug resistant isolate of Haemonchus contortus. Int. J. Parasitol. Drugs Drug Resist. 2012;2:92–97. doi: 10.1016/j.ijpddr.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotze A.C., Hunt P.W., Skuce P., von Samson-Himmelstjerna G., Martin R.J., Sager H., Krucken J., Hodgkinson J., Lespine A., Jex A.R., Gilleard J.S., Beech R.N., Wolstenholme A.J., Demeler J., Robertson A.P., Charvet C.L., Neveu C., Kaminsky R., Rufener L., Alberich M., Menez C., Prichard R.K. Recent advances in candidate-gene and whole-genome approaches to the discovery of anthelmintic resistance markers and the description of drug/receptor interactions. Int. J. Parasitol. Drugs Drug Resist. 2014;4:164–184. doi: 10.1016/j.ijpddr.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwa M.S., Veenstra J.G., Roos M.H. Benzimidazole resistance in Haemonchus contortus is correlated with a conserved mutation at amino acid 200 in beta-tubulin isotype 1. Mol. Biochem. Parasitol. 1994;63:299–303. doi: 10.1016/0166-6851(94)90066-3. [DOI] [PubMed] [Google Scholar]
- Kwa M.S., Veenstra J.G., Van Dijk M., Roos M.H. Beta-tubulin genes from the parasitic nematode Haemonchus contortus modulate drug resistance in Caenorhabditis elegans. J. Mol. Biol. 1995;246:500–510. doi: 10.1006/jmbi.1994.0102. [DOI] [PubMed] [Google Scholar]
- Librado P., Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinforma. Oxf. Engl. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Lichtenfels J.R., Pilitt P.A., Hoberg E.P. New morphological characters for identifying individual specimens of Haemonchus spp. (Nematoda: Trichostrongyloidea) and a key to species in ruminants of North America. J. Parasitol. 1994;80:107–119. [PubMed] [Google Scholar]
- Nikolaou S., Gasser R.B. Prospects for exploring molecular developmental processes in Haemonchus contortus. Int. J. Parasitol. 2006;36:859–868. doi: 10.1016/j.ijpara.2006.04.007. [DOI] [PubMed] [Google Scholar]
- O'Connor L.J., Walkden-Brown S.W., Kahn L.P. Ecology of the free-living stages of major trichostrongylid parasites of sheep. Vet. Parasitol. 2006;142:1–15. doi: 10.1016/j.vetpar.2006.08.035. [DOI] [PubMed] [Google Scholar]
- Pu W.B., Yue C. Detection on the sheep resistance of gastrointestinal nematode to albendazole in Urumqi Area. Xinjiang Agric. Sci. 2009;46:217–221. (In Chinese) [Google Scholar]
- Redman E., Whitelaw F., Tait A., Burgess C., Bartley Y., Skuce P.J., Jackson F., Gilleard J.S. The emergence of resistance to the benzimidazole anthlemintics in parasitic nematodes of livestock is characterised by multiple independent hard and soft selective sweeps. PLoS Negl. Trop. Dis. 2015;9:e0003494. doi: 10.1371/journal.pntd.0003494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rufener L., Kaminsky R., Maser P. In vitro selection of Haemonchus contortus for benzimidazole resistance reveals a mutation at amino acid 198 of beta-tubulin. Mol. Biochem. Parasitol. 2009;168:120–122. doi: 10.1016/j.molbiopara.2009.07.002. [DOI] [PubMed] [Google Scholar]
- Shen J. The brief status of parasite and parasitosis for livestock and poultry in China. Chin. J. Vet. Parasitol. 2005;13:1–4. (In Chinese) [Google Scholar]
- Silvestre A., Cabaret J. Mutation in position 167 of isotype 1 beta-tubulin gene of Trichostrongylid nematodes: role in benzimidazole resistance? Mol. Biochem. Parasitol. 2002;120:297–300. doi: 10.1016/s0166-6851(01)00455-8. [DOI] [PubMed] [Google Scholar]
- Skuce P., Stenhouse L., Jackson F., Hypsa V., Gilleard J. Benzimidazole resistance allele haplotype diversity in United Kingdom isolates of Teladorsagia circumcincta supports a hypothesis of multiple origins of resistance by recurrent mutation. Int. J. Parasitol. 2010;40:1247–1255. doi: 10.1016/j.ijpara.2010.03.016. [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari J., Kumar S., Kolte A.P., Swarnkar C.P., Singh D., Pathak K.M. Detection of benzimidazole resistance in Haemonchus contortus using RFLP-PCR technique. Vet. Parasitol. 2006;138:301–307. doi: 10.1016/j.vetpar.2006.02.003. [DOI] [PubMed] [Google Scholar]
- von Samson-Himmelstjerna G., Blackhall W.J., McCarthy J.S., Skuce P.J. Single nucleotide polymorphism (SNP) markers for benzimidazole resistance in veterinary nematodes. Parasitology. 2007;134:1077–1086. doi: 10.1017/S0031182007000054. [DOI] [PubMed] [Google Scholar]
- von Samson-Himmelstjerna G., Walsh T.K., Donnan A.A., Carriere S., Jackson F., Skuce P.J., Rohn K., Wolstenholme A.J. Molecular detection of benzimidazole resistance in Haemonchus contortus using real-time PCR and pyrosequencing. Parasitology. 2009;136:349–358. doi: 10.1017/S003118200800543X. [DOI] [PubMed] [Google Scholar]
- Waller P.J. Anthelmintic resistance. Vet. Parasitol. 1997;72(3–4):391–405. doi: 10.1016/s0304-4017(97)00107-6. [DOI] [PubMed] [Google Scholar]
- Waller P.J., Rudby-Martin L., Ljungstrom B.L., Rydzik A. The epidemiology of abomasal nematodes of sheep in Sweden, with particular reference to over-winter survival strategies. Vet. Parasitol. 2004;122:207–220. doi: 10.1016/j.vetpar.2004.04.007. [DOI] [PubMed] [Google Scholar]
- Yin F., Gasser R.B., Li F., Bao M., Huang W., Zou F., Zhao G., Wang C., Yang X., Zhou Y., Zhao J.L., Fang R., Hu M. Genetic variablity within and among Haemonchus contortus isolates from goats and sheep in China. Parasit. Vectors. 2013;6:279. doi: 10.1186/1756-3305-6-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin F., Gasser R.B., Li F., Bao M., Huang W., Zou F., Zhao G., Wang C., Yang X., Zhou Y., Zhao J.L., Fang R., Hu M. First microsatellite analysis of Haemonchus contortus populations in China. Parasit. Vectors. 2016 doi: 10.1186/s13071-016-1864-z. under revision (R1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J.S., Pu W.B., Zhan Z.Y., Yue C. Anthelmintic resistance of nematodes in Trichostrongylidae to benzimidazoles. Chin. Vet. Sci. 2010;5:528–531. (In Chinese) [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.