Short abstract
A global quantitative proteomic analysis in a human hepatoma cell line in the presence and absence of interferon treatment using the isotope coded affinity tag method and tandem mass spectrometry identified several novel regulatory components of the interferon response.
Abstract
Background
Interferons (IFNs) play a critical role in the host antiviral defense and are an essential component of current therapies against hepatitis C virus (HCV), a major cause of liver disease worldwide. To examine liver-specific responses to IFN and begin to elucidate the mechanisms of IFN inhibition of virus replication, we performed a global quantitative proteomic analysis in a human hepatoma cell line (Huh7) in the presence and absence of IFN treatment using the isotope-coded affinity tag (ICAT) method and tandem mass spectrometry (MS/MS).
Results
In three subcellular fractions from the Huh7 cells treated with IFN (400 IU/ml, 16 h) or mock-treated, we identified more than 1,364 proteins at a threshold that corresponds to less than 5% false-positive error rate. Among these, 54 were induced by IFN and 24 were repressed by more than two-fold, respectively. These IFN-regulated proteins represented multiple cellular functions including antiviral defense, immune response, cell metabolism, signal transduction, cell growth and cellular organization. To analyze this proteomics dataset, we utilized several systems-biology data-mining tools, including Gene Ontology via the GoMiner program and the Cytoscape bioinformatics platform.
Conclusions
Integration of the quantitative proteomics with global protein interaction data using the Cytoscape platform led to the identification of several novel and liver-specific key regulatory components of the IFN response, which may be important in regulating the interplay between HCV, interferon and the host response to virus infection.
Background
Interferons (IFNs) were originally discovered as antiviral proteins that inhibit virus replication [1]. Upon virus infection, IFNs are induced in mammalian cells and thus mediate cellular homeostatic responses to virus infection. In addition to their antiviral properties, IFNs are involved in many other physiological processes including cell growth and proliferation, cell death, the immune response and other cellular defense mechanisms [2]. The IFN signaling pathway has been extensively studied [3,4]. On binding of IFNs to their cognate receptors, the JAK-STAT signal transduction pathway is triggered, culminating in the transcription of IFN-stimulated genes (ISGs) that mediate IFN function. The proteins encoded by ISGs include, but are not limited to, many antiviral effectors such as the double-stranded RNA-activated protein kinase PKR (which inhibits viral protein synthesis via eIF2α phosphorylation), the 2'-5' oligoadenylate synthetase (2'-5' OAS) (which activates RNase L to degrade viral RNA), and the Mx GTPases (which block viral transport inside the cell). Other ISGs include ISG56 (which inhibits translation via eIF3) and the P200 family (which impairs cell proliferation through cellular factors such as NFκB, E2F, P53, c-Myc, UBF-1, YY1, MyoD) [2].
Owing to its anti-growth and immune-response properties, IFNs have been successfully applied as therapeutics against several types of cancers and infectious diseases including multiple sclerosis, hepatitis and genital warts [2,5]. One of the most prominent clinical applications of IFNs is treatment of patients infected by hepatitis C virus (HCV) [6]. HCV has infected an estimated 3% of the world population [7]. In the absence of a protective vaccine, the only useful therapeutic regimen to date has been treatment using interferon-alpha (IFN-α) together with ribavirin, a broad spectrum antiviral nucleoside [8]. However, more than 50% of HCV-infected patients showed low rates of response to this therapy, in particular patients infected by genotype-1 HCV which is a more infectious sub-genotype among Americans and Europeans. Therefore, further elucidation of the mechanism of IFN response in liver cells could help development of more effective therapeutics against HCV.
Here we describe a quantitative proteomic analysis of the IFN response in human liver carcinoma Huh7 cells using the isotope-coded affinity tag (ICAT) method and mass spectrometry (MS). We compared the global protein expression profile in human liver cells in the presence or absence of IFN-α treatment. Protein identification and quantification were executed and statistically verified using a suite of software tools, including PeptideProphet [9], ProteinProphet [10] and ASAPRatio [11]. By this analysis, we identified more than 1,300 proteins at a threshold that showed a false-positive rate of less than 5%. Of these proteins, 54 were IFN-induced proteins and 24 were IFN-repressed. These include previously well studied IFN-regulated proteins as well as novel proteins. The cellular functions of those proteins were analyzed on the basis of the Gene Ontology [12], using the GoMiner program [13] and the Cytoscape platform [14].
Results and discussion
Global proteomic analysis of IFN- and mock-treated Huh7 cells
To develop a systems-based understanding of the IFN response of liver cells we carried out a global proteomic analysis on human liver carcinoma cells, Huh7, under conditions of IFN or mock treatment. We chose the Huh7 cell line because it is a widely studied cell model and has been used to develop a unique cell system containing the HCV RNA replicon, which is the best cell model system to study HCV propagation and infection [15]. We collected cells 16 hours after treatment with either IFN (400 IU/ml) or mock treatment. The time period and IFN dose were chosen on the basis of the maximal response in gene expression obtained from previous time-course studies on IFN responses using a microarray approach [16,17]. To increase the coverage of protein identification, we fractionated the cell lysates into cytoplasmic, nuclear and membrane fractions and analyzed them individually by the ICAT method and tandem mass spectrometry (MS/MS). For each fraction, the IFN- or mock-treated samples were labeled with either isotopically light (12C) or heavy (13C) acid-cleavable ICAT reagents containing a biotin affinity tag. The light and heavy samples were then combined, proteolyzed to peptides, and fractionated by high-performance liquid chromatography (HPLC) cation-exchange chromatography followed by avidin-affinity chromatography. The purified isotopically labeled peptides were subject to microcapillary HPLC (μLC) followed by electrospray tandem mass spectrometry (ESI-MS/MS) using an ion trap [18,19].
The peptides were identified by matching peptide tandem mass spectra to sequences in the human International Protein Index (IPI) database [20] using SEQUEST software [21]. Each assignment of a MS/MS spectrum to a peptide sequence was statistically validated using the PeptideProphet software tool, which uses the database search scores and peptide end information to compute a probability score for each identified peptide, on a scale of 0 (incorrect identification) to 1 (correct identification) [9]. The peptide identifications were subsequently transformed into protein identifications with overall probability scores using the ProteinProphet software tool [10]. Assigning a probability score to each identified peptide/protein is essential for prioritizing proteins for follow-up analysis, for publication of data that are of consistent quality, and for dataset comparisons. In this study, we identified and quantified 1,190 unique proteins from all three fractions of Huh7 cells with probability scores higher than 0.9 (error rate ≤ 1%) (see Additional data files 1-4 for the original data files) [22]. A reduction of the protein probability scores to 0.4 provided 1,364 protein identifications. This threshold had a sensitivity of 98% and a false-positive error rate of less than 5%. Cellular fractionation improved protein identification, particularly for the nuclear and membrane proteins, which are usually less abundant than the cytoplasmic proteins (Figure 1a).
Figure 1.
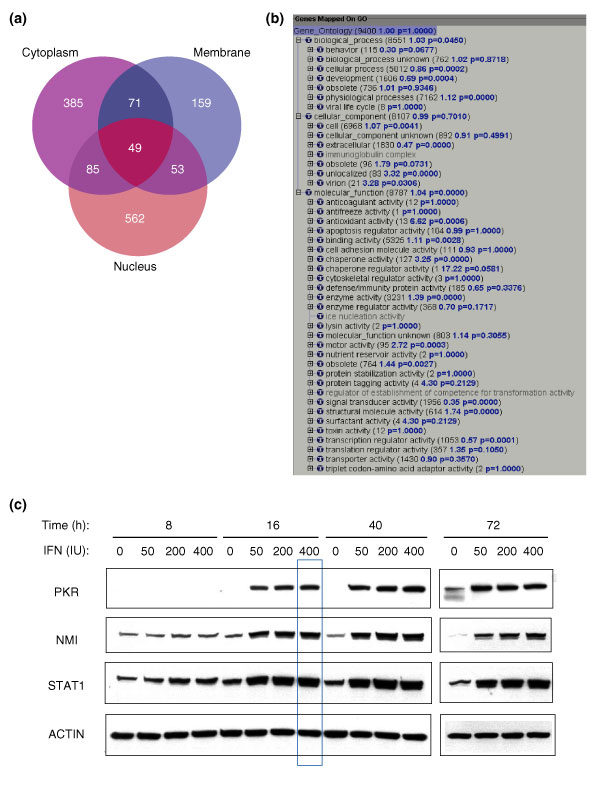
Protein identification. (a) Protein identification from the cytoplasmic (purple), membrane (blue) and nuclear (pink) fractions of the Huh7 cells. Numbers of identified proteins with p ≥ 0.4 are shown. (b) Proteins identified in this study covered most of the 'biological process', 'cellular components' and 'molecular functions' categories at GO level 3. GO analysis was performed following the procedure described in [13]. Briefly, 16,477 human proteins currently carrying human GO nomenclatures were imported into the GoMiner program. Of the 16,477 human genes carrying GO annotations, 9,400 were loaded as Query Gene File in the GoMiner program to build a GO category structure based on the GO annotation database (June 2003). The 1,364 proteins were subsequently loaded as Query Changed Gene File in the GoMiner program to display coverage of the proteomics-identified proteins with respect to GO category. Of the 1,364 proteins, 815 that carry current GO annotations finally contributed to the coverage. (c) For immunoblot analysis, 20 μg of each lysate from IFN-treated Huh7 cells at the indicated time points and dose was subjected to western blot analysis using antibodies as indicated. Data at the 16 h time point and 400 IU/ml IFN concentration, which are the conditions for proteomic analysis, are highlighted with a blue box.
In addition to statistically validating peptide/protein identifications, we also applied statistical analysis to protein quantification. In quantitative proteomics, changes in protein abundance are determined by calculating the ratio of isotopically labeled peptides. The different isotopic peptides were introduced by ICAT labeling (light for IFN and heavy for mock in this study). The abundance ratios were calculated using the ASAPRatio software tool [11]. The ratio for each quantified peptide was calculated from multiple measurements, including measurements of different charge states of the same peptide, measurements of repeat MS analyses of the same peptide from different cation-exchange chromatography fractions, and repeat measurements of the same peptides outside the dynamic exclusion windows of mass spectrometry. Protein abundance was subsequently calculated from the ratios of all the peptides corresponding to the same protein. Therefore, all protein ratios obtained in this study represent the average ± error calculated from numerous independent measurements. Such statistics-based data processing and presentation is particularly critical for global quantitative proteomics analysis, for which the possibility for multiple repeat experiments is technically and economically restricted at present.
Liver proteome representation
The 1,364 proteins identified in the study at a threshold of 5% error rate were examined for their biological association to Gene Ontology (GO) categories [12]. With the help of the recently developed GoMiner program [13], we built a GO category structure based on 9,400 human genes currently carrying human GO annotations (used as Query Gene File). The 1,364 identified proteins from this study were then loaded as Query Changed Gene File into the GoMiner program to examine the distribution of these proteins in the GO category structure. Although only 815 of the 1,364 identified proteins have been currently annotated by the GO consortium, they already covered, at level 3 of the GO ontology, all seven GO 'biological_process' categories, six of the seven 'cellular_component' (except 'immunoglobulin complex') categories, and 27 of the 29 'molecular_function' (except 'ice nucleation activity' and 'regulator of establishment of competence for transformation activity') categories (Figure 1b). Therefore, the proteins identified in this study represent most, if not all, GO-categorized cellular processes. A human liver proteomic dataset consisting of more than 1,300 proteins, which is publicly accessible at [23], is expected to be useful for research on IFN signaling and liver diseases. The dataset, together with its enriched information on human liver protein/peptide identifications, GO categories, cellular fractionations, cation exchange and reverse-phase chromatography elution profiles, and so on, is also expected to be useful as a reference for future human proteomics analyses. In addition, it strongly suggests the feasibility of performing large-scale proteomics analyses on the Huh7 cell-based HCV replicon system to study cellular responses to HCV RNA replication.
Profiling IFN-regulated protein expression
Application of the ICAT methodology in this study allowed us to quantitatively compare protein-expression profiles between IFN- and mock-treated Huh7 cells. The 1,364 identified proteins were quantified by the ratios of protein levels between IFN- and mock-treated samples (Table 1). Upon IFN treatment, 54 of these proteins displayed a more than two-fold increase in level; these included 15 IFN-induced proteins reported previously and 39 novel IFN-induced proteins. Using the GoMiner program, we analyzed the biological classifications of the 54 IFN-induced proteins in the GO categories and found that they were enriched in defense-related cellular pathways such as stress response, immune response, signaling pathways and cell organization (Figure 2a). In contrast, the 24 IFN-repressed proteins were mostly associated with cellular pathways such as cell metabolism, development and RNA transport (Figure 2b) in the GO analysis. The novel IFN-repressed proteins are particularly interesting as little is known about the mechanisms underlying IFN downregulation of gene expression.
Table 1.
List of proteins up- or downregulated by two-fold or more by IFN in this study
| IPI code | Protein* | p† | Ratio‡ | SD | Number§ |
| IFN-induced proteins | |||||
| IPI00164222 | ACACA: acetyl-coenzyme A carboxylase alpha | 1.00 | 3.351 | 0.259 | 2 |
| IPI00033030¶ | ADRM1: adhesion regulating molecule 1 | 0.79 | 9.508c | 1.043 | 1 |
| IPI00012007¶ | AHCY: S-adenosylhomocysteine hydrolase | 0.93 | 6.279n | 0.936 | 1 |
| IPI00032958 | ANLN: anillin, actin binding protein (scraps homolog, Drosophila) | 0.43 | 2.420 | 0.129 | 1 |
| IPI00004656 | B2M: beta-2-microglobulin | 1.00 | 2.768 | 0.583 | 8 |
| IPI00027803 | BAL: B aggressive lymphoma gene | 0.96 | 2.835 | 0.184 | 1 |
| IPI00171487 | CABC1: chaperone, ABC1 activity of bc1 complex like (S. pombe) | 0.99 | 3.533 | 1.659 | 2 |
| IPI00015199 | CD7: CD7 antigen (p41) | 0.57 | 4.523 | 2.204 | 2 |
| IPI00028275 | CH-TOG : Colonic and hepatic tumor overexpressed protein | 1.00 | 2.048 | 0.085 | 4 |
| IPI00011284 | COMT: catechol-O-methyltransferase | 1.00 | 2.614 | 0.239 | 2 |
| IPI00164416 | CSA_PPIasePeptidyl prolyl cis trans isomerase | 1.00 | 8.104 | 1.070 | 55 |
| IPI00029485 | DCTN1: dynactin 1 (p150, glued homolog, Drosophila) | 0.99 | 2.183 | 0.133 | 1 |
| IPI00163553 | DKFZP564C186: DKFZP564C186 protein | 1.00 | 2.108 | 0.505 | 3 |
| IPI00061170 | DNAH11: dynein, axonemal, heavy polypeptide 11 | 0.94 | 9999 | -1 | 2 |
| IPI00015947 | DNAJB1: DnaJ (Hsp40) homolog, subfmaily B, member 1 | 0.89 | 2.571 | 0.223 | 1 |
| IPI00031836 | DRG1: developmentally regulated GTP binding protein 1 | 1.00 | 2.471 | 1.588 | 4 |
| IPI00029536 | EEF1A protein [Fragment] | 1.00 | 3.035 | 0.624 | 5 |
| IPI00010810 | ETFA: electron-transfer-flavoprotein, alpha polypeptide | 0.45 | 2.890 | 0.484 | 1 |
| IPI00065180 | FLJ32915: hypothetical protein FLJ32915 | 0.73 | 6.054 | 4.883 | 2 |
| IPI00031773 | FOXA2: forkhead box A2, hepatic nuclear factor-3-beta | 0.99 | 2.640 | 1.678 | 1 |
| IPI00007241 | G1P2: IFN, alpha-inducible protein (clone IFI-15K) | 0.98 | 4.858 | 0.661 | 1 |
| IPI00028564 | GBP1: guanylate binding protein 1, IFN-inducible | 0.56 | 2.130 | 0.208 | 1 |
| IPI00026268¶ | GNB1:G protein, beta subunit 1 | 1.00 | 5.845n | 0.133 | 2 |
| IPI00171038 | GPR111: G protein-coupled receptor 111 | 0.63 | 21.27 | 4.741 | 2 |
| IPI00007404 | APG7L: ubiquitin activating enzyme E1-like protein, GSA7 | 0.98 | 2.844 | 0.663 | 1 |
| IPI00007853 | IFI30: IFN, gamma-inducible protein 30 | 0.97 | 2.219 | 0.183 | 1 |
| IPI00008613 | IFI35: IFN-induced protein 35 | 1.00 | 2.223 | 0.426 | 2 |
| IPI00018300 | IFIT1: IFN-induced protein with tetratricopeptide repeats 1 | 0.48 | 9999 | -1 | 1 |
| IPI00024254 | IFIT4: IFN-induced protein with tetratricopeptide repeats 4 | 1.00 | 6.230 | 0.794 | 6 |
| IPI00026834 | INPP5E: inositol polyphosphate-5-phosphatase, 72 kDa | 1.00 | 2.470 | 0.399 | 2 |
| IPI00059811 | ISG20: IFN stimulated gene 20 kDa | 0.88 | 2.303 | 0.319 | 1 |
| IPI00032387 | KIAA0186: KIAA0186 gene product | 0.99 | 2.365 | 0.198 | 2 |
| IPI00003499 | KIAA1276: KIAA1276 protein | 0.62 | 3.815 | 0.058 | 1 |
| IPI00020096 | KNS2: kinesin 2 60/70 kDa | 1.00 | 3.140 | 0.335 | 2 |
| IPI00013976 | LAMB1: laminin, beta 1 | 0.94 | 2.693 | 0.180 | 2 |
| IPI00152503 | LOC151636: rhysin 2 | 1.00 | 2.975 | 0.231 | 4 |
| IPI00000421 | M96:mouse metal response element binding transcription factor 2 | 0.98 | 2.923 | 0.390 | 1 |
| IPI00023399 | MTP: microsomal triglyceride transfer protein (88 kDa) | 0.97 | 4.748 | 0.751 | 1 |
| IPI00024681 | MX1: myxovirus resistance 1, IFN-inducible protein p78 | 0.93 | 2.300 | 0.204 | 1 |
| IPI00012450 | NMI: N-Myc (and STAT) interactor | 0.57 | 2.875 | 0.138 | 1 |
| IPI00000022 | NUDT2: nucleoside diphosphate linked moiety X type motif 2 | 0.98 | 3.684 | 0.224 | 2 |
| IPI00002405 | OAS3: 2'-5'-oligoadenylate synthetase 3, 100 kDa | 1.00 | 2.460 | 0.076 | 5 |
| IPI00141040 | PASK PAS domain containing serine/threonine kinase | 0.49 | 12.006 | 1.024 | 2 |
| IPI00024989 | PCMT1: protein-L-isoaspartate (D-aspartate) O-methyltransferase | 0.98 | 2.836 | 0.276 | 1 |
| IPI00016461 | PLCD1: phospholipase C, delta 1 | 0.69 | 4.569 | 0.116 | 1 |
| IPI00019463 | PKR protein kinase, IFN-inducible ds RNA dependent | 1.00 | 3.963 | 0.659 | 3 |
| IPI00008530 | RPLP0: ribosomal protein, large, P0 | 1.00 | 2.044 | 0.198 | 3 |
| IPI00021417 | SART1: T-cell recognized squamous cell carcinoma antigen | 0.57 | 2.616 | 0.093 | 1 |
| IPI00030781¶ | STAT1: large peptide, 91 kDa | 1.00 | 2.153 m | 0.079 | 9 |
| IPI00024723 | UBE2L6: ubiquitin-conjugating enzyme E2L 6 | 0.57 | 9999 | -1 | 1 |
| IPI00146772 | ENSEMBL:ENSP00000295676 Tax_Id = 9606 | 0.78 | 3.240 | 0.955 | 2 |
| IPI00062913 | REFSEQ:XP_062729 hypothetical protein | 0.46 | 3.073 | 2.000 | 1 |
| IPI00045468 | REFSEQ:XP_058770 Hypothetical protein | 0.47 | 2.675 | 0.113 | 1 |
| IPI00145529 | REFSEQ:XP_167245 similar to seven-pass transmembrane receptor protein precursor and cyclophilin type peptidylprolyl isomerase A | 1.00 | 2.29 | 0.448 | 5 |
| IFN-repressed proteins | |||||
| IPI00009137 | CPSF4: cleavage and polyadenylation specific factor 4, 30 kDa | 0.96 | 0.378 | 0.154 | 1 |
| IPI00024549 | FABP: fatty acid binding protein | 0.93 | 0.328 | 0.041 | 1 |
| IPI00004619 | FACL4: fatty-acid-coenzyme A ligase, long-chain 4 | 0.98 | 0.416 | 0.081 | 1 |
| IPI00026781 | FASN: fatty acid synthase | 0.99 | 0.304 | 0.100 | 137 |
| IPI00016250 | FXR2: fragile X mental retardation, autosomal homolog 2 | 0.95 | 0.391 | 0.074 | 1 |
| IPI00011454 | G2AN: alpha glucosidase II alpha subunit | 1.00 | 0.198 | 0.033 | 13 |
| IPI00156282 | GPS1: G protein pathway suppressor 1 | 0.98 | 0.455 | 0.138 | 1 |
| IPI00150148 | HIC1: hypermethylated in cancer 1 | 0.94 | 0.000 | -1 | 1 |
| IPI00015587 | Hypothetical protein FLJ21140 | 0.71 | 0.043 | 0.064 | 3 |
| IPI00166489 | IGLC3: Ig lambda chain C regions | 0.98 | 0.265 | 0.11 | 2 |
| IPI00055954 | KIAA0007: KIAA0007 protein | 1.00 | 0.426 | 0.014 | 2 |
| IPI00166105 | KRT10: keratin 10 | 0.97 | 0.495 | 0.055 | 2 |
| IPI00022980 | KRT6: keratin 6 | 1.00 | 0.003 | 0.008 | 14 |
| IPI00017596 | MAPRE1: RP/EB family, member 1 | 0.98 | 0.339 | 0.016 | 1 |
| IPI00005948 | MGC3207: hypothetical protein MGC3207 | 0.61 | 0.499 | 0.071 | 1 |
| IPI00004399 | MIG-6: Gene 33/Mig-6 | 0.99 | 0.000 | -1 | 2 |
| IPI00022334 | OAT: ornithine aminotransferase | 0.98 | 0.331 | 0.018 | 1 |
| IPI00021794 | PPGB: protective protein for beta-galactosidase (galactosialidosis) | 1.00 | 0.323 | 0.084 | 8 |
| IPI00034308 | SARDH: sarcosine dehydrogenase | 0.98 | 0.484 | 0.008 | 1 |
| IPI00023344 | SYMPK: symplekin | 1.00 | 0.496 | 0.029 | 3 |
| IPI00099730 | SRRM2: serine/arginine repetitive matrix 2 | 0.82 | 0.434 | 0.224 | 1 |
| IPI00027230¶ | TRA1: tumor rejection antigen (gp96) 1 | 1.00 | 0.452 mn | 0.165 | 4 |
| IPI00166768¶ | TUBA6: tubulin alpha 6 | 1.00 | 0.383 m | 0.165 | 14 |
| IPI00024693 | WNT9A: wingless-type MMTV integration site family, member 9A | 0.99 | 0.316 | 0.091 | 2 |
*Genes previously identified in the microarray study by Williams and colleagues [15] are shown in bold. †ProteinProphet probability score. ‡Ratio of IFN/mock. Ratio of 0.000 or 9999 indicates no detection of the IFN or mock signals, respectively. In either case the standard deviation is expressed as -1. §Number of peptides identified. ¶Proteins in which IFN-mediated expression was differentially regulated among the three cellular fractions in this study. Their ratios refer to specific cellular fractions as indicated: n, nuclear; m, membrane; c, cytoplasmic.
Figure 2.
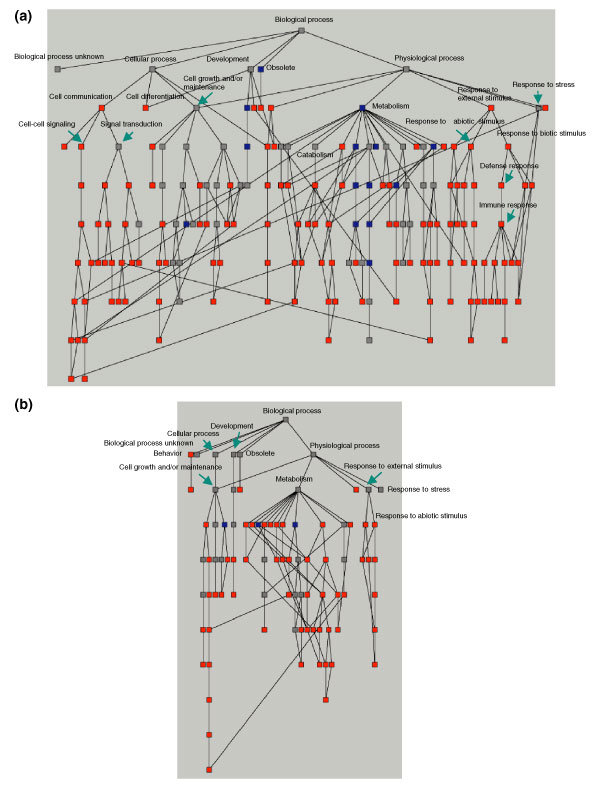
Distribution of IFN-regulated proteins at various levels of GO categories. The 1,364 proteins identified were imported into the GoMiner program to generate a directed acyclic graph (DAG) based on current annotations in the GO database. Each node of the DAG represents one GO category at various levels. (a) Fifty-four IFN-induced and (b) 24 IFN-repressed proteins were used as queries and loaded into GoMiner to classify these proteins into each GO category. The distribution of these IFN-regulated proteins is represented by a different color at each node. The color indicates a 1.5-fold enrichment (red), depletion (blue) or no change (gray) of the IFN-regulated proteins compared with the distribution over all identified proteins at that level. Several critical GO categories are labeled.
The antiviral defense and immune response
We then investigated the individual cellular pathways with which the IFN-regulated proteins were associated. IFNs are best known for their antiviral defense properties. Years of IFN research have discovered several well known IFN-induced proteins such as the protein kinase PKR, 2'-5' OAS, Mx, RNA-specific adenosine deaminase (ADAR) and interferon regulatory factors (IRF) [2-4,24]. From this study, we identified all these classical IFN-induced proteins except the IRFs. Missing the IRFs is possibly due to the very low abundance of these transcription factors inside the liver cell, resulting in a concentration that is below the sensitivity of our MS detection. All these IFN-induced proteins were shown to be induced at least two-fold upon IFN treatment with the exception of ADAR (1.4-fold) (Table 1). Identification of these IFN-induced proteins is consistent with a previous microarray study performed by Williams and colleagues at the Cleveland Clinic Foundation (Table 1, proteins in bold) [17,25]. We also identified/quantified two IFN-induced proteins, STAT1 and NMI, which are involved in the upstream IFN-related JAK/STAT signaling pathways. We performed western blot analysis to confirm the protein expression patterns detected by quantitative proteomics analysis (Figure 1c). We examined the expression patterns of several IFN-induced proteins such as PKR, NMI and STAT1 at multiple doses (0, 50, 200, 400 IU/ml) and multiple time points (8, 16, 40 and 72 hours). These data indicate that the patterns observed by immunoblot analysis are consistent with the data obtained by quantitative mass spectrometry and that the conditions used for the proteomics analysis (16-hour treatment of IFN at 400 IU/ml concentration) represented conditions of which an optimal IFN response was observed (Figure 1c, boxed in blue).
In addition to coverage of many of the well characterized IFN-induced proteins, our proteomic study also identified a significant number of proteins involved in the immune response, which is also critical for IFN's antiviral activities. Applying GO analysis to examine the biological classifications of the 54 IFN-induced proteins, we found that 11 were involved in the immune response, which contributed to a 4.6-fold enrichment at the GO category of 'immune response' (Figure 2a). This is represented by the presence of β2-microglobulin and IFI30, which are involved in MHC class I and MHC class II antigen processing, respectively, as well as CD7, which plays a role in T-cell activation (Figure 3). In addition, we identified several IFN-induced immunomodulatory proteins that function in other cellular pathways and thus may have interesting roles involving cross-talk between the immune response and other cellular regulatory pathways. Examples include IFI35, an IFN inducible protein involved in apoptosis; GBP1, a guanine-nucleotide-binding protein involved in signaling pathways; and G1P2, an ubiquitin cross-reactive protein involved in conjugating and targeting proteins to the cytoskeleton. Interestingly, we also found IFIT1 and IFIT4, two IFN-induced proteins with tetratricopeptide repeats (TPR motif), which are involved in protein-protein interactions. However, their molecular functions and potential interaction targets in the immune response remain to be determined.
Figure 3.
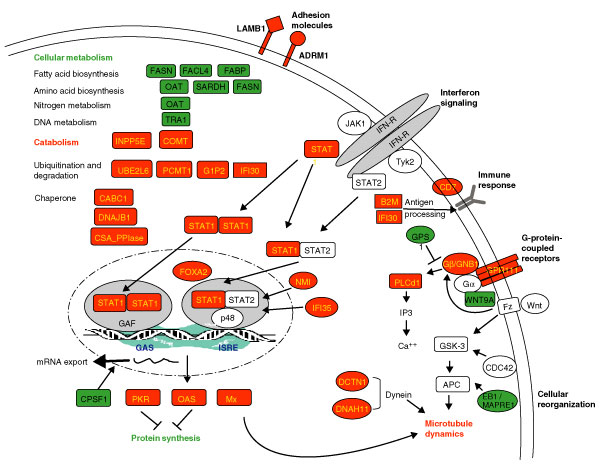
Summary of cellular pathways of IFN-regulated proteins based on data analysis as described in the text. The IFN-induced proteins are shown in red and IFN-repressed proteins in green.
Cell metabolism and growth
IFN treatment has previously been shown to have pleiotropic effects on many aspects of cell physiology, including cell growth, proliferation, reorganization and death [2,4]. The cellular response to stress conditions such as virus infection, which induces high IFN levels, might be to reduce levels of cellular metabolism. In support of this notion, we observed that IFN suppressed expression of proteins involved in metabolic pathways including fatty-acid metabolism (fatty acid synthase (FASN); long-chain fatty-acid-coenzyme A ligase 4 (FACL4); and fatty-acid-binding proteins (FABP)), amino-acid metabolism (ornithine aminotransferase (OAT); sarcosine dehydrogenase (SARDH); and FASN), nitrogen metabolism (OAT), and DNA metabolism (tumor rejection antigen 1 (TRA1) involved in histone acetylation) (Figure 3).
IFN was previously reported to inhibit cell growth, possibly through regulation of cell-cycle control [2]. In support of this, we observed an induction of PKR by IFN treatment; PKR has been reported to negatively regulate cell growth and proliferation [26]. Interestingly, information provided from the GO analysis also revealed repression of protein MAPRE1, which is an EB1 family member binding to APC (adenomatous polyposis coli), and is associated with the mitotic apparatus and cell-cycle regulation [27,28].
Correlation of the cellular pathways
Acquisition of a large global proteomics dataset provides us with a unique opportunity to analyze the IFN-mediated cellular pathways at a systems-based level. The results discussed above suggest cross-talk between activation of the antiviral defense pathways and suppression of pathways involved with cell growth and proliferation. When applying GO analysis to investigate the distribution of the 78 IFN-regulated (induced and repressed) proteins in multiple cellular pathways, we observed that many IFN-induced and repressed proteins may be functionally related and react in a coordinated manner. As shown in Figure 2, at the 'biological process' GO category (level 2) and its subcategories, IFN-induced proteins were enriched in several cellular defense-related pathways including immune response, stress response and signal transductions (Figure 2a, red nodes) and were relatively depleted in the categories of metabolism (Figure 2a, blue nodes). On the other hand, the IFN-repressed proteins are more likely to be enriched in the metabolic pathways (Figure 2b, red nodes) but not in pathways such as those involved in cellular defense mechanisms.
IFN signaling
Taking advantage of the global proteomics approach together with systems-based data mining, we also successfully identified and quantified multiple components of the IFN signaling pathways, including STAT1 and NMI in the JAK-STAT pathway, Mx1 (a GTPase involved in the immune response, cellular organization and apoptosis), PASK (PAS-domain-containing serine/threonine kinase in phosphate metabolism), GPR111 and PLCD1 (G-protein signaling), GNB1 (G-protein signaling and Ras signal transduction), GPS1 (G-protein, MAPK, and JNK signaling), WNT9A (Wnt signaling) (Figure 3). The IFN-mediated differential expression of many signaling molecules within the signal transduction network strongly suggests a crosstalk among multiple signaling pathways.
Of particular interest is the activation of the G-protein-coupled receptor (GPCR) signaling pathway during the IFN response. The GPCR signaling pathway has been widely studied. More than 1,000 GPCRs have been cloned, which regulate, at a minimum, the adenylate cyclase, phospholipase C, and phosphodiesterase-mediated signal transduction pathways [29,30]. However, little is known about the regulation of the IFN-stimulated G-protein signaling pathways. Interestingly, we observed a number of IFN-induced proteins involved in the G-protein signaling pathways: GPR111 (upregulated 21.3-fold), GNB1 (5.8-fold), and PLCD1 (4.6-fold) (Figure 3). GPR111 is the G-protein-coupled receptor 111. GNB1 is the β-subunit of G proteins and has crucial roles in both inositol trisphosphate (IP3)-coupled G-protein signaling and Ras protein signal transduction. The phospholipase C family member PLCD1 is a PIP2 phosphodiesterase (delta subunit) and is also involved in IP3-coupled G-protein signaling. Curiously, a G-protein pathway suppressor, GPS1 was downregulated by IFN treatment (2.2-fold) in our study. Therefore, induction of G-protein-coupled signaling molecules appeared to be coordinated with downregulation of the G-protein pathway suppressor protein. Observation of these multiple signaling molecules involved with the GPCR pathways strongly suggests an IFN-stimulated and GPCR-mediated signal transduction. It is unclear whether this enrichment of the series of G-protein signaling components is reflective of a novel IFN-mediated signaling loop or just involves identification of several components of the G-protein signaling pathways.
In addition to the G-protein signaling, we also observed a decreased expression of the Wnt signaling molecule WNT9A on IFN treatment (downregulated 3.2-fold). The Wnt signaling pathway is essential in development. Whether this downregulation of WNT9A is related to a stress-mediated attenuation of cellular development awaits further study. The Wnt signaling was also reported to regulate cell polarity, in parallel with Cdc42 [31]. The regulation of the cell polarity requires participation of APC, EB1 and activation of cell organization involved with the dynein proteins of the cytoskeleton [32]. Interestingly, we identified multiple components of the cell-polarity signaling pathway using our proteomic approach, including WNT9A, EB1 (MAPRE1) and dynein (DNAH1, a large subunit of dynactin; and DCTN1, ciliary dynein heavy chain 11), suggesting an IFN-mediated regulation of cell polarity.
Novel IFN-regulated proteins and potential targets for future study
Among the IFN-regulated proteins we identified, proteins hitherto not known to be regulated by IFN are of particular interest as they may reveal important new functions of the IFN response. For example, we discovered two new IFN-induced proteins, ADRM1 and LAMB, that are cell adhesion molecules and may be involved in IFN-mediated cell migration, attachment and cell-cell interactions. Interestingly, these two proteins were both recently found to be downregulated in microarray studies of liver biopsies from HCV-infected patients (M. Smith, D. Thomas, and M.G.K., unpublished data). Two keratin proteins, KRT6 (keratin 6) and KRT10 (keratin 10) were found to be repressed by IFN treatment in our study. Keratins are important components of the cytoskeleton and have important roles in cell migration and invasion. Furthermore, one family member, keratin 19, has previously been shown to be correlated with hepatocellular carcinoma metastasis [33].
Three molecular chaperones, CABC1, CSA PPIase, and DNAJB1, were found to be induced by IFN, while another protein with a protein-protective role, PPGB (protective protein for beta-galactosidase), was repressed by IFN. In addition, four IFN-induced proteins - G1P2, IFI30, PCMT1 and UBE2L6 - were involved in ubiquitination and protein degradation in response to stress signals. It is reasonable to speculate that protein degradation might be activated as a cellular survival response.
Importantly, four IFN-regulated proteins were liver specific, including CH-TOG (overexpressed in hepatomas), FOXA2 (also known as hepatocyte nuclear forkhead transcription factor), MTP (lipid transfer to apoB in the endoplasmic reticulum of human hepatoma cells), and FASN (a fatty-acid synthase whose expression is regulated by liver X receptors). Consistent with the antiviral role of IFN, we found that IFN suppressed expression of two proteins, CPSF4 and TRA1, which are known to be regulated during viral infection. CPSF4 is a cleavage and polyadenylation-specific factor which forms a complex with the influenza virus NS1 protein to selectively block the nuclear export of cellular but not viral mRNA [34]. TRA1, a stress-induced tumor rejection antigen (gp96) 1, is involved in histone acetylation and chromatin modification and associates with the RNA polymerase of the hepatitis B virus [35].
Genome analysis of the IFN-regulated proteins
IFN can affect protein expression by directly regulating gene expression or by controlling protein levels through indirect cellular effectors. To learn more about the IFN regulation of gene expression in our study, we examined the cis-acting DNA elements in the promoter regions of the IFN-regulated protein open reading frames (ORFs). One signature of IFN-inducible genes is the presence of IFN-related transcription elements such as the IFN-stimulated response element (ISRE) and IRF-7 binding site [16]. We analyzed 45 IFN-induced proteins with known promoter sequences. As shown in Table 2, 21 of the 45 genes (44%) encoding the IFN-induced proteins contain at least one cis-element of the ISRE, GAS, IRF-3 or IRF-7 binding sites, suggesting possible direct transcriptional regulation of these proteins. They include the well annotated proteins discussed above, such as ACACA, B2M, GBP1, IFIT1, IFIT4, MX1, NMI, OAS3, PKR, STAT1 and UBE2L6. The other 24 genes appeared to lack these known promoter sequences, suggesting an indirect regulation or the presence of other, unknown, promoter elements. In a similar promoter analysis of the IFN-repressed proteins we were unable to identify common promoter regulatory sequence patterns.
Table 2.
Partial list of cis-DNA elements of IFN-induced proteins
| Gene | Position | Strand | Sequence | Binding site |
| ACACA | -1847 | Rev | GTTTCTTTTTC | ISRE |
| -3800 | Rev | AGTTTCGGATTC | IRF-7 | |
| B2M | -167 | GAAAAGGAAAC | ISRE | |
| -161 | GAAACTGAAAAC | IRF-7 | ||
| -2738 | Rev | GCTTTCATTTTC | ISRE/IRF-7 | |
| G1P2 | -316 | GAAAATGAAAC | ISRE/IRF-7 | |
| -114 | GAAAGGGAAACC | ISRE/IRF-3 | ||
| -108 | GAAACCGAAAC(T) | ISRE/IRF-3/IRF-7 | ||
| GBP1 | -128 | Rev | ACTTTCAGTTTC | IRF-7 |
| GSA7 | 185 | Rev | AATTTCATTTTC | IRF-7 |
| IFIT1 | -143 | Rev | (A)GTTTCACTTTC | ISRE/IRF-7 |
| -405 | GAAACTGAAAAT | IRF-7 | ||
| -2096 | Rev | AATTTCATTTTC | IRF-7 | |
| IFIT4 | -173 | Rev | (G)GTTTCATTTTC | ISRE/IRF-7 |
| -139 | Rev | (A)GTTTCACTTTC | ISRE/IRF-7 | |
| ISG20 | -86 | GAAAGGAAAC | ISRE | |
| 140 | GAAACTGAAAC | ISRE | ||
| KIAA0186 | -609 | GAAATGGAAAGC | ISRE | |
| KIAA1276 | -1048 | GAATCCGAAAAC | ISRE | |
| LAMB1 | -3940 | GAAAGGAAAGC | ISRE | |
| LOC151636 | -94 | GAAACTGAAAC(T) | ISRE/IRF-7 | |
| MX1 | -101 | Rev | GTTTCGTTTC | ISRE |
| -56 | Rev | GTTTCATTTC | ISRE | |
| NMI | -78 | GAAAGTGAAATT | IRF-7 | |
| NUDT2 | -746 | TTCCCGGAA | GAS | |
| -2530 | Rev | GTTTCCTTTC | ISRE | |
| OAS3 | -170 | GAAACGAAAC | ISRE | |
| -165 | GAAACTGAAAGC | ISRE/IRF-7 | ||
| -34 | GAAAACGAAACC | ISRE/IRF-7 | ||
| PCMT1 | -1370 | GAATTTGAAAAT | IRF-7 | |
| PRKR | -3617 | GAAAGGAAAC | ISRE | |
| -374 | Rev | GTTTCCTTTC | ISRE | |
| RPLP0 | -1089 | GAAACGGAAAGT | IRF-3 | |
| STAT1 | 39 | Rev | (A)GTTTCGCTTTC | ISRE/IRF-3/IRF-7 |
| UBE2L6 | -1813 | TTCCGGGAA | GAS |
44% of promoter sequences have at least one of the following consensus sequences: GAS, TTCCSGGAA; ISRE, GAAA(N){1,2}GAAA(G){0,1}C [16]; IRF-3, GAAASSGAAANY; IRF-7, GAAWNYGAAANY.
The protein-interaction network of the IFN-regulated proteins
To gain systematic insights into the novel functions of IFN-regulated proteins, we examined the interactions between these proteins using the software package Cytoscape as our bioinformatics analysis platform. Cytoscape provides a unique in silico approach to examine and display protein-interaction networks based on available protein-protein interactions characterized from previous biological studies [36-38]. We loaded the more than 1,300 identified and quantified proteins from our SBEAMS database into Cytoscape, and identified 219 interactions. Of the total 219 interactions, 157 were directly obtained from publicly available protein interaction databases including HPRD [39] and PreBIND [40]. Expanding the search to additional databases such as BIND [41] and a privately curated compiled 'yeast reference network' [40,42-44], an extra 57 interactions were detected as occurring between orthologs (NCBI HomoloGene database [45]) of the reference proteins in Mus musculus, Saccharomyces cerevisiae and Drosophila melanogaster.
Figure 4 shows the proteins that have been implicated in protein-protein interactions and displayed within an interaction network. The proteins' colors portray their IFN-mediated expression changes: red for upregulation and green for downregulation with a threshold of two-fold. This visualization approach allows us to analyze complex biological data at a systems-based level by studying protein expression in conjunction with their interactions. Although the current databases are not nearly complete, a preliminary examination of this network containing only 157 interacting nodes already provides us with useful information that is easily overlooked by traditional approaches. For example, we note an interaction between two IFN-induced proteins, NMI and IFI35. NMI is a Myc-interacting protein that also interacts with STATs and augments STAT-mediated transcription in response to IFN and interleukin-2 (IL-2) [46,47]. Interaction of NMI with IFI35 is induced by IFN [48,49] and dissociation of these two proteins occurs during apoptosis [50]. Interestingly, these two proteins are close to two other IFN-induced proteins, PKR and EEF1A, in the network, mediated only by YY1 (Yin and Yang 1). Both PKR and EEF1A are involved in translational regulation. YY1 is a transcription factor regulating a variety of promoters including the IFN promoter [51]. YY1 was also found to correlate with c-Myc in hepatocarcinogenesis [52]. As the number of known protein interactions expands in the near future, such a Cytoscape-assisted approach for analyzing large genomic/proteomic datasets will dramatically increase our understanding of these systems and could assist in the discovery of many potential targets for biological follow-up studies as well as therapeutic development.
Figure 4.
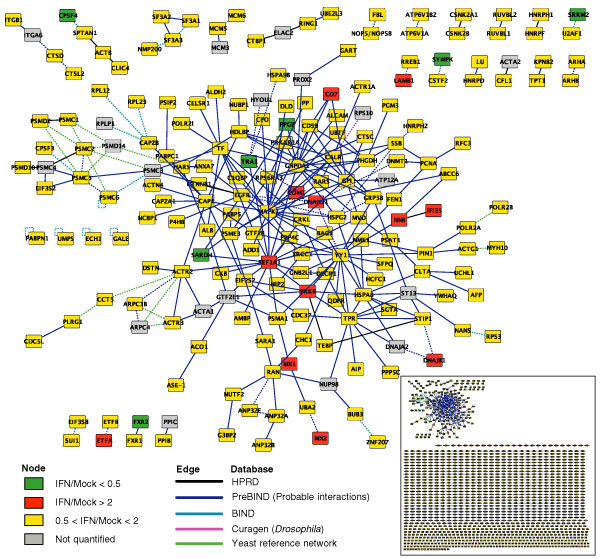
Proteins identified in this study, as visualized by the Cytoscape platform. Orthologs of all significantly expressed proteins were identified in the NCBI HomoloGene database [45], and curated interactions among the proteins and their orthologs were added from the public HPRD [39], BIND [41] and PreBIND [40] databases as well as a privately curated 'yeast reference network' [40,42-44]. ICAT ratios are indicated by node colors as indexed with a two-fold threshold. Interactions are colored according to their source database, and ortholog-inferred interactions are drawn as dashed lines.
Conclusions
In summary, we performed large-scale proteomic analysis on IFN-treated human liver cells and analyzed the large dataset using a systems-biology approach. Data presented here both confirmed earlier work and, more importantly, identified novel IFN-regulated proteins and pathways. We propose a general picture of IFN-mediated regulation involving complex novel pathways and cross-talk between various cellular pathways in responses to multiple environmental signals (Figure 3). Such a systems-based proteomic study will extend our understanding about the IFN-mediated liver physiology and may ultimately lead to development of more efficient therapies against virus infections such as HCV.
Materials and methods
Cell culture and IFN treatment
Human liver carcinoma cells (Huh7) were cultured in DMEM containing 10% FBS, 100 U/ml penicillin and 100 U/ml streptomycin at 37°C. In proteomics analysis, cells were treated for 16 h with 400 IU/ml recombinant IFN-α2b (Intron-A, Schering-Plough). In immunoblot experiments, cells were treated for 8, 16, 40 or 72 h with 0, 50, 200 or 400 IU/ml IFN before harvest.
Cell lysis and fractionation
Cells were collected in a hypotonic lysis buffer (50 mM Tris pH 7.5, 5 mM MgCl2, 5 mM CaCl2, 1 mM DTT, 1 mM EDTA, and 1× Protease Inhibitor Cocktail (Roche) and then lysed by Dounce homogenization followed by centrifugation at 3,000g for 15 min. The pellet obtained (P-3) was washed by buffer containing 600 mM KCl to produce the nucleus fraction. The supernatant (S-3) was subsequently centrifuged at 100,000g for 2 h and generated the cytoplasm fraction (S-100) and the membrane fraction (P-100).
ICAT labeling and mass spectrometry
The cytoplasm, membrane and nuclear fractions were acetone-precipitated and resuspended in ICAT labeling buffer (0.05% SDS, 50 mM Tris pH 8.3, 5 mM EDTA, and 6 M urea). Each fractionated sample was labeled with the second generation ICAT reagents (acid cleavable), either in light (12C for IFN-treated) or in heavy (13C for mock-treated) isotopes (Applied Biosystems). Equal amounts of the two labeled samples were combined and digested into peptides by trypsin (Promega). ICAT-labeled peptides were subsequently purified by cation-exchange chromatography and avidin-affinity chromatography. Peptide mixtures were then analyzed by microcapillary HPLC-electrospray ionization (ESI)-MS/MS using an ion-trap mass spectrometer (LCQ-DecaXP, ThermoFinnigan) as previously described [53].
Immunoblot analysis and antibodies
Immunoblot analysis was performed as previously described [54]. The anti-PKR antibody was a gift from Ara Hovanessian [55]. The anti-NMI antibody was generously provided by Louie Naumovski at Stanford University (California, USA). Antibodies against STAT1 (monoclonal antibody 9H2) and β-actin (ab6276) were purchased from Cell Signaling Technology and Abcam, respectively.
Data analysis and software
SEQUEST matches peptide tandem mass spectra to sequence in the human IPI database [21]; PeptideProphet assigns a probability to the identified peptide sequence [9], and ProteinProphet assigns an overall probability to the protein identification [10]. To quantify protein ratio between IFN- and mock-treated samples, the program ASAPRatio was used to calculate the ICAT ratios [11]. To classify the identified proteins into cellular pathways, GO [12] analysis was performed using the GoMiner program [13]. All proteomics data were stored and systematically analyzed at the Systems Biology Experiment Analytical Management System (SBEAMS) at the Institute for Systems Biology (ISB) [23]. The data were visualized using Cytoscape, which was co-developed by ISB and MIT [14,36].
Additional data files
The following additional data is available with the online version of this paper: the original data files of the ICAT analyses on the cytoplasmic (Additional data 1), membrane (Additional data 2) and nuclear (Additional data 3) fractions of Huh7 cells. Each file contains all proteins identified in individual experiment and only those with ProteinProphet probability score ≥ 0.4 (with an error rate ≤ 5%) are included for data analysis in this article. The descriptions of each column in these three data files are given in Additional data 4; the figure in this file indicates the distribution of the protein ratio (ASAPRatio) of the proteins analyzed in this paper (ProteinProphet probability score ≥ 0.4) (y-axis: ASAPRatio ratio; x-axis: protein number). A normalization factor of 0.8 was obtained from this figure and used to normalize individual protein ratios as shown in columns 'O' and 'P' of the three data files. These data are also available at [23].
Supplementary Material
The original data file of the ICAT analysis on the cytoplasmic fraction of Huh7 cells
The original data file of the ICAT analysis on the membrane fraction of Huh7 cells
The original data file of the ICAT analysis on the nuclear fraction of Huh7 cells
The descriptions of each column in the first three data files
Acknowledgments
Acknowledgements
We thank Kelly Cooke at ISB for technical help. We are grateful to Maria Smith and Eric Chen at the University of Washington for helpful discussions and critical reading of the manuscript and to Maria Smith for sharing unpublished data. This investigation is supported by grant 1P30DA01562501 to M.G.K. and R.A. from the National Institute on Drug Abuse. This project has also been funded in part with Federal funds from the National Heart, Lung and Blood Institute, National Institutes of Health, under contract number N01-HV-28179.
References
- Isaacs A, Lindenmann J. Virus interference. I. The interferon. By A. Isaacs and J. Lindenmann, 1957. J Interferon Res. 1987;7:429–438. doi: 10.1089/jir.1987.7.429. [DOI] [PubMed] [Google Scholar]
- Sen GC. Viruses and interferons. Annu Rev Microbiol. 2001;55:255–281. doi: 10.1146/annurev.micro.55.1.255. [DOI] [PubMed] [Google Scholar]
- Samuel CE. Antiviral actions of interferons. Clin Microbiol Rev. 2001;14:778–809. doi: 10.1128/CMR.14.4.778-809.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katze MG, He Y, Gale M., Jr Viruses and interferon: a fight for supremacy. Nat Rev Immunol. 2002;2:675–687. doi: 10.1038/nri888. [DOI] [PubMed] [Google Scholar]
- Pfeffer LM, Dinarello CA, Herberman RB, Williams BR, Borden EC, Bordens R, Walter MR, Nagabhushan TL, Trotta PP, Pestka S. Biological properties of recombinant alpha-interferons: 40th anniversary of the discovery of interferons. Cancer Res. 1998;58:2489–2499. [PubMed] [Google Scholar]
- Pawlotsky JM. Hepatitis C virus resistance to antiviral therapy. Hepatology. 2000;32:889–896. doi: 10.1053/jhep.2000.19150. [DOI] [PubMed] [Google Scholar]
- Wasley A, Alter MJ. Epidemiology of hepatitis C: geographic differences and temporal trends. Semin Liver Dis. 2000;20:1–16. doi: 10.1055/s-2000-9506. [DOI] [PubMed] [Google Scholar]
- Fried MW, Hoofnagle JH. Therapy of hepatitis C. Semin Liver Dis. 1995;15:82–91. doi: 10.1055/s-2007-1007265. [DOI] [PubMed] [Google Scholar]
- Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- Li XJ, Zhang H, Ranish JA, Aebersold R. Automated statistical analysis of protein abundance ratios from data generated by stable-isotope dilution and tandem mass spectrometry. Anal Chem. 2003;75:6648–6657. doi: 10.1021/ac034633i. [DOI] [PubMed] [Google Scholar]
- Gene Ontology Software and Databases http://www.godatabase.org/dev
- Zeeberg BR, Feng W, Wang G, Wang MD, Fojo AT, Sunshine M, Narasimhan S, Kane DW, Reinhold WC, Lababidi S, et al. GoMiner: a resource for biological interpretation of genomic and proteomic data. Genome Biol. 2003;4:R28. doi: 10.1186/gb-2003-4-4-r28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cytoscape: Analyzing and Visualizing Biological Network Data http://www.cytoscape.org
- Bartenschlager R, Kaul A, Sparacio S. Replication of the hepatitis C virus in cell culture. Antiviral Res. 2003;60:91–102. doi: 10.1016/j.antiviral.2003.08.016. [DOI] [PubMed] [Google Scholar]
- Geiss GK, Carter VS, He Y, Kwieciszewski BK, Holzman T, Korth MJ, Lazaro CA, Fausto N, Bumgarner RE, Katze MG. Gene expression profiling of the cellular transcriptional network regulated by alpha/beta interferon and its partial attenuation by the hepatitis C virus nonstructural 5A protein. J Virol. 2003;77:6367–6375. doi: 10.1128/JVI.77.11.6367-6375.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Veer MJ, Holko M, Frevel M, Walker E, Der S, Paranjape JM, Silverman RH, Williams BR. Functional classification of interferon-stimulated genes identified using microarrays. J Leukoc Biol. 2001;69:912–920. [PubMed] [Google Scholar]
- Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat Biotechnol. 1999;17:994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- Han DK, Eng J, Zhou H, Aebersold R. Quantitative profiling of differentiation-induced microsomal proteins using isotope-coded affinity tags and mass spectrometry. Nat Biotechnol. 2001;19:946–951. doi: 10.1038/nbt1001-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EBI databases: International Protein Index http://www.ebi.ac.uk/IPI/IPIhelp.html
- Eng J, McCormack A, Yates J. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- Von Haller PD, Yi E, Donohoe S, Vaughn K, Keller A, Nesvizhskii AI, Eng J, Li XJ, Goodlett DR, Aebersold R, Watts JD. The application of new software tools to quantitative protein profiling via isotope-coded affinity tag (ICAT) and tandem mass spectrometry: II. Evaluation of tandem mass spectrometry methodologies for large-scale protein analysis, and the application of statistical tools for data analysis and interpretation. Mol Cell Proteomics. 2003;2:428–442. doi: 10.1074/mcp.M300041-MCP200. [DOI] [PubMed] [Google Scholar]
- LC MS/MS Data in PeptideAtlas http://www.peptideatlas.org/repository/
- Levy DE, Garcia-Sastre A. The virus battles: IFN induction of the antiviral state and mechanisms of viral evasion. Cytokine Growth Factor Rev. 2001;12:143–156. doi: 10.1016/S1359-6101(00)00027-7. [DOI] [PubMed] [Google Scholar]
- Der SD, Zhou A, Williams BR, Silverman RH. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc Natl Acad Sci USA. 1998;95:15623–15628. doi: 10.1073/pnas.95.26.15623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proud CG. PKR: a new name and new roles. Trends Biochem Sci. 1995;20:241–246. doi: 10.1016/S0968-0004(00)89025-8. [DOI] [PubMed] [Google Scholar]
- Berrueta L, Kraeft SK, Tirnauer JS, Schuyler SC, Chen LB, Hill DE, Pellman D, Bierer BE. The adenomatous polyposis coli-binding protein EB1 is associated with cytoplasmic and spindle microtubules. Proc Natl Acad Sci USA. 1998;95:10596–10601. doi: 10.1073/pnas.95.18.10596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison EE, Wardleworth BN, Askham JM, Markham AF, Meredith DM. EB1, a protein which interacts with the APC tumour suppressor, is associated with the microtubule cytoskeleton throughout the cell cycle. Oncogene. 1998;17:3471–3477. doi: 10.1038/sj.onc.1202247. [DOI] [PubMed] [Google Scholar]
- Gutkind JS. Cell growth control by G protein-coupled receptors: from signal transduction to signal integration. Oncogene. 1998;17:1331–1342. doi: 10.1038/sj.onc.1202186. [DOI] [PubMed] [Google Scholar]
- Dikic I, Blaukat A. Protein tyrosine kinase-mediated pathways in G protein-coupled receptor signaling. Cell Biochem Biophys. 1999;30:369–387. doi: 10.1007/BF02738120. [DOI] [PubMed] [Google Scholar]
- Ali A, Hoeflich KP, Woodgett JR. Glycogen synthase kinase-3: properties, functions, and regulation. Chem Rev. 2001;101:2527–2540. doi: 10.1021/cr000110o. [DOI] [PubMed] [Google Scholar]
- Harwood A, Braga VM. Cdc42 & GSK-3: signals at the crossroads. Nat Cell Biol. 2003;5:275–277. doi: 10.1038/ncb0403-275. [DOI] [PubMed] [Google Scholar]
- Ding S, Li Y, Tan Y, Jiang M, Tian B, Liu Y, Shao X, Ye SL, Wu J, Zeng R, et al. From proteomic analysis to clinical significance: overexpression of cytokeratin 19 correlates with hepatocellular carcinoma. Mol Cell Proteomics. 2003;3:73–81. doi: 10.1074/mcp.M300094-MCP200. [DOI] [PubMed] [Google Scholar]
- Nemeroff ME, Barabino SM, Li Y, Keller W, Krug RM. Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3' end formation of cellular pre-mRNAs. Mol Cell. 1998;1:991–1000. doi: 10.1016/S1097-2765(00)80099-4. [DOI] [PubMed] [Google Scholar]
- Shin HJ, Kim SS, Cho YH, Lee SG, Rho HM. Host cell proteins binding to the encapsidation signal epsilon in hepatitis B virus RNA. Arch Virol. 2002;147:471–491. doi: 10.1007/s007050200001. [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spirin V, Mirny LA. Protein complexes and functional modules in molecular networks. Proc Natl Acad Sci USA. 2003;100:12123–12128. doi: 10.1073/pnas.2032324100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ideker T, Ozier O, Schwikowski B, Siegel AF. Discovering regulatory and signalling circuits in molecular interaction networks. Bioinformatics. 2002;18 (Suppl 1):S233–S240. doi: 10.1093/bioinformatics/18.suppl_1.s233. [DOI] [PubMed] [Google Scholar]
- Peri S, Navarro JD, Kristiansen TZ, Amanchy R, Surendranath V, Muthusamy B, Gandhi TK, Chandrika KN, Deshpande N, Suresh S, et al. Human protein reference database as a discovery resource for proteomics. Nucleic Acids Res. 2004;32 Database issue:D497–D501. doi: 10.1093/nar/gkh070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson I, Martin J, de Bruijn B, Wolting C, Lay V, Tuekam B, Zhang S, Baskin B, Bader GD, Michalickova K, et al. PreBIND and Textomy - mining the biomedical literature for protein-protein interactions using a support vector machine. BMC Bioinformatics. 2003;4:11. doi: 10.1186/1471-2105-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader GD, Betel D, Hogue CW. BIND: the Biomolecular Interaction Network Database. Nucleic Acids Res. 2003;31:248–250. doi: 10.1093/nar/gkg056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellenberg M, Albermann K, Zollner A, Mewes HW, Hani J. Integrative analysis of protein interaction data. Proc Int Conf Intell Syst Mol Biol. 2000;8:152–161. [PubMed] [Google Scholar]
- Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci USA. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwikowski B, Uetz P, Fields S. A network of protein-protein interactions in yeast. Nat Biotechnol. 2000;18:1257–1261. doi: 10.1038/82360. [DOI] [PubMed] [Google Scholar]
- HomoloGene http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=homologene
- Bao J, Zervos AS. Isolation and characterization of Nmi, a novel partner of Myc proteins. Oncogene. 1996;12:2171–2176. [PubMed] [Google Scholar]
- Zhu M, John S, Berg M, Leonard WJ. Functional association of Nmi with Stat5 and Stat1 in IL-2- and IFNgamma-mediated signaling. Cell. 1999;96:121–130. doi: 10.1016/S0092-8674(00)80965-4. [DOI] [PubMed] [Google Scholar]
- Zhou X, Liao J, Meyerdierks A, Feng L, Naumovski L, Bottger EC, Omary MB. Interferon-alpha induces nmi-IFP35 heterodimeric complex formation that is affected by the phosphorylation of IFP35. J Biol Chem. 2000;275:21364–21371. doi: 10.1074/jbc.M003177200. [DOI] [PubMed] [Google Scholar]
- Chen J, Shpall RL, Meyerdierks A, Hagemeier M, Bottger EC, Naumovski L. Interferon-inducible Myc/STAT-interacting protein Nmi associates with IFP 35 into a high molecular mass complex and inhibits proteasome-mediated degradation of IFP 35. J Biol Chem. 2000;275:36278–36284. doi: 10.1074/jbc.M006975200. [DOI] [PubMed] [Google Scholar]
- Chen J, Naumovski L. Intracellular redistribution of interferon-inducible proteins Nmi and IFP 35 in apoptotic cells. J Interferon Cytokine Res. 2002;22:237–243. doi: 10.1089/107999002753536211. [DOI] [PubMed] [Google Scholar]
- Weill L, Shestakova E, Bonnefoy E. Transcription factor YY1 binds to the murine beta interferon promoter and regulates its transcriptional capacity with a dual activator/repressor role. J Virol. 2003;77:2903–2914. doi: 10.1128/JVI.77.5.2903-2914.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parija T, Das BR. Involvement of YY1 and its correlation with c-Myc in NDEA induced hepatocarcinogenesis, its prevention by d-limonene. Mol Biol Rep. 2003;30:41–46. doi: 10.1023/A:1022207630482. [DOI] [PubMed] [Google Scholar]
- Yi EC, Lee H, Aebersold R, Goodlett DR. A microcapillary trap cartridge-microcapillary high-performance liquid chromatography electrospray ionization emitter device capable of peptide tandem mass spectrometry at the attomole level on an ion trap mass spectrometer with automated routine operation. Rapid Commun Mass Spectrom. 2003;17:2093–2098. doi: 10.1002/rcm.1150. [DOI] [PubMed] [Google Scholar]
- Yan W, Frank CL, Korth MJ, Sopher BL, Novoa I, Ron D, Katze MG. Control of PERK eIF2alpha kinase activity by the endoplasmic reticulum stress-induced molecular chaperone P58IPK. Proc Natl Acad Sci USA. 2002;99:15920–15925. doi: 10.1073/pnas.252341799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent AG, Krust B, Galabru J, Svab J, Hovanessian AG. Monoclonal antibodies to an interferon-induced Mr 68,000 protein and their use for the detection of double-stranded RNA-dependent protein kinase in human cells. Proc Natl Acad Sci USA. 1985;82:4341–4345. doi: 10.1073/pnas.82.13.4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The original data file of the ICAT analysis on the cytoplasmic fraction of Huh7 cells
The original data file of the ICAT analysis on the membrane fraction of Huh7 cells
The original data file of the ICAT analysis on the nuclear fraction of Huh7 cells
The descriptions of each column in the first three data files


