Abstract
We report the case of a 26 year-old patient presenting with a persistent leukocytosis and CML-like marrow but no evidence of a BCR/ABL1 fusion. Molecular cytogenetics revealed that a portion of the ETV6 locus was inserted into the ABL1 locus. An ETV6/ABL1 fusion transcript could subsequently be confirmed. The patient was started on imatinib and went into complete cytomorphological remission. QRT-PCR measurements showed a 4 log reduction of the ETV6/ABL1 fusion. 15 months later, the disease transformed into ALL and the patient expired. Thus, an ETV6/ABL1 fusion positive MPN has the potential to transform very rapidly into ALL.
Keywords: ETV6/ABL1 fusion, CML, ALL, MRD assay, TKI treatment
1. Introduction
The ETV6-ABL1 (TEL-ABL) fusion is a rare aberration found in a range of haematologic malignancies. First described in 1995 [1], only 27 ETV6-ABL1-positive cases diagnosed with atypical chronic myeloid leukemia (aCML), chronic myeloproliferative neoplasm (cMPN), acute lymphoblastic leukemia (ALL) or acute myeloid leukemia (AML) have been reported up to now [2], [3], [4], [5]. The rarity of the ETV6-ABL1 rearrangement is thought to be due to the opposite transcriptional orientations of the ETV6 and the ABL1 genes relative to the telomere-centromere axis, which requires at least three DNA breaks to generate a functional ETV6-ABL1 fusion gene [5]. Thus in most ETV6-ABL1-positive cases, the fusion is generated as the result of complex, often cryptic, chromosomal rearrangements. The frequent cryptic nature of these rearrangements is likely to have resulted in underreporting of ETV6-ABL1-positive cases. Due to breakpoint heterogeneity, two types of ETV6-ABL1 fusion transcripts are found: the ‘type A’ transcript is a fusion of the first 4 ETV6 exons to the second exon of ABL1 and in the ‘type B’ transcript the first 5 exons of ETV6 are fused to the second exon of ABL1. ETV6-ABL1 activates similar cellular pathways like BCR-ABL1. In vitro studies have shown that both fusion proteins lead to the constitutive activation of the ABL1 tyrosine kinase [6]. Like BCR-ABL1, ETV6-ABL1 is sensitive to ABL1 tyrosine kinase inhibitors [7], [8].
Here, we report an ETV6-ABL1 fusion in a young female patient with a myeloproliferative disorder, which rapidly transformed into an acute lymphoblastic leukemia (ALL). The initial diagnostic screening tests for a BCR-ABL fusion did not detect any sign of an ABL1 rearrangement. This case highlights the importance of a detailed cytogenetic work-up in detecting the ETV6-ABL1 fusion and the necessity of careful minimal residual disease monitoring as well as the danger of rapid transformation of ETV6-ABL1 positive MPNs into ALL.
2. Materials and methods
2.1. Cytogenetic analysis
Chromosomes were prepared from short-term bone marrow cultures according to standard protocols. For the confirmation of the cytogenetic findings, fluorescence in situ hybridization (FISH) using commercially available FISH probes was performed following standard protocols (Abott).
2.2. RT-PCR and sequencing
mRNA was isolated from mononuclear cells using the MagnaPure (Roche) kits according to the manufacturer's protocols. Whole RNA was isolated from the mononucleated cells from bone marrow or peripheral blood samples using the RNeasy mini kit (QIAGEN). The RNA was then reverse transcribed to generate cDNA using the Superscript™ III reverse transcriptase (Invitrogen). As a part of the routine diagnostics work up, a reverse transcriptase (RT)-PCR with primers specific for the BCR-ABL1 p210 and p190 fusion transcripts was performed [9]. RT-PCR analysis with the following primers was performed for the detection of the ETV6-ABL1 fusion: ETV6-ABL1-(E4/E2)_F (5′- AAAGCTCTCCTGCTGCTGAC-3′) corresponding to exon 3 of ETV6 and ETV6-ABL1-(E4/E2)_R (5′- TGTTATCTCCACTGGCCACA −3′) corresponding to exon 2 of ABL1 were used for the detection of the ‘type A’ ETV6-ABL1 fusion transcript. Additionally, primers ETV6-ABL1-(E5/E2)_F (5′-AAG CCC ATC AAC CTC TCT CA-3′) from exon 5 of ETV6 and ETV6-ABL1-(E5/E2)_R (5′-ACA CCA TTC CCC ATT GTG AT-3′) from exon 2 of ABL1 were used for the detection of the ‘type B’ ETV6-ABL1 fusion transcript. To detect the reciprocal ABL1 (exon1a/1b)/ ETV6 (exon6) fusion transcript the primers ABL1a_ex1a_F (5′-GTTGGAGATCTGCCTGAAGC−3′), ABL1b_ex1b_F (5′-GGAGCAGGGAAGAAGGAATC-3′) and ETV6_WT_Ex7_R (5′-TCCTGGCTCCTTCCTGATAA-3′) were used. To identify an ABL1-ETV6 fusion transcript, which might be generated by an insertion of the 5′ portion of ETV6 between the exons Ex1a/1b and exon 2 of ABL1, primers ABL1a_ex1a_F, ABL1b_ex1b_F and ETV6_Ex4_R (5′-GGGTGGAAGAATGGTGAAAA-3′) were used. For the mutation detection in the ETV6 transcript, the whole ETV6 transcript was amplified in four overlapping fragments using the primers, ETV6_WT_F1_F (5′-GGGAGAGGAAAGGAAAGTGG-3′), ETV6_WT_F1_R (5’-ACCTCCGGCTGTGT GTGTAT-3’), ETV6_WT__F2_F (5′-CAG CATATTCTGAAGCAGAGGA-3′), ETV6_WT_F2_R (5′-AGGATCAGAGGGTGCATGAT-3′), ETV6_WT__F3_F (5′-CCCATGGAGAATAATCACTGC-3′), ETV6_WT_F3_R (5′-GCAGGGCTCTGGA CATTTT-3′), ETV6_WT__F4_F (5′-CGACTGTGGGGAAACCAT AA-3′) and ETV6_WT_F4_R (5′-AAGTGTCCCTGCCATTTCTG-3′). The RT-PCR products were sequenced with the BigDye terminator v1.1 cycle sequencing kit on an ABI PRISM 310 Genetic analyzer (Applied Biosystems, USA).
2.3. Minimal residual disease (MRD) monitoring
For the monitoring of the minimal residual disease, the level of the ETV6-ABL1 fusion transcript was estimated by reverse transcription quantitative real-time PCR (RQ-PCR). A 150 bp fragment spanning the breakpoint of the ETV6-ABL1 fusion transcript was amplified on a Light-Cycler MxPro3005 instrument (Stratagene) using the primers qPCR_ETV6-ABL1(E5/E2)_F1 (5′-AAGCCCATCAACCTCTCTCA-3′) and qPCR_ETV6-ABL1(E5/E2)_R1 (5′-ACC CTGAGGCTCAAAGTCAG-3′). As a reference transcript, a region from the 5’ portion of ABL1 that is only contained in the non-rearranged ABL1b isoform was amplified using the primers ABL1_qPCR_F (5′-CAGGGAAGAAGGAATCATCG-3′) and ABL1_qPCR_R (5′-GTTGGGGTCATT TTCACTGG-3′). The ETV6-ABL1 fusion transcript was normalized to the ABL1 transcript (ETV6-ABL1/ ABL1 ratio) in the pre- and post treatment samples. Finally, the normalized level of the ETV6-ABL1 transcript at diagnosis was set as 1 and the ETV6-ABL1 fusion transcript levels at the follow-up time points were expressed relative to the level at the initial diagnosis.
3. Results
A 26-year-old women with psoriasis was clinically evaluated with suspicion of psoriatic arthritis of the shoulder. Her psoriasis was not severe and well controlled with stress avoidance and mild topical ointments. Routine blood count showed an increased WBC of 40,000/µl. An RT-PCR assay for a BCR-ABL fusion from peripheral blood was negative. Although her shoulder pain resolved, the leukocytosis persisted. A bone marrow biopsy in December 2012 showed a packed marrow with trilineage hyperplasia and an eosinophilia of 20–30%. The neutrophils and eosinophils showed markedly hypersegmented nuclei. The findings were compatible with a myeloproliferative disease.
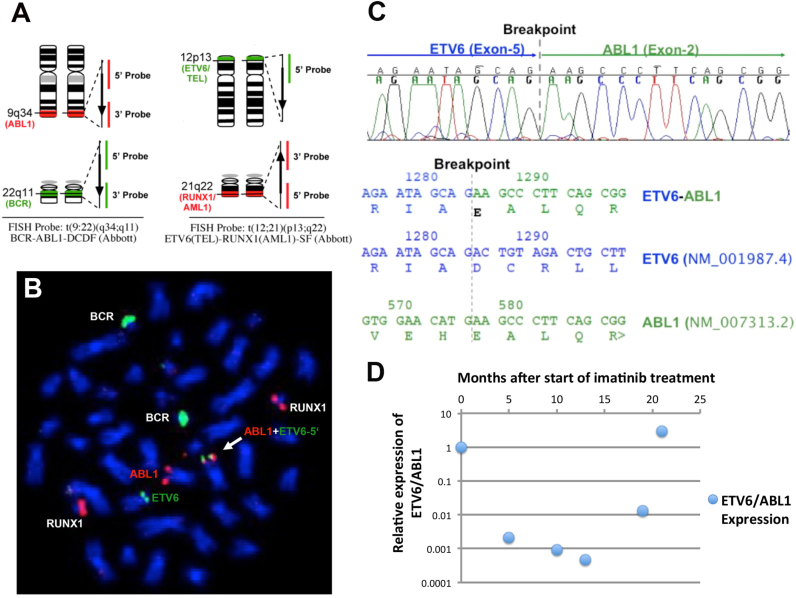
An interphase FISH analysis with BCR-ABL1-DCDF (dual colour dual fusion) probes (Abbott) showed a normal signal pattern. In addition, an RT-PCR screen with primers specific for the BCR-ABL1 p210 and p190 fusion transcripts and a JAK2V617F mutation analysis were both negative. Chromosomal analysis of the bone marrow cultures revealed a 46, XX,t(10;12)(q24;p13) karyotype in 5 of 10 metaphases. The remaining metaphase had a normal karyotype (46, XX,t(10;12)(q24;p13)[5]/46, XX[5]). Whole chromosome painting probes (WCP) specific for chromosomes 10 and 12 were used to confirm the t(10;12) translocation. As band 12p13 was involved in the rearrangement, we assessed the status of the ETV6 locus with an ETV6 break apart probe (Dako). This analysis detected a break in one of the ETV6 loci. Interestingly, instead of on chromosome 10, the probe corresponding to the 5’ genomic portion of ETV6 hybridized, quite unexpectedly, to 9q34 suggesting an involvement of the ABL1 locus. However, when we repeated the BCR/ABL DCDF FISH analysis on metaphase chromosomes there was no alteration of the ABL1 loci visible. Only a combination of the BCR-ABL1-DCDF and an ETV6/AML1-ES (extra signal) dual colour probes (Fig. 1A) revealed a co-localization of the 5’ ETV6 genomic region with the ABL1 locus (Fig. 1B). These findings suggested the presence of an ETV6-ABL1 rearrangement. RT-PCR using ETV6-ABL1 fusion transcript-specific primers indeed detected a ‘type B’ ETV6-ABL1 fusion transcript, which results from the fusion of the first 5 exons of ETV6 to exon 2 of ABL1. Sequence analysis of the RT-PCR product confirmed the presence of the ‘type B’ – ETV6-ABL1 fusion transcript, showing an in-frame fusion between exon 5 of ETV6 and exon 2 of ABL1 (Fig. 1C). These results suggested that an insertion of the 5′ portion of the ETV6 locus up to ETV6 exon 5 into the ABL1 locus had occurred. We also performed a PCR experiment to detect the reciprocal ABL1(exon1a/1b)/ETV6(exon6) fusion and a more 5′ fusion which might result from the insertion (ABL1(exon1a/1b)/ETV6(exon2/3/4)). Neither the reciprocal ABL1(exon1a/1b)/ETV6(exon6) nor evidence for an ABL1(exon1a/1b)/ETV6(exon2/3/4) fusion, as would be expected from an insertion, were found.
Fig. 1.
Detection of the ETV6-ABL1 fusion: A. Schematic diagram of commercial BCR-ABL1-DCDF and the RUNX1-ETV6-ES FISH probe designs. B. Co-hybridization of the BCR-ABL1-DCDF and the RUNX1-ETV6-ES FISH probes on the metaphase chromosomes preparation from the patient sample: RUNX1 and BCR locus were intact. As predicted, one of the green signals for ETV6-5′ region was observed co-localized with one of the ABL1 loci on chromosome 9 (white arrow). C. Electropherogram of the sequence spanning the breakpoint confirming an in-frame fusion between ETV6 exon 5 and ABL1 exon2. (upper panel) and fusion transcript sequence and the corresponding wild type sequences including the amino acid sequence (lower panel). The vertical grey line indicates the position of the breakpoint. D. Overview of the ETV6-ABL1 fusion transcript level during the course of treatment with reference to that at the diagnosis. Imatinib treatment was started 2 months after the diagnosis time point. MRD: minimal residual disease: * Relative Expression of ETV6/ABL1: The expression level of ETV6/ABL1 at the time of diagnosis is set to 1. ETV6-ABL1 expression levels are normalized to the expression levels of ABL1. Relative Expression=2exp(Δ CtETV6/ABL1(follow-up)-Δ CtETV6/ABL1 (diagnosis)).
There is increasing evidence that ETV6 plays the role of a tumor suppressor gene because the non-rearranged ETV6 allele is frequently deleted in childhood ALL cases with an ETV6/RUNX1 fusion [6], [10], [11] and ETV6 point mutations have also been described in hematological malignancies [12]. In addition, ETV6 mutations have been identified in familial leukemia cases [13]. Therefore, we sequenced the complete coding region of ETV6 in our patient. This analysis did not reveal a mutation (data not shown).
Based on the detection of the ETV6-ABL1 fusion transcript, off-label, evidence-based Imatinib therapy (400 mg/d) was commenced [3], [5], [14], [15], [16], [17], [18], [19]. She showed a complete normalization of her blood counts after 4 weeks. We developed a quantitative real time PCR assay (RQ-PCR) for the ETV6-ABL1 fusion transcript. Our minimal residual disease (MRD) monitoring revealed a 4 Log10 fold reduction in the levels of the ETV6-ABL1 fusion transcript over a period of 11 months. There was a moderate increase of MRD levels of about 1.5 Log10 fold 6 months later (Fig. 1D).
The patient presented 3 weeks later at our emergency department because of painful swelling of the right leg in combination with shortness of breath for 1 week. Duplex ultrasonography revealed deep vein thrombosis and CT angiography revealed pulmonary emboli. Full blood count showed 30,000 white blood cells per µl with 48% lymphoid blast cells (normal haematocrit, 80,000 platelets/µl). A bone marrow aspirate showed 70% lymphoid blasts. At this time, the ETV6-ABL1 transcript levels were about three times the level at initial presentation. Sequencing of the tyrosine kinase domain of the ETV6-ABL1 fusion transcript did not reveal any mutations. These results indicate that the ALL clone was ETV6-ABL1 positive. At this time the cytogenetic analysis showed 46, XX, der(9)t(9;12)(q34;p13), del(10)(q24), del(11)(q23), der(12)t(10;12)(q24;p13)[8]/46, XX[2].
The patient was started on standard induction chemotherapy according to the German Multicenter Study Group on Adult Acute Lymphoblastic Leukemia (GMALL) and an allogeneic stem cell donor search was initiated due to the high risk condition. Five days after start of the treatment, the patient fell into a coma due to a cerebral sinus thrombosis with secondary haemorrhage. Despite repeated neurosurgical interventions the patient died two weeks later.
4. Discussion
The ETV6 gene on the short arm of chromosome 12 is transcribed from the telomere to the centromere, whereas ABL1 is transcribed from the centromere to the telomere on the long arm of chromosome 9. The different transcriptional orientations of ETV6 and ABL1 are most likely the reason for the rarity and complexity of ETV6-ABL1 rearrangements. In most of the reported ETV6-ABL1 cases, these so-called cryptic rearrangements were not detected by conventional cytogenetics. FISH analyses showed that the ETV6-ABL1 fusion resulted either from an insertion of the 3′ region of the ABL1 gene into the ETV6 locus or an insertion of the 5′ region of the ETV6 gene within the ABL1 gene locus. FISH screening for the BCR-ABL1 rearrangement is part of the routine diagnostic work-ups for MPD (CML and MPNs) and ALL. Depending on the type of FISH probe used, this assay also allows the detection of unusual rearrangements at the BCR and ABL1 loci. Therefore, cryptic ETV6-ABL1 rearrangements, resulting from an insertion of the 3’ ABL1 region within the ETV6 locus, can be detected during the routine diagnostic work-up as an additional or split ABL1 signal. However, the ETV6-ABL1 fusions resulting from an insertion of the 5’ ETV6 region within the ABL1 locus will be overlooked, unless an ETV6 rearrangement specific FISH assay is performed. In cases with a t(9;12;Var), the involvement of 9q34 or 12p13 is an indication that an ETV6-ABL1 rearrangement might be present, which can then be confirmed by specific FISH assays and ETV6-ABL1 fusion specific RT-PCR. However, cases like the one present here, pose a problem since our initial BCR-ABL1 FISH assay was completely normal and the results of our initial cytogenetics analysis did not indicate an involvement of the ABL1 locus either. The only hint came from the chromosomal rearrangement involving band p13 of chromosome 12 on chromosome 9. With ETV6-ABL1 being a rather rare entity, FISH probes specific for the detection of the ETV6-ABL1 fusion are not commercially available. However, a FISH assay using a mixture of the standard FISH probes for the BCR-ABL1 and the ETV6-RUNX1 fusions on the metaphase chromosomes can detect and characterize even cryptic and rare ETV6-ABL1 rearrangements. Recently, multiplex RT-PCR assays have been shown to be efficient tools for the detection of a wide range of leukemia specific fusion transcripts. Several ETV6-ABL1 cases were detected by such multiplex RT-PCR approach [2], [4]. Table 1 gives an overview of the ETV6-ABL1 positive cases described in the literature so far. This table is an up-date and extension of the table of Zuna et al. [5].
Table 1.
Review of reported ETV6-ABL1 positive cases till date (adapted from [5]).
| Case number | Diagnosisa | Gender | Age at diagnosis (years)b | TKIctreatment | Outcomed | Eosinophilia | Type of transcript | Reference | Cytogenetics: detailed work-up |
|---|---|---|---|---|---|---|---|---|---|
| 1 | ALL | F | 1 (22 M) | No | Died | n.r. | A | [1] | n.r. |
| 2 | AML-M6 | M | 81 | No | Died | n.r. | B | [6] | n.r. |
| 3 | CML (atypical) | n.r. | 49 | No | Died | Yes | B | [22] | n.r. |
| 4 | CML | M | 32 | No | CR (>3Y) | Yes | B | [23] | Yes: Group B |
| 5 | CML | M | 59 | No | Died | Yes | A, B | [24] | n.r. |
| 6 | ALL (T-lineage) | M | 4 | No | Died | Yes | A, B | [24] | n.r. |
| 7 | AML-M6 (or CML-MBC? ) | M | 38 | Yes | Died | n.r. | A, B | [16] | Yes: Group A |
| 8 | CML | M | 53 | No | CR (>6Y) | Yes | A, B | [25] | n.d. (NK, B/FISH-) |
| 9 | CML | F | 44 | No | CR (>6 M) | Yes | n.r. | [26] | Yes: Group B |
| 10 | AML-M2 | M | 29 | No | CR (>20 M post HSCT) | Yes | A | [27] | Yes: Group A |
| 11 | AML-M1(after RAEB) | M | 48 | No | Died | Yes | B | [27] | Yes: Group B |
| 12 | CML-MBC | M | 36 | Yes | Died | Yes | B | [14] | n.a. |
| 13 | CML-LBC | M | 72 | Yes | CR (>12 M) | n.r. | B | [19] | Yes: Group A |
| 14 | cMPN - myeloid sarcoma | F | 65 | No | Died | Yes | A, B | [28] | Yes: Group A |
| 15 | cMPN | M | 57 | No | CR (>15Y) | Yes | n.r. | [29] | Yes: Group A |
| 16 | ALL | M | 30 | n.r. | Died | n.r. | A, B | [30] | Yes: Group A |
| 17 | CML | F | 24 | Yes | CR (>7 M) | Yes Eo: 3920/µl (4%) | A, B | [17] | Yes: Group C |
| 18 | cMPN | F | 61 | Yes | CR (>3Y) | Yes | n.r. | [18] | Yes: Group A |
| 19 | CML (atypical) | M | 79 | Yes | Died | Yes Eo: 2110/µl (6%) | n.r. | [15] | Yes: Group A |
| 20 | ALL | F | 33 | n.r. | Died | No | A, B | [5] | Yes: Group A |
| 21 | ALL | M | 5 | No | CR (>24 M) | Yes | A, B | [5] | Yes: Group A |
| 22 | ALL | M | 0 (8 M) | Yes | Died | No | A, B | [5] | Yes: Group B |
| 23 | ALL | F | 8 | Yes | CR(>10 M) | n.r. | A, B | [21] | Yes: Group A |
| 24 | aCML | M | 36 | Yes | CR (>5Y) | Eo: 1650/µl (3%) | n.r. | [3] | Yes: Group A |
| 25 | B-ALL | M | 58 | No | Died | n.r. | n.r. | [4] | n.r. except t(9;12) + add abn |
| 26 | B-ALL | F | 49 | No | HSCTe | n.r. | n.r. | [4] | n.r. except 46, XX, t(9;12)(q34;p13)[20] |
| 27 | AML | M | 52 | No | Died | Yes | B | [2] | n.r. except 46, XY[20] |
| 28 | B-ALL | F | 25 | No | CR (<15 M) | No | A, B | [2] | Yes: Group A |
| 29 | aCML | F | 52 | Yes | CR (>12 M) | Yes | A, B | [20] | Yes: Group B |
| 30 | MPN -> ALL | F | 26 | Yes | CR (18 M), | Yes | B | This case | Yes: Group B |
n.r., not reported.
Group A: BCR-ABL1- FISH screen: BCR-ABL1 negative, ABL1 split: 12 cases (AML, ALL, MPN);
ETV6-ABL1 fusion on der(12)(p13): 10 cases (AML, ALL, MPN), ETV6-ABL1 fusion on a third chromosome: 2 cases (AML, CML-LBC).
Group B: BCR-ABL1- FISH screen: BCR-ABL1 negative, ABL1 not split, ETV6 rearranged. ETV6-ABL1 fusion on der(9)(q34): 4 cases (AML, ALL, MPN).
Group C: BCR-ABL1- FISH screen: BCR-ABL1 negative, ABL1 not split, but ASS deleted (5′ ABL1), 3 signals for ETV6. Duplication of 5′ETV6 and insertion of 5′ETV6 in der(9)(q34) resulting in ETV6-ABL1 fusion: 1 case (CML).
Diagnosis: ALL, acute lymphoblastic leukemia; AML, acute myeloid leukemia; CML, chronic myeloid leukemia; LBC, lymphoid blast crisis; MBC, myeloid blast crisis; cMPN, chronic myeloproliferative neoplasm; RAEB, refractory anemia with excess blasts.
Age at diagnosis: In the two youngest patients also the age at diagnosis in months is shown in parentheses.
TKI treatment: TKI, tyrosine-kinase inhibitors.
Outcome: CR, complete remission; HSCT, stem cell transplantation, HSCT e: Hematopoietic stemcell transplantation without hematological or molecular CR
The ETV6-ABL1 fusion has been reported as a poor prognostic factor when found in acute leukemia (AML and ALL) and MDS. The prognostic significance of an ETV6/ABL1 chronic myeloproliferative neoplasm is less clear due to the paucity of cases and the lack of controlled clinical trials [5]. There is a significant overlap of molecular targets of ETV6-ABL1 with those of BCR-ABL1, and the ETV6-ABL1 fusion protein triggers similar oncogenic cascades like the BCR-ABL1 fusion protein [3], [10]. Similar to BCR-ABL1 positive cells, ETV6-ABL1 positive cells show in-vitro sensitivity to the tyrosine kinase inhibitor Imatinib [8], [7]. Several authors have also reported a transient [5], [14], [15], [16] or prolonged [3], [17], [18], [19] response to Imatinib or second generation tyrosine kinase inhibitor treatment in ETV6-ABL1-positive neoplasms, with the response to TKI being better and longer in ETV6-ABL1-positive myeloproliferative neoplasms. Thus, the detection of a cryptic ETV6-ABL1 rearrangement has a significant impact on patient care, especially in the case of chronic myeloproliferative neoplasms or chronic myeloid leukemia like phenotypes. However, there are only reports of 8 ETV6-ABL1 positive patients having been treated with Imatinib for various diseases (CML, AML, MPN, ALL) according to different therapy protocols [3], [5], [14], [15], [16], [17], [18], [19], [20], [21]. Of the 7 patients with ETV6-ABL1-positive CML or MPN, who received TKI treatment, 5 responded, 4 with a prolonged remission. In contrast, of the 2 patients with an ETV6/ABL1 positive AML or ALL, who received TKI treatment, none responded with a longer remission duration (Table 1). However, the efficacy of Imatinib and second generation TKIs as well as the optimal treatment schedule for ETV6-ABL1 positive hematological malignancies still needs to be determined.
This case illustrates the importance of a careful cytogenetic work-up for patients with a myeloproliferative neoplasm. If a patient can be treated with a tyrosine kinase inhibitor instead of chemotherapy, a significant improvement in his/her quality of life and survival can be achieved. The benefits of long term TKI treatment in young patients compared to a potentially curative bone marrow transplant need to be carefully assessed. In this situation, careful MRD monitoring is warranted. However despite good response to TKI, even tight MRD monitoring was not able to predict the rapid transformation from a myeloproliferative neoplasm into an acute lymphoblastic leukemia in our patient. Additionally, the clinical presentation of the secondary ETV6/ABL-positive ALL in our case was associated with a fatal paraneoplastic thrombophilic syndrome. We can only speculate as to what caused this rapid transformation from a relatively benign myeloproliferative neoplasm to an aggressive ALL. It is known from cytogenetic studies of CML in blast crisis that additional chromosomal abnormalities arise at blast crisis (e.g. an additional copies of the Philadelphia chromosome). This indicates that additional genetic changes, either additional chromosomal abnormalities or point mutations, are most likely responsible for disease transformation. Also in our patient, chromosomal analysis showed that, in addition to the rearrangements affecting chromosomes 9, 10, and 12 found at presentation, there were additional changes on chromosome 11 in the ALL clone. It is also highly likely that more detailed genomic analyses like exome sequencing, SNP array CGH etc., would detect additional driver mutations or deletion in genes previously shown to be affected in ETV6-ABL1-positive ALL like CDKN2A/B and IKZF1 [31] as well as cryptic chromothripsis events [32]. To the best of our knowledge, this is the first description of an ETV6-ABL1-positive myeloid neoplasm that transformed into an ALL. Therefore, ETV6-ABL1 positive patients, even with a myeloid phenotype and CML-like clinical course, should be viewed as high risk patients.
Acknowledgement
PMK and SKB are supported by the Family of Marijanna Kumerich and Leukaemia & Blood Cancer New Zealand.
References
- 1.Papadopoulos P., Ridge S.A., Boucher C.A., Stocking C., Wiedemann L.M. The novel activation of ABL by fusion to an ets-related gene, TEL. Cancer Res. 1995;55:34–38. [PubMed] [Google Scholar]
- 2.Park J., Kim M., Lim J., Kim Y., Han K., Kim J.S. Variant of ETV6/ABL1 gene is associated with leukemia phenotype. Acta Haematol. 2013;129:78–82. doi: 10.1159/000342490. [DOI] [PubMed] [Google Scholar]
- 3.Perna F., Abdel-Wahab O., Levine R.L., Jhanwar S.C., Imada K., Nimer S.D. ETV6-ABL1-positive “chronic myeloid leukemia”: clinical and molecular response to tyrosine kinase inhibition. Haematologica. 2011;96:342–343. doi: 10.3324/haematol.2010.036673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou M.H., Gao L., Jing Y., Xu Y.Y., Ding Y., Wang N. Detection of ETV6 gene rearrangements in adult acute lymphoblastic leukemia. Ann. Hematol. 2012;91:1235–1243. doi: 10.1007/s00277-012-1431-4. [DOI] [PubMed] [Google Scholar]
- 5.Zuna J., Zaliova M., Muzikova K., Meyer C., Lizcova L., Zemanova Z. Acute leukemias with ETV6/ABL1 (TEL/ABL) fusion: poor prognosis and prenatal origin. Genes Chromosomes Cancer. 2010;49:873–884. doi: 10.1002/gcc.20796. [DOI] [PubMed] [Google Scholar]
- 6.Golub T.R., Goga A., Barker G.F., Afar D.E., McLaughlin J., Bohlander S.K. Oligomerization of the ABL tyrosine kinase by the Ets protein TEL in human leukemia. Mol. Cell Biol. 1996;16:4107–4116. doi: 10.1128/mcb.16.8.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Okuda K., Weisberg E., Gilliland D.G., Griffin J.D. ARG tyrosine kinase activity is inhibited by STI571. Blood. 2001;97:2440–2448. doi: 10.1182/blood.v97.8.2440. [DOI] [PubMed] [Google Scholar]
- 8.Carroll M., Ohno-Jones S., Tamura S., Buchdunger E., Zimmermann J., Lydon N.B. CGP 57148, a tyrosine kinase inhibitor, inhibits the growth of cells expressing BCR-ABL, TEL-ABL, and TEL-PDGFR fusion proteins. Blood. 1997;90:4947–4952. [PubMed] [Google Scholar]
- 9.Kakadia P.M., Tizazu B., Mellert G., Harbott J., Röttgers S., Quentmeier H. A novel ABL1 fusion to the SH2 containing inositol phosphatase-1 (SHIP1) in acute lymphoblastic leukemia (ALL) Leukemia. 2011;25:1645–1649. doi: 10.1038/leu.2011.129. [DOI] [PubMed] [Google Scholar]
- 10.Golub T.R., Barker G.F., Bohlander S.K., Hiebert S.W., Ward D.C., Bray-Ward P. Fusion of the TEL gene on 12p13 to the AML1 gene on 21q22 in acute lymphoblastic leukemia. Proc. Natl. Acad. Sci. USA. 1995;92:4917–4921. doi: 10.1073/pnas.92.11.4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Romana S.P., Mauchauffé M., Le Coniat M., Chumakov I., Le Paslier D., Berger R. The t(12;21) of acute lymphoblastic leukemia results in a tel-AML1 gene fusion. Blood. 1995;85:3662–3670. [PubMed] [Google Scholar]
- 12.Griesinger F., Janke A., Podleschny M., Bohlander S.K. Identification of an ETV6-ABL2 fusion transcript in combination with an ETV6 point mutation in a T-cell acute lymphoblastic leukaemia cell line. Br. J. Haematol. 2002;119:454–458. doi: 10.1046/j.1365-2141.2002.03850.x. [DOI] [PubMed] [Google Scholar]
- 13.Zhang M.Y., Churpek J.E., Keel S.B., Walsh T., Lee M.K., Loeb K.R. Germline ETV6 mutations in familial thrombocytopenia and hematologic malignancy. Nat. Genet. 2015 doi: 10.1038/ng.3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barbouti A., Ahlgren T., Johansson B., Höglund M., Lassen C., Turesson I. Clinical and genetic studies of ETV6/ABL1-positive chronic myeloid leukaemia in blast crisis treated with imatinib mesylate. Br. J. Haematol. 2003;122:85–93. doi: 10.1046/j.1365-2141.2003.04391.x. [DOI] [PubMed] [Google Scholar]
- 15.Kelly J.C., Shahbazi N., Scheerle J., Jahn J., Suchen S., Christacos N.C. Insertion (12;9)(p13;q34q34): a cryptic rearrangement involving ABL1/ETV6 fusion in a patient with Philadelphia-negative chronic myeloid leukemia. Cancer Genet Cytogenet. 2009;192:36–39. doi: 10.1016/j.cancergencyto.2009.02.012. [DOI] [PubMed] [Google Scholar]
- 16.O’Brien S.G., Vieira S.A., Connors S., Bown N., Chang J., Capdeville R. Transient response to imatinib mesylate (STI571) in a patient with the ETV6-ABL t(9;12) translocation. Blood. 2002;99:3465–3467. doi: 10.1182/blood.v99.9.3465. [DOI] [PubMed] [Google Scholar]
- 17.Kawamata N., Dashti A., Lu D., Miller B., Koeffler H.P., Schreck R. Chronic phase of ETV6-ABL1 positive CML responds to imatinib. Genes Chromosomes Cancer. 2008;47:919–921. doi: 10.1002/gcc.20593. [DOI] [PubMed] [Google Scholar]
- 18.Nand R., Bryke C., Kroft S.H., Divgi A., Bredeson C., Atallah E. Myeloproliferative disorder with eosinophilia and ETV6–ABL gene rearrangement: efficacy of second-generation tyrosine kinase inhibitors. Leuk. Res. 2009;33:1144–1146. doi: 10.1016/j.leukres.2009.03.011. [DOI] [PubMed] [Google Scholar]
- 19.Tirado C.A., Sebastian S., Moore J.O., Gong J.Z., Goodman B.K. Molecular and cytogenetic characterization of a novel rearrangement involving chromosomes 9, 12, and 17 resulting in ETV6 (TEL) and ABL fusion. Cancer Genet. Cytogenet. 2005;157:74–77. doi: 10.1016/j.cancergencyto.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 20.Gancheva K., Virchis A., Howard-Reeves J., Cross N.C., Brazma D., Grace C. Myeloproliferative neoplasm with ETV6-ABL1 fusion: a case report and literature review. Mol. Cytogenet. 2013;6:39. doi: 10.1186/1755-8166-6-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malone A., Langabeer S., O’Marcaigh A., Storey L., Bacon C.L., Smith O.P. A doctor(s) dilemma: etv6-abl1 positive acute lymphoblastic leukaemia. Br. J. Haematol. 2010;151:101–102. doi: 10.1111/j.1365-2141.2010.08323.x. [DOI] [PubMed] [Google Scholar]
- 22.Brunel V., Lafage-Pochitaloff M., Alcalay M., Pelicci P.G., Birg F. Variant and masked translocations in acute promyelocytic leukemia. Leuk. Lymphoma. 1996;22:221–228. doi: 10.3109/10428199609051752. [DOI] [PubMed] [Google Scholar]
- 23.Andreasson P., Johansson B., Carlsson M., Jarlsfelt I., Fioretos T., Mitelman F. BCR/ABL-negative chronic myeloid leukemia with ETV6/ABL fusion. Genes Chromosomes Cancer. 1997;20:299–304. [PubMed] [Google Scholar]
- 24.Van Limbergen H., Beverloo H.B., van Drunen E., Janssens A., Hählen K., Poppe B. Molecular cytogenetic and clinical findings in ETV6/ABL1-positive leukemia. Genes Chromosomes Cancer. 2001;30:274–282. [PubMed] [Google Scholar]
- 25.Lin H., Guo J.Q., Andreeff M., Arlinghaus R.B. Detection of dual TEL-ABL transcripts and a Tel-Abl protein containing phosphotyrosine in a chronic myeloid leukemia patient. Leukemia. 2002;16:294–297. doi: 10.1038/sj.leu.2402353. [DOI] [PubMed] [Google Scholar]
- 26.Keung Y.K., Beaty M., Steward W., Jackle B., Pettnati M. Chronic myelocytic leukemia with eosinophilia, t(9;12)(q34;p13), and ETV6-ABL gene rearrangement: case report and review of the literature. Cancer Genet Cytogenet. 2002;138:139–142. doi: 10.1016/s0165-4608(02)00609-x. [DOI] [PubMed] [Google Scholar]
- 27.La Starza R., Trubia M., Testoni N., Ottaviani E., Belloni E., Crescenzi B. Clonal eosinophils are a morphologic hallmark of ETV6/ABL1 positive acute myeloid leukemia. Haematologica. 2002;87:789–794. [PubMed] [Google Scholar]
- 28.Meyer-Monard S., Mühlematter D., Streit A., Chase A.J., Gratwohl A., Cross N.C. Broad molecular screening of an unclassifiable myeloproliferative disorder reveals an unexpected ETV6/ABL1 fusion transcript. Leukemia. 2005;19:1096–1099. doi: 10.1038/sj.leu.2403697. [DOI] [PubMed] [Google Scholar]
- 29.Mozziconacci M.J., Sainty D., Chabannon C. A fifteen-year cytogenetic remission following interferon treatment in a patient with an indolent ETV6-ABL positive myeloproliferative syndrome. Am. J. Hematol. 2007;82:688–689. doi: 10.1002/ajh.20873. [DOI] [PubMed] [Google Scholar]
- 30.Baeumler J., Szuhai K., Falkenburg J.F., van Schie M.L., Ottmann O.G., Nijmeijer B.A. Establishment and cytogenetic characterization of a human acute lymphoblastic leukemia cell line (ALL-VG) with ETV6/ABL1 rearrangement. Cancer Genet. Cytogenet. 2008;185:37–42. doi: 10.1016/j.cancergencyto.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 31.Zaliova M., Moorman A.V., Cazzaniga G., Stanulla M., Harvey R.C., Roberts K.G. Characterization of leukemias with ETV6-ABL1 fusion. Haematologica. 2016;101:1082–1093. doi: 10.3324/haematol.2016.144345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salaverria I., Martín-Garcia D., López C., Clot G., García-Aragonés M., Navarro A. Detection of chromothripsis-like patterns with a custom array platform for chronic lymphocytic leukemia. Genes Chromosomes Cancer. 2015;54:668–680. doi: 10.1002/gcc.22277. [DOI] [PMC free article] [PubMed] [Google Scholar]