Abstract
Elevated serum soluble (s) Suppressor of Tumorigenicity 2 (ST2) is observed during cardiovascular and inflammatory bowel diseases. To ascertain whether modulated ST2 levels signify heart (HTx) or small bowel transplant (SBTx) rejection, we quantified sST2 in serially obtained pediatric HTx (n=41) and SBTx recipient (n=18) sera. At times of biopsy-diagnosed HTx rejection (cellular and/or antibody-mediated), serum sST2 was elevated compared to rejection-free time points (1714±329 vs. 546.5±141.6 pg/ml; P=0.0002). SBTx recipients also displayed increased serum sST2 during incidences of rejection (7536±1561 vs. 2662±543.8 pg/ml; P=0.0347). Receiver operator characteristic (ROC) analysis showed that serum sST2>600 pg/ml could discriminate time points of HTx rejection and non-rejection (Area under the curve (AUC)=0.724±0.053; P=0.0003). ROC analysis of SBTx measures revealed a similar discriminative capacity (AUC=0.6921±0.0820; P=0.0349). Quantitative evaluation of both HTx and SBTx biopsies revealed rejection significantly increased allograft ST2 expression. Pathway and Network Analysis of biopsy data pinpointed ST2 in the dominant pathway modulated by rejection and predicted TNF-α and IL-1β as upstream activators. In total, our data indicate that alloimmune-associated pro-inflammatory cytokines increase ST2 during rejection. They also demonstrate that routine serum sST2 quantification, potentially combined with other biomarkers, should be investigated further to aid in the non-invasive diagnosis of rejection.
INTRODUCTION
Solid organ transplantation prolongs survival and increases quality of life in both adult and pediatric patients with end-stage organ failure (1, 2). Evolving surgical techniques and immunosuppressant therapies have resulted in 1-year graft survival exceeding 90% following kidney transplantation and 80% for those receiving heart, lung, liver or intestine allografts (1, 3, 4). Yet poor long-term patient outcomes and unacceptable transplant (Tx) attrition rates persist in both adult and pediatric recipients (1, 3, 4). Significant innovations in post-Tx therapeutic and diagnostic care are most likely required to make substantial improvements to late post-Tx outcomes.
The current gold standard for establishing allograft rejection is allograft biopsy. Yet, this costly and invasive procedure is acknowledged to be poorly suited for routine monitoring of Tx recipients, particularly children and those recipients needing frequent assessments due to a high risk of rejection (5, 6). Likewise, the limited sample area provided by biopsy, combined with subtle graft pathology found in early rejection, may lead to false-negatives or “indeterminate” diagnosis (7, 8). The subjective nature of pathologist diagnosis and inherent differences between individual pathologist scoring can also cloud biopsy interpretations (9, 10). Given the lack of any reliable alternatives, biopsy remains of paramount importance for rejection diagnosis. However, the establishment of sensitive and reliable serum biomarkers of early allograft rejection would be a significant improvement in our ability to care for organ Tx recipients. Ideally, a rejection biomarker would be alloimmune-driven and graft-derived, thus quantification of its levels in the serum would provide a sensitive and specific readout for rejection diagnosis and also aid monitoring of effective rejection treatment.
Since its identification as an IL-1 family member in 2005 (11), the barrier cell cytokine, IL-33, and its receptor, Suppressor of Tumorigenicity-2 (ST2), have emerged as multifunctional immune regulators, as well as indicators of local inflammation and cell stress (12, 13). Like other IL-1 cytokines, IL-33 is regulated by a decoy receptor, or soluble ST2 (sST2), that is generated through alternative RNA splicing and processing of the ST2 RNA (14-17). In vitro, both the membrane-bound (ST2) and soluble (sST2) isoforms are induced or augmented in both leukocytes and non-hematopoietic cells by pro-inflammatory stimuli such as IL-1β, TNFα, or lipopolysaccharide (LPS) (18-20), as well as mechanical stress (21). Measurement of circulating sST2 is predictive of cardiovascular disease risk and mortality in patients with symptomatic heart failure (22-26) or after myocardial infarction (27-29). Soluble ST2 is also increased in the inflamed bowel and serum in patients with ulcerative colitis (30). High plasma levels of sST2 at the time of diagnosis of graft-versus-host disease (GVHD) are prognostic of treatment resistance and death (31). Thus, accumulating evidence in both experimental models and clinical assessments supports a hypothesis that increasing local and systemic expression of ST2 may serve as valuable measure of pathological inflammation.
Given the identified upregulation of ST2 and sST2 during pathological inflammatory conditions of the vasculature and mucosa, we aimed to define if significant modulations of graft and/or serum ST2 are indicative of acute rejection after solid organ transplantation. We also sought to provide mechanistic insight into the molecular underpinnings of regulation of ST2 expression in transplanted tissues.
Material and Methods
Pediatric Heart Transplant Recipients
We identified 41 children (mean age 8.0±6.3 years; see Table 1) enrolled in IRB-approved (IRB# 0702122, IRB# PRO13050191) observational studies after heart transplantation at Children's Hospital of Pittsburgh of UPMC, who had serial serum samples with an associated pathologist-graded endomyocardial biopsy (EMB) at the time of serum collection. In the clinical management of these patients, all received intravenous thymoglobulin induction therapy (total 7.5 mg/kg) with subsequent tacrolimus-based immunosuppression, plus adjunctive maintenance therapy with oral mycophenolate mofetil or sirolimus. Patients with a positive, donor-specific cytotoxicity crossmatch and/or those with antibody-mediated rejection (AMR) or recurrent acute cellular rejection (ACR) were treated with steroids as part of their maintenance immunosuppression. In the first year post-transplantation, patients underwent routine post-HTx allograft surveillance EMB and serum collection at approximately 1-2 weeks, 2-4 weeks, 2 months, 4 months, 6-7 months, and 10-12 months post-transplantation. Patients also underwent EMB if rejection was suspected and to assess resolution following the anti-rejection treatment. Serum samples were isolated at the time of collection, frozen the same day, and stored at −80°C until use.
Table 1.
Clinical Characteristics of Heart Transplant Recipients
| Total number of patients | 41 | 100% |
| Males | 23 | 56% |
| Females | 18 | 44% |
| Recipient age at transplant | ||
| Mean | 8.0 ±6.3 | |
| Median | 7.0 | |
| Recipient age at transplant <18y | 39 | 95% |
| Recipient age at transplant ≥18y | 2 | 5% |
| Etiology of heart failure | ||
| Congenital heart disease (CHD) | 13 | 31.7% |
| Cardiomyopathy (CM) | 24 | 58.5% |
| Re-Transplantation (ReTx) | 4 | 9.8% |
| Ave. Graft Survival (Years as of 11/18/2013) | 4.53 ±1.4 | |
| Sex Matched Donor (Y) | 51% | |
| Ethnicity/Race | ||
| White | 33 | 80.5% |
| Arabic | 1 | 2.4% |
| White/Asian | 1 | 2.4% |
| White/Arabic | 1 | 2.4% |
| Black | 4 | 9.8% |
| Hispanic | 1 | 2.4% |
Small Bowel Transplant Recipients
Samples from 18 individuals (mean age 9.6±14.5 years; see Table 2) undergoing isolated small bowel or multivisceral transplantation at the University of Nebraska Medical Center from 2004 to 2010 were obtained from an established, IRB-approved tissue collection bank (IRB #417-02). All SBTx recipients received organs from cadaveric donors with identical or compatible ABO blood types. Donors were pretreated with anti-thymocyte globulin and basiliximab. In the clinical management of these SBTx recipients, induction immunosuppression consisted of corticosteroid and basiliximab, while corticosteroid and tacrolimus were used for maintenance therapy. All patients underwent serial post-SBTx allograft surveillance biopsies weekly for the first four weeks. For-cause biopsies were obtained when rejection was suspended or to assess resolution following the treatment of acute rejection. Sequentially obtained material available for experimental assessment included extra biopsy specimens in RNAlater (Applied Biosystems/Ambion, Austin, TX) and serum, both stored at −80°C.
Table 2.
Clinical Characteristics of Small Bowel Transplant Recipients
| Total number of patients | 18 | 100% |
| Males | 7 | 39% |
| Females | 11 | 61% |
| Recipient age at transplant | ||
| Mean | 9.6 ±14.5 | |
| Median | 3.4 | |
| Recipient age at transplant <18y | 15 | 83% |
| Recipient age at transplant ≥18y | 3 | 17% |
| Etiology of intestinal failure | ||
| Necrotizing enterocolitis (NEC) | 1 | 6% |
| Atresia | 3 | 17% |
| Volvulus | 2 | 11% |
| Gastroschisis | 3 | 17% |
| Pseudo-obstruction | 3 | 17% |
| Microvillus inclusion disease (MVID) | 4 | 22% |
| Hisrshsprung | 2 | 11% |
| Isolated bowel transplant | 3 | 17% |
| Multivisceral transplant | 15 | 83% |
Quantum Dot (Qdot) Immunolabeling, High-Resolution Whole-Slide Scanning and Analysis
Paraffin-embedded HTx recipient EMB were sectioned (4 μm) onto slides, deparaffinized, steamed with antigen retrieval buffer (pH 9.0; 30 minutes), and blocked in avidin/biotin (Vector Labs, Burlingame, CA) and Serum-Free Protein Block (Dako, Carpinteria, CA). Slides were labeled overnight with rabbit anti-ST2 antibody (1:30; Sigma Aldrich) washed in phosphate-buffered saline, and incubated with anti-rabbit biotinylated IgG secondary antibody (Vector Labs). After washing and avidin/biotin blocking, streptavidin conjugated Qdot 705 was applied. Washed slides were stained with Hoechst nuclear dye, dehydrated, and coverslipped. Whole Slide Images (WSI) were captured via a Zeiss Mirax MIDI scanner utilizing a Plan-Apochromat 40x/.95N.A. objective lens, AxioCam MRm digital CCD camera (Carl Zeiss, Jena, Germany), and specifically selected excitation/emission Qdot filters (Omega Optical, Brattleboro, VT) as described (32, 33). Pixel-based image analytics were performed on the WSI utilizing the internally developed-IAE-NearCYTE (http://nearcyte.org) imaging software. Fluorophore-labeled WSI were analyzed with IAE-NearCYTE as described (33). Briefly, using IAE-NearCYTE Region of Interest (ROI) tool was used to select satisfactory areas to be analyzed (typically 4-6 fragments) and unacceptable areas (i.e. blood clots, tissue folds, etc.) excluded. Following the establishment of thresholds for fluorophore positivity, the software automatically generated an Area Ratio value (ROI Fluorescence Area normalized to total ROI area). Area Ratio values for each WSI sample were exported for analysis.
ELISA
HTx and SBTx recipient serum sST2 levels were measured by ELISA (DuoSet, R&D Systems, Minneapolis, MN). On the day of assessment, samples were batch thawed and samples were analyzed in triplicate according to manufacturer specifications. Assay limit of detection was 31.25 pg/mL. Absorbance was measured utilizing a Benchmark Plus Reader (Bio Rad) at a wavelength setting of 450 nm.
Quantitative Real-Time (qRT)-polymerase chain reaction (PCR) and Pathway Analysis
Intestinal allograft biopsy tissue mRNA expression for ST2 was measured as part of an Inflammatory Cytokines and Receptors qRT-PCR array (Qiagen, Fredericksburg, MD). A dataset (Appendix 2) containing gene identifiers and corresponding expression values were uploaded into Ingenuity Pathway Analysis (IPA) Software (Ingenuity© Systems, Redwood City, CA; www.ingenuity.com) and analyzed. Up and down regulated genes were considered in the analysis using the Inge nuity Knowledge base. IPA Upstream Regulator Analysis was utilized to rank molecules upstream of modulated networks that potentially explain observed changes.
Statistical Analysis
Data analysis of ELISA and Qdot data was completed with Prism 6 Software (GraphPad Software, La Jolla, CA), except for the linear mixed models (34), which were fit using Stata (StataCorp LP, College Station, TX). All tests were 2-sided and P values of <0.05 was considered significant. Measurement distributions were evaluated using the D'Agostino-Pearson test of normality. In all cases, the measurements were non-normal and differences between groups were therefore transformed (for the mixed model regression analysis) or tested using non-parametric statistics (e.g. the Wilcoxon-Mann-Whitney rank sum test or receiver operating characteristic (ROC) curve analysis). Since there were multiple measurements on some subjects, group effects were tested using a linear mixed model, where log-transformed sST2 was the outcome, the subject ID was the random effect, and the group variable (e.g. rejecters versus non-rejecters) was the time-varying effect of interest. Analyses were also repeated using fold-change in sST2 (adjusted for the baseline value); however, since these results were very similar to analysis of the actual values, they were excluded from the main results. ROC curve analysis was constructed to establish the capacity of sST2 ELISA measures to discriminate rejection relative to non-rejection. Area under the curve (AUC) was calculated as a measure of discriminatory ability; the analysis was repeated using the average sST2 value for a given subject. In assessment of SBTx biopsies by qRT-PCR, fold-Change (2−ΔΔCT) was calculated as normalized gene expression (2−ΔCT) in the Test Sample divided by the normalized gene expression (2−ΔCT) in the Control Sample. The p-values were calculated based on a Student's t-test of the replicate 2−ΔΔCT values for each gene in the control group and treatment groups. In IPA analysis, fold change treated-to-control ≥1.5 and p≤0.05 cutoffs were used to determine significantly modulated networks and Upstream Regulator Analysis outcomes were based on both absolute z-score and p-value.
RESULTS
Local ST2 expression is increased in pediatric HTx patients at the time of rejection
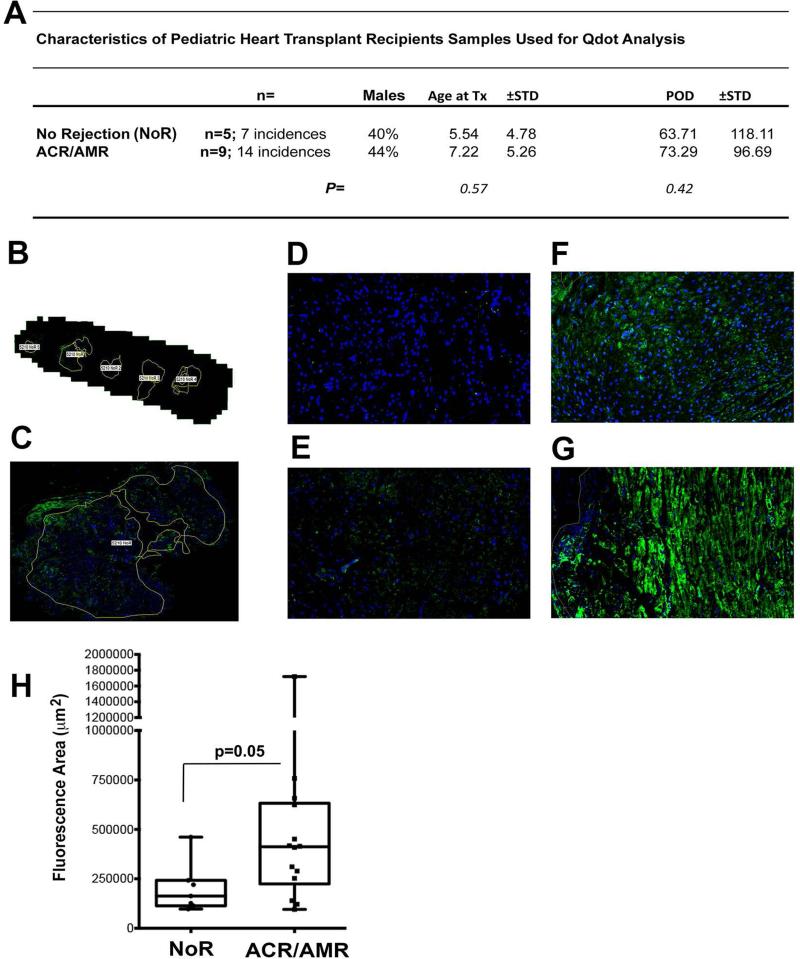
We previously established that ST2 is increased in acutely rejecting heterotopic murine heart transplants, but not in the endogenous recipient hearts (35). These rodent data supported a quantifiable local increase in allograft ST2 during rejection. To define if ST2 is modulated during clinical pediatric HTx rejection, we utilized an antibody recognizing both isoforms of ST2 to investigate staining patterns in EMB obtained from a cohort of pediatric HTx recipients. The samples were from times of pathologist-diagnosis of ACR (ISHLT grade ≥2R) or AMR (ISHLT grade ≥2), as well samples of similar time frame from distinct patients that remained free of both ACR or AMR (No Rejection; NoR) in the first year following transplantation (See Appendix 1: for more details on experimental samples). General characteristics for patients from which EMB were assessed by Qdot-based immunostaining are summarized in Fig. 1A - Table and a represented WSI with ROI indicated in Fig. 1B-C. As depicted in Fig. 1, non-rejecting group samples (representative samples from POD 331; Fig. 1D and POD 26; Fig. 1E) display areas of limited ST2 positive cells. In contrast, EMB of patients at the time of diagnosed AMR (Fig. 1F; POD 11) and ACR (Fig. 1G; POD 25) show marked ST2 staining through-out the allograft. When ST2 Area Ratio measures were generated using IAE-NearCYTE, we found a significant increase (P=0.05) in ST2+ values in AMR and ACR biopsies vs. comparable controls (Fig. 1H). Overall, these data are consistent with our previously described rodent HTx studies (35) and support significant local increases in allograft ST2 protein expression as a result of HTx rejection.
Figure 1. ST2 is augmented in HTx biopsies at the time of rejection.
Quantum dot (Qdot)-based immunostaining for ST2 was completed on ISHLT-graded pediatric heart transplant (HTx) patient endomyocardial biopsies (EMB). (A) The table depicts demographics for pediatric HTx patients whose EMB were stratified into groups based on pathologist classification at the time of biopsy as those suffering ACR, exhibiting evidence of AMR, or those free of both at the time of EMB assessment (No Rejection; NoR). (B) Digital image depicts the capture of a whole slide image (WSI) of a Non-rejecting subject EMB generated utilizing NearCyte software after Mirax MIDI scanning using Qdot-appropriate filters (B; 1X; C; 5X; DAPI (Blue); ST2 (Green). Representative examples of NearCyte image captures (at a magnification of 40X) for EMB at time points diagnosed by a pathologist as: (D-E) No Rejection, (F) AMR (ISHLT Grade 2 and C4d+), or (G) ACR (ISHLT Grade 2R) are also presented. (H) NearCYTE WSI analysis software was further used to assess HTx patient EMB by quantitating ST2 staining. After manual outlining of appropriate EMB regions for each sample (See A-B) was completed, an Area Ratio (Fluorescent+ Area/Total Field Area across all EMB regions) for that subject at that time was automatically calculated for ST2+ staining. The graph depicts box whisker plots, with the median depicted as the line inside each box, the upper box border representing the 75th quartile and the lower box border representing the 25th quartile. Whiskers represent the range and the indicated significance level was calculated through Wilcoxon-Mann-Whitney rank sum test comparison.
Elevated serum sST2 distinguishes HTx rejection
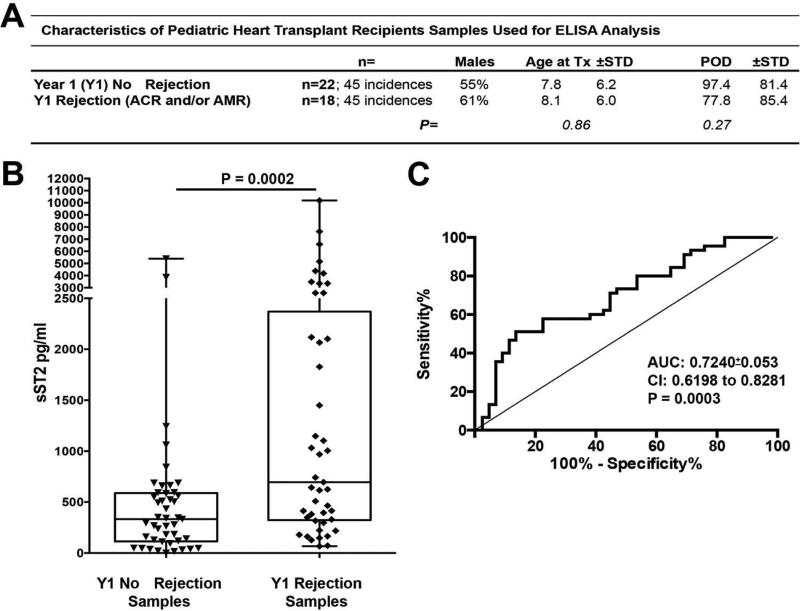
Next, sST2 ELISA measures were determined for all available serum samples for the first year post-HTx for each recipient in our HTx cohort (n=39). Statistical comparisons were completed on these year 1 (Y1) measures and time points classified as rejection-free (NoR; ACR and AMR Grade=0 and C4d−) were compared to measures at times of pathologist diagnosed rejection (ACR Grade>2R and/or histological evidence for pathological AMR (AMR) and C4d+), independent of the recipient. One recipient's samples were abnormally low and excluded from analysis. Fig. 2A-Table provides HTx recipient demographics for serum samples examined and groups analyzed. Additional details on each data point are provided in Appendix 1. Our evaluation revealed a highly significant (P=0.0002) increase in serum sST2 in pediatric HTx patients from the time of diagnosed rejection (Fig. 2B) as compared to times of non-rejection in Y1. ROC analysis depicted in Fig. 2C established that sST2 measures had moderate discriminative capacity for the identification of HTx rejection in Y1 (AUC=0.7240±0.053;P=0.0003); 58% Sensitivity and 79% Specificity at a 600 pg/ml cutoff). Using an estimate of 40% for the incidence of first year HTx rejection (36), Positive Predictive Value of 63% and Negative Predictive Value of 73% were calculated at the above sensitivity and specificity values. To be sure that repeated measures on some subjects were not inflating the AUC and associated P value, ROC analysis was repeated using Y1 mean values and doing so actually increased AUC measures (mean: AUC:0.75±0.08; P=0.0071).
Figure 2. Elevated serum sST2 distinguishes pediatric HTx recipients suffering rejection episodes.
Year 1 (Y1) HTx recipient serum sST2 ELISA measures at time points of pathologist-diagnosed rejection (Y1 Rejection Samples; ACR Grade ≥2R and/or histological evidence for pathological AMR (AMR) and C4d+) were compared to time points where the recipient was classified as rejection-free (Y1 No Rejection Samples; ACR and AMR Grade=0 and C4d-). (A) Table depicts the demographics for the two groups. (B) ELISA values are presented as box whisker plots as in Fig. 1 and indicated significance levels calculated through a Wilcoxon-Mann-Whitney rank sum test comparison. (C) Receiver-operator characteristic (ROC) curve analysis of Y1 No Rejection Samples (Negative Control Group) and Y1 Rejection Samples (Positive Control Group). ACR, acute cellular rejection; AMR, antibody-mediated rejection; HTx, heart transplant.
As indicated in Fig. 2A - Table and Appendix 1, both Y1 Non-Rejection and Rejection measures included samples which were derived from one HTx recipient, potentially during the same rejection episode or, alternatively, rejection free period. Analysis of repeated measures with linear mixed models that account for dependency among measurements from a single subject, and the time-varying nature of rejection status, also found a significant effect of rejection status on sST2 (p=0.003).
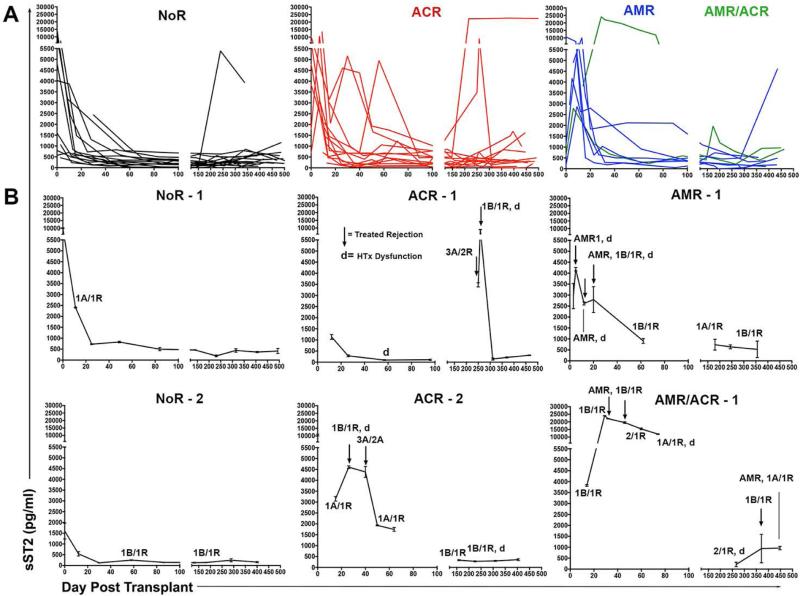
Next, we plotted changes in sST2 serum levels for first year post-HTx serum sST2 levels for 39 recipients. One recipient had only a limited number of samples from isolated time points and was not plotted. All data are summarized in Fig. 3, where data are grouped by Y1 outcomes as: 1. those having at least one or more incidence of diagnosed ACR (ISHLT grade≥2R), 2. those with histologically and immunohistochemistry (C4d+) indicated pathogenic AMR (ISHLT grade≥2) alone or ACR, and 3. recipients that remained free of ACR and AMR in year 1 post-HTx (NoR; Fig. 3A). One or more profiles representative of each group are also depicted in Fig. 3B. Nine of 14 HTx recipients suffering ACR exhibited levels of sST2 >600 pg/ml in the time point before or during diagnosed ACR (Fig. 3). Likewise, 8 of 10 recipients with diagnosed AMR or AMR/ACR displayed sST2 measures >600 pg/ml at the time of diagnosis (Fig. 3). While all the recipients in the NoR did display sST2 levels >600 pg/ml during the first few weeks after transplantation, only 4 of 15 exceeded this level after day 21 post-HTx (Fig. 3). Importantly, in the great majority of recipients (22 of 24) in the ACR or AMR groups, HTx rejection treatment returned and/or maintained sST2 at levels reflective of that of the No Rejection Group (550±142 pg/ml; see Fig. 2).
Figure 3. Serum sST2 is increased during HTx rejection and decreases following recipient treatment.
Circulating sST2 was assessed by ELISA in HTx recipient serum samples obtained serially in the first year post-transplant. (A) Changes of sST2 concentrations are depicted for all patients grouped into cohorts based on Year 1 (Y1) outcomes. Groups include patients suffering one or more episodes of diagnosed ACR (Grade≥2R) and/or histologically and C4d+ indicated pathogenic AMR, or those remaining free from ACR or AMR during Y1 (No Rejection; NoR). (B) Panels depict individual recipients representative of the indicated group. Black arrows indicate times of recipient treatment for rejection; d = points of graft dysfunction; All EMB grades over 0 indicated at appropriate time point. ACR, acute cellular rejection; AMR, antibody-mediated rejection; HTx, heart transplant; sST2, soluble ST2.
Serum sST2 is also elevated in pediatric SBTx recipients with rejection
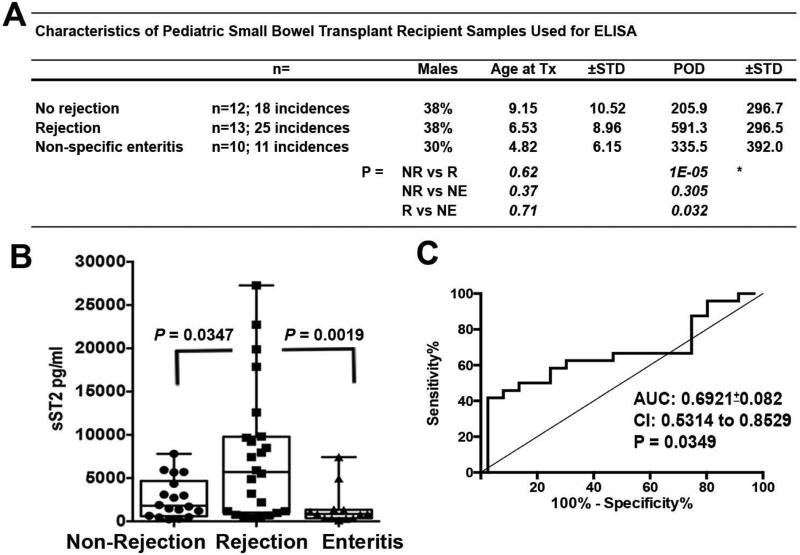
To investigate if sST2 may have the potential to act as a serum biomarker of rejection after transplant of other organs, we assessed sST2 in the circulation of a cohort of predominantly pediatric SBTx recipients time points of non-rejection, rejection, or non-specific enteritis without rejection (Demographics summarized in Fig. 4A-Table and Appendix 1). Given the limited number of subjects available, experimental comparisons were completed across all subjects regardless of day post transplant. Serum levels of sST2 were significantly (P=0.0347) increased at the time of pathologist-diagnosed mild, moderate, or severe rejection of SBTx compared to time periods when rejection was not diagnosed (Non-Rejection; Fig. 4B). Increases in sST2 appeared specific to rejection, as it differentiated rejection from non-specific enteritis in SBTx patients (Fig. 4B). ROC analysis suggested a discriminative capacity for serum ST2 to distinguish rejection in SBTx recipients (AUC:0.6921±0.082; P=0.0349; 62% Sensitivity and 72.2% Specificity at a cutoff of 3,150 pg/ml). As in above HTx recipient serum assessments, both Non-Rejection and Rejection groups included samples derived from one recipient. Due to our limited SBTx recipient sample size, when linear mixed model comparisons or individual mean values were used in ROC analysis, differences between the Non-Rejection and Rejection groups only approached, but no longer reached significance (P=0.07 and AUC=0.72 P=0.06). These data, while not definitive, indicate that serum sST2 is detectably elevated during allograft rejection in SBTx recipients and provide further evidence to support sST2 as a potential biomarker of solid allograft rejection.
Figure 4. Serum sST2 is elevated in pediatric SBTx recipients during diagnosed rejection episodes.
Circulating sST2 determinations were completed using ELISA on the serum collected from a cohort of SBTx recipients during periods of quiescence (No Rejection), rejection (R), or non-specific enteritis. (A) Table depicts the general demographics of these SBTx recipients. (B) ELISA values are presented as box whisker plots as in Figs. 1 and 2. Significance levels indicated were calculated using the Wilcoxon-Mann-Whitney rank sum test. (D) ROC curve analysis of No Rejection Samples (Negative Control Group) and Rejection Samples (Positive Control Group). SBTx, small bowel transplant; sST2, soluble ST2.
Graft expression of ST2 in small bowel transplants
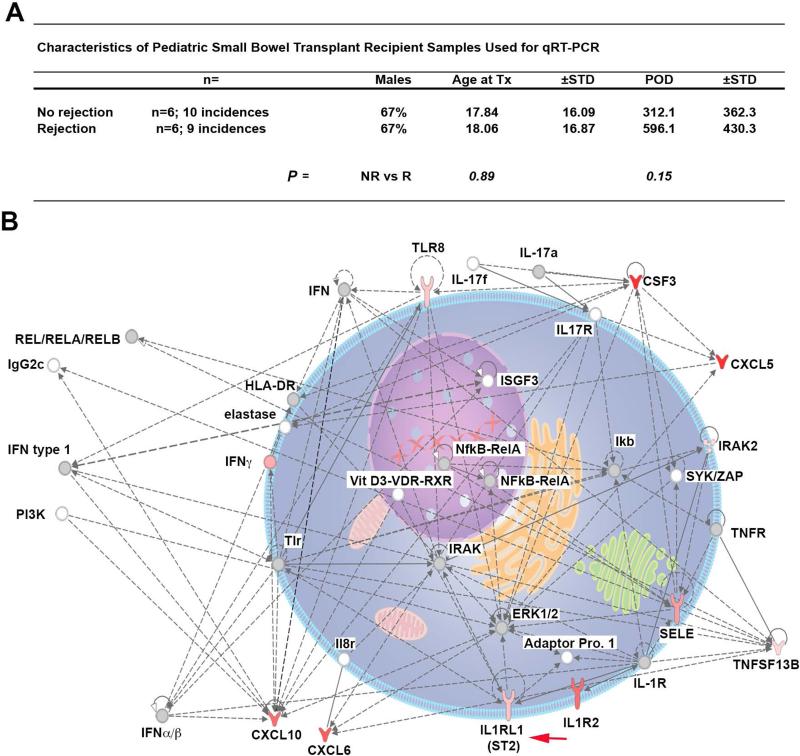
Analysis of mRNA expression of 384 immune-related genes expressed in rejection-free small bowel biopsies from 8 subjects and SBTx samples with pathologist-identified rejection (n=6) showed mean ST2 (IL1RL1) expression was significantly increased during SBTx rejection (3.9-fold increase vs. non-rejection; P=0.02; raw and analyzed data provided in Appendix 2). Thus, examination of biopsy mRNA expression supports distinct modulation of the ST2 axis during SBTx rejection.
Upregulated ST2 (IL1RL1) in network of significantly modulated genes
Of the 384 assessed, in addition to ST2, it was determined that the mRNA message for an additional 35 genes were significantly regulated during SBTx rejection (Table 3). Comparing regulated expression in rejection samples relative to those from non-rejection samples, we observed an increase in 32 gene products and 4 with decreased expression (Table 3 and Appendix 2). To further understand the regulation of genes involved in SBTx rejection, these data were probed using IPA Pathway and Network Analysis that generates associated pathways incorporating sets of upregulated and downregulated genes. Based on Fold Changes and P values, the top modulated network contained ST2 and the gene-gene interaction representation of this network is depicted (Fig. 5). This dataset was further analyzed using IPA to determine upstream transcriptional regulators that could explain the observed gene expression changes. As depicted in Table 4, IPA Upstream Regulator Analytic identified that TNFα, LPS, IL-1β, and IFNγ were the predicted activators of this network containing ST2.
Table 3.
Gene Up-Down Regulation During SBTx Rejection
| Increased | |||
|---|---|---|---|
| Gene | Fold Regulation (Rejection relative to No Rejection) | p-value | |
| 1 | CSF3 | 14.8 | 0.024 |
| 2 | CXCL5 | 13.2 | 0.049 |
| 3 | CXCL6 | 10.4 | 0.019 |
| 4 | CXCL10 | 9.4 | 0.044 |
| 5 | IL1R2 | 9.4 | 0.041 |
| 6 | CSF2 | 8.4 | 0.025 |
| 7 | SELE | 6.3 | 0.015 |
| 8 | CTLA4 | 6.1 | 0.042 |
| 9 | MMP9 | 5.4 | 0.020 |
| 10 | IFNG | 5.2 | 0.008 |
| 11 | MADCAM1 | 4.9 | 0.010 |
| 12 | BDKRB1 | 4.7 | 0.006 |
| 13 | CD80 | 4.6 | 0.020 |
| 14 | PLA2G7 | 4.2 | 0.031 |
| 15 | IL1RL1 | 3.9 | 0.023 |
| 16 | CCR4 | 3.9 | 0.022 |
| 17 | CCL2 | 3.7 | 0.012 |
| 18 | ICAM1 | 3.7 | 0.014 |
| 19 | PTAFR | 3.6 | 0.027 |
| 20 | TLR8 | 3.5 | 0.014 |
| 21 | OAS2 | 3.0 | 0.018 |
| 22 | ITGAM | 2.8 | 0.025 |
| 23 | ITGB2 | 2.6 | 0.017 |
| 24 | CD86 | 2.5 | 0.035 |
| 25 | TNFSF13B | 2.5 | 0.022 |
| 26 | IRAK2 | 2.4 | 0.032 |
| 27 | OASL | 2.3 | 0.033 |
| 28 | BDKRB2 | 2.2 | 0.043 |
| 29 | ISG15 | 2.2 | 0.043 |
| 30 | IFITM1 | 2.2 | 0.022 |
| 31 | RIPK2 | 2.1 | 0.044 |
| 32 | GBP1 | 2.1 | 0.047 |
| Not Significant | IL33 | 1.7 | 0.756 |
| Decreased | |||
| 1 | PPARA | −2.1 | 0.026 |
| 2 | ACE | −3.7 | 0.044 |
| 3 | IL5 | −4.6 | 0.021 |
| 4 | VIPR1 | −7.2 | 0.028 |
Figure 5. Dominant modulated gene network during acute rejection of small bowel transplants includes increased ST2.
Changes in expression of 384 inflammatory cytokines and receptors genes were determined by quantitative RT-PCR (qRT-PCR) in SBTx biopsies harvested at time points subjects were diagnosed as rejection-free (No rejection; n=10) or diagnosed with active rejection (Rejection; n=9). (A) General demographics of the SBTx subjects from which biopsy samples were utilized for qRT-PCR analysis. (B) After determination of fold change and associated P-values for each gene, the data set was further analyzed using Ingenuity Pathway Analysis (IPA; Ingenuity Systems Software). Depicted is a graphical representation of the highest-scored network generated from our data set. Level of upregulation is indicated by intensity of red color at that node. Gray nodes are part of network, but were not significantly modified between rejecting and non-rejecting samples. Solid lines indicate direct relationships, while dashed lines depicted indirect relationships. Red arrow notes ST2. SBTx, small bowel transplant; sST2, soluble ST2.
Table 4.
Predicted Network Upstream Regulator
| Regulator | Molecule Type | Activation z-score | p-value of overlap | |
|---|---|---|---|---|
| 1 | TNF | cytokine | 5.12 | 2.30E-36 |
| 2 | lipopolysaccharide | chemical drug | 4.65 | 7.18E-30 |
| 3 | IL1B | cytokine | 4.19 | 1.75E-23 |
| 4 | IFNG | cytokine | 4.05 | 2.62E-30 |
| 5 | poly rl:rC-RNA | chemical reagent | 4.05 | 1.08E-19 |
| 6 | MYD88 | other | 4.05 | 5.43E-22 |
| 7 | IRF7 | transcription regulator | 4.05 | 1.39E-18 |
| 8 | IL1 | group | 4.05 | 1.09E-19 |
| 9 | TICAM1 | other | 4.05 | 1.25E-17 |
| 10 | Interferon alpha | group | 4.05 | 4.25E-21 |
| 11 | TLR3 | transmembrane receptor | 4.05 | 3.77E-18 |
| 12 | TLR4 | transmembrane receptor | 4.05 | 6.61E-22 |
| 13 | NFkB (complex) | complex | 4.06 | 1.28E-15 |
| 14 | IFNL1 | cytokine | 4.06 | 1.10E-16 |
| 15 | P38 MAPK | group | 4.06 | 4.21E-18 |
| 16 | IL6 | cytokine | 4.06 | 5.01E-16 |
| 17 | E. coli B5 lipopolysaccharide | chemical - endogenous non-mammalian | 4.06 | 4.61E-16 |
| 18 | IL17A | cytokine | 4.06 | 7.38E-15 |
| 19 | E. coli lipopolysaccharide | chemical - endogenous non-mammalian | 4.06 | 3.41E-15 |
| 20 | ERK | group | 4.06 | 1.62E-10 |
DISCUSSION
In this study we make several novel findings that support the development of serum ST2 assessment as a means to potentially aid diagnosis and treatment of transplant rejection, possibly in conjunction with other biomarkers. We recapitulates our findings in rodent HTx studies (35) and provide evidence for increased ST2 expression in both pediatric HTx and SBTx recipients biopsies during allograft rejection. To our knowledge, these data represent the first clinical demonstration of increased ST2 within rejecting allografts and suggest the graft as a source of serum sST2. Consistent with a recent study of adult HTx recipients (37), we also demonstrate that circulating sST2 is elevated during acute rejection in pediatric HTx recipients and serum sST2 measures provide a moderate degree of discrimination in identifying HTx rejection episodes. By establishing a comparable increase in serum sST2 during SBTx rejection, we also extend these finding beyond heart transplantation. These data not only represent the first assessment of ST2 modulation following SBTx, but also support our hypothesis that circulating levels of sST2 increase due to inflammation associated with alloimmunity, and not only as a result of heart damage and myocardial strain and stress as previously advocated (38). That serum sST2 increases do not reflect cardiovascular pathology, but instead indicate local pro-inflammatory events and associated cytokines, is supported by studies reporting serum sST2 increases in other inflammatory diseases, including GVHD, inflammatory bowel disease, ulcerative colitis, sepsis, and juvenile idiopathic arthritis (30, 31, 39-41). Similar to reported serum sST2 decreases with successful anti-TNF therapy for ulcerative colitis (30), we now find that treatment of HTx rejection results in a reduction of sST2 serum levels to those typical of quiescent periods. In total, our data, and that of others, suggest the potential for sST2 to act as a biomarker of immune-mediated pathologies, including alloimmunity.
Qdot-based immunostaining of pediatric HTx EMB revealed that ST2 is minimally expressed in quiescent HTx biopsies, but profoundly increases during ACR and AMR. These data are also consistent with local inflammation acting as the driving force for ST2 augmentation in HTx tissue. Gene expression analysis of SBTx biopsies revealed an analogous upregulation of ST2 message in samples acquired during rejection. Increased ST2 is a member of the most dominant modulated gene network and this network is most likely accounted for by the upstream activity of TNFα, LPS, IL1β, and IFNγ. Our data are in agreement with multiple previous in vitro determinations that found IL-1β, TNFα, or LPS to drive production of sST2 (18-20). In total, local pro-inflammatory cytokines, such as IFNγ from infiltrating T cells or TNFα and IL-1β from inflammatory macrophages, are the likely mediators of the significant increases in circulating sST2 during rejection.
A shortcoming with previously evaluated transplant rejection biomarkers is that they fail to distinguish rejection from infection (42). Our assessment of serum sST2 in SBTx recipients suggests it may be unique in this regard. Our group noted increased sST2 in the serum of SBTx patients during episodes of histologic intestinal allograft rejection, but not enteritis. Features of acute SBTx rejection are well-described (43). Likewise, histologic findings of a mixed inflammatory infiltrate including eosinophils, >6 apoptotic bodies per 10 crypts and a lack of viral inclusions allow the differentiation of rejection from enteritis (44, 45). That said, the exact cause of non-specific enteritis is unknown in the majority of our studied patients, and could be due a number of issues, including viral infection, medication reaction, or bacterial overgrowth. While our initial observation that sST2 levels may distinguish rejection from infection is promising, expanded study including more precise characterization of enteritis etiology as it relates to sST2 levels is needed.
Although our study reveals significant findings, it is not without limitations that may be addressed in future expanded studies. One limitation is that our biopsy-based human assessments do not distinguish between ST2 and sST2, but utilized probes that recognize shared regions of both. The future development of Qdot-based staining compatible antibodies that are able to distinguish ST2 isoforms based on unique C-terminal domains or selective qRT-PCR probes should clarify this issue.
A second limitation results from the sample sizes of both our HTx and SBTx cohorts. Both HTx and SBTx are relatively rare procedures. The limited availability of pediatric HTx and SBTx samples drove our current need to utilize a cross-sectional, retrospective study design. Likewise, in our HTx recipient analyses, we combined patients with ACR and AMR. Although ACR and AMR both represent alloimmune-mediated graft damage, we do appreciate that they represent distinct forms of rejection. Yet the primary goal of our study was to establish if rejection, be it ACR or AMR, modulated ST2, either locally or systemically in pediatric Tx recipients. Despite the sample size limitations and combining of rejection groups, our data examining serum sST2, as well patient HTx expression of this protein, make a convincing case for quantifiable local and systemic increases in ST2 during HTx rejection.
In this report, we evaluated the same group of HTx or SBTx recipients during periods of quiescence, episodes of rejection, and post-rejection treatment. This design allowed us to both leverage cohorts of relatively rare subjects and also provide insight into chronological changes in circulating sST2 relative to rejection diagnosis and treatment. Our serial observations during Y1 pediatric HTx recipient measures extend adult HTx data findings reported by Pascual-Figal et al (37). Together, these two HTx studies demonstrate how sST2 levels are elevated before or during ACR and AMR, as well as return to non-rejection point levels with successful rejection treatment. We also observed in our cohort of SBTx patients that 9 of the 13 SBTx recipients experiencing rejection had sST2 levels exceeding the above-described cutoff value of 3,150 pg/ml during the rejection episode (Supplemental Fig. 1). Likewise, rejection treatment tended to be associated with decreased sST2 levels (Supplemental Fig. 1). While encouraging and supportive of HTx data, these observations are complicated by the fact that sST2 levels decreased even during unsuccessful rejection treatment resulting in graft loss, as all three subjects experiencing graft loss also displayed a subsequent profound decrease in serum sST2 (Supplemental Fig. 1). Thus, while our SBTx data reveal that sST2 increases are associated with rejection episodes, expanded studies in SBTx will be required to define if differences in sST2 levels are acting as a surrogate for successful rejection treatment or result from loss of sST2 producing cells during severe SBTx rejection.
In total, we have completed the first simultaneous biopsy and serum based assessment of ST2 in two distinct pediatric transplant populations. These biopsy analyses establish that significant increases in ST2 occur in the allograft during clinical solid organ rejection. More importantly, our current studies also demonstrate that routine serum sST2 quantification may have value in aiding the non-invasive diagnosis and treatment of solid organ rejection. Most evaluated rejection biomarkers, such as cardiac-specific troponins and B-type natriuretic peptide for HTx rejection, are released from damaged cardiomyocytes, thus only distinguish episodes of severe rejection (46). As sST2 produced by cardiac or other inflamed tissues can be immediately secreted, it is expected that sST2 serum measurements will aide in the detection of early acute Tx rejection. This may be especially true if the sensitivity of sST2 measures can be increased through the use of assay methods more sensitive than the research grade ELISA kits used presently. The high-sensitivity Presage ST2 Assay was recently FDA-cleared for patient evaluation after heart failure (47) and its use for evaluation of solid organ transplant recipients should be particularly informative. In the case of HTx rejection diagnosis, assessment of sST2 measures as part of a biomarker panel including potential HTx rejection biomarkers, such as circulating cell-free donor DNA (48), and indicators of cardiac tissue damage (46, 49, 50) also warrants investigation. Supported by our finding that local inflammatory cytokines are the predicted drivers of allograft ST2 expression, we expect such investigations in larger prospective studies will establish serum sST2 quantification as an effective way to support biopsy-free detection of early allograft rejection and also enable biopsy-free monitoring of anti-rejection therapy to aide effective therapeutic resolution of alloimmune responses.
Supplementary Material
Acknowledgments
We thank Carla Forsythe for her assistance in generation and editing of the manuscript. We also acknowledge the valuable statistical assistance of Kun-Wan Chen, MS. To complete this work H.R.T. was supported by awards from the National Institutes of Health(NIH)/National Heart Lung and Blood Institute (NHLBI; R00 HL097155), Roche Organ Transplant Research Foundation (Grant Award # 5891142), and American Heart Association (14GRNT2040004), as well as an AST/Pfizer Basic Science Faculty Development Grant. B.F. was supported by a grant from NIH/National Center for Advancing Translational Sciences (NCATS; KL2TR000146) and a Children's Hospital of Pittsburgh of UPMC Research Advisory Committee award. Support to R. T. F. was derived from a University of Nebraska Medical Center Lozier Pediatric Research Grant. J.M.L. was supported by NIH/ National Institutes of Allergy and Infectious Diseases (T32AI089443) and a subsequent, individual UNCF/Merck Science Initiative grant.
Abbreviations
- ACR
acute cellular rejection
- AMR
antibody-mediated rejection
- AU
arbitrary units
- AUC
area under the curve
- ELISA
enzyme-linked immunosorbent assay
- EMB
endomyocardial biopsy
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- GVHD
graft-versus-host disease
- HTx
heart transplant
- ISHLT
International Society for Heart and Lung Transplantation
- LPS
lipopolysaccharide
- qRTPCR
quantitative real time polymerase chain reaction
- Qdot
Quantum dot
- ROC
receiver operator characteristic
- ROI
region of interest
- SBTx
small bowel transplant
- sST2
soluble ST2
- ST2
growth stimulation gene-2
- TNF
tumor necrosis factor
- Tx
transplant
- WSI
whole slide images
Footnotes
Disclosure: The authors of this manuscript have no conflicts of interest to disclose as described by the American Journal of Transplantation.
Supporting Information
Additional Supporting Information may be found in the online version of this article.
References
- 1.Lodhi SA, Lamb KE, Meier-Kriesche HU. Solid Organ Allograft Survival Improvement in the United States: The Long-Term Does Not Mirror the Dramatic Short-Term Success. Am J Transplant. 2011;11(6):1226–1235. doi: 10.1111/j.1600-6143.2011.03539.x. [DOI] [PubMed] [Google Scholar]
- 2.LaRosa C, Baluarte HJ, Meyers KE. Outcomes in pediatric solid-organ transplantation. Pediatr Transplant. 2011;15(2):128–141. doi: 10.1111/j.1399-3046.2010.01434.x. [DOI] [PubMed] [Google Scholar]
- 3.Lund LH, Edwards LB, Kucheryavaya AY, Dipchand AI, Benden C, Christie JD, et al. The Registry of the International Society for Heart and Lung Transplantation: thirtieth official adult heart transplant report--2013; focus theme: age. J Heart Lung Transplant. 2013;32(10):951–964. doi: 10.1016/j.healun.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 4.Dipchand AI, Kirk R, Edwards LB, Kucheryavaya AY, Benden C, Christie JD, et al. The Registry of the International Society for Heart and Lung Transplantation: sixteenth official pediatric heart transplantation report--2013; focus theme: age. J Heart Lung Transplant. 2013;32(10):979–988. doi: 10.1016/j.healun.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 5.Evans RW, Williams GE, Baron HM, Deng MC, Eisen HJ, Hunt SA, et al. The economic implications of noninvasive molecular testing for cardiac allograft rejection. Am J Transplant. 2005;5(6):1553–1558. doi: 10.1111/j.1600-6143.2005.00869.x. [DOI] [PubMed] [Google Scholar]
- 6.Hamour IM, Burke MM, Bell AD, Panicker MG, Banerjee R, Banner NR. Limited utility of endomyocardial biopsy in the first year after heart transplantation. Transplantation. 2008;85(7):969–974. doi: 10.1097/TP.0b013e318168d571. [DOI] [PubMed] [Google Scholar]
- 7.Kucirka LM, Maleszewski JJ, Segev DL, Halushka MK. Survey of North American pathologist practices regarding antibody-mediated rejection in cardiac transplant biopsies. Cardiovasc Pathol. 2011;20(3):132–138. doi: 10.1016/j.carpath.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 8.Talmon G, El Behery R, Radio S, Fischer R, Shostrom V, Wisecarver J. Crypt apoptotic count reproducibility in small bowel allograft biopsies. Int J Surg Pathol. 2013;21(3):257–260. doi: 10.1177/1066896912452912. [DOI] [PubMed] [Google Scholar]
- 9.Yang HM, Lai CK, Gjertson DW, Baruch-Oren T, Ra SH, Watts W, et al. Has the 2004 revision of the International Society of Heart and Lung Transplantation grading system improved the reproducibility of the diagnosis and grading of cardiac transplant rejection? Cardiovasc Pathol. 2009;18(4):198–204. doi: 10.1016/j.carpath.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 10.Angelini A, Andersen CB, Bartoloni G, Black F, Bishop P, Doran H, et al. A web-based pilot study of inter-pathologist reproducibility using the ISHLT 2004 working formulation for biopsy diagnosis of cardiac allograft rejection: the European experience. J Heart Lung Transplant. 2011;30(11):1214–1220. doi: 10.1016/j.healun.2011.05.011. [DOI] [PubMed] [Google Scholar]
- 11.Schmitz J, Owyang A, Oldham E, Song Y, Murphy E, McClanahan TK, et al. IL-33, an interleukin-1-like cytokine that signals via the IL-1 receptor-related protein ST2 and induces T helper type 2-associated cytokines. Immunity. 2005;23(5):479–490. doi: 10.1016/j.immuni.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 12.Liew FY, Pitman NI, McInnes IB. Disease-associated functions of IL-33: the new kid in the IL-1 family. Nat Rev Immunol. 2010;10(2):103–110. doi: 10.1038/nri2692. [DOI] [PubMed] [Google Scholar]
- 13.Liu Q, Turnquist HR. Implications for Interleukin-33 in solid organ transplantation. Cytokine. 2013;62(2):183–194. doi: 10.1016/j.cyto.2013.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bergers G, Reikerstorfer A, Braselmann S, Graninger P, Busslinger M. Alternative promoter usage of the Fos-responsive gene Fit-1 generates mRNA isoforms coding for either secreted or membrane-bound proteins related to the IL-1 receptor. EMBO J. 1994;13(5):1176–1188. doi: 10.1002/j.1460-2075.1994.tb06367.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thomassen E, Kothny G, Haas S, Danescu J, Hultner L, Dormer P, et al. Role of cell type-specific promoters in the developmental regulation of T1, an interleukin 1 receptor homologue. Cell growth & differentiation : the molecular biology journal of the American Association for Cancer Research. 1995;6(2):179–184. [PubMed] [Google Scholar]
- 16.Iwahana H, Yanagisawa K, Ito-Kosaka A, Kuroiwa K, Tago K, Komatsu N, et al. Different promoter usage and multiple transcription initiation sites of the interleukin-1 receptor-related human ST2 gene in UT-7 and TM12 cells. Eur J Biochem. 1999;264(2):397–406. doi: 10.1046/j.1432-1327.1999.00615.x. [DOI] [PubMed] [Google Scholar]
- 17.Lipsky BP, Toy DY, Swart DA, Smithgall MD, Smith D. Deletion of the ST2 proximal promoter disrupts fibroblast-specific expression but does not reduce the amount of soluble ST2 in circulation. Eur J Immunol. 2012;42(7):1863–1869. doi: 10.1002/eji.201142274. [DOI] [PubMed] [Google Scholar]
- 18.Kumar S, Tzimas MN, Griswold DE, Young PR. Expression of ST2, an interleukin-1 receptor homologue, is induced by proinflammatory stimuli. Biochem Biophys Res Commun. 1997;235(3):474–478. doi: 10.1006/bbrc.1997.6810. [DOI] [PubMed] [Google Scholar]
- 19.Weinberg EO, Shimpo M, De Keulenaer GW, MacGillivray C, Tominaga S, Solomon SD, et al. Expression and regulation of ST2, an interleukin-1 receptor family member, in cardiomyocytes and myocardial infarction. Circulation. 2002;106(23):2961–2966. doi: 10.1161/01.CIR.0000038705.69871.D9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mildner M, Storka A, Lichtenauer M, Mlitz V, Ghannadan M, Hoetzenecker K, et al. Primary sources and immunological prerequisites for sST2 secretion in humans. Cardiovasc Res. 2010;87(4):769–777. doi: 10.1093/cvr/cvq104. [DOI] [PubMed] [Google Scholar]
- 21.Sanada S, Hakuno D, Higgins LJ, Schreiter ER, McKenzie AN, Lee RT. IL-33 and ST2 comprise a critical biomechanically induced and cardioprotective signaling system. J Clin Invest. 2007;117(6):1538–1549. doi: 10.1172/JCI30634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Januzzi JL, Jr., Peacock WF, Maisel AS, Chae CU, Jesse RL, Baggish AL, et al. Measurement of the interleukin family member ST2 in patients with acute dyspnea: results from the PRIDE (Pro-Brain Natriuretic Peptide Investigation of Dyspnea in the Emergency Department) study. J Am Coll Cardiol. 2007;50(7):607–613. doi: 10.1016/j.jacc.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 23.Mueller T, Dieplinger B, Gegenhuber A, Poelz W, Pacher R, Haltmayer M. Increased plasma concentrations of soluble ST2 are predictive for 1-year mortality in patients with acute destabilized heart failure. Clin Chem. 2008;54(4):752–756. doi: 10.1373/clinchem.2007.096560. [DOI] [PubMed] [Google Scholar]
- 24.Shah RV, Chen-Tournoux AA, Picard MH, van Kimmenade RR, Januzzi JL. Serum levels of the interleukin-1 receptor family member ST2, cardiac structure and function, and long-term mortality in patients with acute dyspnea. Circ Heart Fail. 2009;2(4):311–319. doi: 10.1161/CIRCHEARTFAILURE.108.833707. [DOI] [PubMed] [Google Scholar]
- 25.Ky B, French B, McCloskey K, Rame JE, McIntosh E, Shahi P, et al. High-sensitivity ST2 for prediction of adverse outcomes in chronic heart failure. Circ Heart Fail. 2011;4(2):180–187. doi: 10.1161/CIRCHEARTFAILURE.110.958223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pascual-Figal DA, Ordonez-Llanos J, Tornel PL, Vazquez R, Puig T, Valdes M, et al. Soluble ST2 for predicting sudden cardiac death in patients with chronic heart failure and left ventricular systolic dysfunction. J Am Coll Cardiol. 2009;54(23):2174–2179. doi: 10.1016/j.jacc.2009.07.041. [DOI] [PubMed] [Google Scholar]
- 27.Sabatine MS, Morrow DA, Higgins LJ, MacGillivray C, Guo W, Bode C, et al. Complementary roles for biomarkers of biomechanical strain ST2 and N-terminal prohormone B-type natriuretic peptide in patients with ST-elevation myocardial infarction. Circulation. 2008;117(15):1936–1944. doi: 10.1161/CIRCULATIONAHA.107.728022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shimpo M, Morrow DA, Weinberg EO, Sabatine MS, Murphy SA, Antman EM, et al. Serum levels of the interleukin-1 receptor family member ST2 predict mortality and clinical outcome in acute myocardial infarction. Circulation. 2004;109(18):2186–2190. doi: 10.1161/01.CIR.0000127958.21003.5A. [DOI] [PubMed] [Google Scholar]
- 29.Weinberg EO, Shimpo M, Hurwitz S, Tominaga S, Rouleau JL, Lee RT. Identification of serum soluble ST2 receptor as a novel heart failure biomarker. Circulation. 2003;107(5):721–726. doi: 10.1161/01.cir.0000047274.66749.fe. [DOI] [PubMed] [Google Scholar]
- 30.Pastorelli L, Garg RR, Hoang SB, Spina L, Mattioli B, Scarpa M, et al. Epithelial-derived IL-33 and its receptor ST2 are dysregulated in ulcerative colitis and in experimental Th1/Th2 driven enteritis. Proc Natl Acad Sci U S A. 2010;107(17):8017–8022. doi: 10.1073/pnas.0912678107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vander Lugt MT, Braun TM, Hanash S, Ritz J, Ho VT, Antin JH, et al. ST2 as a marker for risk of therapy-resistant graft-versus-host disease and death. N Engl J Med. 2013;369(6):529–539. doi: 10.1056/NEJMoa1213299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Isse K, Grama K, Abbott IM, Lesniak A, Lunz JG, Lee WM, et al. Adding value to liver (and allograft) biopsy evaluation using a combination of multiplex quantum dot immunostaining, high-resolution whole-slide digital imaging, and automated image analysis. Clin Liver Dis. 2010;14(4):669–685. doi: 10.1016/j.cld.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 33.Isse K, Lesniak A, Grama K, Maier J, Specht S, Castillo-Rama M, et al. Preexisting epithelial diversity in normal human livers: a tissue-tethered cytometric analysis in portal/periportal epithelial cells. Hepatology. 2013;57(4):1632–1643. doi: 10.1002/hep.26131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rabe-Hesketh S, Skrondal A. Multilevel and longitudinal modeling using Stata. Second Edition ed. Strata Press; 2008. [Google Scholar]
- 35.Turnquist HR, Zhao Z, Rosborough BR, Liu Q, Castellaneta A, Isse K, et al. IL-33 Expands Suppressive CD11b+ Gr-1int and Regulatory T Cells, including ST2L+ Foxp3+ Cells, and Mediates Regulatory T Cell-Dependent Promotion of Cardiac Allograft Survival. J Immunol. 2011;187(9):4598–4610. doi: 10.4049/jimmunol.1100519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gossett JG, Canter CE, Zheng J, Schechtman K, Blume ED, Rodgers S, et al. Decline in rejection in the first year after pediatric cardiac transplantation: a multi-institutional study. J Heart Lung Transplant. 2010;29(6):625–632. doi: 10.1016/j.healun.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 37.Pascual-Figal DA, Garrido IP, Blanco R, Minguela A, Lax A, Ordonez-Llanos J, et al. Soluble ST2 is a marker for acute cardiac allograft rejection. The Annals of thoracic surgery. 2011;92(6):2118–2124. doi: 10.1016/j.athoracsur.2011.07.048. [DOI] [PubMed] [Google Scholar]
- 38.Januzzi JL., Jr. ST2 as a cardiovascular risk biomarker: from the bench to the bedside. J Cardiovasc Transl Res. 2013;6(4):493–500. doi: 10.1007/s12265-013-9459-y. [DOI] [PubMed] [Google Scholar]
- 39.Beltran CJ, Nunez LE, Diaz-Jimenez D, Farfan N, Candia E, Heine C, et al. Characterization of the novel ST2/IL-33 system in patients with inflammatory bowel disease. Inflamm Bowel Dis. 2010;16(7):1097–1107. doi: 10.1002/ibd.21175. [DOI] [PubMed] [Google Scholar]
- 40.Hoogerwerf JJ, Tanck MW, van Zoelen MA, Wittebole X, Laterre PF, van der Poll T. Soluble ST2 plasma concentrations predict mortality in severe sepsis. Intensive Care Med. 2010;36(4):630–637. doi: 10.1007/s00134-010-1773-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ishikawa S, Shimizu M, Ueno K, Sugimoto N, Yachie A. Soluble ST2 as a marker of disease activity in systemic juvenile idiopathic arthritis. Cytokine. 2013;62(2):272–277. doi: 10.1016/j.cyto.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 42.Lo DJ, Kaplan B, Kirk AD. Biomarkers for kidney transplant rejection. Nat Rev Nephrol. 2014;10(4):215–225. doi: 10.1038/nrneph.2013.281. [DOI] [PubMed] [Google Scholar]
- 43.Wu T, Abu-Elmagd K, Bond G, Nalesnik MA, Randhawa P, Demetris AJ. A schema for histologic grading of small intestine allograft acute rejection. Transplantation. 2003;75(8):1241–1248. doi: 10.1097/01.TP.0000062840.49159.2F. [DOI] [PubMed] [Google Scholar]
- 44.Adeyi OA, Randhawa PA, Nalesnik MA, Ochoa ER, Abu-Elmagd KM, Demetris AJ, et al. Posttransplant adenoviral enteropathy in patients with small bowel transplantation. Arch Pathol Lab Med. 2008;132(4):703–705. doi: 10.5858/2008-132-703-PAEIPW. [DOI] [PubMed] [Google Scholar]
- 45.Talmon GA. Histologic features of cytomegalovirus enteritis in small bowel allografts. Transplant Proc. 2010;42(7):2671–2675. doi: 10.1016/j.transproceed.2010.04.059. [DOI] [PubMed] [Google Scholar]
- 46.Gleissner CA, Klingenberg R, Nottmeyer W, Zipfel S, Sack FU, Schnabel PA, et al. Diagnostic efficiency of rejection monitoring after heart transplantation with cardiac troponin T is improved in specific patient subgroups. Clin Transplant. 2003;17(3):284–291. doi: 10.1034/j.1399-0012.2003.00050.x. [DOI] [PubMed] [Google Scholar]
- 47.Mueller T, Jaffe AS. Soluble ST2--analytical considerations. Am J Cardiol. 2015;115(7 Suppl):8B–21B. doi: 10.1016/j.amjcard.2015.01.035. [DOI] [PubMed] [Google Scholar]
- 48.De Vlaminck I, Valantine HA, Snyder TM, Strehl C, Cohen G, Luikart H, et al. Circulating Cell-Free DNA Enables Noninvasive Diagnosis of Heart Transplant Rejection. Sci Transl Med. 2014;6(241):241ra277–241ra277. doi: 10.1126/scitranslmed.3007803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pascual-Figal DA, Manzano-Fernandez S, Boronat M, Casas T, Garrido IP, Bonaque JC, et al. Soluble ST2, high-sensitivity troponin T- and N-terminal pro-B-type natriuretic peptide: complementary role for risk stratification in acutely decompensated heart failure. Eur J Heart Fail. 2011;13(7):718–725. doi: 10.1093/eurjhf/hfr047. [DOI] [PubMed] [Google Scholar]
- 50.Bayes-Genis A, de Antonio M, Galán A, Sanz H, Urrutia A, Cabanes R, et al. Combined use of high-sensitivity ST2 and NTproBNP to improve the prediction of death in heart failure. Eur J Heart Fail. 2012;14(1):32–38. doi: 10.1093/eurjhf/hfr156. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.