Abstract
Introduction
With expansion of clinical trials to individuals across the spectrum of Alzheimer disease (AD) from preclinical to symptomatic phases, it is increasingly important to quantify AD severity using methods that capture underlying pathophysiology.
Methods
We derived an AD severity measure based on biomarkers from brain imaging, neuropathology, and cognitive testing using latent variable modeling. We used data from ADNI-1 (N = 822) and applied findings to BIOCARD study (N = 349). We evaluated criterion validity for distinguishing diagnostic groups and construct validity by evaluating rates of change in AD severity.
Results
The AD severity factor cross-sectionally distinguishes cognitively normal participants from MCI (AUC = 0.87) and AD dementia (AUC = 0.94). Among ADNI MCI subjects, worsening scores predict faster progression to AD dementia (HR = 1.17; 95% CI, 1.13–1.22). In ADNI and BIOCARD, the pace of change in AD severity is steepest among progressors, with persisting differences by baseline diagnosis.
Discussion
Our content-valid latent variable measurement model is a reasonable approach for grading AD severity across a broad spectrum beginning at preclinical stages of AD.
Keywords: Alzheimer's disease, Clinical trials, Measurement, Item response theory, Cognitive testing, Imaging, Longitudinal follow-up
1. Introduction
Alzheimer disease (AD) is now recognized to span a spectrum of impairment from normal cognition to dementia, with changes in biomarkers that capture various aspects of the underlying neuropathology [1], [2]. AD develops over decades [3], [4] and has a long prodromal period [5], [6]. The clinical manifestations of AD are often evident only after many years of accumulating neuropathology.
A multitude of AD biomarkers including those derived from brain imaging, cerebrospinal fluid, and neuropsychological testing have been identified which provide distinct information about the pathophysiology and clinical course of AD. A highly influential theoretical model has provided a framework for conceptualizing the pathological cascade of AD [1]. This dynamic biomarkers model proposes that the pathologic cascade of AD begins with abnormal amyloid processing, resulting in build-up of amyloid beta protein (Aβ1–42) in the brain, accelerating tau deposition, which in combination has neurotoxic effects resulting in cellular dysfunction and death, brain atrophy, and impaired neuropsychological function. This process culminates in clinical symptoms and functional disability [1].
Biomarkers in the dynamic biomarkers model are hypothesized to provide information over a spectrum from preclinical to clinical stages of AD. Evidence suggests, however, that no single biomarker provides sufficient information to capture the underlying severity of disease across the entire spectrum. Several efforts are thus underway to objectively and quantitatively combine multiple biomarkers to characterize the clinico-pathophysiological severity of AD [7], [8]. The main goal of this study is to operationalize such a method objectively and quantitatively and to evaluate its validity.
Features of the dynamic biomarkers hypothesis relevant for its operationalization are the prevalence of multiple disease severity markers, thought to represent relationships between disease severity markers and disease stage and the characterization of the phase of disease. First, levels of disease severity markers underlying physiological mechanisms indicate worsening AD severity over time. Different markers worsen from normal to abnormal levels during different phases of AD, ranging from cognitively normal, through mild cognitive impairment (MCI), to clinical dementia. A second feature of the dynamic biomarkers model is the hypothesized sigmoidal (s-shaped) relationship between each disease severity marker and disease stage. In the mid-range of the disease severity marker response, its relationship with underlying disease stage is presumed linear, but the distribution at its tails asymptotes toward normal/abnormal response levels. Different disease severity markers have different dynamic ranges. For example, while deposition of Aβ1–42 is initially occurring, there may be no change in memory. Later, while memory worsens, it is hypothesized that less Aβ1–42 deposition is taking place relative to earlier disease stages. A third key feature of the dynamic biomarkers model is disease stage on the x-axis. Neither time nor age is necessary to describe the advancing disease course, but some quantity (i.e., underlying disease severity) not directly measureable is. Jack et al. [9] suggested a latent variable model, as implemented in this study, may sufficiently represent AD severity and its relationship to disease severity markers.
The dynamic biomarkers model is almost immediately recognizable as a latent variable model. Latent variable models relate item responses on observed variables to a latent, or unobserved, variable using probabilistic models. Severity of underlying pathology is the latent variable. A latent variable is not directly observable but is presumed to causally influence reflective indicators (disease severity markers). The response scale of disease severity markers and sigmoidal response curve shape leads naturally to response variable discretization [10] and graded response variable modeling [11]. Latent variable modeling characterizes aspects of persons (level of latent AD severity) and aspects of latent variable indicators (disease severity markers). This approach quantifies underlying AD pathology in persons without frank impairment.
Our main goal was to operationalize the dynamic biomarkers hypothesis. We present an objective and quantitative method for integrating multiple biomarkers and other disease severity markers of AD into a global measure of AD severity using a latent trait framework based in measures from cerebrospinal fluid, structural neuroimaging, neuropsychological performance, and ratings of functional impairment. We demonstrate the potential utility of the measure of AD severity by using it to describe differences between clinically defined diagnostic groups—normal, MCI, and AD dementia—and to predict future progression to more impaired clinical states. We suggest applications of the model for research and clinical purposes, as well as weaknesses and opportunities for extending the model. We used data from the Alzheimer's Disease Neuroimaging Initiative (ADNI) study to derive the model and applied the findings in the BIOCARD study, both longitudinal studies in which a range of biomarkers was collected.
2. Methods
2.1. Participants
We used data collected in the ADNI and BIOCARD studies. ADNI (adni.loni.usc.edu) was launched in 2003 as a public-private partnership, led by Principal Investigator Michael W. Weiner, MD. The primary goal of ADNI was to test whether biological markers could be used to improve measurement of clinical and neuropsychological progression in clinical trials. In these analyses, we used data from ADNI-1. For up-to-date information, see www.adni-info.org.
The BIOCARD study initially recruited 349 cognitively normal middle-aged persons starting in 1995, most of whom by design had a first-degree relative with dementia. The primary goal of the study was to identify early markers of progression to MCI due to AD in cognitively normal people. Participants were recruited by the Geriatric Psychiatry branch of the Intramural Program of the National Institute of Mental Health. The study was stopped in 2005, and in 2009 was reestablished by a research team at the Johns Hopkins School of Medicine. Clinical assessments and cognitive testing were completed annually; MRI scans, cerebrospinal fluid, and blood specimens were collected approximately every 2 years. Further details are available elsewhere [12]. Importantly, BIOCARD is smaller in sample size and has less heterogeneity in AD severity than ADNI, but BIOCARD has greater longitudinal follow-up.
ADNI data could be characterized as a cross-sectional study with longitudinal follow-up in that participants in diagnostic groups were very different from each other at baseline on cognitive, imaging, and cerebrospinal fluid (CSF) outcomes, and the study has not yet followed people long enough to observe considerable within-person changes in many variables relative to baseline differences [13]. Our reason for using these data is that ADNI provides a wider spread, albeit from cross-sectional information, of AD biomarker levels thank BIOCARD and thus presumably better measurement estimation in the impaired range of AD severity.
2.2. Disease severity markers
We used biomarkers from each dimension proposed by Jack et al. [1]. Concentrations of Aβ1–42 and phosphorylated tau (P-tau) were analyzed from CSF samples in both ADNI and BIOCARD. ADNI and BIOCARD both used the xMAP platform (Luminex Corp, Austin, Texas) and INNO-BIA AlzBio3 research-use-only reagents [14], [15]. In ADNI and BIOCARD, measures of hippocampal volume were available from MRI scans. In analyses, we residualized hippocampal volume by intracranial volume [16].
The ADNI baseline neuropsychological assessment procedures incorporated a wide range of cognitive measures [17]. Tests included the auditory verbal learning test (AVLT) [18] and the digit symbol substitution test [19]. Cognitive assessments in BIOCARD were also extensive [12]; they included the California verbal learning test (CVLT) [20] and the Digit Symbol Substitution Test. The digit symbol substitution test is a multifactorial test shown to be a significant predictor of progression from normal cognition to mild impairment [12]. We used linear equating to scale the CVLT in BIOCARD to the AVLT [21]. To represent clinical functioning, we used the Functional Activities Questionnaire (FAQ) available from both studies [22].
2.3. Scale of the response of the disease severity markers
The disease severity markers used here are not on the same scale. Placing them on a common scale can be accomplished using transformation functions. We broke continuous indicators into nine categories with equally-spaced intervals to preserve floor and ceiling effects if present (Supplementary Table 2). Cut points used to categorize disease severity markers were spaced using their empirical distributions in ADNI and are thus designed to be spread out along the range of the latent trait, unconnected to clinical diagnoses. In a sensitivity analysis, we implemented a normalization transformation function by placing each disease severity marker on a scale from 0 to 1.
2.4. Analysis plan
We developed a latent trait model of AD severity based on disease severity markers guided by the dynamic biomarkers hypothesis [1]. We then empirically validated the model by contrasting the level and change of the derived severity score with clinical diagnosis (Normal, MCI, and AD dementia). We contrasted the score's ability to predict risk of progression to a more impaired clinical state relative to its component disease severity markers. We applied the findings from a cohort that represents a broad range of disease (ADNI) to a cohort designed to focus on the preclinical phase of AD (BIOCARD). We used both samples because although ADNI includes a wide spread of disease severity marker levels, BIOCARD features a younger sample at earlier stages of risk for dementia.
2.4.1. Model derivation
To evaluate dimensionality of the AD severity indicators, we used parallel analysis with scree plots [23]. We then developed the latent variable measurement model of AD severity using a 2-step approach in ADNI and BIOCARD. The approach accounts for variance and covariance among AD signs and symptoms and addresses the challenge of repeated measures. First, we fit a factor analysis measurement model using baseline data in ADNI, evaluating fit by the empirical or pseudo-r2, which is the squared correlation between observed and model-estimated indicator values that represents the proportion of variability explained by the model [24]. The model used statistically appropriate polychoric correlation matrices and is equivalent to a logistic graded response item response theory model [25], [26]. This equation describes the relationship between categorized biomarkers u and the AD severity factor θ:
The expected probability P(θ) of scoring in or above category j of item i is a function of a discrimination parameter ai, a set of item difficulty or threshold parameters bij, and the latent variable θ for AD severity.
Next, we ran the model using all follow-up data in ADNI in which parameters (factor loadings and thresholds) were constrained to their values in the baseline model as a latent variable scoring device. We used the same procedure in ADNI and in BIOCARD. After evaluating model fit in both studies, we scaled the factor in BIOCARD to that in ADNI by fixing model parameters in BIOCARD to values estimated in the baseline ADNI model. We scored BIOCARD to ADNI instead of vice versa because the greater range of disease severity markers in ADNI provided more reliable estimates.
The latent trait, estimated as a factor score for each visit with available disease severity marker data, represents a subject's level of AD severity at a visit, is defined based on the covariance of variables in a way analogous to factor analysis: our approach can be described as a generalized linear item response theory approach [27]. We used Mplus version 7.3 [28].
2.4.2. Model validation
We first contrasted the level and change of the derived severity score with baseline clinical diagnosis from ADNI (AD dementia, MCI, cognitively normal) and risk of progression to a more impaired clinical state.
Second, we determined the degree to which the AD severity factor distinguished diagnostic groups in ADNI and BIOCARD using receiver operator characteristic analyses. We compared area under the curve (AUC) with component disease severity markers.
Third, we determined how well baseline levels of the AD severity factor among cognitively normal individuals predict progression to MCI or to AD. We contrasted these associations with those of components of the AD severity score, to determine whether the score is better than its parts at predicting disease progression.
Fourth, to demonstrate the utility of the AD severity score for predicting future levels of biomarkers, we evaluated the association of changes in biomarker levels with baseline levels of AD severity using regressions of each biomarker on AD severity, time, and their interaction. We included random effects for intercept and slope. We graphed model-estimated levels of each biomarker against time by quintile of baseline AD severity to examine whether the trajectory of the biomarkers varies according to baseline levels of AD severity.
2.4.3. Sensitivity analyses
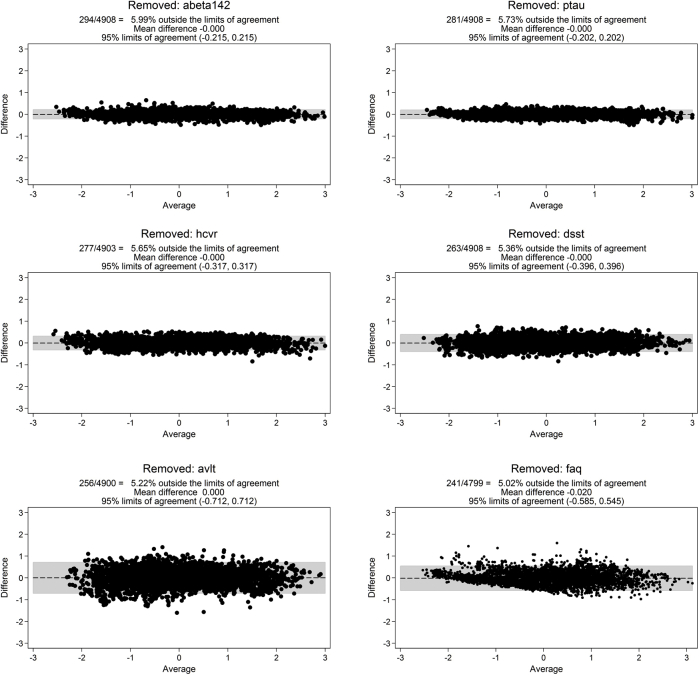
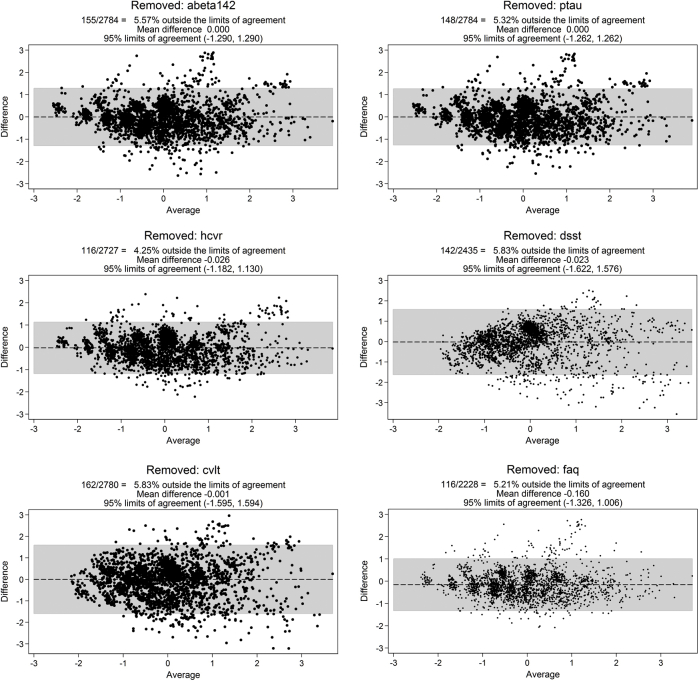
Different uses of an AD severity score (e.g., screening and tracking progression in preclinical cases vs. describing trajectories) place different demands on the level of measurement precision needed at particular ranges of severity that are in turn informed by different measures. Latent trait theory leads to the expectation that the same level of AD severity can be obtained if only a subset of the disease severity markers are observed. Thus, we conducted a series of leave-1-out measurement models which omitted a biomarker and re-estimated the AD severity score. We compared scores with Bland–Altman plots to evaluate precision and bias across the range of underlying AD severity [29]. Results of this sensitivity analysis informed the minimum set of biomarkers that can be assessed to return a reasonably precise estimate of a person's underlying AD severity.
3. Results
ADNI-1 is a diagnostically heterogeneous sample of 822 older adults (mean age, 73 years, range 54–91 years) that at baseline included cognitively normal (N = 229), MCI (N = 400), and AD dementia cases (N = 193) (Table 1). During follow-up on average of 4 years (range 0 to 9 years, median 3 years), N = 73 progressed from cognitively normal to MCI and N = 242 progressed to AD dementia. BIOCARD is a younger sample (58 years, range 20 to 85 years) than ADNI and at baseline included mostly cognitively normal persons (N = 284, 84.8%). During follow-up on average of 11.5 years (range, 0–17.8 years, median 12.1 years), N = 79 participants progressed to MCI and N = 28 to dementia.
Table 1.
Baseline characteristics of the ADNI and BIOCARD samples
| Characteristic | ADNI |
BIOCARD |
||
|---|---|---|---|---|
| Mean (SD) or number (%) | Range | Mean (SD) or number (%) | Range | |
| Sample size | 822 | 349 | ||
| Age at baseline, mean (SD) | 75.3 (6.9) | 54.8–90.9 | 57.6 (10.0) | 20.4–84.8 |
| Race/ethnicity, n (%) | ||||
| White | 748 (91.0) | 336 (96.6) | ||
| Black | 38 (4.6) | 5 (1.4) | ||
| Hispanic | 20 (2.4) | 4 (1.1) | ||
| Other/unknown | 16 (1.9) | 3 (0.9) | ||
| Female sex, n (%) | 344 (41.9) | 201 (57.6) | ||
| Years of education, mean (SD) | 15.5 (3.0) | 4–20 | 17 (2.4) | 12–20 |
| Study visits, n (%) | 6.0 (2.8) | 1–12 | 8.3 (4.0) | 1–18 |
| Years of follow-up, mean (SD) | 3.9 (2.7) | 0–9.1 | 11.5 (4.9) | 0–17.8 |
| Mini-mental state examination score, mean (SD) | 26.7 (2.7) | 18–30 | 29.5 (0.9) | 27–30 |
| Baseline diagnosis, n (%) | ||||
| Normal | 229 (27.9) | 279 (84.5) | ||
| MCI | 400 (48.7) | 30 (9.1) | ||
| Dementia | 193 (23.5) | 21 (6.4) | ||
| Incident MCI, n (%) | 73 (8.9) | 79 (22.4) | ||
| Incident AD dementia, n (%) | 242 (38.5) | 28 (8.0) | ||
| Biomarker levels, mean (SD) | ||||
| Aβ1–42 | 144.4 (56.3) | 1–266.7 | 259.3 (113.5) | 25.1–555.4 |
| Phosphorylated tau | 28.4 (18.6) | 2.0–109.0 | 32.2 (23.2) | 4.5–158.4 |
| Hippocampal volume | 2297.3 (528.2) | 187.5–3772.4 | 765.7 (280.6) | 81.0–1361.7 |
| Digit symbol substitution test | 47.0 (13.1) | 4.0–84.0 | 44.2 (10.8) | 18.0–66.0 |
| Auditory verbal learning test, sum of recall | 43.5 (11.5) | 7.0–76.0 | 36.9 (14.6) | 8.3–69.9 |
| FAQ | 6.0 (6.6) | 1.0–31.0 | 2.8 (5.5) | 1.0–28.0 |
3.1. Measurement model in ADNI and BIOCARD
Measurement models fit the observed data acceptably in ADNI based on empirical r2 statistics (Table 2). Empirical r2 statistics describe the proportion of variance in each indicator explained by the model. In ADNI, factor loadings were highest for the AVLT and FAQ. Owing to missing data by design in BIOCARD, factor loadings and empirical r2 statistics were lower than in ADNI, particularly for hippocampal volume and the FAQ, but the empirical r2 were still acceptable.
Table 2.
Factor loadings for AD Severity: Results from ADNI (N = 822) and BIOCARD (N = 349)
| Biomarker | Standardized loading | Thresholds |
Empirical r2 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |||
| ADNI (N = 822, 4949 observations) | ||||||||||
| Aβ1–42 | 0.54 | −1.63 | −0.92 | −0.59 | −0.45 | 0.01 | 0.57 | 1.00 | 1.78 | 0.85 |
| Phosphorylated tau | 0.48 | −1.35 | −0.67 | −0.16 | 0.19 | 0.65 | 0.88 | 1.33 | 1.71 | 0.78 |
| Hippocampal volume | 0.58 | −2.20 | −1.06 | −0.43 | −0.17 | 0.21 | 0.71 | 1.17 | 1.91 | 0.97 |
| Digit symbol substitution test | 0.64 | −2.59 | −1.69 | −1.06 | −0.32 | 0.40 | 1.02 | 1.64 | 2.11 | 0.97 |
| Auditory Verbal Learning Test, sum of recall | 0.77 | −2.29 | −1.84 | −1.14 | −0.55 | 0.14 | 1.08 | 1.88 | 2.63 | 0.98 |
| FAQ | 0.76 | −0.29 | 0.31 | 0.66 | 0.91 | 1.25 | 1.42 | 1.68 | 1.98 | 0.98 |
| BIOCARD (N = 349, 2773 observations) | ||||||||||
| Aβ1–42 | 0.36 | 0.69 | 0.84 | 0.99 | 1.16 | 1.34 | 1.52 | 1.86 | 2.11 | 0.65 |
| Phosphorylated tau | 0.35 | 0.33 | 0.99 | 1.15 | 1.36 | 1.49 | 1.62 | 2.02 | 2.18 | 0.62 |
| Hippocampal volume | −0.05 | −1.59 | −1.11 | −0.44 | 0.01 | 0.22 | 0.6 | 0.86 | 1.56 | 0.58 |
| Digit symbol substitution test | 0.47 | −1.27 | −0.47 | 0.22 | 0.97 | 1.57 | 1.91 | 2.06 | 2.13 | 0.92 |
| California verbal learning test, sum of recall | 0.56 | −1.27 | −0.61 | 0.05 | 0.58 | 1.04 | 1.51 | 1.94 | 2.35 | 0.96 |
| FAQ | 0.26 | 1.01 | 1.21 | 1.47 | 1.54 | 1.63 | 1.68 | 1.73 | 1.94 | 0.86 |
NOTE. Standardized factor loadings represent correlations between an item and the underlying latent trait. Standardized thresholds represent the location of cut points on the scale of the latent trait. Empirical r2 statistics are the squared correlations between model-estimated values of each indicator and observed values of the indicator in the data and are used here to indicate quality of item-level fit of the model to the data.
3.2. Model validation
In Table 3, the AD severity score in ADNI distinguishes AD dementia from cognitively normal persons (area under the curve, AUC = 0.98), cognitively normal from MCI (AUC = 0.88), and MCI from AD dementia (AUC = 0.79). Results were equally good in BIOCARD. Table 3 also provides classification quality for each disease severity marker. In both ADNI and BIOCARD, the AD severity summary score demonstrated equal or superior sensitivity, specificity, and AUC statistics compared to its component measures for all comparisons.
Table 3.
Criterion validity of the AD severity factor using diagnostic status: Results from ADNI (N = 822) and BIOCARD (N = 349)
| Data set and diagnostic comparison | Predictor | Area under the curve | Sensitivity | Specificity |
|---|---|---|---|---|
| ADNI (N = 822) | ||||
| Normal vs AD | ||||
| AD severity factor | 0.98 | 0.96 | 0.94 | |
| Aβ1–42 | 0.83 | 0.83 | 0.76 | |
| Phosphorylated tau | 0.76 | 0.69 | 0.72 | |
| Hippocampal volume | 0.90 | 0.80 | 0.83 | |
| Digit symbol substitution | 0.91 | 0.83 | 0.85 | |
| Auditory verbal learning | 0.96 | 0.91 | 0.89 | |
| FAQ | 0.97 | 0.93 | 0.95 | |
| Normal vs MCI | ||||
| AD severity factor | 0.88 | 0.79 | 0.85 | |
| Aβ1–42 | 0.73 | 0.69 | 0.72 | |
| Phosphorylated tau | 0.65 | 0.61 | 0.63 | |
| Hippocampal volume | 0.77 | 0.66 | 0.77 | |
| Digit symbol substitution | 0.74 | 0.65 | 0.72 | |
| Auditory verbal learning | 0.84 | 0.75 | 0.79 | |
| FAQ | 0.84 | 0.79 | 0.83 | |
| MCI vs AD | ||||
| AD severity factor | 0.79 | 0.77 | 0.70 | |
| Aβ1–42 | 0.60 | 0.68 | 0.48 | |
| Phosphorylated tau | 0.61 | 0.65 | 0.51 | |
| Hippocampal volume | 0.67 | 0.65 | 0.61 | |
| Digit symbol substitution | 0.73 | 0.69 | 0.64 | |
| Auditory verbal learning | 0.72 | 0.64 | 0.68 | |
| FAQ | 0.78 | 0.79 | 0.67 | |
| BIOCARD (N = 349) | ||||
| Normal vs AD | ||||
| AD severity factor | 0.99 | 0.95 | 0.96 | |
| Aβ1–42 | 0.96 | 1.00 | 0.93 | |
| Phosphorylated tau | 0.86 | 0.82 | 0.88 | |
| Hippocampal volume | 0.67 | 0.71 | 0.68 | |
| Digit symbol substitution | 0.93 | 0.80 | 0.91 | |
| California verbal learning | 0.94 | 0.88 | 0.88 | |
| FAQ | 0.97 | 0.95 | 0.98 | |
| Normal vs MCI | ||||
| AD severity factor | 0.78 | 0.73 | 0.65 | |
| Aβ1–42 | 0.78 | 0.68 | 0.80 | |
| Phosphorylated tau | 0.67 | 0.55 | 0.80 | |
| Hippocampal volume | 0.64 | 0.60 | 0.68 | |
| Digit symbol substitution | 0.76 | 0.67 | 0.73 | |
| California verbal learning | 0.78 | 0.72 | 0.71 | |
| FAQ | 0.63 | 0.30 | 0.96 | |
NOTE. Each row is based on a separate logistic regression of diagnostic status on the predictor (row) of interest. Logistic regressions account for clustering of observations within people over time using a Huber–White variance estimator.
Table 4 shows hazard ratios from survival analyses for associations of the AD severity factor as well as its components and time-to-progression to MCI or to AD dementia. Hazard ratios explain the elevated risk of progression per unit difference in the AD severity factor or its components. Because the variables are in different units, we also present z statistics for the hazard ratios to facilitate comparisons of the relative strength of the hazard ratios. In ADNI data, in which participants contributed 3153 person-years, relative to the individual components of the severity factor, the AD severity factor was most strongly associated with onset of dementia (hazard ratio [HR] = 1.08; 95% confidence interval [CI], 1.07–1.10; Z = 9.94), followed by AVLT and hippocampal volume (both HR = 1.08; 95% CI, 1.6–1.09). In BIOCARD, in which participants contributed 3312 person-years, the AD severity score predicted progression to AD dementia (HR = 1.05; 95% CI, 1.00–1.10; Z = 2.10) but not to MCI (HR = 0.98; 95% CI, 0.95–1.02; Z = −0.88).
Table 4.
Predictive convergent validity of the AD severity factor: Results from ADNI (N = 822) and BIOCARD (N = 349)
| Predictor | Number of progressors | Hazard ratio | 95% confidence interval | Z statistic |
|---|---|---|---|---|
| BIOCARD: Progression to MCI | ||||
| AD severity factor | 32 | 0.98 | 0.94–1.02 | −1.02 |
| Aβ1–42 | 18 | 1.02 | 0.97–1.07 | 0.94 |
| Phosphorylated Tau | 18 | 1.09∗ | 1.03–1.15 | 2.94 |
| Hippocampal volume | 19 | 1.01 | 0.96–1.06 | 0.33 |
| Digit Symbol Substitution | 23 | 1.11∗ | 1.05–1.18 | 3.76 |
| California Verbal Learning | 9 | 1.14∗ | 1.01–1.28 | 2.21 |
| FAQ | 0 | — | ||
| BIOCARD: Progression to Dementia | ||||
| AD Severity factor | 12 | 1.05∗ | 1.00–1.10 | 2.10 |
| Aβ1–42 | 9 | 1.07∗ | 1.01–1.14 | 2.18 |
| Phosphorylated Tau | 9 | 1.12∗ | 1.04–1.20 | 3.10 |
| Hippocampal volume | 10 | 1.07 | 0.97–1.17 | 1.42 |
| Digit Symbol Substitution | 12 | 1.11∗ | 1.03–1.21 | 2.62 |
| California Verbal Learning | 0 | — | ||
| FAQ | 0 | — | ||
| ADNI: Progression to Dementia | ||||
| AD Severity factor | 176 | 1.08∗ | 1.07–1.10 | 9.96 |
| Aβ1–42 | 92 | 1.07∗ | 1.04–1.09 | 5.23 |
| Phosphorylated Tau | 92 | 1.04∗ | 1.02–1.05 | 3.73 |
| Hippocampal volume | 171 | 1.08∗ | 1.07–1.10 | 9.15 |
| Digit symbol substitution | 176 | 1.04∗ | 1.03–1.06 | 5.97 |
| Auditory verbal learning | 176 | 1.08∗ | 1.06–1.10 | 8.95 |
| FAQ | 175 | 1.03∗ | 1.01–1.04 | 4.08 |
NOTE. The AD severity score for BIOCARD was based on a model in which ADNI parameters were applied to the BIOCARD sample. Because the variables are in different units, z statistics facilitate comparisons of the relative strength of the hazard ratios. Each row is based on a separate logistic regression of diagnostic status on baseline levels of the predictor (row) of interest. All models are adjusted for age.
P < .05.
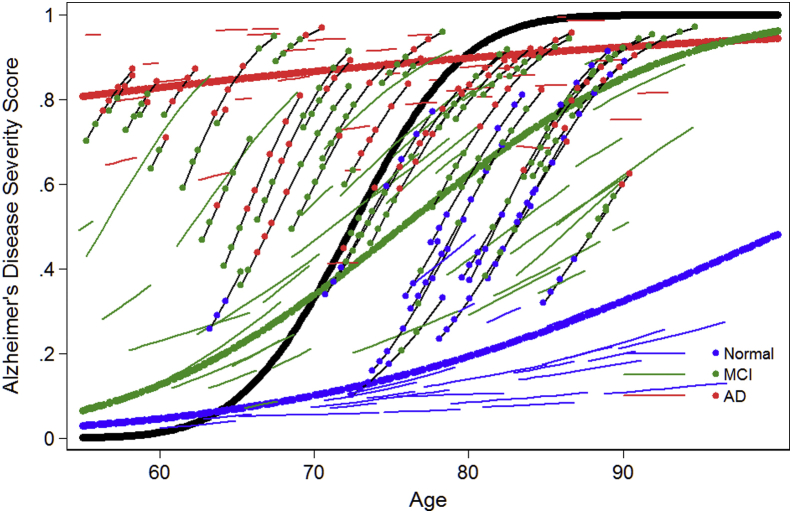
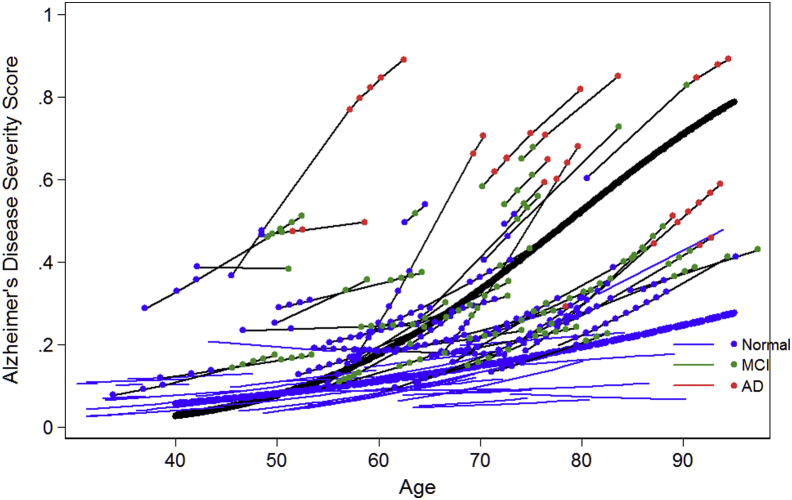
To evaluate construct validity, the power of the AD severity score to distinguish differences in the pace of change in AD severity is illustrated in Figs. 1 (ADNI) and 2 (BIOCARD). These figures summarize the longitudinal trajectory of a random sample of individual persons' AD severity scores over age. Values plotted are best unbiased linear predictors based on a linear mixed effect regression model of predicted AD severity score. Four groups of participant's trajectories are summarized in these figures: those who begin cognitively normal and remain normal (solid blue lines), those who begin with MCI and remain MCI (solid green lines), those who begin with clinical AD dementia and do not worsen (solid red lines), and those who progress to a higher level of severity (solid black lines) over follow-up. The average trajectory for persons who are normal, MCI, and AD dementia at baseline and who progressed are shown with thicker blue, green, red, and black lines, respectively. For persons who progressed, colored dots indicate current (at time of assessment) clinical stage as indicated in the key. In Fig. 2 for BIOCARD, we did not plot average trajectories for AD dementia or MCI cases because, by design, no participants had such a diagnosis at baseline.
Fig. 1.
Longitudinal trends in Alzheimer disease severity in ADNI. This figure shows a random sample of longitudinal trajectories for select ADNI subgroups. Persons who begin the study in the normal group and do not progress over 48M follow-up are shown with solid blue lines without markers (dots). The average trajectory for this group is shown with a heavy blue-dashed line. Persons who begin with clinical AD are shown in solid red lines. The trend line for this group is shown with a heavy dashed red line. Persons who progress (either from normal to mild cognitive impairment (MCI) or AD or from MCI to AD are drawn with black lines, and colored dots indicate their current (at time of assessment) clinical stage as indicated in the key. The average trajectory for this group is shown with a solid black line. The values plotted are best unbiased linear predictors based on a linear mixed effect regression model of predicted Alzheimer disease severity score.
Fig. 2.
Longitudinal trends in Alzheimer disease severity in BIOCARD. This figure shows a random sample of longitudinal trajectories for select BIOCARD subgroups. Persons who begin the study in the normal group and do not progress during follow-up are shown with solid blue lines without markers (dots). The average trajectory for this group is shown with a heavy blue-dashed line. Persons who progress (either from normal to mild cognitive impairment (MCI) or AD or from MCI to AD are drawn with black lines, and colored dots indicate their current (at time of assessment) clinical stage as indicated in the key. The average trajectory for this group is shown with a solid black line. The values plotted are best unbiased linear predictors based on a linear mixed effect regression model of predicted Alzheimer disease severity score.
Fig. 1, Fig. 2 suggest that among older persons who are cognitively normal and remain so over up to 9 years in ADNI and 18 years in BIOCARD, the pace of change in AD severity is slow but positive. Persons initially enrolled with clinical AD dementia begin in a much more impaired range at baseline, yet possibly due to ceiling effects, selection effects, or drop-outs their pace of change is comparable to cognitively normal persons. The pace of change among people with MCI and those who progressed to a higher level of severity is steeper than in other groups.
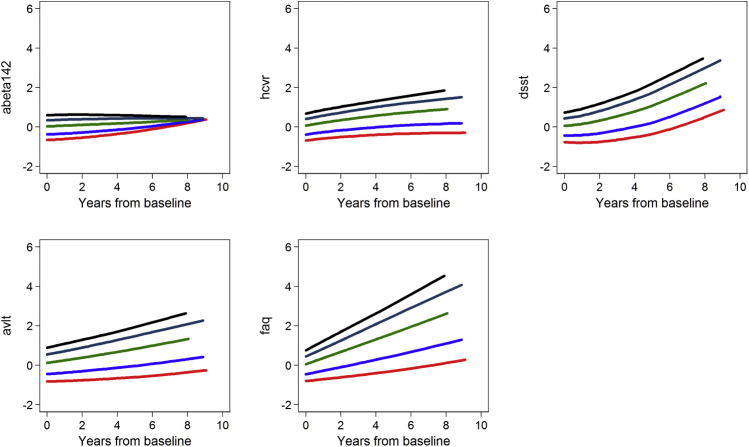
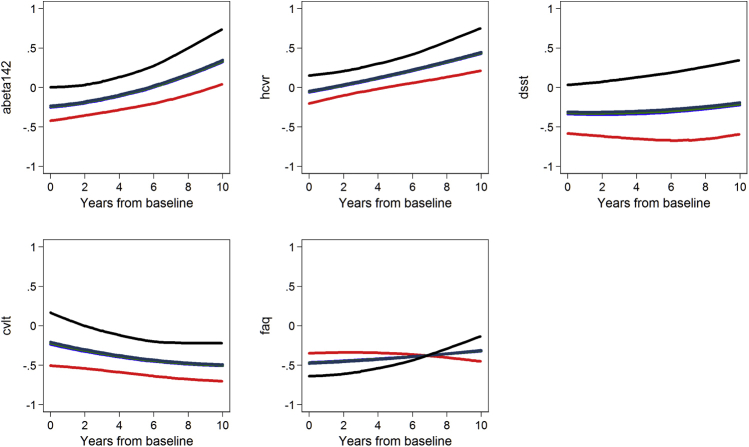
Supplementary Figs. 1 (ADNI) and 2 (BIOCARD) show results of random effects regressions of each biomarker on baseline levels of the AD severity score by quintile of AD severity. The trajectories for each disease severity marker are not parallel, indicating the AD severity score at baseline partially differentiates trajectories of disease severity markers.
3.3. Sensitivity analyses
We evaluated the extent to which an accurate latent trait score can be estimated with subsets of available biomarkers (e.g., if Aβ1–42 is unavailable, if hippocampal atrophy is unavailable). Bland–Altman plots of factor scores from leave-1-out models for each disease severity marker revealed minimal evidence of systematic bias over the range of AD severity (Supplementary Figs. 3 and 4). These results depict that reasonable measurement precision of AD severity for research use can be obtained with any five of the six markers.
In another sensitivity analysis, we estimated factor analyses based on normalized versions of the disease severity markers (Supplementary Table 1). Factor loadings from these models in ADNI and BIOCARD were similar to their counterparts using discretized indicators.
4. Discussion
The overarching goal of this project was to develop an objective and quantitative method for integrating multiple biomarkers of AD into a global measure of AD severity. We incorporated cerebrospinal fluid, structural neuroimaging markers, neuropsychological testing, and functional performance into a latent trait model. The AD severity score is a content-valid measure of AD severity. Results suggest it a reasonable approach for describing individual differences in AD severity and predicts progression from cognitively normal to more impaired clinical states. These findings suggest the approach can characterize underlying severity of disease and may therefore be useful for drug development or evaluating response to treatment.
Our derived AD severity score has potential to advance AD research by providing a method for characterizing the level of severity of AD in both preclinical and symptomatic cases. Because AD is recognized as a disease that develops over decades with a long preclinical period and in the absence of apparent clinical symptoms, a summary severity score based on biomarkers will be essential for screening and targeting of early interventions. Most current clinical trials are using amyloid imaging to determine whether AD pathology is present. However, such a method is not optimal for grading disease severity. An objective statistical measure of AD severity offers a feasible way to shorten clinical trials by providing a method to capture underlying pathology that can be tracked longitudinally.
Our model is based on available biomarkers and is extendible. Future work that identifies new biomarkers can be incorporated into the model and provide greater precision or information regarding AD severity. For example, our model uses CSF Aβ1–42 and P-Tau, but future investigations might instead use the ratio of Aβ1–42/P-Tau. Our study's purpose was not to build a predictive model of AD diagnosis; thus, we did not consider demographic or genetic modifiers of AD risk in measurement models. Age, sex, and APOE status are not part of the dynamic biomarkers hypothesis [1], and AD does not cause these characteristics. Age, sex, and APOE status are important features to include when trying to predict AD progression and conversion, but they are not appropriate factors to include as indicators that define AD severity.
Another future direction for the field, outside the scope of the present analysis, will be to compare the relative strength of the proposed model and alternative biomarker-based models to predict onset of AD and characterize trajectories of AD severity. Such alternative models include the AD signature based on MRI [30], Jednyak et al. [8] disease progression model for AD which bears some resemblance to ours in terms of goal, and a recently proposed spatiotemporal function to simultaneously model the ordering of a multifactorial set of biomarkers in late-onset AD [31].
Strengths of the study include two well-characterized cohorts followed prospectively for years. Strengths of our analysis include the use of readily available standard software, with standard expectations for conduct of analysis and reporting that is an important aspect of reproducibility, thus reducing the likelihood of false findings [32]. Mplus is flexible and provides a framework for testing alternative models (e.g., a nonlinear relationship between underlying AD severity and a biomarker could be tested with a latent variable interaction).
A noteworthy study limitation is that some aspects of the study designs for ADNI and BIOCARD are not ideal for the inferences we sought to make. It would be helpful to have AD severity indicator data for even younger ages, possibly extending back as far as 40 years of age or even younger. It would also have been better if clinically normal participants in ADNI were a random selection from the population. We addressed the issue of younger ages, in part, by applying our findings to the BIOCARD study, but even there participants were not selected to be a random population sample, as it was enriched with people who had a family history of dementia.
Another potential study limitation is our discretization of AD severity markers that are observed quantitatively. This results in some loss of information. In a sensitivity analysis, we reran models using continuous versions of disease severity markers and currently believe the discretization approach is better (Supplementary Table 1). The advantage of the discretization approach is that it does not presume that AD severity increases linearly with increases in the quantitatively observed biomarker. This comes from estimating eight thresholds for a nine-category discretized biomarker: the spacing among the estimated thresholds along the latent trait can vary.
A third limitation is that the extent to which a model-estimated AD severity score corresponds to an assumed underlying reality of an AD pathological process hinges directly on which AD severity indicators are in the model. Although ADNI and BIOCARD collected a multitude of biomarkers, we included a relatively small set. Any number of AD biomarkers could be chosen for a model such as ours, and each might produce some differences in the rank ordering of persons along a continuum of AD severity. For example, Abeta loads determined by PIB, FDG PET-derived measures of metabolism, and size and shape of the corpus callosum are other important markers albeit unavailable in other datasets we used in the present study [33], [34], [35]. Given this, our model can only be seen as a rough empirical guide to the underlying nature of AD severity, and suited for group differences research (e.g., testing the effectiveness of interventions administered to groups; charting the progression of AD in population samples). Application to inference at the individual level should be discouraged. The goal of this study was to introduce the concept, methods, and use of the objective measure of AD severity using biomarkers and other disease severity markers. Future extensions of our model will be to investigate the potential utility of other biomarkers, with an eye toward identifying those with a dynamic range relevant to persons with very early AD.
Research in context.
-
1.
Systematic review: Alzheimer's disease (AD) develops over decades, but underlying disease severity is difficult to measure with individual biomarkers. The dynamic biomarkers hypothesis provides a framework for conceptualizing the pathological cascade of AD that begins with abnormal amyloid processing and culminates in clinical symptoms and functional disability.
-
2.
Interpretation: Recognizing the dynamic biomarkers hypothesis as a latent variable model, we present an objective, quantitative method for integrating multiple biomarkers and other severity indicators of AD into a global measure encompassing preclinical and symptomatic disease stages. The derived AD severity factor distinguishes diagnostic groups as well or better than its component AD biomarkers. Worsening scores predict faster progression to AD dementia among participants with MCI.
-
3.
Future directions: Potential applications of the derived severity score include being an intermediate outcome in clinical trials. The model should additionally facilitate research on novel biomarkers which have a dynamic range relevant to persons with preclinical AD.
Acknowledgments
This work was supported by National Institute on Aging grant R13 AG030995 (PI: Mungas). J-M. S. L.'s work was supported by McGill University, Pfizer Canada, the Government of Canada (Canada Research Chairs program and Canadian Fund for Innovation), the Weston Brain Institute and the Levesque Foundation.
Data collection and sharing for this project was funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer's Association; Alzheimer's Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
The BIOCARD study is supported in part by grants from the National Institutes of Health: U01-AG03365, P50-AG005146 and P41-RR015241. The BIOCARD Study consists of 7 Cores with the following members: (1) the Administrative Core (Marilyn Albert, Barbara Rodzon, Richard Power), (2) the Clinical Core (Ola Selnes, Marilyn Albert, Rebecca Gottesman, Ned Sacktor, Guy McKhann, Scott Turner, Leonie Farrington, Maura Grega, Daniel D'Agostino, Sydney Feagen, David Dolan, Hillary Dolan), (3) the Imaging Core (Michael Miller, Susumu Mori, Tilak Ratnanather, Timothy Brown, Hayan Chi, Anthony Kolasny, Kenichi Oishi, Thomas Reigel, William Schneider, Laurent Younes), (4) the Biospecimen Core (Richard O'Brien, Abhay Moghekar, Richard Meehan), (5) the Informatics Core (Roberta Scherer, Curt Meinert, David Shade, Ann Ervin, Jennifer Jones, Matt Toepfner, Lauren Parlett, April Patterson, Lisa Lassiter), the (6) Biostatistics Core (Mei-Cheng Wang, Yi Lu, Qing Cai), and (7) the Neuropathology Core (Juan Troncoso, Barbara Crain, Olga Pletnikova, Gay Rudow, Karen Fisher).
We would like to acknowledge the contributions to BIOCARD of the Geriatric Psychiatry Branch (GPB) of the intramural program of the NIMH who initiated the study (PI: Dr. Trey Sunderland).
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.dadm.2016.08.005.
Supplementary data
Supplementary Fig. 1.
Baseline AD severity factor predicting changes in disease severity markers: Results from ADNI (N = 822). Average longitudinal trajectories of indicators for quintiles of baseline AD severity show baseline values and rates of change of each disease severity marker are sensitive to AD severity across its full range.
Supplementary Fig. 2.
Baseline AD severity factor predicting changes in disease severity markers: Results from BIOCARD (N = 349). Average longitudinal trajectories of indicators for quintiles of baseline AD severity show baseline values and rates of change of each disease severity marker are sensitive to AD severity across its full range.
Supplementary Fig. 3.
Bland–Altman plots in ADNI for Leave-1-Out sensitivity analyses. Each Bland Altman plot compares factor scores from a full IRT model with factor scores from a model without one of the disease severity markers. See Methods for more details.
Supplementary Fig. 4.
Bland–Altman plots in BIOCARD for Leave-1-Out sensitivity analyses. Each Bland–Altman plot compares factor scores from a full IRT model with factor scores from a model without one of the disease severity markers. See Methods for more details.
References
- 1.Jack C.R., Knopman D.S., Jagust W.J., Shaw L.M., Aisen P.S., Weiner M.W. Hypothetical model of dynamic biomarkers of the alzheimer's pathological cascade. Lancet Neurol. 2010;9:119–128. doi: 10.1016/S1474-4422(09)70299-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McKhann G.M., Albert M.S., Sperling R.A. Changing Diagnostic Concepts of Alzheimer's Disease. In: Hampel H., Carrillo M., editors. Alzheimer's disease - modernizing concept, biological diagnosis and therapy. Karger Publishers; Basel: 2012. [Google Scholar]
- 3.Braak H., Braak E. Neuropathological staging of Alzheimer-related changes. Acta Neuropathol. 1991;82:239–259. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- 4.Rodrigue K., Kennedy K., Devous M., Rieck J., Hebrank A., Diaz-Arrastia R. ß-Amyloid burden in healthy aging: regional distribution and cognitive consequences. Neurology. 2012;78:387–395. doi: 10.1212/WNL.0b013e318245d295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McKhann G.M., Knopman D.S., Chertkow H., Hyman B.T., Jack C.R., Kawas C.H. The diagnosis of dementia due to Alzheimer's disease: recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement. 2011;7:263–269. doi: 10.1016/j.jalz.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sperling R.A., Jack C.R., Aisen P.S. Testing the right target and right drug at the right stage. Sci Transl Med. 2011;3:111cm133. doi: 10.1126/scitranslmed.3002609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Meyer G., Shapiro F., Vanderstichele H., Vanmechelen E., Engelborghs S., De Deyn P.P., Alzheimer's Disease Neuroimaging Initiative Diagnosis-independent alzheimer disease biomarker signature in cognitively normal elderly people. Arch Neurol. 2010;67:949–956. doi: 10.1001/archneurol.2010.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jedynak B.M., Lang A., Liu B., Katz E., Zhang Y., Wyman B.T., Alzheimer's Disease Neuroimaging Initiative A computational neurodegenerative disease progression score: Method and results with the Alzheimer's Disease Neuroimaging Initiative cohort. Neuroimage. 2012;63:1478–1486. doi: 10.1016/j.neuroimage.2012.07.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jack C.R., Knopman D.S., Jagust W.J., Petersen R.C., Weiner M.W., Aisen P.S. Tracking pathophysiological processes in Alzheimer's disease: an updated hypothetical model of dynamic biomarkers. Lancet Neurol. 2013;12:207–216. doi: 10.1016/S1474-4422(12)70291-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kotsiantis S., Kanellopoulos D. Discretization techniques: A recent survey. GESTS Int Trans Computer Sci Eng. 2006;32:47–58. [Google Scholar]
- 11.Samejima F. Estimation of latent ability using a response pattern of graded scores. Psychometrika Monogr Suppl. 1969;34:100. [Google Scholar]
- 12.Albert M., Soldan A., Gottesman R., McKhann G., Sacktor N., Farrington L. Cognitive changes preceding clinical symptom onset of mild cognitive impairment and relationship to ApoE genotype. Curr Alzheimer Res. 2014;11:773–784. doi: 10.2174/156720501108140910121920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Whitwell J.L., Wiste H.J., Weigand S.D., Rocca W.A., Knopman D.S., Roberts R.O., Alzheimer Disease Neuroimaging Initiative Comparison of imaging biomarkers in the Alzheimer Disease Neuroimaging Initiative and the Mayo Clinic Study of Aging. Arch Neurol. 2012;69:614–622. doi: 10.1001/archneurol.2011.3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Olsson A., Vanderstichele H., Andreasen N., De Meyer G., Wallin A., Holmberg B. Simultaneous measurement of beta-amyloid(1-42), total tau, and phosphorylated tau (Thr181) in cerebrospinal fluid by the xMAP technology. Clin Chem. 2005;51:336–345. doi: 10.1373/clinchem.2004.039347. [DOI] [PubMed] [Google Scholar]
- 15.Shaw L.M., Vanderstichele H., Knapik-Czajka M., Clark C.M., Aisen P.S., Petersen R.C., Alzheimer's Disease Neuroimaging Initiative Cerebrospinal fluid biomarker signature in Alzheimer's disease neuroimaging initiative subjects. Ann Neurol. 2009;65:403–413. doi: 10.1002/ana.21610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Voevodskaya O., Simmons A., Nordenskjöld R., Kullberg J., Ahlström H., Lind L., Alzheimer's Disease Neuroimaging Initiative The effects of intracranial volume adjustment approaches on multiple regional MRI volumes in healthy aging and Alzheimer's disease. Front Aging Neurosci. 2014;6:264. doi: 10.3389/fnagi.2014.00264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mueller S.G., Weiner M.W., Thal L.J., Petersen R.C., Jack C.R., Jagust W. Ways toward an early diagnosis in Alzheimer's disease: the Alzheimer's Disease Neuroimaging Initiative (ADNI) Alzheimers Dement. 2005;1:55–66. doi: 10.1016/j.jalz.2005.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rey A. [Clinical examination in psychology]. Presses Univeritaires de France; Paris: 1964. L'examen clinique en psychologie. [Google Scholar]
- 19.Lezak M.D., Howieson D.B., Loring D.W. Oxford University Press; New York: 2004. Neuropsychological assessment; pp. 368–370. [Google Scholar]
- 20.Delis D.C., Kramer J.H., Kaplan E., Ober B.A. The Psychological Corporation; New York: 1986. California Verbal Learning Test. [Google Scholar]
- 21.Gross A.L., Inouye S.K., Rebok G.W., Brandt J., Crane P.K., Parisi J.M. Parallel But Not Equivalent: Challenges and Solutions for Repeated Assessment of Cognition over Time. J Clin Exp Neuropsychol. 2012;34:758–772. doi: 10.1080/13803395.2012.681628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pfeffer R.I., Kurosaki T.T., Harrah C.H., Chance J.M., Filos S. Measurement of functional activities in older adults in the community. J Gerontol. 1982;37:323–329. doi: 10.1093/geronj/37.3.323. [DOI] [PubMed] [Google Scholar]
- 23.Horn J.L. A rationale and test for the number of factors in factor analysis. Psychometrika. 1965;30:179–185. doi: 10.1007/BF02289447. [DOI] [PubMed] [Google Scholar]
- 24.Singer J.D., Willet J. Oxford University Press; New York, NY: 2003. Applied Longitudinal Data Analysis: Modeling Change and Event Occurrence. [Google Scholar]
- 25.Lord F.M. The relation of test score to the trait underlying the test. Educ Psychol Meas. 1953;13:517–549. [Google Scholar]
- 26.Takane Y., de Leeuw J. On the relationship between item response theory and factor analysis of discretized variables. Psychometrika. 1987;52:393–408. [Google Scholar]
- 27.Mellenbergh G.J. Generalized linear item response theory. Psychol Bull. 1994;115:300–307. [Google Scholar]
- 28.Muthen L.K., Muthen B.O. Seventh Edition. Muthen & Muthen; Los Angeles, CA: 1998–2012. Mplus user's guide. [Google Scholar]
- 29.Bland J.M., Altman D.G. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet. 1986;1:307–310. [PubMed] [Google Scholar]
- 30.Dickerson B.C., Bakkour A., Salat D.H., Feczko E., Pacheco J., Greve D.N. The cortical signature of Alzheimer's disease: regionally specific cortical thinning relates to symptom severity in very mild to mild AD dementia and is detecTable in asymptomatic amyloid-positive individuals. Cereb Cortex. 2009;19:497–510. doi: 10.1093/cercor/bhn113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iturria-Medina Y., Sotero R.C., Toussaint P.J., Mateos-Pérez J.M., Evans A.C., Alzheimer's Disease Neuroimaging Initiative Early role of vascular dysregulation on late-onset Alzheimer's disease based on multifactorial data-driven analysis. Nat Commun. 2016;7:11934. doi: 10.1038/ncomms11934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ioannidis J.P. Why most published research findings are false. PLoS Med. 2005;2:e124. doi: 10.1371/journal.pmed.0020124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ardekani B.A., Convit A., Bachman A.H. Analysis of the MIRIAD Data Shows Sex Differences in Hippocampal Atrophy Progression. J Alzheimers Dis. 2016;50:847–857. doi: 10.3233/JAD-150780. [DOI] [PubMed] [Google Scholar]
- 34.Lee S.H., Bachman A.H., Yu D., Lim J., Ardekani B.A. Predicting progression from mild cognitive impairment to Alzheimer's disease using longitudinal callosal atrophy. Alzheimers Dement (Amst) 2016;2:68–74. doi: 10.1016/j.dadm.2016.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bachman A.H., Lee S.H., Sidtis J.J., Ardekani B.A. Corpus callosum shape and size changes in early Alzheimer's disease: a longitudinal MRI study using the OASIS brain database. J Alzheimers Dis. 2014;39:71–78. doi: 10.3233/JAD-131526. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.