Summary
Expansion of a hexanucleotide repeat GGGGCC (G4C2) in C9ORF72 is the most common cause of amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD). Transcripts carrying (G4C2) expansions undergo unconventional, non-ATG-dependent translation, generating toxic dipeptide repeat (DPR) proteins thought to contribute to disease. Here we identify the interactome of all DPRs and find that arginine-containing DPRs, polyGly-Arg (GR) and polyPro-Arg (PR), interact with RNA-binding proteins and proteins with low complexity sequence domains (LCDs) that often mediate the assembly of membrane-less organelles. Indeed, most GR/PR interactors are components of membrane-less organelles such as nucleoli, the nuclear pore complex and stress granules. Genetic analysis in Drosophila demonstrated the functional relevance of these interactions to DPR toxicity. Furthermore, we show that GR and PR altered phase separation of LCD-containing proteins, insinuating into their liquid assemblies and changing their material properties, resulting in perturbed dynamics and/or functions of multiple membrane-less organelles.
Blurb
Arginine-containing dipeptide repeat proteins, which are thought to contribute to amyotrophic lateral sclerosis, disrupt the dynamics of membrane-less organelles
Introduction
Expansion of a hexanucleotide repeat GGGGCC (G4C2) in an intron of chromosome 9 open reading frame 72 (C9ORF72) is the most common cause of ALS and FTD (C9-ALS/FTD) (DeJesus-Hernandez et al., 2011; Renton et al., 2011). Unaffected individuals typically have between 2 and 23 G4C2 repeats, while people with C9-ALS/FTD have hundreds or even thousands of repeats. As this mutation accounts for up to 40% of familial cases of ALS and FTD and ~5–10% of sporadic cases (Renton et al., 2011), there is intense interest in understanding how G4C2 repeat expansions cause disease.
Decreased expression of the C9ORF72 protein may contribute to disease progression in C9-ALS/FTD (O’Rourke et al., 2016), but the primary driver of pathogenesis in C9-ALS/FTD appears to be toxic gain-of-function, for which two non-exclusive mechanisms have been proposed. The first posits that C9ORF72 transcripts sequester RNA-binding proteins (Schweizer Burguete et al., 2015; Xu et al., 2013), since these transcripts accumulate in the brain and spinal cord of C9-ALS/FTD patients (DeJesus-Hernandez et al., 2011; Gendron et al., 2013; Mori et al., 2013; Zu et al., 2013). The second centers on toxicity from dipeptide repeat (DPR) proteins produced by repeat-associated non-ATG (RAN) translation, which occurs in the absence of an initiating AUG codon and produces peptides from all reading frames from both sense and antisense transcripts (Zu et al., 2011). Hexanucleotide repeat expansion in C9ORF72 produces five DPR proteins: glycine-alanine (GA) and glycine-arginine (GR) from sense G4C2-containing transcripts, proline-arginine (PR) and proline-alanine (PA) from antisense C4G2-containing transcripts, and glycine-proline (GP) from both sense and antisense transcripts (Gendron et al., 2013; Mori et al., 2013; Zu et al., 2013). All of these DPRs are produced in C9-ALS/FTD patients and account for cytoplasmic and intranuclear inclusions observed in the brain and spinal cord (Gendron et al., 2013; Zu et al., 2013).
Some DPR species, those containing arginine (GR and PR) in particular, are toxic. GR and PR peptides are taken up by cultured cells when added exogenously and accumulate in nucleoli, causing RNA processing defects and cell death (Kwon et al., 2014). Similarly, GR and PR are toxic when expressed in cultured neurons, while similar levels of PA, GA, and PG are well tolerated (Wen et al., 2014), consistent with findings in Drosophila that GR and PR are toxic in neuronal tissue, whereas GA is modestly toxic and GP and PA are non-toxic (Freibaum et al., 2015; Mizielinska et al., 2014; Wen et al., 2014). A recent series of studies provided converging evidence that at least one downstream consequence of the C9ORF72 mutation is impaired trafficking through the nuclear pore (Freibaum et al., 2015; Jovičić et al., 2015; Zhang et al., 2015). Two of these studies were based on complex disease models expressing expanded G4C2 RNAs and multiple DPR proteins (Freibaum et al., 2015; Zhang et al., 2015), but one used PR overexpression in yeast, suggesting that this DPR species alone is sufficient to impair nuclear pore trafficking (Jovicic et al., 2015). Other studies have reported toxicity with GA expression in cultured cells or AAV-mediated delivery to mouse brain, so multiple DPR species may contribute to disease (Zhang et al., 2016; Zhang et al., 2014). The relative contribution of RNA- versus DPR-mediated toxicity to C9-ALS/FTD pathogenesis, as well as the identities of the most potent DPR species and their mechanisms of toxicity, are pressing questions in the field.
An additional unresolved question is the extent of mechanistic overlap between the many distinct genetic forms of ALS, FTD, and related diseases that share the same histopathology. Accumulation of the RNA-binding protein TDP-43 in ubiquitinated cytoplasmic inclusions is the hallmark pathology of sporadic and most familial ALS and FTD, including C9-ALS/FTD (Taylor et al., 2016). The discovery of ALS/FTD-causing mutations in TDP-43 and related RNA-binding proteins, including FUS and hnRNPA1, focused attention on the role of RNA-binding proteins in ALS/FTD pathogenesis (Kim et al., 2013; Kwiatkowski et al., 2009; Vance et al., 2009). Notably, disease-causing mutations in TDP-43, FUS and hnRNPA1 are most frequently located in low complexity sequence domains (LCDs) of these proteins. LCDs in these and related RNA-binding proteins contribute to the assembly of membrane-less organelles such as RNA granules (Kato et al., 2012). Recently, we and others demonstrated that liquid-liquid phase transition (LLPS) by hnRNPs harboring LCDs, such as hnRNPA1, TIA1 and FUS, contributes to the assembly and liquid properties of stress granules (Lin et al., 2015; Molliex et al., 2015; Murakami et al., 2015; Patel et al., 2015). Importantly, disease mutations in LCDs alter this biophysical property, accelerate the formation of amyloid-like fibrils, and disrupt the dynamics and functions of membrane-less organelles (Taylor et al., 2016).
Here we determined that the two arginine-containing DPRs produced from mutant C9ORF72, GR and PR, interact with RNA-binding proteins and other proteins harboring LCDs, including TDP-43, hnRNPA1 and FUS. We show that GR and PR insinuate into liquid assemblies of LCDs, alter their material properties, and disrupt the dynamics and functions of membrane-less organelles in living cells. These observations suggest a shared pathogenic mechanism for disease initiated by C9ORF72 mutations and mutations in genes encoding RNA-binding proteins.
Results
GR/PR interactomes are enriched in proteins containing LCDs
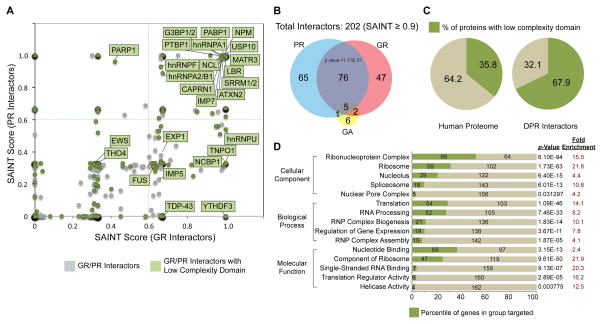
To gain insight into the mechanisms underlying DPR-mediated toxicity, we analyzed the interactomes of DPR proteins in human cells. Green fluorescent protein (GFP)-conjugated 47 or 50 repeat DPR proteins (GFP-GA50, GFP-GP47, GFP-PA50, GFP-GR50 and GFP-PR50) were expressed in human embryonic kidney (HEK293T) cells and immunoprecipitated with anti-GFP nanobody. By gel staining, we found that arginine-containing DPRs, GFP-GR50 and GFP-PR50, interacted with numerous proteins, whereas GFP-GA50, GFP-GP47, and GFP-PA50 showed no enrichment of bound proteins compared to GFP control (Figure S1A and S1B). After analysis of co-purified proteins by liquid chromatography-tandem mass spectrometry (LC-MS/MS), we measured the specificity and reproducibility of individual interactions using the Significance Analysis of INTeractome (SAINT) approach (Figure 1A and S1D) (Choi et al., 2011). Western blotting confirmed a subset of high-scoring interactions (Figure S1C and data not shown). We identified 389 DPR-interacting proteins using a SAINT score cutoff of 0.6 (Figure S1D, Table S1); this number decreased to 202 with a stringent cutoff of 0.9 (Figure 1B, Figure S1D). Of these 202 proteins, 196 were associated with either GFP-GR50 or GFP-PR50, 6 were identified as GFP-GA50-specific interactors, and no proteins were identified as GFP-GP47- or GFP-PA50-specific interactors.
Figure 1. Proteomic analysis showing GR and PR share a common set of interactors enriched in LCDs.
(A) GFP-GR50- and GFP-PR50-interacting proteins displayed by SAINT probabilistic score. Green, interactors with LCDs; gray, interactors without LCDs. (B) Venn diagram illustrating overlap (P < 1.17e-37, hypergeometric test) between GFP-GR50- and GFP-PR50-interacting proteins. (C) Percentage of LCD-containing proteins within the GR/PR interactome compared to percentage of LCD-containing proteins in the human proteome. (D) Gene ontology analysis showing enrichment in proteins that are components of membrane-less organelles. P values were derived via the DAVID algorithm. See also Figure S1.
Among the 196 GFP-GR50 or GFP-PR50 interactors, 81 proteins were associated with both GFP-GR50 and GFP-PR50, far more overlap than expected by chance (p = 1.17E-37). Among these interactors are ALS/FTD-related RNA-binding proteins, including TDP-43, FUS, hnRNPA1, hnRNPA2B1, Matrin-3 and Ataxin-2. Moreover, these common interactors showed significant enrichment in proteins containing LCDs; whereas LCDs are in 35.8% of human proteins, 67.9% of GFP-GR50 and GFP-PR50 common interactors contained LCDs (Figure 1C). Consistent with this observation, a companion manuscript from McKnight and colleagues uses a covalent cross-linking approach yielding similar overlap in PR interactomes (p=9.3 × 10−274; Lin and Mori et al.). Consistent with the emerging role of LCDs in mediating the higher order assembly of membrane-less organelles and related structures, gene ontology analysis of the full GR/PR interactome revealed that GFP-GR50 and GFP-PR50 interactors were significantly enriched in RNA-binding proteins and components of membrane-less organelles, including RNP complexes or granules, nucleoli, spliceosomes, and the nuclear pore complex (Figure 1D), suggesting that membrane-less organelles and related structures might be common targets of arginine-containing DPRs.
Arginine-containing DPR proteins drive C9orf72 G4C2 repeat toxicity in Drosophila
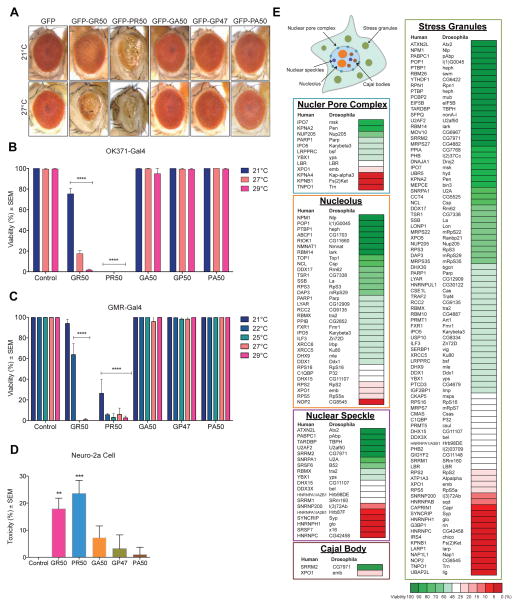
To assess the contribution of each DPR protein to the toxicity associated with C9orf72-mediated neurodegeneration, we used transgenic flies with alternative codons to G4C2 to directly express 47 or 50 repeats of GR, PR, GA, GP, and PA, with an AUG start codon and amino-terminal GFP (Freibaum et al., 2015; Wen et al., 2014). Expression of GFP-GA50, GFP-GP47, or GFP-PA50 in eyes using GMR-GAL4 did not elicit a degenerative eye phenotype (Figure 2A). By contrast, expression of GFP-GR50 or GFP-PR50 resulted in severely degenerated eyes (Figure 2A), consistent with previous reports indicating that arginine-containing dipeptides are toxic in cultured cells and Drosophila (Freibaum et al., 2015; Jovičić et al., 2015; Kwon et al., 2014; Mizielinska et al., 2014; Wen et al., 2014). The toxicity of GR and PR also extends to other tissues; expression of GFP-GR50 or GFP-PR50 in motor neurons (using driver OK371-GAL4) or eyes (using GMR-GAL4, which has some leakiness in the brain) caused pupal lethality, whereas GFP-GA50, GFP-GP47, or GFP-PA50 had no effect on viability (Figure 2B, 2C). Increasing the temperature, which boosts transgene expression, further decreased survival in flies expressing GFP-GR50 or GFP-PR50 (Figure 2B, 2C). We then crossed in deGradFP (Caussinus et al., 2012) to target GFP-GR50 peptides for proteasomal degradation. This rescued the eye phenotype (Figure S2A, S2B), demonstrating that GFP-GR50 caused toxicity at the protein level. Toxicity of GFP-GR50 or GFP-PR50 was also observed in mammalian cells of neuronal origin, Neuro-2a cells, consistent with a prior report of PR and GR toxicity in neurons (Wen et al., 2014)(Figure 2D). Cells expressing GFP-GA50 showed a trend toward increased cell death (Figure 2D). Thus, the highly basic, arginine-containing DPR proteins stand apart as particularly toxic in Drosophila and mammalian cells.
Figure 2. Relevance of the GR/PR interactome to toxicity confirmed in Drosophila.
(A) Eye phenotypes resulting from GMR-GAL4-driven expression of indicated transgenes at indicated temperatures. (B, C) Egg-to-adult viability phenotypes resulting from (B) OK371-GAL4-driven or (C) GMR-GAL4-driven expression of indicated transgenes at indicated temperatures. n=3 biological repeats, ****P < 0.0001 by Two Way ANOVA, Tukey’s post-hoc test. (D) MTT assay following ectopic expression of indicated DPR constructs in Neuro-2a cells. n=5 biological repeats, **P < 0.01, ***P < 0.001 by One Way ANOVA. (E) Schematic of a cell with membrane-less organelles related to the GR/PR interactome. Enhancers (red) and suppressors (green) of GR viability are categorized by membrane-less organelle. Data are represented as mean ± SEM in (B–D). See also Figure S2.
DPR protein interactors are genetic modifiers of GR50-mediated toxicity in Drosophila
We performed an in vivo RNAi screen as an orthologous approach to assess the functional significance of the arginine-containing DPR interactomes. GFP-GR50-expressing flies were crossed with Drosophila lines expressing double-stranded RNA (dsRNA), which use a GMR-GAL4 driver to knock down selected genes and show ~50% viability at 22°C, allowing for detection of enhancer and suppressor effects. A total of 126 GR/PR interactors for which close Drosophila orthologs exist were tested. To minimize variability due to genetic background, we set the basal viability range at 25 – 48%, based on the survival rate of GFP-GR50-expressing flies crossed with two genetically distinct parental controls, V60100 and w1118 (Figure S2C). Remarkably, 84.9% of the GR/PR interactors were genetic modifiers of GFP-GR50 toxicity (Figure S2C). This targeted screen identified 80 suppressors (63%), including 35 (28%) strong suppressors whose depletion increased viability to >70%. The screen also identified 27 enhancers (21%), including 21 that reduced viability to <15%. The high frequency of genetic modifiers in the GR/PR interactome underscores the relevance of these interactions to DPR-related toxicity. Importantly, most of these genetic modifiers encode components of membrane-less organelles, suggesting that disturbance in the function of membrane-less organelles contributes to GR/PR toxicity (Figure 2E, Figure S2D).
Overlap in genetic modifiers of toxicity from GR/PR and expanded-G4C2 repeat
Several genetic modifiers of GR toxicity identified among the GR/PR interactome (e.g. ALYREF, XPO1, TNPO1 and GLE1) were also identified in a prior unbiased, whole-genome screen for modifiers of flies expressing expanded (G4C2) repeats (Freibaum et al., 2015). To assess the extent of overlap between these data sets, we further assessed the 35 strongest suppressors and 21 strongest enhancers in our Drosophila model expressing 58 G4C2 repeats. Twenty-three of 35 suppressors (65.7%) and 13 of 21 (61.9%) enhancers modified the rough eye phenotype in (G4C2)58-expressing flies, similar to their effects on GFP-GR50 fly viability (Figure S2E, S2F). To assess specificity, we tested the same candidate modifiers in a Drosophila model expressing polyglutamine (polyQ) expansion in the androgen receptor (AR(polyQ)52) (Nedelsky et al., 2010), a neurodegenerative model unrelated to C9orf72, and found very little overlap (Figure S2E, S2F). Cluster dendrogram analysis demonstrated that the overlap of shared modifiers of GR and (G4C2)58 toxicities was significantly greater than shared modifiers with AR(polyQ)52 toxicity, suggesting a common pathogenic mechanism by which GR and G4C2 expansion cause toxicity. Thus some portion of the degeneration driven by G4C2 expansion in Drosophila, if not all, may be driven by DPR expression.
Arginine-containing DPRs are recruited to specific nucleolar substructures
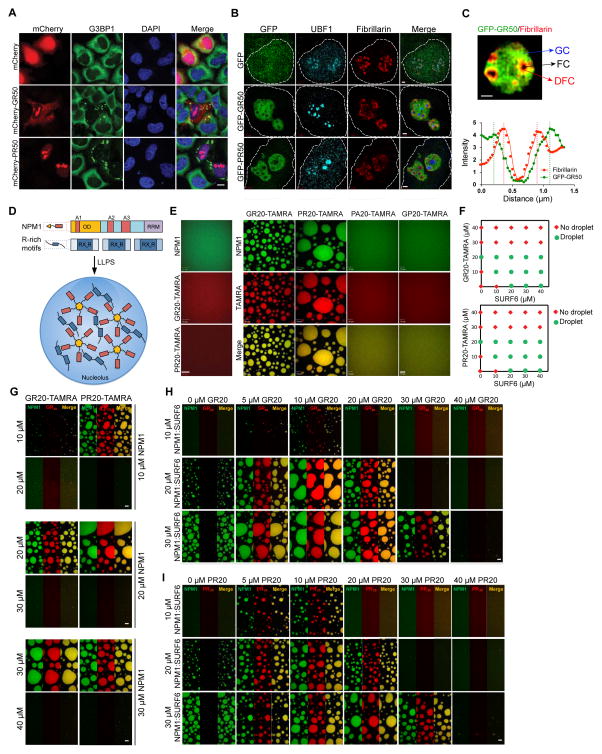
We next examined the distribution of GFP-GR50 and GFP-PR50 in mammalian cells. We found GFP-GR50 in both the cytoplasm and nucleus, GFP-PR50 predominantly in the nucleus, and both GFP-GR50 and GFP-PR50 in the nucleolus, all consistent with prior reports (Kwon et al., 2014; Wen et al., 2014) (Figure S3A, S3B). Similar localization patterns were observed with mCherry-GR50 and -PR50 (Figure 3A), and with fluorescein amidite-labeled peptides of GR20 and PR20 added to the culture media (Figure S3C).
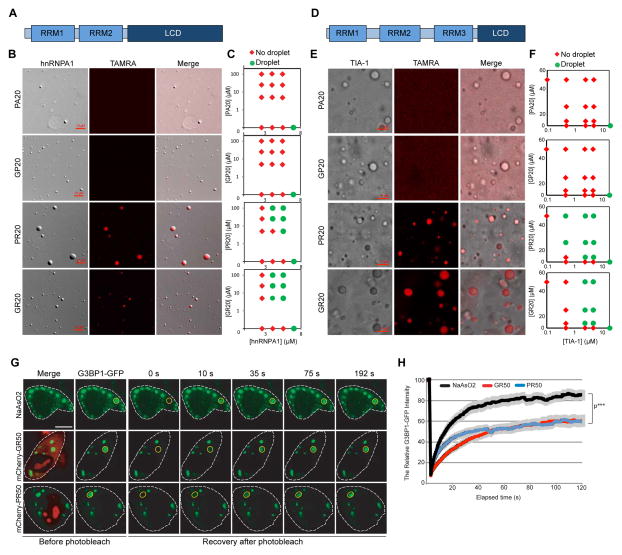
Figure 3. GR and PR dipeptides localize to nucleolar substructures and impair biophysical properties of NPM1 in vitro.
(A) Cellular localization of mCherry- GR50 or -PR50 (red) and endogenous G3BP1 (green) in HeLa cells. DAPI is shown in blue. Scale bar, 10 μm. (B) Structured illumination microscopy showing sub-nucleolar localization of GFP-GR50 and -PR50 (green), UBF1 (cyan), and Fibrillarin (red) in HeLa cells. Nuclei were outlined based on DAPI stain (not shown). Scale bar, 1 μm. (C) GR localizes within GC and DFC, but is excluded from the FC. A representative line scan intensity graph of Fibrillarin and GFP-GR50 is displayed. Scale bar, 1 μm. (D) Schematic representation of LLPS mediated by NPM1 and R-rich motifs. NPM1 has N-terminal oligomerization domain (OD, gold), three acidic tracts (A1, A2, and A3, red) and one C-terminal RNA recognition motif (RRM, purple). (E) Poly-arginine peptides phase separate in vitro with NPM1 at 1:1 stoichiometry. Shown are confocal microscopy images of 20 μM NPM1, GR20 and PR20 (left panel) and binary mixtures of 20 μM NPM1 with 20 μM of the indicated peptide (right panels). 1 μM of the total NPM1 and each peptide was labeled with Alexa488 or TAMRA, respectively. Scale bar, 10 μm. (F) Phase diagrams for ternary mixtures of NPM1 (20 μM) with the indicated concentrations of SURF6 and GR20 (top) or SURF6 and PR20 (bottom). Single-phase regime (red diamonds) and two-phase regime with the formation of liquid droplets (green circles) are indicated. (G) GR20 (left) and PR20 (right) phase separate with NPM1 at 1:1 stoichiometry (DPR:NPM1) but not at higher stoichiometries. Alexa488-NPM1 was detected using confocal fluorescence microscopy. Scale bar, 10 μm. (H, I). Titrations of GR20 (H) and PR20 (I) (concentrations at top) into liquid-phase droplets comprised of 1:1 NPM1:SURF6 peptide prepared at different concentrations (indicated at left). Alexa488-NPM1 (left in each panel) and TAMRA-DPR peptides (middle) were detected using confocal fluorescence microscopy. Scale bar, 10 μm. See also Figures S3 and S4.
Nucleoli consist of three morphologically distinct regions: fibrillar centers (FCs) surrounded by a dense fibrillar component (DFC), both embedded in a liquid-like granular component (GC) (Boisvert et al., 2007; Brangwynne et al., 2011). These regions host different aspects of ribosome biogenesis and have different biophysical properties (Boisvert et al., 2007). Super-resolution microscopy using structured illumination (SIM) revealed that GR and PR are recruited to the NPM1-positive GC, but excluded from the upstream binding factor (UBF)-positive FC (Figure 3B), and GR but not PR was enriched in the Fibrillarin-positive DFC (Figure 3C).
Recruitment of GR and PR to the GC is consistent with the observed physical interaction of both DPRs with GC component NPM1 (Figure 1A). None of the non-arginine DPRs showed accumulation in the nucleolus or its individual compartments (Figure S3A and S3B). We further assessed association of GR and PR with nucleolar components by fluorescent recovery after photobleaching (FRAP). GFP-GR50 and GFP-PR50 showed prolonged recovery time after photobleaching compared to GFP control (Figure S3D, S3E), consistent with tighter physical interaction with a nucleolar constituent. Despite the comparable recovery times of GFP-GR50 and GFP-PR50, GFP-GR50 showed a smaller mobile fraction, indicating a more stable interaction with one or more nucleolar constituents (Figure S3D, S3E), possibly due to its tight association with less mobile DFC components (Feric et al., 2016) in the nucleolus (Figure 3C).
In contrast to the dynamic association of GFP-GR50 and GFP-PR50 with the nucleolus, GFP-GA50 forms prominent cytoplasmic aggregates that are non-dynamic and show no fluorescence recovery after photobleaching. Moreover, when cells were treated with the aliphatic alcohol 1,6-hexanediol, which can disturb labile, phase-separation assemblies (Kroschwald et al., 2015; Molliex et al., 2015), GFP-GR50 and GFP-PR50 are washed away from accumulation in nucleoli, whereas GFP-GA50 cytoplasmic aggregates persist (Figure S3F). Finally, when cells expressing GFP-GR50, GFP-PR50 or GFP-GA50 were subjected to serial detergent extraction, GFP-GR50 and GFP-PR50 were mostly in the RIPA-soluble fraction (containing 0.1% SDS), whereas GFP-GA50 was more soluble in the urea fraction (Figure S3G). This difference in solubility reflects their distinct physicochemical properties; GR and PR peptides (and the non-toxic GP and PA dipeptides) form flexible, expanded coils, which are expected to remain soluble (Figure S4A), while the GA dipeptide, composed of small hydrophobic residues and devoid of charge, is expected to collapse into poorly soluble globules and form aggregates through inter- and intramolecular interactions.
Arginine-containing DPRs impair biophysical properties of NPM1 in vitro
NPM1 is abundant in the GC and helps organize this liquid-like component of the nucleolus (Mitrea et al., 2016). It is a multivalent protein containing three acidic LCD tracts permitting interaction with many nucleolar binding partners with multivalent arginine-rich motifs (R-motifs) (Mitrea et al., 2016). Along with R-motif-containing proteins, NPM1 undergoes LLPS to form liquid-like droplets believed to reflect the liquid-like properties of the GC (Mitrea et al., 2016) (Figure 3D). Due to the high arginine content of GR and PR, we hypothesized that these dipeptides might interfere with nucleolar function by directly interacting with NPM1 and altering its interactions with R-motif-containing partners. Indeed, FLAG-GR20 and -PR20 peptides directly interacted with purified NPM1 in vitro (Figure S4A, S4B). Remarkably, although neither NPM1 nor GR20-FLAG/PR20-FLAG peptides underwent LLPS in isolation, LLPS was observed when NPM1 was added, indicating that interaction of arginine-containing DPRs with NPM1 is sufficient to induce LLPS (Figure 3E). LLPS occurred at a NPM1:DPR stoichiometric ratio of 1:1 and was reduced or absent with excess GR20 or PR20 (Figure 3G). These observations suggest that the multivalent R-motifs in DPRs, at equimolar ratios, form dynamic crosslinks between NPM1 pentamers, mediating LLPS as previously described for other multivalent R-rich peptides derived from nucleolar proteins (Mitrea et al., 2016).
We next investigated whether adding GR20 or PR20 peptides would influence the ability of NPM1 to undergo LLPS with one of its native nucleolar binding partners, SURF6. Using a fixed concentration of NPM1 (20 μM), we titrated increasing amounts of SURF6 peptide and observed LLPS only when SURF6 concentrations exceeded 20 μM (Figure 3F). Co-addition of increasing concentrations of GR20 or PR20 peptides substantially reduced the critical concentration of SURF6 necessary to induce LLPS (Figure 3F), suggesting cooperation between the two peptides in mediating LLPS. However, concentrations of GR20 and PR20 peptides in excess of that of NPM1 inhibited LLPS at any SURF6 concentration (Figure 3F). Moreover, pre-formed NPM1/SURF6 droplets were dissolved by addition of excess GR20 or PR20, suggesting that GR20 or PR20 peptides are able to outcompete SURF6 for interaction with NPM1 (Figure 3H, 3I) and adopt a bound conformation incompatible with LLPS. In contrast, non-arginine-containing DPR peptides (GP20 and PA20) did not undergo LLPS with NPM1 and failed to influence pre-formed NPM1/SURF6 droplets (Figures 3E and S4C).
Arginine-containing DPRs impair nucleolar dynamics and function in live cells
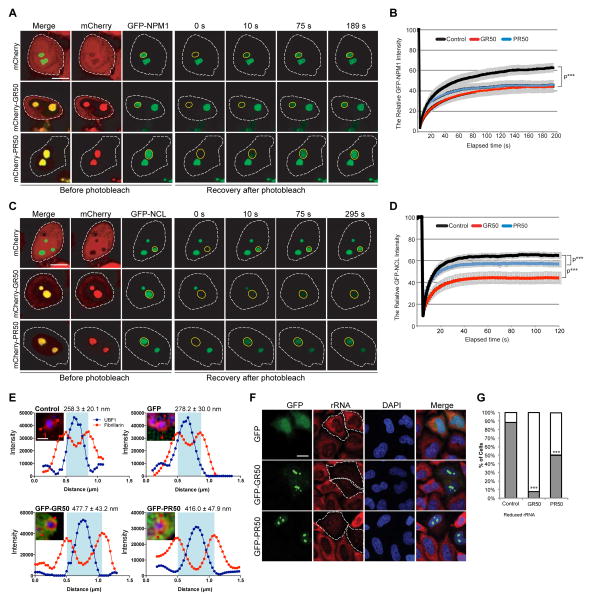
To test the in vivo consequences of GR and PR interaction with nucleolar constituents, we assessed the impact of these DPRs on the mobility of NPM1 and Nucleolin (NCL) in living cells with FRAP. NPM1 and NCL both physically interact with GR/PR (Figure 1A and Table S1) and genetically suppress GR toxicity in vivo (Figure 2E and S2C). After photobleaching a single nucleolus labeled by GFP-NPM1, ~60% of fluorescence signal recovered within ~120s, indicating a mobile fraction in dynamic equilibrium with the surrounding nucleoplasm (Figure 4A, 4B). Expression of mCherry-GR50 or mCherry-PR50 slowed NPM1 recovery after photobleaching and significantly increased the NPM1 immobile fraction, consistent with strong interactions between GR/PR and NPM1 (Figure 4A, 4B).
Figure 4. GR and PR dipeptides disrupt nucleolar dynamics and function in live cells.
(A–D) HeLa cells transfected with (A, B) GFP-NPM1 or (C, D) GFP-NCL and mCherry-GR50 or -PR50 constructs were analyzed using FRAP. The yellow circle marks the photobleached region. Representative images of the same area before and after photobleaching are shown. Scale bar, 10 μm. Fluorescence of (B) GFP-NPM1 or (C) GFP-NCL in the photobleached region over time. n=10–12 cells per sample for GFP-NPM1; n=7–8 cells per sample for GFP-NCL, ***P < 0.001 by Student t-test, paired. (E) Average diameter of the nucleolar fibrillar center (FC) is shown following expression of GFP-GR50 or PR50 dipeptides or controls. P < 0.01, ANOVA. Scale bar, 1 μm. At least 12 cells were analyzed for each condition (n=2 biological repeats). Representative line scan intensity graphs of FC surrounded by DFC are displayed. (F) rRNA-specific antibody (red) shows rRNA levels in HeLa cells transiently transfected with GFP-GR50 or -PR50 or GFP alone. (G) Quantification of cells showing the percentage of normal (gray) and reduced (white) immunostaining of rRNA relative to neighboring untransfected cells. n=3 biological repeats, ***P < 0.001 by One Way ANOVA. Scale bar, 10 μm. Data are represented as mean ± SEM in (B), (D) and (E). See also Figure S4.
NCL is primarily located in the DFC (Ginisty et al., 1999) and directly interacts with synthetic GR20-FLAG and PR20-FLAG peptides, showing greater association with GR than PR (Figure S4B). When we photobleached a single nucleolus labeled by GFP-NCL, approximately 60% of fluorescence signal recovered within ~10s (Figure 4C, 4D). PR50 or GR50 reduced the mobile fraction of NCL, with GR50 showing a more potent effect, consistent with the binding data (Figures 3C, 4C, 4D, S4B). This phenomenon is specific, as the mobility of DDX47, a nucleolar protein that neither contains an LCD nor interacts with GR50 or PR50, was unaffected by GR50 or PR50 expression (Figure S4D, S4E)
We also examined the impact of GR50 and PR50 accumulation on the morphology of nucleolar substructures with super-resolution SIM microscopy, using UBF1 staining to identify the FC and Fibrillarin staining to identify the DFC. The diameter of the FC in cells expressing GFP did not significantly differ from that of untransfected cells, but was significantly enlarged in cells expressing GR50 and PR50 (Figure 4E). Total rRNA in cells expressing GFP-GR50 or GFP-PR50 was significantly reduced compared to neighboring untransfected or GFP-expressing cells (Figure 4F, 4G), consistent with prior reports of decreased processing and maturation of rRNA in cells treated with PR20 peptide (Kwon et al., 2014) and in C9-ALS/FTD patient-derived cells and tissue (Haeusler et al., 2014). Together, these results indicate that interaction of arginine-containing DPRs with nucleolar proteins impacts their biophysical properties, alters nucleolar dynamics, and impairs nucleolar function.
GR and PR interact with stress granule proteins
Cytoplasmic stress granules are membrane-less cytosolic bodies assembled by mRNAs and proteins when translation initiation is limiting (Anderson and Kedersha, 2009). Many GR/PR physical and genetic interactors are components of these structures and are essential for their assembly and dynamics (Figures 1A, 2E and Table S1). The LCD-containing G3BP1 and G3BP2 (hereafter G3BP) and its binding partner Caprin1 both promote stress granule assembly (Kedersha et al., 2016) and were identified as strong enhancers of GR-mediated toxicity in our RNAi screen (Figure 2E). USP10, which also binds to G3BP but inhibits stress granule assembly (Kedersha et al., 2016) was identified as a suppressor. These observations suggest that stress granule biology is tightly associated with DPR-mediated toxicity.
GR and PR induce spontaneous assembly of poorly dynamic stress granules and impair translation
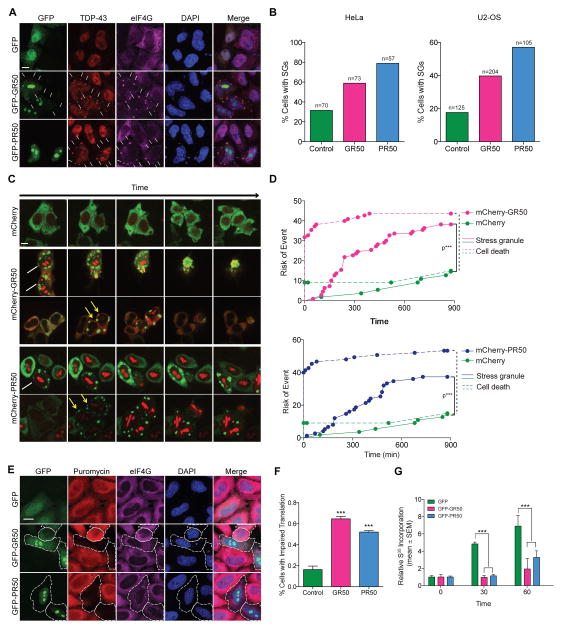
Cytosolic GR50 showed frequent colocalization with stress granule markers (Figure 5A, S4). Although GFP-PR50 was not detectable in stress granules, expression of GR50 or PR50 was associated with spontaneous stress granule assembly in both HeLa and U2-OS cells (Figure 5A, 5B), and numerous stress granule proteins were detected in PR/GR-induced stress granules (Figure S5A–S5E). Live imaging of HeLa cells for 14 hours revealed that these stress granules were poorly dynamic, very rarely disassembled, and their frequency increased with time (Figure 5C, Movie S1–3). Hazard analysis revealed that the risk of death in GR50-positive or PR50-positive cells forming stress granules approached 87.5% and 70.0%, respectively, over the 14-hour window (Figure 5D).
Figure 5. GR and PR dipeptides promote spontaneous assembly of poorly dynamic stress granules and inhibit cellular translation.
(A) Cellular localization of GFP-GR50, GFP-PR50, or GFP and endogenous TDP-43 (red) and eIF4G (purple) in HeLa cells. DAPI is shown in blue. Scale bar, 10 μm. (B) Percentage of cells containing stress granules in HeLa or U2-OS cells following expression of GFP, GFP-GR50 or GFP-PR50. n=2 biological repeats. (C) Stress granule dynamics in HeLa cells as visualized by G3BP1-GFP (green) and mCherry, mCherry-GR50 or mCherry-PR50 (red). Live cells were imaged continuously over 14 hours (also see Movies S1–S3). White arrows, cells with stress granules at the beginning of imaging; yellow arrows, cells spontaneously forming stress granules during imaging. Scale bar, 10 μm. (D) Hazard analysis demonstrating risk of stress granule formation and cell death. ***P < 0.0001 by Log-rank test. (E) Translation efficiency in HeLa cells as visualized by measuring puromycin (red) incorporation into nascent peptides; GFP (green), EIF4G (purple). Scale bar, 10 μm. (F) Quantification of translation in HeLa cells as visualized in (E). n=3 biological repeats, ***P < 0.001 by One Way ANOVA. (G) Translational inhibition as assessed by metabolic assay monitoring incorporation of S35 into newly translated proteins. n=5 biological repeats, ***P < 0.001, **P < 0.01, *P < 0.05, Two Way ANOVA, Dunnett’s post-hoc test. Data are represented as mean ± SEM in (F) and (G). See also Figure S5 and Movies S1–S3.
Stress granules are thought to represent a pool of mRNPs stalled in the process of translation initiation (Anderson and Kedersha, 2009). Therefore, we assessed the impact of GR50/PR50-induced stress granules on translation. Results from two independent methods (puromycin incorporation (Schmidt et al., 2009), Figure 5E, 5F; 35S-Met/Cys metabolic labeling, Figure 5G, S5F) showed that mRNA translation was dramatically reduced in cells expressing GFP-GR50 or GFP-PR50.
GR and PR directly interact with LCD-containing RNA-binding proteins and alter their biophysical properties
RNA-binding proteins with LCDs undergo concentration-dependent LLPS to form liquid droplets, a property believed to contribute to the assembly and liquid properties of stress granules (Lin et al., 2015; Molliex et al., 2015; Murakami et al., 2015; Patel et al., 2015). hnRNPA1 and TIA-1 are prototypical hnRNPs consisting of two or three folded RNA recognition motifs (RRMs), respectively, with C-terminal LCDs (Figures 6A, 6D, S6A, S6D). Under conditions in which hnRNPA1 and TIA-1 formed liquid droplets, tetramethylrhodamine (TAMRA)-labeled GR20 (TAMRA-GR20) and PR20 (TAMRA-PR20) peptides were recruited into these protein-dense droplets, whereas TAMRA-PA20 and TAMRA-GP20 peptides were not (Figure 6B, 6E). Importantly, the presence of GR20 or PR20 substantially decreased the critical concentration of hnRNPA1 or TIA-1 required for LLPS, whereas neither PA20 nor GP20 impacted LLPS by hnRNPA1 or TIA-1 (Figure 6C, F). Consistent with these observations, FRAP analysis showed that GR20 or PR20 significantly altered dynamic exchange of hnRNPA1 (Figure S6B, S6C) and TIA-1 (Figure S6E, S6F) between the dense droplet phase and the surrounding mono-disperse phase, suggesting that these peptides strengthen the multivalent interactions that comprise the liquid phase of hnRNPA1 and TIA-1. Moreover, incorporation of GR20 or PR20 changed the material properties of hnRNPA1 droplets, which are normally highly fusible and readily wet the coverslip (Movie S4), but with addition of GR20 or PR20 fail to wet the surface of the coverslip or to fuse over time (Movie S5), suggesting that these droplets have higher surface tension (Figure S6).
Figure 6. GR and PR dipeptides alter biophysical properties of hnRNPA1 and TIA-1 and alter stress granule dynamics in live cells.
(A) Schematic of hnRNPA1. RRM, RNA-recognition motif; LCD, low-complexity sequence domain. (B) Incorporation of DPRs into phase-separated liquid droplets of hnRNPA1. Fluorescence images of hnRNPA1 (DIC) mixed with TAMRA-labeled polypeptides. Scale bar, 10 μm. (C) Phase diagram of hnRNPA1 as a function of concentration of protein concentration and polypeptides concentration. GR20 and PR20 significantly shift the phase diagram. Red and green symbols indicate that the samples are shown as being in either was in the one-phase (red) or the two-phase (green) regime, respectively. (D) Schematic of TIA-1. (E) Incorporation of DPRs into phase-separated liquid droplets of TIA-1. Fluorescence images of TIA-1 (DIC) mixed with TAMRA-labeled polypeptides. Scale bar, 10 μm. (F) Phase diagram of TIA-1 using the same analysis as for (C). (G) G3BP1-GFP-transfected HeLa cells were either treated with NaAsO2 to induce stress granules or co-transfected with mCherry-GR50, followed by photobleaching (yellow circle) and monitoring for fluorescence recovery. Scale bar, 10 μm. (H) Fluorescence of the photobleached region over time. Data are represented as mean ± SEM for n=7–8, ***P < 0.001 by Student t-test, paired. See also Figure S6 and Movies S4–S5.
GR and PR alter stress granule dynamics in live cells
We next monitored G3BP1, which directly interacts with GR and PR (Figure S4B), to assess the impact of GR and PR on stress granule dynamics in living cells. G3BP1-GFP was distributed diffusely in the cytoplasm of untreated HeLa cells (Figure 5C), but rapidly assembles into stress granules upon NaAsO2 treatment (Figure 6G). FRAP analysis showed G3BP1-GFP in stress granules is in rapid dynamic equilibrium with the cytoplasm (Figure 6G, 6H). mCherry-GR50 or mCherry-PR50 expression induced spontaneous assembly of stress granules and impaired the dynamic exchange of G3BP1-GFP between the stress granule and the cytoplasm. GR or PR reduced the rate of fluorescence recovery and decreased the mobile fraction after photobleaching of G3BP1-GFP within stress granules (Figure 6G, 6H), suggesting that these peptides strengthen the multivalent interactions that comprise the liquid phase of stress granules in living cells.
GR and PR alter nuclear speckle dynamics and Cajal body assembly in live cells
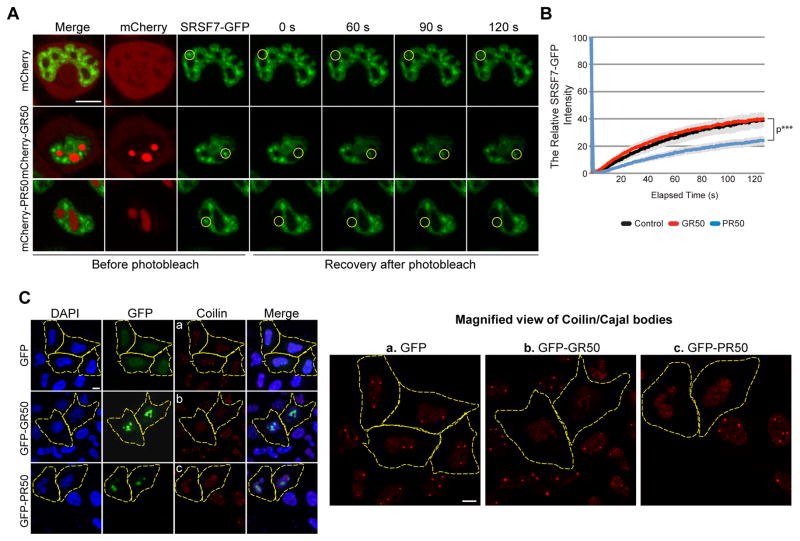
Nuclear speckles, also known as interchromatin granule clusters, are membrane-less nuclear organelles enriched in pre-mRNA splicing and transcription factors (Wansink et al., 1993) and recently found to have dynamic, liquid-like properties (Marzahn et al., 2016). Cajal bodies are another type of membrane-less nuclear organelle, enriched in snRNPs and serving as a site of spliceosome assembly (Machyna et al., 2015). Components of both of these membrane-less organelles were identified in the GR/PR interactome (Figure 1A and Table S1) and as genetic modifiers of GR toxicity in Drosophila (Figure 2E, S2C, S2D). For example, the splicing factor and nuclear speckle component SRSF7 interacted strongly with GFP-PR50 but weakly with GFP-GR50 (Figure S1C). FRAP analysis showed that fluorescence recovery of SRSF7-GFP in nuclear speckles was significantly reduced by mCherry-PR50 but not mCherry-GR50 (Figure 7A, 7B). The splicing factor SRSF2 (also called SC-35) is another nuclear speckle component that was not identified in the GR/PR interactome. In contrast to SRSF7, SRSF2 mobility was not influenced either by mCherry-PR50 or mCherry-GR50, suggesting that GR and PR specifically impact the dynamics of proteins they interact with (Figure S7). To examine Cajal bodies, we stained for their principal structural component, Coilin (Machyna et al., 2015). In untransfected and GFP-expressing GFP cells, we detected multiple Cajal bodies in most nuclei, but cells expressing GFP-PR50 or GFP-GR50 were often devoid of Cajal bodies (Figure 7C). Thus nuclear speckles and Cajal bodies are additional membrane-less organelles whose dynamics and/or assembly are impaired by GR and PR.
Figure 7. GR and/or PR dipeptides impair the assembly or dynamics of other membrane-less organelles.
(A) Nuclear speckle dynamics in HeLa cells transfected with SRSF7-GFP and mCherry-GR50 or -PR50 constructs were analyzed using FRAP. The yellow circle marks the photobleached region. Representative images of the same area before and after photobleaching are shown. Scale bar, 10 μm. (B) Fluorescence of SRSF7-GFP in the photobleached region over time. Data are represented as mean ± SEM for n=10, ***P < 0.001 by Student t-test, paired. (C) Assembly of Cajal bodies in HeLa cells transfected with mCherry-PR50 or -GR50 were immunostained with anti-Coilin antibody. For panels a – c (Coilin staining), magnified images are shown on the right. Scale bar, 10 μm. See also Figure S7.
Discussion
Despite efforts to define the basis of repeat expansion toxicity in C9-ALS/FTD, the field has struggled to integrate seemingly disparate findings, such as evidence of impaired nucleolar function (Kwon et al., 2014; Wen et al., 2014), disturbances in nucleocytoplasmic transport (Freibaum et al., 2015; Jovičić et al., 2015; Zhang et al., 2015), altered RNA splicing (Prudencio et al., 2015), impaired trafficking of RNA granules (Alami et al., 2014; Schweizer Burguete et al., 2015) and impaired translation (Kanekura et al., 2016). Furthermore, despite extensive clinicopathological overlap in ALS/FTD caused by mutation in C9ORF72 and RNA-binding proteins, a cohesive view of pathogenesis has been lacking. Here we unite these pathogenic mechanisms, demonstrating that DPRs, in particular GR and PR, disturb phase transitions mediated by LCDs. This finding mirrors the defects in phase transitions observed with disease-causing mutations LCDs in TDP-43, FUS, and hnRNP1 (Lin et al., 2015; Molliex et al., 2015; Murakami et al., 2015; Patel et al., 2015). Furthermore, the altered assembly, dynamics and function of membrane-less organelles that result from disturbed phase transitions fully account for the widespread cellular abnormalities observed in ALS/FTD.
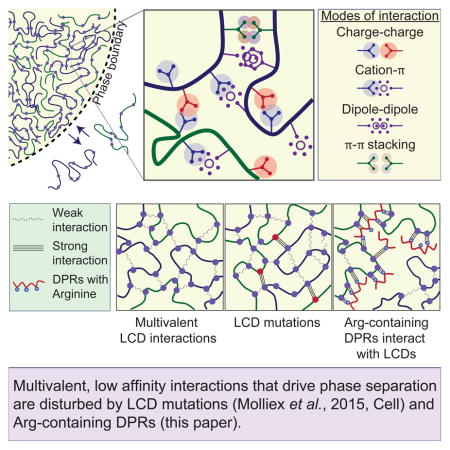
We are only beginning to understand the molecular details that mediate LLPS in living cells, but a common underlying principle seems to be the formation of a network of relatively weak, multivalent interactions (Li et al., 2012). Many of the proteins that drive LLPS belong to the general class of intrinsically disordered proteins that make up about one-third of the human proteome (Oldfield and Dunker, 2014). Examination of the LCDs that underlie LLPS reveals a sequence bias that provides a biophysical basis for the weak and dynamic interactions that occur between the macromolecules concentrated within the dense, liquid phase. Within these long stretches of low overall amino acid diversity is an enrichment in specific amino acids, such as glycine and those with polar, positively or negatively charged, or aromatic sidechains (Brangwynne et al., 2015). Often these residues are present within LCDs as reiterated short motifs, providing a potential basis for multivalency. While the nature of the interactions mediated by LCDs that permit LLPS remains unclear, certain clues are beginning to emerge. For example, we previously showed that LLPS of hnRNPA1, a component of stress granules, is enthalpy-driven and that aromatic and electrostatic interactions are driving forces (Molliex et al., 2015). Indeed, hnRNPA1 is enriched in the aromatic residues phenylalanine and tyrosine and the positively charged residue arginine relative to the overall eukaryotic proteome (Hormoz, 2013). Moreover, the LCD of hnRNPA1 is patterned; aromatics and positively charged residues, mainly arginines, are well distributed. Thus, these features may represent reiterated interaction motifs in the background of a polar polymer and enable multivalent, weak interactions that drive LLPS during stress granule assembly. Analogously, NPM1, containing multivalent acidic LCDs, undergoes LLPS with nucleolar proteins that contain multivalent, linear arginine-rich motifs (Mitrea et al., 2016). The importance of arginine-rich motifs in both of these examples likely relates to the ability of this residue to participate in charge-charge and Pi-cation interactions within the multivalent network (Brangwynne et al., 2015). However, this feature also creates an exquisite vulnerability to the inappropriate expression of the C9ORF72 DPRs GR and PR. Indeed, nature would be hard-pressed to engineer a more potent toxin to membrane-less organelles than polymers of arginine.
As discussed earlier, an appreciation for the importance of the RNA-binding protein TDP-43 in ALS and FTD pathogenesis (Neumann et al., 2006; Kim et al., 2013; Kwiatkowski et al., 2009; Vance et al., 2009), as well as inclusion body myopathy (IBM) (Salajegheh et al., 2009), has emerged. Disease-causing mutations in two familial forms of IBM were also identified in the RNA-binding proteins hnRNPDL and TIA-1 (Hackman et al., 2013; Klar et al., 2013; Vieira et al., 2014), with the pathogenic TIA-1 mutations impairing stress granule dynamics (Hackman et al., 2013). Disease-causing mutations in these RNA-binding proteins are most frequently located in the LCDs (Taylor et al., 2016) and can alter their biophysical properties, accelerate the formation of amyloid-like fibrils, and disrupt the dynamics and functions of membrane-less organelles (Kim et al., 2013; Lin et al., 2015; Molliex et al., 2015; Murakami et al., 2015; Patel et al., 2015). We propose a common mechanism of pathogenesis for genetically overlapping forms of ALS, FTD and IBM, namely disturbances in the biophysical properties of LCDs that disrupt the dynamics and functions of multiple membrane-less organelles.
STAR★METHODS
Detailed methods are provided in the online version of this paper and include the following:
KEY RESOURCES TABLE
-
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Generation of Drosophila Lines and Drosophila Stocks
Cell Culture
CONTACT FOR REAGENT AND RESOURCE SHARING
-
METHODS DETAILS
Proteomic Analysis
Define Proteins with Low Complexity Sequence Domain
Drosophila Eye and Viability Analysis
Transient Transfection
Immunofluorescence and Microscopy
Structured Illumination Microscopy (SIM)
Fluorescence Recovery after Photo Bleaching (FRAP)
Live Cell Imaging and Hazard Analysis
Cell Toxicity Analysis
Immunoblotting
Puromycin Incorporation Assay and [35S]-Methionine Labeling
Peptide Synthesis
Circular Dichroism
Peptide Treatment in Cells
Protein Expression and Purification
In vitro FLAG pull-down assay
In vitro NPM1 Phase Separation Assays
In vitro Determination of hnRNPA1 and TIA-1 Phase Diagrams
QUANTIFICATION AND STATISTICAL ANALYSIS
-
DATA AND SOFTWARE AVAILABILITY
Data Resources
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for reagents may be directed to, and will be fulfilled by the corresponding author J. Paul Taylor (JPaul.Taylor@STJUDE.ORG).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Generation of Drosophila Lines and Drosophila Stocks
The GFP-GP47, GFP-GA50, and GFP-GR50 Drosophila stocks used were previously described (Freibaum et al., 2015). The GFP-PA50 and GFP-PR50 stocks were kindly provided by Udai Pandey (Wen et al., 2014). Flies were raised at 25°C on a standard diet. RNAi stocks were obtained from either the Bloomington Drosophila Stock Center or the Vienna Drosophila RNAi Center. DPR expression Drosophila lines RNAi lines were obtained from either the Bloomington Drosophila Stock Center or the Vienna Drosophila RNAi center. Drosophila lines used are listed in Table S2.
Cell Culture
HEK 293T, HeLa, U2OS, and Neuro2a cells were cultured in Dulbecco’s modified Eagle’s medium (HyClone) supplemented with 10% fetal bovine serum and 1% antibiotics, and maintained at 37°C in a humified incubator with 95% air and 5% CO2.
METHODS DETAILS
Proteomic Analysis
pEGFP-C3, pEGFP-GR50, pEGFP-PR50, pEGFP-GA50, pEGFP-GP47 or pEGFP-PA50 plasmid was transfected into HEK 293T cells by using lipofectamine 2000 (Invitrogen) as manufacturer’s instruction. Forty-eight hours post-transfection, cells were lysed in lysis buffer [10mM Tris/Cl pH 7.5; 150mM NaCl; 0.5mM EDTA; 0.5% NP-40] with protease inhibitor (cOmplete™ Mini EDTA-free protease inhibitor Cocktail, Roche). 1mg of cell extracts were incubated with GFP-Trap®_MA beads, and incubated for 1 hour at 4°C. Magnetically separated samples were resuspended and boiled in 2x LDS sample buffer (NuPAGE® LDS sample buffer, Life technologies), and then were analyzed by mass-spectrometry. Mass-spectrometry results were analyzed by SAINT scoring method, CRAPome (Mellacheruvu et al., 2013), and DPR interacting proteins showing over 0.9 SAINT score were analyzed by DAVID bioinformatics tool. The overlapping probability between GR and PR interactors (SAINT >= 0.9) was calculated based on the hypergeometric distribution. The population size consisted of all unfiltered GR or PR interactors (SAINT >= 0.0).
Define Proteins with Low Complexity Sequence Domain
Swiss-Prot annotated human proteome was downloaded from the UniProt database (UniProt, 2015). SEG with default parameters (Wootton and Federhen, 1996) was used to search for the low complexity sequence region (LCR) in the human proteome. After examining the LCR length distribution, protein region with a span of 20 LCR residues was defined as a low complexity sequence domain.
Drosophila Eye and Viability Analysis
Phenotypic analysis of DPR expression in the Drosophila eye was assessed by crossing GFP, GR50, PR50, GA50, GP47 and PA50 lines to GMR-GAL4 or OK371-GAL4 at different temperatures. Eye phenotypes are representative images resulting from Drosophila crosses, the phenotypes were validated by 4 independent crosses. GFP-GR50 expression Drosophila lines were crossed with RNAi lines targeting GR50 or PR50 interacting proteins. Viability was checked by counting adults and dead pupae.
Transient Transfection
For transient transfection, lipofectamine 2000 (Invtrogen) for 293T and FuGENE 6 (Promega) for HeLa were used as per the manufacturer’s instructions.
Immunofluorescence and Microscopy
Cells were seeded in 8-well chamber slides (Millipore), and grown for 24h, prior to being transfected with DPR expressing plasmids, and incubated for an additional 48h. Cells were then fixed with 4% paraformaldehyde (Electron Microscopy Science), and then permeabilized with 0.2% Triton X-100. Mouse anti-G3BP1 (BD Transduction Lab), mouse anti-EIF4G (1:200, Santa Cruz Biotechnology), mouse anti-5.8s rRNA oligonucleotides specific antibody (Y10b, Novus Biologicals), goat anti-eIF3η (1:200, Santa Cruz Biotechnology), rabbit anti- Coilin (1:400, Abcam), rabbit anti-TDP43 (1:350, Proteintech), mouse anti-NPM1 (1:200, Thermo), rabbit anti-PABPC1 (1:400, Abcam), rabbit anti-PTBP1 (1:200, Millipore), goat anti-IPO7 (1:400, Abcam), and goat anti-TIA-1 (1:400, Santa Cruz Biotechnology) were used as primary antibodies for immunofluorescence. For visualization, the appropriate host-specific Alexa Fluor 488, 555, or 647 (Life Technologies) secondary was used.
For immunofluorescence microscopic imaging, slides were mounted with ProLong® Diamond Antifade Mountant with DAPI (Invitrogen). Images were captured using LSM 780 NLO (Zeiss) or Leica TCS SP8 STED 3X confocal microscope with a 63x objective.
Structured Illumination Microscopy (SIM)
HeLa cells were seeded and grown on sterilized 18mm round #1.5 glass coverslips (Warner Instruments). 24 hours after seeding, the cells were transfected with DPR expressing plasmids and incubated for 48 hours. Then the cells were fixed with 4% paraformaldehyde (Electron Microscopy Science), permeabilized with 0.5% Triton X-100 (Fisher Scientific), and blocked with 3% BSA (Sigma). Rabbit anti-Fibrillarin (1:1000, abcam) and mouse anti-UBF (1:150, Santa Cruz Biotechnology) were used as the primary antibodies. Alexa Fluor 555 anti-rabbit and Alexa Fluor 647 anti-mouse (Life Technologies) were used as the secondary antibodies. The coverslips were mounted on slides with ProLong® Gold Antifade Mountant with DAPI (Invitrogen). Super resolved three-dimensional images of fixed cells were taken on a Zeiss Elyra PS.1 microscope (Carl Zeiss Microscopy Inc., Thornwood, NY) with Structured Illumination Microscopy (SIM) technique using an Apochromat 63×1.4 NA oil objective lens. SIM images were taken at five rotations of the excitation grid with five phases per rotation. Raw images were acquired with 32nm pixel resolution in XY dimension and 90nm spacing between Z-planes. Images were processed with the SIM module of Zen software package (Carl Zeiss Microscopy, Thornwood, NY). In order to determine the diameter of the Fibrillarin cores, a line scan analysis was applied to the processed SIM images using the Zen software. Only fibrillarin cores, which displayed a positive UBF signal within them, indicating that the fibrillarin was surrounding a fibrillar center, were measured. For each construct the nucleoli of at least 12 cells were analyzed, and were only considered if there were at least 10 measurable fibrillarin cores within the nucleoli of the cell.
Fluorescence Recovery after Photo Bleaching (FRAP)
For the FRAP assay, HeLa cells were seeded onto 4-well chamber slides (Nunc), and incubated for 24h. After 24h, cells were transfected with G3BP1-GFP, or co-transfected with G3BP1-GFP and mCherry DPR expressing plasmids. After 48h incubation, G3BP1-GFP transfected cells were treated with sodium arsenite for 15 min, to induce stress granules. Co-transfected cells with G3BP1-GFP and mCherry-DPR were not treated with sodium arsenite. G3BP1-GFP positive stress granules in the transfected and cotransfected cells were photobleached and the GFP signal intensity was measured before and after photobleaching. For NPM1, NCL, SRSF7, SRSF2 or DDX47 FRAP, GFP-tagged NPM1, NCL, SRSF7, SRSF2 or DDX4 expressing plasmids were co-transfected with mCherry or mCherry-tagged DPR expressing plasmids, and incubated for 48h. The signal intensity of GFP in the nucleoli was measured before and after photobleaching.
Live Cell Imaging and Hazard Analysis
The G3BP1-GFP plasmid was constructed by inserting the G3BP1 (HsCD00042033) in the peGFP-N1 (Clontech) with primers containing the restriction enzyme sites EcoRI and BamHI in the 5′ and 3′ site of G3BP1. All live-cell imaging experiments were performed using a Marianas 2 spinning disk confocal (SDC) imaging system on a Zeiss Axio Observer inverted microscope platform. HeLa cells were plated in 4-well lab-Tek chambered coverglass and transfected with indicated plasmids. Before imaging, change the medium to FluoroBrite medium. When imaging, cells were maintained in a humidified 5% CO2 incubator at 37 °C with an environmental control chamber. Multi-positions and definite focus were used during the imaging. Time-lapse images were collected for 870 min using a Zeiss Plan-Apochromat 40 1.3 NA oil objective and Evolve 512 EMCCD camera. Images were analyzed with SlideBook 6 software. For statistical analysis, the imaging time points at which a cell formed stress granules and a cell died were defined. Kaplan–Meier curves were used to estimate hazard functions for stress granule formation and cell death with commercially available software (Prism 6). Differences in Kaplan–Meier curves were assessed with the log-rank (Mantel-Cox) test.
Cell Toxicity Analysis
Cell toxicity of Neuro-2a cells was assayed using Vybrant® MTT Cell Proliferation Assay Kit according to the manufacturer’s instructions. Briefly, Neuro-2a cells were seeded at a density of 5000 cells per well into 96-well plates, and after 24h incubation, were transfected with DPR expressing plasmids. 48h after transfection, the media was replaced with fresh culture media and MTT reagent was added at a 1:10 volumetric ratio to the media. After a 4h incubation at 37°C, 100μL SDS-HCL solution was added, and the plate was incubated at 37°C for another 4h. Absorbance of the samples was read at 570 nm and DPR toxicity was determined from the growth relative to the control.
Immunoblotting
For Drosophila samples, adult flies were frozen with dry ice and vortexed to remove the head or thorax. Samples from each genotype were homogenized in RIPA buffer with proteinase inhibitor cocktail added. For cultured cells, cells were lysed in RIPA buffer with proteinase inhibitor cocktail added. Sample was mixed with 4X sample buffer (1 M Tris-HCl (pH 6.8), 8% SDS, 40% glycerol, 0.1% bromophenol blue) and boiled for 5 min, separated on an SDS gel and transferred to a membrane. The membrane was blocked, probed with primary antibody, and incubated with secondary antibody. The signal was visualized with either chemiluminescent substrate (SuperSignal West Pico; Pierce) or by using an Odyssey Fc (Li-Cor). Primary western blot antibodies were anti-GFP (AB3080, Millipore or SC-9996 Santa Cruz Biotechnology), anti-β-actin antibody (4967; Cell Signaling Technology or sc-1616, Santa Cruz Biotechnology), anti-KPNA2 (ab70160, Abcam), anti-LBR (ab32535, Abcam), anti-Nucleolin (ab22758, Abcam), anti-NPM1 (MA5-12508, Thermo), anti-C1QBP (#5734, Cell signaling), anti-PABPC1 (Ab21060, Abcam), anti-PTBP1 (MABE623, Millipore), anti-G3BP1 (6111126, BD biosciences), anti-SRSF7 (11044-1-AP, Proteintech), anti-Flag (F7425, Sigma), anti-His (ab18184, Abcam), and anti-GST (71-7500 or MA4-004, Thermo). Species-specific IRDye® secondary antibodies were used and visualized using Odyssey® FC imaging system, and the acquired images were analyzed using Image Studio (LI-COR) software according to the manufacturer’s instructions.
Puromycin Incorporation Assay and [35S]-Methionine Labeling
HeLa cells were incubated for 48 hours after DPR expressing plasmids transfection. Then the media was removed and replaced with fresh media containing 1μM puromycin and incubated for 1 hour. Following incubation, the cells were fixed, permeabilized, and stained with anti-puromycin (3RH11, Kerafast) and anti-EIF4G (Santa Cruz Biotechnology). Alexa Fluors 555 and 647 were used as secondaries for visualization. Images were captured using a LSM 780 NLO (Zeiss) confocal microscope with a 63x objective. The percentage of DPR expressing cells with impaired translation was determined from the absence of puromycin signal. For [35S]-Methionine labeling, DPR expressing plasmids were transfected into HEK 293T, and incubated for 48h. Medium was exchanged to Met-/Cys- medium for 15 min, then Met-/Cys- medium was replaced to [35S]Met/Cys medium. After 30min incubation, cells were washed with PBS and harvested. Cells were lysised by RIPA buffer, then protein concentration and cpm/ul was measured. The [35S] signal was normalized to protein concentration.
Peptide Synthesis
Flag, TAMRA or FAM labeled DPR peptides (GR20, PR20 GP20, and PA20) and the SURF6 peptide (299RRAQRQRRWEKRTAGVVEKMQQRQDRRR326) were synthesized by the Hartwell Center for Bioinformatics and Biotechnology at St. Jude Children’s Research Hospital, Molecular Synthesis Resource, using standard solid phase peptide synthesis chemistry. The peptides were reconstituted from lyophilized form into the assay buffer, the pH was re-adjusted accordingly and the concentration was determined by absorbance at 280 nm.
Circular Dichroism
200 μM peptide stocks in 10 mM Tris, 150 mM NaCl, pH 7.5 were diluted to 10 μM final concentration in 10 mM Tris pH 7.5 or 10 mM Tris, 150 mM NaCl, pH 7.5 buffers to yield the low ionic strength and physiological ionic strength samples, respectively. CD spectra were recorded at 25 °C, in an Aviv 202 circular dichoism spectrometer (Aviv Biomedical, Inc, Lakewood, NJ), in a 1 mm path length cuvette, with a signal averaging of 15 s.
Peptide Treatment in Cells
Prior to peptide treatment, cells were seeded onto 8-well chamber slides. After 24h incubation, DMSO solubilized 10μM of FAM labeled peptides were added to the medium for 60 minutes. Then cells were subjected to fixation and permeabilization step, and anti-NPM1 antibody was used to validate nucleolus localization.
Protein Expression and Purification
The expression and purification of human hnRNPA1 protein were previously described (Freibaum et al., 2015). The human TIA-1 gene was subcloned into the pETite N-His SUMO Kan expression vector (Lucigen). Human TIA-1 was expressed as His-SUMO-tagged fusion protein in HI-Control™ BL21(DE3) Chemically competent cells (SOLOS) (Lucigen) in LB medium. Cells were lysed in 50 mM HEPES (pH 7.5), 250 mM NaCl, 30 mM imidazole, 2 mM β-mercaptoethanol, and complete protease inhibitor cocktail without EDTA (Roche) with a microfluidizer. The cleared lysate was loaded onto a gravity NiNTA column, washed with lysis buffer, and eluted in 50 mM HEPES (pH 7.5), 250 mM NaCl, 300 mM imidazole, and 2 mM β-mercaptoethanol. The proteins were treated with 0.1 mg/ml RNase A (Roche) for 5 min at 37°C. The protein was purified by linear gradient elution on ion exchange chromatography with a HiTrap Q column (GE Healthcare). The fractions were analyzed by SDS-PAGE, pooled, and concentrated. They were then subjected to size exclusion chromatography on a Superdex 200 16/60 column (GE Healthcare) equilibrated in 50 mM HEPES (pH 7.5), 400 mM NaCl, and 5 mM DTT. The fractions were analyzed by SDS-PAGE, pooled, concentrated, and stored at −80 °C freezer for further use.
The human NPM1 gene was subcloned into the pET28 vector (Novagen, Darmstadt, Germany) with an N-terminal hexa-histidine tag and a TEV protease cleavage site. NPM1 was expressed in E. Coli BL21 (DE3) strain (Novagen, Darmstadt, Germany) in Luria broth, and expression was induced at OD600 ~ 0.6 with 100 mg/mL IPTG. The cultures were incubated with shaking at 20 °C overnight. The bacteria were harvested by centrifugation and lysed by sonication in buffer A (25 mM TRIS, 300 mM NaCl, 10 mM β-mercaptoethanol, pH 7.5), supplemented with EDTA-free protease inhibitors (SigmaFAST, Sigma-Aldrich, St. Louis, MO). The soluble fraction of the lysate was loaded onto a NiNTA column and the bound protein eluted with a linear gradient of buffer B (25 mM TRIS, 300 mM NaCl, 10 mM β-mercaptoethanol, 500 mM Imidazole, pH 7.5). The hexa-histidine tag was removed by cleavage with TEV protease in an overnight dialysis step, against 4 L of Buffer C (10 mM TRIS, 150 mM NaCl, 2 mM DTT, pH 7.5) at 4 °C. The cleaved product was subsequently purified on a C4 HPLC column, loaded in 0.1% trifluoroacetic acid (TFA) in H2O and eluted with a linear gradient of 0.1% TFA in acetonitrile. Fractions containing NPM1, confirmed by SDS-PAGE, were flash frozen and lyophilized. The lyophilized protein was solubilized in 6 M guanidinium hydrochloride buffer at a NPM1 monomer concentration of 100 μM and refolded by dialysis against 3 changes of a 100-fold excess of Buffer C at 4°C. Fluorescent labeling of NPM1 with AlexaFluor 488 was performed as previously described (Mitrea et al., 2016).
In vitro FLAG pull-down assay
Incubated the GR20-FLAG and PR20-FLAG peptides with anti-FLAG M2 antibody-conjugated magnetic beads (Sigma) in IP buffer (150 mM NaCl, 50 mM Tris pH 7.5, 1 mM EDTA, 0.5% NP40, 10% Glycerol) for 1h at 4°C, then added purified His-NPM1, GST-NCL, His-SUMO-hnRNPA1, His-SUMO-TIA-1, or GST-G3BP1. Washed the beads for 3 times with washing buffer (150 mM NaCl, 50 mM Tris pH 7.5, 1 mM EDTA, 0.5% NP40, 10% Glycerol), followed by elution with FLAG peptides in elution buffer (150 mM NaCl, 50 mM Tris pH 7.5, 1 mM EDTA, 0.05% NP40, 10% Glycerol, FLAG peptide: 500 μg/ml).
In vitro NPM1 Phase Separation Assays
All in vitro assays were performed in Buffer C. The ternary mixtures were prepared by first forming the complexes or liquid-like droplets comprised of NPM1 (containing 1 μM AlexaFluor 488-labeled NPM1) with unlabeled SURF6 peptide, followed by the addition of the poly-xR peptides. 1 μM Alexa488-NPM1 or 1 μM TAMRA-poly-xR peptides were included in the samples for fluorescence microscopy detection. The samples were incubated at room temperature for 1 hour and transferred to 16-well chambered slides (Grace Bio-Labs, Bend, OR), pre-coated as described (Mitrea DM et al, eLife (2016)). Fluorescence confocal microscopy images were collected using a Zeiss Axio Observer (Carl Zeiss Microscopy, Jena, Germany) microscope with a 63X 1.40 NA oil magnification objective. Images were processed in Slidebook 6.0 (Intelligent Imaging Innovations, Gottingen, Germany).
In vitro Determination of hnRNPA1 and TIA-1 Phase Diagrams
For the phase separation of hnRNPA1 with DPRs, 50 μM hnRNPA1 were mixed with 50 μM TAMRA labeled polypeptides and DIC images were taken at RT. 100 mg/ml ficoll was added to the samples. For the phase diagram of hnRNPA1, experiment was performed in 50 mM HEPES, 150 mM NaCl, 5 mM DTT at 16 °C. For the phase separation of TIA-1 with DPRs, 5 μM TIA-1 were mixed with 5 μM TAMRA labeled polypeptides at RT. 100 mg/ml ficoll was added to the samples. For the phase diagram of TIA-1, experiment was performed in 50 mM HEPES, 150 mM NaCl, 5 mM DTT at 22 °C. The presence or absence of droplets was scored using a Linkam PE100 thermal stage mounted on a Marianas spinning disk confocal (SDC) imaging system on a Zeiss Axio Observer inverted microscope platform. Sealed sample chambers containing protein solutions comprised coverslips sandwiching Secure-Seal™ Imaging Spacers (0.12 mm depth) were taped on the PE100 silver heating/cooling block.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical parameters including the definitions and exact values of n (e.g., number of biological repeats, number of flies, number of cells, etc), distributions and deviations are reported in the Figures and corresponding Figure Legends. Data of Drosophila viability analysis, MTT assay, fibrillar center measurement, translation assay and FRAP assay were expressed as mean ± SEM. p > 0.05, not significant, *p < 0.05, **p < 0.01 and ***p < 0.001 by Two Way ANOVA, One Way ANOVA, Log-rank test or Student’s t-test. Statistical analysis was performed in GraphPad Prism or Excel.
DATA AND SOFTWARE AVAILABILITY
Data Resources
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD004938.
Supplementary Material
(A) HEK 293T cells were transfected with GFP or the indicated GFP-tagged dipeptide and immunoprecipitated with an antibody targeting the GFP tag. The samples were separated by SDS-PAGE, and proteins were visualized with SYPRO-Ruby. (B) Western blot performed with an anti-GFP antibody was used to visualize transfected dipeptides (or GFP control) in both input and immunoprecipitated samples. (C) Western blot analysis of immunoprecipitated proteins with indicated antibodies to validate results from proteomic analysis. (D) Venn diagrams illustrating the overlap among DPR-interacting proteins at various confidence levels as indicated by SAINT analysis.
Live HeLa cells co-transfected with G3BP1-GFP and mCherry constructs were imaged continuously over 14 hours. Related to Figure 5.
Live HeLa cells co-transfected with G3BP1-GFP and mCherry-GR50 constructs were imaged continuously over 14 hours. Related to Figure 5.
Live HeLa cells co-transfected with G3BP1-GFP and mCherry-PR50 constructs were imaged continuously over 14 hours. Related to Figure 5.
Wetting and fusion of hnRNPA1 droplets. Related to Figure 6.
300 μM protein undergoes LLPS, and droplets of hnRNPA1 exhibit wetting when they encounter the surface of the coverslip as shown in Molliex et al. (Molliex et al., 2015).
Arginine-containing peptides increase the surface tension of hnRNPA1 droplets. Related Figure 6.
150μM hnRNPA1 protein mixed with 50 μM PR20 peptide undergoes LLPS, however hnRNPA1 droplets do not wet the surface or fuse over time due to high surface tension.
(A) GFP-GR50 protein levels, but not mRNA levels, were decreased in a dose dependent manner in both pupae and adult Drosophila by expression of an intrabody (deGradFP) targeted against the GFP sequence. (B) Expression of the deGradFP intrabody rescues the rough eye phenotype caused by GFP-GR50 dipeptide in a dose dependent manner when protein expression is restricted to the Drosophila eye using the GMR driver (C) A genetic screen of RNAi lines targeting DPR interacting proteins in Drosophila using egg-to-adult viability as a read out. Genetic suppressors GR50 toxicity are labeled in green whereas enhancers of GR50 toxicity are labeled in red, as indicated in the key. W1118 and v60100 lines were used as controls. (D) Integration of the genetic screen with gene ontology results depicting GR50 suppressors in green and enhancers in red. (E) Strong suppressors of GR50 toxicity were predominantly found to suppress expanded G4C2-mediated toxicity using the (G4C2)58 Drosophila model (Freibaum et al., 2015). Common suppressors were largely specific as most of these RNAi lines failed to suppress non-specific toxicity caused by expression of the androgen receptor polyQ expansion (ARpolyQ)52. Cluster dendrogram analysis demonstrated that the overlap of shared modifiers of GR and (G4C2)58 toxicities was significantly greater than shared modifiers with AR(polyQ)52 toxicity. (F) Strong enhancers of GR50 toxicity were predominantly found to enhance C9orf72 mediated toxicity using a (G4C2)58 model.
(A) HeLa cells expressing either transfected GFP or GFP-tagged DPR were immunostained with NPM1 (red) and G3BP1 (purple) specific antibodies. DAPI was used to visualize the nucleus. GR50 and PR50 but not other DPRs or GFP were found to colocalize with NPM1 and induce the formation of G3BP1 positive cytoplasmic stress granules. Scale bar, 10 μm. (B) The percentage of cells in which the transfected DPR was found to localize to nucleoli. (C) FAM-labeled GR20 or PR20 peptides (green, 10μM) were incubated in HeLa cell culture media for 60 minutes and their nucleolar localization was determined using an NPM1 specific antibody (red). Scale bar, 10 μm. (D) HeLa cells were transfected with GFP, GFP-GR50, GFP-PR50 or GFP-GA50. GFP-positive nucleoli were analyzed by FRAP. The yellow circle marks the photobleached region. Representative images of the same area before and after photobleaching at different times are shown. Scale bar, 10 μm. (E) Signal intensity of GFP fluorescence in the photobleached yellow circle region was plotted over time. The average fluorescence before photobleaching was counted as 100%. Data are represented as mean +/− SEM. n=10 cells per sample, P values for GFP-GR50, GFP-PR50 or GFP-GA50 (over GFP control) < 0.001 by Student t-test, paired. (F) HeLa cells were transfected with GFP-GR50, GFP-PR50, or GFP-GA50 for 48 hours, then changed the media with 3.5% 1,6-hexanediol and imaged the cells continuously for 1 hour. (G) HEK293T cells were transfected with GFP, GFP-GR50, GFP-PR50, GFP-GA50, GFP-GP47, or GFP-PA50, incubated for 48 hours, and then sequentially extracted with RIPA buffer and urea buffer. Immunoblotting was conducted with anti-GFP antibody.
(A) Circular dichroism spectra of 10 μM peptides at 25 °C, in 10 mM Tris pH 7.5 buffer, under low ionic strength (red) and physiological ionic strength (black) conditions. (B) Flag pull-down assay reveals that GR20 and PR20 peptides directly interact with NPM1, NCL, hnRNPA1, G3BP1, and TIA-1 in vitro. (C) Titrations of GP20, PA20, GR20 and PR20 (concentrations indicated at the top) into liquid-phase droplets comprised of 1:1 NPM1:SURF6 peptide prepared at 20 μM. The GR20 and PR20 peptides entered into the NPM1:SURF6 peptide droplets at concentrations at and below 1:1 stoichiometry with NPM1 (DPR:NPM1) but the droplets were disrupted at higher stoichiometries. The GP20 and PA20 peptides neither entered into nor disrupted the NPM1:SURF6 peptide droplets at any concentrations tested. Alexa488-NPM1 (left in each panel) and TAMRA-DPR peptides (middle) were detected using confocal fluorescence microscopy. These colors were merged on the right of each panel. Scale bar, 10 μm. (D) HeLa cells transfected with DDX47-GFP either with mCherry-GR50 or mCherry-PR50 constructs were analyzed using FRAP. The yellow circle marks the photobleached region. Representative images of the same area before and after photobleaching at different times are shown. Scale bar, 5 μm. (E) Signal intensity of fluorescence of DDX47-GFP in the photobleached yellow circle region was plotted over time. The average fluorescence before photobleaching was counted as 100%. Data are represented as mean +/− SEM. n=15 cells per sample. P > 0.05 by Student t-test, paired.
(A–E) Stress granules in HeLa cells expressing either transfected GFP-GR50 or GFP-PR50 (GFP was used as a control) were visualized using immunofluorescence. Antibodies specific to (A) G3BP1 (red) and eIF3η (purple), (B) PABPC1 (red) and G3BP1 (purple), (C) PTBP1 (red) and EIF4G (purple), (D) IPO7 (red) and eIF4G (purple), or (E) TIA-1 (red) were used to visualize stress granules. DAPI was used to visualize the nucleus. Scale bar, 10 μm. (F) Western blot revealing total protein levels of GFP, GFP-GR50 or GFP-PR50 in transfected and untransfected cells treated with S35 from Figure 5G.
(A) Schematic diagram of hnRNPA1 domain structure and amino acid sequence of the LCD domain. Aromatic amino acids (phenylalanine and tyrosine) are highlighted with purple color, positively charged amino acids are highlighted with blue, and negatively charged amino acids are highlighted with red color. (B) Oregon-green labeled hnRNPA1 droplet was analyzed by FRAP. The yellow circle marks the photobleached region. Representative images of the same area before and after photobleaching at different times are shown. Scale bar, 10 μm. (C) Signal intensity of fluorescence of Oregon-green hnRNPA1 in the photobleached yellow circle region was plotted over time. The average fluorescence before photobleaching was counted as 100%. Data are represented as mean +/− SEM. n=10 droplets per sample, ***P < 0.001 by Student’s t-test, paired. (D) Schematic diagram of TIA-1 domain structure and amino acid sequence of the LCD domain. Amino acids are highlighted with different colors as described in (A). (E) Oregon-green labeled TIA-1 droplet was analyzed by FRAP. The yellow circle marks the photobleached region. Representative images of the same area before and after photobleaching at different times are shown. Scale bar, 10 μm. (F) Signal intensity of fluorescence of Oregon-green TIA-1 in the photobleached yellow circle region was plotted over time. The average fluorescence before photobleaching was counted as 100%. Data are represented as mean +/− SEM. n=9–10 droplets per sample, ***P < 0.001 by Student’s t-test, paired.
(A) HeLa cells transfected with SRSF2-GFP either with mCherry-GR50 or mCherry-PR50 constructs were analyzed using FRAP. The yellow circle marks the photobleached region. Representative images of the same area before and after photobleaching at different times are shown. Scale bar, 5 μm. (B) Signal intensity of fluorescence of SRSF2-GFP in the photobleached yellow circle region was plotted over time. The average fluorescence before photobleaching was counted as 100%. Data are represented as mean +/− SEM. n=6 cells per sample, P > 0.05 by Student’s t-test, paired.
Table S1. List of GR/PR Interactors, Related to Figure 1.
Table S2. List of Drosophila lines, Related to Figure 2.
Highlights.
Arginine-containing DPRs bind to proteins harboring low complexity domains (LCDs)
The GR and PR-interacting proteins are components of membrane-less organelles
GR and PR alter phase separation by LCD-containing proteins
GR and PR alter the assembly, dynamics and function of membrane-less organelles
Acknowledgments
We thank the Bloomington Drosophila Stock Center, the VDRC Stock Center for fly lines, the Proteomics and Cell and Tissue Imaging Centers at St Jude Children’s Research Hospital for assistance. We also thank Natalia Nedelsky for editorial assistance. This work was supported by grants from Target ALS, The Packard Center for ALS Research at the Johns Hopkins University, the ALS Association, the American-Lebanese-Syrian Associated Charities, NIH grant R35NS097974, and HHMI to J.P.T.
Footnotes
Author Contributions
H.J.K., and J.P.T. conceived and supervised the project. K.-H.L. and B.B. performed the proteomic analysis with input from J.P. K.-H.L. and P.Z. validated the proteomic data. K.-H.L. and T.S. analyzed the proteomic data. K.-H.L., B.F., and M.C. performed the genetic screen and validation. K.-H.L., B.F., N.C.K., and M.C. characterized Drosophila phenotypes. K.-H.L., P.Z., B.F., R.-M.K, J.M., J. Moore and J.T. performed cell based assay. M.S., D.M., J.C., A.M., B.M, and J.T. performed in vitro phase separation assays. H.J.K. and J.P.T. wrote the manuscript with input from R.K.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alami NH, Smith RB, Carrasco MA, Williams LA, Winborn CS, Han SS, Kiskinis E, Winborn B, Freibaum BD, Kanagaraj A, et al. Axonal transport of TDP-43 mRNA granules is impaired by ALS-causing mutations. Neuron. 2014;81:536–543. doi: 10.1016/j.neuron.2013.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P, Kedersha N. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat Rev Mol Cell Biol. 2009;10:430–436. doi: 10.1038/nrm2694. [DOI] [PubMed] [Google Scholar]
- Boisvert FM, van Koningsbruggen S, Navascues J, Lamond AI. The multifunctional nucleolus. Nat Rev Mol Cell Biol. 2007;8:574–585. doi: 10.1038/nrm2184. [DOI] [PubMed] [Google Scholar]
- Brangwynne C, Tompa P, Pappu R. Polymer physics of intracellular phase transitions. NATURE PHYSICS. 2015;11:899–904. [Google Scholar]
- Brangwynne CP, Mitchison TJ, Hyman AA. Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc Natl Acad Sci U S A. 2011;108:4334–4339. doi: 10.1073/pnas.1017150108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caussinus E, Kanca O, Affolter M. Fluorescent fusion protein knockout mediated by anti-GFP nanobody. Nat Struct Mol Biol. 2012;19:117–121. doi: 10.1038/nsmb.2180. [DOI] [PubMed] [Google Scholar]
- Choi H, Larsen B, Lin ZY, Breitkreutz A, Mellacheruvu D, Fermin D, Qin ZS, Tyers M, Gingras AC, Nesvizhskii AI. SAINT: probabilistic scoring of affinity purification-mass spectrometry data. Nat Methods. 2011;8:70–73. doi: 10.1038/nmeth.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feric M, Vaidya N, Harmon TS, Mitrea DM, Zhu L, Richardson TM, Kriwacki RW, Pappu RV, Brangwynne CP. Coexisting Liquid Phases Underlie Nucleolar Subcompartments. Cell. 2016 doi: 10.1016/j.cell.2016.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freibaum BD, Lu Y, Lopez-Gonzalez R, Kim NC, Almeida S, Lee KH, Badders N, Valentine M, Miller BL, Wong PC, et al. GGGGCC repeat expansion in C9orf72 compromises nucleocytoplasmic transport. Nature. 2015 doi: 10.1038/nature14974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron TF, Bieniek KF, Zhang YJ, Jansen-West K, Ash PE, Caulfield T, Daughrity L, Dunmore JH, Castanedes-Casey M, Chew J, et al. Antisense transcripts of the expanded C9ORF72 hexanucleotide repeat form nuclear RNA foci and undergo repeat-associated non-ATG translation in c9FTD/ALS. Acta Neuropathol. 2013;126:829–844. doi: 10.1007/s00401-013-1192-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginisty H, Sicard H, Roger B, Bouvet P. Structure and functions of nucleolin. J Cell Sci. 1999;112(Pt 6):761–772. doi: 10.1242/jcs.112.6.761. [DOI] [PubMed] [Google Scholar]
- Hackman P, Sarparanta J, Lehtinen S, Vihola A, Evila A, Jonson PH, Luque H, Kere J, Screen M, Chinnery PF, et al. Welander distal myopathy is caused by a mutation in the RNA-binding protein TIA1. Ann Neurol. 2013;73:500–509. doi: 10.1002/ana.23831. [DOI] [PubMed] [Google Scholar]
- Hormoz S. Amino acid composition of proteins reduces deleterious impact of mutations. Sci Rep. 2013;3:2919. doi: 10.1038/srep02919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovičić A, Mertens J, Boeynaems S, Bogaert E, Chai N, Yamada SB, Paul JW, 3rd, Sun S, Herdy JR, Bieri G, et al. Modifiers of C9orf72 dipeptide repeat toxicity connect nucleocytoplasmic transport defects to FTD/ALS. Nat Neurosci. 2015;18:1226–1229. doi: 10.1038/nn.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanekura K, Yagi T, Cammack AJ, Mahadevan J, Kuroda M, Harms MB, Miller TM, Urano F. Poly-dipeptides encoded by the C9ORF72 repeats block global protein translation. Hum Mol Genet. 2016 doi: 10.1093/hmg/ddw052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J, et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Panas MD, Achorn CA, Lyons S, Tisdale S, Hickman T, Thomas M, Lieberman J, McInerney GM, Ivanov P, et al. G3BP-Caprin1-USP10 complexes mediate stress granule condensation and associate with 40S subunits. J Cell Biol. 2016;212:845–860. doi: 10.1083/jcb.201508028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Kim NC, Wang YD, Scarborough EA, Moore J, Diaz Z, MacLea KS, Freibaum B, Li S, Molliex A, et al. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature. 2013;495:467–473. doi: 10.1038/nature11922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klar J, Sobol M, Melberg A, Mabert K, Ameur A, Johansson AC, Feuk L, Entesarian M, Orlen H, Casar-Borota O, et al. Welander distal myopathy caused by an ancient founder mutation in TIA1 associated with perturbed splicing. Hum Mutat. 2013;34:572–577. doi: 10.1002/humu.22282. [DOI] [PubMed] [Google Scholar]
- Kroschwald S, Maharana S, Mateju D, Malinovska L, Nuske E, Poser I, Richter D, Alberti S. Promiscuous interactions and protein disaggregases determine the material state of stress-inducible RNP granules. Elife. 2015;4 doi: 10.7554/eLife.06807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski TJ, Jr, Bosco DA, Leclerc AL, Tamrazian E, Vanderburg CR, Russ C, Davis A, Gilchrist J, Kasarskis EJ, Munsat T, et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323:1205–1208. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- Kwon I, Xiang S, Kato M, Wu L, Theodoropoulos P, Wang T, Kim J, Yun J, Xie Y, McKnight SL. Poly-dipeptides encoded by the C9orf72 repeats bind nucleoli, impede RNA biogenesis, and kill cells. Science. 2014;345:1139–1145. doi: 10.1126/science.1254917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Banjade S, Cheng HC, Kim S, Chen B, Guo L, Llaguno M, Hollingsworth JV, King DS, Banani SF, et al. Phase transitions in the assembly of multivalent signalling proteins. Nature. 2012;483:336–340. doi: 10.1038/nature10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Protter DS, Rosen MK, Parker R. Formation and Maturation of Phase-Separated Liquid Droplets by RNA-Binding Proteins. Mol Cell. 2015;60:208–219. doi: 10.1016/j.molcel.2015.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machyna M, Neugebauer KM, Stanek D. Coilin: The first 25 years. RNA Biol. 2015;12:590–596. doi: 10.1080/15476286.2015.1034923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzahn MR, Marada S, Lee J, Nourse A, Kenrick S, Zhao H, Ben-Nissan G, Kolaitis RM, Peters JL, Pounds S, et al. Higher-order oligomerization promotes localization of SPOP to liquid nuclear speckles. EMBO J. 2016 doi: 10.15252/embj.201593169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellacheruvu D, Wright Z, Couzens AL, Lambert JP, St-Denis NA, Li T, Miteva YV, Hauri S, Sardiu ME, Low TY, et al. The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nat Methods. 2013;10:730–736. doi: 10.1038/nmeth.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitrea DM, Cika JA, Guy CS, Ban D, Banerjee PR, Stanley CB, Nourse A, Deniz AA, Kriwacki RW. Nucleophosmin integrates within the nucleolus via multi-modal interactions with proteins displaying R-rich linear motifs and rRNA. Elife. 2016;5 doi: 10.7554/eLife.13571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizielinska S, Gronke S, Niccoli T, Ridler CE, Clayton EL, Devoy A, Moens T, Norona FE, Woollacott IO, Pietrzyk J, et al. C9orf72 repeat expansions cause neurodegeneration in Drosophila through arginine-rich proteins. Science. 2014;345:1192–1194. doi: 10.1126/science.1256800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, Mittag T, Taylor JP. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell. 2015;163:123–133. doi: 10.1016/j.cell.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K, Arzberger T, Grasser FA, Gijselinck I, May S, Rentzsch K, Weng SM, Schludi MH, van der Zee J, Cruts M, et al. Bidirectional transcripts of the expanded C9orf72 hexanucleotide repeat are translated into aggregating dipeptide repeat proteins. Acta Neuropathol. 2013;126:881–893. doi: 10.1007/s00401-013-1189-3. [DOI] [PubMed] [Google Scholar]
- Murakami T, Qamar S, Lin JQ, Schierle GS, Rees E, Miyashita A, Costa AR, Dodd RB, Chan FT, Michel CH, et al. ALS/FTD Mutation-Induced Phase Transition of FUS Liquid Droplets and Reversible Hydrogels into Irreversible Hydrogels Impairs RNP Granule Function. Neuron. 2015;88:678–690. doi: 10.1016/j.neuron.2015.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedelsky NB, Pennuto M, Smith RB, Palazzolo I, Moore J, Nie Z, Neale G, Taylor JP. Native functions of the androgen receptor are essential to pathogenesis in a Drosophila model of spinobulbar muscular atrophy. Neuron. 2010;67:936–952. doi: 10.1016/j.neuron.2010.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- O’Rourke JG, Bogdanik L, Yanez A, Lall D, Wolf AJ, Muhammad AK, Ho R, Carmona S, Vit JP, Zarrow J, et al. C9orf72 is required for proper macrophage and microglial function in mice. Science. 2016;351:1324–1329. doi: 10.1126/science.aaf1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldfield CJ, Dunker AK. Intrinsically disordered proteins and intrinsically disordered protein regions. Annu Rev Biochem. 2014;83:553–584. doi: 10.1146/annurev-biochem-072711-164947. [DOI] [PubMed] [Google Scholar]
- Patel A, Lee HO, Jawerth L, Maharana S, Jahnel M, Hein MY, Stoynov S, Mahamid J, Saha S, Franzmann TM, et al. A Liquid-to-Solid Phase Transition of the ALS Protein FUS Accelerated by Disease Mutation. Cell. 2015;162:1066–1077. doi: 10.1016/j.cell.2015.07.047. [DOI] [PubMed] [Google Scholar]
- Prudencio M, Belzil VV, Batra R, Ross CA, Gendron TF, Pregent LJ, Murray ME, Overstreet KK, Piazza-Johnston AE, Desaro P, et al. Distinct brain transcriptome profiles in C9orf72-associated and sporadic ALS. Nat Neurosci. 2015;18:1175–1182. doi: 10.1038/nn.4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renton AE, Majounie E, Waite A, Simon-Sanchez J, Rollinson S, Gibbs JR, Schymick JC, Laaksovirta H, van Swieten JC, Myllykangas L, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salajegheh M, Pinkus JL, Taylor JP, Amato AA, Nazareno R, Baloh RH, Greenberg SA. Sarcoplasmic redistribution of nuclear TDP-43 in inclusion body myositis. Muscle Nerve. 2009;40:19–31. doi: 10.1002/mus.21386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt EK, Clavarino G, Ceppi M, Pierre P. SUnSET, a nonradioactive method to monitor protein synthesis. Nat Methods. 2009;6:275–277. doi: 10.1038/nmeth.1314. [DOI] [PubMed] [Google Scholar]
- Schweizer Burguete A, Almeida S, Gao FB, Kalb R, Akins MR, Bonini NM. GGGGCC microsatellite RNA is neuritically localized, induces branching defects, and perturbs transport granule function. Elife. 2015;4:e08881. doi: 10.7554/eLife.08881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JP, Brown RJ, Cleveland DW. Decoding ALS from Genes to Mechanism. Nature. 2016 doi: 10.1038/nature20413. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt C. UniProt: a hub for protein information. Nucleic Acids Res. 2015;43:D204–212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance C, Rogelj B, Hortobagyi T, De Vos KJ, Nishimura AL, Sreedharan J, Hu X, Smith B, Ruddy D, Wright P, et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–1211. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira NM, Naslavsky MS, Licinio L, Kok F, Schlesinger D, Vainzof M, Sanchez N, Kitajima JP, Gal L, Cavacana N, et al. A defect in the RNA-processing protein HNRPDL causes limb-girdle muscular dystrophy 1G (LGMD1G) Hum Mol Genet. 2014;23:4103–4110. doi: 10.1093/hmg/ddu127. [DOI] [PubMed] [Google Scholar]
- Wansink DG, Schul W, van der Kraan I, van Steensel B, van Driel R, de Jong L. Fluorescent labeling of nascent RNA reveals transcription by RNA polymerase II in domains scattered throughout the nucleus. J Cell Biol. 1993;122:283–293. doi: 10.1083/jcb.122.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen X, Tan W, Westergard T, Krishnamurthy K, Markandaiah SS, Shi Y, Lin S, Shneider NA, Monaghan J, Pandey UB, et al. Antisense proline-arginine RAN dipeptides linked to C9ORF72-ALS/FTD form toxic nuclear aggregates that initiate in vitro and in vivo neuronal death. Neuron. 2014;84:1213–1225. doi: 10.1016/j.neuron.2014.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wootton JC, Federhen S. Analysis of compositionally biased regions in sequence databases. Methods Enzymol. 1996;266:554–571. doi: 10.1016/s0076-6879(96)66035-2. [DOI] [PubMed] [Google Scholar]
- Xu Z, Poidevin M, Li X, Li Y, Shu L, Nelson DL, Li H, Hales CM, Gearing M, Wingo TS, et al. Expanded GGGGCC repeat RNA associated with amyotrophic lateral sclerosis and frontotemporal dementia causes neurodegeneration. Proc Natl Acad Sci U S A. 2013;110:7778–7783. doi: 10.1073/pnas.1219643110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Donnelly CJ, Haeusler AR, Grima JC, Machamer JB, Steinwald P, Daley EL, Miller SJ, Cunningham KM, Vidensky S, et al. The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature. 2015;525:56–61. doi: 10.1038/nature14973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YJ, Gendron TF, Grima JC, Sasaguri H, Jansen-West K, Xu YF, Katzman RB, Gass J, Murray ME, Shinohara M, et al. C9ORF72 poly(GA) aggregates sequester and impair HR23 and nucleocytoplasmic transport proteins. Nat Neurosci. 2016 doi: 10.1038/nn.4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YJ, Jansen-West K, Xu YF, Gendron TF, Bieniek KF, Lin WL, Sasaguri H, Caulfield T, Hubbard J, Daughrity L, et al. Aggregation-prone c9FTD/ALS poly(GA) RAN-translated proteins cause neurotoxicity by inducing ER stress. Acta Neuropathol. 2014;128:505–524. doi: 10.1007/s00401-014-1336-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu T, Gibbens B, Doty NS, Gomes-Pereira M, Huguet A, Stone MD, Margolis J, Peterson M, Markowski TW, Ingram MA, et al. Non-ATG-initiated translation directed by microsatellite expansions. Proc Natl Acad Sci U S A. 2011;108:260–265. doi: 10.1073/pnas.1013343108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu T, Liu Y, Banez-Coronel M, Reid T, Pletnikova O, Lewis J, Miller TM, Harms MB, Falchook AE, Subramony SH, et al. RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc Natl Acad Sci U S A. 2013;110:E4968–4977. doi: 10.1073/pnas.1315438110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) HEK 293T cells were transfected with GFP or the indicated GFP-tagged dipeptide and immunoprecipitated with an antibody targeting the GFP tag. The samples were separated by SDS-PAGE, and proteins were visualized with SYPRO-Ruby. (B) Western blot performed with an anti-GFP antibody was used to visualize transfected dipeptides (or GFP control) in both input and immunoprecipitated samples. (C) Western blot analysis of immunoprecipitated proteins with indicated antibodies to validate results from proteomic analysis. (D) Venn diagrams illustrating the overlap among DPR-interacting proteins at various confidence levels as indicated by SAINT analysis.
Live HeLa cells co-transfected with G3BP1-GFP and mCherry constructs were imaged continuously over 14 hours. Related to Figure 5.
Live HeLa cells co-transfected with G3BP1-GFP and mCherry-GR50 constructs were imaged continuously over 14 hours. Related to Figure 5.
Live HeLa cells co-transfected with G3BP1-GFP and mCherry-PR50 constructs were imaged continuously over 14 hours. Related to Figure 5.
Wetting and fusion of hnRNPA1 droplets. Related to Figure 6.
300 μM protein undergoes LLPS, and droplets of hnRNPA1 exhibit wetting when they encounter the surface of the coverslip as shown in Molliex et al. (Molliex et al., 2015).
Arginine-containing peptides increase the surface tension of hnRNPA1 droplets. Related Figure 6.
150μM hnRNPA1 protein mixed with 50 μM PR20 peptide undergoes LLPS, however hnRNPA1 droplets do not wet the surface or fuse over time due to high surface tension.
(A) GFP-GR50 protein levels, but not mRNA levels, were decreased in a dose dependent manner in both pupae and adult Drosophila by expression of an intrabody (deGradFP) targeted against the GFP sequence. (B) Expression of the deGradFP intrabody rescues the rough eye phenotype caused by GFP-GR50 dipeptide in a dose dependent manner when protein expression is restricted to the Drosophila eye using the GMR driver (C) A genetic screen of RNAi lines targeting DPR interacting proteins in Drosophila using egg-to-adult viability as a read out. Genetic suppressors GR50 toxicity are labeled in green whereas enhancers of GR50 toxicity are labeled in red, as indicated in the key. W1118 and v60100 lines were used as controls. (D) Integration of the genetic screen with gene ontology results depicting GR50 suppressors in green and enhancers in red. (E) Strong suppressors of GR50 toxicity were predominantly found to suppress expanded G4C2-mediated toxicity using the (G4C2)58 Drosophila model (Freibaum et al., 2015). Common suppressors were largely specific as most of these RNAi lines failed to suppress non-specific toxicity caused by expression of the androgen receptor polyQ expansion (ARpolyQ)52. Cluster dendrogram analysis demonstrated that the overlap of shared modifiers of GR and (G4C2)58 toxicities was significantly greater than shared modifiers with AR(polyQ)52 toxicity. (F) Strong enhancers of GR50 toxicity were predominantly found to enhance C9orf72 mediated toxicity using a (G4C2)58 model.
(A) HeLa cells expressing either transfected GFP or GFP-tagged DPR were immunostained with NPM1 (red) and G3BP1 (purple) specific antibodies. DAPI was used to visualize the nucleus. GR50 and PR50 but not other DPRs or GFP were found to colocalize with NPM1 and induce the formation of G3BP1 positive cytoplasmic stress granules. Scale bar, 10 μm. (B) The percentage of cells in which the transfected DPR was found to localize to nucleoli. (C) FAM-labeled GR20 or PR20 peptides (green, 10μM) were incubated in HeLa cell culture media for 60 minutes and their nucleolar localization was determined using an NPM1 specific antibody (red). Scale bar, 10 μm. (D) HeLa cells were transfected with GFP, GFP-GR50, GFP-PR50 or GFP-GA50. GFP-positive nucleoli were analyzed by FRAP. The yellow circle marks the photobleached region. Representative images of the same area before and after photobleaching at different times are shown. Scale bar, 10 μm. (E) Signal intensity of GFP fluorescence in the photobleached yellow circle region was plotted over time. The average fluorescence before photobleaching was counted as 100%. Data are represented as mean +/− SEM. n=10 cells per sample, P values for GFP-GR50, GFP-PR50 or GFP-GA50 (over GFP control) < 0.001 by Student t-test, paired. (F) HeLa cells were transfected with GFP-GR50, GFP-PR50, or GFP-GA50 for 48 hours, then changed the media with 3.5% 1,6-hexanediol and imaged the cells continuously for 1 hour. (G) HEK293T cells were transfected with GFP, GFP-GR50, GFP-PR50, GFP-GA50, GFP-GP47, or GFP-PA50, incubated for 48 hours, and then sequentially extracted with RIPA buffer and urea buffer. Immunoblotting was conducted with anti-GFP antibody.
(A) Circular dichroism spectra of 10 μM peptides at 25 °C, in 10 mM Tris pH 7.5 buffer, under low ionic strength (red) and physiological ionic strength (black) conditions. (B) Flag pull-down assay reveals that GR20 and PR20 peptides directly interact with NPM1, NCL, hnRNPA1, G3BP1, and TIA-1 in vitro. (C) Titrations of GP20, PA20, GR20 and PR20 (concentrations indicated at the top) into liquid-phase droplets comprised of 1:1 NPM1:SURF6 peptide prepared at 20 μM. The GR20 and PR20 peptides entered into the NPM1:SURF6 peptide droplets at concentrations at and below 1:1 stoichiometry with NPM1 (DPR:NPM1) but the droplets were disrupted at higher stoichiometries. The GP20 and PA20 peptides neither entered into nor disrupted the NPM1:SURF6 peptide droplets at any concentrations tested. Alexa488-NPM1 (left in each panel) and TAMRA-DPR peptides (middle) were detected using confocal fluorescence microscopy. These colors were merged on the right of each panel. Scale bar, 10 μm. (D) HeLa cells transfected with DDX47-GFP either with mCherry-GR50 or mCherry-PR50 constructs were analyzed using FRAP. The yellow circle marks the photobleached region. Representative images of the same area before and after photobleaching at different times are shown. Scale bar, 5 μm. (E) Signal intensity of fluorescence of DDX47-GFP in the photobleached yellow circle region was plotted over time. The average fluorescence before photobleaching was counted as 100%. Data are represented as mean +/− SEM. n=15 cells per sample. P > 0.05 by Student t-test, paired.
(A–E) Stress granules in HeLa cells expressing either transfected GFP-GR50 or GFP-PR50 (GFP was used as a control) were visualized using immunofluorescence. Antibodies specific to (A) G3BP1 (red) and eIF3η (purple), (B) PABPC1 (red) and G3BP1 (purple), (C) PTBP1 (red) and EIF4G (purple), (D) IPO7 (red) and eIF4G (purple), or (E) TIA-1 (red) were used to visualize stress granules. DAPI was used to visualize the nucleus. Scale bar, 10 μm. (F) Western blot revealing total protein levels of GFP, GFP-GR50 or GFP-PR50 in transfected and untransfected cells treated with S35 from Figure 5G.
(A) Schematic diagram of hnRNPA1 domain structure and amino acid sequence of the LCD domain. Aromatic amino acids (phenylalanine and tyrosine) are highlighted with purple color, positively charged amino acids are highlighted with blue, and negatively charged amino acids are highlighted with red color. (B) Oregon-green labeled hnRNPA1 droplet was analyzed by FRAP. The yellow circle marks the photobleached region. Representative images of the same area before and after photobleaching at different times are shown. Scale bar, 10 μm. (C) Signal intensity of fluorescence of Oregon-green hnRNPA1 in the photobleached yellow circle region was plotted over time. The average fluorescence before photobleaching was counted as 100%. Data are represented as mean +/− SEM. n=10 droplets per sample, ***P < 0.001 by Student’s t-test, paired. (D) Schematic diagram of TIA-1 domain structure and amino acid sequence of the LCD domain. Amino acids are highlighted with different colors as described in (A). (E) Oregon-green labeled TIA-1 droplet was analyzed by FRAP. The yellow circle marks the photobleached region. Representative images of the same area before and after photobleaching at different times are shown. Scale bar, 10 μm. (F) Signal intensity of fluorescence of Oregon-green TIA-1 in the photobleached yellow circle region was plotted over time. The average fluorescence before photobleaching was counted as 100%. Data are represented as mean +/− SEM. n=9–10 droplets per sample, ***P < 0.001 by Student’s t-test, paired.
(A) HeLa cells transfected with SRSF2-GFP either with mCherry-GR50 or mCherry-PR50 constructs were analyzed using FRAP. The yellow circle marks the photobleached region. Representative images of the same area before and after photobleaching at different times are shown. Scale bar, 5 μm. (B) Signal intensity of fluorescence of SRSF2-GFP in the photobleached yellow circle region was plotted over time. The average fluorescence before photobleaching was counted as 100%. Data are represented as mean +/− SEM. n=6 cells per sample, P > 0.05 by Student’s t-test, paired.
Table S1. List of GR/PR Interactors, Related to Figure 1.
Table S2. List of Drosophila lines, Related to Figure 2.