Abstract
MicroRNAs (miRNAs) are an evolutionarily conserved class of small, regulatory non-coding RNAs that negatively regulate protein coding gene and other non-coding transcripts expression. miRNAs have been established as master regulators of cellular processes, and they play a vital role in tumor initiation, progression and metastasis. Further, widespread deregulation of microRNAs have been reported in several cancers, with several microRNAs playing oncogenic and tumor suppressive roles. Based on these, miRNAs have emerged as promising therapeutic tools for cancer management. In this review, we have focused on the roles of miRNAs in tumorigenesis, the miRNA-based therapeutic strategies currently being evaluated for use in cancer, and the advantages and current challenges to their use in the clinic.
Keywords: microRNA, Non-coding RNA, Cancer, Therapeutics, Clinical trials
Highlights
-
•
miRNAs can act as oncogenes or tumor suppressors depending on the specific tissue/cancer targets.
-
•
miRNAs can be used as drugs or can be targets for drugs.
-
•
Clinical trials using miRNA mimetics or anti-miRNAs as therapeutic targets are currently underway and show promising results.
MiRNAs were originally identified as small non-coding RNAs that control the timing of larval development in Caenorhabditis elegans (Lee et al., 1993). MiRNAs are short, single stranded RNA molecules that serve as master regulators of gene expression. They have been widely implicated in pathogenesis of several human diseases, including cancers (Berindan-Neagoe et al., 2014). Their abnormal levels in tumors have important pathogenetic consequences: miRNAs overexpressed in tumors contribute to oncogenesis by downregulating tumor suppressors. For example, miR17–92 cluster reduces tumorigenic levels of E2F1 transcription factor in lymphomas (Ji et al., 2011), or miR-21 represses PTEN tumor suppressor in hepatocellular carcinomas (Meng et al., 2007). On the other hand, miRNAs lost by malignant cells generally result in oncogene overexpression. For example, let-7 family represses RAS, HMGA2 and MYC in lung cancers (Wang et al., 2012), or miR-15a and miR-16-1 downregulate BCL2 in chronic lymphocytic leukemias and cyclin D1 in prostate cancer and mantle cell lymphoma (Calin and Croce, 2006a). However, several studies have shown that miRNAs' roles in cancer are tissue and tumor specific: for example, in breast cancer models, miR-200 family has been shown to work as an oncogene and enhance distant metastasis (Korpal et al., 2011), whereas in ovarian, renal and lung tumors low expression of miR-200 family members significantly associated with worse overall survival and also inhibited angiogenesis (Pecot et al., 2013).
1. MiRNA Biogenesis and Mechanism of Action
miRNAs are short (19 to 24 nucleotides) non-coding RNAs that are processed from longer primary transcripts by successive endonuclease enzymatic maturation steps (by Drosha in the nucleus and Dicer in the cytoplasm) (Fig. 1). Functionally, miRNAs regulate gene expression in a sequence specific manner. Following incorporation into the ribonucleoprotein (RNP) complex RISC (RNA induced silencing complex (comprising of proteins like Dicer and members of the Argonaute (AGO) family), miRNAs bind messenger RNAs (mRNAs) primarily at their 3′UTRs, via partial complementarity with their “seed” sequence (the first 2 to 8 nts at the miRNA's 5′ end, which defines miRNA families and is important for proper target recognition). Consequently, mRNA translation and/or stability are impaired (Filipowicz et al., 2008, Valencia-Sanchez et al., 2006) with an ultimate reduction in protein expression levels (Bartel, 2004, Kim, 2005).
Fig. 1.
miRNA mechanism and modulation. Canonical biogenesis and processing of miRNAs and mechanism of RNAi-regulated gene silencing is presented. Additionally, the several mechanisms of delivery of miRNA and therapeutic agents are also presented.
In addition to conventional 3′-UTR mechanism of action, we now know that miRNAs can function in multiple ways. For example, miR-363 and let-7 can activate mRNA expression of proteins they normally repress during cell proliferation via recruitment of specific micro-RNPs (like AGO2 and FXR1) to AU-rich elements inside mRNA 3′UTRs (Vasudevan et al., 2007). It has also been shown that miRNAs are able to target to 5′UTR and 3′UTR sequences alike. miR-10a can bind to the 5′UTR of ribosomal proteins following starvation and enhance their translation (Vasudevan et al., 2007, Orom et al., 2008). In addition, miRNA dependent mRNA repression can also occur via binding sites located inside mRNA coding sequences, as shown for miRNAs regulating embryonic stem cell differentiation (Tay et al., 2008). Some studies have suggested non-cytoplasmic functions of miRNAs in different subcellular compartments. miR-29b, for example, carries a distinct hexanucleotide terminal motif that allows its nuclear translocation and subsequent enrichment in the nucleus (Hwang et al., 2007). miRNAs in the nucleus have been shown to act at the promoter level affecting transcription. For example, miR-551b-3p directly upregulates STAT3 expression by binding to a complementary sequence on the STAT3 promoter, and recruiting RNA polymerase II and the TWIST1 transcription factor to activate STAT3 transcription (Chaluvally-Raghavan et al., 2016). miRNAs have also been detected in membrane-bound compartments, such as secreted vesicles (Zhang et al., 2010) and mitochondria (Das et al., 2012). Interestingly, muscle-specific miR-1 is able to stimulate mitochondrial translation of multiple mitochondrial DNA-encoded transcripts, while repressing its nuclear DNA-encoded targets in the cytoplasm (Zhang et al., 2014). Few miRNAs act as decoys, by binding directly to RNA-binding proteins, and inhibiting the interaction with their target RNA (Eiring et al., 2010). Moreover, miRNAs can also regulate gene expression at the transcriptional level (Kim et al., 2008), by binding directly to the DNA regulatory elements. Thus miRNA-mediated regulation of gene expression is a complex science and is still an evolving concept.
2. Mechanisms of miRNA Deregulation in Cancer
The widespread differential expression of miRNA genes between malignant and normal cells is a complex phenomenon, which requires simultaneous combination of several factors, including miRNA expression control by oncogenes, tumor suppressor genes, epigenetic mechanisms and preferential genomic location of miRNAs within cancer-associated regions (Lujambio et al., 2007, Calin et al., 2004). As a paradigm of this sophistication, the tumor suppressor miR-34a is positively controlled by TP53 (Chang et al., 2007), repressed by MYC (Chang et al., 2008), silenced by aberrant CpG methylation (Lodygin et al., 2008) and is located at 1p36, a chromosomal region frequently lost in neuroblastomas (Wei et al., 2008). Accordingly, numerous genetic studies allowed the identification of miRNA abnormalities in human cancer by dissecting their transcriptional regulators (Chang et al., 2008, Calin and Croce, 2006b). Cancer associated miRNAs have been located downstream of major oncogenic and tumor suppressive transcription factors: for example, TP53 promotes the transcription of all the members of the miR-34 family, while MYC can both positively and negatively regulate transcription of different miRNAs (e.g. miR-17–92 cluster and let-7 family, respectively). Additionally, a miRNA hypermethylation profile characteristic of human metastasis was identified (Lujambio et al., 2007). It was identified that somatic mutations in DICER1 and DROSHA impaired biogenesis of tumor suppressive miRNA, including let-7 family, in Wilms tumor (Rakheja et al., 2014). Methylation of miR-9 family genes (miR-9-1, miR-9-2 and miR-9-3) has been identified in several metastatic cancer cell lines (Lujambio et al., 2008) miR-9 family genes are simultaneously methylated in gastric cancer (Tsai et al., 2011). Methylation of miR-9-1 is associated with lymph node metastasis in CRC (Bandres et al., 2009), and methylation of miR-9-1 and miR-9-3 is correlated with metastatic recurrence of renal cell carcinoma (Hildebrandt et al., 2010). These are only initial steps toward the understanding of the causes of miRNA deregulation during metastases, and newer mechanisms will continue to be identified as the field evolves.
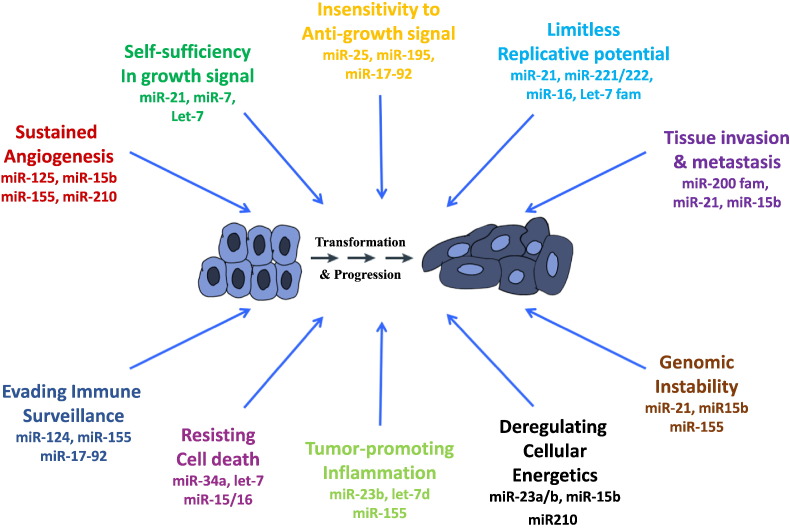
Deregulation of miRNA expression in cancer subsequently leads to altered functionality of these miRNAs. Upregulated miRNAs often act as oncogenes, as exemplified by miR-21 or miR-155 overexpression which causes acute B cell leukemia in transgenic mice models (Medina et al., 2010, Costinean et al., 2006). Consequently, downregulated miRNAs act as tumor suppressor, such as miR-15a/16–1 cluster, for which the knock-out mice develop chronic lymphocytic leukemia (Klein et al., 2010). In certain cases, the same miRNA that acts like an oncogene in one type of cell and as a suppressor in another. This is primarily due to different targets and mechanisms of action (for example miR-222 is overexpressed in liver cancers and targets PTEN suppressor, while it is downregulated in erythroblastic leukemia where it targets c-KIT oncogene). Below, we provide a few examples of miRNAs that affect the emerging hallmarks of cancer as described by Hanahan and Weinberg (Hanahan and Weinberg, 2000, Hanahan and Weinberg, 2011): sustaining proliferative signaling (miR-21, let-7 family) (Dalmay and Edwards, 2006); evading growth receptors (miR-17-92 cluster) (Dalmay and Edwards, 2006); resisting cell death (miR-15/16, miR-34 cluster) (Dalmay and Edwards, 2006); enabling replicative immortality (miR-34a, miR-372/373 cluster) (Dalmay and Edwards, 2006); inducing angiogenesis (miR-210) (Dalmay and Edwards, 2006); Activating invasion and metastasis (miR-10b) (Nicoloso et al., 2009); avoiding immune destruction (miR-520d) (Stern-Ginossar et al., 2008); deregulating cellular genetics (miR-122, miR-210) (Esau et al., 2006, Chan et al., 2009); tumor-promoting inflammation (miR-146, miR-155) (Schetter et al., 2010); and genome instability and mutation (miR-155) (Valeri et al., 2010). The role of miRNAs in other hallmarks of cancer have been discussed in detail in (Negrini et al., 2009) (Fig. 2).
Fig. 2.
microRNAs and cancer hallmarks. Specific examples of miRNAs involved in the hallmarks of cancer are presented.
3. miRNAs and Tumor Microenvironment
While miRNAs have been well established to play an important role in intracellular processes, more recent evidence also support an extracellular role of miRNAs that are produced by microenvironment cells. As hormones, miRNAs are released by a donor cell as ‘free’ molecules or in various forms of vesicles secreted by active mechanisms (Shah and Calin, 2013). These miRNAs are then taken up by cells located in other parts of the body, regulating the protein expression profile in these recipient cells. It was shown that miR-181c released from cancer-derived extracellular vesicles triggered brain metastasis by inducing the breakdown of blood brain barrier via the downregulation of its target gene PDPK1 (Tominaga et al., 2015).
A role of circulating miRNAs in transforming fibroblasts, major constituents of the extracellular matrix and involved in several cellular mechanisms including wound repair, into cancer-associated fibroblasts (CAFs) has been reported (Carstens et al., 2014). Low expression of miR-31 and miR-214 and high expression of miR-155 have been found to be involved in reprograming quiescent fibroblasts to CAFs in ovarian cancers. miR-214 was found to directly target the CCL5 (C-C motif ligand 5) chemokine important for CAF function (Mitra et al., 2012). Furthermore, miR-31 was reported to be the most downregulated miRNA in CAFs isolated from endometrial cancer when compared to normal endometrial fibroblasts. miR-31 directly targets the homeobox gene SATB2, which is significantly elevated in CAFs and is responsible for chromatin remodeling and regulation of gene expression (Aprelikova et al., 2010).
It was shown that endogenous miRNAs, miR-155 and miR-146a are transferred between primary bone-marrow derived dendritic cells. Administration of miR-155 or miR-146a-containing exosomes modulates the endogenous immune response to endotoxins in vivo (Alexander et al., 2015). Essentially, these studies highlight that secreted miRNAs represent a novel regulatory mechanism by which donor cells can influence the gene expression of recipient cells, and thus impact physiological and pathological processes.
4. miRNAs and Immune Response
Circulating miRNAs have been reported to modulate immune responses. For example, it was shown that antigen-dependent transfer of miR-335 from T-cells to antigen-presenting cells is important during immune synapse formation (Mittelbrunn et al., 2011). Recent studies have demonstrated that miRNAs in placenta-derived exosomes from trophoblasts function as immune regulators in fetal–maternal crosstalk (Luo et al., 2009). miR-517a is secreted into maternal circulation where it improves maternal acclimatization to pregnancy and promotes fetal allograft survival.
Furthermore, miRNAs have been shown to be important in the development, differentiation and modulation of the immune cell repertoire, as well as of the innate and adaptive immune responses (Chou et al., 2013, Tili et al., 2013). miRNAs have been identified to regulate T cell response, maturation, differentiation and function, such as activation, proliferation and apoptosis (Liu et al., 2013). These include, for example, the oncogenic members of the miR-17–92 cluster involved in apoptosis of CD4 + T cells (Molitoris et al., 2011), while miR-222 and miR-339 promote resistance of cancer cells to cytotoxic T lymphocytes (CTL) by downregulation of ICAM-1 (Ueda et al., 2009). In addition, it was reported that p53 regulated PDL1 expression via miR-34 in non-small cell lung cancer (NSCLC). Administration of miR-34a mimics, alone or in combination with radiotherapy, reduced PDL1 expression in the tumor and antagonized T-cell exhaustion (Cortez et al., 2016).
5. Various Strategies to use miRNAs Therapeutics
Despite considerable advances in our understanding of the molecular carcinogenesis of human cancers and the extensive research on combined and targeted therapies, there is still a constant need for the development of novel therapeutic tools. Additionally, treatment with any individual therapy confines them to the ‘one-drug-one-target’ paradigm and renders them susceptible to resistance in due course. RNA molecules are now at the center of molecular oncology, with applications for diagnosis and therapy starting to be proposed. The ability of miRNAs to regulate important cellular processes by concurrently regulating multiple targets illustrates their potential as a viable therapeutic tool. There are currently two strategies described for the treatment of cancer using RNAi-based therapy.
5.1. Sandwich RNAi Inhibition Strategy
One strategy involves using multiple agents to target one specific molecular defect linked with cancer pathogenesis – a ‘sandwich RNAi inhibition’ strategy (Calin and Croce, 2009). We exemplified this strategy by targeting an important ovarian cancer oncogene, EphA2, using a combination of EphA2-targeting siRNAs and miR-520d-3p (an EphA2-targeting miRNA) mimics (Nishimura et al., 2013). Dual targeting of EphA2 exhibited synergistic anti-tumor efficiency than either monotherapy alone, both in vitro and in vivo. Combined miRNA-siRNA therapy prominently decreased EphA2 protein levels, suppressed tumor growth, and inhibited migration and invasion. Thus regimens using a cocktail of RNAi-based therapeutics to target dominant oncogenes might achieve better therapeutic outcomes in human cancers.
5.2. Multiplex RNAi Inhibition Strategy
In this strategy, multiple molecular defects accumulated in the multistep pathway of a specific cancer can be targeted – a ‘multiplex RNAi inhibition’ strategy (Calin and Croce, 2009). For example, it was demonstrated the applicability of ‘sensor’ siRNAs, a universal platform for the combination RNAi therapeutics, in targeting the complete RAF node (KRAS + PIK3CA/B) to treat KRAS-mutant colorectal cancer (Yuan et al., 2014). Using in vitro and in vivo modes they showed that siRNA-mediated inhibition of KRAS as well as RAF or PI3K combinations could impair KRAS-mutant colorectal cancer in xenograft models. These studies highlight the efficiency and applicability of RNAi-based therapeutic strategies in management of cancers.
Current approaches for miRNA therapy involve either a) the restoration of tumor suppressive genes by inhibition of oncogenic miRNA using ‘anti-miRNAs’; or b) inhibition of oncogenic genes by treatment with ‘miRNA-mimics’ (Fig. 1, Table 1).
Table 1.
Types of RNA therapeutic drugs.
| Agent | Definition | Mechanism of action | Preclinical or clinical applications | |
|---|---|---|---|---|
| miRNA inhibition | AMOs | Antisense oligonucleotides targeting miRNAs | The miRNA/AMO – duplexes induce degradation of the miRNA and recycling of the antagomir (Krutzfeldt et al., 2005). | Preclinical studies |
| LNA anti-miRs | The LNAs anti miRNAs represent LNA modified ASOs. LNAs are bicyclic RNA analogues where the ribose is locked in a C3’-endo conformation by the introduction of a 2’-O,4′-C methylene bridge (Elmen et al., 2008) | The miRNA/LNA – duplexes induce degradation of the miRNA and recycling of the antagomir. | Phase I and 2a (for HCV) (Janssen et al., 2013, Lieberman and Sarnow, 2013) | |
| Antagomirs | Single-stranded 23 nt RNA molecules complementary to the targeted miRNA that have been modified to increase the stability of the RNA and protect it from degradation. The modifications included a partial phosphorothioate backbone in addition to 2′-O-methoxyethyl (Krutzfeldt et al., 2005). | The miRNA/antagomir – duplexes induce degradation of the miRNA and recycling of the antagomir (Krutzfeldt et al., 2005). | Preclinical studies | |
| miRNA sponges | RNAs containing multiple tandem binding sites to a miRNA of interest and are transcribed from expression vectors (Ebert et al., 2007). | miRNA sponges compete with the native targets of miRNAs, reducing miRNA's effects, and thus result in increased expression of the miRNA's native targets (Ebert et al., 2007). | Preclinical studies | |
| SMIRs | Small molecule chemical compounds | Block activities of specific miRNAs by structure-based docking onto the precursor or mature from of miRNA structure. | Preclinical studies | |
| miRNA Restoration | Small molecules | Hypomethylating agents (Decitabine or 5-azacytidine) and enoxacin | Non-specific induction of miRNA expression | Preclinical studies |
| miRNA mimics | Double stranded synthetic RNAs that mimic endogenous miRNAs | Restore the expression and function of a specific miRNA | Phase I | |
| miRNA expression vectors | Vectors expressing a specific type of miRNA | Restore the expression and function of a specific miRNA | Preclinical studies |
Note: nt – nucleotide; AMO – anti-microRNA antisense oligodeoxyribonucleotide; LNA – locked nucleic acids; SMIRs: small-molecule inhibitors of miRNAs (SMIRs).
5.3. miRNA Inhibition Therapy
Oncogenic miRNAs that are frequently overexpressed in human cancers and need to be inhibited to help restore the normal expression and function of its target tumor suppressive genes. miRNA inhibitors are essentially complementary single stranded oligonucleotides that sequester the endogenous miRNA in an unrecognized conformation. As a result, the mature miRNA cannot be processed by the RISC, and is thus excluded from the RISC. These include antisense anti-miR oligonucleotides (AMOs), locked nucleic acid (LNA) anti-miRNAs, antagomirs, miRNA sponges, miRNA masks and small molecule inhibitors of miRNAs.
Anti-miRNA oligonucleotides (AMOs) are single-stranded, chemically modified anti-sense oligonucleotides (ASOs), that are 17 to 22 nt in length and designed to be complementary to a selected miRNA (Garzon et al., 2010). They work as competitive inhibitors of miRNAs by annealing to the mature miRNA and inhibiting the interaction of that miRNA with its target mRNAs. Thus targeted inhibition of a specific miRNA and subsequent upregulation of its target mRNAs can be achieved. Mechanistically, they produce ASO-miRNA duplex through Watson-Crick binding, leading to RNAse-H-mediated cleavage of the target miRNA gene. Important for the potential clinical use, AMOs harboring a complete 2′-O-methoxyethyl and phosphorothioate modification have been demonstrated to silence in vivo miR-122 in mouse liver (Hutvagner et al., 2004). In contrast, unmodified AMOs are unable to inhibit miRNA function in vitro.
The antagomirs are chemically modified and cholesterol-conjugated single-stranded 23-nt RNA molecules complementary to the targeted miRNAs. The modifications were introduced to increase the stability of the RNA and protect it from degradation. When intravenously administered to mice, antagomir-122 induced a marked, specific, and persistent (up to 23 days) reduction of endogenous miR-122 gene expression in liver, lung, kidney, heart, intestine, fat, skin, bone marrow, muscle, ovaries and adrenals (Krutzfeldt et al., 2005). The same was true for antagomir-16, targeting the ubiquitously expressed miR-16 (Krutzfeldt et al., 2005). Silencing of miRNAs by these new agents also produced other positive physiologic effects, for example the decrease in plasma cholesterol levels after antagomir-122 administration. One clear advantage with respect to siRNA technology is that antagomirs did not induce an immune response.
Another example of modified AMOs are the Locked Nucleic Acid (LNA) anti-miRs, in which an extra methylene bridge connecting the 2′-O atom and the 4′-C atom ‘locks’ the ribose ring in a C3′-endo or C2′-endo conformation (Vester and Wengel, 2004, Elmen et al., 2008). LNA-modified oligonucleotides exhibit higher thermal stability and high-affinity Watson-Crick hybridization with their RNA target molecules, with improved mismatch discrimination. Furthermore, they display higher aqueous solubility and increased metabolic stability for in vivo delivery. miR-21, shown to be strongly overexpressed in glioblastomas, was silenced in vitro by using LNA-modified antisense oligonucleotides leading to a significant reduction in cell viability and elevated intracellular levels of caspases (Griveau et al., 2013). In a recent study (Gallo et al., 2016), the authors evaluated the pharmacokinetic and pharmacodynamic properties of LNA-anti-miR-221 in NOD.SCID mice and Cynomolgus monkeys. They reported that LNA-anti-miR-221 have a short half-life, optimal tissue bioavailability minimal urine excretion in both mice and monkeys and was detectable in mice vital organs and in xenografted tumors for up to 3 weeks after treatment. These studies highlight the suitability of LNA-anti-miRNAs for clinical use.
‘miRNA sponges’ or ‘miRNA decoys’ contain multiple artificial miRNA binding sites that compete with the endogenous miRNA targets for miRNA binding (Ebert et al., 2007). Inhibition of miR-9, which is upregulated in breast cancer cells and directly targets CDH1, using a ‘miRNA sponge’ inhibited metastasis formation (Ma et al., 2010). ‘miRNA masks’ are novel gene-specific anti-miRNAs that can selectively inhibit the interaction of the target miRNA with a specific mRNA. These effectively mask the specific mRNA from the endogenous miRNA and thus prevent its repression (Xiao et al., 2007).
5.4. miRNA Mimetic Agents
‘miRNA mimics’ are an effective alternative to restore the normal function of tumor suppressive miRNAs by replacing or substituting the lost miRNA using synthetic miRNA-like molecules. These are small, chemically modified (2′-O′methoxy) RNA duplexes that can be loaded into RISC and achieve the downstream inhibition of the target mRNAs. Numerous studies have validated the efficiency of miRNA replacement therapy in in vitro and in vivo models. For example, introduction of miRNA mimics for miR-15a in prostate cancer cell lines induced marked apoptosis and blocked proliferation (Bonci et al., 2008). Intranasal administration of let-7 in a K-ras mutant mouse effectively restrained the growth of the tumors by repression of proliferation and cell cycle pathways (Esquela-Kerscher et al., 2008, Trang et al., 2010). More recently, a new RNA polymerase II driven expression vector for miR-155 has been shown to effectively increase miR-155 expression levels in vitro and in in vivo xenograft models (Chung et al., 2006). An aptamer-miRNA conjugate of tumor suppressor let-7 g miRNA and the GL21. T aptamer, was engineered and demonstrated target specific delivery of the conjugate (Esposito et al., 2014). let-7g:GL21. T conjugate can successfully inhibit cell survival and migration in vitro and in vivo in lung cancer model.
Thus administration of miRNA-mimetic agents in patients might be a new avenue for clinical cancer management. Although all of these strategies have been truly exciting, there are still challenges involved in the delivery of these agents. Several new delivery agents are currently being explored to afford safe, effective and efficient delivery of miRNAs. One example is the chitosan based delivery system that was reported to be quite versatile for delivery into multiple tumor and stromal compartments (Han et al., 2010), as well as for miRNAs (Gaur et al., 2015). However, we are still in the need of finding new alternative therapeutic approaches to inhibit oncomiRs, and decrease their activity.
5.5. SMIRs — Small Molecules Inhibitors of miRNAs
Small molecule inhibitors of miRNAs (SMIRs) (Monroig et al., 2015) are small molecules that primarily function by inhibiting miRNA biogenesis or by actively impeding miRNA-target interaction. The SMIR-approach is an appealing one, specifically because it is a way of taking the “fast-track lane” in the drug-developing race, reducing time of production/approval and therefore the cost of it. Gumireddy et al. (Gumireddy et al., 2008) reported a cellular screen for miRNA-pathway inhibitors and found the first small-molecule inhibitor of miRNA function. The mode of action of the small molecules is mainly through the transcriptional regulation of miR-21 rather than inhibition of target recognition by miR-21. It was reported that the small molecule enoxacin, an antibacterial fluoroquinolone, binds to the miRNA biosynthesis protein TAR RNA-binding protein 2 (TRBP) and enhances the production of tumor suppressor miRNAs (Melo et al., 2011). Conversely two compounds, polylysine (PLL) and trypaflavine (TPF) were identified that suppressed miRNA-RISC activity and exhibited anti-tumor activity in vitro (Watashi et al., 2010). Thus, small molecule modulators of miRNAs represent a unique strategy for restoring dysregulated miRNAs in cancer.
5.6. Targeting miRNAs From Microvesicles and Exosomes
As miRNAs are often transferred between various types of cells within a tumor or between the tumor and the metastatic sites, one way to perturb this mode of transport is to block extracellular miRNAs in exosomes. It has been shown that the small molecule GW4869, an inhibitor of neutral sphingomyelinase that is also known to inhibit miRNA and exosome secretion, can be effectively used to interrupt miRNA-mediated aberrant cross-talk between cancer cells and surrounding immune cells within the tumor microenvironment (Fabbri et al., 2012, Kosaka et al., 2010). MiR-21 and miR-29a can be released by cancer cells within exosomes and are engulfed by macrophages in the tumor microenvironment expressing TLRs. It has also been shown in mice that extracellular let-7 can activate TLR7 and induce neurodegeneration through neuronal TLR7 (Lehmann et al., 2012). Intriguingly, let-7b levels are higher in the cerebrospinal fluid (CSF) of patients with Alzheimer's disease, indicating that miRNA-mediated activation of TLRs may have implications beyond cancer.
The use of molecules that block the functions of specific miRNAs (such as LNA anti-miR-21 and LNA anti-miR-29a) in tumor cells could reduce miRNA levels in exosomes released by cancer cells and effectively decrease miRNA-mediated TLR activation (Fabbri et al., 2012). Likewise, it can be postulated that miR-21 or miR-29a could be mutated in such a way that they retain the ability to bind to TLRs but fail to activate them, thereby offsetting the cross talk between cancer-released miRNAs and TLRs. Moreover, genetically engineered TLR decoy molecules could be designed to bind and sequester miRNAs released by cancer cells in the tumor microenvironment, without triggering TLR activated signaling transduction pathways.
An additional strategy targeting miRNA transport involves the use of antibodies that recognize tumor specific antigens expressed by cancer-released exosomes. The advantage of this approach could be that some of the antigens most likely have reduced antigenic properties and permit the production of cancer-released exosomes without any obvious stimulation of the immune system. Finally, we can envision a futuristic therapeutic strategy where cells are stimulated to secrete oncogenic miRNA-loaded nanovesicles and the cancer patient is subsequently treated with dialysis, as a way to “wash-out” oncogenes from cancer cells.
6. First Clinical Trials With miRNAs
MiRNAs represent promising therapeutic agents and several pharmaceutical companies already have miRNA therapeutics in their developmental pipelines. Current strategies for inhibitory-miRNA therapies are based on antisense antimiRs, (LNA), LNA-antimiR constructs, antagomirs, and miRNA sponges. Some of these have proven to be effective not only in vitro, but also in vivo. For instance, Regulus Therapeutics is actively exploring the value of anti-miRs in the treatment of diseases such as fibrosis, hepatitis C virus (HCV) infection, atherosclerosis and cancer. MIRagen Therapeutics is using chemically modified structures of miRNA (including miR-15/195, miR-29, and others) in work that has reached preclinical development in pathologies such as metabolic and cardiovascular diseases. MRX34, a liposome-formulated mimic of the tumor suppressor, miR-34, developed by Mirna Therapeutics, produced complete tumor regression in orthotopic mouse models of liver cancer, with no observed immunostimulatory activity or toxicity to normal tissues. In a Phase I clinical trial with patients with advanced solid tumors (N = 99), a standard dose escalation trial of MRX34 infused IV on a biweekly or daily schedule were given. Phase 1 results, as reported at ASCO 2016 meeting, showed that MRX34 has a manageable toxicity profile and strong evidence of activity in hepatocellular carcinoma, renal cell carcinoma and melanoma. Analysis of RNA from WBCs showed dose-dependent repression of miR-34a target oncogenes, including FOXP1, BCL2, HDAC1, and CTNNB1 in these patients. Unfortunately, recently Mirna Therapeutics halted the Phase I clinical trial due to multiple immune-related severe adverse events (SAE) (http://www.businesswire.com). Therefore, strategies as the ones presented above should be used to reduce the dosages of miR mimetics/anti-miRs and consequently their potential adverse reactions.
Miravirsen (SPC3649) is an LNA against miR-122 developed by Santaris Pharma A/S for the treatment of hepatitis C (HCV), a viral infection known to predispose patients to hepatocellular carcinoma. A phase I clinical trial demonstrated that antagomiR-122 has dose-dependent pharmacology and is well tolerated. When investigated in a phase II clinical trial, Miravirsen was found to be well tolerated in patients with HCV, with mild side effects including light coryza, diarrhea, and headache. Importantly, the administration of Miravirsen in patients with chronic HCV-1 displayed extended dose-dependent diminutions in HCV RNA levels without any manifestation of viral resistance (Janssen et al., 2013). However, it was found that long term treatment with Miravirsen in vitro induces resistance due to mutations in the viral genome (Ottosen et al., 2015, Li et al., 2016).
miR-16 mimics are also currently under Phase I clinical trials for patients with Malignant Pleural Mesothelioma (MPM) and Advanced Non-Small Cell Lung Cancer (NSCLC) that have failed standard therapy. These miR-16 mimics were delivered intravenously, using EnGeneIC Delivery Vehicle (EDV)-Packaging, and were conjugated with an EGFR-targeting antibody. Preliminary data presented by Van Zandwijk et al. (Van Zandwijk et al., 2015) show manageable safety profile in 5 patients. In addition, MIRagen Therapeutics recently announced phase I clinical trials for two candidate miRNA-based candidates: MRG-201, a synthetic microRNA mimic to microRNA-29b, will be tested for patients with scleroderma, and MRG-106, a synthetic microRNA antagonist of microRNA-155, will be tested for patients with cutaneous T-cell lymphoma of the mycosis fungoides (MF) sub-type. The estimated primary completion date for both the studies is late 2016.
7. Conclusion
microRNAs represent critical regulators of tumor initiation, progression, and dissemination. Extensive evidence suggests that inhibition of overexpressed oncogenic miRNAs or substitution of tumor suppressive miRNAs could become a robust strategy for cancer therapy. The optimization of miRNA delivery systems, improvements in the stability of miRNAs, and a detailed understanding of the off-target effects of miRNA therapeutics are several challenges that need to be resolved for successful translation of miRNA therapeutics from bench to bedside.
8. Outstanding Questions
miRNA-based therapeutics hold great promise as highly specific, targeted therapies for cancer treatment. However, to achieve superior sensitivity and specificity, and accelerate their adoption in the clinic, there still exists the need to improve their chemical designs, develop better delivery options, show prolonged therapeutic efficiency, and evaluate the long-term safety of these agents in vivo. Furthermore, it is imperative to understand the underlying intricate network of interactions between miRNAs and the human genome, transcriptome and proteome before their transition into clinical use. In addition, a full assessment of their toxicities need to be performed, and low-toxicity strategies such as combining miRNAs and siRNAs at low doses, or using miRNA therapy as an additive to established chemotherapy regimens, should be evaluated. Overall, miRNA-based therapy can potentially bring an exciting new facet to personalized medicine for cancer treatment; however, a deeper and clearer understanding of its biology is required.
9. Search Strategy and Selection Criteria
Data for this Review were identified by searches of PubMed and references from relevant articles using the search terms “microRNA”, “cancer”, and “therapeutics”. Abstracts and reports from meetings were included only when they related directly to previously published work. Only articles published in English between 1993 and 2016 were included.
Author Contribution
All authors contributed equally to the design and writing of this manuscript.
Acknowledgements
Dr. Calin is The Alan M. Gewirtz Leukemia & Lymphoma Society Scholar. Work in Dr. Calin's laboratory is supported in part by the NIH/NCI grants 1UH2TR00943-01 and 1 R01 CA182905-01, the UT MD Anderson Cancer Center SPORE in Melanoma grant from NCI (P50 CA093459), Aim at Melanoma Foundation and the Miriam and Jim Mulva research funds, the UT MD Anderson Cancer Center Brain SPORE (2P50CA127001), a Developmental Research award from Leukemia SPORE, a CLL Moonshot Flagship project, a 2015 Knowledge GAP MDACC grant, an Owens Foundation grant, and the RGK Foundation. The funding agencies had no role in the design, data collection, data analysis, interpretation, writing of this paper.
References
- Alexander M., Hu R., Runtsch M.C., Kagele D.A., Mosbruger T.L., Tolmachova T., Seabra M.C., Round J.L., Ward D.M., O'connell R.M. Exosome-delivered microRNAs modulate the inflammatory response to endotoxin. Nat. Commun. 2015;6:7321. doi: 10.1038/ncomms8321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aprelikova O., Yu X., Palla J., Wei B.R., John S., Yi M., Stephens R., Simpson R.M., Risinger J.I., Jazaeri A., Niederhuber J. The role of miR-31 and its target gene SATB2 in cancer-associated fibroblasts. Cell Cycle. 2010;9:4387–4398. doi: 10.4161/cc.9.21.13674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandres E., Agirre X., Bitarte N., Ramirez N., Zarate R., Roman-Gomez J., Prosper F., Garcia-Foncillas J. Epigenetic regulation of microRNA expression in colorectal cancer. Int. J. Cancer. 2009;125:2737–2743. doi: 10.1002/ijc.24638. [DOI] [PubMed] [Google Scholar]
- Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Berindan-Neagoe I., Monroig Pdel C., Pasculli B., Calin G.A. MicroRNAome genome: a treasure for cancer diagnosis and therapy. CA Cancer J. Clin. 2014;64:311–336. doi: 10.3322/caac.21244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonci D., Coppola V., Musumeci M., Addario A., Giuffrida R., Memeo L., D'urso L., Pagliuca A., Biffoni M., Labbaye C., Bartucci M., Muto G., Peschle C., de Maria R. The miR-15a-miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat. Med. 2008;14:1271–1277. doi: 10.1038/nm.1880. [DOI] [PubMed] [Google Scholar]
- Calin G.A., Croce C.M. Genomics of chronic lymphocytic leukemia microRNAs as new players with clinical significance. Semin. Oncol. 2006;33:167–173. doi: 10.1053/j.seminoncol.2006.01.010. [DOI] [PubMed] [Google Scholar]
- Calin G.A., Croce C.M. MicroRNA signatures in human cancers. Nat. Rev. Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- Calin G.A., Croce C.M. Chronic lymphocytic leukemia: interplay between noncoding RNAs and protein-coding genes. Blood. 2009;114:4761–4770. doi: 10.1182/blood-2009-07-192740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin G.A., Sevignani C., Dumitru C.D., Hyslop T., Noch E., Yendamuri S., Shimizu M., Rattan S., Bullrich F., Negrini M., Croce C.M. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl. Acad. Sci. U. S. A. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carstens J.L., Lovisa S., Kalluri R. Microenvironment-dependent cues trigger miRNA-regulated feedback loop to facilitate the EMT/MET switch. J. Clin. Invest. 2014;124:1458–1460. doi: 10.1172/JCI75239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaluvally-Raghavan P., Jeong K.J., Pradeep S., Silva A.M., Yu S., Liu W., Moss T., Rodriguez-Aguayo C., Zhang D., Ram P., Liu J., Lu Y., Lopez-Berestein G., Calin G.A., Sood A.K., Mills G.B. Direct upregulation of STAT3 by MicroRNA-551b-3p deregulates growth and metastasis of ovarian cancer. Cell Rep. 2016;15:1493–1504. doi: 10.1016/j.celrep.2016.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan S.Y., Zhang Y.Y., Hemann C., Mahoney C.E., Zweier J.L., Loscalzo J. MicroRNA-210 controls mitochondrial metabolism during hypoxia by repressing the iron-sulfur cluster assembly proteins ISCU1/2. Cell Metab. 2009;10:273–284. doi: 10.1016/j.cmet.2009.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang T.C., Wentzel E.A., Kent O.A., Ramachandran K., Mullendore M., Lee K.H., Feldmann G., Yamakuchi M., Ferlito M., Lowenstein C.J., Arking D.E., Beer M.A., Maitra A., Mendell J.T. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol. Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang T.C., Yu D., Lee Y.S., Wentzel E.A., Arking D.E., West K.M., Dang C.V., Thomas-Tikhonenko A., Mendell J.T. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat. Genet. 2008;40:43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou J., Shahi P., Werb Z. microRNA-mediated regulation of the tumor microenvironment. Cell Cycle. 2013;12:3262–3271. doi: 10.4161/cc.26087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung K.H., Hart C.C., Al-Bassam S., Avery A., Taylor J., Patel P.D., Vojtek A.B., Turner D.L. Polycistronic RNA polymerase II expression vectors for RNA interference based on BIC/miR-155. Nucleic Acids Res. 2006;34 doi: 10.1093/nar/gkl143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez M.A., Ivan C., Valdecanas D., Wang X., Peltier H.J., Ye Y., Araujo L., Carbone D.P., Shilo K., Giri D.K., Kelnar K., Martin D., Komaki R., Gomez D.R., Krishnan S., Calin G.A., Bader A.G., Welsh J.W. PDL1 regulation by p53 via miR-34. J. Natl. Cancer Inst. 2016;108 doi: 10.1093/jnci/djv303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costinean S., Zanesi N., Pekarsky Y., Tili E., Volinia S., Heerema N., Croce C.M. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice; Proceedings of the National Academy of Sciences of the United States of America; 2006. pp. 7024–7029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalmay T., Edwards D.R. MicroRNAs and the hallmarks of cancer. Oncogene. 2006;25:6170–6175. doi: 10.1038/sj.onc.1209911. [DOI] [PubMed] [Google Scholar]
- Das S., Ferlito M., Kent O.A., Fox-Talbot K., Wang R., Liu D., Raghavachari N., Yang Y., Wheelan S.J., Murphy E., Steenbergen C. Nuclear miRNA regulates the mitochondrial genome in the heart. Circ. Res. 2012;110:1596–1603. doi: 10.1161/CIRCRESAHA.112.267732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert M.S., Neilson J.R., Sharp P.A. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat. Methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiring A.M., Harb J.G., Neviani P., Garton C., Oaks J.J., Spizzo R., Liu S., Schwind S., Santhanam R., Hickey C.J., Becker H., Chandler J.C., Andino R., Cortes J., Hokland P., Huettner C.S., Bhatia R., Roy D.C., Liebhaber S.A., Caligiuri M.A., Marcucci G., Garzon R., Croce C.M., Calin G.A., Perrotti D. miR-328 functions as an RNA decoy to modulate hnRNP E2 regulation of mRNA translation in leukemic blasts. Cell. 2010;140:652–665. doi: 10.1016/j.cell.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmen J., Lindow M., Schutz S., Lawrence M., Petri A., Obad S., Lindholm M., Hedtjarn M., Hansen H.F., Berger U., Gullans S., Kearney P., Sarnow P., Straarup E.M., Kauppinen S. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- Esau C., Davis S., Murray S.F., Yu X.X., Pandey S.K., Pear M., Watts L., Booten S.L., Graham M., Mckay R., Subramaniam A., Propp S., Lollo B.A., Freier S., Bennett C.F., Bhanot S., Monia B.P. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Esposito C.L., Cerchia L., Catuogno S., De Vita G., Dassie J.P., Santamaria G., Swiderski P., Condorelli G., Giangrande P.H., De Franciscis V. Multifunctional aptamer-miRNA conjugates for targeted cancer therapy. Mol. Ther. 2014;22:1151–1163. doi: 10.1038/mt.2014.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esquela-Kerscher A., Trang P., Wiggins J.F., Patrawala L., Cheng A., Ford L., Weidhaas J.B., Brown D., Bader A.G., Slack F.J. The let-7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle. 2008;7:759–764. doi: 10.4161/cc.7.6.5834. [DOI] [PubMed] [Google Scholar]
- Fabbri M., Paone A., Calore F., Galli R., Gaudio E., Santhanam R., Lovat F., Fadda P., Mao C., Nuovo G.J., Zanesi N., Crawford M., Ozer G.H., Wernicke D., Alder H., Caligiuri M.A., Nana-Sinkam P., Perrotti D., Croce C.M. MicroRNAs bind to toll-like receptors to induce prometastatic inflammatory response. Proc. Natl. Acad. Sci. U. S. A. 2012;109:E2110–E2116. doi: 10.1073/pnas.1209414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W., Bhattacharyya S.N., Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Gallo Cantafio M.E., Nielsen B.S., Mignogna C., Arbitrio M., Botta C., Frandsen N.M., Rolfo C., Tagliaferri P., Tassone P., Di Martino M.T. Pharmacokinetics and pharmacodynamics of a 13-mer LNA-inhibitor-miR-221 in mice and non-human primates. Mol. Ther.–Nucleic Acids. 2016;5 doi: 10.1038/mtna.2016.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzon R., Marcucci G., Croce C.M. Targeting microRNAs in cancer: rationale, strategies and challenges. Nat. Rev. Drug Discov. 2010;9:775–789. doi: 10.1038/nrd3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaur S., Wen Y., Song J.H., Parikh N.U., Mangala L.S., Blessing A.M., Ivan C., Wu S.Y., Varkaris A., Shi Y., Lopez-Berestein G., Frigo D.E., Sood A.K., Gallick G.E. Chitosan nanoparticle-mediated delivery of miRNA-34a decreases prostate tumor growth in the bone and its expression induces non-canonical autophagy. Oncotarget. 2015;6:29161–29177. doi: 10.18632/oncotarget.4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griveau A., Bejaud J., Anthiya S., Avril S., Autret D., Garcion E. Silencing of miR-21 by locked nucleic acid-lipid nanocapsule complexes sensitize human glioblastoma cells to radiation-induced cell death. Int. J. Pharm. 2013;454:765–774. doi: 10.1016/j.ijpharm.2013.05.049. [DOI] [PubMed] [Google Scholar]
- Gumireddy K., Young D.D., Xiong X., Hogenesch J.B., Huang Q., Deiters A. Small-molecule inhibitors of microrna miR-21 function. Angew. Chem. 2008;47:7482–7484. doi: 10.1002/anie.200801555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H.D., Mangala L.S., Lee J.W., Shahzad M.M., Kim H.S., Shen D., Nam E.J., Mora E.M., Stone R.L., Lu C., Lee S.J., Roh J.W., Nick A.M., Lopez-Berestein G., Sood A.K. Targeted gene silencing using RGD-labeled chitosan nanoparticles. Clin. Cancer Res. 2010;16:3910–3922. doi: 10.1158/1078-0432.CCR-10-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D., Weinberg R.A. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Hildebrandt M.A., Gu J., Lin J., Ye Y., Tan W., Tamboli P., Wood C.G., Wu X. Hsa-miR-9 methylation status is associated with cancer development and metastatic recurrence in patients with clear cell renal cell carcinoma. Oncogene. 2010;29:5724–5728. doi: 10.1038/onc.2010.305. [DOI] [PubMed] [Google Scholar]
- Hutvagner G., Simard M.J., Mello C.C., Zamore P.D. Sequence-specific inhibition of small RNA function. PLoS Biol. 2004;2 doi: 10.1371/journal.pbio.0020098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang H.W., Wentzel E.A., Mendell J.T. A hexanucleotide element directs microRNA nuclear import. Science. 2007;315:97–100. doi: 10.1126/science.1136235. [DOI] [PubMed] [Google Scholar]
- Janssen H.L., Reesink H.W., Lawitz E.J., Zeuzem S., Rodriguez-Torres M., Patel K., Van Der Meer A.J., Patick A.K., Chen A., Zhou Y., Persson R., King B.D., Kauppinen S., Levin A.A., Hodges M.R. Treatment of HCV infection by targeting microRNA. N. Engl. J. Med. 2013;368:1685–1694. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- Ji M., Rao E., Ramachandrareddy H., Shen Y., Jiang C., Chen J., Hu Y., Rizzino A., Chan W.C., Fu K., Mckeithan T.W. The miR-17-92 microRNA cluster is regulated by multiple mechanisms in B-cell malignancies. Am. J. Pathol. 2011;179:1645–1656. doi: 10.1016/j.ajpath.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim V.N. MicroRNA biogenesis: coordinated cropping and dicing. Nat. Rev. Mol. Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- Kim D.H., Saetrom P., Snove O., Jr., Rossi J.J. MicroRNA-directed transcriptional gene silencing in mammalian cells. Proc. Natl. Acad. Sci. U. S. A. 2008;105:16230–16235. doi: 10.1073/pnas.0808830105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein U., Lia M., Crespo M., Siegel R., Shen Q., Mo T., Ambesi-Impiombato A., Califano A., Migliazza A., Bhagat G., Dalla-Favera R. The DLEU2/miR-15a/16-1 cluster controls B cell proliferation and its deletion leads to chronic lymphocytic leukemia. Cancer Cell. 2010;17:28–40. doi: 10.1016/j.ccr.2009.11.019. [DOI] [PubMed] [Google Scholar]
- Korpal M., Ell B.J., Buffa F.M., Ibrahim T., Blanco M.A., Celia-Terrassa T., Mercatali L., Khan Z., Goodarzi H., Hua Y., Wei Y., Hu G., Garcia B.A., Ragoussis J., Amadori D., Harris A.L., Kang Y. Direct targeting of Sec23a by miR-200s influences cancer cell secretome and promotes metastatic colonization. Nat. Med. 2011;17:1101–1108. doi: 10.1038/nm.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosaka N., Iguchi H., Yoshioka Y., Takeshita F., Matsuki Y., Ochiya T. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J. Biol. Chem. 2010;285:17442–17452. doi: 10.1074/jbc.M110.107821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krutzfeldt J., Rajewsky N., Braich R., Rajeev K.G., Tuschl T., Manoharan M., Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- Lee R.C., Feinbaum R.L., Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lehmann S.M., Kruger C., Park B., Derkow K., Rosenberger K., Baumgart J., Trimbuch T., Eom G., Hinz M., Kaul D., Habbel P., Kalin R., Franzoni E., Rybak A., Nguyen D., Veh R., Ninnemann O., Peters O., Nitsch R., Heppner F.L., Golenbock D., Schott E., Ploegh H.L., Wulczyn F.G., Lehnardt S. An unconventional role for miRNA: let-7 activates toll-like receptor 7 and causes neurodegeneration. Nat. Neurosci. 2012;15:827–835. doi: 10.1038/nn.3113. [DOI] [PubMed] [Google Scholar]
- Li Y.P., Van Pham L., Uzcategui N., Bukh J. Functional analysis of microRNA-122 binding sequences of hepatitis C virus and identification of variants with high resistance against a specific antagomir. J. Gen. Virol. 2016;97:1381–1394. doi: 10.1099/jgv.0.000445. [DOI] [PubMed] [Google Scholar]
- Lieberman J., Sarnow P. Micromanaging hepatitis C virus. N. Engl. J. Med. 2013;368:1741–1743. doi: 10.1056/NEJMe1301348. [DOI] [PubMed] [Google Scholar]
- Liu J., Wu C.P., Lu B.F., Jiang J.T. Mechanism of T cell regulation by microRNAs. Cancer Biol. Med. 2013;10:131–137. doi: 10.7497/j.issn.2095-3941.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodygin D., Tarasov V., Epanchintsev A., Berking C., Knyazeva T., Korner H., Knyazev P., Diebold J., Hermeking H. Inactivation of miR-34a by aberrant CpG methylation in multiple types of cancer. Cell Cycle. 2008;7:2591–2600. doi: 10.4161/cc.7.16.6533. [DOI] [PubMed] [Google Scholar]
- Lujambio A., Ropero S., Ballestar E., Fraga M.F., Cerrato C., Setien F., Casado S., Suarez-Gauthier A., Sanchez-Cespedes M., Git A., Spiteri I., Das P.P., Caldas C., Miska E., Esteller M. Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res. 2007;67:1424–1429. doi: 10.1158/0008-5472.CAN-06-4218. [DOI] [PubMed] [Google Scholar]
- Lujambio A., Calin G.A., Villanueva A., Ropero S., Sanchez-Cespedes M., Blanco D., Montuenga L.M., Rossi S., Nicoloso M.S., Faller W.J., Gallagher W.M., Eccles S.A., Croce C.M., Esteller M. A microRNA DNA methylation signature for human cancer metastasis. Proc. Natl. Acad. Sci. U. S. A. 2008;105:13556–13561. doi: 10.1073/pnas.0803055105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S.S., Ishibashi O., Ishikawa G., Ishikawa T., Katayama A., Mishima T., Takizawa T., Shigihara T., Goto T., Izumi A., Ohkuchi A., Matsubara S., Takeshita T., Takizawa T. Human villous trophoblasts express and secrete placenta-specific microRNAs into maternal circulation via exosomes. Biol. Reprod. 2009;81:717–729. doi: 10.1095/biolreprod.108.075481. [DOI] [PubMed] [Google Scholar]
- Ma L., Young J., Prabhala H., Pan E., Mestdagh P., Muth D., Teruya-Feldstein J., Reinhardt F., Onder T.T., Valastyan S., Westermann F., Speleman F., Vandesompele J., Weinberg R.A. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat. Cell Biol. 2010;12:247–256. doi: 10.1038/ncb2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina P.P., Nolde M., Slack F.J. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature. 2010;467:86–90. doi: 10.1038/nature09284. [DOI] [PubMed] [Google Scholar]
- Melo S., Villanueva A., Moutinho C., Davalos V., Spizzo R., Ivan C., Rossi S., Setien F., Casanovas O., Simo-Riudalbas L., Carmona J., Carrere J., Vidal A., Aytes A., Puertas S., Ropero S., Kalluri R., Croce C.M., Calin G.A., Esteller M. Small molecule enoxacin is a cancer-specific growth inhibitor that acts by enhancing TAR RNA-binding protein 2-mediated microRNA processing. Proc. Natl. Acad. Sci. U. S. A. 2011;108:4394–4399. doi: 10.1073/pnas.1014720108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng F., Henson R., Wehbe-Janek H., Ghoshal K., Jacob S.T., Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra A.K., Zillhardt M., Hua Y., Tiwari P., Murmann A.E., Peter M.E., Lengyel E. MicroRNAs reprogram normal fibroblasts into cancer-associated fibroblasts in ovarian cancer. Cancer Discov. 2012;2:1100–1108. doi: 10.1158/2159-8290.CD-12-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittelbrunn M., Gutierrez-Vazquez C., Villarroya-Beltri C., Gonzalez S., Sanchez-Cabo F., Gonzalez M.A., Bernad A., Sanchez-Madrid F. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat. Commun. 2011;2:282. doi: 10.1038/ncomms1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molitoris J.K., Mccoll K.S., Distelhorst C.W. Glucocorticoid-mediated repression of the oncogenic microRNA cluster miR-17 ~ 92 contributes to the induction of Bim and initiation of apoptosis. Mol. Endocrinol. 2011;25:409–420. doi: 10.1210/me.2010-0402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroig Pdel C., Chen L., Zhang S., Calin G.A. Small molecule compounds targeting miRNAs for cancer therapy. Adv. Drug Deliv. Rev. 2015;81:104–116. doi: 10.1016/j.addr.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrini M., Nicoloso M.S., Calin G.A. MicroRNAs and cancer—new paradigms in molecular oncology. Curr. Opin. Cell Biol. 2009;21:470–479. doi: 10.1016/j.ceb.2009.03.002. [DOI] [PubMed] [Google Scholar]
- Nicoloso M.S., Spizzo R., Shimizu M., Rossi S., Calin G.A. MicroRNAs—the micro steering wheel of tumour metastases. Nat. Rev. Cancer. 2009;9:293–302. doi: 10.1038/nrc2619. [DOI] [PubMed] [Google Scholar]
- Nishimura M., Jung E.J., Shah M.Y., Lu C., Spizzo R., Shimizu M., Han H.D., Ivan C., Rossi S., Zhang X., Nicoloso M.S., Wu S.Y., Almeida M.I., Bottsford-Miller J., Pecot C.V., Zand B., Matsuo K., Shahzad M.M., Jennings N.B., Rodriguez-Aguayo C., Lopez-Berestein G., Sood A.K., Calin G.A. Therapeutic synergy between microRNA and siRNA in ovarian cancer treatment. Cancer Discov. 2013;3:1302–1315. doi: 10.1158/2159-8290.CD-13-0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orom U.A., Nielsen F.C., Lund A.H. MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and enhances their translation. Mol. Cell. 2008;30:460–471. doi: 10.1016/j.molcel.2008.05.001. [DOI] [PubMed] [Google Scholar]
- Ottosen S., Parsley T.B., Yang L., Zeh K., van Doorn L.J., van der Veer E., Raney A.K., Hodges M.R., Patick A.K. In vitro antiviral activity and preclinical and clinical resistance profile of miravirsen, a novel anti-hepatitis C virus therapeutic targeting the human factor miR-122. Antimicrob. Agents Chemother. 2015;59:599–608. doi: 10.1128/AAC.04220-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecot C.V., Rupaimoole R., Yang D., Akbani R., Ivan C., Lu C., Wu S., Han H.D., Shah M.Y., Rodriguez-Aguayo C., Bottsford-Miller J., Liu Y., Kim S.B., Unruh A., Gonzalez-Villasana V., Huang L., Zand B., Moreno-Smith M., Mangala L.S., Taylor M., Dalton H.J., Sehgal V., Wen Y., Kang Y., Baggerly K.A., Lee J.S., Ram P.T., Ravoori M.K., Kundra V., Zhang X., Ali-Fehmi R., Gonzalez-Angulo A.M., Massion P.P., Calin G.A., Lopez-Berestein G., Zhang W., Sood A.K. Tumour angiogenesis regulation by the miR-200 family. Nat. Commun. 2013;4:2427. doi: 10.1038/ncomms3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakheja D., Chen K.S., Liu Y., Shukla A.A., Schmid V., Chang T.C., Khokhar S., Wickiser J.E., Karandikar N.J., Malter J.S., Mendell J.T., Amatruda J.F. Somatic mutations in DROSHA and DICER1 impair microRNA biogenesis through distinct mechanisms in Wilms tumours. Nat. Commun. 2014;2:4802. doi: 10.1038/ncomms5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schetter A.J., Heegaard N.H., Harris C.C. Inflammation and cancer: interweaving microRNA, free radical, cytokine and p53 pathways. Carcinogenesis. 2010;31:37–49. doi: 10.1093/carcin/bgp272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah M.Y., Calin G.A. The mix of two worlds: non-coding RNAs and hormones. Nucleic Acid Ther. 2013;23:2–8. doi: 10.1089/nat.2012.0375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern-Ginossar N., Gur C., Biton M., Horwitz E., Elboim M., Stanietsky N., Mandelboim M., Mandelboim O. Human microRNAs regulate stress-induced immune responses mediated by the receptor NKG2D. Nat. Immunol. 2008;9:1065–1073. doi: 10.1038/ni.1642. [DOI] [PubMed] [Google Scholar]
- Tay Y., Zhang J., Thomson A.M., Lim B., Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455:1124–1128. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- Tili E., Michaille J.J., Croce C.M. MicroRNAs play a central role in molecular dysfunctions linking inflammation with cancer. Immunol. Rev. 2013;253:167–184. doi: 10.1111/imr.12050. [DOI] [PubMed] [Google Scholar]
- Tominaga N., Kosaka N., Ono M., Katsuda T., Yoshioka Y., Tamura K., Lotvall J., Nakagama H., Ochiya T. Brain metastatic cancer cells release microRNA-181c-containing extracellular vesicles capable of destructing blood-brain barrier. Nat. Commun. 2015;6:6716. doi: 10.1038/ncomms7716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trang P., Medina P.P., Wiggins J.F., Ruffino L., Kelnar K., Omotola M., Homer R., Brown D., Bader A.G., Weidhaas J.B., Slack F.J. Regression of murine lung tumors by the let-7 microRNA. Oncogene. 2010;29:1580–1587. doi: 10.1038/onc.2009.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai K.W., Liao Y.L., Wu C.W., Hu L.Y., Li S.C., Chan W.C., Ho M.R., Lai C.H., Kao H.W., Fang W.L., Huang K.H., Lin W.C. Aberrant hypermethylation of miR-9 genes in gastric cancer. Epigenetics. 2011;6:1189–1197. doi: 10.4161/epi.6.10.16535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda R., Kohanbash G., Sasaki K., Fujita M., Zhu X., Kastenhuber E.R., Mcdonald H.A., Potter D.M., Hamilton R.L., Lotze M.T., Khan S.A., Sobol R.W., Okada H. Dicer-regulated microRNAs 222 and 339 promote resistance of cancer cells to cytotoxic T-lymphocytes by down-regulation of ICAM-1. Proc. Natl. Acad. Sci. U. S. A. 2009;106:10746–10751. doi: 10.1073/pnas.0811817106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valencia-Sanchez M.A., Liu J., Hannon G.J., Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- Valeri N., Gasparini P., Fabbri M., Braconi C., Veronese A., Lovat F., Adair B., Vannini I., Fanini F., Bottoni A., Costinean S., Sandhu S.K., Nuovo G.J., Alder H., Gafa R., Calore F., Ferracin M., Lanza G., Volinia S., Negrini M., Mcilhatton M.A., Amadori D., Fishel R., Croce C.M. Modulation of mismatch repair and genomic stability by miR-155; Proceedings of the National Academy of Sciences of the United States of America; 2010. pp. 6982–6987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Zandwijk N., Pavlakis N., Kao S., Clarke S., Lee A., Brahmbhatt H., Macdiarmid J., Pattison S., Leslie F., Huynh Y., Linton A., Reid G. P1.02, MesomiR 1: a phase I study of TargomiRs in patients with refractory malignant pleural mesothelioma (MPM) and lung cancer (NSCLC) Ann. Oncol. 2015;26(suppl 2) [Google Scholar]
- Vasudevan S., Tong Y., Steitz J.A. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- Vester B., Wengel J. LNA (locked nucleic acid): high-affinity targeting of complementary RNA and DNA. Biochemistry. 2004;43:13233–13241. doi: 10.1021/bi0485732. [DOI] [PubMed] [Google Scholar]
- Wang X., Cao L., Wang Y., Wang X., Liu N., You Y. Regulation of let-7 and its target oncogenes (review) Oncol. Lett. 2012;3:955–960. doi: 10.3892/ol.2012.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watashi K., Yeung M.L., Starost M.F., Hosmane R.S., Jeang K.T. Identification of small molecules that suppress microRNA function and reverse tumorigenesis. J. Biol. Chem. 2010;285:24707–24716. doi: 10.1074/jbc.M109.062976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J.S., Song Y.K., Durinck S., Chen Q.R., Cheuk A.T., Tsang P., Zhang Q., Thiele C.J., Slack A., Shohet J., Khan J. The MYCN oncogene is a direct target of miR-34a. Oncogene. 2008;27:5204–5213. doi: 10.1038/onc.2008.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J., Yang B., Lin H., Lu Y., Luo X., Wang Z. Novel approaches for gene-specific interference via manipulating actions of microRNAs: examination on the pacemaker channel genes HCN2 and HCN4. J. Cell. Physiol. 2007;212:285–292. doi: 10.1002/jcp.21062. [DOI] [PubMed] [Google Scholar]
- Yuan T.L., Fellmann C., Lee C.S., Ritchie C.D., Thapar V., Lee L.C., Hsu D.J., Grace D., Carver J.O., Zuber J., Luo J., Mccormick F., Lowe S.W. Development of siRNA payloads to target KRAS-mutant cancer. Cancer Discov. 2014;4:1182–1197. doi: 10.1158/2159-8290.CD-13-0900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Liu D., Chen X., Li J., Li L., Bian Z., Sun F., Lu J., Yin Y., Cai X., Sun Q., Wang K., Ba Y., Wang Q., Wang D., Yang J., Liu P., Xu T., Yan Q., Zhang J., Zen K., Zhang C.Y. Secreted monocytic miR-150 enhances targeted endothelial cell migration. Mol. Cell. 2010;39:133–144. doi: 10.1016/j.molcel.2010.06.010. [DOI] [PubMed] [Google Scholar]
- Zhang X., Zuo X., Yang B., Li Z., Xue Y., Zhou Y., Huang J., Zhao X., Zhou J., Yan Y., Zhang H., Guo P., Sun H., Guo L., Zhang Y., Fu X.D. MicroRNA directly enhances mitochondrial translation during muscle differentiation. Cell. 2014;158:607–619. doi: 10.1016/j.cell.2014.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]