Abstract
Background
It has been suggested that bladder cancer can be divided into two molecular subtypes referred to as luminal and basal with distinct clinical behaviors and sensitivities to chemotherapy. We aimed to validate these subtypes in several clinical cohorts and identify signature immunohistochemical markers that would permit simple and cost-effective classification of the disease in primary care centers.
Methods
We analyzed genomic expression profiles of bladder cancer in three cohorts of fresh frozen tumor samples: MD Anderson (n = 132), Lund (n = 308), and The Cancer Genome Atlas (TCGA) (n = 408) to validate the expression signatures of luminal and basal subtypes and relate them to clinical follow-up data. We also used an MD Anderson cohort of archival bladder tumor samples (n = 89) and a parallel tissue microarray to identify immunohistochemical markers that permitted the molecular classification of bladder cancer.
Findings
Bladder cancers could be assigned to two candidate intrinsic molecular subtypes referred to here as luminal and basal in all of the datasets analyzed. Luminal tumors were characterized by the expression signature similar to the intermediate/superficial layers of normal urothelium. They showed the upregulation of PPARγ target genes and the enrichment for FGFR3, ELF3, CDKN1A, and TSC1 mutations. In addition, luminal tumors were characterized by the overexpression of E-Cadherin, HER2/3, Rab-25, and Src. Basal tumors showed the expression signature similar to the basal layer of normal urothelium. They showed the upregulation of p63 target genes, the enrichment for TP53 and RB1 mutations, and overexpression of CD49, Cyclin B1, and EGFR. Survival analyses showed that the muscle-invasive basal bladder cancers were more aggressive when compared to luminal cancers. The immunohistochemical expressions of only two markers, luminal (GATA3) and basal (KRT5/6), were sufficient to identify the molecular subtypes of bladder cancer with over 90% accuracy.
Interpretation
The molecular subtypes of bladder cancer have distinct clinical behaviors and sensitivities to chemotherapy, and a simple two-marker immunohistochemical classifier can be used for prognostic and therapeutic stratification.
Funding
U.S. National Cancer Institute and National Institute of Health.
Keywords: Bladder cancer, Biomarker
Highlights
-
•
Bladder cancer can be assigned to two intrinsic molecular subtypes referred to as luminal and basal.
-
•
Luminal tumors are characterized by the expression signature of intermediate/superficial layers of normal urothelium and they show up-regulation of PARγ target genes with enrichment for FGFR3, ELF3, CDKN1A, and TSC1 mutations.
-
•
Basal tumors show the expression signature similar to the basal layer of normal urothelium and they show the up-regulation of p63 target genes with the enrichment of TP53 and RB1 mutations.
-
•
Muscle invasive tumors of basal type are more aggressive when compared to luminal cancers.
-
•
The immunohistochemical expression of only two markers, luminal (GATA3) and basal (KRT5/6) is sufficient to identify the molecular subtypes of bladder cancer with over 90% accuracy.
Our analyses of several large cohorts comprising of a total of 937 bladder cancer tumors samples with annotated clinical data showed that bladder cancers can be classified into molecular subtypes referred to as luminal and basal. Luminal tumors are similar in their gene expression pattern to more differentiated intermediate and superficial layers of normal urothelium. Basal cancers recapitulate the gene expression signature of the less differentiated basal layer of the urothelium. These molecular subtypes have distinct up-regulation of transcription factors that regulate the expression pattern of their target genes and are enriched for specific mutations. Basal tumors which deeply invade the bladder wall are more aggressive when compared to luminal types and they show distinct sensitivities to frontline chemotherapy. The classification of bladder cancer into its intrinsic molecular subtypes can be accomplished with two immunohistochemical markers such as GATA3 for luminal tumors and KRT5/6 for basal tumors which can be performed in primary care centers.
1. Introduction
Recent genomic investigations of bladder cancer have revealed complex alterations with heavy mutational load and frequent involvement of chromatin remodeling genes (Cancer Genome Atlas Research N, 2014, Gui et al., 2011, Lawrence et al., 2013). Other studies have identified distinct genomic signatures associated with cancer progression, metastasis and response to therapeutic manipulations (Takata et al., 2005, Puzio-Kuter et al., 2009, Cheng et al., 2013, Van Allen et al., 2014, Groenendijk et al., 2016, Dyrskjot et al., 2003). Several groups used whole genome expression profiling to classify bladder cancer into various distinct subtypes (Cancer Genome Atlas Research N, 2014, Damrauer et al., 2014, Choi et al., 2014a, Lindgren et al., 2010, Sjodahl et al., 2012). Although the names for the respective classes used by these groups were different, they showed striking similarities to the intrinsic basal and luminal subtypes identified in human breast cancers (Damrauer et al., 2014, Choi et al., 2014b, Perou et al., 2000). In general, the markers that are used to classify bladder cancers into the two major groups reflect an expression signature of normal basal and intermediate/luminal urothelial cell layers (Choi et al., 2014b). Most importantly, the two intrinsic subtypes of bladder cancer show distinct clinical behaviors and responses to frontline chemotherapy (Choi et al., 2014a, Choi et al., 2014b, McConkey et al., 2015). In the chemotherapy naive setting, the muscle-invasive bladder cancers of the basal subtype were more aggressive with shorter survival when compared to luminal cancers (Choi et al., 2014a, Choi et al., 2014b). On the other hand, basal bladder cancers were more sensitive to cisplatinum based chemotherapy and the patients with this form of the disease appeared to gain more benefits from frontline chemotherapy when compared to luminal subtypes (Choi et al., 2014a, Choi et al., 2014b).
Since the classification of bladder cancer into intrinsic molecular subtypes provides prognostic information and may help to identify a subgroup of patients with increased sensitivity to chemotherapy, we performed a meta-analysis of the luminal and basal subtypes of bladder cancer in several MD Anderson and publicly available cohorts. We also validated the signature profiles of luminal and basal cancers on retrospectively collected paraffin-embedded tumor samples, as these are the types of tissue on which the standard of clinical care is based. Finally, in order to identify a minimal set of clinically applicable biomarkers permitting simple classification of bladder cancers into luminal and basal subtypes, we performed image assisted analysis of selected immunohistochemical markers on parallel tissue microarrays.
2. Methods
2.1. Patients and Study Design
The expression profiling studies of molecular subtypes of bladder cancer were conducted on four cohorts: (Cancer Genome Atlas Research N, 2014) the MD Anderson cohort of fresh frozen bladder tumor tissue (n = 132); (Gui et al., 2011) the cohort of fresh frozen bladder tumor tissue from Lund University in Sweden (n = 308) referred to as the Lund cohort; (Lawrence et al., 2013) the Cancer Genome Atlas (TCGA) cohort of fresh frozen bladder tumor tissue (n = 408); and (Takata et al., 2005) the MD Anderson cohort of formalin-fixed paraffin-embedded bladder tumor tissue samples (n = 89) (Table 1).
Table 1.
Summary of clinical data: the MD Anderson, TCGA#, Lund,⁎ and FFPE MD Anderson cohorts.
| Stage | Subtype | Gender F/M | Total | Age, yr, mean ± SD | Med. sur. mo | 95% CI, mo |
|---|---|---|---|---|---|---|
| MD Anderson cohort | ||||||
| Superficial(Ta-Tis) | Luminal | 8/26 | 34 | 65.3 ± 11.3 | NA | 110.7 - NA |
| Invasive | Luminal | 11/49 | 60 | 68.3 ± 10.6 | 87.9 | 45.1 - NA |
| (T1 and higher) | Basal | 12/22 | 34 | 67.9 ± 16.1 | 11 | 7.2 - NA |
| Invasive | Luminal-non p53 | 9/36 | 45 | 66.7 ± 11.3 | 91.4 | 41.6 - NA |
| (T1 and higher) | Luminal-p53 | 2/13 | 15 | 73.1 ± 6.2 | 80.8 | 26.3 - NA |
| Basal-non p53 | 8/8 | 16 | 73.1 ± 8.8 | 10.6 | 6.5 - NA | |
| Basal-p53 | 4/14 | 18 | 63.2 ± 19.7 | 13.7 | 6.2 - NA | |
| Double negative | 1/3 | 4 | 62.7 ± 8.1 | NA | 14.5 - NA | |
| TCGA cohort | ||||||
| Invasive | Luminal | 48/164 | 212 | 68.3 ± 11.0 | 46.8 | 31.2 - 97.1 |
| (T2 and higher) | Basal | 56/123 | 179 | 68.0 ± 10.1 | 27.1 | 20.7 - 51.2 |
| Invasive | Luminal-non p53 | 27/106 | 133 | 66.2 ± 11.3 | NA | 56.5 - NA |
| (T2 and higher) | Luminal-p53 | 21/58 | 79 | 71.9 ± 9.5 | 28.2 | 22.4 - 46.8 |
| Basal-non p53 | 29/69 | 98 | 67.6 ± 11.0 | 29.7 | 20.2 - NA | |
| Basal-p53 | 27/54 | 81 | 68.5 ± 9.0 | 24.1 | 16.8 - 104.6 | |
| Double negative | 3/14 | 17 | 65.6 ± 10.2 | 18.6 | 7.3 - NA | |
| Lund cohort | ||||||
| Superficial | Luminal | 36/80 | 116 | 69.7 ± 12.8 | NA | NA - NA |
| (Ta-pTis) | ||||||
| Invasive | Luminal | 27/112 | 139 | 70.3 ± 11.3 | NA | NA - NA |
| (T1 and higher) | Basal | 15/23 | 38 | 75.6 ± 11.1 | NA | 24.2 - NA |
| Invasive | Luminal-non p53 | 18/73 | 91 | 69.1 ± 11.6 | NA | NA - NA |
| (T1 and higher) | Luminal-p53 | 9/39 | 48 | 72.6 ± 10.3 | NA | NA - NA |
| Basal-non p53 | 13/14 | 27 | 76.3 ± 12.1 | 34.8 | 13.4 - NA | |
| Basal-p53 | 2/9 | 11 | 73.7 ± 8.3 | NA | 24.2 - NA | |
| Double negative | 2/11 | 13 | 66.8 ± 7.3 | NA | NA - NA | |
| MD Anderson paraffin-embedded formalin-fixed tissue cohort | ||||||
| Invasive | Luminal | 8/38 | 46 | 70.2 ± 11.6 | 57.3 | 23.4 - NA |
| T2 and higher) | Basal | 11/18 | 29 | 69.2 ± 11.0 | 22.7 | 15.4 - NA |
| Invasive | Luminal-non p53 | 3/15 | 18 | 72.6 ± 13.7 | 20.4 | 17 - NA |
| (T2 and higher) | Lum-p53 | 5/23 | 28 | 68.7 ± 10.1 | NA | 36 - NA |
| Basal-non p53 | 6/12 | 18 | 68.9 ± 11.7 | 37.9 | 12.1 - NA | |
| Basal-p53 | 5/6 | 11 | 69.7 ± 10.3 | 19.1 | 15.4 - NA | |
| Double Neg | 3/11 | 14 | 68.0 ± 8.4 | 41.8 | 24.9 - NA | |
F, female; M, male; yr, year; SD, standard deviation; Med., median; sur., survival; mo, months; CI, Confidence Interval. #Stage is unknown in three cases. *Stage is unknown in two cases.
The MD Anderson cohort of fresh frozen bladder tumor samples was from 100 men and 32 women (mean age 67.2 years ± 12.3 SD). The tumors were classified according to the World Health Organization histologic grading system into low-grade (n = 25) and high-grade (n = 107) (Moch et al., 2016). According to the TNM staging system the tumors were divided into superficial (stage Ta-Tis; n = 34) and invasive (stage T1and higher; n = 98) categories (Sobin et al., 2009).
The Lund cohort mRNA expression and clinical data were retrieved from GEO (GSE32894) as per the original publication (Sjodahl et al., 2012). This cohort consisted of fresh frozen bladder tumor tissue samples from 80 women and 228 men (mean age 70.6 years ± 11.8 SD). The tumors were divided into non-invasive (Ta and Tis; n = 116) and invasive (T1 and higher; n = 190) according to the TNM staging system. The tumors were considered low-grade (n = 151) if they were originally reported as grade 1–2 and high-grade (n = 155) if they were reported as grade 3 in the original publication (Sjodahl et al., 2012). In two cases the tumor staging and in another two cases grading information were not available.
The TCGA cohort was comprised of 408 bladder tumors, most of which were muscle-invasive tumors (stage T2 and higher; n = 405). The level of invasion was unknown for three cases. The vast majority of tumors were of high histologic grade (n = 384) and only a few tumors were of low histologic grade (n = 21). The histologic grade was unknown for three cases. The publicly available whole transcriptome data (RNA-seq) and mutations of tumor samples with annotated clinical and survival data were downloaded from the TCGA website (https://tcga-data.nci.nih.gov/tcga/). In addition, the reverse phase protein array (RPPA) data available for 126 samples comprising the expression levels for 190 proteins were downloaded from the same website.
To assemble the MD Anderson formalin-fixed paraffin-embedded tissue sample cohort we searched the pathology files and randomly selected 89 tumor samples (67 men and 22 women; mean age 69.6 years ± 0.9 SD) from cystectomy specimens with invasive bladder cancer. The clinical information, demographic data and follow-up outcomes were obtained from the patient's medical records. Similar to the MD Anderson cohort of fresh frozen bladder tumor samples, the tumors were graded according to World Health Organization histologic grading system and staged according to the TNM staging system (Moch et al., 2016, Sobin et al., 2009). All tumors in this cohort were high-grade and muscle-invasive (stage T2 and higher). The histologic slides were reviewed by two independent pathologists and well-preserved areas rich in tumor cells were identified. The corresponding areas were marked on paraffin blocks and two parallel tissue cores were obtained with a 2 mm biopsy punch (Miltex, York, PA). One core was submitted for RNA extraction and the other core was used for tissue microarray preparation. The use of human samples and related clinical data for this study was approved by the institutional IRB.
2.2. Procedures
2.2.1. Microarray Experiments
RNAs from fresh frozen tissue were extracted using the mirvana™miRNA isolation kit (Ambion, Inc.) and the microarray experiments were performed by direct hybridization on the Illumina HumanHT-12 v3 Expression BeadChip platform as previously described (Choi et al., 2014a). In brief, intact RNAs from samples were converted to double-stranded cDNA, followed by in vitro transcription to generate biotin-labeled cRNA using Ambion Illumina TotalPrep RNA Amplification Kit (Illumina). The labeled cRNA was hybridized containing complimentary gene-specific sequences. After washing the BeadChips, Cy3-Streptavidin was added to bind to analytical probes hybridized on the BeadChips. The chips were then scanned on Illumina BeadArray Reader to measure the fluorescence intensities. Data from the array images were analyzed using Illumina's GenomeStudio Gene Expression Module.
RNA from formalin-fixed paraffin-embedded samples was extracted using the MasterPure Complete DNA and RNA Purification Kit (Epicenter Biotechnologies, Madison, WI, USA). The microarray experiments were performed on Illumina's WG-DASL platform as previously described (Singh et al., 2015, Guo et al., 2016). In brief, the RNA samples (0.25–1.0 μg) were reverse transcribed to biotinylated cDNAs using the Master Mix cDNA Synthesis kit. The biotinylated cDNAs were annealed to assay specific chimeric-oligos containing universal PCR priming sites and a gene-specific sequence. The cDNA-oligo complexes were captured by strepavidin-conjugated paramagnetic particles. The captured cDNA templates were amplified with a pair of fluorophore labeled universal PCR primers. The labeled single stranded PCR products were hybridized to Illumina HumanHT-12 DASL Expression BeadChips containing the gene-specific complimentary sequences. After hybridization, fluorescence intensities were measured at each bead location on the array using Illumina BeadArray Reader. Array data export, processing and analyses were performed with Illumina BeadStudio v3.1.3 (Gene Expression Module V3.3.8).
2.2.2. Tissue Microarrays and Immunohistochemistry
Parallel tissue microarrays comprising 76 cases obtained from MD Anderson formalin-fixed and paraffin-embedded cohort were designed and prepared as previously described (Wang et al., 2002, Kim et al., 2005). Expression of selected genes was measured by immunohistochemical staining. Briefly, tissue microarrays were created using a tissue arrayer (Beecher Instruments, Silver Spring, MD). Based on gene expression profiles, a set of antibodies was selected for immunohistochemical analyses of luminal, basal, and p53-like subtypes. The luminal markers included mouse monoclonal antibody against human GATA3 (HG3-31 clone, 1:100 dilution; Santa Cruz Biotechnology Inc., Santa Cruz, CA), KRT18 (DC10, 1:50 dilution, Dako, Carpinteria, CA), KRT20 (Ks20.8 clone, 1:400 dilution, Dako), uroplakin 2 (BC21 clone, 1:100 dilution; Biocare Medical, Concord, CA), cyclin D1 (SP4 clone, 1:40 dilution, Lab Vision Corp, Fremont, CA), and ERBB2/HER2 (e2-4001 clone. 1:300 dilution, Lab Vision Corp, Fremont, CA); KRT5/6 (D5/16B4 clone, 1:50 dilution, Dako), KRT14 (LL002 clone, 1:50 dilution; BioGenex, Fremont, CA), and p63 (4A4 clone, 1:1000 dilution, BioCare Medical, Concord, CA) antibodies were selected as candidate basal markers and p16 (E6H4 clone, 1:3 dilution, Ventana Medical, Tucson, AZ), BCL2 (100 clone, 1:200 dilution, Leica Microsystems, Newcastle Upon Tyne, UK), smooth muscle actin (1A4 clone, 1:80,000 dilution, Sigma-Aldrich, St. Louis, MO), myosin (MY32 clone, 1:1000 dilution, Invitrogen, Waltham, MA), calponin (CALP clone, 1:2000 dilution, Dako) and desmin (D33 clone, 1:200 dilution, Dako) as p53-like markers. Immunohistochemical stains were performed using the Bond-Max Autostainer (Leica Biosystems, Buffalo Grove, IL). The bound primary antibodies were detected with the visualization reagent linked to a dextran polymer backbone with DAB (3, 3-diaminobenzidine) as a chromogen solution. Then, the slides were counterstained with Mayer's hematoxylin. The immunohistochemical results were initially inspected visually under the microscope and semi-quantitatively assessed and scored by two pathologists as follows: 0, negative; 1 +, weak; 2 +, moderate; and 3 +, strong. Then, for selected markers we performed a quantitative image analysis using an automated digital image analyzer, GenoMx (BioGenex). The proportions of tumor cell nuclei positive for GATA3 staining and the proportion of positive tumor tissue for all remaining markers were measured.
2.3. Statistical Analysis
Data processing and statistical analyses were performed using R package available from the Bioconductor website (http://www.Bioconductor.org). The gene signal values from the array data for the MD Anderson cohorts of fresh and formalin-fixed paraffin-embedded samples were transformed to logarithmic scale and normalized by the sample-wise medians. The data were subjected to hierarchical clustering analyses using Euclidean distance and average linkage algorithm as distance metric. Samples were classified into luminal and basal molecular subtypes as previously described (Choi et al., 2014a). In addition, subsets of tumors characterized by the upregulation of p53 target genes were identified (Choi et al., 2014a). Similar clustering analyses were performed for the genome-wide expression data on the Lund and The Cancer Genome Atlas cohorts and tumors were assigned to specific subtypes by applying the set of luminal, basal and p53-like markers described previously (Choi et al., 2014a). In addition, gene set enrichment analyses (GSEA) were performed to evaluate the significance of gene expression signature enrichment in molecular subsets of bladder cancer (Subramanian et al., 2005). Mutational data were downloaded from the Cancer Genome Atlas portal (https://tcga-data.nci.nih.gov/tcga/). MutSigCV (version 1.4; https://www.broadinstitute.org/cancer/cga/mutsig) and used to identify genes that were mutated more often than expected by chance given the background mutation processes (Lawrence et al., 2013). The significant gene list was obtained using a cutoff of false discovery rate < 0.05. The statistical significance of association between the mutations and the molecular subtypes were assessed by Fisher's exact test. RPPA data of 126 samples comprising of 190 proteins from the TCGA cohort were processed using R package in Bioconductor (http://www.bioconductor.org) and ANOVA analysis was used to identify proteins with the expression levels associated with molecular subtypes of bladder cancer. By performing pair-wise t-tests, subsets of proteins up-regulated in specific molecular subtypes were identified. Linear discrimination and support vector machine analyses were used to determine the best combination of immunohistochemical biomarkers to predict molecular subtypes. Leave-one-out cross-validation was employed to evaluate the performance of the classifiers. Survival outcomes of different molecular subtypes were analyzed using log-rank test and Kaplan-Meier curves. The associations of molecular types of bladder cancer with survival were also analyzed in a multivariate fashion with histologic grade, stage, age, and gender as covariates.
3. Results
To visualize the intrinsic luminal and basal molecular subtypes of bladder cancer, we performed supervised hierarchical clustering of basal and luminal biomarkers using whole transcriptome expression data from four independent cohorts of bladder cancer samples (Fig. 1). Three of these cohorts were composed of fresh frozen tumor samples: the MD Anderson cohort (n = 132); the TCGA cohort (n = 408); and the Lund cohort (n = 308). The fourth MD Anderson cohort contained retrospectively collected formalin-fixed and paraffin-embedded bladder tumor samples (n = 89).
Fig. 1.
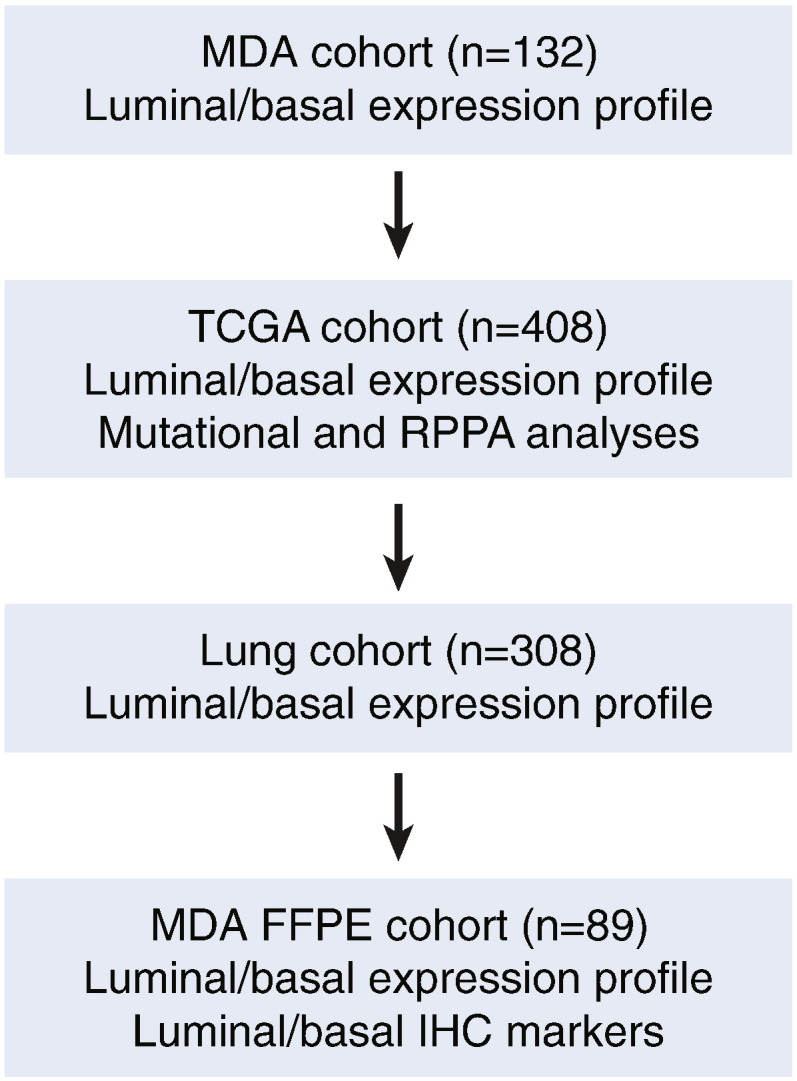
Organizational flow-chart of meta-analysis in four cohorts of bladder cancer samples.
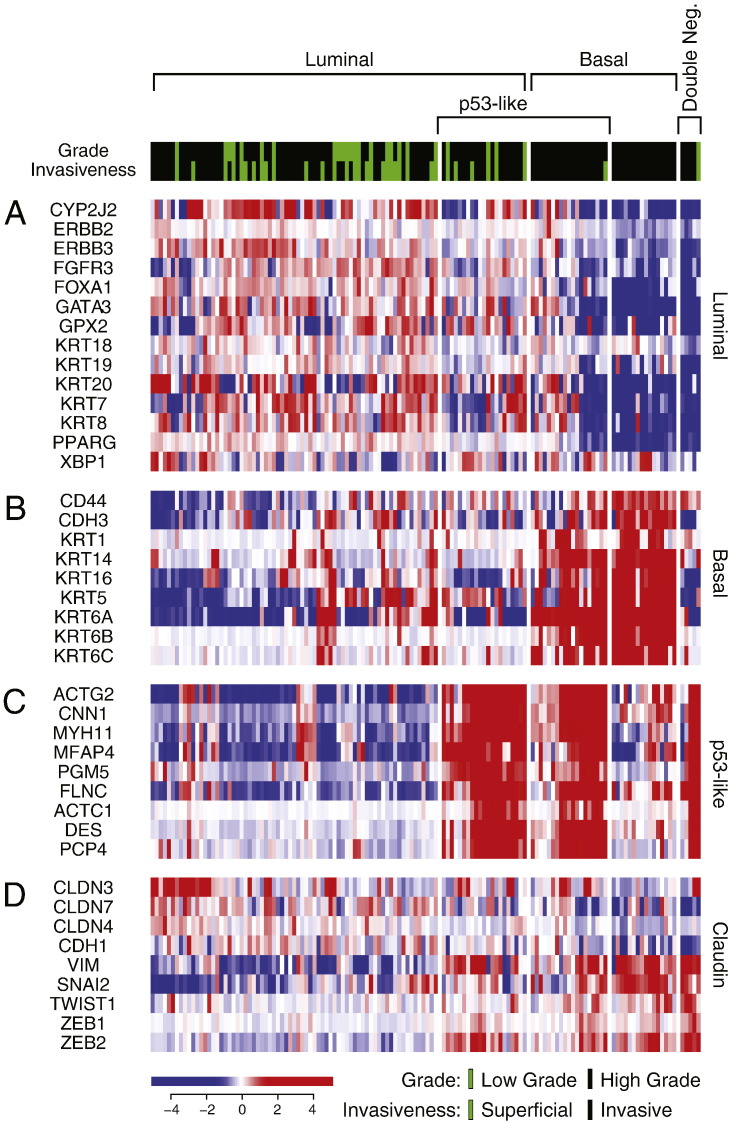
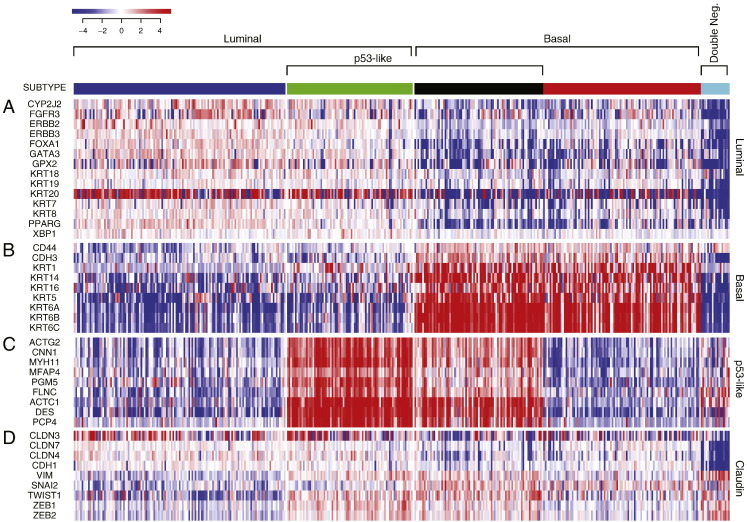
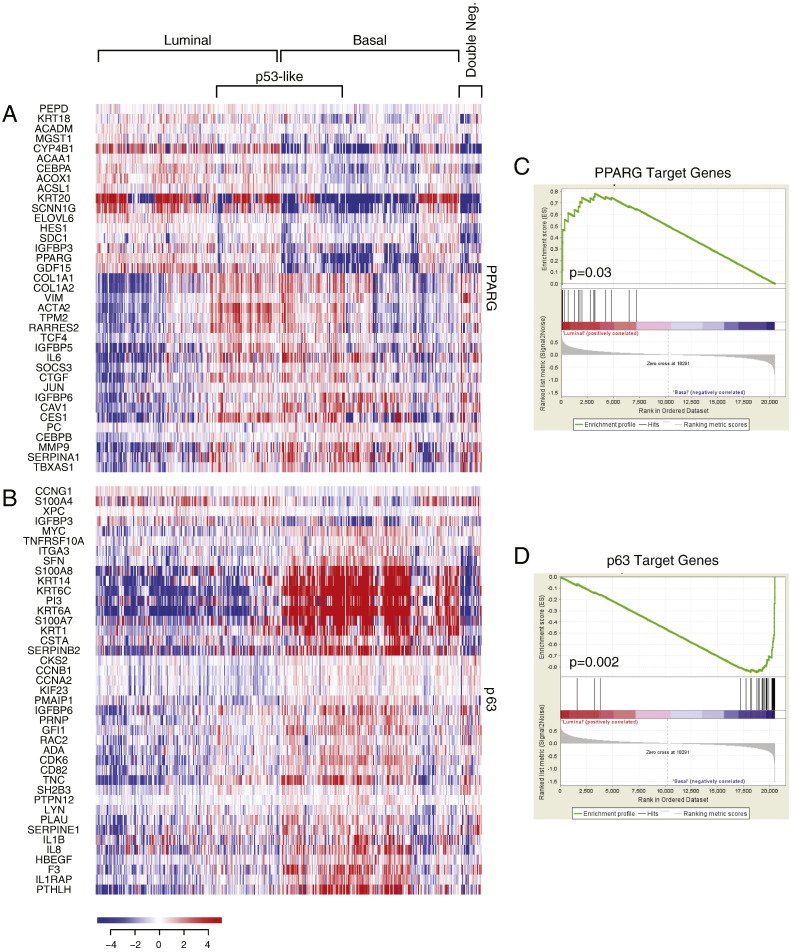
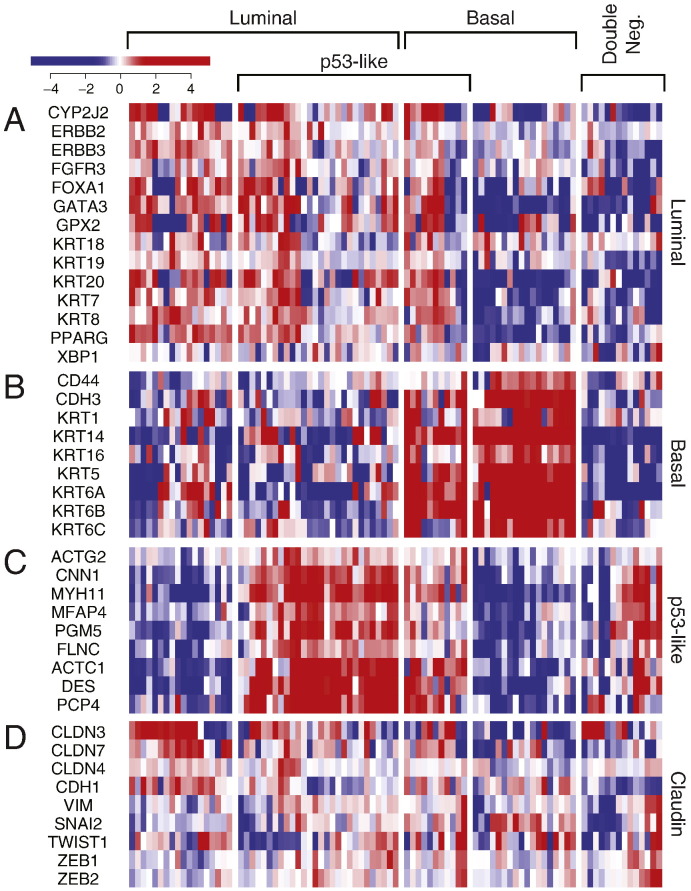
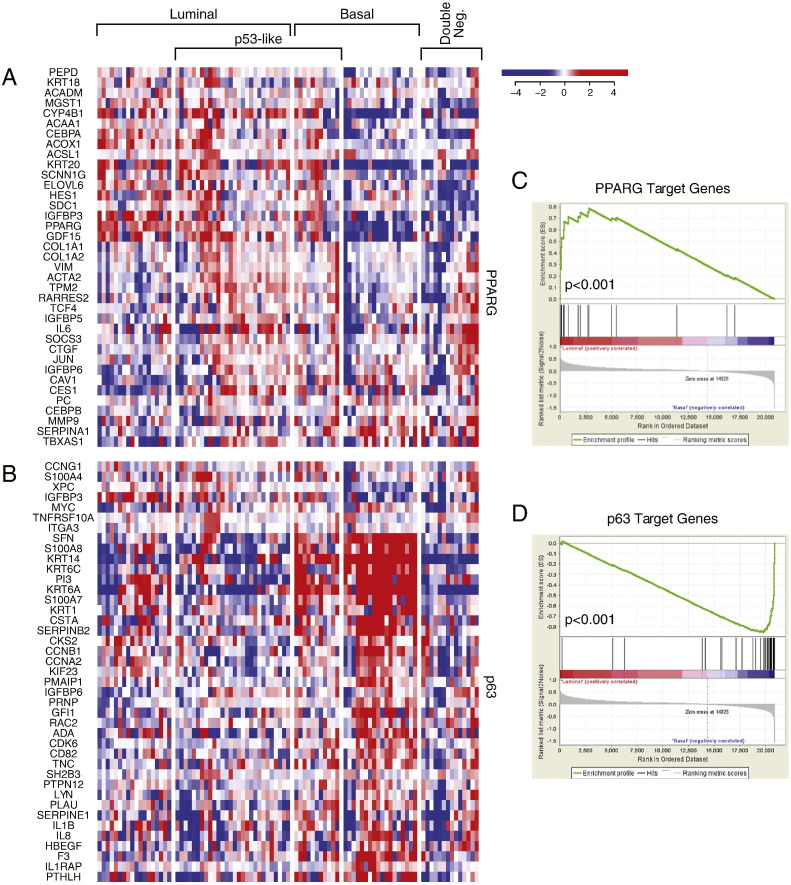
First, we analyzed the MD Anderson cohort which included 34 superficial (Ta-Tis) and 98 invasive (T1 and higher) bladder cancers further classified into low- and high-grade histologic categories. We identified two major groups of samples. The first group was characterized by the strong expression of luminal markers such as KRT20, GATA3, FOXA1, XBP1 and CD24 among others signifying intermediate and terminal urothelial differentiation referred to as luminal type (Fig. 2A). This group consisted of 92 (70%) of the samples and included 24 low-grade tumors and 68 high-grade invasive tumors. Another subset (n = 35; 26%) of the tumors displayed strong expression of high molecular weight keratins (KRT5, KRT6 and KRT14) and co-expressed CDH3 and CD44 characteristic of the basal urothelial cell layer and were referred to as basal-type (Fig. 2B). The luminal tumors were characterized by the upregulation of PPARγ target genes while the basal tumors were characterized by the upregulation of p63 target genes, all of which were confirmed by GSEA analyses (Fig. S1A–D). In addition, a subset of tumors in both luminal and basal groups showed the upregulation of p53 target genes that was referred to as p53-like subtype (Fig. 2C). This subtype consisted of 40 tumors and 21 of them were luminal and 19 were basal. A small fraction of tumors (n = 5; 4%) did not express either luminal or basal markers and were referred to as “double-negative” (Fig. 2A, B). Since subsets of tumors with such characteristics were identified in human breast and bladder cancer before and showed low expression signature of claudin related genes, we tested the expression levels of claudin target genes in a double-negative subset of bladder tumors (Choi et al., 2014a, McConkey et al., 2015). This showed that our double-negative tumors were indeed characterized by low expression signature of claudin related genes (Fig. 2D).
Fig. 2.
Whole-genome messenger RNA profiling of fresh frozen bladder tumor samples in the MD Anderson cohort (n = 132). Unsupervised hierarchical clustering was performed with luminal, p53-like, basal, and claudin markers. A. Expression pattern of signature luminal markers. B. Expression pattern of signature basal markers. C. Expression pattern of signature p53 target genes. D. Expression pattern of signature claudin target genes.
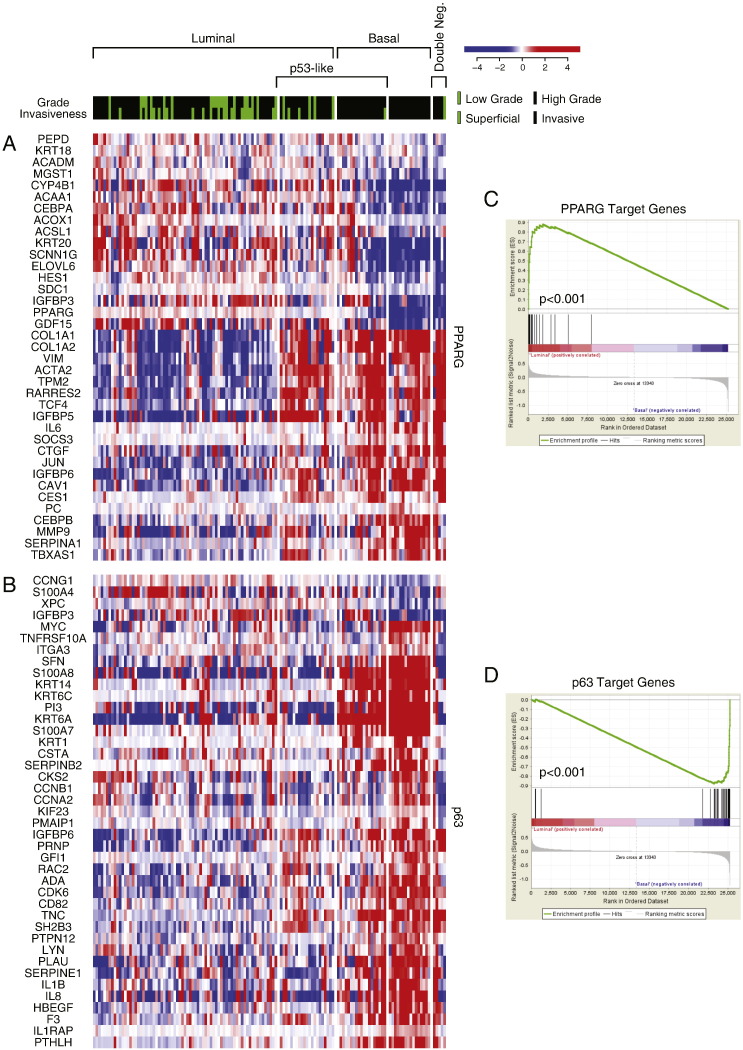
Fig. S1.
The expression pattern of signature transcriptional regulators in molecular subtypes of bladder cancer in fresh frozen bladder tumor samples of the MD Anderson cohort (n = 132). A. The expression pattern of signature PPARγ target genes. B. The expression pattern of signature p63 target genes. C. PPARγ expression signature in luminal and basal subtypes compared by gene set enrichment analysis (GSEA). D. p63 expression signature in luminal and basal subtypes compared by GSEA.
Next we evaluated the cohort of 308 cases from Lund for the presence of intrinsic molecular subtypes of bladder cancer. Like the MD Anderson cohort, this cohort included not only invasive bladder tumors, but also contained a significant proportion of superficial non-invasive lesions. Similar to the two other cohorts, two groups of bladder cancer samples, referred to as luminal (n = 251) and basal (n = 41), with upregulation of PPARγ and p63 signature genes, respectively, were identified (Fig. S2A–C and Fig. S3A–D). Again, a subset of tumors in both luminal (n = 76) and basal (n = 12) subtypes showed overexpression of p53 pathway signature genes (Fig. SC). Additionally, a small subset of double-negative tumors (n = 16; 5%) with claudin-low expression signature were identified (Fig. S2A, B and D).
Fig. S2.
Whole-genome messenger RNA profiling of fresh frozen bladder tumor samples in the Lund cohort (n = 308). Unsupervised hierarchical clustering was performed with luminal, p53-like, basal, and claudin markers. A. The expression pattern of signature luminal markers. B. The expression pattern of signature basal markers. C. The expression pattern of signature p53 target genes. D. The expression pattern of signature claudin target genes.
Fig. S3.
The expression pattern of signature transcription regulators in molecular subtypes of bladder cancer in fresh frozen bladder tumor samples of the Lund cohort (n = 308). A. The expression pattern of PPARγ target genes. B. The expression pattern of p63 target genes. C. PPARγ expression signature in luminal and basal subtypes compared by gene set enrichment analysis (GSEA). D. p63 expression signature in luminal and basal subtypes compared by GSEA.
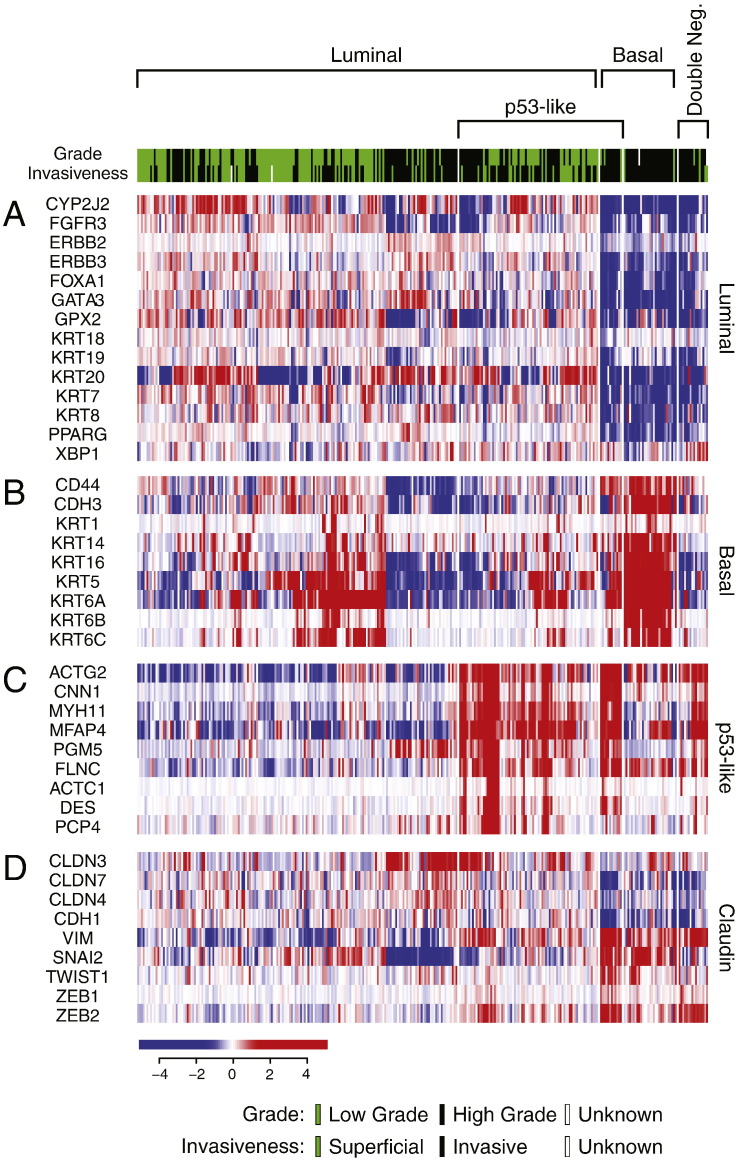
We then validated the presence of the same intrinsic molecular subtypes on 408 cases of high-grade invasive bladder cancers from the publicly available TCGA cohort. By applying our classification algorithm, similar to our MD Anderson cohort, tumors could be segregated into two major groups. One group of tumors (n = 212; 52%) exhibited the luminal expression signature while the other group (n = 189; 44%) expressed basal markers (Fig. S4A, B). Moreover, a subset of tumors (n = 160) exhibited p53 target expression signature and of these 79 were luminal and 81 were basal (Fig. 4C). Similarly, luminal tumors were characterized by the enrichment of PPARγ target genes while basal tumors showed the expression signature of p63 target genes (Fig. S5A–D). In addition, a small subset of double-negative tumors (n = 17; 4%) which did not show expression signatures of either luminal or basal cancers and was characterized by the downregulation of claudin target genes, was also identified (Fig. S4A, B, and D).
Fig. S4.
Whole-genome messenger RNA profiling and mutational analysis of fresh frozen bladder tumor samples in the TCGA cohort (n = 408). Unsupervised hierarchical clustering was performed with luminal, p53-like, basal, and claudin markers. A. Expression pattern of signature luminal markers. B. Expression pattern of signature basal markers. C. Expression pattern of signature p53 target genes. D. Expression pattern of signature claudin target genes.
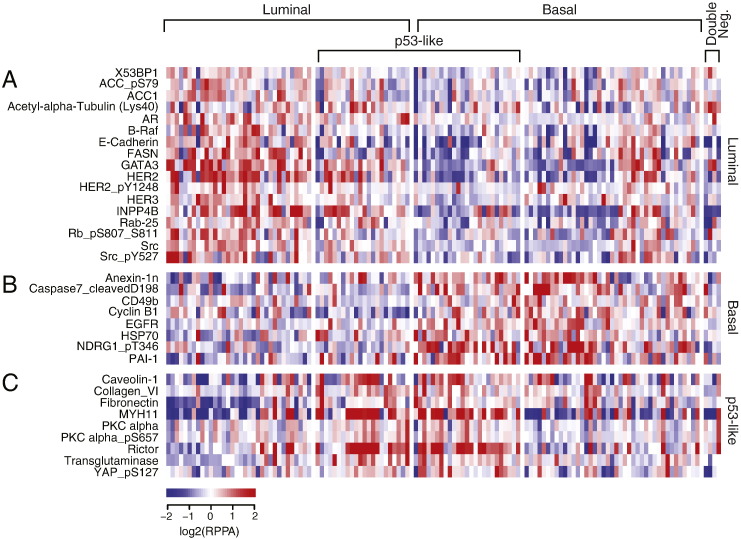
Fig. 4.
RPPA heat map showing the expression level of proteins in the TCGA cohort (n = 127). A. Expression patterns of signature proteins in luminal subtypes. B. Expression patterns of signature proteins in basal subtypes. C. Expression patterns of signature proteins in p53-like subtypes.
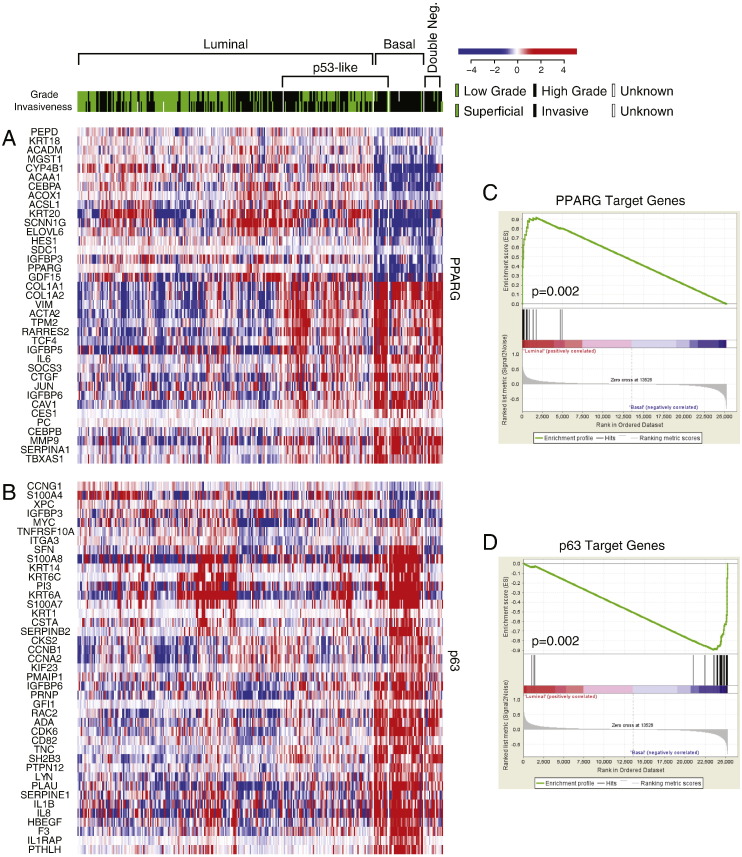
Fig. S5.
The expression pattern of signature transcription regulators in molecular subtypes of bladder cancer in fresh frozen bladder tumor samples of the TCGA cohort (n = 408). A. The expression pattern of PPARγ target genes. B. The expression pattern of p63 target genes. C. The PPARγ expression signature in luminal and basal subtypes compared by gene set enrichment analysis (GSEA). D. The p63 expression signature in luminal and basal subtypes compared by GSEA.
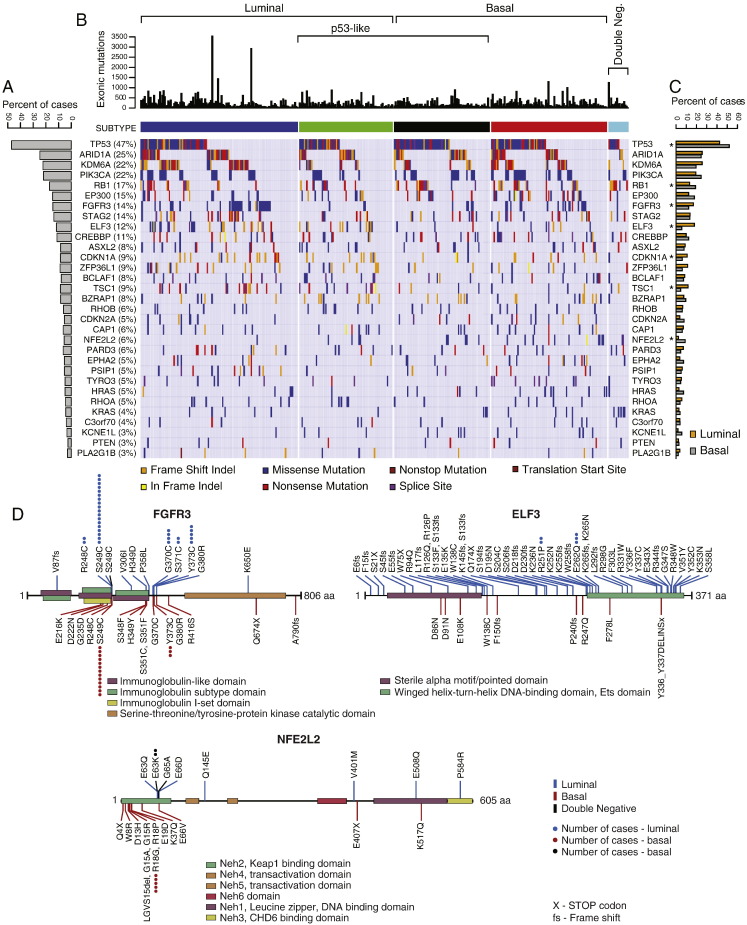
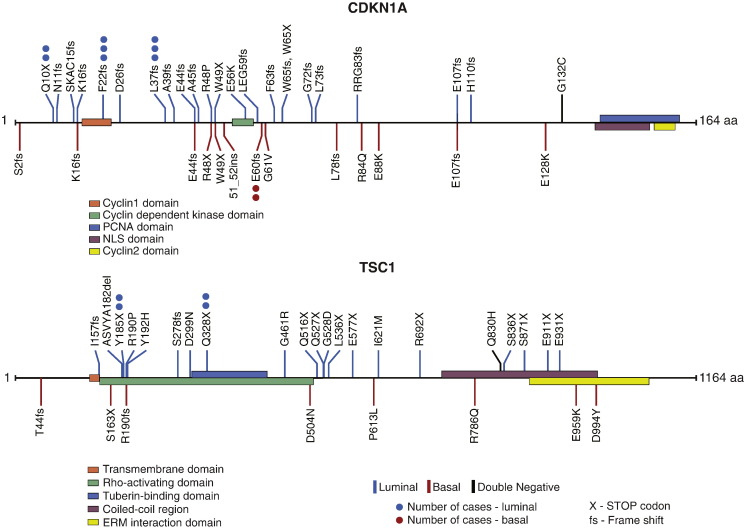
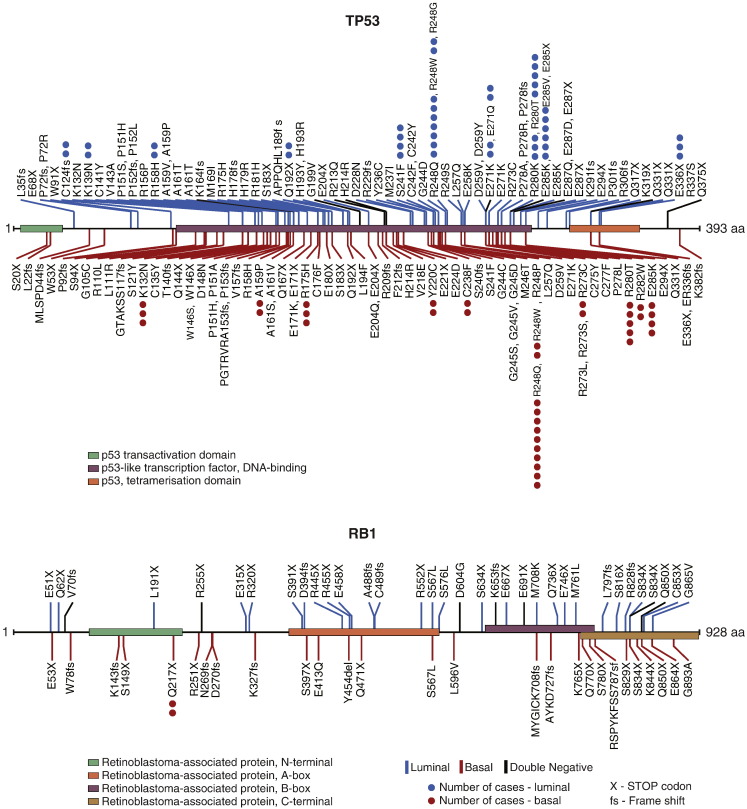
The TCGA cohort provided an opportunity to analyze the mutational and protein expression patterns of the molecular subtypes of bladder cancer (Fig. 3A–C). First we analyzed the mutations in the two intrinsic luminal and basal subtypes. Since there were only 17 cases classified as double negative, their mutations were recorded in figures and tables but the quantitative mutational analyses of these cases were not performed. Whole genome sequencing of 408 tumors identified 99,983 non-synonymous somatic mutations including 97,685 point mutations and 2298 insertions or deletions, yielding the mean and median somatic mutation load per case of 244 and 170, respectively. A small portion of cases showed a high mutational rate with 1.5% of cases having > 1000 non-synonymous mutations and 8.8% of cases having > 500 non-synonymous mutations. Among the luminal cancers, the mean and median numbers of mutations per case were 258 and 177 respectively. A similar mutational rate was found in basal cancers with the mean and the median number of mutations per case being 225 and 160 respectively. MutSigCV (v1.4) identified 31 genes which showed statistically significant levels of recurrent somatic mutations. The five most frequently mutated genes included TP53 (47%), ARID1A (25%), KDM6A (22%), PIK3CA (22%), and RB1 (17%). The genes identified as statistically significantly mutated included, in general, multiple genes involved in cell cycle regulation, chromatin remodeling and kinase signaling pathways as reported previously (Cancer Genome Atlas Research N, 2014). The overall mutational landscape of luminal and basal bladder cancers was similar but several genes were distinctively enriched for their mutations in specific molecular subtypes. The mutations of four genes (FGFR3, ELF3, CDKN1A, and TSC1) were enriched in luminal tumors (Supplemental Table 1, Supplemental Table 2, Supplemental Table 3, Supplemental Table 4) while the basal tumors were enriched for the mutations of TP53, RB1, and NFE2L2 genes (Supplemental Table 5, Supplemental Table 6, Supplemental Table 7). For some of these genes, their mutational enrichments, in specific, molecular subtypes, were particularly evident when the mutational patterns of the functional domains of their encoded protein were analyzed (Fig. 3D, Fig. S6, Fig. S7). For FGFR3, the mutations clustered in the extracellular receptor domain and were enriched in luminal subtype. A particularly strong enrichment was observed in the ETS encoding domain of the ELF3 gene where 20 of 22 mutations clustered in the luminal subtype. On the other hand, 15 of 17 deleterious mutations of the NFE2L2 gene involved the Neh2 domain and affected the basal subtype.
Fig. 3.
The mutational landscape of molecular subtypes of bladder cancer of fresh frozen bladder tumor samples in the TCGA cohort (n = 408). A. The proportion of bladder cancer cases with statistically significant levels of mutations (MutSig, false discovery rate < 0.05). B. Mutation types in molecular subtypes of bladder cancer classified according to their expression profiles of luminal and basal markers. C. The proportion of mutations for individual genes in luminal and basal subtypes of bladder cancer. The genes which show statistically significant difference (p < 0.05) in molecular subtypes are indicated by an asterisk. D. Mutation patterns of the FGFR3, ELF3, and NFE2L2 genes in luminal and basal cancers.
Fig. S6.
Mutation patterns of the CDKN1A and TSC1 genes in luminal and basal cancers.
Fig. S7.
Mutation patterns of the TP53 and RB1 genes in luminal and basal cancers.
The RPPA expression levels of 190 proteins were available for 127 TCGA bladder tumor samples. By using the molecular subtype identifications based on cDNA expression levels from the same cohort, we performed analyses of variance (ANOVA) and identified 46 proteins that were significantly (p < 0.05) differentially expressed among the molecular subtypes. By performing t-tests we found that 34 of these proteins were significantly up-regulated in one of the molecular subtypes of bladder cancer (Fig. 4A–C). This approach showed that 17 of the proteins were up-regulated in the luminal subtypes and included GATA3, E-Cadherin, HER2/3, Rab-25, and Src among others (Fig. 4A). There were eight proteins up-regulated in the basal subtype, which included Anexin-1, CD49, Cyclin B1, and EGFR (Fig. 4B). The nine proteins identified as overexpressed in the p53-like tumors included Caveolin-1, Collagen VI, Fibronectin, and PKC alpha among others (Fig. 4C).
Because the analyses of the three independent bladder cancer cohorts of frozen fresh tumor samples confirmed the presence of the intrinsic luminal and basal types of the disease, we proceeded to analyze the formalin-fixed and paraffin-embedded cohort as well as the parallel tissue microarray. The goal of this analysis was to address the question of whether the genome profiling with the identification of intrinsic molecular subtypes of bladder cancer can be successfully performed on formalin-fixed and paraffin-embedded tissue, which is routinely used for clinical management. In addition, we wanted to identify a limited set of biomarkers that could be used to subclassify bladder cancer into subtypes by simple immunohistochemical tests that can be applied to the management of bladder cancer patients in primary care centers around the world. Similar to the three fresh frozen bladder cancer tissue cohorts, the formalin-fixed and paraffin-embedded samples could be divided into luminal (n = 46; 52%) and basal (n = 29; 32%) groups (Fig. 5A, B). In addition, a subset of tumors with the wild-type p53 gene expression signature was identified (n = 39; 44%); 28 of which were luminal and 11 were basal (Fig. 5C). Analogous to the fresh frozen tumor sample cohorts, the luminal paraffin-embedded tumor samples exhibited PPARγ pathway activation and basal tumors showed the upregulation of the p63 transcription factor pathway genes (Fig. S8A–D). As in the other cohorts, a small group of double-negative tumors (n = 14, 16%) which did not express either luminal or basal markers with claudin-low expression signature was identified (Fig. 5A, B, and D).
Fig. 5.
Whole-genome messenger RNA profiling and semi-quantitative immunohistochemical analysis of signature basal and luminal markers in formalin-fixed and paraffin-embedded tumor samples (n = 89) and parallel tissue microarray (n = 76) in the MD Anderson cohort. A. Expression pattern of signature luminal markers. B. Expression pattern of signature basal markers. C. Expression signature of p53 target genes. D. Expression pattern of signature claudin target genes.
Fig. S8.
The expression pattern of signature transcription regulators in molecular subtypes of bladder cancer in the formalin-fixed and paraffin-embedded MD Anderson cohort (n = 89). A. The expression pattern of PPARγ target genes. B. The expression pattern of p63 target genes. C. PPARγ expression signature in luminal and basal subtypes compared by gene set enrichment analysis (GSEA). D. p63 expression signature in luminal and basal subtypes compared by GSEA.
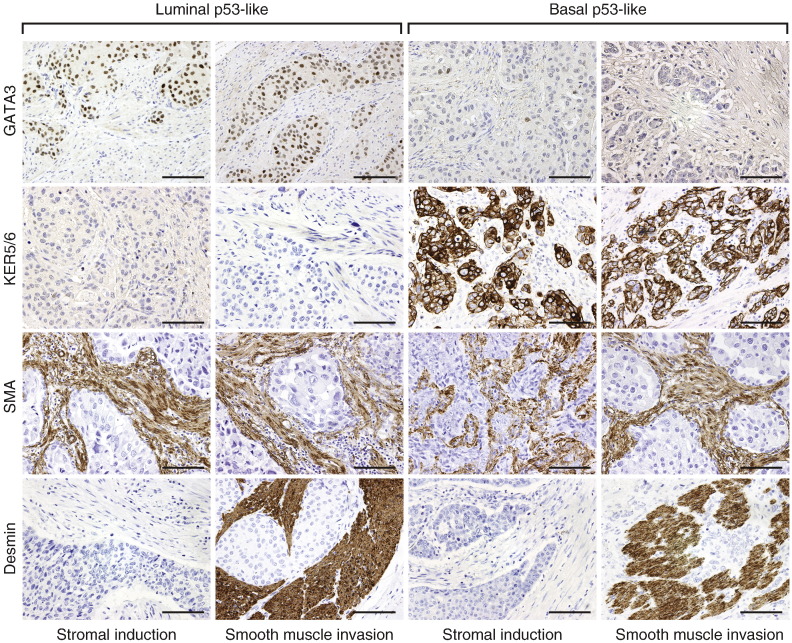
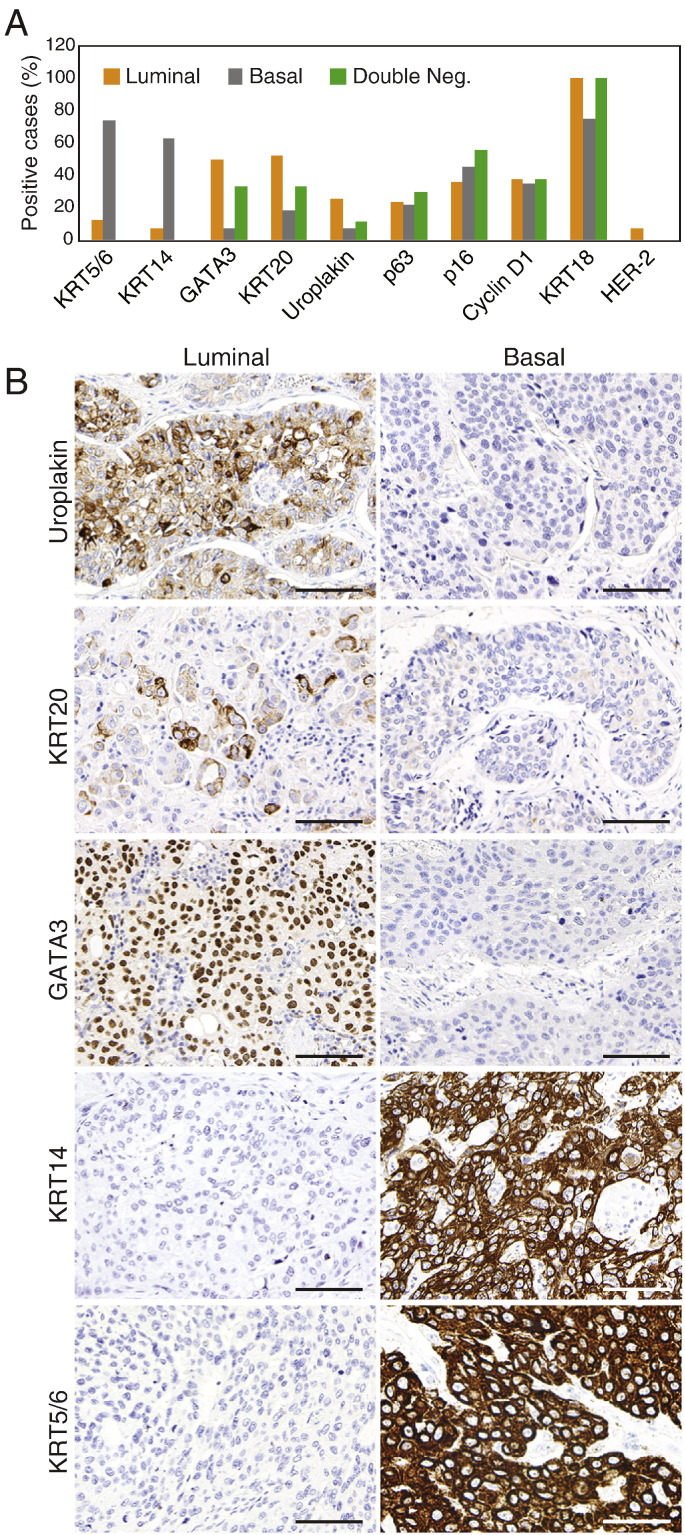
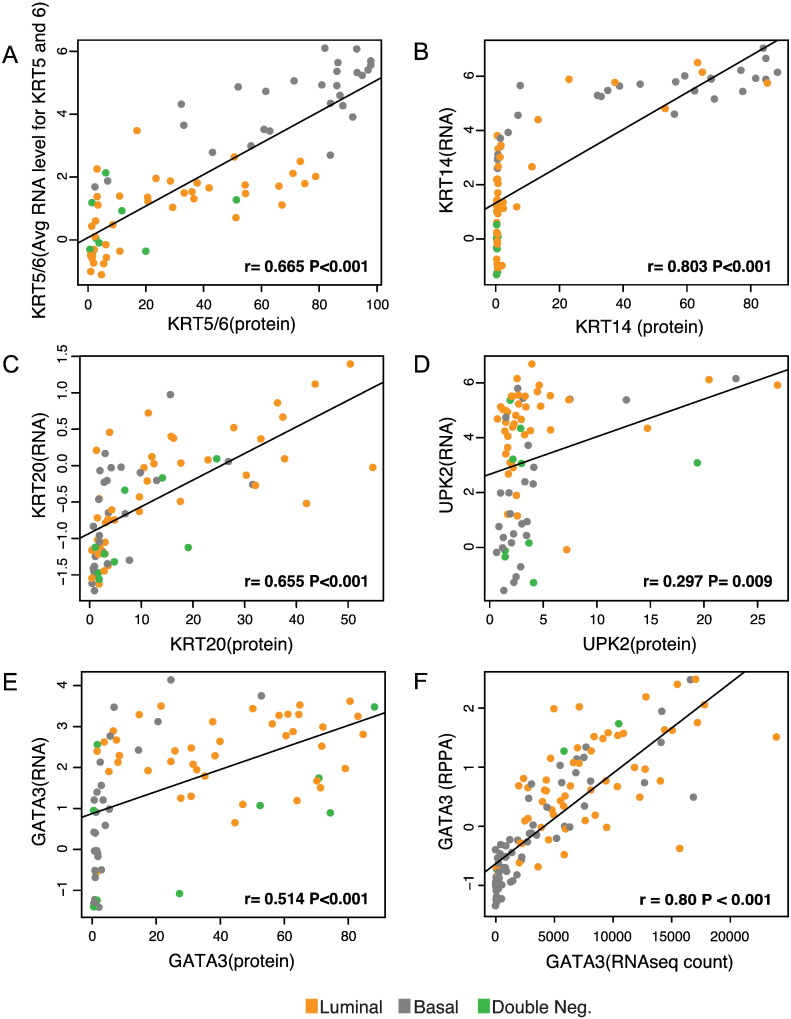
In order to identify immunohistochemical markers to classify bladder cancers into basic intrinsic molecular categories we used genomic expression profiling to select a set of markers that are routinely used in the pathologic diagnostic workup of human tumor samples. These included the GATA3 transcription factor, KRT18, KRT20, uroplakin 2, cyclin D1, and ERBB2/HER2 as candidate luminal markers and KRT5/6, KRT14, and p63 as candidate markers of basal tumors. In addition, we selected p16, BCL2, smooth muscle actin, myosin, calponin, and desmin for p53-like subtype. The initial microscopic inspection of tissue microarray stained for these markers showed that several of the markers (smooth muscle actin, myosin, calponin, and desmin) that were selected for the p53-like subset of tumors stained only the stromal tissue intervening with the nests of cancer cells, and none of these markers were convincingly positively stained in tumor cells. The stromal components comprising either smooth muscle infiltrated by tumor cells or the so called stromal induction composed of florid proliferations of myofibroblastic cells intermixed with the nests of tumor cells were positive for the mesenchymal markers that, in fact, represented the signature of stromal smooth muscle or myofibroblastic differentiation (Fig. 6). In addition, staining for another p53-like marker, BCL2, an anti-apoptotic protein, showed that inflammatory cells infiltrating tumors were positive while the tumor cells were negative. Therefore, the microscopic analysis of the immunohistochemical stains of the so called p53-like tumors provided evidence that most of their markers were not distinctively positive in tumor cells and the p53-like signature profile resulted from stromal contamination. The remaining p-53 marker (p16) did not show significant difference in expression between p53-like and non-p53-like tumors. In addition, the ERBB2/HER2 stains were only positive in 3 cases of luminal p53-like tumors. Therefore it was not considered to represent an effective differentiation marker and was not further analyzed. The immunohistochemical expression levels of the remaining luminal and basal markers were semi-quantitatively scored and their results are summarized in Fig. 7A. The expression patterns of some of these markers (p63, cyclin D1, KRT18) showed clear overlap in the luminal and basal subtypes, but five of them (KRT5/6 and KRT14 for basal tumors and GATA3, KRT20, and uroplakin 2 for luminal tumors) showed promising differential expression patterns and they were selected for further analyses (Fig. 7B). First we addressed the question whether the immunohistochemical expression levels in tissue microarrays could be correlated with their cDNA expression levels used as the original classifiers. These analyses were performed on the parallel sample sets from the formalin-fixed and paraffin-embedded MD Anderson cohort (Fig. S9A–E). For GATA3 in addition to comparing its cDNA and protein expression levels in the MD Anderson cohort, we used the RNASeq and RPPA expression levels from the TCGA cohort (Fig. S9F). All of the proteins analyzed showed statistically significant correlation between their immunohistochemical expression, quantitated by image analysis, and their cDNA expression levels.
Fig. 6.
Immunohistochemical analysis of p53 target markers in molecular subtypes of bladder cancer. Note that in both luminal (GATA3 +; KRT5/6-) and basal (GATA3-; KRT5/6 +) subtypes, the p53-like tumors are showing the expression of SMA or desmin in the stromal component of the tumor while the tumor cells do not express these markers. Scale bars indicate 100 μM.
Fig. 7.
Immunohistochemical analysis of luminal and basal markers in molecular subtypes of bladder cancer. A. Summary of semi-quantitative scoring of immunohistochemical expression levels of signature luminal and basal markers in tissue microarrays. The proportion of positive cases with score 2 + or higher is shown. B. The immunohistochemical expression of signature luminal and basal markers in representative luminal and basal bladder cancers. Scale bars indicate 100 μ.
Fig. S9.
Correlation between mRNA expression levels and their encoded proteins detected by immunohistochemistry and RPPA. A–E. Correlation between mRNA expression level in cDNA microarray and their respective encoded protein detected by immunohistochemistry and quantitative image analysis in tissue microarrays for signature basal and luminal markers (MD Anderson FFPE cohort). F. Correlation between GATA3 mRNA quantitated by RNASeq and its protein quantitated by RPPA (TCGA cohort).
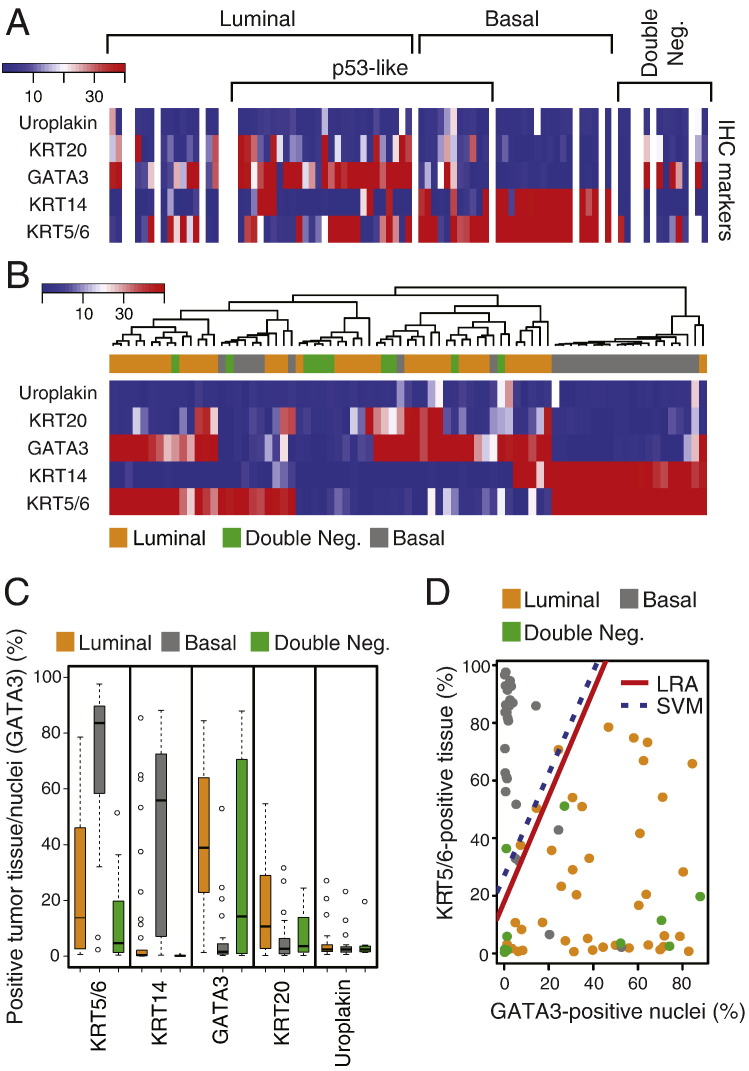
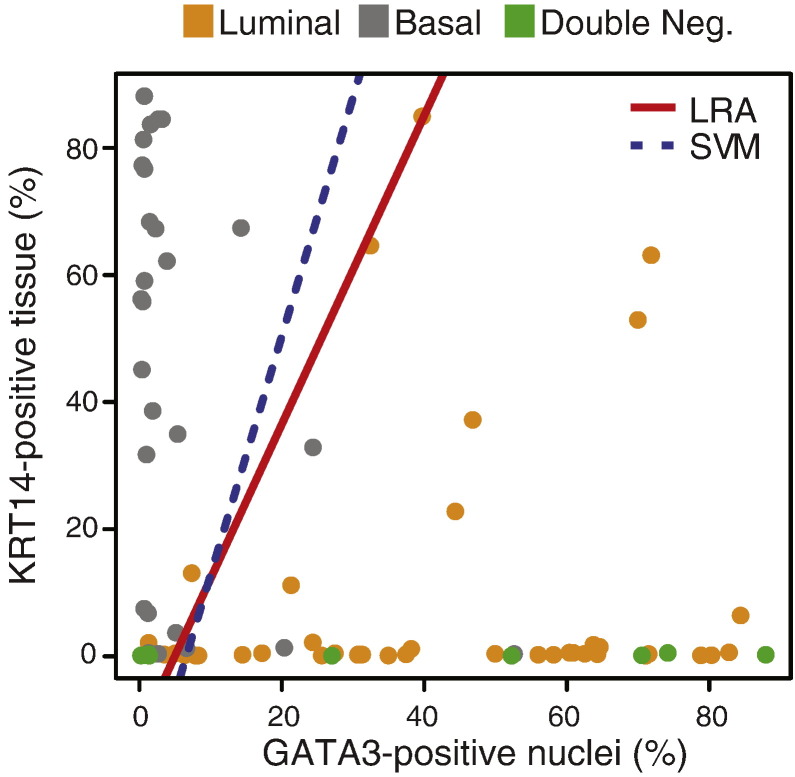
When the results from image analyses were compared with mRNA-based tumor subtype assignments, it became evident that the luminal tumors were clearly enriched for the immunohistochemically detectable protein expression levels of luminal markers while the basal tumors showed clear enrichment for expressions of basal markers (Fig. 8A). Although, in general, tumors classified as luminal based on genome expression profiling were positive for luminal markers (GATA3, KRT20, and uroplakin) and basal tumors were positive for basal makers (KRT5/6 and KRT14), there was significant overlap between these two sets of markers, particularly evident in the luminal category. When tumors were clustered according to quantitative immunohistochemical expression levels, they were classified into two distinct groups designated as clusters A and B corresponding to luminal and basal subtypes. We performed this clustering using different cut-off expression levels corresponding to 10, 20, 30, and 40% of tumor tissue positivity for KRT5/6, KRT14, KRT20, and uroplakin. The cut-off expression levels of 10, 20, 30, and 40% of tumor nuclei positivity were used for GATA3. Regardless of the cut-off positivity level, the tumors were segregated into two major groups referred to as clusters A and B corresponding to luminal and basal categories. The best segregation of specific molecular subtypes into their respective clusters was accomplished with 20% tumor tissue positivity and 20% tumor nuclei positivity cut-off levels (Fig. 8B). At these cut-off levels, all but one of the tumors classified as luminal by genome expression profiling were in cluster A. Cluster B contained almost exclusively basal tumors. Unfortunately, due to the overlapping expression between immunohistochemical luminal and basal markers, several basal tumors (8; 28%) co-clustered with luminal tumors in cluster A. In addition, all double-negative tumors were classified by immunohistochemical expression levels as luminal as a part of cluster A. In order to find the best immunohistochemical classifier of molecular subtypes of bladder cancer, we performed box-type analyses of the marker expression levels which showed that KRT5/6, KRT14 and GATA3 may be the most efficient markers with minimal overlap to classify the tumors into luminal and basal subtypes (Fig. 8C). We verified this by performing logistic regression and support vector machine analyses in which we analyzed models of all combinations of marker expression levels and showed that KRT5/6 and GATA3 represent the best pair of markers classifying the tumor into molecular categories with a 91% accuracy (Fig. 8D). The second most effective pair of markers was keratin 14 and GATA3, which classified the tumors into luminal and basal subtypes with 89% accuracy (Fig. S10). In these analyses the support vector machine performed in a similar fashion to logistic linear regression and did not improve the accuracy of the classifier. In both of the analytical models the majority of double-negative tumors were in the luminal category.
Fig. 8.
Quantitative image-based immunohistochemical analysis of selected luminal and basal markers in formalin-fixed and paraffin-embedded tumor samples (n = 76) in the MD Anderson cohort. A. Expression levels of selected luminal and basal markers in tissue microarray analyzed by quantitative image analysis. The cases were classified and clustered on the basis of cDNA microarray analysis as shown in Fig. 3A. B. Hierarchical clustering analysis using 20% of tumor tissue positivity and 20% of tumor nuclei positivity as cut-off expression levels of signature luminal and basal markers revealed by immunohistochemistry and analyzed by quantitative image analysis. C. Mean expression levels of signature luminal and basal markers revealed by immunohistochemistry and quantitated by image analysis. D. Logistic regression (LRA) and support vector machine (SVM) analyses using the immunohistochemical levels of two signature markers: luminal (GATA3) and basal (KRT5/6).
Fig. S10.
Logistic regression and support vector machine analyses using the immunohistochemical levels of two signature luminal (GATA3) and basal (KRT14) markers.
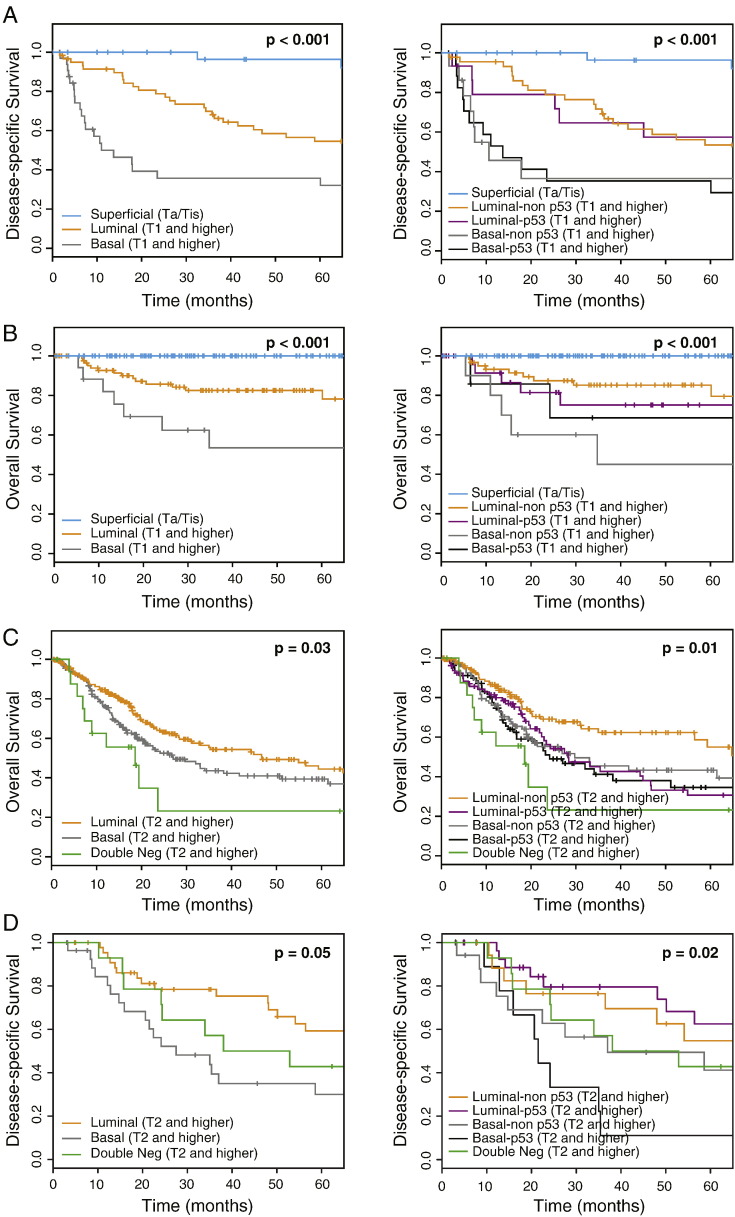
Since the intrinsic molecular subtypes of bladder cancer were reported previously to have differences in clinical behavior with basal tumors being more aggressive when compared to luminal tumors, we performed survival analyses on all four cohorts (Fig. 9A–D, Table 1). Disease specific survival analyses were performed on both fresh frozen and formalin-fixed paraffin-embedded MD Anderson cohorts while for the two publically available cohorts, i.e., Lund and TCGA, only overall survival analyses could be performed. In addition, two cohorts (fresh frozen tissue MD Anderson and the Lund cohorts) included superficial non-invasive tumors and these groups were separately analyzed and compared to invasive tumors. Initially the invasive tumors were separated into three sub-groups based on their expression profiles and classified as luminal, basal and double-negative. Since the number of cases with follow-up for the double-negative group was insufficient in some of the cohorts, the survival analyses for this group was not performed on the fresh frozen tissue MD Anderson and Lund cohorts. In all four cohorts, the invasive basal tumors were more aggressive as compared to invasive luminal tumors and were associated with a significantly shorter survival. The difference was particularly striking when the disease specific-survival was analyzed in the two MD Anderson cohorts. The double-negative tumors were more aggressive in the TCGA cohort when compared to basal tumors in the TCGA cohort but the Kaplan-Meier curve of the double-negative tumors in the MD Anderson cohort of archival paraffin-embedded samples overlapped with that of basal tumors and the differences in survival between double-negative and basal tumors were statistically insignificant.
Fig. 9.
Survival analyses in four independent cohorts. A. Fresh frozen tumor samples in the MD Anderson cohort (n = 132). B. Fresh frozen tumor samples in the Lund cohort (n = 308). C. Fresh frozen tumor samples in the TCGA cohort (n = 408). D. Formalin-fixed and paraffin-embedded samples in the MD Anderson cohort (n = 89). Left panels in each cohort show the Kaplan-Meier plots of luminal, basal, and double-negative subtypes. Right panels in each cohort show the Kaplan-Meier plots of luminal and basal cancers divided into non-p53-like and p53-like subtypes. The superficial non-invasive tumors in the fresh frozen tumor samples of the MD Anderson and the Lund cohorts were analyzed as a separate group. The survival analyses for the double-negative group were not performed in the fresh frozen tumor samples of the MD Anderson and the Lund cohorts because of the insufficient number of cases for follow-up data.
Next, we included in the analyses the p53-like tumors and separated the luminal and basal tumors into two subgroups defined as luminal or basal non-p53-like and luminal or basal with p53-like signature. In these analyses we addressed the question whether the p53-like tumors have distinct clinical behavior. These analyses did not disclose significant differences in clinical behavior of p53-like tumors across the cohorts. The survival curves of the tumors with p53-like signatures either overlapped with the survival curves of the tumors exhibiting luminal or basal phenotypes or were between them. The p53-like basal tumors displayed significantly more aggressive behavior in the formalin-fixed and paraffin-embedded MD Anderson cohort only.
Finally, we addressed the issue of whether the molecular subtypes are associated with clinical outcome when analyzed together with other parameters such as histologic grade and stage of the tumor stage as well as the patient's age and gender in a multivariate analysis. In both MD Anderson cohorts as well as in the TCGA cohort the molecular subtypes were significantly associated with outcomes in the multivariate analysis together with tumor grade, stage, as well as the patient's age and gender. In particular, it was evident that luminal subtype was associated with more favorable outcome when compared to basal subtype. These associations were not, however, evident in the Lund cohort in which the molecular subtypes were not independent predictors of the outcome in the multivariate analysis.
4. Discussion
Our meta-analysis of the genome expression profiles on several large cohorts, comprising a total of 937 bladder cancer tumor samples with annotated clinical data, showed that bladder cancers can be consistently and reliably classified into two intrinsic molecular types, luminal and basal. These classifications can be successfully performed, not only on fresh frozen tumors but also on formalin-fixed and paraffin-embedded archival samples. Moreover, parallel analyses of genomic expression profiles and immunohistochemical expression patterns in tissue microarrays identified a limited number of biomarkers that can be used to classify bladder cancer into its intrinsic molecular types. The immunohistochemical expression levels of just two signatures markers, one luminal and the other basal, specifically GATA3 and KRT5/6 respectively, are sufficient to classify bladder cancers into luminal and basal categories with over 90% accuracy but the performance of this classifier should be further validated on larger independent cohorts. Survival analyses confirmed that invasive luminal and basal bladder cancers had distinctively different clinical behaviors. The differences were particularly evident when disease specific survival was analyzed. The basal tumors were consistently more aggressive when compared to luminal tumors and were associated with significantly shorter survival than luminal tumors.
A small fraction (4–16%) of double-negative cases characterized by the low level of expression of either luminal or basal markers was identified in all cohorts. These cases appear similar to a previously identified subcategory of bladder and breast cancers with down regulated claudin target genes (Damrauer et al., 2014, Prat et al., 2010). The previously identified category of so called p53-like tumors was identified as a subgroup of both luminal and basal tumors. In addition, a fraction of double-negative tumors were also characterized by the expression signature of p53 target genes. The microscopic inspection of the immunohistochemical stains of tumors classified by expression profiling as p53-like, disclosed that the markers used for their identification were expressed in the stromal component while the tumor cells were, in general, negative. This indicated that the so called p53 phenotype resulted from the contamination of the tumor tissue with stromal components. The RPPA data from the TCGA cohort provides additional clues by showing that the proteins overexpressed in the p53-like subtypes in general belong to the category of the so-called mesenchymal markers. This was particularly evident when the tumor infiltrated the smooth muscle of the bladder or in the areas of the so called stromal induction which contained florid proliferations of myofibroblastic cells. Such samples showed strong stromal overexpression of smooth muscle and myofibroblastic markers in the stromal tissue surrounding the nests of tumor cells. This data strongly suggests that stromal contaminations might have contributed to the identification of the so-called p53-like phenotype and indicate that further analyses should be performed with purified tumor tissue such as laser microdissection. Previous studies demonstrated that membership in the p53-like subtype was unstable and could be enhanced by exposure to neoadjuvant chemotherapy (Choi et al., 2014a). Therefore, the p53-like subtype does not appear to be an “intrinsic” subtype of bladder cancer. Nevertheless, these infiltrated tumors tended to be chemoresistant (Choi et al., 2014a), and data from a recently completed Phase II clinical trial of the blocking anti-PDL1 antibody atezolizumab suggest that patients with infiltrated (TCGA cluster II) tumors obtained the most clinical benefit from the drug (Rosenberg et al., 2016). Therefore, it may be useful to distinguish these infiltrated basal and luminal tumors from the others when selecting patients for neoadjuvant or adjuvant therapy.
The overall mutational pattern of luminal and basal subtypes was similar but there were several genes that have their mutations enriched in specific molecular subtypes. The mutations of FGFR3, ELF3, CDKN1A, and TSC1 were enriched in luminal subtypes. The mutations of TP53, RB1, and NFE2L2 were enriched in basal tumors. However, the frequency of the involvement of these genes was too low and there was also an overlap in their involvement in molecular subtypes precluding their use as effective molecular classifiers. The protein expression pattern of molecular subtypes revealed by RPPA provided a roster of proteins distinctively overexpressed in luminal and basal subtypes. They included E-Cadherin, HER2/3, Rab-25, and Src as markers of luminal tumors and CD49, Cyclin B1 and EGFR as markers of basal tumors. These proteins in addition to contributing to a list of potential markers of molecular subtypes may represent attractive therapeutic targets.
Molecular subtypes of bladder cancer resemble those originally identified in human breast carcinomas that can also be divided into luminal and basal subtypes using a similar set of markers (Perou et al., 2000). Several groups independently classified bladder cancer into distinct subtypes by using genomic expression profiling (Cancer Genome Atlas Research N, 2014, Damrauer et al., 2014, Choi et al., 2014a, Lindgren et al., 2010, Sjodahl et al., 2012). One of these groups divided bladder cancers into two subtypes, (Damrauer et al., 2014) another classified them into three subtypes, (Choi et al., 2014a) and the third concluded that there were four distinct molecular categories of bladder cancer, (Cancer Genome Atlas Research N, 2014) while the fourth proposed the classification scheme based on five subcategories (Sjodahl et al., 2012). Although these groups used different names for their respective categories they used overlapping sets of markers that were originally used to subclassify breast cancers (Perou et al., 2000). The subtypes of bladder cancer identified by these groups show overlapping expression signatures with at least three of the previously identified molecular subtypes referred to as squamous, genomically unstable, and infiltrated (Sjodahl et al., 2012). From these investigations it is evident that bladder cancer is not only clinically and pathologically, but also molecularly, a heterogeneous disease. However, there is a general consensus that the top-level separation occurs at the basal and luminal differentiation checkpoint proposed by the group at the University of North Carolina (Damrauer et al., 2014). Although the well-known study of bladder cancer molecular subtypes performed by the group at the University of Lund, Sweden identified five distinct molecular subtypes, termed urobasal A, genomically unstable, infiltrated, urobasal B and squamous cell carcinoma-like, more recently the same group reconciled their subtypes with the basal and luminal phenotypes identified by the University of North Carolina (Aine et al., 2015). Specifically they showed that the urobasal A and genomically unstable tumors were contained within the luminal subtype cluster whereas the infiltrated, urobasal B, and squamous cell carcinoma-like tumors were contained within the basal subtype cluster (Aine et al., 2015). Similarly, even though the Cancer Genome Atlas group concluded that bladder cancers could be subdivided into four clusters, it recognized that clusters I and II were enriched with luminal markers and their gene expression signatures whereas clusters III and IV contained basal or mesenchymal markers characteristic of the basal subtype identified by the University of North Carolina (Cancer Genome Atlas Research N, 2014).
In summary, our studies confirmed the existence of two distinct molecular subtypes of bladder cancer referred to as luminal and basal. The luminal cancers appear to evolve through the papillary track while basal forms were nonpapillary. The superficial papillary tumors were exclusively luminal while the invasive bladder cancers can be almost equally divided into luminal and basal types. In this scenario, those invasive tumors that show luminal expression signatures most likely evolve from the preexisting papillary disease and likely represent a progression of superficial papillary tumors.
Our unpublished genomic whole-organ studies suggest that luminal and basal tumors develop from preexisting diffuse mucosal field effects which retain the expression signature of their respective luminal and basal tumors. Further support for early dichotomy of bladder carcinogenesis is provided by recent animal lineage tracing studies in the BBN mouse model (Van Batavia et al., 2014, Shin et al., 2014a, Shin et al., 2014b, Czerniak et al., 2016). These studies implicate that basal and luminal forms of bladder cancers are derived from distinct progenitor cells. Basal cancers originate from KRT5 and Sonic hedge-hog-positive basal uroprogenitor cells while papillary luminal tumors are derived from the intermediate cells (Shin et al., 2014b). In addition, the molecular subtypes of bladder cancer show distinct mutation signature and involvement of transcription factors which may represent therapeutic targets (Cancer Genome Atlas Research N, 2014). Our meta-analysis of bladder cancer subtypes has important implication for management of patients with bladder cancer by providing tools for prognostication and selection for specific therapies.
Contributors
DM and BC conceived the project ideas. BC was responsible for the project final design, supervision, interpretation of the data, and he wrote the manuscript. VD, MZ, and LZ performed data analysis and VD drafted the original text. JB and TM performed gene expression experiments and were involved in their data analysis. BC, VD, MZ, and CG performed pathological reviews and immunohistochemical analysis. DEC, SZ, SL, and JGL contributed to the organization of the pathological material and performed confirmatory testing. LZ, JNW, and KB were responsible for design and statistical analysis of genomic expression profiles. VD and LZ performed the mutational analysis of the TCGA cohort. ASR and CD provided and analyzed clinical data.
Declaration of Interest
We declare no competing interest.
The following are the supplementary data related to this article.
List of mutations of FGFR3 in molecular subtypes of bladder cancer in the TCGA cohort.
List of mutations of ELF3 in molecular subtypes of bladder cancer in the TCGA cohort.
List of mutations of CDKN1A in molecular subtypes of bladder cancer in the TCGA cohort.
List of mutations of TSC1 in molecular subtypes of bladder cancer in the TCGA cohort.
List of mutations of TP53 in molecular subtypes of bladder cancer in the TCGA cohort.
List of mutations of RB1 in molecular subtypes of bladder cancer in the TCGA cohort.
List of mutations of NFE2L2 in molecular subtypes of bladder cancer in the TCGA cohort.
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.ebiom.2016.08.036.
Acknowledgements
This project was supported by National Cancer Institute grants R01 CA151489 and P50 CA91846 (Project 1 and Core C) to B.C. V.D. is supported by T32 CA163185 grants. The content is solely the responsibility of the authors and does not necessarily represent the views of the National Cancer Institute or the National Institutes of Health. The study funders had no role in the design of the study, the collection, analysis, or interpretation of the data, the writing of the manuscript, or the decision to submit the manuscript for publication. The authors thank Virginia Hurley and Stephanie Garza for administrative assistance as well as Kim Anh Vu for graphical design.
References
- Aine M., Eriksson P., Liedberg F., Sjodahl G., Hoglund M. Biological determinants of bladder cancer gene expression subtypes. Sci. Rep. 2015;5:10957. doi: 10.1038/srep10957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research N Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014;507(7492):315–322. doi: 10.1038/nature12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng T., Roth B., Choi W., Black P.C., Dinney C., McConkey D.J. Fibroblast growth factor receptors-1 and -3 play distinct roles in the regulation of bladder cancer growth and metastasis: implications for therapeutic targeting. PLoS One. 2013;8(2) doi: 10.1371/journal.pone.0057284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi W., Czerniak B., Ochoa A. Intrinsic basal and luminal subtypes of muscle-invasive bladder cancer. Nat. Rev. Urol. 2014;11(7):400–410. doi: 10.1038/nrurol.2014.129. [DOI] [PubMed] [Google Scholar]
- Choi W., Porten S., Kim S. Identification of distinct basal and luminal subtypes of muscle-invasive bladder cancer with different sensitivities to frontline chemotherapy. Cancer Cell. 2014;25(2):152–165. doi: 10.1016/j.ccr.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czerniak B., Dinney C., McConkey D. Origins of bladder cancer. Annu. Rev. Pathol. 2016;11:149–174. doi: 10.1146/annurev-pathol-012513-104703. [DOI] [PubMed] [Google Scholar]
- Damrauer J.S., Hoadley K.A., Chism D.D. Intrinsic subtypes of high-grade bladder cancer reflect the hallmarks of breast cancer biology. Proc. Natl. Acad. Sci. U. S. A. 2014;111(8):3110–3115. doi: 10.1073/pnas.1318376111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyrskjot L., Thykjaer T., Kruhoffer M. Identifying distinct classes of bladder carcinoma using microarrays. Nat. Genet. 2003;33(1):90–96. doi: 10.1038/ng1061. [DOI] [PubMed] [Google Scholar]
- Groenendijk F.H., de Jong J., van de Putte EE F. ERBB2 mutations characterize a subgroup of muscle-invasive bladder cancers with excellent response to neoadjuvant chemotherapy. Eur. Urol. 2016;69(3):384–388. doi: 10.1016/j.eururo.2015.01.014. [DOI] [PubMed] [Google Scholar]
- Gui Y., Guo G., Huang Y. Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nat. Genet. 2011;43(9):875–878. doi: 10.1038/ng.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C.C., Dadhania V., Zhang L. Gene expression profile of the clinically aggressive micropapillary variant of bladder cancer. Eur. Urol. 2016 doi: 10.1016/j.eururo.2016.02.056. (March 15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.H., Tuziak T., Hu L. Alterations in transcription clusters underlie development of bladder cancer along papillary and nonpapillary pathways. Lab. Investig. 2005;85(4):532–549. doi: 10.1038/labinvest.3700250. [DOI] [PubMed] [Google Scholar]
- Lawrence M.S., Stojanov P., Polak P. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499(7457):214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindgren D., Frigyesi A., Gudjonsson S. Combined gene expression and genomic profiling define two intrinsic molecular subtypes of urothelial carcinoma and gene signatures for molecular grading and outcome. Cancer Res. 2010;70(9):3463–3472. doi: 10.1158/0008-5472.CAN-09-4213. [DOI] [PubMed] [Google Scholar]
- McConkey D.J., Choi W., Dinney C.P. Genetic subtypes of invasive bladder cancer. Curr. Opin. Urol. 2015;25(5):449–458. doi: 10.1097/MOU.0000000000000200. [DOI] [PubMed] [Google Scholar]
- Moch H., Humphrey P., Ulbright T., Reuter V., WHO . 4th ed. International Agency for Research on Cancer; Lyon: 2016. Classification of Tumours of the Urinary System and Male Genital Organs. [Google Scholar]
- Perou C.M., Sorlie T., Eisen M.B. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- Prat A., Parker J.S., Karginova O. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12(5):R68. doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puzio-Kuter A.M., Castillo-Martin M., Kinkade C.W. Inactivation of p53 and Pten promotes invasive bladder cancer. Genes Dev. 2009;23(6):675–680. doi: 10.1101/gad.1772909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg J.E., Hoffman-Censits J., Powles T. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: a single-arm, multicentre, phase 2 trial. Lancet. 2016;387(10031):1909–1920. doi: 10.1016/S0140-6736(16)00561-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin K., Lim A., Odegaard J.I. Cellular origin of bladder neoplasia and tissue dynamics of its progression to invasive carcinoma. Nat. Cell Biol. 2014;16(5):469–478. doi: 10.1038/ncb2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin K., Lim A., Zhao C. Hedgehog signaling restrains bladder cancer progression by eliciting stromal production of urothelial differentiation factors. Cancer Cell. 2014;26(4):521–533. doi: 10.1016/j.ccell.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R.R., Murugan P., Patel L.R. Intratumoral morphologic and molecular heterogeneity of rhabdoid renal cell carcinoma: challenges for personalized therapy. Mod. Pathol. 2015;28(9):1225–1235. doi: 10.1038/modpathol.2015.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjodahl G., Lauss M., Lovgren K. A molecular taxonomy for urothelial carcinoma. Clin. Cancer Res. 2012;18(12):3377–3386. doi: 10.1158/1078-0432.CCR-12-0077-T. [DOI] [PubMed] [Google Scholar]
- Sobin L., Gospodarowicz M., Wittekind C. 7th ed. Wiley-Blackwell; Hoboken: 2009. TNM Classification of Malignant Tumours. [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata R., Katagiri T., Kanehira M. Predicting response to methotrexate, vinblastine, doxorubicin, and cisplatin neoadjuvant chemotherapy for bladder cancers through genome-wide gene expression profiling. Clin. Cancer Res. 2005;11(7):2625–2636. doi: 10.1158/1078-0432.CCR-04-1988. [DOI] [PubMed] [Google Scholar]
- Van Allen E.M., Mouw K.W., Kim P. Somatic ERCC2 mutations correlate with cisplatin sensitivity in muscle-invasive urothelial carcinoma. Cancer Discov. 2014;4(10):1140–1153. doi: 10.1158/2159-8290.CD-14-0623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Batavia J., Yamany T., Molotkov A. Bladder cancers arise from distinct urothelial sub-populations. Nat. Cell Biol. 2014;16(10):982–991. doi: 10.1038/ncb3038. (1–5) [DOI] [PubMed] [Google Scholar]
- Wang H., Wang H., Zhang W., Fuller G.N. Tissue microarrays: applications in neuropathology research, diagnosis, and education. Brain Pathol. 2002;12(1):95–107. doi: 10.1111/j.1750-3639.2002.tb00426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of mutations of FGFR3 in molecular subtypes of bladder cancer in the TCGA cohort.
List of mutations of ELF3 in molecular subtypes of bladder cancer in the TCGA cohort.
List of mutations of CDKN1A in molecular subtypes of bladder cancer in the TCGA cohort.
List of mutations of TSC1 in molecular subtypes of bladder cancer in the TCGA cohort.
List of mutations of TP53 in molecular subtypes of bladder cancer in the TCGA cohort.
List of mutations of RB1 in molecular subtypes of bladder cancer in the TCGA cohort.
List of mutations of NFE2L2 in molecular subtypes of bladder cancer in the TCGA cohort.