Abstract
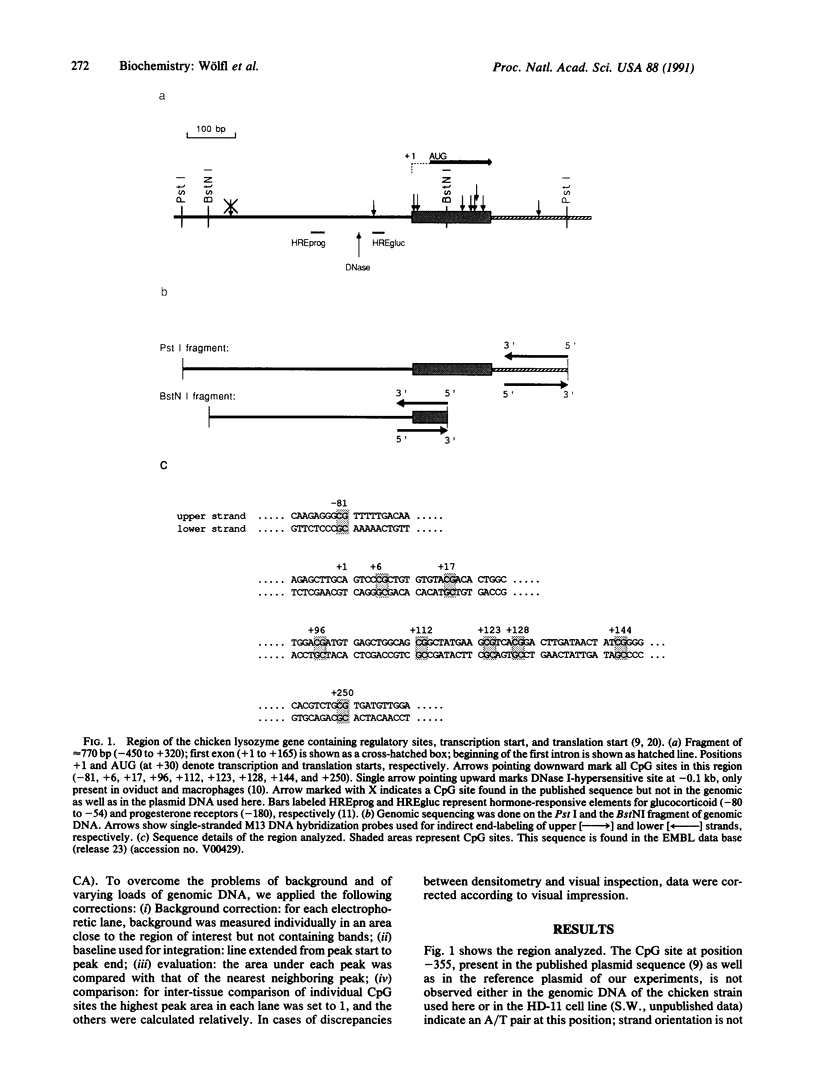
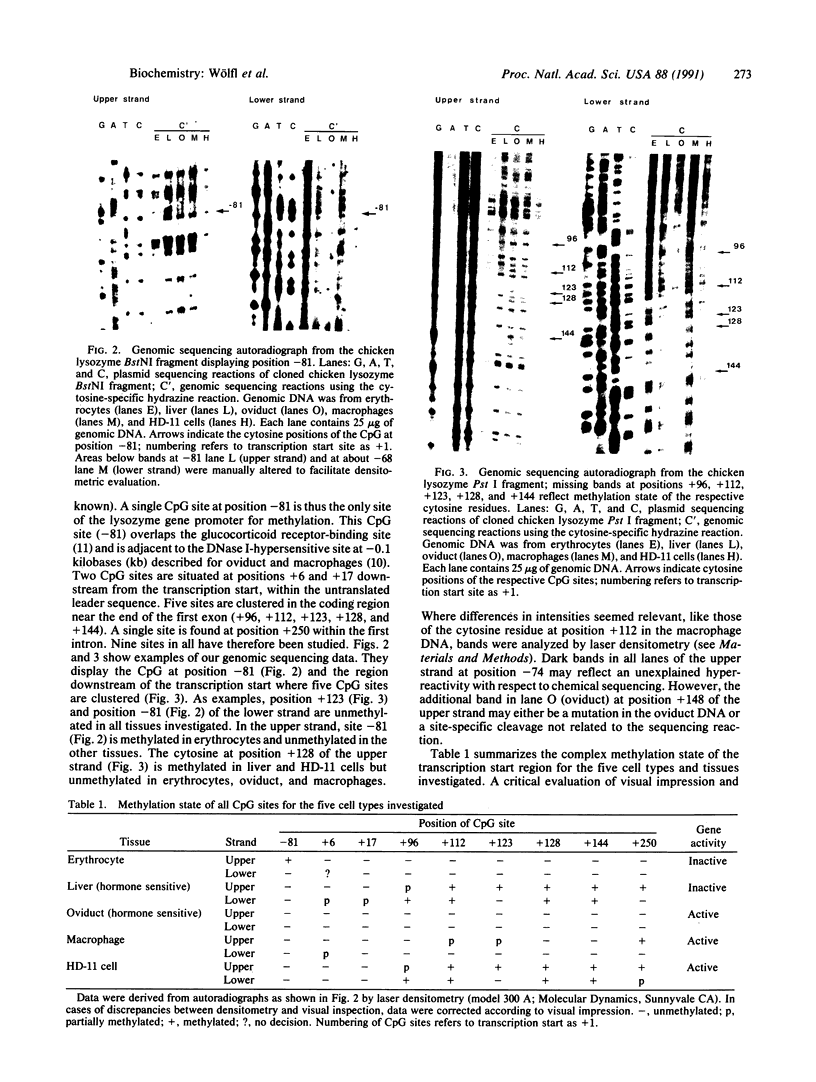
We have analyzed the methylation state of all nine CpG sites in the transcription start region (-420 to +250 base pairs) of the chicken lysozyme gene by genomic sequencing. One of these sites, at -81, lies within the promoter, seven are clustered within the first exon, and the last is in the first intron. Five cell types and tissues have been investigated to study the relationship between methylation level and gene expression. For each cell type used, the majority of CpG sites showed a similar level of methylation. Of two gene-nonexpressing tissues, erythrocytes are hypomethylated, whereas liver is methylated at most of its CpG sites. For gene-expressing tissues, oviduct is completely unmethylated, whereas HD-11 culture cells are methylated. Thus no correlation is observed between degree of CpG methylation and level of expression of the lysozyme gene. The observed methylation patterns are discussed in terms of possible features of the local chromatin structure.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Becker P. B., Ruppert S., Schütz G. Genomic footprinting reveals cell type-specific DNA binding of ubiquitous factors. Cell. 1987 Nov 6;51(3):435–443. doi: 10.1016/0092-8674(87)90639-8. [DOI] [PubMed] [Google Scholar]
- Ben-Hattar J., Beard P., Jiricny J. Cytosine methylation in CTF and Sp1 recognition sites of an HSV tk promoter: effects on transcription in vivo and on factor binding in vitro. Nucleic Acids Res. 1989 Dec 25;17(24):10179–10190. doi: 10.1093/nar/17.24.10179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Hattar J., Jiricny J. Methylation of single CpG dinucleotides within a promoter element of the Herpes simplex virus tk gene reduces its transcription in vivo. Gene. 1988 May 30;65(2):219–227. doi: 10.1016/0378-1119(88)90458-1. [DOI] [PubMed] [Google Scholar]
- Beug H., von Kirchbach A., Döderlein G., Conscience J. F., Graf T. Chicken hematopoietic cells transformed by seven strains of defective avian leukemia viruses display three distinct phenotypes of differentiation. Cell. 1979 Oct;18(2):375–390. doi: 10.1016/0092-8674(79)90057-6. [DOI] [PubMed] [Google Scholar]
- Bird A. P. CpG-rich islands and the function of DNA methylation. Nature. 1986 May 15;321(6067):209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- Buschhausen G., Wittig B., Graessmann M., Graessmann A. Chromatin structure is required to block transcription of the methylated herpes simplex virus thymidine kinase gene. Proc Natl Acad Sci U S A. 1987 Mar;84(5):1177–1181. doi: 10.1073/pnas.84.5.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cedar H. DNA methylation and gene activity. Cell. 1988 Apr 8;53(1):3–4. doi: 10.1016/0092-8674(88)90479-5. [DOI] [PubMed] [Google Scholar]
- Church G. M., Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dierich A., Gaub M. P., LePennec J. P., Astinotti D., Chambon P. Cell-specificity of the chicken ovalbumin and conalbumin promoters. EMBO J. 1987 Aug;6(8):2305–2312. doi: 10.1002/j.1460-2075.1987.tb02505.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorbic T., Wittig B. Chromatin from transcribed genes contains HMG17 only downstream from the starting point of transcription. EMBO J. 1987 Aug;6(8):2393–2399. doi: 10.1002/j.1460-2075.1987.tb02517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dynan W. S. Understanding the molecular mechanism by which methylation influences gene expression. Trends Genet. 1989 Feb;5(2):35–36. doi: 10.1016/0168-9525(89)90016-4. [DOI] [PubMed] [Google Scholar]
- Fritton H. P., Igo-Kemenes T., Nowock J., Strech-Jurk U., Theisen M., Sippel A. E. Alternative sets of DNase I-hypersensitive sites characterize the various functional states of the chicken lysozyme gene. Nature. 1984 Sep 13;311(5982):163–165. doi: 10.1038/311163a0. [DOI] [PubMed] [Google Scholar]
- Grez M., Land H., Giesecke K., Schütz G., Jung A., Sippel A. E. Multiple mRNAs are generated from the chicken lysozyme gene. Cell. 1981 Sep;25(3):743–752. doi: 10.1016/0092-8674(81)90182-3. [DOI] [PubMed] [Google Scholar]
- Harrington M. A., Jones P. A., Imagawa M., Karin M. Cytosine methylation does not affect binding of transcription factor Sp1. Proc Natl Acad Sci U S A. 1988 Apr;85(7):2066–2070. doi: 10.1073/pnas.85.7.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser H., Graf T., Beug H., Greiser-Wilke I., Lindenmaier W., Grez M., Land H., Giesecke K., Schütz G. Structure of the lysozyme gene and expression in the oviduct and macrophages. Haematol Blood Transfus. 1981;26:175–178. doi: 10.1007/978-3-642-67984-1_29. [DOI] [PubMed] [Google Scholar]
- Höller M., Westin G., Jiricny J., Schaffner W. Sp1 transcription factor binds DNA and activates transcription even when the binding site is CpG methylated. Genes Dev. 1988 Sep;2(9):1127–1135. doi: 10.1101/gad.2.9.1127. [DOI] [PubMed] [Google Scholar]
- Jung A., Sippel A. E., Grez M., Schütz G. Exons encode functional and structural units of chicken lysozyme. Proc Natl Acad Sci U S A. 1980 Oct;77(10):5759–5763. doi: 10.1073/pnas.77.10.5759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlmann I., Achten S., Rudolph R., Doerfler W. Tumor induction by human adenovirus type 12 in hamsters: loss of the viral genome from adenovirus type 12-induced tumor cells is compatible with tumor formation. EMBO J. 1982;1(1):79–86. doi: 10.1002/j.1460-2075.1982.tb01128.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leutz A., Beug H., Graf T. Purification and characterization of cMGF, a novel chicken myelomonocytic growth factor. EMBO J. 1984 Dec 20;3(13):3191–3197. doi: 10.1002/j.1460-2075.1984.tb02278.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Palmiter R. D. Regulation of protein synthesis in chick oviduct. I. Independent regulation of ovalbumin, conalbumin, ovomucoid, and lysozyme induction. J Biol Chem. 1972 Oct 25;247(20):6450–6461. [PubMed] [Google Scholar]
- Razin A., Riggs A. D. DNA methylation and gene function. Science. 1980 Nov 7;210(4470):604–610. doi: 10.1126/science.6254144. [DOI] [PubMed] [Google Scholar]
- Rossi G., Himmelhoch S. Immunocytochemical localization of the mannose receptor on ultrathin cryosections of chicken macrophages. Eur J Cell Biol. 1985 Sep;38(2):280–291. [PubMed] [Google Scholar]
- Saluz H. P., Feavers I. M., Jiricny J., Jost J. P. Genomic sequencing and in vivo footprinting of an expression-specific DNase I-hypersensitive site of avian vitellogenin II promoter reveal a demethylation of a mCpG and a change in specific interactions of proteins with DNA. Proc Natl Acad Sci U S A. 1988 Sep;85(18):6697–6700. doi: 10.1073/pnas.85.18.6697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saluz H. P., Jiricny J., Jost J. P. Genomic sequencing reveals a positive correlation between the kinetics of strand-specific DNA demethylation of the overlapping estradiol/glucocorticoid-receptor binding sites and the rate of avian vitellogenin mRNA synthesis. Proc Natl Acad Sci U S A. 1986 Oct;83(19):7167–7171. doi: 10.1073/pnas.83.19.7167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner C., Muller M., Baniahmad A., Renkawitz R. Lysozyme gene activity in chicken macrophages is controlled by positive and negative regulatory elements. Nucleic Acids Res. 1987 May 26;15(10):4163–4178. doi: 10.1093/nar/15.10.4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth M., Lichtenberg U., Doerfler W. Genomic sequencing reveals a 5-methylcytosine-free domain in active promoters and the spreading of preimposed methylation patterns. Proc Natl Acad Sci U S A. 1989 May;86(10):3728–3732. doi: 10.1073/pnas.86.10.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von der Ahe D., Renoir J. M., Buchou T., Baulieu E. E., Beato M. Receptors for glucocorticosteroid and progesterone recognize distinct features of a DNA regulatory element. Proc Natl Acad Sci U S A. 1986 May;83(9):2817–2821. doi: 10.1073/pnas.83.9.2817. [DOI] [PMC free article] [PubMed] [Google Scholar]






