Abstract
Streptomyces sp. TP-A0867 (=NBRC 109436) produces structurally complex polyketides designated alchivemycins A and B. Here, we report the draft genome sequence of this strain together with features of the organism and assembly, annotation, and analysis of the genome sequence. The 9.9 Mb genome of Streptomyces sp. TP-A0867 encodes 8,385 putative ORFs, of which 7,232 were assigned with COG categories. We successfully identified a hybrid polyketide synthase (PKS)/ nonribosomal peptide synthetase (NRPS) gene cluster that could be responsible for alchivemycin biosynthesis, and propose the biosynthetic pathway. The alchivemycin biosynthetic gene cluster is also present in Streptomyces rapamycinicus NRRL 5491T, Streptomyces hygroscopicus subsp. hygroscopicus NBRC 16556, and Streptomyces ascomycinicus NBRC 13981T, which are taxonomically highly close to strain TP-A0867. This study shows a representative example that distribution of secondary metabolite genes is correlated with evolution within the genus Streptomyces.
Keywords: Alchivemycin, Biosynthetic gene cluster, Genome mining, Polyketide synthase, Streptomyces, Taxonomy
Introduction
Actinomycetes are known for their ability of producing a variety of secondary metabolites with useful pharmacological potency such as antimicrobial, antitumor, and immunosuppressive activities. In particular, the genus Streptomyces is one of the most prolific sources of chemically diverse small molecules [1]. Terrestrial surface soil is the well-known habitat of this genus, but, since Streptomyces have been extensively searched for several decades, discovery of strains producing novel compounds becomes difficult from easily accessible soil samples. Therefore, untapped sources such as plants have recently attracted attention to obtain new strains for new secondary metabolites [2, 3]. In our continuing search for structurally rare metabolites from Streptomyces, alchivemycins A and B, which have potent antimicrobial activity and inhibitory effects on tumor cell invasion, were discovered from a plant-derived Streptomyces strain TP-A0867. These compounds are novel polycyclic polyketides with an unprecedented carbon backbone [4, 5], however the biosynthetic gene cluster has not been known to date. In this study, we performed whole genome shotgun sequencing of the strain TP-A0867 to elucidate the biosynthetic pathway of alchivemycins. We herein present the draft genome sequence of Streptomyces sp. TP-A0867, together with the taxonomical identification of the strain, description of its genome properties, and annotation for secondary metabolite genes. The putative alchivemycin biosynthetic gene cluster and the plausible biosynthetic pathway are also described.
Organism information
Classification and features
In the course of screening for new bioactive compounds produced by plant-derived actinomycetes, Streptomyces sp. TP-A0867 was isolated from a leaf of a Chinese chive (Allium tuberosum) collected in Toyama, Japan [2] and two new polyketides, alchivemycins A and B, were found from its culture broth [4, 5]. The characteristics of Streptomyces sp. TP-A0867 were examined by the same manner of our previous report [6]. This strain grew well on ISP 2, ISP 4, and ISP 6 agar media, but poorly on ISP 5 and ISP 7. Colors of aerial mycelia were determined using the Japanese Industrial Standard Common Color Names (JIS Z 8102: 2001). The color of aerial mycelia was light gray and that of the reverse side was pale yellow on ISP 2 agar medium. No diffusible pigment was observed on ISP 2, ISP 3, ISP 4, ISP 5, ISP 6, and ISP 7 agar media. A scanning electron micrograph of this strain (Fig. 1) shows that spore chains were spiral and contained 2–3 helixes and 5–8 spores per chain; spores were cylindrical and 0.9 × 1.8 μm in size, and had a rugose ornamentation. Motile cells were not observed in hanging drops under a light microscope. Growth occurred at 15–45 °C (optimum 40 °C) on ISP 2 agar medium. Strain TP-A0867 exhibited growth with 0–5 % (w/v) NaCl (optimum 0 % NaCl) at 28 °C on ISP 2 agar medium and pH 4–10 (optimum pH 7) at 28 °C in ISP 2 liquid medium. Carbohydrate utilization was determined on Pridham-Gottlieb carbon utilization (ISP 9) agar medium supplemented with 1 % (w/v) of carbon sources sterilized by filtration. Strain TP-A0867 utilized fructose, glucose, rhamnose, sucrose, and xylose for growth. These results are summarized in Table 1. The genes encoding 16S rRNA were amplified by PCR using two universal primers, 9 F (5′-GAGTTTGATCCTGGCTCAG-3′) and 1541R (5′-AAGGAGGTGATCCAGCC-3′) [7]. KOD FX (Toyobo Co., Ltd., Tokyo, Japan) was used as described by the manufacturer for the PCR. The reaction was started with denaturation at 94 °C for 1 min followed by a total 30 cycles that consisted of denaturation at 98 °C for 10 s, annealing at 55.5 °C for 30 s, and extension at 68 °C for 1.5 min. The amplicon size was 1.5 kb. After purification of the PCR product by AMPure (Beckman Coulter), sequencing was carried out according to an established method [7]. The sequence was deposited into DDBJ under the accession number LC150789. BLAST search of the sequence by EzTaxon-e [8] indicated the highest similarities to those of Streptomyces hygroscopicus subsp. hygroscopicus NRRL 2387 T (AB231803, 100 %, 1456/1456), Streptomyces endus NRRL 2339 T (AY999911, 100 %, 1456/1456), and Streptomyces sporocinereus NBRC 100766 T (AB249933, 100 %, 1456/1456). A phylogenetic tree was reconstructed on the basis of the 16S rRNA gene sequence together with Streptomyces type strains showing over 98.5 % similarities and S. hygroscopicus subsp. hygroscopicus NBRC 16556 using ClustalX2 [9] and NJPlot [10] as shown in Fig. 2. The phylogenetic analysis confirmed that the strain TP-A0867 belongs to the genus Streptomyces.
Fig. 1.

Scanning electron micrograph of Streptomyces sp. TP-A 0867 grown on double-diluted ISP 2 agar for 7 days at 28 °C. Bar, 2 μm
Table 1.
Classification and general features of Streptomyces sp. TP-A0867 [12]
| MIGS ID | Property | Term | Evidence codea |
|---|---|---|---|
| Classification | Domain Bacteria | TAS [34] | |
| Phylum Actinobacteria | TAS [35] | ||
| Class Actinobacteria | TAS [36] | ||
| Order Actinomycetales | TAS [36–39] | ||
| Suborder Streptomycineae | TAS [36, 39] | ||
| Family Streptomycetaceae | TAS [36, 38–41] | ||
| Genus Streptomyces | TAS [38, 41–43] | ||
| Species Streptomyces hygroscopicus | IDA | ||
| Subspecies Streptomyces hygroscopicus subsp. hygroscopicus | IDA | ||
| Strain TP-A0867 | [4] | ||
| Gram stain | Not tested, likely positive | NAS | |
| Cell shape | Branched mycelia | IDA | |
| Motility | Not observed | IDA | |
| Sporulation | Sporulating | IDA | |
| Temperature range | Grows from 15 °C to 45 °C | IDA | |
| Optimum temperature | 40 °C | IDA | |
| pH range; Optimum | 4 to 10; 7 | IDA | |
| Carbon source | Fructose, glucose, rhamnose, sucrose, xylose | IDA | |
| MIGS-6 | Habitat | Chinese chive (Allium tuberosum) | TAS [2, 4] |
| MIGS-6.3 | Salinity | Grows from 0 % to 7 % NaCl | IDA |
| MIGS-22 | Oxygen requirement | Aerobic | IDA |
| MIGS-15 | Biotic relationship | Free-living | IDA |
| MIGS-14 | Pathogenicity | Not reported | |
| MIGS-4 | Geographic location | Toyama, Japan | TAS [2] |
| MIGS-5 | Sample collection | from April to June in 1998 | TAS [2] |
| MIGS-4.1 | Latitude | Not reported | |
| MIGS-4.2 | Longitude | Not reported | |
| MIGS-4.4 | Altitude | Not reported |
aEvidence codes - IDA Inferred from Direct Assay, TAS Traceable Author Statement (i.e., a direct report exists in the literature), NAS Non-traceable Author Statement (i.e., not directly observed for the living, isolated sample, but based on a generally accepted property for the species, or anecdotal evidence). These evidence codes are from the Gene Ontology project [44]
Fig. 2.
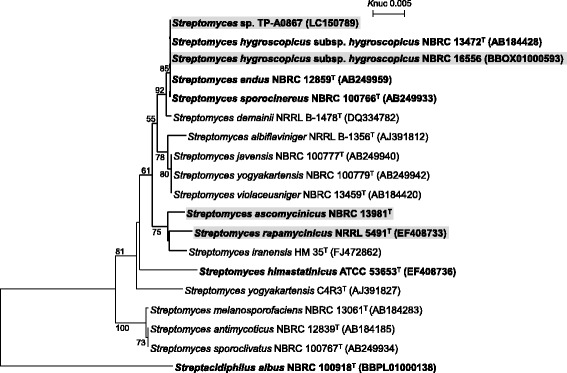
Phylogenetic tree of Streptomyces strains based on 16S rRNA gene sequences. The 16S rRNA sequences were obtained from GenBank, whose accession numbers are shown in parentheses, whereas that of Streptomyces ascomycinicus NBRC 13981T was downloaded from ‘Sequence Information’ of the NBRC Culture Catalog Search (www.nbrc.nite.go.jp/NBRC2/SequencSearchServlet?ID=NBRC&CAT=00013981&DNA=2). The tree was constructed by the neighbor-joining method [45] using sequences aligned by ClustalX2 [9]. All positions containing gaps were eliminated. The building of the tree also involves a bootstrapping process repeated 1,000 times to generate a majority consensus tree, and only bootstrap values above 50 % are shown at branching points. Streptacidiphilus albus NBRC 100918T was used as an outgroup. Strains whose genome were sequenced are boldfaced. Among the genome-sequenced strains, those harboring the putative alchivemycin biosynthetic gene cluster are shadowed in gray
Chemotaxonomic data
Biomass for chemotaxonomic studies was obtained by cultivating strain TP-A0867 in shake flasks of ISP 2 broth for 2 days at 28 °C at 100 r.p.m. The isomer of diaminopimelic acid in the whole-cell hydrolysate was analyzed according to the method described by Hasegawa et al. [11]. Isoprenoid quinones and cellular fatty acids were analyzed as described previously [7]. The whole-cell hydrolysate of strain TP-A0867 contained ll-diaminopimelic acid as its diagnostic peptidoglycan diamino acid. The predominant menaquinones were identified as MK-9(H2) (33 %), MK-9(H4) (40 %) and MK-9(H6) (23 %). The major cellular fatty acids were found to be C16:0 (27 %), anteiso-C15:0 (18 %) and iso-C15:0 (12 %).
Genome sequencing information
Genome project history
In collaboration between Toyama Prefectural University and NBRC, the organism was selected for genome sequencing to elucidate the alchivemycin biosynthetic pathway. We successfully accomplished the genome project of Streptomyces sp. TP-A0867 as reported in this paper. The draft genome sequences have been deposited in the INSDC database under the accession numbers BBON01000001 to BBON01000259. The project information and its association with MIGS version 2.0 compliance are summarized in Table 2 [12].
Table 2.
Project information
| MIGS ID | Property | Term |
|---|---|---|
| MIGS 31 | Finishing quality | Improved-high-quality draft |
| MIGS-28 | Libraries used | Illumina paired-end library |
| MIGS 29 | Sequencing platforms | Illumina MiSeq |
| MIGS 31.2 | Fold coverage | 98 x |
| MIGS 30 | Assemblers | Newbler v2.6, GenoFinisher, Sequencher v5.1 |
| MIGS 32 | Gene calling method | Prodigal |
| Locus tag | TPA0867 | |
| GenBank ID | BBON00000000 | |
| GenBank date of release | March 24, 2016 | |
| GOLD ID | Not registered | |
| BioProject | PRJDB3206 | |
| MIGS 13 | Source material identifier | NBRC 109436 |
| Project relevance | Industrial |
Growth conditions and genomic DNA preparation
Streptomyces sp. TP-A0867 was deposited in the NBRC culture collection with the registration number of NBRC 109436. Its monoisolate was grown on polycarbonate membrane filter (Advantec) on double diluted NBRC 227 agar medium (0.2 % yeast extract, 0.5 % malt extract, 0.2 % glucose, 2 % agar, pH 7.3) at 28 °C. High quality genomic DNA for sequencing was extracted and isolated from the mycelia with an EZ1 DNA Tissue Kit and a Bio Robot EZ1 (Qiagen) according to the manufacturer’s protocol for extraction of nucleic acid from Gram-positive bacteria. The size, purity, and double-strand DNA concentration of the genomic DNA were measured by agarose gel electrophoresis, ratio of absorbance values at 260 nm and 280 nm, and Quant-iT PicoGreen dsDNA Assay Kit (Life Technologies) to assess the quality. Two hundreds fifty ng of the genomic DNA were used for the preparations of Illumina paired-end library.
Genome sequencing and assembly
A paired-end library with 500 bp insert was constructed and 130 bp from each end was sequenced using MiSeq (Illumina K.K., Tokyo, Japan) according to manufacturer’s protocols (Table 2). The 799 Mb paired-end sequences were assembled into 259 scaffolds larger than 500 bp using Newbler v2.6 (Roche Applied Science, Branford, CT, USA) with the default parameters. Subsequently, each sequence gap in scaffolds was checked and re-assembled using sequence reads belonging to gap extremes by GenoFinisher [13]. Branching contigs, one connected to multiple other contigs, were also examined and misassembled linkages were corrected. The sequences of the alchivemycin biosynthetic gene cluster were further checked manually by Sequencher v.5.1 (Gene Codes Corporation, Ann Arbor, MI, USA)
Genome annotation
Coding sequences were predicted with Prodigal [14] and tRNA-scanSE [15]. The gene functions were assigned using an in-house genome annotation pipeline, and searched for domains related to polyketide synthase (PKS) and nonribosomal peptide synthetase (NRPS) using the SMART and PFAM domain databases [16, 17]. PKS and NRPS gene clusters and their domain organizations were determined as reported previously [18]. Similarity search results against the NCBI non-redundant database were also used for predicting function of genes in the biosynthetic gene clusters.
Genome properties
The total size of the genome is 9,889,163 bp and the GC content is 71.9 % (Table 3), similar to other genome-sequenced Streptomyces members. Of the total 8,453 genes, 8,385 are protein-coding genes and 68 are RNA genes. The classification of genes into COGs functional categories is shown in Table 4. As for the synthesis of secondary metabolites such as polyketides and nonribosomal peptides, this genome encodes at least five type I PKS gene clusters, one type II PKS gene cluster, four NRPS gene clusters, and two hybrid PKS/NRPS gene clusters. This suggests the potential to produce diverse polyketide- and nonribosomal peptide-compounds as the secondary metabolites. Two type I PKS gene clusters are putatively identified for syntheses of nigericin and geldanamycin, respectively, and one hybrid PKS/NRPS gene cluster could be responsible for alchivemycin synthesis as stated below. The others are orphan gene clusters at present.
Table 3.
Genome statistics
| Attribute | Value | % of Total |
|---|---|---|
| Genome size (bp) | 9,889,163 | 100.0 |
| DNA coding (bp) | 8,515,958 | 85.2 |
| DNA G + C (bp) | 7,107,274 | 71.8 |
| DNA scaffolds | 259 | - |
| Total genes | 8,453 | 100.0 |
| Protein coding genes | 8,385 | 99.2 |
| RNA genes | 68 | 0.8 |
| Pseudogenes | - | - |
| Genes in internal clusters | 3,697 | 44.1 |
| Genes with function prediction | 5,588 | 66.1 |
| Genes assigned to COGs | 7,232 | 86.2 |
| Genes with Pfam domains | 6,077 | 71.9 |
| Genes with signal peptides | 625 | 7.4 |
| Genes with transmembrane helices | 1,629 | 19.3 |
| CRISPR repeats | 3 | - |
Table 4.
Number of genes associated with general COG functional categories
| Code | Value | % age | Description |
|---|---|---|---|
| J | 289 | 3.4 | Translation, ribosomal structure and biogenesis |
| A | 4 | 0.04 | RNA processing and modification |
| K | 1,036 | 12.3 | Transcription |
| L | 345 | 4.1 | Replication, recombination and repair |
| B | 3 | 0.03 | Chromatin structure and dynamics |
| D | 52 | 0.6 | Cell cycle control, Cell division, chromosome partitioning |
| V | 133 | 1.58 | Defense mechanisms |
| T | 506 | 6.03 | Signal transduction mechanisms |
| M | 337 | 4.02 | Cell wall/membrane biogenesis |
| N | 43 | 0.51 | Cell motility |
| U | 85 | 1.01 | Intracellular trafficking and secretion |
| O | 211 | 2.52 | Posttranslational modification, protein turnover, chaperones |
| C | 543 | 6.48 | Energy production and conversion |
| G | 751 | 8.96 | Carbohydrate transport and metabolism |
| E | 811 | 9.67 | Amino acid transport and metabolism |
| F | 134 | 1.60 | Nucleotide transport and metabolism |
| H | 286 | 3.41 | Coenzyme transport and metabolism |
| I | 431 | 5.14 | Lipid transport and metabolism |
| P | 462 | 5.51 | Inorganic ion transport and metabolism |
| Q | 529 | 6.31 | Secondary metabolites biosynthesis, transport and catabolism |
| R | 1,383 | 16.5 | General function prediction only |
| S | 471 | 5.62 | Function unknown |
| - | 1,153 | 13.8 | Not in COGs |
The total is based on the total number of protein coding genes in the genome
Insights from the genome sequence
Taxonomic identification of Streptomyces sp. TP-A0867
The 16S rRNA gene sequence of Streptomyces sp. TP-A0867 was identical to those of S. hygroscopicus subsp. hygroscopicus NBRC 13472 T (AB184428), S. hygroscopicus subsp. hygroscopicus NBRC 16556 (BBOX01000593), S. endus NBRC 12859 T (AB249959), and S. sporocinereus NBRC 100766 T (AB249933). To determine the scientific name of the strain TP-A0867, we calculated average nucleotide identity based on BLAST values between strain TP-A0867 and the three type strains using their genome sequences (NBRC 13472, BBOX00000000; NBRC 12859, BBOY00000000; NBRC 100766, BCAN00000000) using JSpecies [19]. The ANIb values between Streptomyces sp. TP-A0867 and the type strains of S. hygroscopicus subsp. hygroscopicus, S. endus, and S. sporocinereus were 97.16 %, 97.10 %, and 98.54 %, respectively. Since these values are above the threshold (95–96 %) corresponding to DNA relatedness value of 70 % recommended as the cut-off point for the assignment of bacterial strains to the same species [19, 20], strain TP-A0867 can be classified into these three taxa. We also analyzed the in silico DNA-DNA hybridization values using these genome sequences with a different and quickly method provided from the DSMZ website [21]. The analysis estimated that the DDH values between Streptomyces sp. TP-A0867 and the three type strains were 76.2 %, 76.2 %, and 87.6 %, respectively, supporting our results clearly. Once this strain was reported to be S. endus [22], however S. endus and S. sporocinereus were reported as the later heterotypic synonyms of S. hygroscopicus subsp. hygroscopicus in 2012 [23], although the taxonomic proposal has not been validated. Therefore, we classified strain TP-A0867 into S. hygroscopicus subsp. hygroscopicus as shown in Table 1.
Proposal of alchivemycin biosynthetic pathway
Our previous study suggested that the carbon backbone of alchivemycins is assembled from five methylmalonyl-CoA, nine malonyl-CoA and one glycine molecules by a hybrid PKS/NRPS pathway [5]. We therefore searched for a hybrid PKS/NRPS gene cluster consisting of fourteen PKS modules and one NRPS module and, indeed, a hybrid PKS/NRPS gene cluster was found in scaffold00155 (Table 5, Fig. 3) that consisted of fourteen PKS modules and one NRPS module (Fig. 4), while no other such gene clusters are present in the genome. Almost all domains in each module conserved active residues and/or signature sequences defined in the previous report [24], but the first ketosynthase (KS) domain in TPA0867_155_00340 had glutamine substituted for the active site cysteine residue, suggesting this domain is KSQ [25, 26] and this module is for loading starter molecule in this assembly line. The acyltransferase (AT) domains of modules 1, 4, 7, 10, and 11 were predicted to load a methylmalonyl-CoA in the elongating polyketide chain, because they have YASHS as signature amino-acid residues specific for methylmalonyl-CoA [27, 28]. In contrast, the remaining nine AT domains were predicted to load a malonyl-CoA since the diagnostic residues HAFHS, specific for malonyl-CoA, were found; although that of module 2 is not HAFHS but RAFHS. These results suggest that the PKS assembly line synthesizes a polyketide chain by sequential incorporation of C2-C3-C2-C2-C3-C2-C2-C3-C2-C2-C3-C3-C2-C2 units, consistent with our previous 13C-labeled precursor feeding experiments [5]. In the PKS assembly line, combination of optional domains such as ketoreductase (KR), dehydratase (DH) and enoylreductase (ER) between AT and acyl carrier protein in each module determines reduction of the ketone group, dehydration of the resulting hydroxyl group and subsequent reduction of the double bond, respectively [29]. PKS modules in the PKS/NRPS gene cluster have three KRs, five DH/KR pairs and four DH/ER/KR trios, corresponding to hydroxyl group, double bond, and saturated carbon, respectively, as the optional domains. We also analyzed signature sequences of KR and ER domains to predict absolute configuration of secondary hydroxyl groups derived from acyl keto groups and methyl branches derived from methylmalonyl-CoA based on the fingerprinting and flowchart reported previously [30, 31]. Based on these experimental and bioinfomatic analyses, a putative linear polyketide precursor of alchivemycin for macrocyclization is shown under module 13 (m13) in Fig. 4, which is in good accordance with the carbon backbone of alchivemycins. Alchivemycin contains an unusual heterocyclic system tetrahydrooxazine ring that derives from glycine-incorporation [5]. A gene encoding NRPS (TPA0867_155_00310) is present upstream the PKS genes (Fig. 3), and the substrate of its adenylation (A) domain was predicted to be glycine by the PKS/NRPS Analysis Web-site (http://nrps.igs.umaryland.edu/nrps/) [32]. This strongly supports the idea that this NRPS is involved in the glycine uptake into the tetrahydrooxazine ring: Kim et al. found that the 13C-labeled glycine was actually incorporated into the heterocyclic part of alchivemycin A [5]. After the tetrahydrooxazine ring formation, modifications such as cyclization, epoxidation, and oxidation may take place as shown in Fig. 4. Three monooxygenases (TPA0867_155_00270, TPA0867_155_00280 and TPA0867_155_00420) and a cytochrome P450 (TPA0867_155_00320) are encoded in this cluster, but it was unable to determine which enzymes catalyze the epoxidation at two positions and oxidation at C-24 only by bioinformatic analyses. On the basis of the above-mentioned bioinfomatic evidences, we propose that this PKS/NRPS gene cluster could be responsible for the synthesis of alchivemycins. Further experiments including gene-disruption to prove this proposal are currently in progress.
Table 5.
ORFs in the putative alchivemycin-biosynthetic gene cluster of Streptomyces sp. TP-A0867
| TPA0867_155_ (locus tag) | Length (aa) | Deduced function | Protein homolog [origin] |
Identity/similarity (%) | Accession number |
|---|---|---|---|---|---|
| 00270 a | 526 | monooxygenase | hypothetical protein M271_21675 [S. rapamycinicus NRRL 5491] |
96/97 | AGP55859 |
| 00280 | 510 | monooxygenase | hypothetical protein M271_21670 [S. rapamycinicus NRRL 5491] |
95/96 | AGP55858 |
| 00290 | 197 | unknown | hypothetical protein M271_21665 [S. rapamycinicus NRRL 5491] |
97/99 | AGP55857 |
| 00300 | 270 | unknown | hypothetical protein M271_21660 [S. rapamycinicus NRRL 5491] |
95/96 | AGP55856 |
| 00310 | 1,117 | NRPS | hypothetical protein M271_21655 [S. rapamycinicus NRRL 5491] |
88/89 | AGP55855 |
| 00320 | 405 | cytochrome P450 | hypothetical protein M271_21650 [S. rapamycinicus NRRL 5491] |
96/97 | AGP55854 |
| 00330a | 293 | oxidoreductase | hypothetical protein M271_21645 [S. rapamycinicus NRRL 5491] |
98/99 | AGP55853 |
| 00340 | 2,516 | PKS | hypothetical protein M271_21640 [S. rapamycinicus NRRL 5491] |
87/90 | AGP55852 |
| 00350 | 578 | PKS | hypothetical protein M271_21640 [S. rapamycinicus NRRL 5491] | 90/92 | AGP55852 |
| 00360 | 2,890 | PKS | type I polyketide synthase AVES 4 [Streptomyces avermitilis MA-4680] |
54/63 | NP_822118 |
| 00370 | 3,731 | PKS | hypothetical protein M271_21625, partial [S. rapamycinicus NRRL 5491] |
89/91 | AGP55849 |
| 00380 | 7,654 | PKS | AmphC [Streptomyces nodosus] | 53/64 | AAK73514 |
| 00390 | 4,354 | PKS | hypothetical protein M271_21600, partial [S. rapamycinicus NRRL 5491] |
87/90 | AGP55844 |
| 00400 | 3,637 | PKS | beta-ketoacyl synthase [S. violaceusniger Tu 4113] |
54/64 | YP_004817601 |
| 00410a | 309 | phytanoyl-CoA dioxygenase | hypothetical protein M271_21580 [S. rapamycinicus NRRL 5491] |
93/96 | AGP55840 |
| 00420 | 608 | monooxygenase | hypothetical protein M271_21575 [S. rapamycinicus NRRL 5491] |
91/93 | AGP55839 |
| 00430 | 426 | transcriptional regulator | helix-turn-helix domain-containing protein [S. violaceusniger Tu 4113] |
93/95 | YP_004817135 |
| 00440 | 199 | unknown | hypothetical protein [S. violaceusniger Tu 4113] |
75/80 | YP_004812903 |
| 00450 | 157 | unknown | hypothetical protein M271_33560 [S. rapamycinicus NRRL 5491] |
56/64 | AGP58126 |
| 00460 | 295 | phosphotransferase | aminoglycoside phosphotransferase [S. violaceusniger Tu 4113] |
71/78 | YP_004817724 |
aencoded in complementary strand. Genes shown in Fig. 4 are bold-faced
Fig. 3.
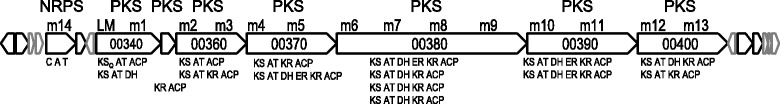
Genetic map of the putative alchivemycin biosynthetic gene cluster (TPA0867_155_00270 to TPA0867_155_00460) of Streptomyces sp. TP-A0867
Fig. 4.
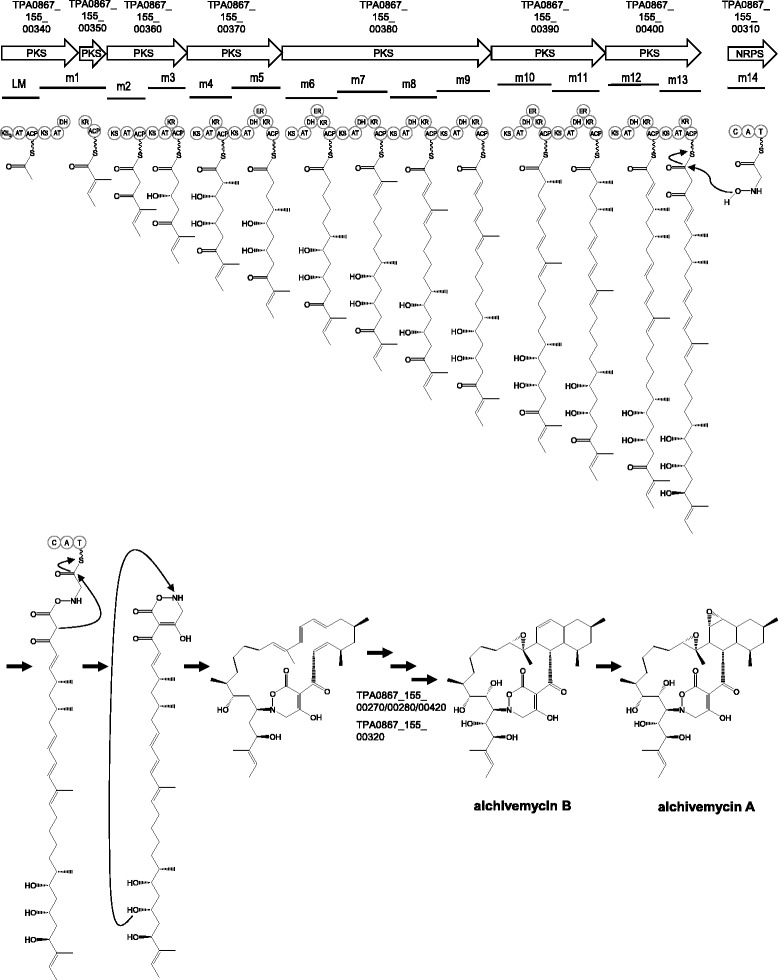
Proposed alchivemycin biosynthetic pathway
Distribution of putative alchivemycin biosynthetic gene clusters in other strains
BLAST search of ORFs in the putative alchivemycin gene cluster within the NCBI database suggested that a similar gene cluster is present in Streptomyces rapamycinicus NRRL 5491 T because this strain has several protein homologues with high sequence homology (Table 5). Analysis of secondary metabolite gene clusters in the genome of strain NRRL 5491 T revealed that a gene cluster from M271_21585 to M271_21655 and the PKS/NRPS domain organizations are identical between Streptomyces sp. TP-A0867 (Fig. 3) and S. rapamycinicus NRRL 5491 T (Fig. 5a), although the genome sequence of the strain NRRL 5491 T is incomplete and its cluster sequence contains several undetermined DNA sequence regions. This finding prompted us to investigate distribution of putative alchivemycin biosynthetic gene clusters in other Streptomyces strains. Further BLAST search of putative alchivemycin-biosynthetic genes indicated that the gene cluster is also present in S. hygroscopicus subsp. hygroscopicus NBRC 16556 (Fig. 5b) and Streptomyces ascomycinicus NBRC 13981 T (Fig. 5c). These strains are phylogenetically close to strain TP-A0867 (Fig. 2, shaded in gray), suggesting that putative alchivemycin-biosynthetic pathway is likely specific in this taxonomical group highlighted by bold lines, although it is unclear whether strains whose genome sequences are unavailable, not boldfaced in the phylogenetic tree, harbor the pathway at present. All the four clusters of Streptomyces sp. TP-A0867, S. rapamycinicus NRRL 5491 T, S. hygroscopicus subsp. hygroscopicus NBRC 16556, and Streptomyces ascomycinicus NBRC 13981 T show conserved synteny, and encode all essential enzymes such as PKSs, an NRPS, and P450/monooxygenases likely for alchivemycin synthesis (Figs. 3 and 5). These results suggest that these strains might also have potential to produce alchivemycins.
Fig. 5.
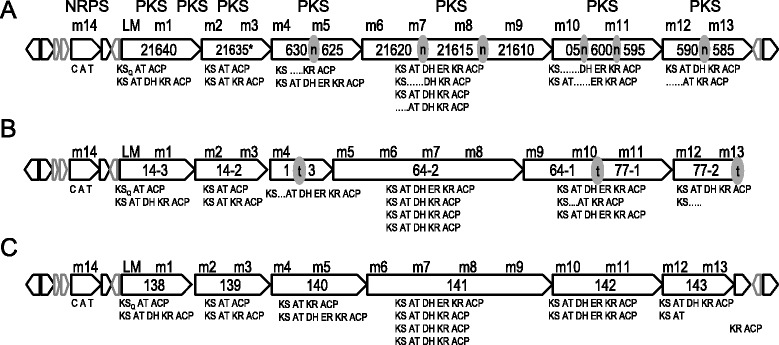
Genetic maps of putative alchivemycin biosynthetic gene clusters of S. rapamycinicus NRRL 5491 (a M271_21675 to M271_21575), S. hygroscopicus subsp. hygroscopicus NBRC 16556 (b orf10 to orf1 in scaffold14, orf3 to orf1 in scaffold64, and orf1 to orf2 in scaffold77), and S. ascomycinicus NBRC 13981T (c orf131 to orf145 of scaffold16). n (in grey circle), these parts contained many undetermined DNA sequences; t (in grey circle), scaffold terminal because b was not obtained as single scaffold. *We manually annotated the ORF, which were longer than registered in GenBank/EMBL/DDBJ
Alchivemycin production by S. ascomycinicusNBRC 13981T
We examined alchivemycin production of S. hygroscopicus subsp. hygroscopicus NBRC 16556 and S. ascomycinicus NBRC 13981 T, both of which are available from the NBRC culture collection. However, the production was not reproducibly observed in some liquid culture conditions tested in this study. Then, we attempted to obtain mutants that can stably produce alchivemycins. S. ascomycinicus NBRC 13981 T was inoculated and cultured on potato dextrose agar (PDA) medium (Merck & Co.) to obtain single colonies, and then the subculture was continuously performed using PDA medium. Within five generations of the subculture, bald mutants were observed. The bald mutants were isolated and maintained on PDA medium to check bald phenotype. Each mutant was cultured using PDA medium for 7 days at 30 °C. The mycelial cells were harvested by steel spatula, and the cells were extracted by equal volume of methanol (MeOH). After centrifugation to remove insoluble materials, the MeOH extracts were analyzed by HPLC coupled with ESI-MS to detect alchivemycins. The alchivemycin production was observed in the MeOH extract of a mutant strain designated as T3. Since loss of morphological differentiation leads to loss of secondary metabolite production in Streptomyces [33], it is generally recognized that bald mutants lose their ability to produce secondary metabolites. Our result differs from such an empirical recognition. We also deposited the bald mutant to the NBRC culture collection and the comparative genome analysis is in progress.
Conclusions
The 9.9 Mb draft genome of Streptomyces sp. TP-A0867, a producer of alchivemycins isolated from a leaf of a Chinese chive, has been deposited at GenBank/ENA/DDBJ under the accession number BBON00000000. This strain was identified to be S. hygroscopicus subsp. hygroscopicus. We successfully identified a putative PKS/NRPS hybrid gene cluster that could be for alchivemycin synthesis and proposed the plausible biosynthetic pathway. Alchivemycin biosynthetic gene clusters are also present in the genomes of taxonomically close strains, one of which was able to produce alchivemycins. The genome sequence information disclosed in this study will be utilized for the investigation of additional new bioactive compounds and will also serve as a valuable reference for evaluation of the metabolic potential in plant-derived Streptomyces.
Acknowledgements
This research was supported by a Grant-in-aid for Scientific Research from the Ministry of Education, Culture, Sports, and Technology of Japan to Y.I. We would like to thank Ms. Machi Sasagawa and Ms. Yuko Kitahashi for assistance with searching and completely sequencing of the alchivemycin biosynthetic gene cluster, respectively. We also thank Ms. Tomoko Hanamaki, Ms. Chiyo Shibata and Ms. Satomi Saitou for technical assistance with whole genome sequencing, chemotaxonomic analyses and SEM observation, respectively.
Authors’ contributions
HK elucidated alchivemycin-biosynthetic pathway and drafted the manuscript. NI annotated the genome sequences. AO carried out the genome sequencing and sequence alignment. MH performed chemotaxonomic study. EH examined the features of the strain. SK isolated an alchivemycin-producing mutant. NF participated in coordination of genome sequencing. YI designed this study and edited the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors’ declare that they have no competing Interests.
Abbreviations
- A
Adenylation
- ACP
Acyl carrier protein
- ANI
Average nucleotide identity
- ANIb
ANI based on BLAST
- AT
Acyltransferase
- BLAST
Basic Local Alignment Search Tool
- C
Condensation
- CoA
Coenzyme A
- DDBJ
DNA Data Bank of Japan
- DDH
DNA-DNA hybridization
- DH
Dehydratase
- ER
Enoylreductase
- ESI
Electrospray ionization
- HPLC
High-performance liquid chromatography
- INSDC
International Nucleotide Sequence Database Collaboration
- ISP
International Streptomyces Project
- KR
Ketoreductase
- KS
Ketosynthase
- KSQ
KS-like domain with glutamine substituted for the active site cysteine residue
- LM
Loading module
- m
Module
- MeOH
Methanol
- MIGS
The minimum information about a genome sequence
- MS
Mass spectrometer
- NBRC
Biological Resource Center, National Institute of Technology and Evaluation
- NCBI
National Center for Biotechnology Information
- NRPS
Nonribosomal peptide synthetase
- PDA
Potato dextrose agar
- PKS
Polyketide synthase
- T
Thiolation
References
- 1.Berdy J. Bioactive microbial metabolites. J Antibiot. 2005;58(1):1–26. doi: 10.1038/ja.2005.1. [DOI] [PubMed] [Google Scholar]
- 2.Igarashi Y, Iida T, Sasaki T, Saito N, Yoshida R, Furumai T. Isolation of actinomycetes from live plants and evaluation of antiphytopathogenic activity of their metabolites. Actinomycetologica. 2002;16(1):9–13. doi: 10.3209/saj.16_9. [DOI] [Google Scholar]
- 3.Igarashi Y. Screening of novel bioactive compounds from plant-associated actinomycetes. Actinomycetologica. 2004;18(2):63–66. doi: 10.3209/saj.18_63. [DOI] [Google Scholar]
- 4.Igarashi Y, Kim Y, In Y, Ishida T, Kan Y, Fujita T, Iwashita T, Tabata H, Onaka H, Furumai T, Alchivemycin A, A bioactive polycyclic polyketide with an unprecedented skeleton from Streptomyces sp. Org Lett. 2010;12(15):3402–3405. doi: 10.1021/ol1012982. [DOI] [PubMed] [Google Scholar]
- 5.Kim Y, In Y, Ishida T, Onaka H, Igarashi Y. Biosynthetic origin of alchivemycin A, a new polyketide from Streptomyces and absolute configuration of alchivemycin B. Org Lett. 2013;15(14):3514–3517. doi: 10.1021/ol401071j. [DOI] [PubMed] [Google Scholar]
- 6.Harunari E, Hamada M, Shibata C, Tamura T, Komaki H, Imada C, Igarashi Y. Streptomyces hyaluromycini sp. nov., isolated from a tunicate (Molgula manhattensis) J Antibiot. 2016;69(3):159–163. doi: 10.1038/ja.2015.110. [DOI] [PubMed] [Google Scholar]
- 7.Hamada M, Yamamura H, Komukai C, Tamura T, Suzuki K, Hayakawa M. Luteimicrobium album sp. nov., a novel actinobacterium isolated from a lichen collected in Japan, and emended description of the genus Luteimicrobium. J Antibiot. 2012;65(8):427–431. doi: 10.1038/ja.2012.45. [DOI] [PubMed] [Google Scholar]
- 8.Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, Park SC, Jeon YS, Lee JH, Yi H, et al. Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol. 2012;62(Pt 3):716–721. doi: 10.1099/ijs.0.038075-0. [DOI] [PubMed] [Google Scholar]
- 9.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 10.Perriere G, Gouy M. WWW-query: an on-line retrieval system for biological sequence banks. Biochimie. 1996;78(5):364–369. doi: 10.1016/0300-9084(96)84768-7. [DOI] [PubMed] [Google Scholar]
- 11.Hasegawa T, Takizawa M, Tanida S. A rapid analysis for chemical grouping of aerobic actinomycetes. J Gen Appl Microbiol. 1983;29(4):319–322. doi: 10.2323/jgam.29.319. [DOI] [Google Scholar]
- 12.Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, Thomson N, Allen MJ, Angiuoli SV, et al. The minimum information about a genome sequence (MIGS) specification. Nat Biotechnol. 2008;26(5):541–547. doi: 10.1038/nbt1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ohtsubo Y, Maruyama F, Mitsui H, Nagata Y, Tsuda M. Complete genome sequence of Acidovorax sp. strain KKS102, a polychlorinated-biphenyl degrader. J Bacteriol. 2012;194(24):6970–6971. doi: 10.1128/JB.01848-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25(5):955–964. doi: 10.1093/nar/25.5.0955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador-Vegas A, et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016;44(D1):D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Letunic I, Doerks T, Bork P. SMART: recent updates, new developments and status in 2015. Nucleic Acids Res. 2015;43(Database issue):D257–D260. doi: 10.1093/nar/gku949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Komaki H, Ichikawa N, Hosoyama A, Fujita N, Igarashi Y. Draft genome sequence of marine-derived Streptomyces sp. TP-A0598, a producer of anti-MRSA antibiotic lydicamycins. Stand Genomic Sci. 2015;10:58. doi: 10.1186/s40793-015-0046-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Richter M, Rossello-Mora R. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci U S A. 2009;106(45):19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol. 2007;57(Pt 1):81–91. doi: 10.1099/ijs.0.64483-0. [DOI] [PubMed] [Google Scholar]
- 21.Meier-Kolthoff JP, Auch AF, Klenk HP, Goker M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics. 2013;14:60. doi: 10.1186/1471-2105-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Onaka H, Mori Y, Igarashi Y, Furumai T. Mycolic acid-containing bacteria induce natural-product biosynthesis in Streptomyces species. Appl Environ Microbiol. 2011;77(2):400–406. doi: 10.1128/AEM.01337-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rong X, Huang Y. Taxonomic evaluation of the Streptomyces hygroscopicus clade using multilocus sequence analysis and DNA-DNA hybridization, validating the MLSA scheme for systematics of the whole genus. Syst Appl Microbiol. 2012;35(1):7–18. doi: 10.1016/j.syapm.2011.10.004. [DOI] [PubMed] [Google Scholar]
- 24.Keatinge-Clay AT. The structures of type I polyketide synthases. Nat Prod Rep. 2012;29(10):1050–1073. doi: 10.1039/c2np20019h. [DOI] [PubMed] [Google Scholar]
- 25.Long PF, Wilkinson CJ, Bisang CP, Cortes J, Dunster N, Oliynyk M, McCormick E, McArthur H, Mendez C, Salas JA, et al. Engineering specificity of starter unit selection by the erythromycin-producing polyketide synthase. Mol Microbiol. 2002;43(5):1215–1225. doi: 10.1046/j.1365-2958.2002.02815.x. [DOI] [PubMed] [Google Scholar]
- 26.Wilkinson CJ, Frost EJ, Staunton J, Leadlay PF. Chain initiation on the soraphen-producing modular polyketide synthase from Sorangium cellulosum. Chem Biol. 2001;8(12):1197–1208. doi: 10.1016/S1074-5521(01)00087-4. [DOI] [PubMed] [Google Scholar]
- 27.Del Vecchio F, Petkovic H, Kendrew SG, Low L, Wilkinson B, Lill R, Cortes J, Rudd BA, Staunton J, Leadlay PF. Active-site residue, domain and module swaps in modular polyketide synthases. J Ind Microbiol Biotechnol. 2003;30(8):489–494. doi: 10.1007/s10295-003-0062-0. [DOI] [PubMed] [Google Scholar]
- 28.Kakavas SJ, Katz L, Stassi D. Identification and characterization of the niddamycin polyketide synthase genes from Streptomyces caelestis. J Bacteriol. 1997;179(23):7515–7522. doi: 10.1128/jb.179.23.7515-7522.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fischbach MA, Walsh CT. Assembly-line enzymology for polyketide and nonribosomal peptide antibiotics: logic, machinery, and mechanisms. Chem Rev. 2006;106(8):3468–3496. doi: 10.1021/cr0503097. [DOI] [PubMed] [Google Scholar]
- 30.Keatinge-Clay AT. A tylosin ketoreductase reveals how chirality is determined in polyketides. Chem Biol. 2007;14(8):898–908. doi: 10.1016/j.chembiol.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 31.Kwan DH, Sun Y, Schulz F, Hong H, Popovic B, Sim-Stark JC, Haydock SF, Leadlay PF. Prediction and manipulation of the stereochemistry of enoylreduction in modular polyketide synthases. Chem Biol. 2008;15(11):1231–1240. doi: 10.1016/j.chembiol.2008.09.012. [DOI] [PubMed] [Google Scholar]
- 32.Bachmann BO, Ravel J. Chapter 8. Methods for in silico prediction of microbial polyketide and nonribosomal peptide biosynthetic pathways from DNA sequence data. Methods Enzymol. 2009;458:181–217. doi: 10.1016/S0076-6879(09)04808-3. [DOI] [PubMed] [Google Scholar]
- 33.Horinouchi S, Beppu T. Hormonal control by A-factor of morphological development and secondary metabolism in Streptomyces. Proc Jpn Acad Ser B Phys Biol Sci. 2007;83(9–10):277–295. doi: 10.2183/pjab.83.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci U S A. 1990;87(12):4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goodfellow M. Phylum XXVI. Actinobacteria phyl. nov. In: Goodfellow M, Kämpfer P, Busse H-J, Trujillo ME, Suzuki K-I, Ludwig W, Whitman WB, editors. Bergey’s manual of systematic bacteriology. 2. New York: Springer; 2012. p. 33. [Google Scholar]
- 36.Stackebrandt E, Rainey FA, Ward-Rainey NL. Proposal for a new hierarchic classification system, Actinobacteria classis nov. Int J Syst Bacteriol. 1997;47:479–491. doi: 10.1099/00207713-47-2-479. [DOI] [Google Scholar]
- 37.Buchanan RE. Studies in the Nomenclature and Classification of the Bacteria: II. The Primary Subdivisions of the Schizomycetes. Journal of Bacteriology. 1917;2(2):155–164. doi: 10.1128/jb.2.2.155-164.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Skerman VBD, McGowan V, Sneath PHA. Approved lists of bacterial names. Int J Syst Bacteriol. 1980;30:225–420. doi: 10.1099/00207713-30-1-225. [DOI] [PubMed] [Google Scholar]
- 39.Zhi XY, Li WJ, Stackebrandt E. An update of the structure and 16S rRNA gene sequence-based definition of higher ranks of the class Actinobacteria, with the proposal of two new suborders and four new families and emended descriptions of the existing higher taxa. Int J Syst Evol Microbiol. 2009;59(Pt 3):589–608. doi: 10.1099/ijs.0.65780-0. [DOI] [PubMed] [Google Scholar]
- 40.Kim SB, Lonsdale J, Seong CN, Goodfellow M. Streptacidiphilus gen. nov., acidophilic actinomycetes with wall chemotype I and emendation of the family Streptomycetaceae (Waksman and Henrici (1943)AL) emend. Rainey et al. 1997. Antonie van Leeuwenhoek. 2003;83(2):107–116. doi: 10.1023/A:1023397724023. [DOI] [PubMed] [Google Scholar]
- 41.Wellington EM, Stackebrandt E, Sanders D, Wolstrup J, Jorgensen NO. Taxonomic status of Kitasatosporia, and proposed unification with Streptomyces on the basis of phenotypic and 16S rRNA analysis and emendation of Streptomyces Waksman and Henrici 1943, 339AL. Int J Syst Bacteriol. 1992;42(1):156–160. doi: 10.1099/00207713-42-1-156. [DOI] [PubMed] [Google Scholar]
- 42.Waksman SA, Henrici AT. The nomenclature and classification of the Actinomycetes. J Bacteriol. 1943;46(4):337–341. doi: 10.1128/jb.46.4.337-341.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Witt D, Stackebrandt E. Unification of the genera Streptoverticillium and Streptomyces, and amendation of Streptomyces Waksman and Henrici 1943, 339 AL. Syst Appl Microbiol. 1990;13:361–371. doi: 10.1016/S0723-2020(11)80234-1. [DOI] [Google Scholar]
- 44.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]


