Abstract
Nodulation, nodule development and senescence directly affects nitrogen fixation efficiency, and previous studies have shown that inhibition of some cysteine proteases delay nodule senescence, so their nature inhibitors, cystatin genes, are very important in nodulation, nodule development, and senescence. Although several cystatins are actively transcribed in soybean nodules, their exact roles and functional diversities in legume have not been well explored in genome-wide survey studies. In this report, we performed a genome-wide survey of cystatin family genes to explore their relationship to nodulation and nodule development in soybean and identified 20 cystatin genes that encode peptides with 97–245 amino acid residues, different isoelectric points (pI) and structure characteristics, and various putative plant regulatory elements in 3000 bp putative promoter fragments upstream of the 20 soybean cystatins in response to different abiotic/biotic stresses, hormone signals, and symbiosis signals. The expression profiles of these cystatin genes in soybean symbiosis with rhizobium strain Bradyrhizobium japonicum strain 113-2 revealed that 7 cystatin family genes play different roles in nodulation as well as nodule development and senescence. However, these genes were not root nodule symbiosis (RNS)—specific and did not encode special clade cystatin protein with structures related to nodulation and nodule development. Besides, only two of these soybean cystatins were not upregulated in symbiosis after ABA treatment. The functional analysis showed that a candidate gene Glyma.15G227500 (GmCYS16) was likely to play a positive role in soybean nodulation. Besides, evolutionary relationships analysis divided the cystatin genes from Arabidopsis thaliana, Nicotiana tabacum, rice, barley and four legume plants into three groups. Interestingly, Group A cystatins are special in legume plants, but only include one of the above-mentioned 7 cystatin genes related to nodulation and nodule development. Overall, our results provide useful information or clues for our understanding of the functional diversity of legume cystatin family proteins in soybean nodulation and nodule development and for finding nodule-specific cysteine proteases in soybean.
Keywords: soybean, cystatin, genome-wide survey, gene expression, root nodule symbiosis
Introduction
Nitrogen fixation efficiency plays important roles in plant cultivation and fertilizer application and is closely related to nodulation, nodule development, and senescence (Biswas and Gresshoff, 2014). Previous studies have shown that cysteine proteases are important in nodule development and senescence in several legume plants. For example, silencing of AsNODF32 delays root nodule development and bacteroid senescence and prolongs nodule lifespan in Astragalus sinicus (Li et al., 2008). Inhibition of cysteine protease CYP15A delays nodule senescence in Medicago trunctula (Sheokand et al., 2005) and protease PsCyp15A is activated at the onset of senescence in the indeterminate nodules of pea (Vincent and Brewin, 2000). In soybean, since the first report of cysteine protease expression during nodule senescence in 1983 (Pfeiffer et al., 1983), 18 soybean cysteine proteases have been found actively transcribed during nodule development and senescence (van Wyk et al., 2014). However, it is not well understood whether these cysteine proteases play specific roles in nodule development and senescence and why so many soybean cysteine proteases are involved in regulation of nodule symbiosis. Studies on their nature inhibitors could help us better understand these questions.
Cystatins are a group of small proteins known to reversibly inhibit the activity of cysteine proteases in families of papain C1A and legumain C13 peptidase (Martinez et al., 2007). Their inhibitory mechanism involves a tripartite wedge formed by the partially flexible N terminus containing a Gly residue and two hairpin loops that carry a conserved QxVxG motif and a conserved tryptophan (Trp) residue (Stubbs et al., 1990). Besides, a conserved LARFAV motif ([LVI]-[AGT]-[RKE]-[FY]-[AS]-[VI]) is also common to plant cystatins (Margis et al., 1998). Disturbance of the equilibrium between cystatins and their cysteine proteases in plant is a key event in endogenous protein turnover (Martinez et al., 2009), accumulation and mobilization of storage proteins (Benchabane et al., 2010), seed germination and maturation (Arai et al., 2002), programmed cell death (Belenghi et al., 2003), abiotic environmental stresses (Hwang et al., 2010), and protection of plants against attack by mites (Carrillo et al., 2011), fungi (Martinez et al., 2003; Popovic et al., 2013), and viruses (Gutierrez-Campos et al., 1999). Although previous studies have well described the genome-wide characteristics of cystatin family genes in four different species, Hordeum vulgare (Martinez et al., 2009), Nicotiana tabacum (Zhao et al., 2014), Apple (Tan et al., 2014), and Oryza sativa L. (Wang et al., 2015), the genome-wide studies of cystatin family genes in legume are quite limited.
Soybean (Glycine max) is an important oil crop, food and feed material with planting area >1734 million acres worldwide in 2015. Twenty soybean cystatins have already been identified by releasing of the complete soybean genome sequence (Schmutz et al., 2010) and the RNA-seq atlas showed they were expressed in 14 different soybean tissues, especially in nodules (Severin et al., 2010). In addition, the inhibitory activities of these cystatins were analyzed and seven of them were actively transcribed in nodules (van Wyk et al., 2014). Ectopic expression of rice phytocystatin positively regulates nodulation and nitrogen deficiency of soybean (Quain et al., 2015). However, almost all of these studies were focused on individual cystatin members. Which soybean cystatins play roles in symbiotic signal transduction, nodulation, nodule development, and senescence is still incompletely understood.
In this report, the detail information of genome-wide identification and characterization of 20 soybean cystatins were shown to reveal the special characteristics of soybean cystatin family genes. Expression profiles of cystatin genes in soybean symbiosis with Bradyrhizobium japonicum strain 113-2 were used to explore their putative roles in nodulation and nodule development. And the function analysis of Glyma.15G227500 (GmCYS16) was used to confirm the expression of these soybean cystatin genes in symbiosis. These investigations and analyses could provide useful information for the research of the functional diversity of legume cystatin family proteins, and helpful clues for investigation of nodule-specific cysteine proteases in soybean.
Materials and methods
Database searches for the identification of cystatin family genes from soybean, L. japonicus, M. truncatula, common bean, A. thaliana, N. tabacum, rice and barely
To identify cystatin family genes in soybean, the Soybean Genome Database (http://soybase.org/), Phytozome Database (http://www.phytozome.net/soybean), and NCBI-BLAST (http://blast.ncbi.nlm.nih.gov/) online resources were searched to identify the entire family of cystatins in G. max. Soybean cystatins were applied as model for identification of cystatin homologs in Lotus japonicus, Medicago truncatula, common bean, Arabidopsis thaliana, rice and barely at LIS (Legume Information System) (http://legumeinfo.org/lis_gene_families/chado/msa/phytozome_10_2.59086800-consensus/). N. tabacum cystatin proteins were identified in NCBI-BLAST (http://blast.ncbi.nlm.nih.gov/) online resources.
Identification of conserved motifs and phylogenetic tree constructions
Multiple alignments of amino acid sequences of soybean cystatin family genes were performed with DNAMAN software to retrieve the conserved motifs of soybean cystatins. Phylogenetic tree analysis was performed using MEGA6 software (Tamura et al., 2013). Multiple alignments of the full-length deduced amino acid sequences of cystatin family genes were conducted with Clustal W program. The Poisson substitution model, uniform rates, pairwise deletion and 1000 Bootstrapping replicates were applied in Neighbor-joining method to perform phylogenetic tree analysis.
The different cystatins including 20 cystatin proteins from soybean, 34 cystatin proteins from M. trunctula, 7 cystatin proteins from L. japonicus, 10 cystatin proteins from common bean, 7 cystatin proteins from A. thaliana, 10 cystatin proteins from N. tabacum, 18 cystatin proteins from rice, and 13 cystatin proteins from barley were applied for phylogenetic and molecular evolutionary analyses using the programs RAxML and MEGA version 6.0 (http://www.megasoftware.net; Guindon and Gascuel, 2003; Tamura et al., 2013). Maximum likelihood (ML) analysis was performed using RAxML 8.0.0 (Stamatakis, 2014) and rapid bootstrap analysis was performed with the bootstrap convergence test using the extended majority-rule consensus tree criterion (autoMRE) in RAxML. GenBank accession numbers are listed in Table S1.
Soybean cystatin genes sequence and structure analysis
The genomic DNA and cDNA sequences corresponding to each predicted soybean cystatin genes were downloaded from Phytozome Database (http://www.phytozome.net/soybean) and Soybean Genome Database (http://soybase.org/). The gene structure display server program (http://gsds.cbi.pku.edu.cn) (Guo et al., 2007) was used to analyze the exon/intron structures of these 20 soybean cystatin genes. The SignalP version 3.0 program (http://www.cbs.dtu.dk/services/SignalP) (Bendtsen et al., 2004) was used to perform the signal peptide analysis. The secondary structures (a-helix and b-sheets) were predicted as previously reported (http://ps2.life.nctu.edu.tw/) (Chen et al., 2009). The automated SWISS-MODEL program (http://swissmodel.expasy.org/interactive) (Peitsch, 1996) was used to model the three-dimensional structures of soybean cystatins, and the known crystal structure of rice oryzacystatin I (OC-I) (Nagata et al., 2000), SiCYS (Hu et al., 2015), and PMC (Green et al., 2013) were used to construct the homology-based models. The Plant CARE database (http://bioinformatics.psb. ugent.be/ webtools/ plantcare/html/) (Lescot et al., 2002) was used to analyze the promoter sequences of soybean cystatins.
Plant materials and treatments
Seeds of soybean Tianlong No.1 (stored in our lab) were surface-sterilized, germinated and grown on moistened filter paper in a greenhouse with a 16/8 h day/night cycle and 70% relative humidity (RH) for 2–3 d at 28°C (Yuan et al., 2016). For nodulation experiments, the germinated soybean seeds were grown in pots filled with sterilized vermiculite and sand (1:1) supplemented with half-strength B&D medium in a chamber with a 16/8 h day/night cycle at 28°C for 4–5 d before inoculation with rhizobium strain 113-2 (stored in our lab). After inoculation, plants were kept under the same growth conditions.
For RNA isolation, soybean roots from CK and 113-2 inoculated groups were collected with three biological replicates at five different time points [0.5 h, 7–24 h (mixture of 7 h and 24 h), 5, 16, and 21 d of post inoculation] (Yuan et al., 2016) and frozen at −80°C, and soybean nodules from CK and 113-2 inoculated groups were collected with three biological replicates at five important nodule development time points (12, 30, 42, 60, and 84 d of post inoculation) and frozen at −80°C.
For ABA treatment, nodules-containing, strain 113-2 inoculated soybean roots were collected at 6, 33, and 62 d of post inoculation, transferred into containers with half-strength B&D medium supplemented with 200 μM ABA, and cultured for 5, 10, and 9 d, respectively. The collected soybean roots samples were collected with three biological replicates and were frozen at −80°C for RNA isolation.
RNA extraction, semi-quantitative RT-PCR, and qPCR
Total RNA was isolated using TRIzol reagent (Invitrogen, USA) and stored in a −80°C freezer for downstream gene-expression experiments. Potential genomic DNA were removed using RNeasy plant mini kit (QIAGEN, Germany) and RNA quantity and quality were measured using an Epoch Multi-Volume Spectrophotometer system, NanoDrop and Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA). Glyma.15G227500 expression in L. japonicus were detected using semi-quantitative RT-PCR at the following conditions: an initial denaturation at 94°C for 5 min, followed by 31 cycles of 30 s at 94°C, 30 s at 55°C, and 30 s at 72°C, as well as a final extension at 72°C for 10 min. PCR products were detected by 1% agarose gel analysis. Expressions of other genes in L. japonicus were determined with qPCR using primer sets listed in Table S2, and following cycling conditions: 30 s at 95°C, followed by 40 cycles of 5 s at 95°C, 15 s at 60°C and 12 s at 72°C and final 5 s at 72°C. The soybean QACT and L. japonicus polyubiquitin transcript were used as the internal, endogenous control to normalize the samples, and the 2−ΔΔCT method (Livak and Schmittgen, 2001) was used to analyze the relative changes in gene expression in qPCR experiments. Three biological replica samples were used to detect the expression of soybean cystatins in symbiosis with B. japonicum strain 113-2, and three or four replicate reactions per sample were used to ensure statistical credibility. Student's t-test was conducted to analyze the differences in qPCR experiments using software SPSS Statistics 17.0.
Overexpression of Glyma.15G227500 in L. japonicus by hairy root transformation
The full-length coding sequence of Glyma.15G227500 was cloned into the KpnI/ BamHI site of pU1301 using primer set of 5′-AGGGTACCATGAGAGCATTAACC TCTTC-3′ and 5′-CAGGATCCTTAGGAATGATCTTGTTCC-3′ to generate pMUb: Glyma.15G227500. A. rhizogenes strain LBA1334 cells carrying pMUb: Glyma.15G227500 were used to induce hairy root formation in wild-type L. japonicus “MG-20” using an A. rhizogenes-mediated procedure as described previously (Yuan et al., 2012). Briefly, seedlings were cut at the base of the hypocotyls, placed in suspension of A. rhizogenes LBA1334 cells containing plasmids for 30 min, and transferred onto agar plates of Murashige and Skoog (MS; Sigma) medium containing 1.5% (w/v) Suc and cocultivated for 5 d. The plants were then transferred onto agar plates containing 250 mg/ml of cefotaxime and grown until hairy roots (a few cm long) developed from the base of hypocotyls. Each hairy root was properly labeled and a short root tip (1- to 3-mm long) was removed to test for GUS activity in staining solution overnight at 37°C in the dark. Hairy roots with GUS-positive tips were preserved and allowed to continue to grow. Each seedling was allowed to have 1–2 transgenic hairy roots. Plants with transgenic hairy roots were transferred to pots filled one-half-strength Broughton&Dilworth (B&D) medium supplemented with vermiculite and sand (1:1) (Broughton and Dilworth, 1971) and grown in a chamber in a 16-/8-h day/night cycle at 23°C. After a week of adaptation, plants were inoculated with Mesorhizobium loti strain MAFF303099 and grown in the same medium without ammonium nitrate. Nodulation phenotypes of the transgenic hairy roots were scored at 32 days of post inoculation with M. loti. The transgenic hairy roots expressing the empty vector (pU1301) were used as the control.
Results
Characterization of the soybean cystatin genes
The Soybean Genome Databases were searched to identify the entire family of cystatins in G. max. Twenty full-length soybean cystatin cDNAs containing complete ORF were obtained and named as GmCYS1- GmCYS20 according to their positions in chromosomes. Table 1 lists their detailed information. The identified soybean cystatin genes encode peptides with 97–245 amino acid residues and isoelectric point (pI) of 4.98–9.88. SignalIP was used to find signal peptides. Among the 20 soybean cystatins, 14 soybean cystatins have signal peptides of 17–33 amino acid residues and the other six cystatins (GmCYS9, GmCYS12, GmCYS13, GmCYS14, GmCYS18, and GmCYS20) has no signal peptide. The cDNA sequences of each cystatin were compared with their genomic sequences to analyze their exon/intron structures. The results revealed that 12 soybean cystatins are single-exon cystatins without intron, 3 soybean cystatins (GmCYS9, GmCYS12, and GmCYS20) contain one intron, 3 (GmCYS13, GmCYS14, and GmCYS17) contain two introns and 2 (GmCYS10 and GmCYS16) contain three introns. The secondary structures (α-helix and β-strand) of cystatin genes were predicted in http://ps2.life.nctu.edu.tw/, and the results are shown in Table 1. Among the 20 soybean cystatins, 14 have two α-helixes, 3 (GmCYS9, GmCYS12, and GmCYS18) have only one α-helix, one (GmCYS14) has three α-helixes, and 2 (GmCYS10 and GmCYS16) have six α-helixes. The number of β-strand is divergent from 4 to 11 in these 20 cystatins.
Table 1.
Detailed information of soybean cystatin family genes.
| Name | Soybean gene ID | Chromosome location | No. of amino acid residues | PI | Signal peptide | α-Helix | β-Strand | Exon | Intron |
|---|---|---|---|---|---|---|---|---|---|
| GmCYS1 | Glyma.04G096400 | Chr04 8659349–8659856 | 114 | 9.88 | 17 | 2 | 4 | 1 | 0 |
| GmCYS2 | Glyma.05G149800 | Chr05 34385547–34386483 | 130 | 9.23 | 26 | 2 | 4 | 1 | 0 |
| GmCYS3 | Glyma.07G266000 | Chr07 43976589–43977195 | 114 | 7.77 | 20 | 2 | 4 | 1 | 0 |
| GmCYS4 | Glyma.08G168600 | Chr08 13381885–13382591 | 125 | 9.41 | 19 | 2 | 6 | 1 | 0 |
| GmCYS5 | Glyma.09G010900 | Chr09 837176–837835 | 114 | 9.88 | 17 | 2 | 4 | 1 | 0 |
| GmCYS6 | Glyma.09G110600 | Chr09 21896029–21896583 | 114 | 9.88 | 17 | 2 | 4 | 1 | 0 |
| GmCYS7 | Glyma.11G064200 | Chr11 4842312–4842656 | 114 | 9.83 | 17 | 2 | 4 | 1 | 0 |
| GmCYS8 | Glyma.11G253300 | Chr11 34413583–34414431 | 126 | 9.52 | 20 | 2 | 4 | 1 | 0 |
| GmCYS9 | Glyma.13G071800 | Chr13 17264906–17266886 | 97 | 5.83 | 1 | 6 | 2 | 1 | |
| GmCYS10 | Glyma.13G189500 | Chr13 30296749–30300668 | 245 | 6.56 | 33 | 6 | 11 | 4 | 3 |
| GmCYS11 | Glyma.13G209000 | Chr13 32301762–32302169 | 124 | 5.73 | 19 | 2 | 4 | 1 | 0 |
| GmCYS12 | Glyma.14G038200 | Chr14 2854471–2855966 | 103 | 5.8 | 1 | 5 | 2 | 1 | |
| GmCYS13 | Glyma.14G038300 | Chr14 2859830–2861347 | 200 | 5.26 | 2 | 9 | 3 | 2 | |
| GmCYS14 | Glyma.14G038500 | Chr14 2876239–2877988 | 184 | 4.98 | 3 | 9 | 3 | 2 | |
| GmCYS15 | Glyma.15G115300 | Chr15 9076848–9077830 | 112 | 9.07 | 17 | 2 | 5 | 1 | 0 |
| GmCYS16 | Glyma.15G227500 | Chr15 42044582–42049151 | 245 | 7.27 | 33 | 6 | 9 | 4 | 3 |
| GmCYS17 | Glyma.18G003700 | Chr18 304856–305625 | 142 | 9.56 | 20 | 2 | 4 | 2 | 2 |
| GmCYS18 | Glyma.18G103700 | Chr18 11309282–11311702 | 101 | 5.64 | 1 | 5 | 1 | 0 | |
| GmCYS19 | Glyma.19G206500 | Chr19 46202432–46203385 | 114 | 9.88 | 17 | 2 | 4 | 1 | 0 |
| GmCYS20 | Glyma.20G045500 | Chr20 8475315–8478532 | 97 | 5.83 | 2 | 6 | 2 | 1 |
Phylogenetic analysis and structural predication of the soybean cystatins
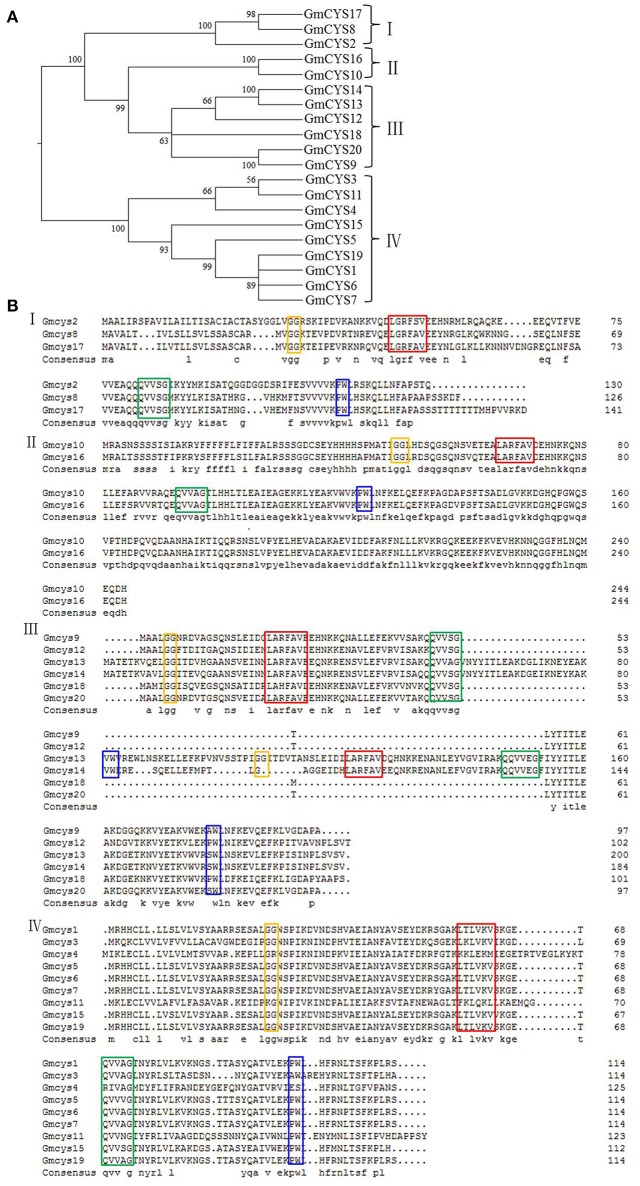
A phylogenetic analysis based on alignments of the 20 full-length cystatin protein sequences was performed to determine the phylogenetic relationships among the different members of the soybean cystatins, and the results are revealed in Figure 1A.
Figure 1.
Phylogenetic analysis and amino acid residue alignment of soybean cystatins. (A) Phylogenetic analysis of soybean cystatins. Phylogenetic tree construction of soybean cystatins is based on the full-length deduced amino acid sequences using MEGA 6 by the neighbor-joining method with 1000 bootstrap replicates. The tree shows four major phylogenetic clades (clades I to IV) indicated with braces. (B) Alignments of conserved motifs of different clades of the soybean cystatins. The main cystatin conserved motifs in four phylogenetic clades (clades I to IV) are in orange, red, green, and blue boxes.
Twenty soybean cystatins were divided into four clades (I, II, III, and IV) in the neighbor-joining phylogenetic tree, and the members within each clade show similar gene structures (Table 1).
Alignments of cystatin sequences in different soybean clades were used to search for amino acid variants that could affect their inhibitory capability on cysteine proteases. The results are shown in Figure 1B. All protein signatures responsible for the cysteine proteinase inhibitory properties were conserved along the sequence of Clades I, II, and III, with exception of the alanine in the second position of the conserved LARFAV motif (A/G) in Clade I. Clade IV comprises 9 members (the largest number of members) with different amino acid residues in the conserved LARFAV motif. Besides, Clade III contains variable numbers of the four conserved motifs (Figure 1B). In addition, the sequence length of soybean cystatins in different clades also varies (Figure 1B).
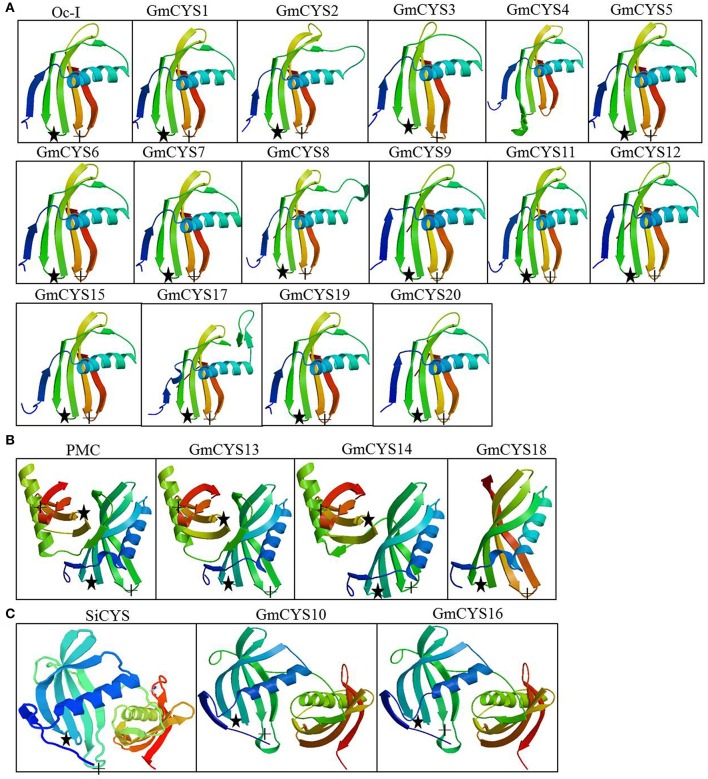
The predicted three-dimensional structures of the soybean cystatins were established using the automated SWISS-MODEL program with the known crystal structure of OC-I (Nagata et al., 2000), SiCYS (Hu et al., 2015), and PMC (Green et al., 2013) as templates (Figure 2). Although these structures were predicted with variable degrees of accuracy, GmCYS1 to GmCYS9, GmCYS11, GmCYS12, GmCYS15, GmCYS17, GmCYS19, and GmCYS20 share similar protein structures with rice OC-I (Figure 2A) and GmCYS13, GmCYS14 and GmCYS18 share similar protein structure with PMC (Figure 2B). Besides, GmCYS10 and GmCYS16 show significant variations in their predicted three-dimensional structures, and share similar protein structure with SiCYS (Figure 2C). Two important motifs (the conserved QxVxG motif and the conserved tryptophan) involved in the interaction with the cysteine proteinases were shown in Figure 2. The predicted structure of GmCYS4 shows some distortions in the region of QxVxG motif and W residue, probably due to the glutamine in the first position of the conserved QxVxG motif (R/Q) and change of the conserved tryptophan to phenylalanine (W/F) in this cystatin (Figure 1B).
Figure 2.
The three-dimensional structure prediction of soybean cystatins. (A) The three-dimensional structures of 15 cystatins were predicted using the automated SWISS-MODEL program with OC-I as a template. (B) The three-dimensional structures of GmCYS13, GmCYS14, and GmCYS18 were predicted using the automated SWISS-MODEL program with PMC as a template. (C) The three-dimensional structures of GmCYS10 and GmCYS16 were predicted using the automated SWISS-MODEL program with SiCYS as a template. Two important motifs involved in the interaction with the target enzymes are indicated the reactive site (asterisks) and W residue (crosses).
Evolutionary relationships among the cystatin family in rice, barley, Arabidopsis thaliana, Nicotiana tabacum and four legume plants
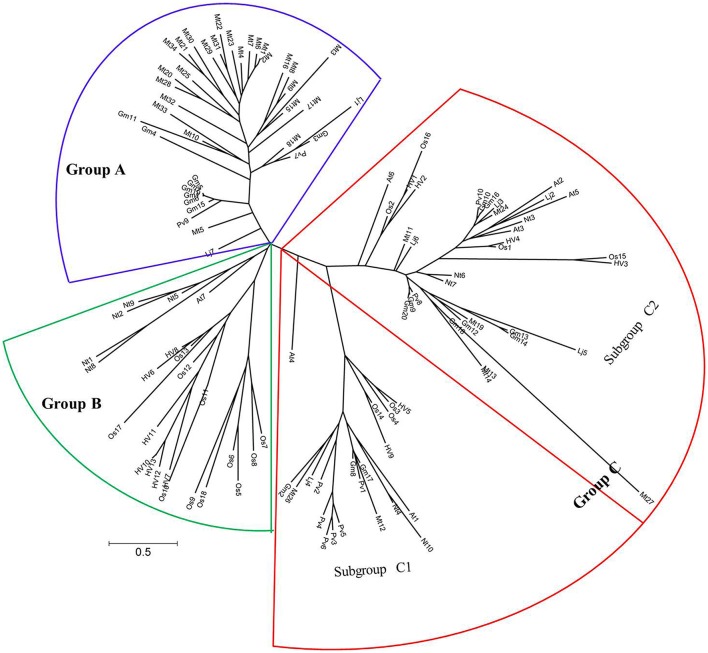
To estimate the phylogenetic relationships of cystatins in legume plants to other known cystatins in rice, barley, A. thaliana and N. tabacum, a multiple sequence alignment of soybean cystatin protein sequences to the sequences from other three legume plants (M. trunctula, L. japonicus, and common bean) and four non-legume plants (A. thaliana, N. tabacum, rice, and barley) was conducted to establish the phylogenetic tree using the programs RAxML and MEGA version 6.0 (Figure 3). The accession numbers of cystatins in M. trunctula, L. japonicus, common bean, A. thaliana, N. tabacum, rice, and barley are shown in Table S1. These cystatin proteins from eight different species are distributed in three major groups (Group A, Group B, and Group C). Among them, Group A was formed by 39 cystatins with only legume plant cystatin, 26 M. trunctula cystatins, 9 soybean cystatins, 2 L. japonicus cystatins, and 2 common bean cystatin. Group B was the smallest group composed of 24 cystatins with only non-legume plant cystatin, 11 rice cystatins, 7 barley cystatins, 5 N. tabacum cystatins, and 1 A. thaliana cystatin. Group C was the largest group composed of 56 cystatins with 32 legume plant cystatins and 24 non-legume plant cystatins, and was subdivided in two subgroups: subgroup C1 and subgroup C2. Cystatins from soybean almost equally fall into Group A and Group C. Cystatins from M. trunctula mainly fall into Group A, and cystatins from common bean mainly fall into subgroup C1, while most cystatins from of L. japonicus are in subgroup C2. The phylogenetic relationship may reflect some distinction between legume plant cystatins and four non-legume plants cystatins (2 monocotyledon plants and 2 non-legume dicotyledonous plants), and indicate that the potential biological functions of some cystatins are conservative in legume plants and in some special developmental processes.
Figure 3.
Phylogenetic tree of cystatin proteins from soybean, M. trunctula, L. japonicus, common bean, A. thaliana, N. tabacum, rice and barley. Twenty cystatin proteins from soybean, 34 cystatin proteins from M. trunctula, 7 cystatin proteins from L. japonicus, 10 cystatin proteins from common bean, 7 cystatin proteins from A. thaliana, 10 cystatin proteins from N. tabacum, 18 cystatin proteins from rice, and 13 cystatin proteins from barley were used to construct the phylogenetic tree using the programs RAxML and MEGA6.0. These cystatin proteins were classified into three groups, namely Group A, Group B, and Group C. Group C was subdivided in two subgroups: subgroup C1 and subgroup C2. GenBank accession numbers of these cystatin proteins are listed in Table S1.
Promoter sequence analysis of the soybean cystatins
Based on soybean Genome Database (http://www.phytozome.net/soybean), 3000 bp putative promoter fragments upstream of the soybean cystatins were obtained and putative cis-acting elements were analyzed with the Plant CARE database (http://bioinformatics.psb.ugent.be/ webtools/ plantcare/html/). Various putative plant regulatory elements in these promoters are shown in Table 2, including elements in response to environmental cues and hormone signals, such as temperature responsive elements (HSE and LTR), drought-inducibility elements (MBS), defense and stress responsiveness elements (TC-rich repeats), wound-responsive element (WUN-motif), fungal elicitor responsive element (Box-W1), circadian-regulated elements (circadian) and elements in response to hormones including abscisic acid (ABRE and CE3), gibberellin acid (GARE-motif, P-box, and TATC-box), salicylic acid (TCA-element), auxin (TGA-element, TGA-box, and AuxRR-core) and jasmonic acid (TGACG-motif), and 7 other putative regulatory elements including flavonoid biosynthetic genes regulation elements (MBSI and MBSII), nodule specific factor binding site element (Nodule-site2), MYBHv1 binding site element (CCAAT-box), anaerobic induction element (ARE), ethylene-responsive element (ERE), and elicitor-responsive element (ELI-box3). The statistical analyses of data in Table 2 showed that 95% soybean cystatins have TC-rich repeats element, 90% have HSE element, TGACG-motif and circadian elements, 80% have TCA-element, ABRE element and gibberellin acid elements, and so on. These results indicated that the expressions of these 20 soybean cystatins might be regulated by various environmental stresses and hormone signals.
Table 2.
Distribution of relative elements in upstream 3000 bp sequences of soybean CYS genes.
| Gene CYS | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ABRE | 4 | 2 | 3 | 2 | 1 | 2 | 2 | 3 | 4 | 1 | 1 | 3 | 2 | 3 | 1 | 2 | ||||
| ARE | 1 | 1 | 2 | 4 | 4 | 4 | 2 | 3 | 3 | 4 | 3 | 4 | 3 | 1 | 6 | |||||
| GARE-motif | 2 | 5 | 3 | 1 | 2 | 1 | 1 | 1 | 3 | 1 | 2 | 1 | ||||||||
| P-box | 1 | 1 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 3 | 1 | |||||||||
| HSE | 2 | 3 | 2 | 1 | 2 | 2 | 3 | 2 | 5 | 7 | 3 | 2 | 2 | 4 | 1 | 2 | 3 | 7 | ||
| LTR | 1 | 1 | 1 | 1 | 1 | 1 | ||||||||||||||
| TC-rich repeats | 4 | 3 | 7 | 2 | 3 | 3 | 3 | 2 | 2 | 5 | 2 | 6 | 4 | 3 | 4 | 5 | 1 | 5 | 5 | |
| TGA-element | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |||||||||
| TGACG-motif | 3 | 2 | 3 | 3 | 3 | 3 | 4 | 2 | 7 | 3 | 3 | 1 | 1 | 1 | 1 | 2 | 3 | 4 | ||
| Circadian | 4 | 5 | 5 | 1 | 1 | 3 | 5 | 5 | 6 | 2 | 4 | 4 | 4 | 4 | 2 | 1 | 3 | 4 | ||
| TCA-element | 1 | 1 | 1 | 1 | 3 | 2 | 3 | 4 | 1 | 1 | 2 | 1 | 5 | 2 | 3 | 5 | ||||
| MBS | 6 | 2 | 2 | 1 | 3 | 1 | 3 | 3 | 1 | 1 | 4 | 1 | 2 | 1 | ||||||
| MBSI | 1 | |||||||||||||||||||
| MBSII | 1 | |||||||||||||||||||
| Nodule-site2 | 1 | |||||||||||||||||||
| Box-W1 | 1 | 1 | 1 | 1 | 2 | 1 | 2 | 2 | 1 | 1 | ||||||||||
| ERE | 1 | 1 | 3 | 3 | 1 | 1 | 3 | |||||||||||||
| CCAAT-box | 1 | 2 | 1 | 1 | 1 | 1 | ||||||||||||||
| WUN-motif | 1 | 1 | 1 | 2 | 2 | 1 | 2 | |||||||||||||
| AuxRR-core | 1 | 1 | ||||||||||||||||||
| TATC-box | 1 | 1 | 1 | 1 | ||||||||||||||||
| CE3 | 1 | 1 | ||||||||||||||||||
| ELI-box3 | 1 | 1 | ||||||||||||||||||
| TGA-box | 1 |
Expression profiles of soybean cystatin genes in symbiosis with B. japonicum strain 113-2
Based on plant Phytozome database (http://www.phytozome.net/soybean), the expression profiles of the 20 soybean cystatins were investigated in various tissues, including nodules. symbiotic condition, root. symbiotic condition, flower, leaves, nodules, pod, root, root hairs, seed, and stem. The results showed that the soybean cystatin genes were expressed in distinct patterns and most of them were expressed in multiple tissues (Table 3). Among them, 10 soybean cystatins (GmCYS2, GmCYS3, GmCYS9, GmCYS10, GmCYS12, GmCYS15, GmCYS16, GmCYS17, GmCYS18, and GmCYS20) showed relative high expression in nodules and/or nodules at symbiotic condition, while other 6 soybean cystatins (GmCYS1, GmCYS4, GmCYS5, GmCYS6, GmCYS7, and GmCYS19) showed no expression in these two tissues. GmCYS8 had no expression in nodules and very low expression in nodules at symbiotic condition while GmCYS11 was not expressed in nodules at symbiotic condition, but had very low expression in nodules. In addition, 13 soybean cystatins (GmCYS2, GmCYS3, GmCYS8, GmCYS9, GmCYS10, GmCYS12, GmCYS13, GmCYS14, GmCYS15, GmCYS16, GmCYS17, GmCYS18, and GmCYS20) were expressed in root, root hairs and roots at symbiotic condition, and GmCYS5 had low expression in roots at symbiotic condition. GmCYS1, GmCYS5, GmCYS6, GmCYS7, and GmCYS19 were mainly expressed in flower, and GmCYS4 was mainly expressed in pod and seed.
Table 3.
Expression analysis of 20 soybean cystatins in different tissues in phytozome database.
| PKM (Phytozome database) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Tissue | Flower | Leaves | Nodules | Pod | Root | Root Hairs | Seed | Stem | Nodules. symbiotic condition | Root. symbiotic condition |
| GmCYS1 | 2.223 | 0 | 0 | 0.36 | 0 | 0 | 0.328 | 0 | 0 | 0 |
| GmCYS2 | 65.448 | 5.741 | 14.347 | 9.69 | 7.617 | 14.376 | 299.668 | 2.31 | 106.506 | 104.914 |
| GmCYS3 | 3.529 | 5.776 | 40.961 | 50.279 | 16.214 | 29.289 | 90.18 | 3.426 | 3.518 | 0.701 |
| GmCYS4 | 0.261 | 0 | 0 | 31.197 | 0 | 0 | 78.61 | 0 | 0 | 0 |
| GmCYS5 | 11.236 | 0 | 0 | 1.629 | 0 | 0 | 0.818 | 0 | 0 | 0.626 |
| GmCYS6 | 1.973 | 0 | 0 | 0.107 | 0 | 0 | 0.041 | 0 | 0 | 0 |
| GmCYS7 | 0.261 | 0 | 0 | 0.17 | 0 | 0 | 0.04 | 0 | 0 | 0 |
| GmCYS8 | 0 | 0 | 0 | 0 | 3.17 | 2.938 | 243.649 | 0 | 0.048 | 0.173 |
| GmCYS9 | 49.973 | 62.288 | 46.363 | 112.498 | 72.678 | 45.571 | 88.956 | 101.283 | 13.656 | 31.935 |
| GmCYS10 | 98.534 | 57.723 | 55.677 | 65.267 | 95.346 | 71.81 | 229.767 | 63.73 | 31.794 | 30.9 |
| GmCYS11 | 0 | 0 | 0.366 | 0.436 | 0 | 0 | 0.065 | 0 | 0 | 0 |
| GmCYS12 | 215.075 | 67.664 | 9.519 | 500.369 | 41.927 | 25.159 | 62.095 | 95.536 | 0.487 | 8.882 |
| GmCYS13 | 24.323 | 3.608 | 0.001 | 14.707 | 9.976 | 0.063 | 4.731 | 4.187 | 0.023 | 0.185 |
| GmCYS14 | 312.627 | 24.379 | 0.289 | 323.377 | 24.549 | 1.237 | 1.185 | 55.414 | 0.005 | 0.109 |
| GmCYS15 | 82.558 | 67.571 | 145.753 | 50.291 | 91.941 | 140.95 | 67.492 | 213.426 | 718.33 | 897.133 |
| GmCYS16 | 45.481 | 21.699 | 19.798 | 35.242 | 32.294 | 24.917 | 48.012 | 29.017 | 16.074 | 21.96 |
| GmCYS17 | 1.125 | 0 | 0 | 0.018 | 7.901 | 4.054 | 2.032 | 0 | 38.085 | 15.773 |
| GmCYS18 | 0 | 0.032 | 3.491 | 0 | 26.299 | 3.067 | 0 | 0 | 40.862 | 205.927 |
| GmCYS19 | 2.639 | 0 | 0 | 0.09 | 0 | 0 | 0.086 | 0 | 0 | 0 |
| GmCYS20 | 94.949 | 71.153 | 64.095 | 111.068 | 73.98 | 57.858 | 192.429 | 108.603 | 13.033 | 8.785 |
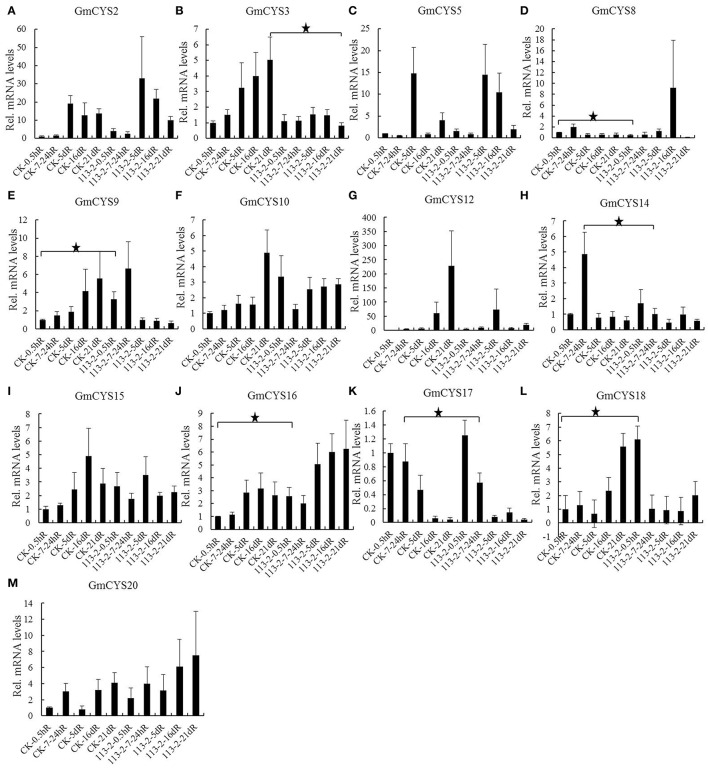
To determine whether cystatins play roles in symbiotic signal transduction, nodulation and early nodule development, 14 soybean cystatins that were expressed in root, root hairs and/or root at symbiotic condition were analyzed, and their expression levels in roots (CK and inoculated with B. japonicum strain 113-2) at 0.5 h, 7–24 h, 5, 16, and 21 d of post-inoculation were compared using t-tests to show statistical difference between CK and inoculated roots. Because no specific sequence was suitable for GmCYS13 qPCR (Figure S1), only 13 soybean cystatins were detected (Figure 4). Inoculation of B. japonicum strain 113-2 significantly increased the expression of GmCYS2, GmCYS9, GmCYS10, GmCYS12, GmCYS15, GmCYS16, GmCYS18, and GmCYS20 (>2 fold) at 0.5 hR of post-inoculation (Figures 4A,E,F,G,I,J,L,M), but significantly decreased the expression of GmCYS8 (>2 fold) at 0.5 hR of post-inoculation (Figure 4D). The expression levels of GmCYS8 and GmCYS14 decreased (>2 fold) at 7–24 hR of post-inoculation (Figures 4D,H), while the expression level of GmCYS9 increased (>2 fold) at 7–24 hR of post-inoculation (Figure 4E). The expression levels of GmCYS17 decreased (>2 fold) at 5 dR of post-inoculation (Figure 4K), while the expression level of GmCYS20 increased (>2 fold) at 5 dR of post-inoculation (Figure 4M). The expression of GmCYS5 and GmCYS8 in inoculated roots were increased (>2 fold) at 16 dR of post-inoculation, but decreased (>2 fold) at 21 dR of post-inoculation (Figures 4C,D), while for other 5 soybean cystatins (GmCYS3, GmCYS9, GmCYS12, GmCYS15, and GmCYS18), the expression levels were decreased (>2 fold) at 16 dR and/or 21 dR of post-inoculation (Figures 4B,E,G,I,L).
Figure 4.
Expression profile of cystatins family genes in soybean roots with and without inoculation of B. japonicum strain 113-2 at five post-inoculation time points. Three biological replica samples were used to detect the expression levels of 13 cystatins. Relative expression levels of each cystatin gene were obtained by qPCR and normalized to the average expression level of soybean reference gene QACT, and calculated using the expression level in CK-0.5 hR as control. Student's t-test was done using software SPSS Statistics 17.0 and data are presented as the mean ± SE of 9 or 10 independent replicate experiments. “★” indicates statistical difference between the inoculated roots and CK roots (t-test, p < 0.05). (A) GmCYS2. (B) GmCYS3. (C) GmCYS5. (D) GmCYS8. (E) GmCYS9. (F) GmCYS10. (G) GmCYS12. (H) GmCYS14. (I) GmCYS15. (J) GmCYS16. (K) GmCYS17. (L) GmCYS18. (M) GmCYS20.
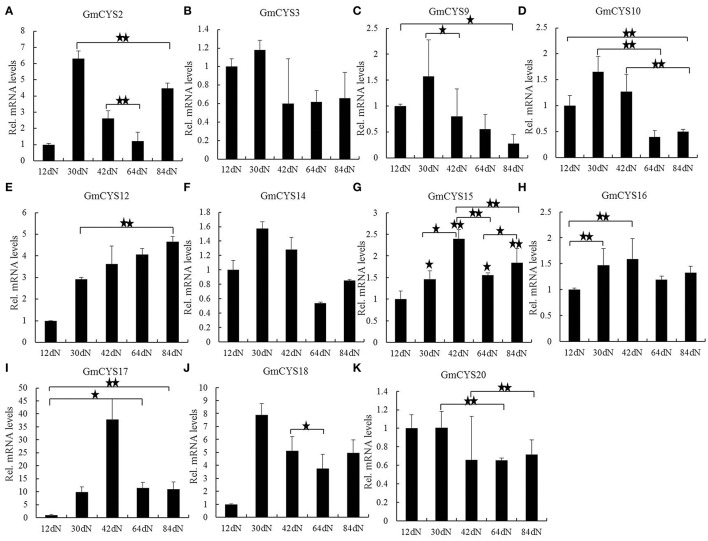
Previous studies showed that some members of soybean cystatins are actively transcribed during nodule development and senescence (van Wyk et al., 2014). To determine which soybean cystatins play roles in nodule development and/or senescence, soybean cystatins that were expressed in nodule and/or nodules at symbiotic condition were analyzed using t-tests to show statistical difference between different nodule development time points. The results showed that 11 soybean cystatins (GmCYS2, GmCYS3, GmCYS9, GmCYS10, GmCYS12, GmCYS14-18, and GmCYS20) were differentially expressed in the nodules at five important nodule development time points (Figure 5). Among them, 4 soybean cystatins (GmCYS3, GmCYS14, GmCYS16, and GmCYS20) showed <2-fold expression variation in different nodules, suggesting that they were not regulated at the transcription level during soybean nodule development (Figures 5B,F,H,K). GmCYS12, GmCYS17, and GmCYS18 were upregulated (>2 fold) between 12 dN and others (Figures 5E,I,J), GmCYS2 was up-regulated (>2 fold) between 12 dN and other three nodules (30, 42, and 84 dN) and between 60 and 84 dN (Figure 5A), indicating that it may play a role in nodule senescence. The expressions of GmCYS9 and GmCYS10 were reduced (most were <2 fold) during the nodule development (Figures 5C,D). The expression levels of GmCYS15 and GmCYS17 reached their peaks at 42 dN (the nitrogen-fixation rate are relatively high; Figures 5G,I), suggesting that they may be regulated at the transcription level in the nitrogen-fixation process. The expression analysis results of 4 soybean cystatins (GmCYS9, GmCYS10, GmCYS12, and GmCYS15) agreed with transcriptional profile data (van Wyk et al., 2014). GmCYS2 was downregulated between 30 and 64 dN (Figure 5A) and GmCYS16 and GmCYS20 showed no change at the transcription level during nodule development and senescence (Figures 5H,K).
Figure 5.
Expression profile of cystatins family genes in soybean nodules at five important nodule development time points. Three biological replica samples and three replicate reactions per sample were used to calculate the expression level of each cystatin during the nodule development. Relative expression levels of each cystatin gene were obtained by qPCR and normalized to the average expression level of soybean reference gene QACT, and calculated using the expression level in 12 dN as a starting point control. Data represent the mean ± SE of nine independent replicate experiments. “★”and “★★” indicate statistical difference between different nodule development time points (t-test, p < 0.05 or p < 0.01, respectively). “★”and “★★” without wire frame marker in (G), indicate statistical difference compared to the 12 dN (starting point control). (A) GmCYS2. (B) GmCYS3. (C) GmCYS9. (D) GmCYS10. (E) GmCYS12. (F) GmCYS14. (G) GmCYS15. (H) GmCYS16. (I) GmCYS17. (J) GmCYS18. (K) GmCYS20.
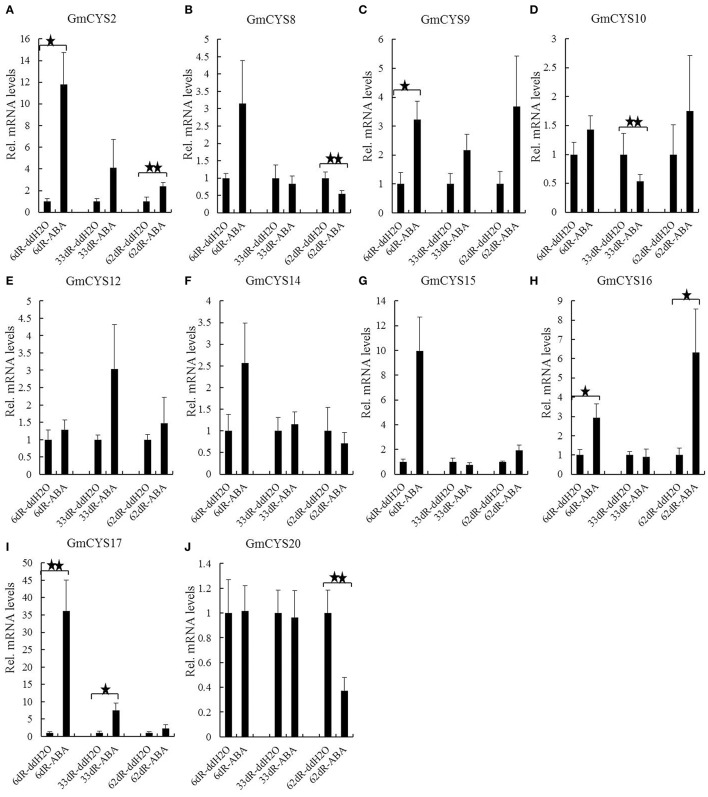
Expression analysis of soybean cystatin genes in response to ABA treatment in symbiosis with B. japonicum strain 113-2
As Table 2 described, 80% soybean cystatin genes have abscisic acid responsive elements (ABRE or CE3), suggesting that their expressions are induced by ABA, a negative regulator of symbiotic nitrogen fixation (Tominaga et al., 2009, 2010) that decreases nodule number (Phillips, 1971; Suzuki et al., 2004; Ding et al., 2008; Biswas et al., 2009) and negatively regulates nitrogen fixation (González et al., 2001). To confirm the response of soybean cystatins to ABA in symbiosis, the transcriptional levels of 10 soybean cystatins that contain ABRE and/or CE3 elements, and are expressed in inoculated roots (Tables 2, 3) at 6, 33, and 62 d of post-inoculation were compared using t-tests to show statistical difference among different treatment groups (with nodules, ddH2O and ABA treatment) (Figure 6). Three soybean cystatins (GmCYS2, GmCYS9 and GmCYS17) were upregulated (>2 fold) after ABA treatment in inoculated roots at 6, 33, and 62 d of post-inoculation (Figures 6A,C,I). The expression of GmCYS16 was increased (>2 fold) after ABA treatment in inoculated roots at 6 and 62 d of post-inoculation (Figure 6H), the expression level of GmCYS8, GmCYS14 and GmCYS15 was increased (>2 fold) after ABA treatment in inoculated roots at 6 d of post-inoculation (Figures 6B,F,G), and the expression level of GmCYS12 was increased (>2 fold) after ABA treatment in inoculated roots at 33 d of post-inoculation (Figure 6E). The expression of GmCYS10 was not changes (<2 fold) at the transcription level in soybean inoculated roots after ABA treatment (Figure 6D), suggesting that its expressions were not induced by ABA in soybean symbiosis with B. japonicum strain 113-2. Whether its expressions were induced by ABA in non-inoculated roots and/or other tissues need to be further explored.
Figure 6.
Expression profile of soybean cystatin genes in response to ABA treatment in symbiosis with B. japonicum strain 113-2. Three biological replica samples and three replicate reactions per sample were used to calculate the expression level of 10 cystatins in inoculated roots (with nodules, ddH2O, and ABA treatment) at 6, 33, and 62 days of post inoculation. Relative expression levels of each cystatin gene were obtained by qPCR and normalized to the average expression level of soybean reference gene QACT, and calculated using the expression level in ddH2O treatment samples as controls. Data are presented as mean ± SE of nine independent replicate experiments. “★”and “★★” indicate statistical difference between different treatment inoculated roots (t-test, p < 0.05 or p < 0.01, respectively). (A) GmCYS2. (B) GmCYS8. (C) GmCYS9. (D) GmCYS10. (E) GmCYS12. (F) GmCYS14. (G) GmCYS15. (H) GmCYS16. (I) GmCYS17. (J) GmCYS20.
Functional analysis of a candidate gene Glyma.15G227500 (GmCYS16) in nodulation
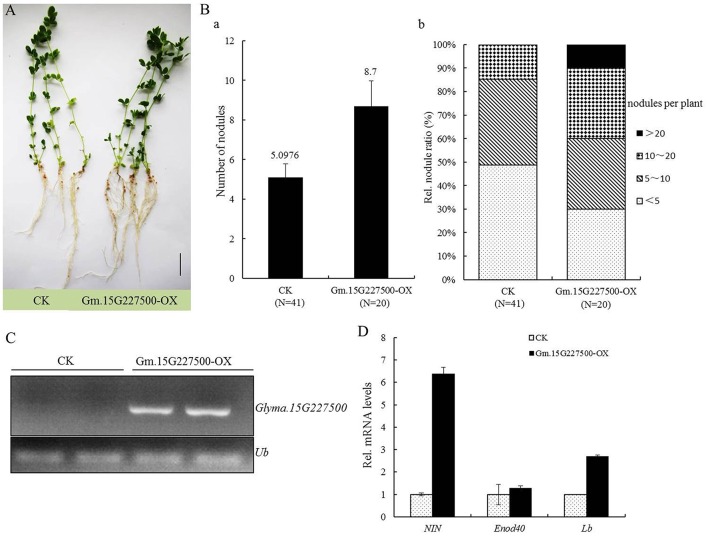
As described above, when inoculated with B. japonicum strain 113-2, the expression of GmCYS16 in soybean roots was significantly increased (Figure 4J), suggesting that GmCYS16 may plays a role in nodulation. To confirm this result, Glyma.15G227500 (GmCYS16) was expressed under the control of the maize (Zea mays) ubiquitin promoter (Glyma.15G227500-OX) in transgenic hairy roots of L. japonicus (Kumagai and Kouchi, 2003; Yuan et al., 2012). The phenotypes of nodulation were scored at 32 days of post-inoculation with M. loti MAFF303099, which expresses β-galactosidase (lacZ) as a constitutive marker for the presence of rhizobial cells (Tansengco et al., 2003). As shown in Figures 7A,B, more nodules were produced in Glyma.15G227500-OX hairy roots. The nodule number per root system was increased from 5.0976 in the control to 8.7 in Glyma.15G227500-OX hairy roots (Figure 7Ba). Analysis of the distribution of nodule number per plant revealed that 40% of Glyma.15G227500-OX hairy roots developed more than 10 nodules. In contrast, only 15% of the control hairy roots produced more than 10 nodules, and no hairy roots produced more than 20 nodules (Figure 7Bb). Semi-quantitative RT-PCR results showed that the expression level of Glyma.15G227500 was abundant in Glyma.15G227500-OX hairy roots (Figure 7C). qPCR analyses of expressions of NIN and ENOD40, two early nodulin genes implicated in the processes of rhizobial entrance, nodule initiation, and subsequent organogenesis (Schauser et al., 1999; Kumagai et al., 2006), as well as Lb (leghemoglobin), a typical nodulin gene required for nitrogen fixation (Ott et al., 2005) showed that the expression levels of these three nodulin genes, especially NIN and Lb, were increased in Glyma.15G227500-OX hairy roots, as compared with those in the control hairy roots, (Figure 7D). These results suggest that Glyma.15G227500 plays a positive role in nodulation of L. japonicus.
Figure 7.
Effect of Glyma.15G227500 overexpression on symbiosis in L. japonicus. (A) Symbiotic phenotypes of transgenic plants at 32 days of post inoculation with M. loti. MAFF303099. Hairy roots expressing vector pU1301 served as a control. Independent transgenic plants were chosen for photography. Bars = 5 mm. (B) Nodule numbers of transgenic hairy roots with altered Glyma.15G227500 transcript levels. Transgenic hairy roots expressing vector (pU1301) served as control. (A) Mean numbers of nodules per plant with standard deviation (SD) of L. japonicus expressing pMUb: Glyma.15G227500 (Gm.15G227500-OX) or the empty vector pU1301 (CK) at 32 days of post inoculation with M. loti. (B) Plants were divided into different groups on the basis of nodules per plant, and the relative ratios of various groups were calculated. (C) Semi-quantitative RT-PCR analysis of the transcript levels of Glyma.15G227500 in the control and Gm.15G227500-OX hairy roots. (D) qPCR analysis of the transcript levels of NIN, Enod40, and Lb in the control and Gm.15G227500-OX hairy roots. Total RNA isolated from root system was used for qPCR. Relative expression levels of NIN, Enod40, and Lb transcripts in Gm.15G227500-OX hairy roots were calculated with reference to that of the control hairy roots.
Discussion
Nodulation, nodule development and senescence directly affect nitrogen fixation efficiency. Previous studies have shown that inhibition of some cysteine proteases delays nodule senescence (Sheokand et al., 2005; Li et al., 2008). Thus, their nature inhibitors, cystatins, are very important in nodule development and senescence. Soybean is an important oil crop, food and feed material, providing large amounts of dietary proteins and edible oil and an excellent model for studying nitrogen fixation and nodule development. The inhibitory activities of 20 soybean cystatins including seven that were actively transcribed in nodules were analyzed (van Wyk et al., 2014). However, the exact roles of these cystatin genes in nodulation, nodule development, and senescence remain poorly characterized. To search for nodulation and nodule development-related cystatin genes in soybean, the detail information of genome-wide identification and characterization of 20 soybean cystatins were shown in this study to explore their functional diversities. Our study for the first time conducted the genome-wide survey of soybean cystatin family genes in nodulation and nodule development, and the resultant expression patterns of soybean cystatin family genes in symbiosis provide insights into their putative roles in nodulation and nodule development.
Potential roles of soybean cystatin family genes in nodulation, nodule development, and senescence
Previous studies showed that cystatins could regulate nodulation and nodule development (Quain et al., 2015) and may play a role in regulating proteolysis in the process of nodules senescence and programmed cell death (van Wyk et al., 2014). However, which soybean cystatins play roles in nodulation, nodule development, and senescence is still largely unknown. Therefore, the expression profiles of soybean cystatins in five tissues (nodules. symbiotic condition, root. symbiotic condition, root, root hairs, and nodules) were comprehensively analyzed (Table 3) in present work, respectively (Figures 4, 5). The results showed that the expression levels of three soybean cystatins (GmCYS2, GmCYS16, and GmCYS20) were significantly increased in inoculated roots (Figures 4A,J,M), suggesting that they may play roles in nodulation. GmCYS15 and GmCYS17 showed highest expression at the stage with relatively high nitrogen-fixation rate (42 dN), indicating that they may play roles in the nitrogen-fixation process. For nodule senescence, previous transcriptional profile data has showed that expression of GmCYS2, GmCYS15, and GmCYS16 were particularly increased during the onset of senescence (van Wyk et al., 2014). In this study, expression of GmCYS15 and GmCYS16 barely changed between 60 and 84 dN or between 42 and 84 dN (Figures 5G,H). Besides, the expression levels of GmCYS12 and GmCYS18 were significantly increased in roots at 0.5 h of post inoculation and during nodule developments (Figures 4G,L, 5E,J), suggesting that these two cystatins may play roles in nodulation and nodule development. These data indicated that these 7 soybean cystatin proteins (GmCYS2, GmCYS12, GmCYS15, GmCYS16, GmCYS17, GmCYS18, and GmCYS20) might play different roles in soybean nodule symbiosis. Their specific regulatory roles and distinct functions are interesting and worthy to be explored in the future.
Characteristics of soybean nodulation and nodule development-related cystatin genes
As above described in the present work, the identified 20 soybean cystatin genes encode peptides with 97–245 amino acid residues, and different isoelectric points (pI) and structure characteristics (Table 1, Figures 1, 2). Three typical motifs for cystatin proteins, “QxVxG,” “G” in N-terminal and “W” residue were detected in most cystatins with only one exception (GmCYS4), and the other conserved “LARFAV” motif varied among the four Clades of soybean cystatin genes (Figure 1B). Five soybean cystatins (GmCYS10, GmCYS13, GmCYS14, GmCYS16, and GmCYS18) showed significant variations with the other 15 cystatins in their predicted three-dimensional structures (Figure 2), mainly due to these soybean cystatins have relatively complicated motifs and longer coding sequences than the others (except GmCYS18). Among them, 7 nodulation and nodule development-related cystatin proteins fall into different Clades (Figure 1) and have different structures (Table 1, Figure 2), indicating that there are no special clade cystatin protein or special structure in nodulation and nodule development-related cystatins.
Previous reports demonstrated that cystatin could inhibit activity of cysteine proteases in the process of plant seed germination (Martinez et al., 2009) and host immunity (Koiwa et al., 2000; van der Linde et al., 2012), and also play a key role in hypersensitive cell death and the regulation of defense against pathogens and insects (Bobek and Levine, 1992; Belenghi et al., 2003; Gholizadeh et al., 2005; Pogány et al., 2015). Besides, their expression is also responsive to abiotic stresses and hormone signals (Zhang et al., 2008; Wang et al., 2015). Similar to this study, various putative plant regulatory elements in response to different abiotic/biotic stresses (various environmental stresses) and hormone signals were shown in 3000 bp putative promoter fragments upstream of the 20 soybean cystatins (Table 2). The 7 nodulation and nodule development-related cystatin proteins have inconsistent plant regulatory elements in their promoters (Table 2). For hormones responsive elements, GmCYS2 has elements in response to 5 different hormones, including abscisic acid, gibberellin acid, salicylic acid, auxin and jasmonic acid, GmCYS12, GmCYS15, GmCYS16, and GmCYS17 have no auxin responsive elements, GmCYS15 has no jasmonic acid responsive element, GmCYS18 has no abscisic acid and gibberellin acid responsive elements, and GmCYS20 has no gibberellin acid responsive elements. For environmental responsive elements, GmCYS15, GmCYS16, and GmCYS18 have no MBS element, GmCYS12 has no TC-rich repeats element, and GmCYS15 has no circadian-regulated elements. Besides, GmCYS2, GmCYS15, GmCYS17, and GmCYS18 have fungal elicitor responsive element, GmCYS15 and GmCYS17 have wound-responsive element, and GmCYS15, GmCYS17, and GmCYS20 have ethylene-responsive element. Interesting, GmCYS2 has a nodule specific factor binding site element (Nodule-site2, Table 2), which also exists in the promoter fragment of soybean leghemoglobin (LBC3), a typical nodulin gene required for nitrogen fixation (She et al., 1993). However, whether GmCYS2 plays an important role in the soybean nitrogen fixation needs to be further studied. These results indicated that these 7 soybean cystatin proteins are not root nodule symbiosis (RNS)—specific cystatins, and have different putative biological functions.
A special branch for legume plant cystatins containing only one soybean cystatin related to nodulation and nodule development
As described in the evolutionary relationships among the cystatin family in rice, barley, A. thaliana, N. tabacum and four legume plants, Group A cystatins are special branch for legume plant, Group B cystatins only contain non-legume plant cystatins (non-legume plant dicotyledon and monocotyledon), and Group C cystatins are both legume plants and non-legume plant related (Figure 3). Of the 7 cystatins related to nodulation and nodule development, only GmCYS15 fall into Group A, the other 6 cystatins fall into Group C (Figure 3). These results indicated that cystatins in the special branch are not special for legume property-nodulation and nitrogen fixation and may possess other biological functions.
In summary, 20 cystatin family genes were identified in soybean genome through a genome-wide survey with the aim to search for nodulation and nodule development-related cystatin genes in soybean. Characterization of their gene sequences, structures, and promoter sequences as well as phylogenetic analysis revealed special characteristics of cystatin family genes in soybean. Expression profiling of the cystatin genes in soybean symbiosis with B. japonicum 113-2 indicated that 7 cystatin family genes might play different roles in nodulation and nodule development. The functional analysis showed that the candidate gene Glyma.15G227500 (GmCYS16) likely to play a positive role in soybean nodulation. These investigations and analyses shed new light on the research of the functional diversity of all members in the soybean cystatin family proteins, and could increase our knowledge on their functions in regulating soybean nodulation and nodule development, and provide helpful clues for investigation of nodule-specific cysteine proteases in soybean.
Author contributions
SY and XZho designed this work. SY wrote the manuscript. SY and RL performed most of the experiments. HC, CZ, LC, QH, ZS, XZha, SC, ZY, DQ, and LW contributed substantially to the completion of this work. All authors read and approved the final manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by funds from the China Agriculture Research System (CAAS-04-PS08), the National Transgenic Project of China (2016ZX08004-005), the Agricultural Science and Technology Innovation Program of CAAS, and the Scientific and Technological Support Project of Soybean Breeding and Propagation (2016BAD35B06).
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01595
References
- Arai S., Matsumoto I., Emori Y., Abe K. (2002). Plant seed cystatins and their target enzymes of endogenous and exogenous origin. J. Agric. Food Chem. 50, 6612–6617. 10.1021/jf0201935 [DOI] [PubMed] [Google Scholar]
- Belenghi B., Acconcia F., Trovato M., Perazzolli M., Bocedi A., Polticelli F., et al. (2003). AtCYS1, a cystatin from Arabidopsis thaliana, suppresses hypersensitive cell death. Eur. J. Biochem. 270, 2593–2604. 10.1046/j.1432-1033.2003.03630.x [DOI] [PubMed] [Google Scholar]
- Benchabane M., Schlüter U., Vorster J., Goulet M. C., Michaud D. (2010). Plant cystatins. Biochimie 92, 1657–1666. 10.1016/j.biochi.2010.06.006 [DOI] [PubMed] [Google Scholar]
- Bendtsen J. D., Nielsen H., von Heijne G., Brunak S. (2004). Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340, 783–795. 10.1016/j.jmb.2004.05.028 [DOI] [PubMed] [Google Scholar]
- Biswas B., Chan P. K., Gresshoff P. M. (2009). A novel ABA insensitive mutant of Lotus japonicus with a wilty phenotype displays unaltered nodulation regulation. Mol. Plant 2, 487–499. 10.1093/mp/ssp009 [DOI] [PubMed] [Google Scholar]
- Biswas B., Gresshoff P. M. (2014). The role of symbiotic nitrogen fixation in sustainable production of biofuels. Int. J. Mol. Sci. 15, 7380–7397. 10.3390/ijms15057380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobek L. A., Levine M. J. (1992). Cystatins–inhibitors of cysteine proteinases. Crit. Rev. Oral Biol. Med. 3, 307–332. [DOI] [PubMed] [Google Scholar]
- Broughton W. J., Dilworth M. J. (1971). Control of leghaemoglobin synthesis in snake beans. Biochem. J. 125, 1075–1080. 10.1042/bj1251075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo L., Martinez M., Ramessar K., Cambra I., Castañera P., Ortego F., et al. (2011). Expression of a barley cystatin gene in maize enhances resistance against phytophagous mites by altering their cysteine-proteases. Plant Cell Rep. 30, 101–112. 10.1007/s00299-010-0948-z [DOI] [PubMed] [Google Scholar]
- Chen C. C., Hwang J. K., Yang J. M. (2009). (PS)2-v2: template-based protein structure prediction server. BMC Bioinformatics 10:366. 10.1186/1471-2105-10-366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y., Kalo P., Yendrek C., Sun J., Liang Y., Marsh J. F., et al. (2008). Abscisic acid coordinates nod factor and cytokinin signaling during the regulation of nodulation in Medicago truncatula. Plant Cell 20, 2681–2695. 10.1105/tpc.108.061739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gholizadeh A., Santha I. M., Kohnehrouz B. B., Lodha M. L., Kapoor H. C. (2005). Cystatins may confer viral resistance in plants by inhibition of a virus-induced cell death phenomenon in which cysteine proteinases are active: cloning and molecular characterization of a cDNA encoding cysteine-proteinase inhibitor (celostatin) from Celosia cristata (crested cock's comb). Biotechnol. Appl. Biochem. 42, 197–204. 10.1042/BA20050029 [DOI] [PubMed] [Google Scholar]
- González E. M., Gálvez L., Arrese-Igor C. (2001). Abscisic acid induces a decline in nitrogen fixation that involves leghaemoglobin, but is independent of sucrose synthase activity. J. Exp. Bot. 52, 285–293. 10.1093/jexbot/52.355.285 [DOI] [PubMed] [Google Scholar]
- Green A. R., Nissen M. S., Kumar G. N., Knowles N. R., Kang C. (2013). Characterization of Solanum tuberosum multicystatin and the significance of core domains. Plant Cell 25, 5043–5052. 10.1105/tpc.113.121004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S., Gascuel O. (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696–704. 10.1080/10635150390235520 [DOI] [PubMed] [Google Scholar]
- Guo A. Y., Zhu Q. H., Chen X., Luo J. C. (2007). [GSDS: a gene structure display server]. Yi Chuan 29, 1023–1026. 10.1360/yc-007-1023 [DOI] [PubMed] [Google Scholar]
- Gutierrez-Campos R., Torres-Acosta J. A., Saucedo-Arias L. J., Gomez-Lim M. A. (1999). The use of cysteine proteinase inhibitors to engineer resistance against potyviruses in transgenic tobacco plants. Nat. Biotechnol. 17, 1223–1226. 10.1038/70781 [DOI] [PubMed] [Google Scholar]
- Hu Y. J., Irene D., Lo C. J., Cai Y. L., Tzen T. C., Lin T. H., et al. (2015). Resonance assignments and secondary structure of a phytocystatin from Sesamum indicum. Biomol. NMR Assign. 9, 309–311. 10.1007/s12104-015-9598-y [DOI] [PubMed] [Google Scholar]
- Hwang J. E., Hong J. K., Lim C. J., Chen H., Je J., Yang K. A., et al. (2010). Distinct expression patterns of two Arabidopsis phytocystatin genes, AtCYS1 and AtCYS2, during development and abiotic stresses. Plant Cell Rep. 29, 905–915. 10.1007/s00299-010-0876-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koiwa H., Shade R. E., Zhu-Salzman K., D'Urzo M. P., Murdock L. L., Bressan R. A., et al. (2000). A plant defensive cystatin (soyacystatin) targets cathepsin L-like digestive cysteine proteinases (DvCALs) in the larval midgut of western corn rootworm (Diabrotica virgifera virgifera). FEBS Lett. 471, 67–70. 10.1016/S0014-5793(00)01368-5 [DOI] [PubMed] [Google Scholar]
- Kumagai H., Kinoshita E., Ridge R. W., Kouchi H. (2006). RNAi knock-down of ENOD40s leads to significant suppression of nodule formation in Lotus japonicus. Plant Cell Physiol. 47, 1102–1111. 10.1093/pcp/pcj081 [DOI] [PubMed] [Google Scholar]
- Kumagai H., Kouchi H. (2003). Gene silencing by expression of hairpin RNA in Lotus japonicus roots and root nodules. Mol. Plant Microbe Interact. 16, 663–668. 10.1094/MPMI.2003.16.8.663 [DOI] [PubMed] [Google Scholar]
- Lescot M., Déhais P., Thijs G., Marchal K., Moreau Y., Van de Peer Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. 10.1093/nar/30.1.325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Zhou L., Li Y., Chen D., Tan X., Lei L., et al. (2008). A nodule-specific plant cysteine proteinase, AsNODF32, is involved in nodule senescence and nitrogen fixation activity of the green manure legume Astragalus sinicus. New Phytol. 180, 185–192. 10.1111/j.1469-8137.2008.02562.x [DOI] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Margis R., Reis E. M., Villeret V. (1998). Structural and phylogenetic relationships among plant and animal cystatins. Arch. Biochem. Biophys. 359, 24–30. 10.1006/abbi.1998.0875 [DOI] [PubMed] [Google Scholar]
- Martinez M., Cambra I., Carrillo L., Diaz-Mendoza M., Diaz I. (2009). Characterization of the entire cystatin gene family in barley and their target cathepsin L-like cysteine-proteases, partners in the hordein mobilization during seed germination. Plant Physiol. 151, 1531–1545. 10.1104/pp.109.146019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez M., Diaz-Mendoza M., Carrillo L., Diaz I. (2007). Carboxy terminal extended phytocystatins are bifunctional inhibitors of papain and legumain cysteine proteinases. FEBS Lett. 581, 2914–2918. 10.1016/j.febslet.2007.05.042 [DOI] [PubMed] [Google Scholar]
- Martinez M., Lopez-Solanilla E., Rodriguez-Palenzuela P., Carbonero P., Diaz I. (2003). Inhibition of plant-pathogenic fungi by the barley cystatin Hv-CPI (gene Icy) is not associated with its cysteine-proteinase inhibitory properties. Mol. Plant Microbe Interact. 16, 876–883. 10.1094/MPMI.2003.16.10.876 [DOI] [PubMed] [Google Scholar]
- Nagata K., Kudo N., Abe K., Arai S., Tanokura M. (2000). Three-dimensional solution structure of oryzacystatin-I, a cysteine proteinase inhibitor of the rice, Oryza sativa L. japonica. Biochemistry 39, 14753–14760. 10.1021/bi0006971 [DOI] [PubMed] [Google Scholar]
- Ott T., van Dongen J. T., Gunther C., Krusell L., Desbrosses G., Vigeolas H., et al. (2005). Symbiotic leghemoglobins are crucial for nitrogen fixation in legume root nodules but not for general plant growth and development. Curr. Biol. 15, 531–535. 10.1016/j.cub.2005.01.042 [DOI] [PubMed] [Google Scholar]
- Peitsch M. C. (1996). ProMod and Swiss-model: internet-based tools for automated comparative protein modelling. Biochem. Soc. Trans. 24, 274–279. 10.1042/bst0240274 [DOI] [PubMed] [Google Scholar]
- Pfeiffer N. E., Torres C. M., Wagner F. W. (1983). Proteolytic Activity in soybean root nodules : activity in host cell cytosol and bacteroids throughout physiological development and senescence. Plant Physiol. 71, 797–802. 10.1104/pp.71.4.797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips D. A. (1971). Abscisic acid inhibition of root nodule initiation in Pisum sativum. Planta 100, 181–190. 10.1007/BF00387034 [DOI] [PubMed] [Google Scholar]
- Pogány M., Dankó T., Kámán-Tóth E., Schwarczinger I., Bozsó Z. (2015). Regulatory proteolysis in arabidopsis-pathogen interactions. Int. J. Mol. Sci. 16, 23177–23194. 10.3390/ijms161023177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovic M., Andjelkovic U., Burazer L., Lindner B., Petersen A., Gavrovic-Jankulovic M. (2013). Biochemical and immunological characterization of a recombinantly-produced antifungal cysteine proteinase inhibitor from green kiwifruit (Actinidia deliciosa). Phytochemistry 94, 53–59. 10.1016/j.phytochem.2013.06.006 [DOI] [PubMed] [Google Scholar]
- Quain M. D., Makgopa M. E., Cooper J. W., Kunert K. J., Foyer C. H. (2015). Ectopic phytocystatin expression increases nodule numbers and influences the responses of soybean (Glycine max) to nitrogen deficiency. Phytochemistry 112, 179–187. 10.1016/j.phytochem.2014.12.027 [DOI] [PubMed] [Google Scholar]
- Schauser L., Roussis A., Stiller J., Stougaard J. (1999). A plant regulator controlling development of symbiotic root nodules. Nature 402, 191–195. 10.1038/46058 [DOI] [PubMed] [Google Scholar]
- Schmutz J., Cannon S. B., Schlueter J., Ma J., Mitros T., Nelson W., et al. (2010). Genome sequence of the palaeopolyploid soybean. Nature 463, 178–183. 10.1038/nature08670 [DOI] [PubMed] [Google Scholar]
- Severin A. J., Woody J. L., Bolon Y. T., Joseph B., Diers B. W., Farmer A. D., et al. (2010). RNA-Seq Atlas of Glycine max: a guide to the soybean transcriptome. BMC Plant Biol. 10:160. 10.1186/1471-2229-10-160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- She Q., Sandal N. N., Stougaard J., Marcker K. A. (1993). Comparative sequence analysis of cis elements present in Glycine max L. leghemoglobin lba and lbc3 genes. Plant Mol. Biol. 22, 931–935. 10.1007/BF00027380 [DOI] [PubMed] [Google Scholar]
- Sheokand S., Dahiya P., Vincent J. L., Brewin N. J. (2005). Modified expression of cysteine protease affects seed germination, vegetative growth and nodule development in transgenic lines of Medicago truncatula. Plant Sci. 169, 966–975. 10.1016/j.plantsci.2005.07.003 [DOI] [Google Scholar]
- Stamatakis A. (2014). RAxML version 8: a tool for phylogenetic analysis and post- analysis of large phylogenies. Bioinformatics 30, 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbs M. T., Laber B., Bode W., Huber R., Jerala R., Lenarcic B., et al. (1990). The refined 2.4 A X-ray crystal structure of recombinant human stefin B in complex with the cysteine proteinase papain: a novel type of proteinase inhibitor interaction. EMBO J. 9, 1939–1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A., Akune M., Kogiso M., Imagama Y., Osuki K., Uchiumi T., et al. (2004). Control of nodule number by the phytohormone abscisic Acid in the roots of two leguminous species. Plant Cell Physiol. 45, 914–922. 10.1093/pcp/pch107 [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y., Wang S., Liang D., Li M., Ma F. (2014). Genome-wide identification and expression profiling of the cystatin gene family in apple (Malus x domestica Borkh.). Plant Physiol. Biochem. 79, 88–97. 10.1016/j.plaphy.2014.03.011 [DOI] [PubMed] [Google Scholar]
- Tansengco M. L., Hayashi M., Kawaguchi M., Imaizumi-Anraku H., Murooka Y. (2003). Crinkle, a novel symbiotic mutant that affects the infection thread growth and alters the root hair, trichome, and seed development in Lotus japonicus. Plant Physiol. 131, 1054–1063. 10.1104/pp.102.017020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga A., Nagata M., Futsuki K., Abe H., Uchiumi T., Abe M., et al. (2009). Enhanced nodulation and nitrogen fixation in the abscisic acid low-sensitive mutant enhanced nitrogen fixation1 of Lotus japonicus. Plant Physiol. 151, 1965–1976. 10.1104/pp.109.142638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga A., Nagata M., Futsuki K., Abe H., Uchiumi T., Abe M., et al. (2010). Effect of abscisic acid on symbiotic nitrogen fixation activity in the root nodules of Lotus japonicus. Plant Signal. Behav. 5, 440–443. 10.4161/psb.5.4.10849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Linde K., Hemetsberger C., Kastner C., Kaschani F., Van der Hoorn R. A., Kumlehn J., et al. (2012). A maize cystatin suppresses host immunity by inhibiting apoplastic cysteine proteases. Plant Cell 24, 1285–1300. 10.1105/tpc.111.093732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wyk S. G., Du Plessis M., Cullis C. A., Kunert K. J., Vorster B. J. (2014). Cysteine protease and cystatin expression and activity during soybean nodule development and senescence. BMC Plant Biol. 14:294. 10.1186/s12870-014-0294-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent J. L., Brewin N. J. (2000). Immunolocalization of a cysteine protease in vacuoles, vesicles, and symbiosomes of pea nodule cells. Plant Physiol. 123, 521–530. 10.1104/pp.123.2.521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Zhao P., Zhou X. M., Xiong H. X., Sun M. X. (2015). Genome-wide identification and characterization of cystatin family genes in rice (Oryza sativa L.). Plant Cell Rep. 34, 1579–1592. 10.1007/s00299-015-1810-0 [DOI] [PubMed] [Google Scholar]
- Yuan S., Li R., Chen S., Chen H., Zhang C., Chen L., et al. (2016). RNA-Seq analysis of differential gene expression responding to different rhizobium strains in soybean (Glycine max) roots. Front. Plant Sci. 7:721. 10.3389/fpls.2016.00721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S., Zhu H., Gou H., Fu W., Liu L., Chen T., et al. (2012). A ubiquitin ligase of symbiosis receptor kinase involved in nodule organogenesis. Plant Physiol. 160, 106–117. 10.1104/pp.112.199000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Liu S., Takano T. (2008). Two cysteine proteinase inhibitors from Arabidopsis thaliana, AtCYSa and AtCYSb, increasing the salt, drought, oxidation and cold tolerance. Plant Mol. Biol. 68, 131–143. 10.1007/s11103-008-9357-x [DOI] [PubMed] [Google Scholar]
- Zhao P., Zhou X. M., Zou J., Wang W., Wang L., Peng X. B., et al. (2014). Comprehensive analysis of cystatin family genes suggests their putative functions in sexual reproduction, embryogenesis, and seed formation. J. Exp. Bot. 65, 5093–5107. 10.1093/jxb/eru274 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.