Abstract
Most HIV-1 vaccines elicit neutralizing antibodies that are active against highly sensitive (tier-1) viruses or rare cases of vaccine-matched neutralization-resistant (tier-2) viruses, but no vaccine has induced antibodies that can broadly neutralize heterologous tier-2 viruses. In this study, we isolated antibodies from an HIV-1-infected individual that targeted the gp41 membrane-proximal external region (MPER) that may have selected single-residue changes in viral variants in the MPER that resulted in neutralization sensitivity to antibodies targeting distal epitopes on the HIV-1 Env. Similarly, a single change in the MPER in a second virus from another infected-individual also conferred enhanced neutralization sensitivity. These gp41 single-residue changes thus transformed tier-2 viruses into tier-1 viruses that were sensitive to vaccine-elicited tier-1 neutralizing antibodies. These data demonstrate that Env amino acid changes within the MPER bnAb epitope of naturally-selected escape viruses can increase neutralization sensitivity to multiple types of neutralizing antibodies, and underscore the critical importance of the MPER for maintaining the integrity of the tier-2 HIV-1 trimer.
Highlights
-
•
Amino acid changes in the HIV gp41 MPER can regulate neutralization sensitivity of distal epitopes.
-
•
MPER antibodies isolated early are resistant to MPER changes that enhance neutralization sensitivity.
-
•
HIV gp41 MPER is critical for determining overall HIV envelope conformations.
The HIV-1 envelope protein (Env) is the primary target for neutralizing antibodies. Most HIV-1 vaccines elicit neutralizing antibodies that are active against highly neutralization-sensitive (tier-1) or rare vaccine-matched more neutralization-resistant (tier-2) viruses, but no vaccine has induced antibodies that can broadly neutralize heterologous tier-2 viruses. In this study, we identified changes that occurred in two HIV-1-infected individuals in the membrane proximal region of the HIV-1 Env that resulted in neutralization sensitivity to antibodies targeting distal epitopes on the HIV Env. These single-residue changes thus transformed tier-2 viruses into tier-1 viruses, highlighting the importance of MPER residues in maintaining neutralization-resistant virus.
Graphical Abstract
1. Introduction
HIV-1 neutralization sensitivity occurs on a continuum, with some virus strains highly neutralization sensitive (tier-1 viruses) while others are more neutralization-resistant (tier-2 viruses). Transmitted/founder (T/F) HIV-1 strains are uniformly tier-2 viruses (Seaman et al., 2010, Derdeyn et al., 2014, Haim et al., 2011). The sensitivity of T/F viruses is shaped by easy-to-induce antibodies that can neutralize tier-1 viruses, thus selecting for neutralization-resistant founder viruses (Moody et al., 2015, Moore et al., 2009, Richman et al., 2003, Wei et al., 2003). The structural correlate of neutralization sensitivity is the exposure of epitopes such as the second (V2) and third (V3) variable loops on tier-1 viruses that are not exposed on tier-2 viruses (Mascola and Montefiori, 2010, Mccoy and Weiss, 2013). FRET analysis has demonstrated that the HIV-1 envelope can oscillate between an “open” neutralization-sensitive state (tier-1) and a “closed” more neutralization-resistant (tier-2) state (Munro et al., 2014). However, our understanding of the contribution of specific Env sequences to overall trimer conformation is incomplete.
HIV-1 Env is comprised of three gp120 monomers, each non-covalently linked with a transmembrane gp41 subunit. The gp120 subunits interact with target cell surface CD4 and a co-receptor to mediate viral entry (Berger, 1997, Wyatt and Sodroski, 1998). The membrane-proximal external region (MPER) is a highly conserved 23 amino acid stretch that is located in the heptad region-2 (HR-2) at the base of gp41, proximal to the transmembrane domain. The MPER plays a critical role in membrane fusion during viral entry into the host cell (Montero et al., 2008). The high sequence diversity and glycan shield of HIV-1 Env limit the breadth of most neutralizing antibodies, but broadly neutralizing antibodies (bnAbs) targeting the gp120, MPER and gp120-gp41 bridging regions have been identified (de Taeye et al., 2016, Burton and Mascola, 2015). BnAbs take years to develop during natural HIV-1 infection, have attributes that are disfavored by the host immune system, and have yet to be generated by any vaccine strategy (Haynes et al., 2012, Mascola and Haynes, 2013). One fundamental challenge has been the development of native-like Env trimers capable of expressing bnAb epitopes while shielding non-neutralizing epitopes. Recently, examples of native-like Env trimers, stabilized soluble gp140 SOSIP trimers that have truncated gp41 ectodomains (Sanders et al., 2015), have been developed and these trimers are able to induce autologous tier-2 neutralizing antibodies but have yet to elicit bnAbs. SOSIP trimers do not include the MPER (Sanders et al., 2015), and therefore would not be expected to elicit antibodies against that epitope.
CAP206 is a South African CAPRISA 002 cohort participant, who was infected by a clade C virus and at 81 weeks post-infection developed neutralization breadth that was mediated by MPER-reactive antibodies in the plasma (Gray et al., 2009). We previously isolated an MPER-reactive neutralizing mAb from CAP206 by single memory B cell sorting (Morris et al., 2011). This mAb, CAP206-CH12 utilized the same VH and VΚ gene segments, VH1–69 and VΚ3–20, and had a similar binding footprint as bnAb 4E10. Study of Env sequences in CAP206 from soon after infection to over 2.5 years revealed accumulation of amino acid changes within the CAP206 MPER (K677 N, W680R, and K683Q). We have also identified a similar gp41 MPER change in another clade C African individual, CH505, who developed bnAbs against the CD4-binding site (CD4bs) of gp120 (Liao et al., 2013b). Here, we show that these MPER changes determine HIV-1 neutralization sensitivity in both infected individuals. MPER antibodies isolated early during infection from CAP206 did not neutralize the viruses with MPER changes that displayed enhanced neutralization sensitivity, indicating that early autologous MPER-targeting antibodies could have selected for the MPER amino acid changes.
2. Materials and Methods
2.1. Study Subjects
Plasma and PBMCs were isolated from serial blood samples that were collected from subtype C HIV-1 infected, antiretroviral therapy-naïve individuals CAP206 and CH505 (Liao et al., 2013b, Gray et al., 2009, Morris et al., 2011). Plasma and PBMC samples were stored at − 80 °C and in liquid nitrogen tanks, respectively. HIV-1 viral envelope sequences were obtained from plasma over the course of infection (L. Morris, Unpublished). Ethical approval for studies using CAP206 specimens was obtained from the Universities of KwaZulu-Natal and the Witwatersrand. All work related to human subjects was in compliance with Institutional Review Board protocols approved by the Duke University Institutional Review Board and the local ethics boards where the individuals were recruited.
2.2. Site-Directed Mutagenesis
Specific amino acid changes to HIV-1 envelopes were introduced using QuikChange Site-Directed Mutagenesis Kit and Quik Change Lightning Site-Directed Mutagenesis Kit (Aligent Technologies, Santa Clara, CA). Mutations were confirmed by sequence analysis.
2.3. Neutralization Assays
Neutralizing antibody assays in TZM-bl cells were performed as described (Montefiori, 2005). Recombinant monoclonal antibodies were tested against autologous and heterologous HIV-1 Env-pseudotyped viruses in eight serial threefold dilutions starting at 100 μg/mL or 50 μg/mL as described (Montefiori, 2005, Seaman et al., 2010). IC50 values were calculated using the five-parameter logistic nonlinear regression model. The virus subtypes in the panel were selected to be consistent with previous publications (Huang et al., 2012, Seaman et al., 2010, Wu et al., 2010). Rhesus plasma neutralization titers were tested against the same HIV-1 Env-pseudotyped viruses in serial dilutions using non-heat inactivated plasma and titer was calculated as the reciprocal plasma dilutions causing a 50% reduction of relative light units (ID50).
2.4. Isolation and Expression of VHDHJH and VLJL Genes
The VHDHJH and VLJL gene-segment pairs of the isolated antibodies were amplified by reverse transcription followed by semi-nested PCR (RT-PCR) (Liao et al., 2009, Tiller et al., 2008) performed on flow-sorted or limited dilution memory B cell cultures (Bonsignori et al., 2011). Antigen-specific flow sorting was performed using HIV-1 Env CON-S gp140, an HIV-1 envelope known to react with all clades of HIV-1 positive sera (Tomaras et al., 2008), or MPR.03 peptide (KKKNEQELLELDKWASLWNWFDITNWLWYIRKKK) tetramers as described (Morris et al., 2011). Initial screening of memory B cell cultures was performed with CAP206 T/F gp140 Env. Antibodies were produced in bulk cultures by transient transfection of Expi293F cells (Life technologies, Grand Island, NY) with 1 mg of each heavy- and light-chain genes synthesized in pcDNA plasmids (GeneScript, Piscataway, NJ) per 1 L transfection as described (Liao et al., 2009). Rhesus macaque memory B cell sorting, gene amplification and antibody production was performed as described (Bradley et al., 2016, Wiehe et al., 2014).
2.5. Single Cell PCR Sequencing, Next-Generation Sequencing and Sequence Annotation
A PCR purification kit (Qiagen, Valencia, CA) was used to purify all single cell PCR products of Ig VHDHJH and VLJL genes. PCR products were sequenced in forward and reverse directions using ABI 3700 instrument and BigDye sequencing kit (Applied Biosystems).
Base calling for each sequence was performed using Phred (Ewing and Green, 1998). Forward and reverse strands of the Ig genes were assembled into one final nucleotide sequence based on quality scores at each base position and genetic information was inferred by using SoDA (Munshaw and Kepler, 2010).
For high throughput DNA sequencing of Ig V(D)J genes, genomic DNA samples were isolated from 9 serial aliquots of PBMCs from CAP206 sampled from the following weeks post HIV-1 transmission: 4, 15, 22, 33, 68, 120, 146, 198 and 254 weeks using Using QIAamp DNA mini kit. Heavy chain V gene segment family specific primers and a consensus J gene segment primer were multiplexed and used to amplify the rearranged VHDHJH Ig heavy chain sequences as previously described (Boyd et al., 2009). Six barcoded V(D)J libraries from independent aliquots of DNA template from each time pont were amplified, pooled and sequenced using the 454 platform with Titanium chemistry (Roche) (Boyd et al., 2009).
2.6. Identification of Clone Members and Inference of UCA
Clonal relatedness of VHDHJH and VLJL sequences was determined using an algorithm and the UCAs were inferred as described (Kepler et al., 2014, Munshaw and Kepler, 2010).
2.7. CH82 Antibody Blocking Assay
Antibody blocking assays were performed with CH82_UCA, CH82, CH133, PG9, PG16, PGT121, PGT125, PGT128 and 2G12. Three hundred eight-four-well ELISA plates (Costar #3700) were coated with CAP206 month 0 T/F gp140 overnight at 4 degree C and blocked with assay diluent (PBS containing 4% (weight/volume) whey protein/ 15% Normal Goat Serum/0.5% Tween20/ 0.05% Sodium Azide) for 1 h at room temperature (RT). Antibodies CH82_UCA, CH82, CH133, PG9, PG16, PGT121, PGT125, PGT128 and 2G12, starting at 100 μg/mL and diluted two-fold, were incubated in triplicate wells. Biotinylated mAb CH82 was added at (0.1 μg/mL), the EC50 determined by a direct binding of biotinylated-CH82, for 1 h at RT. Biotin-CH82 binding was detected with streptavidin-HRP (Thermo Scientific; Waltham, MA) at 1:30,000 (1 hour RT) followed by SureBlue Reserve TMB Microwell Peroxidase Substrate (Kirkegaard & Perry Laboratories, Inc.; Gaithersburg, MD). Reaction was stopped with 0.33 N HCL and plates were read at 450 nm. After background subtractions, percent inhibition was calculated as follows: 100-(mAb triplicate mean / no inhibition control mean) ∗ 100.
2.8. Antibody Autoreactivity
The polyreactivity of MPER-reactive antibodies was assayed in the AtheNA multi-lyte system (Zeus Scientific).
2.9. Recombinant HIV-1 Proteins
HIV-1 Envs for ELISA and SPR assays included HIV-1 MN recombinant gp41 (Immunodiagnostics), HIV-1 group M consensus gp120 (CON-S) (Liao et al., 2006), HIV-1 clade C consensus (ConC) gp120, ConC gp120 N332A mutant, Env immunodominant region peptide sp400 (RVLAVERYLRD-QQLLGIWGCSG-KLICTTAVPWN-ASWSNKSLNK), gp41 MPER region peptide SP62 (QQEKNEQELLELDKWASLWN) and GCN4 gp41 Inter (Frey et al., 2010). CAP206 autologous transmitted/founder env and 6 additional envs from the first 30 months of infection were obtained from serial blood samples by single genome amplification (Keele et al., 2008), codon optimized (Andre et al., 1998) and de novo synthesized (GeneScript) as gp140 or gp120, and cloned into pcDNA3.1/hygromycin (Invitrogen). Recombinant Env glycoproteins were produced in 293F cells in serum-free media transfected with HIV-1 gp140 or gp120 expressing pcDNA3.1 plasmids, purified from the supernatant of transfected 293F cells using Galanthus nivalis lectin-agarose (Vector Labs) column chromatography, and stored at − 80 °C. ELISA was performed as described (Liao et al., 2011, Liao et al., 2013a).
2.10. Rhesus Macaque Immunizations
12 Indian origin Macaca mulatta were immunized every 6 weeks either sequentially (n = 6) or with a swarm (n = 6) of 100 μg of 7 gp140 Envs isolated from the first 30 months of infection from CAP206 (T/F, 2 month, 6 month, 12 month, 21 month, 24 month and 30 month Envs) formulated with adjuvant MF59 (Novartis) in a 1:1 ratio in 1 mL. Blood samples were collected 2 weeks after each immunization. All rhesus macaques were housed at Bioqual. All rhesus macaques were maintained in accordance with the Association for Assessment and Accreditation of Laboratory Animals with the approval of the Animal Care and Use Committees of the NIH and Harvard Medical School. Research was conducted in compliance with the Animal Welfare Act and other federal statutes and regulations relating to animals and experiments involving animals and adheres to principles stated in the Guide for the Care and Use of Laboratory Animals, NRC Publication, 2011 edition.
3. Results
3.1. Amino Acid Changes in the gp41 MPER Increase Global Neutralization Sensitivity
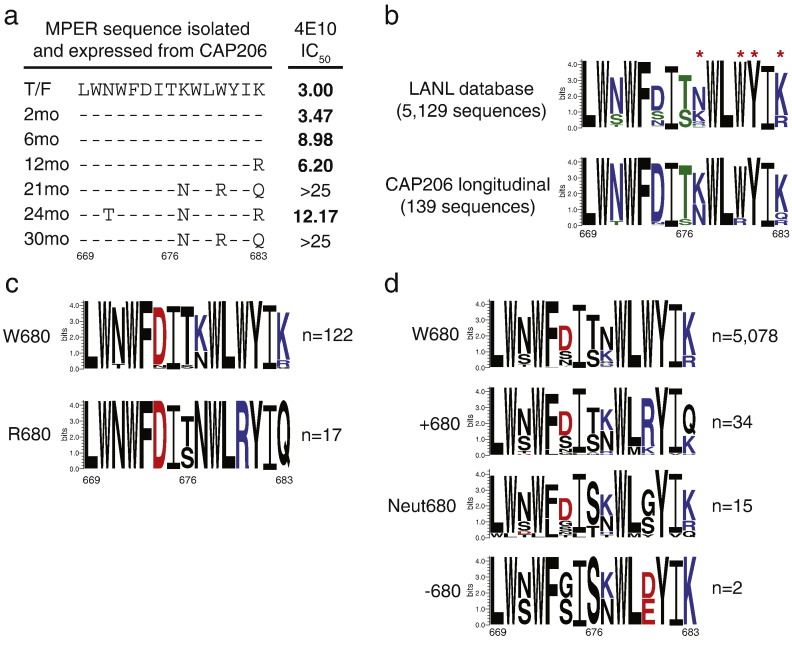
We performed sequencing of HIV-1 env soon after infection through 2.5 years in CAP206 using single-genome amplification (SGA) (Bradley et al., 2016). Examination of the C-terminal MPER sequences revealed three amino acid changes (K677 N, W680R, and K683Q) in HIV-1 Envs isolated at 12 months and beyond (Fig. 1A). The month 21 and month 30 viruses with the W680R mutation were resistant to neutralization by the MPER bnAb 4E10 (Fig. 1A). Analysis of 5129 HIV-1 Env sequences from the Los Alamos National laboratory (LANL) HIV-1 sequence database revealed limited variability at positions 677 and 683, and conservation of the tryptophan at MPER position 680 with 99.4% of the sequences having W680 (Fig. 1B). In contrast, this position varied in sequences isolated longitudinally from CAP206, with tryptophan or arginine at position 680; only 87.8% of CAP206 sequences had W680 (Fig. 1B; lower panel). In all CAP206 sequences that had the W680R change, there were also coincidental changes of positions 677 and 683 to neutral amino acids, indicating that preserving the charge of the C-terminus of the MPER may be important for interactions with the viral membrane (Fig. 1C). Although rare, there were 51 Env sequences in the LANL database that had W680 changes. Like the change in CAP206, the majority of changes at position 680 were to positively charged amino acids (34 of 51), but there were select examples of changes to neutral or negatively charged residues (Fig. 1D). In the MPER sequences that changed to a positive residue at position 680 there was also an increase in a neutral change at position 683, but variation at positions 677 and 683 was observed (Fig. 1D).
Fig. 1.
Amino acid changes in the gp41 MPER of CAP206 Envs after infection. (A) Amino acid alignment of the MPER region (669–683, HXB2) of the CAP206 T/F and 6 mutant Envs isolated at months 2, 6, 12, 21, 24 and 30 months post-infection. Neutralization of each virus in the TZM-bl assay by MPER bnAb 4E10 measured as IC50 μg/ml. (B) Graphical representation of amino acid sequences of MPER positions 669–683 from all HIV sequences in the LANL database and CAP206 longitudinal sequences sequenced by SGA. The size of each letter indicates the relative proportion of sequences that utilize that amino acid at that position. The colour of each letter indicates the hydrophobicity property of the amino acid; hydrophilic, blue; neutral, green; hydrophobic, black. n = number of sequences in the analyzed in the logoplot. Red asterisks are mutation sites studied. (C) Logo plots of CAP206 MPER amino acid sequences with W680 (top) and R680 (bottom) with amino acids colored by charge. Positive amino acids, blue; Negative amino acids, red; and Neutral, black. n = number of sequences in the analyzed in the logoplot. (D) Logo plots of the LANL MPER amino acid sequences with W680, position 680 with a positive amino acid, position 680 with a neutral amino acid and position 680 with a negative amino acid change with amino acids colored by charge. Positive amino acids, blue; Negative amino acids, red; and Neutral, black. n = number of sequences in the analyzed in the logoplot.
Changes in the MPER have been shown to alter virus sensitivity to neutralizing antibodies and fusion inhibitors (Blish et al., 2008, Nakamura et al., 2010, Shen et al., 2010, Ringe and Bhattacharya, 2012), so to examine the role of these changes in HIV-1 immune escape, we tested sensitivity of all 7 CAP206 pseudoviruses to neutralization by purified IgG from 5 HIV-1 clade C infected individuals (Table 1). All 7 CAP206 viruses, including viruses with the W680 changes, were resistant to neutralization with geometric mean titers > 100 μg/mL and remained classified as tier-2, difficult-to-neutralize, viruses (Table 1). We used site-directed mutagenesis to introduce the 3 individual naturally occurring changes alone and in combination (NRQ) into the CAP206 T/F virus. We also altered the highly conserved tyrosine in this region (Y681D) to examine its role in antibody escape; this residue was not a natural viral variant present in CAP206. All of the MPER mutant viruses demonstrated increased neutralization sensitivity to a panel of 5 HIV-1-infected plasma IgG samples, and all MPER mutant viruses, with the exception of CAP206 T/F W680R, exhibited a transition from a tier-2 to a tier-1 phenotype (Table 1). Although the CAP206 T/F W680R virus remained classified as tier-2, this virus demonstrated over a 3-fold enhancement in neutralization sensitivity as measured by the geometric mean titer of neutralization compared to the CAP206 T/F virus (Table 1). Relative infectivity of the CAP206 T/F and 5 mutant viruses was assessed by determining the tissue culture infectious dose 50 (TCID50) which is the viral stock dilution that results in approximately 150,000 RLU in TZM-bl cells. Only the CAP206 T/F Y681D mutant virus showed reduced infectivity by this measure, having a 2-log reduction in TCID50 (Fig. S1).
Table 1.
Neutralization sensitivity of CAP206 and CH505 viruses to a panel of purified IgG from clade C plasma (Tier phenotyping).
| IC50 (μg/mL) in TZM-bl cells | |||||||
|---|---|---|---|---|---|---|---|
| Virus ID | |||||||
| CAP206 viruses | SA-C2 | SA-C8 | SA-C62 | SA-C72 | SA-C74 | GMT | Tier |
| CAP206 T/F | 647 | 333 | 142 | 549 | 5000 | 609 | 2 |
| CAP206.6.mo | 316 | 88 | 264 | 520 | 499 | 286 | 2 |
| CAP206.12mo | 294 | 304 | 70 | 466 | 589 | 280 | 2 |
| CAP206.21mo | 584 | 461 | 269 | 1203 | 5000 | 847 | 2 |
| CAP206.24mo | 448 | 287 | 178 | 434 | 555 | 353 | 2 |
| CAP206.30mo | 667 | 514 | 154 | 765 | 5000 | 726 | 2 |
| CAP206 mutant viruses | |||||||
| CAP206 T/F K677N | 9 | 6 | 3 | 154 | 18 | 13 | 1B |
| CAP206 T/F W680R | 165 | 229 | 100 | 202 | 235 | 178 | 2 |
| CAP206 T/F Y681D | 17 | 41 | 71 | 37 | 5 | 24 | 1B |
| CAP206 T/F K683Q | 12 | 8 | 4 | 300 | 18 | 18 | 1B |
| CAP206 T/F NRQ | 11 | 7 | 4 | 151 | 18 | 13 | 1B |
| Control viruses | |||||||
| MW965.26 | 1.14 | 1.70 | 1.40 | 1.14 | 1.14 | 1.29 | 1 A |
| Q23.17 | 25 | 16 | 432 | 151 | 19 | 55 | 1B |
| CAP45.2.00.G3 | 71 | 209 | 337 | 56 | 662 | 179 | 2 |
| C.DU156.12 | 352 | 278 | 811 | 207 | 393 | 365 | 2 |
| Virus ID | |||||||
| CH505 viruses | HIVIG-C | SA-C102 | SA-C82 | SA-C36 | SA-C8 | GMT | Tier |
| CH505 T/F | 556 | 2222 | 790 | 118 | 1110 | 663 | 2 |
| CH505 w4.3 | 62.2 | 9 | 36 | 5 | 2 | 12 | 1B |
| Control viruses | |||||||
| C.MW965.26 | 2.99 | 1 | 7 | 2 | 3 | 3 | 1A |
| C.DU156.12 | 46.7 | 432 | 352 | 119 | 110 | 156 | 2 |
We sequenced the transmitted/founder virus and evolved Envs of another clade C HIV-1-infected individual, CH505, who produced bnAbs targeting the CD4 binding site. We identified a viral variant of the CH505 T/F virus with a single change (W680G) four weeks after transmission (w4.3) with no other amino acid changes when compared to the predominant T/F virus (Gao et al., 2014, Liao et al., 2013b). This was a single variant identified from 53 SGA sequences from week 4 post-transmission. We determined that the CH505 T/F virus was a tier-2 virus, whereas the CH505 w4.3 mutant with W680G was a more neutralization-sensitive tier-1b virus (Table 1). These data demonstrate that MPER mutations within CAP206 and CH505 T/F virus backbones enhanced the global neutralization sensitivity and transform the tier-2 T/F virus into an easy-to-neutralize tier-1 virus.
3.2. Isolation of HIV Env-Targeting Antibodies during Infection from CAP206
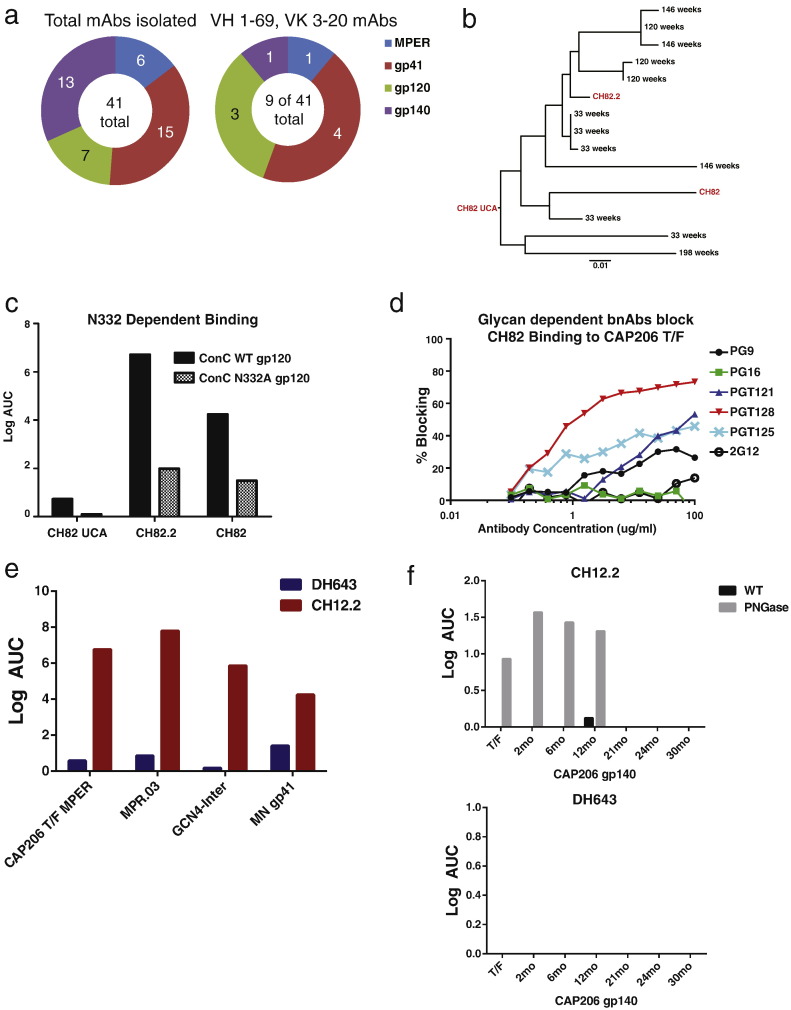
We previously isolated a neutralizing MPER-reactive antibody, CAP206-CH12, from CAP206 (Morris et al., 2011). To identify additional antibodies that targeted the HIV-1 Env in CAP206, we performed single-cell PCR on HIV-1-specific memory B cells sorted from peripheral blood memory B cells 4–254 weeks post transmission using MPR.03 tetramers and group M consensus Env CON-S. We also utilized limiting dilution cultures (Bonsignori et al., 2011) of single memory B cells and sequenced wells that exhibited CAP206 T/F gp140 reactivity. Using these methods, we isolated 41 monoclonal antibodies (mAbs) from CAP206 and confirmed HIV-1 Env reactivity by ELISA (Table S1). Fifteen of the 41 mAbs utilized the heavy chain variable gene segment (VH) 1–69 and 9 also used the kappa chain variable gene segment (Vκ) 3–20; these are the same V-gene segments utilized by the neutralizing MPER-reactive mAb CAP206-CH12 and the broadly neutralizing MPER antibody 4E10 (Table S1; Fig. 2A). The reactivity of other mAbs that used VH1–69 and Vκ3–20 was not limited to MPER but included additional gp41 and gp120 epitopes (Fig. 2A).
Fig. 2.
Isolation of V3 and MPER-targeting antibodies from CAP206. (A) Total number of isolated antibodies that bind the MPER, gp41, gp120 and gp140 subunits of the HIV-1 Env determined by ELISA and number of antibodies that utilize VH 1–69 and Vκ3–20 genes that target HIV Env. (B) The phylogenetic tree of the CAP206-CH82 antibody lineage heavy chain sequences, rooted on the unmutated common ancestor (CH82 UCA). The nodes in black are lineage members isolated by next-generation 454 sequencing and are labeled with the weeks post transmission the sequence was isolated from. CH82 and CH82.2 were natural VHDHJH and VLJL gene pairs isolated by single-cell PCR. The antibodies in red were selected for further studies. (C) ELISA binding of CH82 antibody lineage members to a consensus C (ConC) gp120 protein and ConC protein with asparagine (N) at position 332 mutated to alanine (A). Values graphed are Log area under the curve (Log AUC) of 3-fold antibody dilutions starting with 100 μg of antibody. (D) Competitive ELISA blocking of CH82 binding to CAP206 T/F gp140 protein by glycan dependent bnAbs PG9, PG16, PGT121, PGT128, PGT125 and 2G12. (E) ELISA binding of MPER mAbs (DH643 and CH12.2) to gp41 and MPER epitope antigens. The CAP206 T/F MPER peptide (KKKNEKDLLALDSWKNLWNWFDITKWLWYIRKKK), MPR.03 peptide (KKKNEQELLELDKWASLWNWFDITNWLWYIRKKK), GCN4-inter (construct that mimics the gp41-intermediate state), and clade B MN gp41 protein subunit. Binding measured in Log area under curve (Log AUC). (F) ELISA binding of DH643 and CH12.2 to the 7 CAP206 Envs isolated at various timepoints from CAP206 untreated (WT) or treated with PNGase enzyme to digest Env glycans. Graphed as Log Area Under the Curve (LOG AUC) using a titration of antibody starting at 100 μg.
Two of the mAbs isolated by single-cell PCR that utilized VH 1–69 and VK 3–20, CH82 and CH82.2, have clonally related VHDHJH and VKJK sequences (Table S1). We pyrosequenced genomic DNA isolated from 9 PBMC samples over the first 3 years of infection of CAP206, and identified additional members of this clonal lineage from multiple time points 33–146 weeks post transmission, from which we inferred the unmutated common ancestor (UCA, CH82 UCA (Fig. 2B). CH82 UCA, CH82.2 and CH82 all bound the consensus C (ConC) gp120 protein and binding was reduced when the critical glycan site within the V3 loop at position 332 was mutated (N332A; Fig. 2C). To compare the binding epitope of CH82 to glycan dependent bnAbs, we tested variable loop 2 (V2)-reactive bnAbs, PG9, PG16, and V3-glycan bnAbs PGT121, PGT125, and PGT128 for their ability to block CH82 binding to the CAP206 T/F gp140 (Fig. 2D). V3-glycan dependent bnAb PGT128 blocked 73% of CH82 binding at 100 μg/mL, binding was also blocked by other V3-glycan dependent bnAbs, PGT121 and PGT126, 53% and 46%, respectively (Fig. 2D). V1/V2-glycan dependent mAbs PG9 and PG16 did not block CH82 binding by > 35%.
The CAP206 evolved viruses with the W680R change were detected 21 months post-infection. For this reason, we sought to isolate MPER antibodies at earlier time points that may have provided selective pressure in this region. Two antibodies (DH643, CH12.2) that reacted with gp41, GCN4-inter (a gp41 intermediate state mimic), and MPER peptides were isolated 17 weeks post infection from CAP206 by single-cell PCR after sorting memory B cells from CAP206 that were positive for MPR.03 peptides (Fig. 2E). CH12.2 used the same antibody gene segments (VH1–69, Vκ3–20) utilized by MPER-targeting bnAbs CAP206-CH12 and 4E10, and was determined to be an early member of the CH12 bnAb clonal lineage (Morris et al., 2011). Antibody DH643 used unrelated gene segments (Table S1). CH12.2 and DH643 had limited somatic hypermutation (SHM), with heavy chain mutation frequencies of 2.7% and 3.6%, respectively. Both antibodies interacted weakly with CAP206 gp140 recombinant Envs, but CH12.2 had increased reactivity with CAP206 Envs that were natively deglycosylated with PNGase treatment (Fig. 2F). DH643 did not bind to any forms of the CAP206 21, 24 or 30 month Envs (Fig. 2F).
3.3. CAP206 Viruses with MPER Changes that Confer Neutralization Sensitivity Are Resistant to Neutralization by Early MPER-Targeting Antibodies
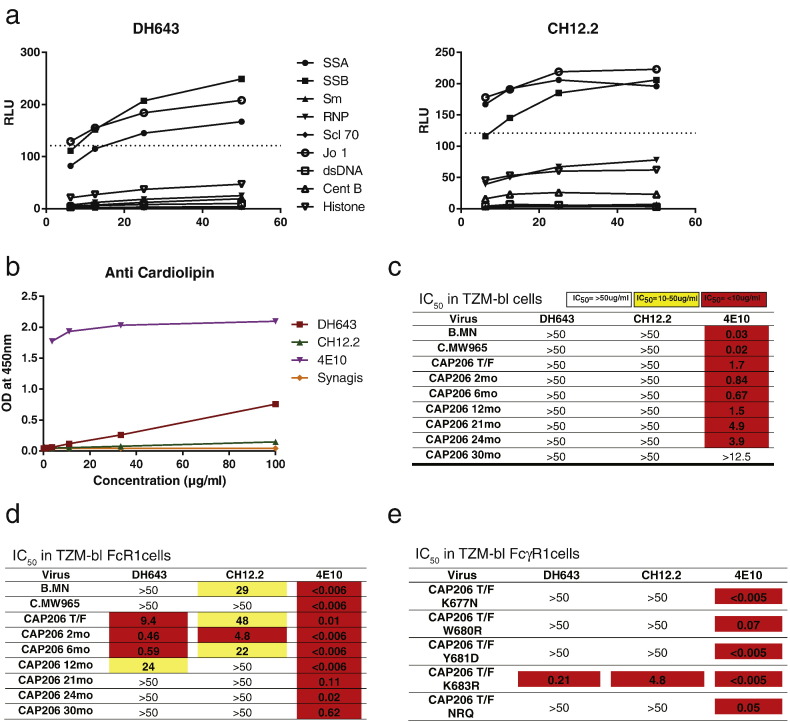
Like the MPER bnAbs 4E10 and CH12, CH12.2 and DH643, isolated early after infection, were cross-reactive with host proteins that are common autoantigens in autoimmune disease (SSA, SSB and Jo1), but only DH643 reacted weakly with cardiolipin (Fig. 3A & B). To assess neutralization by MPER antibodies, we used the TZM-bl assay and the TZM-bl/FcγRI assay that employs TZM-bl epithelial cells transfected with CD64 (FcγRI), thus enabling the cells to bind MPER bnAbs and augment their ability to associate with the virion prior to receptor-mediated activation (Perez et al., 2009). By using a cell line that provides antibodies a kinetic advantage for interaction with the transiently-exposed MPER epitope, this assay has enhanced sensitivity for MPER antibodies (Perez et al., 2009). Antibodies DH643 and CH12.2 did not neutralize any viruses in the TZM-bl neutralization assay, but DH643 neutralized 4 autologous viruses and CH12.2 neutralized 3 autologous viruses and 1 heterologous virus in TZM-bl cells that expressed FcγR1/CD64 (Fig. 3C & D). Both antibodies neutralized the CAP206 T/F virus but could not neutralize any of the evolved variants after month 21 that contained W680 changes; these antibodies did not neutralize the CAP206 T/F variants with the K677N, W680R, Y681D, and the NRQ changes (Fig. 3E). These observations raise the possibility that the observed changes in the MPER that conferred enhanced neutralization sensitivity were escape mutants from these early MPER-targeting antibodies.
Fig. 3.
MPER antibodies isolated early after infection were autoreactive and could not neutralize late viruses with MPER amino acid changes. (A) Intensity of binding autoantigens in the AtheNA multilyte assay by DH643 and CH12.2 and 4E10 at increasing antibody concentrations (x-axis). Dashed lined indicates assay positivity cutoff. (B) Binding of DH643 and CH12.2 to cardiolipin at increasing antibody concentrations (x-axis) measured by ELISA using antibodies 4E10 and Synagis as positive and negative control, respectively. (C–D) Neutralization of 7 autologous Tier-2 CAP206 viruses and 2 heterologous Tier-1 viruses (B.MN and C.MW965) by DH643 and CH12.2 and MPER-targeting bnAb 4E10 in TZM-bl cells (C) and TZM-bl FcγR1 expressing cells (D) measured as IC50. (E) Neutralization by DH643 and CH12.2 and MPER-targeting bnAb 4E10 of CAP206 T/F virus with MPER mutations introduced observed in later viruses in the TZM-bl FcγR1 neutralization assay measured as IC50.
3.4. Enhanced Neutralization of MPER Mutant Viruses by gp120-Targeted Antibodies from CAP206 and CAP206 Env-Immunized Macaques
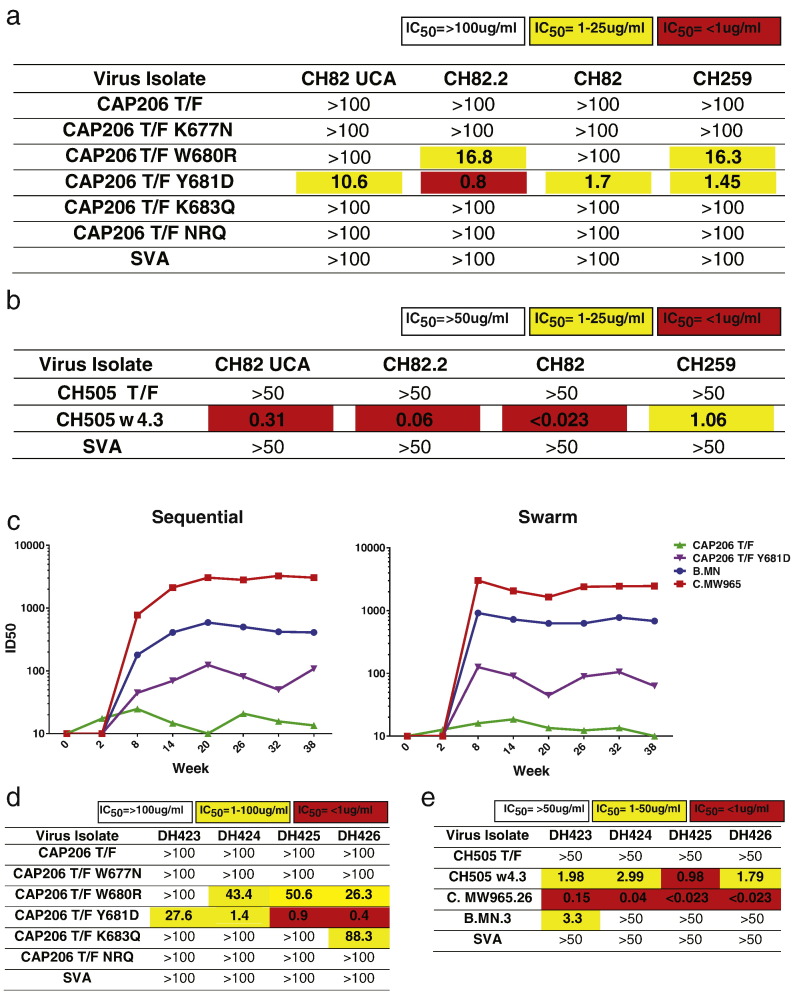
V3-targeting CH82 lineage antibodies did not mediate any tier-2 autologous or heterologous neutralization and CH82 and CH82.2 only could neutralize tier-1 viruses (Table S2).Given the observed global increase in neutralization sensitivity for viruses with changes in the MPER, we tested the CH82 clonal antibody lineage and an additional V3-targeting antibody isolated from CAP206, CH259, for their ability to neutralize the CAP206 MPER mutant viruses. Clonal lineage members CH82 and CH82.2, along with the CH82 lineage unmutated common ancestor (UCA), and CH259 did not neutralize CAP206 T/F virus. CH82.2, CH82 and CH259 all weakly neutralized the virus with a W680R mutation, potently neutralized the CAP206 T/F Y681D virus (Fig. 4A & S2). Similar to the CAP206 viruses, the CH505 T/F virus was resistant to neutralization by CH82 UCA, CH82.2, CH82, and CH259, but the CH505 w4.3 virus was sensitive to neutralization by all 3 CH82 clonal lineage mAbs and CH259 (Fig. 4B).
Fig. 4.
Tier-1 neutralizing gp120-targeting antibodies from CAP206 and CAP206 Env-immunized rhesus macaques neutralized viruses with MPER changes. (A–B) CH82 lineage and CH259 neutralization of (A) CAP206 T/F and CAP206 T/F MPER mutant viruses and (B) of heterologous CH505 T/F and variant w4.3 that has a W680 mutation in the MPER in the TZM-bl neutralization assay displayed as IC50 values. Simian virus amphotropic murine leukemia virus (SVA) was tested as a negative control. (C) Plasma neutralization at 8 timepoints post-immunization of 12 rhesus macaques immunized with a sequential (group 1) or swarm (group 2) of CAP206 T/F virus, CPA206 T/F Y681D virus and heterologous viruses B.MN and C. MW965 in the TZM-bl assay measured by ID50 (y-axis). Animals were immunized at weeks 0,6,12,18,24,30 and 36. (D–E) Neutralization profiles of mAbs isolated from CAP206 Env immunized rhesus macaques (DH423-DH424) against (D) CAP206 T/F, CAP206 T/F MPER mutants, (E) CH505 T/F and CH505 w4.3 viruses. Measured as IC50 in the TZM-bl neutralization assay. Simian virus amphotropic murine leukemia virus (SVA) was tested as a negative control.
Next, we tested if vaccine-elicited antibodies from CAP206-immunized rhesus macaques neutralized viruses with MPER changes that conferred enhanced neutralization sensitivity. Two groups of rhesus macaques were immunized with 7 CAP206 Env proteins sequentially or in a swarm every 6 weeks (Fig. S3A). We have previously demonstrated that the swarm immunized group elicited tier-1 neutralizing antibodies in all animals and a single animal had tier-2 neutralizing antibodies to the CAP206 virus isolated at 6 months (Bradley et al., 2016). After two immunizations, plasma neutralization was detected against heterologous tier-1 viruses MN and MW965 for both the sequential and swarm immunized animals (Fig. 4C). We tested plasma neutralization of the autologous CAP206 T/F virus and the CAP206 Y681D mutant virus that displayed enhanced neutralization sensitivity. Only sporadic weak neutralization of the tier-2 CAP206 T/F could be detected in the immunized animals, but much higher titers of neutralizing antibodies to the tier-1 CAP206 Y681D virus was detected in all animals (Fig. 4C). We isolated 3 gp120 reactive mAbs DH423-DH425 from macaque 5096 2 weeks after the 4th immunization, and a 4th gp120 reactive mAb, DH426, was isolated from animal 5160 after the 3rd immunization. All four mAbs were IgG isotype and used a VH gene segment from VH family 4 (Table S3). DH424–426 reacted with a linear epitope in V3 and DH423 recognized a conformation-dependent epitope in gp120 (Fig. S3B).
The isolated antibodies were unable to neutralize the CAP206 T/F virus, but DH424, DH425 and DH426 neutralized the CAP206 T/F W680R virus, and all 4 isolated mAbs neutralized the CAP206 T/F Y681D virus in the TZM-bl neutralization assay (Fig. 4D). DH423-DH426 also neutralized the heterologous CH505 w4.3 virus, but lacked the ability to neutralize the CH505 T/F virus (Fig. 4E). Additionally, these antibodies neutralized the heterologous tier-1 virus C. MW965 and DH423 could also neutralize B. MN (Fig. 4E). These data demonstrated that viruses with MPER changes are sensitive to autologous CAP206 tier-1 neutralizing antibodies and easy-to-induce vaccine-elicited tier-1 gp120-targeting antibodies.
3.5. MPER Amino Acid Changes Increase Viral Neutralization Sensitivity to Antibodies against Multiple Epitopes and Alter gp120 Conformational Preference
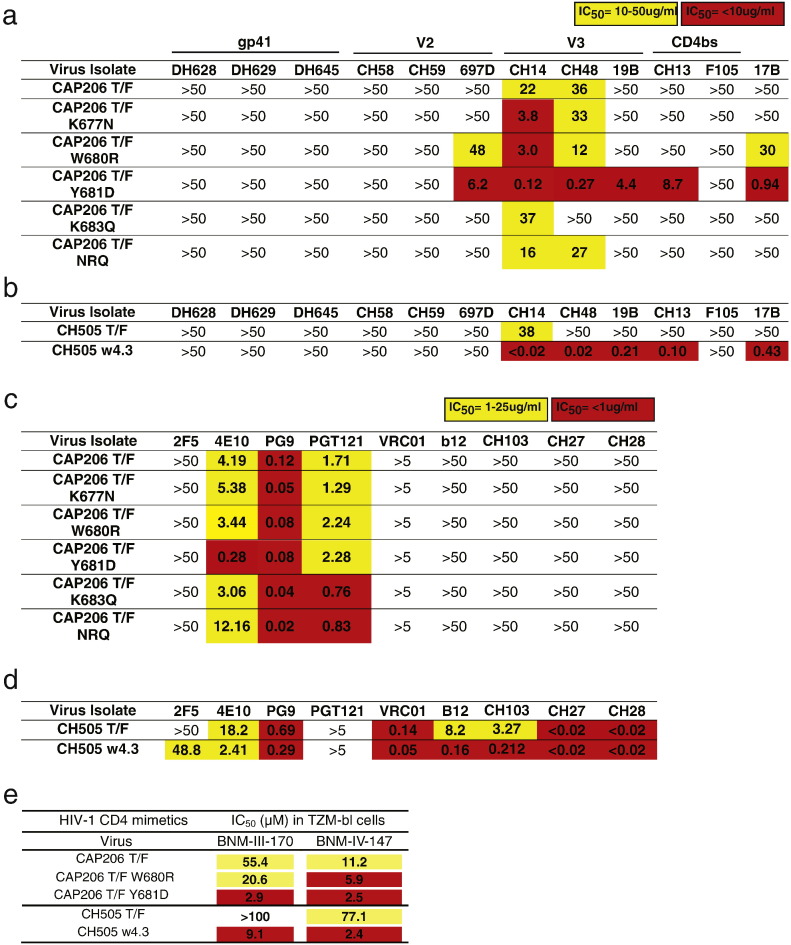
Next, we tested neutralization of the CAP206 T/F and MPER mutant viruses by antibodies that targeted different Env epitopes and that were known to neutralize heterologous tier-1 or limited numbers of tier-2 pseudoviruses (Fig. 5A). Antibodies that targeted gp41 outside of the MPER did not neutralize the CAP206 T/F virus or any of the MPER mutants (DH628, DH629, and DH645). V2-targeting antibodies CH58 and CH59 isolated from RV144 vaccines also did not neutralize CAP206 T/F or any mutants; in contrast, the V2 antibody 697D was able to neutralize the CAP206 W680R and Y681D mutant viruses but it failed to neutralize the CAP206 T/F virus. Antibodies that target the V3 loop (CH14, CH48 and 19B) also exhibited enhanced neutralization of the MPER mutant viruses—when compared with the CAP206 T/F virus, CH14 more potently neutralized K677N, W680R, and Y681D mutant viruses; CH48 more potently neutralized W680R and Y681D mutant viruses; and 19B was only able to neutralize the Y681D mutant virus. The CD4-binding site targeting mAb F105 was unable to neutralize any of the viruses, but CH13, which also targets the CD4-binding site, potently neutralized the CAP206 Y681D mutant virus. Lastly, antibody 17B, which binds preferentially to the CD4-induced, CCR5 co-receptor binding site epitopes on Env, could neutralize the W680R virus, and even more potently, the Y681D mutant virus. Thus, changes at positions 680 and 681 result in greater binding of antibodies that have been shown to bind to more open Env conformations, suggesting that these changes in the MPER allow Env to sample more open states that resemble the CD4-bound conformation where the CCR5 binding site is exposed (Fig. 5A). Similar enhanced neutralization sensitivity to V3 (CH14, CH48 and 19B), CD4bs (CH13) and CD4-induced coreceptor (17B) targeting antibodies were observed for the CH505 w4.3 virus that had a W680G change in the MPER (Fig. 5B). We also tested neutralization of the MPER mutant viruses by bnAbs that target gp41 and gp120 epitopes on HIV Env. Among the CAP206 mutant viruses, only the Y681D mutant showed enhanced neutralization sensitivity to MPER bnAb 4E10 when compared to neutralization of the T/F virus (Fig. 5C). The CH505 w4.3 virus also exhibited enhanced neutralization sensitivity to 4E10, and was more sensitive to the CD4bs bnAbs b12 and CH103 (Fig. 5D). These data demonstrate that the CAP206 W680R and Y681D viruses and the CH505 w4.3 virus, which has a W680G change, exhibit enhanced neutralization sensitivity to weakly-neutralizing heterologous antibodies that target distal epitopes (V2, V3 and CD4bs) in gp120. In contrast, there was minimal impact of these mutations on sensitivity to bnAbs.
Fig. 5.
MPER changes enhanced neutralization sensitivity to heterologous HIV antibodies and altered Env conformational preference. (A–D) Neutralization profiles of non-bnAbs (gp41 mAbs (DH628, DH629, DH645), V2 mAbs (CH58, CH59, 697D), V3 mAbs (CH14, CH18, 19B), CD4bs mAbs (CH13, F105) and conformational epitope mAb 17B; A–B) and bnAbs (C–D), measured as IC50 in TZM-bl neutralization assays, against transmitted founder viruses isolated from clade C infected individuals CAP206 and CH505, compared to T/F viruses containing MPER mutations that confer enhanced neutralization sensitivity (A,C) CAP206 viruses. (B,D) CH505 viruses. (E) Neutralization by two compounds the mimic CD4 and induce Env conformations similar the CD4-bound state (BNM-III-170 and BNM-IV-147) measured in the TZM-bl neutralization assay displayed as IC50 concentration (Yellow, < 100; Red, < 10).
Next, we tested the ability of small molecules that mimic CD4 to inhibit infection of the viruses (Fig. 5E) (Melillo et al., 2016). The CAP206 T/F virus was inhibited by CD4 mimetics BNM-III-170 and BNM-IV-147 at IC50s of 55.4 μM and 11.2 μM, respectively. The CAP206 T/F Y681D mutant virus exhibited increased sensitivity to both molecules with IC50 values of 2.9 μM and 2.5 μM. The CAP206 T/F W680R mutant had a more modest two-fold increase in sensitivity to both molecules. The CH505 T/F virus was resistant to BNM-III-170 but was inhibited by BNM-IV-147 with an IC50 of 77.1 μM. The CH505 w4.3 virus with a single W680G change became sensitive to BNM-III-170 and was over 30 times more sensitive to BNM-IV-147 (Fig. 5E). These results suggest that changes in the gp41 MPER can confer enhanced Env reactivity to CD4-mimetic compounds, consistent with a greater propensity for Envs with these mutations to sample more open trimer conformations, which results in enhanced neutralization sensitivity.
4. Discussion
In this study, we identified amino acid changes in the gp41 MPER of CAP206 Envs that modulate neutralization sensitivity by antibodies that target multiple HIV-1 Env epitopes. Changes in the MPER of the CAP206 Env can be detected as early as 12 months after infection, and when introduced into the CAP206 T/F virus, they converted the virus neutralization phenotype from a neutralization-resistant tier-2 virus to a more easy-to-neutralize tier-1 virus. Additionally, we isolated MPER-targeting antibodies during the first 6 months of infection that could have selected these viral escape mutants. This includes antibody CH12.2 which was an early member of the CH12 bnAb lineage (Morris et al., 2011). Similarly, a T/F viral variant (W680G) from another bnAb individual, CH505, also conferred enhanced antibody neutralization sensitivity. Using bnAbs and narrow neutralizing antibodies, we demonstrated that antibodies that target the V2, V3 and CD4bs displayed enhanced neutralization of MPER-mutant viruses.
Changes in the gp41 and cytoplasmic domain outside of the MPER have been demonstrated to increase resistance to neutralization by gp120 antibodies (Watkins et al., 1996, Back et al., 1993, Haim et al., 2011) and modulate sensitivity to MPER-reactive bnAbs (Shen et al., 2010, Blish et al., 2008, Shen et al., 2009, Ringe and Bhattacharya, 2012). Natural polymorphisms are extremely rare for W680; however naturally occurring variants resistant to the MPER bnAb 4E10 have been isolated and shown to be capable of mother-to-child transmission (Blish et al., 2008, Nakamura et al., 2010). The precise mechanisms of how the MPER changes enhance neutralization sensitivity remains unclear. Prolonged epitope exposure due to changes in fusion kinetics, Env dissociation, Env expression and changes in infectivity may play a role in global enhancement of neutralization sensitivity, but were not sole factors for neutralization sensitivity in previous studies (Nakamura et al., 2010, Blish et al., 2008, Ringe and Bhattacharya, 2012, Vishwanathan and Hunter, 2008, Munoz-Barroso et al., 1999). MPER residue W680R and Y681D changes in CAP206 and W680G change in CH505 increase neutralization sensitivity to CD4-induced antibody 17b indicated that these two mutations acted by exposing the coreceptor binding site and other gp120 epitopes.
Consistent with previous studies we found that the MPER polymorphisms have different neutralization sensitivities depending on the genetic background of the virus. The CAP206 month 21 and month 30 viruses with the NRQ mutations remained difficult-to-neutralize tier-2 viruses, but when this mutation was introduced in the T/F virus it converted it to tier-1 (Nakamura et al., 2010). Moreover, we showed that two clade C T/F viruses have varying degrees of sensitivity to MPER changes; these changes in sensitivity are likely due in part to the fact that the conserved amino acid tryptophan is large and hydrophobic. Replacement of this tryptophan at position 680 in CH505 T/F with the less hydrophobic residue glycine confers greater neutralization sensitivity than we observed with the larger charged arginine residue as we observed in the CAP206 T/F. These results indicated that the properties of the amino acid changes in the MPER and compensatory mutations elsewhere in the Env can determine the magnitude of the neutralization phenotype change.
Changes at amino acid position 680 in CAP206 were always associated with changes at positions 677 and 683, and in the LANL database, when position 680 is mutated to a positively charged residue there is higher prevalence of a neutral (Q) change at position 683. These 3 residues are solvent exposed within the second amphipathic helix and the coordinated changes of these positions may be important for interactions with distal Env regions or fusion dynamics and this mutation pattern has been observed in other viral sequences (Nakamura et al., 2010, Sun et al., 2008). Furthermore, amino acid residues 679–683 of the HIV-1 gp41 MPER have a cholesterol recognition amino acid consensus motif and disruption of this motif may inhibit viral fusion or stabilization of the Env within the viral membrane (Vishwanathan and Hunter, 2008). The motif WLWYIK overlaps with epitopes recognized by MHC class I alleles, but WLGYIK or WLRYIK do not. Thus, in addition to neutralizing antibody escape, changes at W680 could result in escape from cytotoxic T-lymphocyte responses early in infection before a potent neutralizing antibody response (Kundu et al., 1998, Colleton et al., 2009, Reinis et al., 2007).
DH643 and CH12.2 are MPER-targeting mAbs isolated 17 weeks post-infection and only neutralized viruses in TZM-bl cells that expressed FcγR1/CD64. MPER nAbs interact with the viral membrane in order to have the kinetic advantage to access the MPER after viral docking (Dennison et al., 2009). DH643 and CH12.2 that could not neutralize in the TZM-bl assay were autoreactive, but had little to no reactivity with lipids; this suggests that further somatic hypermutation would be required to gain lipid reactivity and the ability to neutralize viruses without the assistance of FcγR1 on the cell surface. That CH12.2, an early CH12 clonal lineage member, required FcγR1-TZM-bl cells to demonstrate neutralization, indicates the usefulness of using this cell line to identify MPER-targeted bnAb precursors (Zhang et al., 2016).
The enhanced sensitivity of these MPER mutant viruses to gp120 antibodies and CD4-mimetic molecules demonstrate the importance of gp41 MPER residues near the viral membrane in maintaining the stability of the closed native trimer state. Structural studies of viruses containing these changes may provide insights into how these gp41 alterations affect Env conformations and present conserved epitopes more effectively. Moreover, eliciting MPER-targeting antibodies, like DH463 and CH12.2 that arose early and did not require high somatic hypermutation, that selected MPER changes that induce more open, easy-to-neutralize, trimer conformations by vaccination may contribute to viral clearance and protection. In particular, this scenario could be important in the setting of maternal-to-child transmission (MTCT) of HIV-1 where inducing these types of MPER-targeting antibodies that select MPER-changes that transform tier-2 viruses to tier-1 viruses in pregnant women could lead to lower transmission rates. Indeed, a previous study characterized a W680R mutation that was transmitted by MTCT that was resistant to the bnAb 4E10 (Nakamura et al., 2010), and a recent analysis has shown that a correlate of decreased MTCT transmission risk was high plasma tier-1 neutralizing antibodies (Permar et al., 2015). Future studies will be required to determine if antibodies can be elicited by vaccination that can select viral mutants that induce transition of neutralization-resistant viruses to neutralization-sensitive.
Conflicts of Interest
The authors declare no conflicts of interest.
Author Contributions
T.B. and A.T. isolated and characterized antibodies, designed assays, analyzed and interpreted data and wrote and edited the manuscript. N.T., E.G. S.S.A. and L.M. provided CAP206 samples and sequences and edited the manuscript. X.L., S.M.A and H.X.L. produced recombinant proteins and characterized their antigenicity. N.M., F.J., B.M., A.B.S·III and J.S. provided and tested CD4 mimetic compounds. A.E., S.X. and D.M. performed neutralization assays. R.P. and K.E.L. performed antibody binding assays. L.L.S., R.M.S., C.M.B. and S.S. assisted with rhesus macaque immunizations and sample processing. S.B. provided vaccine adjuvant. S. D.B. performed NGS sequencing. T.B.K. designed software and performed analysis of antibody sequences. F.G. performed analysis of viral sequences. M.B. led memory B cell culture work. M.A.M. produced fluorophor-labeled Env proteins for memory B cell staining and oversaw flow cytometry and edited the manuscript. B.F.H. designed the study, oversaw all experiments, analyzed all data and wrote and edited the manuscript.
Acknowledgments
We would like to acknowledge Dawn J. Marshall, John F. Whitesides, Joshua D. Amos, Tarra A. Von Holle, Thaddeus C. Gurley, Lawrence C. Armand, Andrew Foulger, Annie Hogan, Daniel M. Kozink, Florence Perrin, and Kwan-Ki Hwang for expert technical assistance. We also would like to acknowledge Kelly Soderberg and Celia C. LaBranche for project coordination and management. This work was supported by the Center for HIV/AIDS Vaccine Immunology-Immunogen Discovery (CHAVI-ID; UMI-AI100645) grant from NIH/NIAID/DAIDS. Our funding sources had no role in data collection, analysis or interpretation, and were not involved in the writing of this manuscript.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.ebiom.2016.08.045.
Contributor Information
Todd Bradley, Email: todd.bradley@duke.edu.
Barton F. Haynes, Email: barton.haynes@duke.edu.
Appendix A. Supplementary data
Supplementary material.
References
- Andre S., Seed B., Eberle J., Schraut W., Bultmann A., Haas J. Increased immune response elicited by DNA vaccination with a synthetic gp120 sequence with optimized codon usage. J. Virol. 1998;72:1497–1503. doi: 10.1128/jvi.72.2.1497-1503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Back N.K., Smit L., Schutten M., Nara P.L., Tersmette M., Goudsmit J. Mutations in human immunodeficiency virus type 1 gp41 affect sensitivity to neutralization by gp120 antibodies. J. Virol. 1993;67:6897–6902. doi: 10.1128/jvi.67.11.6897-6902.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger E.A. HIV entry and tropism: the chemokine receptor connection. AIDS. 1997;11(Suppl A):S3–16. [PubMed] [Google Scholar]
- Blish C.A., Nguyen M.A., Overbaugh J. Enhancing exposure of HIV-1 neutralization epitopes through mutations in gp41. PLoS Med. 2008;5 doi: 10.1371/journal.pmed.0050009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonsignori M., Hwang K.K., Chen X., Tsao C.Y., Morris L., Gray E., Marshall D.J., Crump J.A., Kapiga S.H., Sam N.E., Sinangil F., Pancera M., Yongping Y., Zhang B., Zhu J., Kwong P.D., O'dell S., Mascola J.R., Wu L., Nabel G.J., Phogat S., Seaman M.S., Whitesides J.F., Moody M.A., Kelsoe G., Yang X., Sodroski J., Shaw G.M., Montefiori D.C., Kepler T.B., Tomaras G.D., Alam S.M., Liao H.X., Haynes B.F. Analysis of a clonal lineage of HIV-1 envelope V2/V3 conformational epitope-specific broadly neutralizing antibodies and their inferred unmutated common ancestors. J. Virol. 2011;85:9998–10009. doi: 10.1128/JVI.05045-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd S.D., Marshall E.L., Merker J.D., Maniar J.M., Zhang L.N., Sahaf B., Jones C.D., Simen B.B., Hanczaruk B., Nguyen K.D., Nadeau K.C., Egholm M., Miklos D.B., Zehnder J.L., Fire A.Z. Measurement and clinical monitoring of human lymphocyte clonality by massively parallel VDJ pyrosequencing. Sci. Transl. Med. 2009;1:12ra23. doi: 10.1126/scitranslmed.3000540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley T., Fera D., Bhiman J., Eslamizar L., Lu X.Z., Anasti K., Zhang R.J., Sutherland L.L., Scearce R.M., Bowman C.M., Stolarchuk C., Lloyd K.E., Parks R., Eaton A., Foulger A., Nie X.Y., Karim S.S.A., Barnett S., Kelsoe G., Kepler T.B., Alam S.M., Montefiori D.C., Moody M.A., Liao H.X., Morris L., Santra S., Harrison S.C., Haynes B.F. Structural constraints of vaccine-induced tier-2 autologous HIV neutralizing antibodies targeting the receptor-binding site. Cell Rep. 2016;14:43–54. doi: 10.1016/j.celrep.2015.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton D.R., Mascola J.R. Antibody responses to envelope glycoproteins in HIV-1 infection. Nat. Immunol. 2015;16:571–576. doi: 10.1038/ni.3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colleton B.A., Huang X.L., Melhem N.M., Fan Z., Borowski L., Rappocciolo G., Rinaldo C.R. Primary human immunodeficiency virus type 1-specific CD8 + T-cell responses induced by myeloid dendritic cells. J. Virol. 2009;83:6288–6299. doi: 10.1128/JVI.02611-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Taeye S.W., Moore J.P., Sanders R.W. 2016. HIV-1 Envelope Trimer Design and Immunization Strategies to Induce Broadly Neutralizing Antibodies. Trends Immunol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennison S.M., Stewart S.M., Stempel K.C., Liao H.X., Haynes B.F., Alam S.M. Stable docking of neutralizing human immunodeficiency virus type 1 gp41 membrane-proximal external region monoclonal antibodies 2F5 and 4E10 is dependent on the membrane immersion depth of their epitope regions. J. Virol. 2009;83:10211–10223. doi: 10.1128/JVI.00571-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derdeyn C.A., Moore P.L., Morris L. Development of broadly neutralizing antibodies from autologous neutralizing antibody responses in HIV infection. Curr. Opin. HIV AIDS. 2014;9:210–216. doi: 10.1097/COH.0000000000000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing B., Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- Frey G., Chen J., Rits-Volloch S., Freeman M.M., Zolla-Pazner S., Chen B. Distinct conformational states of HIV-1 gp41 are recognized by neutralizing and non-neutralizing antibodies. Nat. Struct. Mol. Biol. 2010;17:1486–1491. doi: 10.1038/nsmb.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F., Bonsignori M., Liao H.X., Kumar A., Xia S.M., Lu X., Cai F., Hwang K.K., Song H., Zhou T., Lynch R.M., Alam S.M., Moody M.A., Ferrari G., Berrong M., Kelsoe G., Shaw G.M., Hahn B.H., Montefiori D.C., Kamanga G., Cohen M.S., Hraber P., Kwong P.D., Korber B.T., Mascola J.R., Kepler T.B., Haynes B.F. Cooperation of B cell lineages in induction of HIV-1-broadly neutralizing antibodies. Cell. 2014;158:481–491. doi: 10.1016/j.cell.2014.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray E.S., Madiga M.C., Moore P.L., Mlisana K., Abdool Karim S.S., Binley J.M., Shaw G.M., Mascola J.R., Morris L. Broad neutralization of human immunodeficiency virus type 1 mediated by plasma antibodies against the gp41 membrane proximal external region. J. Virol. 2009;83:11265–11274. doi: 10.1128/JVI.01359-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haim H., Strack B., Kassa A., Madani N., Wang L., Courter J.R., Princiotto A., Mcgee K., Pacheco B., Seaman M.S., Smith A.B., III, Sodroski J. Contribution of intrinsic reactivity of the HIV-1 envelope glycoproteins to CD4-independent infection and global inhibitor sensitivity. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1002101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes B.F., Kelsoe G., Harrison S.C., Kepler T.B. B-cell-lineage immunogen design in vaccine development with HIV-1 as a case study. Nat. Biotechnol. 2012;30:423–433. doi: 10.1038/nbt.2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Ofek G., Laub L., Louder M.K., Doria-Rose N.A., Longo N.S., Imamichi H., Bailer R.T., Chakrabarti B., Sharma S.K., Alam S.M., Wang T., Yang Y., Zhang B., Migueles S.A., Wyatt R., Haynes B.F., Kwong P.D., Mascola J.R., Connors M. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature. 2012;491:406–412. doi: 10.1038/nature11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keele B.F., Giorgi E.E., Salazar-Gonzalez J.F., Decker J.M., Pham K.T., Salazar M.G., Sun C., Grayson T., Wang S., Li H., Wei X., Jiang C., Kirchherr J.L., Gao F., Anderson J.A., Ping L.H., Swanstrom R., Tomaras G.D., Blattner W.A., Goepfert P.A., Kilby J.M., Saag M.S., Delwart E.L., Busch M.P., Cohen M.S., Montefiori D.C., Haynes B.F., Gaschen B., Athreya G.S., Lee H.Y., Wood N., Seoighe C., Perelson A.S., Bhattacharya T., Korber B.T., Hahn B.H., Shaw G.M. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc. Natl. Acad. Sci. U. S. A. 2008;105:7552–7557. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepler T.B., Munshaw S., Wiehe K., Zhang R.J., Yu J.S., Woods C.W., Denny T.N., Tomaras G.D., Alam S.M., Moody M.A., Kelsoe G., Liao H.X., Haynes B.F. Reconstructing a B-cell clonal lineage. II. Mutation, selection, and affinity maturation. Front. Immunol. 2014;5 doi: 10.3389/fimmu.2014.00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu S.K., Dupuis M., Sette A., Celis E., Dorner F., Eibl M., Merigan T.C. Role of preimmunization virus sequences in cellular immunity in HIV-infected patients during HIV type 1 MN recombinant gp160 immunization. AIDS Res. Hum. Retrovir. 1998;14:1669–1678. doi: 10.1089/aid.1998.14.1669. [DOI] [PubMed] [Google Scholar]
- Liao H.X., Sutherland L.L., Xia S.M., Brock M.E., Scearce R.M., Vanleeuwen S., Alam S.M., Mcadams M., Weaver E.A., Camacho Z., Ma B.J., Li Y., Decker J.M., Nabel G.J., Montefiori D.C., Hahn B.H., Korber B.T., Gao F., Haynes B.F. A group M consensus envelope glycoprotein induces antibodies that neutralize subsets of subtype B and C HIV-1 primary viruses. Virology. 2006;353:268–282. doi: 10.1016/j.virol.2006.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao H.X., Levesque M.C., Nagel A., Dixon A., Zhang R., Walter E., Parks R., Whitesides J., Marshall D.J., Hwang K.K., Yang Y., Chen X., Gao F., Munshaw S., Kepler T.B., Denny T., Moody M.A., Haynes B.F. High-throughput isolation of immunoglobulin genes from single human B cells and expression as monoclonal antibodies. J. Virol. Methods. 2009;158:171–179. doi: 10.1016/j.jviromet.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao H.X., Chen X., Munshaw S., Zhang R., Marshall D.J., Vandergrift N., Whitesides J.F., Lu X., Yu J.S., Hwang K.K., Gao F., Markowitz M., Heath S.L., Bar K.J., Goepfert P.A., Montefiori D.C., Shaw G.C., Alam S.M., Margolis D.M., Denny T.N., Boyd S.D., Marshal E., Egholm M., Simen B.B., Hanczaruk B., Fire A.Z., Voss G., Kelsoe G., Tomaras G.D., Moody M.A., Kepler T.B., Haynes B.F. Initial antibodies binding to HIV-1 gp41 in acutely infected subjects are polyreactive and highly mutated. J. Exp. Med. 2011;208:2237–2249. doi: 10.1084/jem.20110363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao H.X., Bonsignori M., Alam S.M., Mclellan J.S., Tomaras G.D., Moody M.A., Kozink D.M., Hwang K.K., Chen X., Tsao C.Y., Liu P., Lu X., Parks R.J., Montefiori D.C., Ferrari G., Pollara J., Rao M., Peachman K.K., Santra S., Letvin N.L., Karasavvas N., Yang Z.Y., Dai K., Pancera M., Gorman J., Wiehe K., Nicely N.I., Rerks-Ngarm S., Nitayaphan S., Kaewkungwal J., Pitisuttithum P., Tartaglia J., Sinangil F., Kim J.H., Michael N.L., Kepler T.B., Kwong P.D., Mascola J.R., Nabel G.J., Pinter A., Zolla-Pazner S., Haynes B.F. Vaccine induction of antibodies against a structurally heterogeneous site of immune pressure within HIV-1 envelope protein variable regions 1 and 2. Immunity. 2013;38:176–186. doi: 10.1016/j.immuni.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao H.X., Lynch R., Zhou T., Gao F., Alam S.M., Boyd S.D., Fire A.Z., Roskin K.M., Schramm C.A., Zhang Z., Zhu J., Shapiro L., Program N.C.S., Mullikin J.C., Gnanakaran S., Hraber P., Wiehe K., Kelsoe G., Yang G., Xia S.M., Montefiori D.C., Parks R., Lloyd K.E., Scearce R.M., Soderberg K.A., Cohen M., Kamanga G., Louder M.K., Tran L.M., Chen Y., Cai F., Chen S., Moquin S., Du X., Joyce M.G., Srivatsan S., Zhang B., Zheng A., Shaw G.M., Hahn B.H., Kepler T.B., Korber B.T., Kwong P.D., Mascola J.R., Haynes B.F. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature. 2013;496:469–476. doi: 10.1038/nature12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascola J.R., Haynes B.F. HIV-1 neutralizing antibodies: understanding nature's pathways. Immunol. Rev. 2013;254:225–244. doi: 10.1111/imr.12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascola J.R., Montefiori D.C. The role of antibodies in HIV vaccines. Annu. Rev. Immunol. 2010;28:413–444. doi: 10.1146/annurev-immunol-030409-101256. [DOI] [PubMed] [Google Scholar]
- Mccoy L.E., Weiss R.A. Neutralizing antibodies to HIV-1 induced by immunization. J. Exp. Med. 2013;210:209–223. doi: 10.1084/jem.20121827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melillo B., Liang S., Park J., Schon A., Courter J.R., Lalonde J.M., Wendler D.J., Princiotto A.M., Seaman M.S., Freire E., Sodroski J., Madani N., Hendrickson W.A., Smith A.B., III Small-Molecule CD4-Mimics: Structure-Based Optimization of HIV-1 Entry Inhibition. ACS Med. Chem. Lett. 2016;7:330–334. doi: 10.1021/acsmedchemlett.5b00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montefiori D.C. Evaluating neutralizing antibodies against HIV, SIV, and SHIV in luciferase reporter gene assays. Curr. Protoc. Immunol. 2005;Chapter 12(Unit 12):11. doi: 10.1002/0471142735.im1211s64. [DOI] [PubMed] [Google Scholar]
- Montero M., Van Houten N.E., Wang X., Scott J.K. The membrane-proximal external region of the human immunodeficiency virus type 1 envelope: dominant site of antibody neutralization and target for vaccine design. Microbiol. Mol. Biol. Rev. 2008;72:54–84. doi: 10.1128/MMBR.00020-07. (table of contents) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody M.A., Gao F., Gurley T.C., Amos J.D., Kumar A., Hora B., Marshall D.J., Whitesides J.F., Xia S.M., Parks R., Lloyd K.E., Hwang K.K., Lu X., Bonsignori M., Finzi A., Vandergrift N.A., Alam S.M., Ferrari G., Shen X., Tomaras G.D., Kamanga G., Cohen M.S., Sam N.E., Kapiga S., Gray E.S., Tumba N.L., Morris L., Zolla-Pazner S., Gorny M.K., Mascola J.R., Hahn B.H., Shaw G.M., Sodroski J.G., Liao H.X., Montefiori D.C., Hraber P.T., Korber B.T., Haynes B.F. Strain-specific V3 and CD4 binding site autologous HIV-1 neutralizing antibodies select neutralization-resistant viruses. Cell Host Microbe. 2015;18:354–362. doi: 10.1016/j.chom.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore P.L., Ranchobe N., Lambson B.E., Gray E.S., Cave E., Abrahams M.R., Bandawe G., Mlisana K., Abdool Karim S.S., Williamson C., Morris L., STUDY, C., IMMUNOLOGY, N. C. F. H. A. V Limited neutralizing antibody specificities drive neutralization escape in early HIV-1 subtype C infection. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris L., Chen X., Alam M., Tomaras G., Zhang R., Marshall D.J., Chen B., Parks R., Foulger A., Jaeger F., Donathan M., Bilska M., Gray E.S., Abdool Karim S.S., Kepler T.B., Whitesides J., Montefiori D., Moody M.A., Liao H.X., Haynes B.F. Isolation of a human anti-HIV gp41 membrane proximal region neutralizing antibody by antigen-specific single B cell sorting. PLoS One. 2011;6 doi: 10.1371/journal.pone.0023532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Barroso I., Salzwedel K., Hunter E., Blumenthal R. Role of the membrane-proximal domain in the initial stages of human immunodeficiency virus type 1 envelope glycoprotein-mediated membrane fusion. J. Virol. 1999;73:6089–6092. doi: 10.1128/jvi.73.7.6089-6092.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro J.B., Gorman J., Ma X., Zhou Z., Arthos J., Burton D.R., Koff W.C., Courter J.R., Smith A.B., III, Kwong P.D., Blanchard S.C., Mothes W. Conformational dynamics of single HIV-1 envelope trimers on the surface of native virions. Science. 2014;346:759–763. doi: 10.1126/science.1254426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munshaw S., Kepler T.B. SoDA2: a hidden Markov model approach for identification of immunoglobulin rearrangements. Bioinformatics. 2010;26:867–872. doi: 10.1093/bioinformatics/btq056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K.J., Gach J.S., Jones L., Semrau K., Walter J., Bibollet-Ruche F., Decker J.M., Heath L., Decker W.D., Sinkala M., Kankasa C., Thea D., Mullins J., Kuhn L., Zwick M.B., Aldrovandi G.M. 4E10-resistant HIV-1 isolated from four subjects with rare membrane-proximal external region polymorphisms. PLoS One. 2010;5 doi: 10.1371/journal.pone.0009786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez L.G., Costa M.R., Todd C.A., Haynes B.F., Montefiori D.C. Utilization of immunoglobulin G Fc receptors by human immunodeficiency virus type 1: a specific role for antibodies against the membrane-proximal external region of gp41. J. Virol. 2009;83:7397–7410. doi: 10.1128/JVI.00656-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Permar S.R., Fong Y., Vandergrift N., Fouda G.G., Gilbert P., Parks R., Jaeger F.H., Pollara J., Martelli A., Liebl B.E., Lloyd K., Yates N.L., Overman R.G., Shen X., Whitaker K., Chen H., Pritchett J., Solomon E., Friberg E., Marshall D.J., Whitesides J.F., Gurley T.C., Von Holle T., Martinez D.R., Cai F., Kumar A., Xia S.M., Lu X., Louzao R., Wilkes S., Datta S., Sarzotti-Kelsoe M., Liao H.X., Ferrari G., Alam S.M., Montefiori D.C., Denny T.N., Moody M.A., Tomaras G.D., Gao F., Haynes B.F. Maternal HIV-1 envelope-specific antibody responses and reduced risk of perinatal transmission. J. Clin. Invest. 2015;125:2702–2706. doi: 10.1172/JCI81593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinis M., Weiser B., Kuiken C., Dong T., Lang D., Nachman S., Zhang Y., Rowland-Jones S., Burger H. Genomic analysis of HIV type 1 strains derived from a mother and child pair of long-term nonprogressors. AIDS Res. Hum. Retrovir. 2007;23:309–315. doi: 10.1089/aid.2006.0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richman D.D., Wrin T., Little S.J., Petropoulos C.J. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc. Natl. Acad. Sci. U. S. A. 2003;100:4144–4149. doi: 10.1073/pnas.0630530100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringe R., Bhattacharya J. Association of enhanced HIV-1 neutralization by a single Y681H substitution in gp41 with increased gp120-CD4 interaction and macrophage infectivity. PLoS One. 2012;7 doi: 10.1371/journal.pone.0037157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders R.W., Van Gils M.J., Derking R., Sok D., Ketas T.J., Burger J.A., Ozorowski G., Cupo A., Simonich C., Goo L., Arendt H., Kim H.J., Lee J.H., Pugach P., Williams M., Debnath G., Moldt B., Van Breemen M.J., ISIK G., Medina-Ramirez M., Back J.W., Koff W.C., Julien J.P., Rakasz E.G., Seaman M.S., Guttman M., Lee K.K., Klasse P.J., Labranche C., Schief W.R., Wilson I.A., Overbaugh J., Burton D.R., Ward A.B., Montefiori D.C., Dean H., Moore J.P. HIV-1 VACCINES. HIV-1 neutralizing antibodies induced by native-like envelope trimers. Science. 2015;349:aac4223. doi: 10.1126/science.aac4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman M.S., Janes H., Hawkins N., Grandpre L.E., Devoy C., Giri A., Coffey R.T., Harris L., Wood B., Daniels M.G., Bhattacharya T., Lapedes A., Polonis V.R., Mccutchan F.E., Gilbert P.B., Self S.G., Korber B.T., Montefiori D.C., Mascola J.R. Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies. J. Virol. 2010;84:1439–1452. doi: 10.1128/JVI.02108-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X., Parks R.J., Montefiori D.C., Kirchherr J.L., Keele B.F., Decker J.M., Blattner W.A., Gao F., Weinhold K.J., Hicks C.B., Greenberg M.L., Hahn B.H., Shaw G.M., Haynes B.F., Tomaras G.D. In vivo gp41 antibodies targeting the 2F5 monoclonal antibody epitope mediate human immunodeficiency virus type 1 neutralization breadth. J. Virol. 2009;83:3617–3625. doi: 10.1128/JVI.02631-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X., Dennison S.M., Liu P., Gao F., Jaeger F., Montefiori D.C., Verkoczy L., Haynes B.F., Alam S.M., Tomaras G.D. Prolonged exposure of the HIV-1 gp41 membrane proximal region with L669S substitution. Proc. Natl. Acad. Sci. U. S. A. 2010;107:5972–5977. doi: 10.1073/pnas.0912381107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z.Y., Oh K.J., Kim M., Yu J., Brusic V., Song L., Qiao Z., Wang J.H., Wagner G., Reinherz E.L. HIV-1 broadly neutralizing antibody extracts its epitope from a kinked gp41 ectodomain region on the viral membrane. Immunity. 2008;28:52–63. doi: 10.1016/j.immuni.2007.11.018. [DOI] [PubMed] [Google Scholar]
- Tiller T., Meffre E., Yurasov S., Tsuiji M., Nussenzweig M.C., Wardemann H. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J. Immunol. Methods. 2008;329:112–124. doi: 10.1016/j.jim.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomaras G.D., Yates N.L., Liu P., Qin L., Fouda G.G., Chavez L.L., Decamp A.C., Parks R.J., Ashley V.C., Lucas J.T., Cohen M., Eron J., Hicks C.B., Liao H.X., Self S.G., Landucci G., Forthal D.N., Weinhold K.J., Keele B.F., Hahn B.H., Greenberg M.L., Morris L., Karim S.S., Blattner W.A., Montefiori D.C., Shaw G.M., Perelson A.S., Haynes B.F. Initial B-cell responses to transmitted human immunodeficiency virus type 1: virion-binding immunoglobulin M (IgM) and IgG antibodies followed by plasma anti-gp41 antibodies with ineffective control of initial viremia. J. Virol. 2008;82:12449–12463. doi: 10.1128/JVI.01708-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishwanathan S.A., Hunter E. Importance of the membrane-perturbing properties of the membrane-proximal external region of human immunodeficiency virus type 1 gp41 to viral fusion. J. Virol. 2008;82:5118–5126. doi: 10.1128/JVI.00305-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins B.A., Buge S., Aldrich K., Davis A.E., Robinson J., Reitz M.S., JR., Robert-Guroff M. Resistance of human immunodeficiency virus type 1 to neutralization by natural antisera occurs through single amino acid substitutions that cause changes in antibody binding at multiple sites. J. Virol. 1996;70:8431–8437. doi: 10.1128/jvi.70.12.8431-8437.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X., Decker J.M., Wang S., Hui H., Kappes J.C., Wu X., Salazar-Gonzalez J.F., Salazar M.G., Kilby J.M., Saag M.S., Komarova N.L., Nowak M.A., Hahn B.H., Kwong P.D., Shaw G.M. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- Wiehe K., Easterhoff D., Luo K., Nicely N.I., Bradley T., Jaeger F.H., Dennison S.M., Zhang R.J., Lloyd K.E., Stolarchuk C., Parks R., Sutherland L.L., Scearce R.M., Morris L., Kaewkungwal J., Nitayaphan S., Pitisuttithum P., Rerks-Ngarm S., Sinangil F., Phogat S., Michael N.L., Kim J.H., Kelsoe G., Montefiori D.C., Tomaras G.D., Bonsignori M., Santra S., Kepler T.B., Alam S.M., Moody M.A., Liao H.X., Haynes B.F. Antibody light-chain-restricted recognition of the site of immune pressure in the RV144 HIV-1 vaccine trial is phylogenetically conserved. Immunity. 2014;41:909–918. doi: 10.1016/j.immuni.2014.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Yang Z.Y., Li Y., Hogerkorp C.M., Schief W.R., seaman M.S., Zhou T., Schmidt S.D., Wu L., Xu L., Longo N.S., Mckee K., O'dell S., Louder M.K., Wycuff D.L., Feng Y., Nason M., Doria-Rose N., Connors M., Kwong P.D., Roederer M., Wyatt R.T., Nabel G.J., Mascola J.R. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 2010;329:856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt R., Sodroski J. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science. 1998;280:1884–1888. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- Zhang R., Verkoczy L., Wiehe K., Munir Alam S., Nicely N.I., Santra S., Bradley T., Pemble C.W.T., Zhang J., Gao F., Montefiori D.C., Bouton-Verville H., Kelsoe G., Larimore K., Greenberg P.D., Parks R., Foulger A., Peel J.N., Luo K., Lu X., Trama A.M., Vandergrift N., Tomaras G.D., Kepler T.B., Moody M.A., Liao H.X., Haynes B.F. Initiation of immune tolerance-controlled HIV gp41 neutralizing B cell lineages. Sci. Transl. Med. 2016;8:336ra62. doi: 10.1126/scitranslmed.aaf0618. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.