Abstract
The aberrant expression of microRNAs (miRNAs) has emerged as an important hallmark of cancer. However, the molecular mechanisms underlying the changes in miRNA expression remain unclear. In this study, we discovered a novel epigenetic mechanism of miR-506 regulation and investigated its functional significance in pancreatic cancer. Sequencing analysis revealed that the miR-506 promoter is highly methylated in pancreatic cancer tissues compared with non-cancerous tissues. Reduced miR-506 expression was significantly associated with clinical stage, pathologic tumor status, distant metastasis and decreased survival of pancreatic cancer patients. miR-506 inhibited cell proliferation, induced cell cycle arrest at the G1/S transition and enhanced apoptosis and chemosensitivity of pancreatic cancer cells. Furthermore, we identified sphingosine kinase 1 (SPHK1) as a novel target of miR-506, the expression of which inhibited the SPHK1/Akt/NF-κB signaling pathway, which is activated in pancreatic cancer. High SPHK1 expression was significantly associated with poor survival in a large cohort of pancreatic cancer specimens. Our data suggest that miR-506 acts as a tumor suppressor miRNA and is epigenetically silenced in pancreatic cancer. The newly identified miR-506/SPHK1 axis represents a novel therapeutic strategy for future pancreatic cancer treatment.
Introduction
Pancreatic cancer remains one of the deadliest cancers, with a 5-year overall survival rate of less than 7%.1 This poor prognosis is due to its late presentation, early metastasis and unresponsiveness to most treatment options.2 Surgical resection is the only curative treatment; however, only ~15–20% of tumors are resectable at the time of diagnosis.3 A large proportion of patients are diagnosed with locally advanced or metastatic disease at the time of presentation,1 and these patients receive chemoradiation therapy. Nevertheless, pancreatic cancer responds poorly to both chemotherapy and radiation.4 Although gemcitabine-based chemotherapy is standard treatment for advanced pancreatic cancer and has improved patient prognosis, its effect is limited because of high drug resistance.5 Therefore, the development of new diagnostic and treatment strategies for pancreatic cancer is urgently needed.
MicroRNAs (miRNAs) are a class of small noncoding RNAs (~22 nt) that function in post-transcriptional regulation mainly by binding to the 3′-untranslated region (3′-UTR) of target mRNAs, resulting in mRNA degradation or inhibition of translation.6 Altered miRNA expression has been reported in almost all types of human cancers. miRNAs can function as oncogenes or tumor suppressor genes, involving multiple pathways and cell functions in cancer development and progression, such as proliferation, apoptosis, invasion and resistance to therapy.7 To date, a series of miRNAs (including miR-21, miR-34a, miR-30d, miR-155 and miR-203) has been shown to be associated with tumor progression and overall survival in patients with pancreatic cancer.8, 9 In an integrated data analysis, Frampton et al.10 identified that miR-21, miR-23a and miR-27a functioned as cooperative repressors of a network of tumor suppressor genes, and high levels of a combination of these miRNAs were correlated with shorter survival times in pancreatic ductal adenocarcinoma (PDAC) patients. Previous studies from our group demonstrated that miR-15a inhibits cell proliferation and the epithelial-to-mesenchymal transition in PDAC by downregulating Bmi-1 expression.11 These studies indicate that several miRNAs are involved in the tumorigenesis of PDAC. Recently, miR-506, a member of an X chromosome-linked miRNA cluster in the primate,12 was found to have pivotal roles in several cancer types.13, 14, 15 However, the role of miR-506 is complicated, even contradictory, in the reported cancer types. For instance, miR-506 acts as a tumor suppressor in ovarian13 and lung14 cancer, but functions as an oncogene in melanomas,15 which indicates that miR-506 may function in a tissue type-dependent manner. Nevertheless, whether and how miR-506 is involved in the pathogenesis of pancreatic cancer is yet to be investigated.
In this study, we discovered that miR-506 acts as a novel tumor suppressor in pancreatic cancer. The tumor-suppressive function of miR-506 is mediated in part by the suppression of sphingosine kinase 1 (SPHK1) expression, thereby inhibiting Akt/NF-κB signaling. We found that miR-506 suppressed pancreatic cancer cell growth both in vitro and in vivo. In addition, the ectopic expression of miR-506 caused cell cycle arrest and enhanced pancreatic cancer cell apoptosis and chemosensitivity. We further revealed the clinical relevance of miR-506 in pancreatic cancer, as the expression of miR-506 was significantly reduced in pancreatic cancer due to hypermethylation of its promoter. Reduced miR-506 expression significantly correlated with a poor prognosis of pancreatic cancer patients.
Results
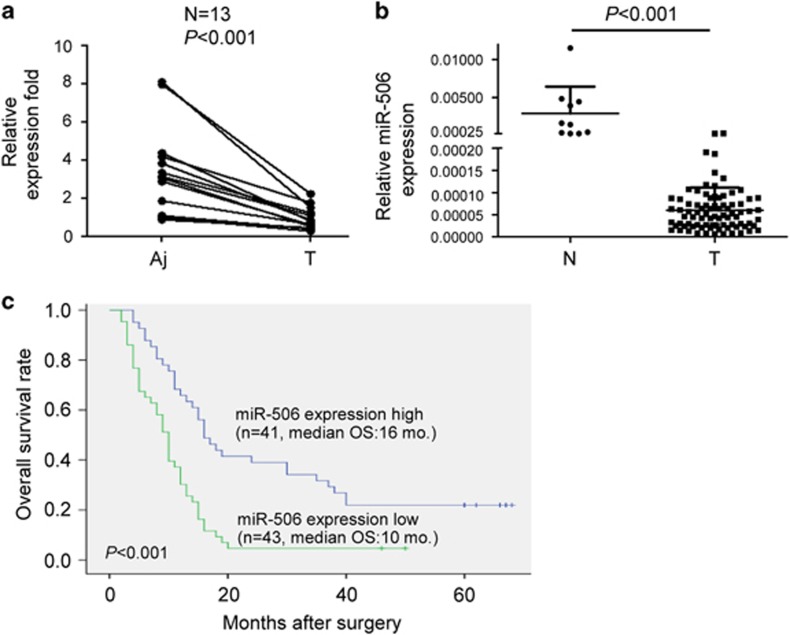
miR-506 is aberrantly downregulated in pancreatic cancer tissues and correlates with disease progression
To investigate the expression level and significance of miR-506 in pancreatic cancer, we first performed quantitative real-time PCR (qRT–PCR) to evaluate miR-506 expression in 13 matched pairs of pancreatic cancer and adjacent non-cancerous tissues. The results showed that miR-506 expression was significantly downregulated in pancreatic cancer tissues compared with adjacent non-cancerous tissues (Figure 1a). Consistently, the expression of miR-506 was clearly reduced in pancreatic cancer tissues compared with normal pancreatic tissues (Figure 1b). Clinicopathological analyses of 84 pancreatic cancer patients showed that a decrease in miR-506 expression was significantly correlated with clinical stage, pathologic tumor status and distant metastasis (Supplementary Table 1). In addition, Kaplan–Meier survival analysis showed that patients with lower miR-506 expression levels had significantly reduced 5-year overall survival rates (Figure 1c). A multivariate Cox regression analysis showed that low miR-506 expression was an independent poor prognostic factor in pancreatic cancer (hazard ratio, 1.880; 95% confidence interval, 1.048–3.026; P=0.033; Supplementary Table 2). These results suggest that the downregulation of miR-506 might promote the progression of pancreatic cancer.
Figure 1.
miR-506 is aberrantly downregulated in pancreatic cancer tissues and correlates with disease progression. (a) Comparison of miR-506 expression in 13 paired pancreatic cancer tissues and adjacent non-cancer tissues via qRT–PCR. U6 was used as an internal control. Aj, adjacent non-cancerous tissue; T, pancreatic cancer. (b) miR-506 expression was determined via qRT–PCR in 10 normal pancreatic tissues and 84 cancer tissues. miR-506 expression levels were calculated by the miR-506/U6 expression ratio (2−ΔΔCt). N: normal pancreas. (c) Kaplan–Meier curves for the survival time of patients with pancreatic cancer divided according to miR-506 expression. The low and high miR-506 expression levels are based on the average value of miR-506 expression.
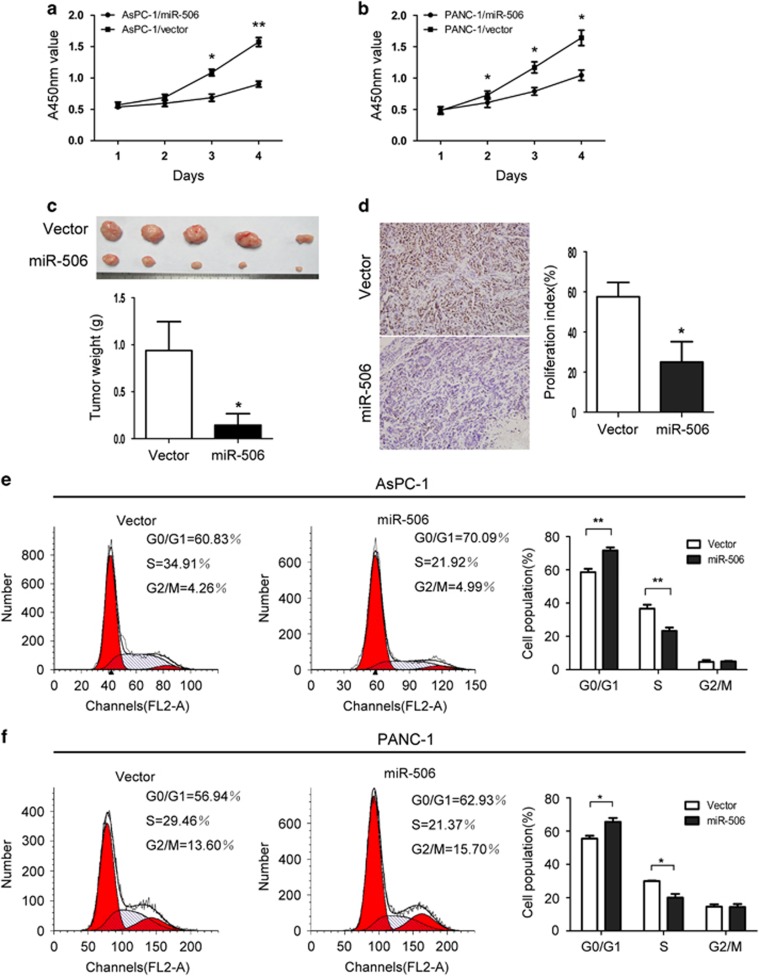
miR-506 inhibits pancreatic cancer cell proliferation in vitro and in vivo and induces cell cycle arrest
To investigate its biological function in pancreatic cancer, miR-506 was ectopically expressed via a lentiviral infection system to establish two stable pancreatic cancer cell lines, AsPC-1 and PANC-1, both of which have low levels of miR-506 (Supplementary Figure 1a). Cell proliferation assays showed that miR-506 overexpression significantly suppressed pancreatic cancer cell proliferation (Figures 2a and b). In contrast, the proliferation rate of the pancreatic cancer cell lines PANC-1 and BxPC-3 that were transiently transfected with miR-506 inhibitors was significantly increased compared with negative control cells (Supplementary Figures 1b, 2a and b). To determine whether miR-506 upregulation inhibits pancreatic cancer cell growth in vivo, AsPC-1 cell lines that were transfected with a control vector or miR-506 were implanted subcutaneously into nude mice. The tumors in the nude mice inoculated with AsPC-1/miR-506 cells grew more slowly than the tumors in the nude mice inoculated with AsPC-1/vector cells (Supplementary Figure 3). After 30 days, the weights of the tumors derived from the AsPC-1/miR-506 cells were significantly lower than the weights of those from the control cells (Figure 2c). Immunohistochemistry analysis revealed lower Ki-67 levels in tumors from the AsPC-1/miR-506 group compared with those from the AsPC-1/vector group (Figure 2d). Collectively, our results indicate that miR-506 suppresses pancreatic cancer cell growth both in vitro and in vivo. Moreover, the functional impact of miR-506 on the cell cycle was analyzed using flow cytometry. Compared with the control, miR-506 significantly decreased the fraction of cells in the S phase and increased the population in the G0/G1 phase in both AsPC-1 and PANC-1 cells (Figures 2e and f). Consistent with this result, miR-506 inhibition noticeably decreased the populations of both PANC-1 and BxPC-3 cells in the G0/G1 phase (Supplementary Figures 2c and d).
Figure 2.
miR-506 inhibits pancreatic cancer cell proliferation in vitro and in vivo and induces cell cycle arrest. (a, b) The effects of miR-506 on pancreatic cancer cell growth were measured via a CCK-8 assay. The results are presented as the means±s.d. of the values obtained in three independent experiments. Significance was calculated using Student's t-test. (c) Representative graph of tumor growth and the mean tumor weights 30 days after inoculation. (d) Representative immunohistochemical staining of Ki-67-stained cells from the indicated tumors. (e, f) On the basis of flow cytometric analysis, miR-506 overexpression induced via stable lentiviral transfection resulted in a significant increase in the cellular population in the G0/G1 phase but a decrease in those in the S phase in both AsPC-1 and PANC-1 cells. *P<0.05; **P<0.01.
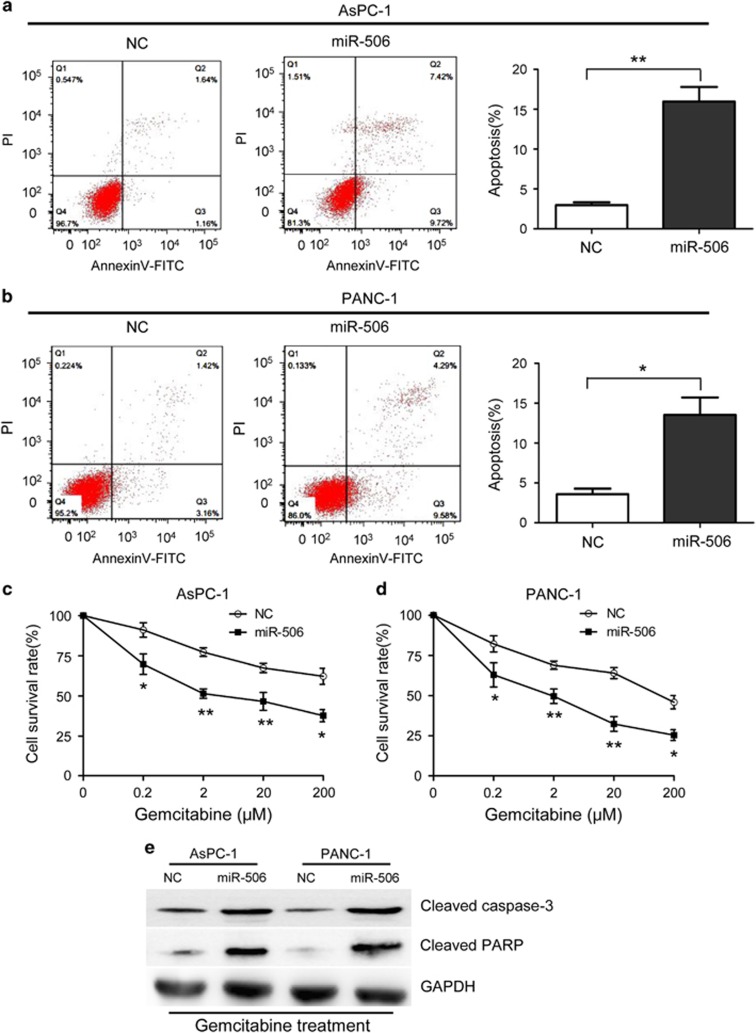
miR-506 induces apoptosis and enhances the chemosensitivity of pancreatic cancer cells
We next measured the effect of miR-506 on apoptosis and chemosensitivity of pancreatic cancer cells. For the apoptosis assay, pancreatic cancer cells were harvested after 48 h of incubation, stained with 50 μg/ml propidium iodide and Annexin V-fluorescein isothiocyanate and subsequently analyzed via flow cytometry. miR-506-mimic transfection resulted in a significant increase in the percentages of cells undergoing apoptosis (Figures 3a and b; Supplementary Figure 1c), whereas miR-506 inhibition noticeably prevented apoptosis of PANC-1 and BxPC-3 cells (Supplementary Figures 2e and f). To study the effects of miR-506 on the chemosensitivity of pancreatic cancer cells, various gemcitabine concentrations were used. AsPC-1 and PANC-1 cells were transfected with miR-506 mimics 24 h before gemcitabine treatment, and the antiproliferative effects of gemcitabine were measured 48 h after treatment using the CCK-8 assay. The results revealed that the miR-506 mimics noticeably enhanced the chemosensitivity of the cells to gemcitabine (Figures 3c and d), whereas miR-506 inhibitors significantly conferred chemoresistance to PANC-1 and BxPC-3 cells (Supplementary Figures 2g and h). Moreover, the noted effect of miR-506 on apoptosis was confirmed, as increased activating cleavage of PARP and caspase-3 was induced by gemcitabine in the pancreatic tumor cells that overexpressed miR-506 (Figure 3e).
Figure 3.
miR-506 induces apoptosis and enhances chemosensitivity in pancreatic cancer cells. (a, b) Induction of apoptosis in AsPC-1 and PANC-1 cells transfected with miR-506 mimics or negative control. Compared with the negative control, miR-506 overexpression significantly increased the apoptosis rate in both cell types. (c, d) miR-506 enhanced the chemosensitivity of AsPC-1 and PANC-1 cells to gemcitabine. (e) Western blotting for cleavage of caspase-3 and PARP in NC- or miR-506-transduced AsPC-1 and PANC-1 cells after gemcitabine (20 μM) for 48 h using glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as a loading control. *P<0.05; **P<0.01.
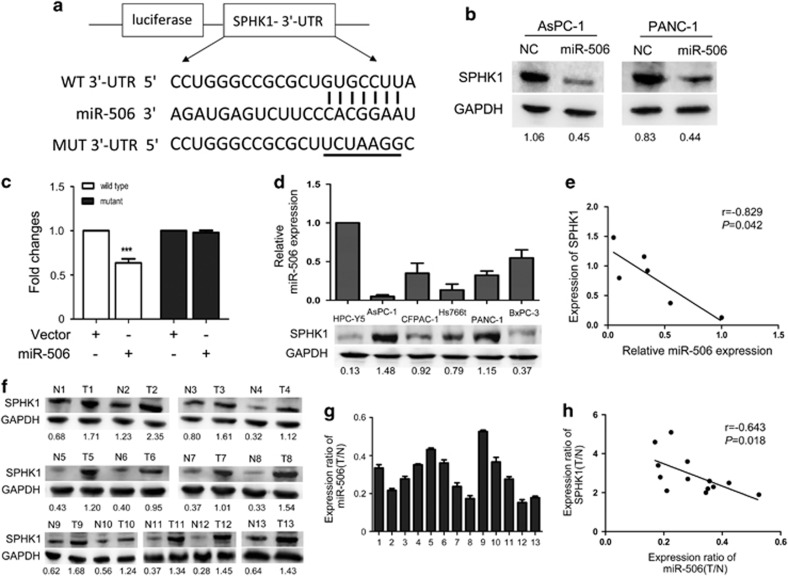
miR-506 targets SPHK1 in pancreatic cancer
On the basis of miRNA target analysis algorithms (miRanda and TargetScan), SPHK1 is a potential target mRNA of miR-506 (Figure 4a). We chose SPHK1 as a validation target, as it has been shown to be involved in various relevant biological processes, such as cell proliferation, apoptosis and chemoresistance.16 Western blot analysis revealed that miR-506 overexpression significantly reduced the expression of SPHK1 in AsPC-1 and PANC-1 pancreatic cancer cells (Figure 4b), whereas miR-506 inhibitors noticeably increased the expression of SPHK1 in PANC-1 and BxPC-3 cells (Supplementary Figure 4a). However, qRT–PCR analysis revealed that the mRNA expression of SPHK1 did not notably change (Supplementary Figure 4b), suggesting that the suppressive effect of miR-506 on SPHK1 was mainly via translational suppression. Using a dual-luciferase reporter system, we showed that the coexpression of miR-506 significantly inhibited the firefly luciferase reporter activity of the wild-type SPHK1 3′-UTR but not the mutant 3′-UTR (Figure 4c), indicating that SPHK1 is a direct target of miR-506.
Figure 4.
miR-506 targets SPHK1. (a) The predicted miR-506-binding sequence in the 3′-UTR of SPHK1 mRNA is shown. Mutations were generated in the SPHK1 3′-UTR sequence at the complementary sites for the seed regions in miR-506. (b) Western blot analysis of SPHK1 expression in AsPC-1 and PANC-1 cells transfected with NC or miR-506 mimic after 48 h; GAPDH was used as a loading control. (c) Analysis of luciferase activity. 293 T cells were transfected with pGL3-wild-SPHK1 3′-UTR or pGL3-mutant-SPHK1 3′-UTR using a vector control or miR-506. The Renilla luciferase vector was used as an internal control. *P<0.05. (d) miR-506 and SPHK1 expression in pancreatic cancer cell lines. Top: qRT–PCR analysis of miR-506 levels; bottom: western blot analysis of SPHK1 protein levels. (e) An inverse relationship between miR-506 and SPHK1 protein levels was shown in the pancreatic cancer cell lines and in the immortalized pancreatic cell line HPC-Y5 based on Spearman's correlation. (f) Western blot analysis of SPHK1 in 13 pairs of pancreatic cancer tissues. (g) The related expression levels of miR-506 in the 13 pairs of tissue are indicated. (h) An inverse relationship between miR-506 and SPHK1 protein was demonstrated in pancreatic cancer tissues based on Spearman's correlation.
To further confirm that SPHK1 is a direct target of miR-506, we detected the endogenous expression levels of miR-506 and SPHK1 in five pancreatic cancer cell lines and an immortalized human pancreatic ductal epithelial cell line to investigate the relationship between the expression levels of miR-506 and SPHK1. qRT–PCR and western blot analysis showed a significant inverse correlation between the levels of miR-506 and SPHK1 proteins (Figures 4d and e). We also examined the expression levels of miR-506 and SPHK1 in 13 pairs of matched pancreatic cancer tissues and adjacent non-cancerous tissues. Among the 13 pairs of pancreatic cancer tissues, we observed a significant inverse correlation between the expression of miR-506 and SPHK1 (Figures 4f–h). Moreover, we subsequently examined SPHK1 expression in subcutaneous xenotransplanted tumors of nude mice via immunohistochemistry. Lower SPHK1 levels were observed in tumors from the AsPC-1/miR-506 cells compared with those from the AsPC-1/vector group (Supplementary Figures 5a and b). These results indicate that miR-506 downregulation by promoter methylation may be the primary mechanism of SPHK1 upregulation in pancreatic cancer.
Increased SPHK1 expression is associated with a poor prognosis in pancreatic cancer
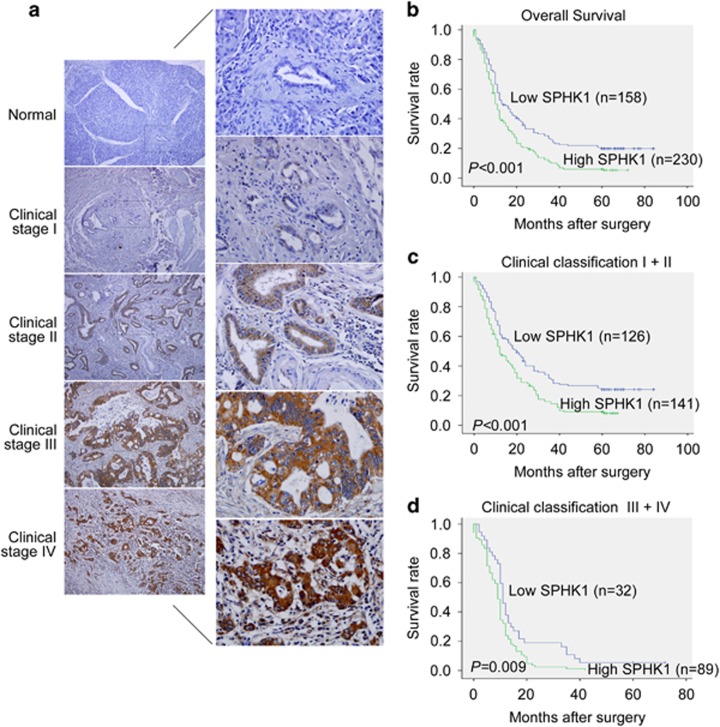
The protein expression status of SPHK1 was further investigated in an additional 388 archival formalin-fixed paraffin-embedded pancreatic cancer tissues and 20 normal pancreatic tissues using immunohistochemistry. SPHK1 was markedly upregulated in pancreatic cancer tissues but was nearly undetectable in normal pancreatic tissues (Figure 5a). Statistical analyses revealed that SPHK1 expression was significantly associated with the clinical stage, the pathologic tumor status, lymph node status and distant metastasis of patients with pancreatic cancer (Supplementary Table 3). A Kaplan–Meier survival analysis revealed that patients with high SPHK1 expression had significantly lower 5-year overall survival rates (P<0.001; Figure 5b). Notably, SPHK1 expression also significantly correlated with overall survival in both the early clinical subgroups (stages I and II) and the advanced-disease group (stages III and IV; Figures 5c and d), indicating that SPHK1 could be a valuable prognostic marker for pancreatic cancer patients at all disease stages. In addition, a multivariate Cox regression analysis showed that high expression of SPHK1 was an independent poor prognostic factor in pancreatic cancer (hazard ratio, 1.395; 95% confidence interval, 1.107–1.758; P=0.005; Supplementary Table 4).
Figure 5.
SPHK1 expression in pancreatic cancer and normal pancreatic tissues via immunohistochemistry. (a) IHC staining indicated that SPHK1 expression is upregulated in human pancreatic cancer tissues (clinical stages I–IV) compared with normal pancreatic tissues (left panel: magnification × 100; right panel: magnification × 400). (b) Kaplan–Meier curves of pancreatic cancer patients with low versus high expression levels of SPHK1 (n=388; P<0.001). (c, d) Significance of the difference between the curves of SPHK1 high-expressing and low-expressing patients was compared in clinical stages I−II (c) and clinical stages III−IV (d) patient subgroups. P-values were calculated based on the log-rank test.
SPHK1 silencing recapitulates the effects of miR-506 in pancreatic cancer cells
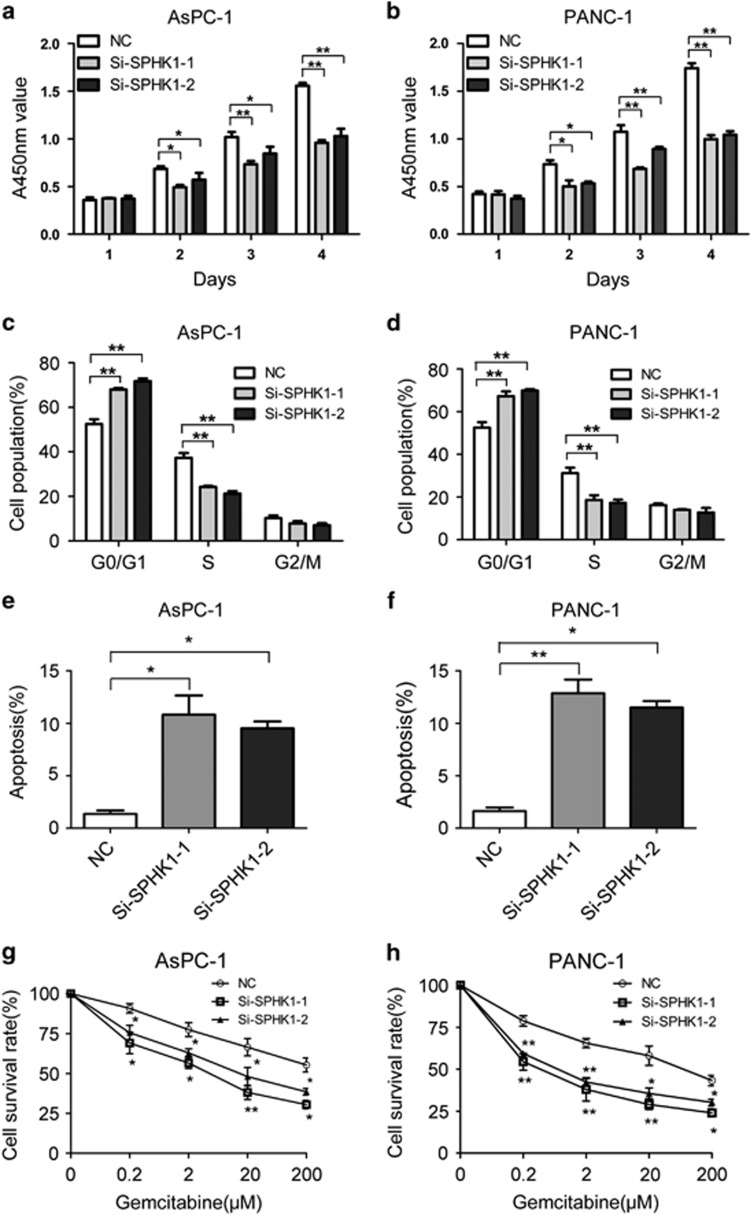
To determine the functional impacts of SPHK1 on pancreatic cancer cells, two specific small interfering RNAs against SPHK1 mRNA, si-SPHK1-1 and si-SPHK1-2, significantly reduced the expression of SPHK1 protein (Supplementary Figure 6a) and clearly inhibited the proliferation of both AsPC-1 and PANC-1 cells (Figures 6a and b). Moreover, we conducted a knockdown approach using a lentiviral infection system to establish AsPC-1 and PANC-1 transfectants that stably express SPHK1 short hairpin RNA (Supplementary Figure 6b). Compared with control cells, SPHK1-RNAi inhibited pancreatic cancer cell proliferation both in vitro (Supplementary Figure 6c) and in vivo (Supplementary Figures 6d and e). Both si-SPHK1-1 and si-SPHK1-2 induced G1−S arrest (Figures 6c and d) and apoptosis (Figures 6e and f) of AsPC-1 and PANC-1 cells. In addition, the chemosensitivity of AsPC-1 and PANC-1 cells to gemcitabine was significantly increased because of si-SPHK1 treatment (Figures 6g and h). These results suggest that SPHK1 is one of the key functional mediators for miR-506 in pancreatic cancer cells. The suppression of proliferation, the blockade of cell cycle progression, the enhancement of apoptosis and the chemosensitivity observed due to the silencing of SPHK1 were similar to the phenotype induced by the restored expression of miR-506 in pancreatic cancer cells. These results suggest that miR-506-mediated downregulation of SPHK1 is one of the major mechanisms that inhibit pancreatic cancer growth and progression.
Figure 6.
SPHK1 silencing recapitulates the effects of miR-506 in pancreatic cancer cells. (a, b) The proliferation of AsPC-1 and PANC-1 cells was significantly inhibited by transient transfection with SPHK1 small interfering RNAs (siRNAs). Cell growth was determined by the Cell Counting Kit-8 assay. (c, d) Silencing of SPHK1 induced G1 arrest of AsPC-1 and PANC-1 cells. (e, f) Apoptosis of AsPC-1 and PANC-1 cells was noticeably accelerated following transient transfection with SPHK1 siRNA. (g, h) Cells were treated for 48 h with gemcitabine (0–200 μM), and chemosensitivity was measured via cell viability. The chemosensitivities of the AsPC-1 and PANC-1 cells to gemcitabine were significantly increased subsequent to transient transfection with SPHK1 siRNA, compared with NC. *P<0.05; **P<0.01.
Re-introduction of SPHK1 abrogates the miR-506-induced effects on cell proliferation and apoptosis
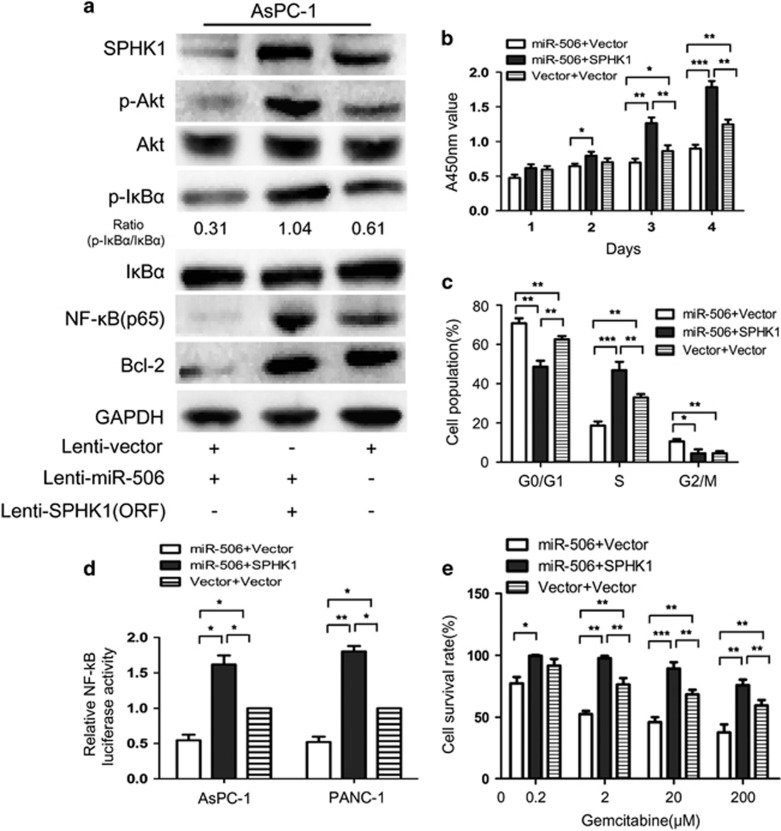
To further confirm that the functional impact of miR-506 in pancreatic cancer is mediated by the suppression of SPHK1, we re-introduced SPHK1 expression in miR-506-expressing cells to observe the direct antagonizing effect of miR-506. We constructed a lentiviral expression vector of SPHK1 without the 3′-UTR and infected miR-506-expressing cells. As shown in Figure 7a, SPHK1 expression was restored after SPHK1 lentiviral infection. Functional studies showed that the re-introduction of SPHK1 enhanced the proliferation of miR-506-expressing cells (Figure 7b). In addition, the inhibitory effect of miR-506 on the G1/S phase transition of the cell cycle was also rescued by enforced SPHK1 expression (Figure 7c). Moreover, the enforced SPHK1 expression significantly counteracted the chemosensitivity induced by miR-506 (Figure 7e). Similar results were obtained in PANC-1 cells (data not shown). Collectively, these results show that SPHK1 re-introduction could abrogate miR-506-induced cell growth suppression and apoptosis, suggesting that SPHK1 mediates the tumor-suppressive function of miR-506 in pancreatic cancer.
Figure 7.
Re-introduction of SPHK1 abrogates the miR-506-induced effects on cell proliferation and apoptosis. AsPC-1 cells stably expressing miR-506 or the vector were infected with lenti-SPHK1 or the corresponding vector. The following experiments were conducted with the above cells: (a) western blot analysis of SPHK1, (p)Akt, (p)IKBα, NF-κB(p65) and Bcl-2; (b) cell proliferation analyses; (c) fluorescence-activated cell sorting assays for cell cycle distribution; (d) NF-κB reporter assays; and (e) cell viability after gemcitabine treatment. *P<0.05, **P<0.01, ***P<0.001.
miR-506 induces pancreatic cancer cell growth suppression and apoptosis via the SPHK1/Akt/NF-κB signaling pathway
SPHK1 is associated with the promotion of Akt/NF-κB-dependent cell proliferation and apoptosis.17 Therefore, we next examined whether the Akt/NF-κB pathway is involved in miR-506-SPHK1-mediated cell growth and apoptosis in pancreatic cancer. Interestingly, western blot analysis showed that miR-506 overexpression decreased Akt/NF-κB signaling, whereas the ectopic expression of SPHK1 blocked the miR-506-mediated inactivation of the Akt/NF-κB pathway (Figure 7a). Moreover, an NF-κB reporter assay showed that SPHK1 upregulation could abrogate miR-506-induced suppression of NF-κB transcriptional transactivating activity in both AsPC-1 and PANC-1 cells (Figure 7d).
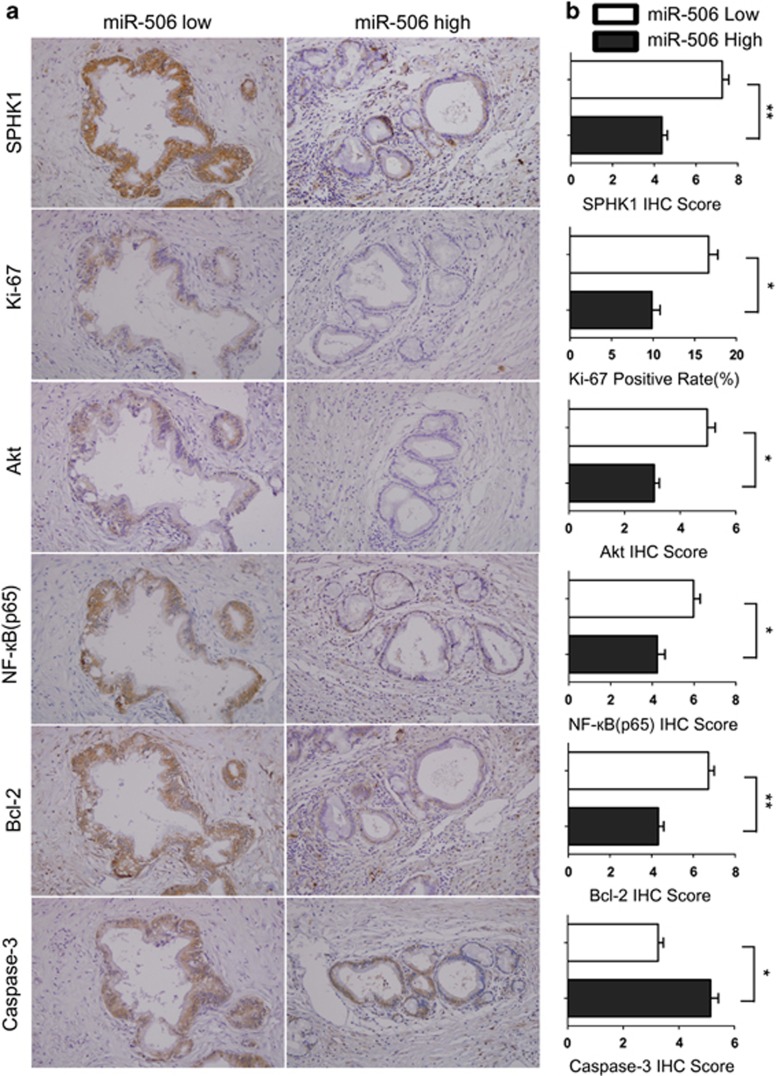
To further determine whether the Akt/NF-κB pathway has a role in miR-506-SPHK1-mediated tumor progression, we performed immunohistochemistry in human pancreatic cancer tissues for SPHK1, Akt, NF-κB(p65), Bcl-2, caspase-3 and Ki-67 and analyzed their correlation with miR-506 expression (determined via qRT−PCR). As shown in Figure 8, miR-506 expression inversely correlated with SPHK1, Akt, NF-κB(p65) and Bcl-2 protein expression in the 84 cases of pancreatic cancer (P<0.05 for all), which was consistent with the in vitro results. Moreover, high miR-506 expression was significantly associated with low Ki-67 and high caspase-3 expression (P<0.05). Altogether, our results from the in vitro and human tumor samples showed that miR-506 suppressed the SPHK1/Akt/NF-κB signaling pathway and consequently reduced cell proliferation and promoted apoptosis in pancreatic cancer.
Figure 8.
miR-506 expression is inversely associated with the SPHK1/Akt/NF-κB pathway in human pancreatic cancer. (a) Representative images (× 200) of IHC staining for SPHK1, Ki-67, Akt, NF-κB(p65), Bcl-2 and caspase-3 in low- or high-miR-506 expression groups are shown. (b) Bar charts show the association between miR-506 expression and the expression levels of SPHK1, Ki-67, Akt, NF-κB(p65), Bcl-2 and caspase-3. The x axes represent the relative expression levels of SPHK1, Ki-67, Akt, NF-κB(p65), Bcl-2 and caspase-3, as indicated by IHC. The results are presented as the means±s.e.m. *P<0.05; **P<0.01.
Microarray analysis identified additional potential target genes and pathways of miR-506, further elucidating the mechanism behind its tumor-suppressive effect
It should be noted that any given miRNA has the capacity to exert its function on dozens, if not hundreds, of target genes,18 and we cannot exclude the possibility that other target genes are involved in the tumor-suppressive role of miR-506. To identify additional miR-506 target genes and the mechanisms behind its tumor-suppressive effect, we performed global microarray analysis of mRNA expression in the miR-506-overexpressing AsPC-1 pancreatic cancer cell line and a negative control. The array results are listed on the website http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE76280, and the gene expression omnibus accession number is GSE76280. We identified that miR-506 could potentially enhance the expression of 144 genes and downregulate the expression of 78 genes, respectively (Supplementary Figure 7a). Collectively, miR-506 could potentially target ~10 pathways including the tumor-related PI3K-Akt pathway that was intensively investigated in this study (Supplementary Figure 7b). In addition, a Venn diagram demonstrated the overlap between the genes downregulated by the miR-506 and the common miR-506 target genes in three Databases (miRanda, miRDB and Targetscan; Supplementary Figure 7c). The use of three different bioinformatics predicted that target algorithm will increase the probability of true miR-506-mediated targets. These results provide a better understanding of the global impact of miR-506-mediated targets and pathways in pancreatic cancer, in addition to the key target SPHK1 and the major PI3K-Akt pathway.
Hypermethylation mediates miR-506 silencing in pancreatic cancer
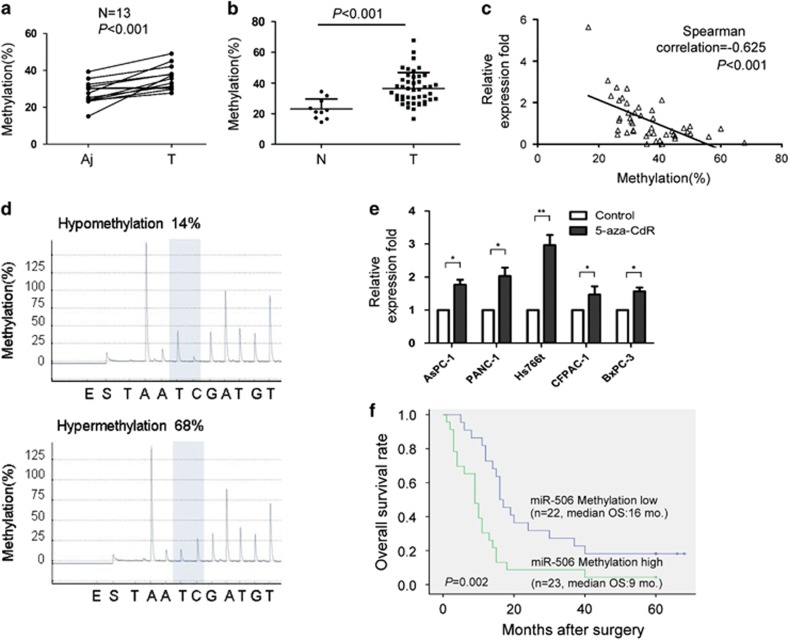
Because miR-506 has a key role in pancreatic cancer progression and chemoresistance, we further investigated the mechanism responsible for its downregulation in cancer tissues. We identified five CpG sites in the promoter region of the miR-506 gene in pancreatic cancer tissues (Supplementary Figure 8a). Sequencing analysis revealed that, among the five methylation sites of the miR-506 promoter, the fifth site displayed hypermethylation in pancreatic cancer tissues compared with adjacent non-cancerous tissues (Figure 9a), whereas there were no differences in methylation of the other four sites between cancer tissues and adjacent non-cancerous tissues (Supplementary Figures 8b−e). The hypermethylation of the miR-506 promoter in pancreatic cancer tissues was confirmed using an additional 45 cancer tissues and 10 normal pancreatic tissues (Figure 9b). The impact of hypermethylation of the miR-506 promoter on miR-506 expression was quantified via qRT−PCR using total RNA isolated from paired normal and pancreatic cancer tissues. A high level of promoter methylation in cancer tissues was significantly associated with reduced miR-506 expression compared with adjacent non-cancerous tissues (Figure 1a). Furthermore, a significant inverse correlation between miR-506 promoter methylation and miR-506 expression was observed (Figures 9c and d). In addition, 5-aza-2'-deoxycitidine (5-aza-CdR), a demethylating agent, markedly increased miRNA-506 expression, indicating that DNA methylation was responsible for miR-506 gene silencing (Figure 9e). We also evaluated the association between DNA methylation and patient survival time in 45 pancreatic cancer patients. We found that miR-506 hypermethylation was associated with a shorter survival time (Figure 9f). Taken together, our results suggest that hypermethylation at least partially mediates miR-506 silencing in pancreatic cancer.
Figure 9.
Hypermethylation mediates miR-506 silencing in pancreatic cancer. (a) The DNA methylation levels of site five were detected in 13 paired pancreatic cancer tissues and adjacent non-cancer tissues by pyrosequencing. The differences in DNA methylation levels between cancer tissues and adjacent non-cancer tissues were calculated using Student's t-test. Aj: adjacent non-cancerous tissue; T: pancreatic cancer. (b) The DNA methylation levels of site five were detected in 10 normal pancreatic tissues and 45 cancer tissues by pyrosequencing. N: normal pancreas. (c) The association between miR-506 expression levels and the DNA methylation status of miR-506 was evaluated using a Spearman's correlation analysis. (d) Representative output of bisulfite pyrosequencing. (e) Pancreatic cancer cell lines were treated with 5-aza-CdR (3 μM) for 72 h, and miR-506 expression levels were determined via qRT−PCR and normalized to U6. The results are presented as the means±s.d. of the values obtained in three independent experiments. *P<0.05; **P<0.01. (f) The impact of miR-506 DNA methylation status on overall survival in pancreatic cancer patients. The low and high levels of miR-506 methylation are separated according to the median value. The P-values were determined using a log-rank test.
Discussion
Dysregulation of miRNAs, which can result from aberrant DNA methylation, has been demonstrated to contribute to pancreatic cancer tumorigenesis.19, 20 In this study, we found that miR-506 expression was reduced in pancreatic cancer, mediated by hypermethylation of the promoter region of the miR-506 gene and significantly associated with poor prognosis. miR-506 inhibited pancreatic cancer cell growth in vitro and in vivo and enhanced apoptosis and chemosensitivity through downregulating SPHK1. Silencing of SPHK1 recapitulated the effects of miR-506 overexpression, whereas enforced expression of SPHK1 reversed the suppressive effects of miR-506. This functional impact of miR-506 was mediated directly through SPHK1 to inhibit Akt/NF-κB signaling. These findings support the important roles of miR-506 in suppressing the tumorigenesis of pancreatic cancer.
Altered expression of miRNA is strongly implicated in cancer, and recent studies have shown that certain miRNA genes may be silenced in human tumors by aberrant CpG island hypermethylation.21, 22 Aberrant hypermethylation has also been reported to regulate the expression of specific miRNAs in pancreatic cancer, including miR-124, miR-132 and miR-615-5p.19, 20, 23 Moreover, in a recent report, Yang et al. demonstrated that promoter DNA methylation attenuated miR-506 expression in human ovarian cancer. However, the methylation status of miR-506 in pancreatic cancer has remained unclear. In this study, we first performed pyrosequencing to evaluate the detailed methylation patterns of five CpG islands in the promoter region of miR-506 and found that the miR-506 promoter was more highly methylated in pancreatic cancer tissues compared with adjacent non-cancerous tissues and normal controls. In addition, we showed that promoter hypermethylation resulted in miR-506 silencing in pancreatic cancer and that miR-506 hypermethylation was associated with reduced survival time. Our study suggests that miR-506 may be involved in pancreatic cancer pathogenesis and progression; furthermore, it may serve as a prognostic biomarker of pancreatic cancer.
The reported expression patterns and roles of miR-506 markedly differ among various malignancies. In ovarian cancer, miR-506 was identified as a key epithelial-to-mesenchymal transition inhibitor that inhibited cell migration and invasion; thus, the clinically downregulated miR-506 expression in tumor tissues was significantly correlated with poor prognosis.13 Similar results have been demonstrated in cervical cancer,24 breast cancer25 and gastric cancer,26 indicating that miR-506 also acts as a tumor suppressor in these cancer types. However, miR-506 is upregulated and functions as an oncogene to promote proliferation in melanomas15 and confers chemoresistance in colon cancer.27 Moreover, miR-506 has been shown to be associated with diverse biological behaviors via the regulation of different target genes. Recent studies have shown that miR-506 prevents TGFβ-induced epithelial-to-mesenchymal transition by targeting SNAI213 and can also suppress the expression of vimentin and N-cadherin in ovarian cancer.28 In addition, miR-506 has been involved in antiproliferative functions through the regulation of its target genes, CDK629 and Gli3.24 These various observations for the roles of miR-506 may not be surprising because the function of certain miRNAs and their target preference may be highly dependent on the cellular and disease context. Although these mechanisms mediated by miR-506 may also be involved in pancreatic cancer, we provide strong evidence that a novel miR-506 epigenetic silencing alteration and targeting SPHK1 promote pancreatic cancer cell proliferation in vitro and in vivo. The restoration of miR-506 induces cell cycle arrest at the G1/S transition and enhances apoptosis and the chemosensitivity of pancreatic cancer cells. Therefore, reduced miR-506 expression promotes the tumorigenesis of pancreatic cancer.
SPHK1, the rate-limiting enzyme of sphingosine 1 phosphate synthesis, has been demonstrated to have a crucial role in oncogenesis.30 Accumulating evidence suggests that SPHK1 expression correlates with various cellular functions in tumors, such as proliferation, apoptosis, chemoresistance and angiogenesis.31, 32, 33 In addition, SPHK1 activation correlates with the progression of several types of cancer.17, 34, 35 Moreover, SPHK1 downregulation enhances chemosensitivity to gemcitabine via the targeting of sphingolipid metabolism.36 We reveal for the first time the key roles of SPHK1 in pancreatic cancer. In this study, we demonstrated that SPHK1 was overexpressed and correlated with poor prognosis in a large cohort of pancreatic cancer specimens. In addition, we revealed that miR-506 bound to the 3′-UTR of SPHK1, resulting in a dramatic reduction in SPHK1 protein levels. On the basis of the observations that SPHK1 silencing recapitulated the effects of miR-506 overexpression, and enforced expression of SPHK1 reversed the suppressive effects of miR-506, our results indicate that the functional roles of miR-506 are mediated by its direct targeting of SPHK1 in pancreatic cancer. It has been reported that SPHK1 is associated with Akt/NF-κB-dependent cell proliferation and apoptosis.17 Consistent with this observation, we found that miR-506 upregulation significantly inhibited levels of the activated forms of Akt and IKBα. Moreover, we performed immunohistochemistry on serial sections of human pancreatic cancer tissues with antibodies specific for SPHK1, p-Akt, NF-κB and Bcl-2 and showed that the expression of these proteins inversely correlated with miR-506 expression. Thus, our current study suggests that activation of the Akt/NF-κB signaling pathway by SPHK1 may have a pivotal role in miR-506-mediated tumor suppression and enhanced chemosensitivity in pancreatic cancer.
It has been recently demonstrated that both miRNA mimics and anti-miRs are potential novel anticancer therapeutics. Modified anti-miRs, such as peptide nucleic acids, cholesterol-conjugated ‘antagomirs' and anti-miRNA oligonucleotides, have shown effectiveness in vivo.37, 38, 39 Notably, MRX34 is entering a Phase 1 trial as the first miRNA mimic for cancer therapy.40 In this study, our primary data demonstrated that the miR-506 mimics effectively suppress pancreatic cancer cell growth in vitro, revealing its potential therapeutic significance in pancreatic cancer based on an miRNA ‘replacement' strategy. Moreover, miRNAs may be promising candidates as biomarkers for early cancer diagnosis and prognosis and for the modification of responses to anticancer treatment.41, 42, 43, 44 Because the hypermethylation and downregulation of miR-506 were associated with poor prognoses, miR-506 has a strong potential for use as a biomarker for improved management of pancreatic cancer.
In conclusion, we demonstrated that promoter hypermethylation mediated the silencing of miR-506 in pancreatic cancer. Transcriptional repression of miR-506 results in the upregulation of the key target SPHK1, which has an important role in the progression of pancreatic cancer by regulating SPHK1/Akt/NF-κB signaling. The newly identified miR-506/SPHK1 axis provides crucial mechanistic insights and provides a foundation for miR-506-based therapeutics and biomarkers in pancreatic cancer.
Materials and methods
Cell culture
The human pancreatic cancer cell lines AsPC-1, PANC-1, BxPC-3, Hs766t and CFPAC-1 (ATCC, Manassas, VA, USA) and an immortalized human pancreatic ductal epithelial cell line (HPC-Y5, Chinese Academy of Sciences) were cultured in complete growth medium, as recommended by the manufacturer. Cultured cells were maintained in a humidified 5% CO2 atmosphere at 37 °C.
Tissue samples
Eighty-four fresh-frozen pancreatic cancer samples (13 with adjacent non-cancerous tissues) and 20 normal pancreatic tissues were obtained from the Institute of Hepatopancreatobiliary Surgery, Southwest Hospital, Third Military Medical University. The normal pancreatic samples were obtained from organ donors. Another 388 formalin-fixed paraffin-embedded tissues were acquired from the archival collections of the Southwest Hospital, Third Military Medical University (262), Union Hospital (57) and Tongji Hospital (32), Tongji Medical College, Huazhong University of Science and Technology, the First Affiliated Hospital of Anhui Medical University (37). None of the patients had received radiotherapy or chemotherapy before surgery. The use of human tissues was approved by the ethics committee of Southwest Hospital.
DNA isolation, sodium bisulfite conversion and pyrosequencing analysis
Genomic DNA was isolated using the TIANamp Genomic DNA Kit (TIANGEN Biotech, Beijing, China). Sodium bisulfite modification of the DNA was performed using an EZ DNA Methylation-Gold Kit (ZYMO, Beijing, China) according to the manufacturer's protocol. The five CpG sites of the miR-506 promoter region were amplified via PCR using the bisulfite-modified DNA template, forward primers and biotinylated reverse primers according to the established procedures.13 The primer sequences are listed in Supplementary Table 5. The PCR products were then analyzed via pyrosequencing technology using a PyroMark Gold Q96 Reagent and a PyroMark ID system (Qiagen, Hilton, Germany). Pyro Q-CpG software v. 1.0.9 was used to determine the optimal order of nucleotide addition when designing the assays. The pyrosequencing analysis was performed at Sangon Biotech Co., Ltd. (Shanghai, China).
Quantitative real-time PCR
Total RNA from the tumor cells or tissues was extracted using RNAiso (TaKaRa, Dalian, China). For miR-506 detection, complementary DNA synthesis was performed using a PrimeScript RT reagent kit (TaKaRa), and qRT–PCR was performed with SYBR Premix Ex Taq II (TaKaRa) using a Stratagene Mx3000P real-time PCR system (Agilent Technologies, Santa Clara, CA, USA). The expression levels were normalized to the endogenous small nuclear RNA U6 control. For the mRNA analyses, PrimeScript RT Master Mix (TaKaRa) and SYBR Premix Ex Taq II (TaKaRa) were used. β-actin was used as an endogenous control. The 2−ΔΔCt method was used to calculate expression relative to the endogenous control. The primer sequences are listed in Supplementary Table 5.
Western blotting
Protein extracts were separated by electrophoresis in a sodium dodecyl sulfate-polyacrylamide gel (Invitrogen, Camarillo, CA, USA) and transferred onto polyvinylidene fluoride membranes (Millipore, Billerica, MA, USA) for immunoblotting. The membranes were hybridized with a primary antibody at 4 °C overnight followed by incubation with a secondary antibody for 1 h at room temperature. Glyceraldehyde-3-phosphate dehydrogenase was used as the loading control. Signals were visualized using a chemiluminescent detection system (Thermo Scientific, Rockford, IL, USA) and exposure to the film. The antibodies used for western blotting are described in Supplementary Table 6.
Immunohistochemistry
Immunohistochemistry was performed as previously described.45 Briefly, after blocking, the sections were incubated with primary antibodies overnight, followed by incubation with secondary antibodies and further incubation with the streptavidin–biotin complex (Maixin, Fuzhou, China). The antibodies used for immunohistochemistry are described in Supplementary Table 6. Ki-67-positive cells were defined as those with brown staining in the nucleus, and the expression was evaluated based on the percentage of positive tumor cells out of 1000 tumor cells. SPHK1-, Akt-, NF-κB(p65)-, BCL-2- and caspase-3-positive cells were defined as those with immunoreactivity in both the cytoplasm and the nucleus, and were quantified using a composite score obtained by multiplying the values of staining intensities (0, no staining; 1, weak staining; 2, moderate staining; 3, strong staining) and the percentage of positive cells (0, 0% 1, <10% 2, 10–50% 3, >50%). For statistical analyses, the tumor sample cohort was grouped into those with low expression (⩽4) and high expression (⩾6).
Transfection and vector construction
miRNA mimics and inhibitors (Qiagen) and small interfering RNAs (sequences listed in Supplementary Table 5) were transiently transfected using Lipofectamine 2000 (Invitrogen) at a final concentration of 100 nM, according to the manufacturer's instructions. Lentiviral miR-506 expression constructs and the negative control lentiviruses were purchased from GeneChem (Shanghai, China). The SPHK1 expression lentivirus was constructed by inserting the SPHK1 open reading frame into the pLenti6.3 vector (Invitrogen). For SPHK1 depletion, human SPHK1-targeting small interfering RNA sequences (GGCTGAAATCTCCTTCACG) were cloned into pENTR vectors to generate pENTR-SPHK1-RNAi lentiviruses (Invitrogen). Lentiviral infection was performed as previously described.46
Luciferase reporter assay
The wild-type or mutant 3′-UTR sequences of SPHK1 were ligated into the XbaI and FseI sites of the pGL3 vector (GeneChem). HEK293T cells infected with the miR-506 lentivirus or the negative control lentivirus were seeded into 96-well plates. The cells were co-transfected with 50 ng of the pGL3 vector and 10 ng of the pRL-TK vector using Lipofectamine LTX (Invitrogen). Twenty-four hours after transfection, the cells were harvested according to the manufacturer's protocol (Promega, Madison, WI, USA), and firefly and Renilla luciferase activities were measured using a Dual Luciferase Reporter System (Promega) with a Victor X machine (Perkin-Elmer, Boston, MA, USA). For the NF-κB transcriptional activity assay, the pGL4.32[luc2P/NF-κB-RE/Hygro] plasmid or control luciferase plasmid pGL4.74[hRluc/TK] (Promega) was transfected using the Lipofectamine 2000 reagent (Invitrogen), and luciferase and Renilla signals were measured using a Dual Luciferase Reporter Assay kit (Promega) according to the manufacturer's protocol.
Cell proliferation and chemosensitivity assays
Cell proliferation was measured via WST-8 staining using a Cell Counting Kit-8 (Dojinodo, Shanghai, China) according to the manufacturer's instructions. For the chemosensitivity assays, the cells were treated for 48 h with gemcitabine (Gemzar, Lilly, France), and cell viability was measured using a Cell Counting Kit-8.
Cell cycle and apoptosis assays
For the cell cycle analysis, the transfected cells were collected and fixed in 70% ethanol overnight at −20 °C and stained with propidium iodide (Kaiji, Nanjing, China) in a phosphate-buffered saline solution containing RNase. For the apoptosis assay, the cells were stained with 50 μg/ml propidium iodide and Annexin V-fluorescein isothiocyanate (Kaiji) following the manufacturer's instructions. The data were analyzed using the ModFit 3.3 software (BD Bioscience, Sparks, MD, USA).
Animal experiments
Five-week-old female BALB/c-nu mice were purchased from the Peking University Animal Center (Beijing, China). After 5 days of acclimatization, a total of 2 × 106 AsPC-1 cells stably transfected with either miR-506(SPHK1-RNAi) or negative control were injected subcutaneously into the right oxter of each mouse. Tumor volume was calculated using the equation (L × W2)/2, and the tumor weights were measured and recorded in grams. The mice were killed on the 30th day after injection. All animal studies were approved by the Institutional Animal Care and Use Committee of the Third Military Medical University.
Complementary DNA microarray analysis
Total RNA was isolated from three independent cultures of AsPC-1/miR-506 cells or AsPC-1/vector cells. RNA quantity and purity were determined using the Nanodrop ND-1000 (NanoDrop Technologies, Wilmington, DE, USA) and denaturing gel electrophoresis. Microarray analysis of mRNA profiles using Agilent Array platforms and the identification of differentially expressed genes were performed by KangChen Bio-tech (Shanghai, China) as previously described.47
Statistical analysis
All data were analyzed using the SPSS 17.0 statistical software (version 17.0, Chicago, IL, USA). The data are presented as the means±s.d. of three independent experiments; Student's t-test was used for comparisons. P<0.05 was considered significant.
Acknowledgments
We thank Professor Renyi Qin from the Department of Biliary-Pancreatic Surgery (Affiliated Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan) and Professor Yijun Zhao from the Department of Hepatobiliary Surgery (the First Affiliated Hospital of Anhui Medical University, Hefei) for providing human pancreatic cancer samples. This work was supported by the following grants: the National Natural Science Foundation of China (No. 81372242); the National High Technology Research and Development Program of China (Program 863; No. 2012AA021105); and the Research Special Fund for Public Welfare Industry of Health (No. 201202007).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
Supplementary Material
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin 2015; 65: 5–29. [DOI] [PubMed] [Google Scholar]
- Maitra A, Hruban RH. Pancreatic cancer. Annu Rev Pathol 2008; 3: 157–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Xie K, Wolff R, Abbruzzese JL. Pancreatic cancer. Lancet 2004; 363: 1049–1057. [DOI] [PubMed] [Google Scholar]
- Garrido-Laguna I, Hidalgo M. Pancreatic cancer: from state-of-the-art treatments to promising novel therapies. Nat Rev Clin Oncol 2015; 12: 319–334. [DOI] [PubMed] [Google Scholar]
- Oettle H, Neuhaus P, Hochhaus A, Hartmann JT, Gellert K, Ridwelski K et al. Adjuvant chemotherapy with gemcitabine and long-term outcomes among patients with resected pancreatic cancer: the CONKO-001 randomized trial. JAMA 2013; 310: 1473–1481. [DOI] [PubMed] [Google Scholar]
- Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, Eachus R et al. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell 2005; 122: 553–563. [DOI] [PubMed] [Google Scholar]
- Chen CZ. MicroRNAs as oncogenes and tumor suppressors. N Engl J Med 2005; 353: 1768–1771. [DOI] [PubMed] [Google Scholar]
- Jamieson NB, Morran DC, Morton JP, Ali A, Dickson EJ, Carter CR et al. MicroRNA molecular profiles associated with diagnosis, clinicopathologic criteria, and overall survival in patients with resectable pancreatic ductal adenocarcinoma. Clin Cancer Res 2012; 18: 534–545. [DOI] [PubMed] [Google Scholar]
- Frampton AE, Krell J, Jamieson NB, Gall TM, Giovannetti E, Funel N et al. MicroRNAs with prognostic significance in pancreatic ductal adenocarcinoma: a meta-analysis. Eur J Cancer 2015; 51: 1389–1404. [DOI] [PubMed] [Google Scholar]
- Frampton AE, Castellano L, Colombo T, Giovannetti E, Krell J, Jacob J et al. MicroRNAs cooperatively inhibit a network of tumor suppressor genes to promote pancreatic tumor growth and progression. Gastroenterology 2014; 146: 268–277 e218. [DOI] [PubMed] [Google Scholar]
- Guo S, Xu X, Tang Y, Zhang C, Li J, Ouyang Y et al. miR-15a inhibits cell proliferation and epithelial to mesenchymal transition in pancreatic ductal adenocarcinoma by down-regulating Bmi-1 expression. Cancer Lett 2014; 344: 40–46. [DOI] [PubMed] [Google Scholar]
- Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet 2005; 37: 766–770. [DOI] [PubMed] [Google Scholar]
- Yang D, Sun Y, Hu L, Zheng H, Ji P, Pecot CV et al. Integrated analyses identify a master microRNA regulatory network for the mesenchymal subtype in serous ovarian cancer. Cancer Cell 2013; 23: 186–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin M, Ren X, Zhang X, Luo Y, Wang G, Huang K et al. Selective killing of lung cancer cells by miRNA-506 molecule through inhibiting NF-(kappa)B p65 to evoke reactive oxygen species generation and p53 activation. Oncogene 2014; 34: 691–703. [DOI] [PubMed] [Google Scholar]
- Streicher KL, Zhu W, Lehmann KP, Georgantas RW, Morehouse CA, Brohawn P et al. A novel oncogenic role for the miRNA-506-514 cluster in initiating melanocyte transformation and promoting melanoma growth. Oncogene 2012; 31: 1558–1570. [DOI] [PubMed] [Google Scholar]
- Shida D, Takabe K, Kapitonov D, Milstien S, Spiegel S. Targeting SphK1 as a new strategy against cancer. Curr Drug Targets 2008; 9: 662–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Xiong H, Li J, Liao W, Wang L, Wu J et al. Sphingosine kinase-1 enhances resistance to apoptosis through activation of PI3K/Akt/NF-kappaB pathway in human non-small cell lung cancer. Clin Cancer Res 2011; 17: 1839–1849. [DOI] [PubMed] [Google Scholar]
- Bracken CP, Khew-Goodall Y, Goodall GJ. Network-based approaches to understand the roles of miR-200 and other microRNAs in cancer. Cancer Res 2015; 75: 2594–2599. [DOI] [PubMed] [Google Scholar]
- Wang P, Chen L, Zhang J, Chen H, Fan J, Wang K et al. Methylation-mediated silencing of the miR-124 genes facilitates pancreatic cancer progression and metastasis by targeting Rac1. Oncogene 2014; 33: 514–524. [DOI] [PubMed] [Google Scholar]
- Gao W, Gu Y, Li Z, Cai H, Peng Q, Tu M et al. miR-615-5p is epigenetically inactivated and functions as a tumor suppressor in pancreatic ductal adenocarcinoma. Oncogene 2015; 34: 1629–1640. [DOI] [PubMed] [Google Scholar]
- Bueno MJ, Perez de Castro I, Gomez de Cedron M, Santos J, Calin GA, Cigudosa JC et al. Genetic and epigenetic silencing of microRNA-203 enhances ABL1 and BCR-ABL1 oncogene expression. Cancer Cell 2008; 13: 496–506. [DOI] [PubMed] [Google Scholar]
- Kong KL, Kwong DL, Chan TH, Law SY, Chen L, Li Y et al. MicroRNA-375 inhibits tumour growth and metastasis in oesophageal squamous cell carcinoma through repressing insulin-like growth factor 1 receptor. Gut 2012; 61: 33–42. [DOI] [PubMed] [Google Scholar]
- Zhang S, Hao J, Xie F, Hu X, Liu C, Tong J et al. Downregulation of miR-132 by promoter methylation contributes to pancreatic cancer development. Carcinogenesis 2011; 32: 1183–1189. [DOI] [PubMed] [Google Scholar]
- Wen SY, Lin Y, Yu YQ, Cao SJ, Zhang R, Yang XM et al. miR-506 acts as a tumor suppressor by directly targeting the hedgehog pathway transcription factor Gli3 in human cervical cancer. Oncogene 2014; 34: 717–725. [DOI] [PubMed] [Google Scholar]
- Arora H, Qureshi R, Park WY. miR-506 regulates epithelial mesenchymal transition in breast cancer cell lines. PLoS One 2013; 8: e64273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakimura S, Sugimachi K, Kurashige J, Ueda M, Hirata H, Nambara S et al. The miR-506-induced epithelial-mesenchymal transition is involved in poor prognosis for patients with gastric cancer. Ann Surg Oncol 2015; 22: 1436–1443. [DOI] [PubMed] [Google Scholar]
- Tong JL, Zhang CP, Nie F, Xu XT, Zhu MM, Xiao SD et al. MicroRNA 506 regulates expression of PPAR alpha in hydroxycamptothecin-resistant human colon cancer cells. FEBS Lett 2011; 585: 3560–3568. [DOI] [PubMed] [Google Scholar]
- Sun Y, Hu L, Zheng H, Bagnoli M, Guo Y, Rupaimoole R et al. MiR-506 inhibits multiple targets in the epithelial-to-mesenchymal transition network and is associated with good prognosis in epithelial ovarian cancer. J Pathol 2015; 235: 25–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Sun Y, Ji P, Li X, Cogdell D, Yang D et al. MiR-506 suppresses proliferation and induces senescence by directly targeting the CDK4/6-FOXM1 axis in ovarian cancer. J Pathol 2014; 233: 308–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamori T, Kaneshiro T, Okumura M, Maalouf S, Uflacker A, Bielawski J et al. Role for sphingosine kinase 1 in colon carcinogenesis. FASEB J 2009; 23: 405–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitonov D, Allegood JC, Mitchell C, Hait NC, Almenara JA, Adams JK et al. Targeting sphingosine kinase 1 inhibits Akt signaling, induces apoptosis, and suppresses growth of human glioblastoma cells and xenografts. Cancer Res 2009; 69: 6915–6923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyne NJ, Pyne S. Sphingosine 1-phosphate and cancer. Nat Rev Cancer 2010; 10: 489–503. [DOI] [PubMed] [Google Scholar]
- Nagahashi M, Ramachandran S, Kim EY, Allegood JC, Rashid OM, Yamada A et al. Sphingosine-1-phosphate produced by sphingosine kinase 1 promotes breast cancer progression by stimulating angiogenesis and lymphangiogenesis. Cancer Res 2012; 72: 726–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Guan HY, Gong LY, Song LB, Zhang N, Wu J et al. Clinical significance of sphingosine kinase-1 expression in human astrocytomas progression and overall patient survival. Clin Cancer Res 2008; 14: 6996–7003. [DOI] [PubMed] [Google Scholar]
- Li W, Yu CP, Xia JT, Zhang L, Weng GX, Zheng HQ et al. Sphingosine kinase 1 is associated with gastric cancer progression and poor survival of patients. Clin Cancer Res 2009; 15: 1393–1399. [DOI] [PubMed] [Google Scholar]
- Guillermet-Guibert J, Davenne L, Pchejetski D, Saint-Laurent N, Brizuela L, Guilbeau-Frugier C et al. Targeting the sphingolipid metabolism to defeat pancreatic cancer cell resistance to the chemotherapeutic gemcitabine drug. Mol Cancer Ther 2009; 8: 809–820. [DOI] [PubMed] [Google Scholar]
- Kasinski AL, Slack FJ. Epigenetics and genetics. MicroRNAs en route to the clinic: progress in validating and targeting microRNAs for cancer therapy. Nat Rev Cancer 2011; 11: 849–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babar IA, Cheng CJ, Booth CJ, Liang X, Weidhaas JB, Saltzman WM et al. Nanoparticle-based therapy in an in vivo microRNA-155 (miR-155)-dependent mouse model of lymphoma. Proc Natl Acad Sci USA 2012; 109: E1695–E1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennox KA, Behlke MA. Chemical modification and design of anti-miRNA oligonucleotides. Gene Ther 2011; 18: 1111–1120. [DOI] [PubMed] [Google Scholar]
- Bouchie A. First microRNA mimic enters clinic. Nat Biotechnol 2013; 31: 577. [DOI] [PubMed] [Google Scholar]
- Xi Y, Nakajima G, Gavin E, Morris CG, Kudo K, Hayashi K et al. Systematic analysis of microRNA expression of RNA extracted from fresh frozen and formalin-fixed paraffin-embedded samples. RNA 2007; 13: 1668–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai NS, Wu DG, Fang XG, Lin YC, Chen SS, Li ZB et al. Serum microRNA-210 as a potential noninvasive biomarker for the diagnosis and prognosis of glioma. Br J Cancer 2015; 112: 1241–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Kong W, He L, Zhao JJ, O'Donnell JD, Wang J et al. MicroRNA expression profiling in human ovarian cancer: miR-214 induces cell survival and cisplatin resistance by targeting PTEN. Cancer Res 2008; 68: 425–433. [DOI] [PubMed] [Google Scholar]
- Drayton RM, Dudziec E, Peter S, Bertz S, Hartmann A, Bryant HE et al. Reduced expression of miRNA-27a modulates cisplatin resistance in bladder cancer by targeting the cystine/glutamate exchanger SLC7A11. Clin Cancer Res 2014; 20: 1990–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Xu X, Guo S, Zhang C, Tang Y, Tian Y et al. An increased abundance of tumor-infiltrating regulatory T cells is correlated with the progression and prognosis of pancreatic ductal adenocarcinoma. PLoS One 2014; 9: e91551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Hilaire C, Ziegler SG, Markello TC, Brusco A, Groden C, Gill F et al. NT5E mutations and arterial calcifications. N Engl J Med 2011; 364: 432–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su J, You P, Li WL, Tao XR, Zhu HY, Yao YC et al. The existence of multipotent stem cells with epithelial-mesenchymal transition features in the human liver bud. Int J Biochem Cell Biol 2010; 42: 2047–2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.