Abstract
We classified the carboxylic ester hydrolases (CEHs) into families and clans by use of multiple sequence alignments, secondary structure analysis, and tertiary structure superpositions. Our work for the first time has fully established their systematic structural classification. Family members have similar primary, secondary, and tertiary structures, and their active sites and reaction mechanisms are conserved. Families may be gathered into clans by their having similar secondary and tertiary structures, even though primary structures of members of different families are not similar. CEHs were gathered from public databases by use of Basic Local Alignment Search Tool (BLAST) and divided into 91 families, with 36 families being grouped into five clans. Members of one clan have standard α/β‐hydrolase folds, while those of other two clans have similar folds but with different sequences of their β‐strands. The other two clans have members with six‐bladed β‐propeller and three‐α‐helix bundle tertiary structures. Those families not in clans have a large variety of structures or have no members with known structures. At the time of writing, the 91 families contained 321,830 primary structures and 1378 tertiary structures. From these data, we constructed an accessible database: CASTLE (CArboxylic eSTer hydroLasEs, http://www.castle.cbe.iastate.edu).
Keywords: carboxylesterases, cholinesterases, cocaine esterases, cutinases, lysopholipases, phospholipases, triacylglycerol lipases
Introduction
Carboxylic ester hydrolases (CEHs) catalyze the hydrolysis of ester bonds into alcohols and carboxylic acids, and they are ubiquitous throughout life. Members of this enzyme group that attack different substrates, form different products, and have different names are listed as EC 3.1.1.1 to EC 3.1.1.98, with seven deleted entries.1 Carboxylesterases (EC 3.1.1.1), triacylglycerol lipases (EC 3.1.1.3), phospholipase A2s (EC 3.1.1.4), lysophospholipases (EC 3.1.1.5), acetylcholinesterases (EC 3.1.1.7), butyrylcholinesterases (EC 3.1.1.8), aminoacyl‐tRNA hydrolases (EC 3.1.1.29), and cocaine esterases (EC 3.1.1.84) are the most extensively researched CEHs.
According to the CATH database,2 many CEHs have standard α/β hydrolase folds, which are composed of three α/β/α layers, with the second β‐strand being antiparallel to generally seven others in the β‐sheet.3, 4 Other CEHs with other types of α/β hydrolase folds have different arrangements of their α‐helices and β‐strands. Some CEHs have six‐propeller folds, which consist of a six‐bladed β‐sheet with a central axis. Others have four‐layer β‐sandwich folds, where several antiparallel β‐strands are arranged in two β‐sheets. Three‐solenoid folds are also found in CEH structures; they consist of many parallel β‐strands arranged into three β‐sheets. The outer‐membrane CEHs are commonly found in β‐barrel folds.
To this point, there is no systematic structural classification of the CEHs. Such a classification is likely to yield very different results than found with Enzyme Commission (EC) numbering, because enzymes with different EC numbers and different names may have very similar amino acid sequences (primary structures) and three‐dimensional (tertiary) structures. This has been demonstrated in the Carbohydrate‐Active EnzYme (ThYme) databases and Thioester‐Active EnzYme databases,5, 6 as well as elsewhere.
Earlier research has partially covered this topic, but with many fewer primary structures than amassed in this project. The ESTerases and α/β‐Hydrolase Enzymes and Relatives (ESTHERs) database7, 8 covers some CEHs, focusing on α/β hydrolase structures. It is not limited to CEHs, but includes other enzymes such as peptidases and thioesterases that have this fold. ESTHER classifies sequences into three levels: blocks, rank 1 families, and rank 2 families, where each block is based on conserved and characteristic parts of sequences. At the time of writing, it had over 40,000 primary structures classified into four blocks, 94 rank 1 families, and 93 rank 2 families further divided from 11 rank 1 families.
The CAZy database has classified 15 homologous families of carbohydrate esterases, which catalyze the de‐O‐ or de‐N‐acylation of substituted saccharides, by their primary structures. Twelve of these families contain CEHs, mainly acetyl xylan esterases, and mainly with α/β hydrolase folds. At the time of writing, about 32,000 primary structures of carbohydrate esterases were included, of which almost 21,000 were CEHs.
The lipase engineering database (LED)9 classified three classes and 38 superfamilies with α/β hydrolase folds, of which 16 included lipases and 10 covered other CEHs, by their functions, sequences, and crystal structures. Its founders employed much smaller E‐values in their use of Basic Local Alignment Search Tool (BLAST)10 to gather primary structures than used in this work, implying that the family members in LED were more similar to each other than here, perhaps leading to more families. It encompassed 112 homologous families and almost 25,000 primary and over 1100 tertiary structures. However, it has not been maintained since 2009.
The MELDB database sorted microbial carboxylesterases and triacylglycerol lipases by their primary structure similarities.11 It corresponded to parts of the LED database, but it appears to be no longer available.
The research reported in this article systematically classifies the CEHs by their primary, secondary, and tertiary structure similarities, as opposed to classifying them by their EC numbers. This will cast light on the various ways that CEHs with different structures catalyze the hydrolysis of ester bonds to yield carboxylic acids and alcohols.
Potential Family Identification
CEH families were generally identified by the techniques used for classifying fatty acid synthesis enzymes.6, 12, 13 All the primary structures of CEHs, chosen by their EC numbers, with evidence at protein level in the UniProt database14 were collected. These totaled 752 sequences. The criterion of evidence at protein level is to ensure that wet laboratory experiments had been conducted on these proteins to verify their functions as CEHs. This criterion ruled out most available CEH sequences, mainly those obtained from whole‐genome projects, whose functions are putative because their sequences have been compared only with those of known CEHs rather than being verified experimentally.
The collected primary structures were checked on the Pfam database15 to obtain their catalytic domains only. BLAST was used consecutively to find primary structures similar to these catalytic domains (query sequences) from the National Center for Biotechnology Information's up‐to‐date nr database,16 which gathers nonredundant protein sequences from various sources such as the GenBank,17 Protein Data Bank (PDB),18 Protein Information Resource,19 Protein Research Foundation,20 RefSeq,16 and Swiss‐Prot14 databases. The threshold E‐value in BLAST was set to 0.001. Protein sequences with E‐values lower than 0.001 were regarded as similar enough to the query sequence to be included in one potential family.10 In‐house Python and shell scripts were implemented to automate the process of obtaining catalytic domains of query sequences in Pfam, to conduct BLAST consecutively, and to further analyze the results of structure comparison. All the scripts were run on the Google cloud platform with Linux Cent OS7 installed.
Family Verification
Multiple sequence alignment (MSA), secondary structure comparison, and tertiary structure superposition are the three main techniques to verify membership in the potential families, possibly merging or splitting them. It is assumed that all members of a family have the same protein ancestor.
A random sample of sequences in each potential family was used to perform MSA with ClustalX 2.1.21 The alignment is to ensure that these sequences are similar enough, with several positions of amino acid residues conserved along the entire sample. Different potential families gathered by BLAST were subjected to joint MSA to ascertain whether they could be merged into one family. Conversely, if no amino acid residue is conserved and if clear differences are observed in the MSA result, then the potential family was split into two or more families. Occasionally no residue is conserved in what clearly is a family because of a sequence error in one or a few of its members.
Up to 50 tertiary structures from each potential family, if available in the PDB, were superimposed by MultiProt.22 The monomer of each tertiary structure was extracted and compared. The root mean square deviation (RMSD) of the α‐carbon atoms of the different tertiary structures was calculated, together with the P avg, a measure of the percentage of these atoms that are close enough (<4.0 Å) to be compared.12, 13
A further criterion to verify family membership is that active sites of potential members should remain in similar positions within each family. Also, secondary structure elements, based on the DSSP database23, 24 embedded in the PDB, were compared and analyzed to ensure that potential members of each family have almost the same elements.
Finally, memberships of potential families were manually inspected to confirm that they held a significant number of entries with names and EC numbers specific to CEHs.
Clan Identification and Verification
Clans are composed of two or more different families, where their active sites, reaction mechanisms, and secondary and tertiary structures are conserved from family to family, although their primary structures may not be significantly similar from one family to the next. It is assumed that family members of different clans are more distantly derived from the same protein ancestor than are members of the same family.
We used CATH‐defined folds to first divide available tertiary structures in different families into separate groups. We then used two separate procedures to determine membership of families in clans. In the first, tertiary structure representatives from different families with similar tertiary structures were superimposed by MultiProt. Varying from previous methods to calculate RMSD and P avg values of members of all potential families in a clan, pairwise RMSD and P avg values after overlapping representative tertiary structures from pairs of families were calculated. This variation was caused by the large number of families with similar folds, making it difficult to visually distinguish them by PyMOL.25 The MultiProt superposition, along with RMSD and P avg calculations, were implemented by Python scripts, and RMSD and P avg values were recorded in matrices. To cluster similar structures into potential clans, the pairwise RMSD matrices were imported into MEGA 6.06,26 and neighbor‐joining trees were produced as curved and circular trees. Although MEGA was intended to produce phylogenetic trees for the study of molecular evolution, the pairwise distance matrix used in MEGA is similar enough to be used for RMSD matrices. Potential clans were proposed according to the trees. Then the structures of potential clan members were superimposed and inspected in PyMOL.
In the second procedure, similar secondary and tertiary structures from different families were grouped roughly into potential clans, and then their structures were superimposed by MultiProt and visually inspected by PyMOL. If the superposition were satisfactory, RMSD and P avg values for single representatives of all the potential family members of a potential clan were calculated. The proposed classification was tuned until the structures superimposed in PyMOL were in good alignment and RMSD and P avg values were minimized. Active sites were checked, if available, to see whether the catalytic residues are in similar positions to act on the substrates, and whether they share the same mechanism in each clan.
Interestingly, the initial pairwise alignment procedure did not perform as well as the initial visual inspection procedure. Therefore, the latter technique was chosen to assign families to clans.
Results
BLAST searches using the 752 query sequences yielded 480,148 primary structures of CEHs and other enzymes. In addition, 2101 tertiary structures were gathered from the PDB. The primary structures were classified into 130 potential families.
The membership of each potential family was verified by three methods: MSA of primary structures, secondary structure analysis, and tertiary structure superpositions. The potential 130 families obtained by BLAST became 91 families after MSA using ClustalX, secondary structure analysis, and tertiary structure superposition by MultiProt and PyMOL, and after noting that some potential families had no or very few CEH members (Table 1). After these operations, 321,830 primary structures, 1490 sequences with evidence at protein level, and 1378 tertiary structures remained.
Table 1.
Clans and Families of Carboxylic Ester Hydrolases
| Family | Number of sequences | Number of sequences with evidence at protein level | Number of known tertiary structures (representative PDB structures) | Producing organismsa | Dominant enzyme names | Dominant EC numbers |
|---|---|---|---|---|---|---|
| Clan A (α/β‐hydrolase, three‐layer α/β/α sandwich, Rossmann fold, second β‐strand antiparallel with sequence 1, 2, 4, 3, 5, 6, 7, and 8) | ||||||
| 1 | 1216 | 5 | 1 (3QIT) | B, Ea | α/β‐Hydrolase, esterase, thioesterase | 3.1.1.– |
| 2 | 31,202 | 41 | 131 (5ALM) | A, B, E | α/β‐Hydrolase, 3‐oxoadipate enol‐lactonase | 3.1.1.24 |
| 3 | 26,277 | 69 | 53 (3L1J) | A, B, E | α/β‐Hydrolase, acetylesterase, esterase/lipase | 3.1.1.– |
| 4 | 497 | 5 | 10 (4CG1) | B | Lipase, triacylglycerol lipase | 3.1.1.3 |
| 5 | 2191 | 5 | 14 (3FYU) | B | Acetyl xylan esterase | 3.1.1.72 |
| 6 | 1181 | 10 | 2 (3C5V) | B, E | Protein phosphatase methylesterase, peroxidase | 3.1.1.89, 1.11.1.– |
| 7 | 1437 | 8 | 6 (3D59) | B, E | Carboxylic ester hydrolase, platelet‐activating factor acetylhydrolase | 3.1.1.–, 3.1.1.47 |
| 8 | 730 | 2 | 4 (4G4G) | B, E | Acetyl xylan esterase | 3.1.1.72 |
| 9 | 2432 | 13 | 2 (1K8Q) | E | Lysomal acid lipase, lipase member M | 3.1.1.– |
| 10 | 3896 | 3 | 2 (3HXK) | B, E | Xylanase, pectin acetylesterase, esterase | 3.2.1.8, 3.1.1.– |
| 11 | 1359 | 4 | 14 (4UYU) | B, E | Pectin acetylesterase, protein notum homolog | 3.1.1.– |
| 12 | 24,560 | 169 | 280 (2JGJ) | A, B, E | Carboxylesterase, carboxylic ester hydrolase, acetylcholinesterase, cholinesterase, cocaine esterase | 3.1.1.8, 3.1.1.84 |
| 13 | 4538 | 6 | 19 (3ZI7) | B, E | Esterase | 3.1.1.– |
| Clan B (α/β‐hydrolase, three‐layer α/β/α sandwich, Rossmann fold, all β‐strands parallel with sequence 2, 1, 3, 4, and 5) | ||||||
| 14 | 410 | 5 | 2 (1YQE) | A, E | D‐Aminoacyl‐tRNA deacylase | 3.1.1.96 |
| 15 | 5264 | 4 | 7 (1U8U) | B | GDSL‐like lipase, arylesterase, acyl‐CoA thioesterase | 3.1.1.–, 3.1.1.2, 3.1.2.2 |
| 16 | 1869 | 2 | 13 (1R50) | B, E | Lipase | 3.1.1.– |
| 17 | 2960 | 12 | 60 (1XZG) | B, E | Cutinase, acetyl xylan esterase | 3.1.1.74, 3.1.1.72 |
| 18 | 1463 | 14 | 10 (1BWQ) | B, E | GDSL‐like lipase, acetylhydrolase | 3.1.1.– |
| 19 | 2262 | 6 | 6 (1DEO) | A, B, E | Rhamnogalacturonan acetylesterase, GDSL family lipase, carbohydrate esterase family 12 protein | 3.1.1.86, 3.1.1.– |
| 20 | 1717 | 15 | 43 (1EB8) | B, E | α/β‐Hydrolase, esterase | 3.1.1.– |
| 21 | 1985 | 5 | 4 (1ESD) | B, E | GDSL family lipase, triacylglycerol lipase | 3.1.1.–, 3.1.1.3 |
| 22 | 2374 | 9 | 18 (1CVL) | B, E | Lipase, lactonizing lipase | 3.1.1.3 |
| 23 | 498 | 8 | 7 (4X92) | B, E | Phospholipase A2, lecithin‐cholesterol acyltransferase | 3.1.1.4, 2.3.1.43 |
| 24 | 1537 | 17 | 15 (4X71) | B, E | Lipase, triacylglycerol lipase | 3.1.1.–, 3.1.1.3 |
| Clan C (α/β‐hydrolase, three‐layer α/β/α sandwich, Rossmann fold, first β‐strand antiparallel with sequence 1, 3, 2, 4, 5, 6, and 7) | ||||||
| 25 | 6115 | 6 | 19 (1ZJ5) | A, B, E | Carboxymethylenebutenolidase, dienelactone hydrolase | 3.1.1.45 |
| 26 | 8649 | 9 | 14 (4ETW) | A, B, E | α/β‐Hydrolase, hydrolase (biotin biosynthesis), carboxylesterase | 3.1.1.1 |
| 27 | 4643 | 15 | 5 (4UUQ) | A, B, E | α/β‐Hydrolase, lysophospholipase | 3.1.1.5 |
| 28 | 5655 | 30 | 9 (3CN9) | B, E | Carboxylesterase, phospholipase | 3.1.1.1, 3.1.1.– |
| 29 | 4262 | 3 | 18 (1TQH) | A, B | Esterase, carboxylesterase | 3.1.1.–, 3.1.1.1 |
| 30 | 1818 | 4 | 2 (3BF8) | A, B, E | α/β‐Hydrolase, 3‐oxoadipate enol lactonase, 2‐succinyl‐6‐hydroxy‐2,4‐cyclohexadiene‐1‐carboxylate synthase | 3.1.1.24, 4.2.99.20 |
| 31 | 314 | 1 | 14 (1LBS) | B, E | Lipase | 3.1.1.– |
| Clan D (six‐bladed β‐propeller) | ||||||
| 32 | 8864 | 16 | 42 (4GN9) | A, B, E | Gluconolactonase, SMP‐30/gluconolactone/LRE‐like region | 3.1.1.17 |
| 33 | 672 | 18 | 7 (3SRE) | B, E | Serum paraoxonase/arylesterase 2 | 3.1.1.2, 3.1.1.81 |
| Clan E (three α‐helix bundle) | ||||||
| 34 | 2791 | 325 | 288 (1TG1) | E | Phospholipase A2 | 3.1.1.4 |
| 35 | 738 | 14 | 1 (1POC) | B, E | Phospholipase A2 | 3.1.1.4 |
| 36 | 269 | 5 | 3 (2WG7) | B, E | Phospholipase A2 | 3.1.1.4 |
| Not part of a clan | ||||||
| (α/β‐Hydrolase, three‐layer α/β/α sandwich, Rossmann fold, various β‐strand arrangements) | ||||||
| 37 | 1715 | 15 | 8 (3ERJ) | A, B, E | Peptidyl‐tRNA hydrolase | 3.1.1.29 |
| 38 | 7996 | 29 | 15 (3EB9) | B, E | 6‐Phosphogluconolactonase | 3.1.1.31 |
| 39 | 4994 | 56 | 12 (1LPB) | B, E | Pancreatic glycerol lipase, phospholipase A1 | 3.1.1.3, 3.1.1.32 |
| 40 | 2593 | 32 | 35 (4GBG) | B, E | Lipase | 3.1.1.– |
| 41 | 1493 | 11 | 1 (2YIJ) | B, E | Phospholipase A1, lipase | 3.1.1.32 |
| 42 | 1059 | 11 | 1 (1CJY) | E | Lysophospholipase | 3.1.1.5 |
| 43 | 688 | 2 | 1 (3KVN) | B | Esterase | 3.1.1.– |
| 44 | 869 | 6 | 4 (1UWC) | E | Diacylglycerol lipase | 3.1.1.– |
| 45 | 11,783 | 17 | 43 (4HOY) | B, E | Peptidyl‐tRNA hydrolase | 3.1.1.29 |
| 46 | 43 | 1 | 1 (1TIA) | E | Lipase | 3.1.1.– |
| 47 | 12,511 | 8 | 4 (1CHD) | A, B | Chemotaxis‐specific regulator protein, protein‐glutamate methylesterase | 3.1.1.61 |
| Patatin‐like fold | ||||||
| 48 | 11,787 | 40 | 4 (4PKB) | B, E | Patatin, patatin‐like phospholipase family | 3.1.1.– |
| α/β TIM barrel | ||||||
| 49 | 8906 | 10 | 15 (3CL6) | A, B, E | Polysaccharide deacetylase | 3.1.1.58 |
| 50 | 3765 | 7 | 7 (4DI9) | B, E | 2‐Pyrone‐4,6‐dicarboxylate hydrolase, amidohydrolase | 3.1.1.57, 3.5.1.–, 3.5.2.– |
| Seven‐bladed β‐propeller | ||||||
| 51 | 8410 | 8 | 5 (3FGB) | A, B, E | 6‐Phosphogluconolactonase, 3‐carboxy‐muconate cyclase | 3.1.1.31, 5.5.1.5 |
| Three‐solenoid fold | ||||||
| 52 | 8166 | 22 | 13 (2NSP) | A, B, E | Pectinesterase, pectin methylesterase | 3.1.1.11 |
| Four‐layer α/β/β/α fold | ||||||
| 53 | 1325 | 2 | 3 (2WYM) | A, B | L‐Ascorbate 6‐phosphate lactonase, β‐lactamase | 3.1.1.–, 3.5.2.6 |
| Three‐layer β/β/α fold | ||||||
| 54 | 8877 | 14 | 19 (3KO9) | B, E | D‐Tyrosyl‐tRNA(Tyr) deacylase, D‐aminoacyl‐tRNA deacylase | 3.1.–.– |
| β‐barrel | ||||||
| 55 | 1024 | 2 | 1 (2ERV) | B | Outer membrane enzyme, deacylase, lipid A 3‐O‐deacylase | 3.1.1.77 |
| 56 | 3296 | 2 | 7 (1ILZ) | B, E | Phospholipase A1 | 3.1.1.32 |
| Two‐layer sandwich fold | ||||||
| 57 | 3136 | 94 | 4 (1J26) | B, E | Peptidyl‐tRNA hydrolase, peptide chain release factor I | 3.1.1.29 |
| β‐sandwich | ||||||
| 58 | 2129 | 16 | 3 (1BCI) | B, E | Cytosolic phospholipase A2 | 3.1.1.4 |
| Not described | ||||||
| 59 | 5037 | 4 | 5 (2RTX) | B, E | Peptide chain release factor 1, peptidyl‐tRNA hydrolase | 3.1.1.29 |
| 60 | 395 | 10 | 9 (4C7W) | V | Hemagglutinin‐esterase, E3 glycoprotein | 3.1.1.53 |
| 61 | 521 | 3 | 9 (2JZ7) | B | Lipase, polyurethanase, hemolysin E | 3.1.1.– |
| 62 | 386 | 4 | 1 (4NFU) | E | Senescence‐associated carboxylesterase | 3.1.1.1 |
| 63 | 3312 | 5 | 1 (3WMT) | B, E | Feruloyl esterase, tannase | 3.1.1.20, 3.1.1.73 |
| 64 | 944 | 14 | 5 (3FBX) | B, E | Phospholipase B‐like 2 | 3.1.1.– |
| No known tertiary structure | ||||||
| 65 | 13 | 1 | 0 | E | Phospholipase A2 | 3.1.1.4 |
| 66 | 438 | 7 | 0 | E | Putative peptidyl‐tRNA hydrolase | 3.1.1.29 |
| 67 | 11 | 11 | 0 | E | Phospholipase A1, phospholipase A2 | 3.1.1.4, 3.1.1.32 |
| 68 | 728 | 8 | 0 | E | Groups XIIA and XIIB secretory phospholipase A2 | 3.1.1.4 |
| 69 | 1234 | 4 | 0 | B, E | Poly(3‐hydroxybutyrate) depolymerase, feruloyl esterase | 3.1.1.75, 3.1.1.73 |
| 70 | 1334 | 1 | 0 | B, E | Carboxymethylenebutenolidase, dienelactone hydrolase | 3.1.1.45 |
| 71 | 34 | 1 | 0 | B | Poly(3‐hydroxyoctanoate) depolymerase | 3.1.1.76 |
| 72 | 2686 | 10 | 0 | A, B, E | Polyhydroxybutyrate depolymerase family, acetylxylan esterase, feruloyl esterase, carbohydrate esterase family 1 | 3.1.1.75, 3.1.1.72, 3.1.1.73 |
| 73 | 335 | 1 | 0 | B | Phospholipase A1 | 3.1.1.32 |
| 74 | 3091 | 1 | 0 | B, E | Lysophospholipase L2 | 3.1.1.5 |
| 75 | 7331 | 14 | 0 | B, E | GDSL family esterase/lipase | 3.1.1.– |
| 76 | 261 | 4 | 0 | B, E | Chlorophyllase | 3.1.1.14 |
| 77 | 284 | 1 | 0 | E | Triacylglycerol lipase | 3.1.1.3 |
| 78 | 688 | 2 | 0 | B, E | Acetylhydrolase, esterase, lipase, α/β‐hydrolase | 3.1.1.– |
| 79 | 514 | 3 | 0 | E | Triglyceride lipase, cholesterol esterase, lysosomal acid lipase | 3.1.1.–, 3.1.1.13 |
| 80 | 1828 | 10 | 0 | B, E | Patatin‐like phospholipase domain | 3.1.1.– |
| 81 | 212 | 1 | 0 | E | ATG15 protein, triacylglycerol lipase | 3.1.1.3 |
| 82 | 192 | 2 | 0 | E | Steroyl esterase, YEH1p, YEH2p | 3.1.1.13 |
| 83 | 2953 | 3 | 0 | B, E | Sialate O‐acetylesterase, 9‐O‐acetylesterase | 3.1.1.53 |
| 84 | 332 | 3 | 0 | E | Acyloxyacyl hydrolase | 3.1.1.77 |
| 85 | 55 | 3 | 0 | B | EstP (carboxylesterase) | 3.1.1.1 |
| 86 | 799 | 1 | 0 | B, E | Lipase | 3.1.1.– |
| 87 | 439 | 2 | 0 | B | Lipase | 3.1.1.– |
| 88 | 2378 | 34 | 0 | E | Phospholipase DDHD | 3.1.1.– |
| 89 | 465 | 3 | 0 | B | D‐(−)−3‐hydroxybutyrate oligomer hydrolase, cytochrome C1 | 3.1.1.22 |
| 90 | 1666 | 17 | 0 | B, E | Patatin‐like phospholipase domain protein, triacylglycerol lipase | 3.1.1.–, 3.1.1.3 |
| 91 | 862 | 14 | 0 | B, E | Phospholipase B1 | 3.1.1.– |
Most prevalent producers are bolded.
A, archaea; B, bacteria; E, eukaryota; V, virus.
The ClustalX sequence alignment files, using 50 representative sequences of each family (or all that were available, if <50), are in Supporting Information Figure S1. The conserved amino acid counts in each family are summarized in Supporting Information Table SI. In the great majority of cases, at least one and usually substantially more amino acid residues are totally conserved over all primary structures. A roughly equal number of residues of chemically similar but not identical amino acid residues are also conserved. The secondary structures of a representative of each family with a known tertiary structure are shown in Supporting Information Figure S2. Each family member has similar secondary structures in its core, but some members have either extra or missing α‐helices or β‐strands. RMSD and P avg values obtained by tertiary structure superposition also appear in Supporting Information Table SI. In a large fraction of all cases where multiple tertiary structures in a single family are available, RMSD < 1.5 Å and P avg > 90%.
A large majority of families have >1000 primary structures, and most of the rest have 100–1000 sequences (Table 1). As expected, all families have at least one sequence with evidence at protein level. Among them, 64 of the 91 families have known tertiary structures, with 27 families having none.
Many families have members produced by organisms in all three life kingdoms (Table 1). Members of other families are produced by organisms in two kingdoms, usually bacteria and eukaryota, or in a single kingdom (Table 1).
A total of 36 families were grouped into five clans by having secondary and tertiary structures that could be closely superimposed (Table 1), even though their primary structures may not be significantly similar. Supporting Information Table SII shows RMSD and P avg values and protein folds of each clan. In all cases, RMSD < 2.5 Å. However, P avg values cover a wide range. Tertiary structures of families within clans are more variable than those of family members, as the former do not share similar primary structures while the latter do.
Each clan has characteristic tertiary folds (Fig. 1). Clan A CEHs all have standard α/β hydrolase folds, in which the second β‐strand is antiparallel to the others in the β‐sheet.3, 4 The β‐sheet generally has eight β‐strands, which are found in the order 1, 2, 4, 3, 5, 6, 7, and 8, based on their amino acid residue numbers. Clan B members have similar tertiary structures as those in clan A; however, all their β‐strands are parallel to each other, proceeding in the same direction, and they are arranged in the order 2, 1, 3, 4, and 5. A sixth β‐strand, if present, may be found before or after the fifth one. Clan C enzymes have α/β hydrolase‐like tertiary structures as well, but their first rather than their second β‐strands are antiparallel to the others on the same β‐sheet. The β‐strand order is 1, 3, 2, 4, 5, 6, and 7. The tertiary structures of clan D members are six‐bladed β‐propeller folds, where each blade, consisting of four β‐sheets, shares a central axis. Clan E enzymes have three‐α‐helix up‐down bundle tertiary structures.
Figure 1.
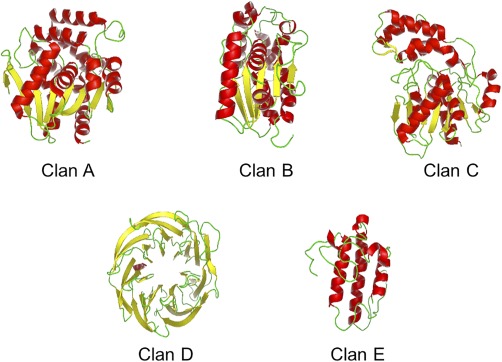
Tertiary structures of representative families from the five CEH clans. Clan A: family 2, Burkholderia xenovorans 3‐oxoadipate enol‐lactonase, PDB 2XUA. Clan B: family 17, Trichoderma reesei cutinase, PDB 4PSD. Clan C: family 30, Escherichia coli esterase, PDB 3BF8. Clan D: family 32, mouse SMP30/GNL hydrolase, PDB 4GN9. Clan E: family 34, Daboia russelii phospholipase A2, PDB 1TG1.
At present, over 60% of the CEH families are not part of clans, either because they have no known tertiary structures or because their members cannot be superimposed well on the tertiary structures of the members of existing clans or with each other, even when they share much the same fold. A total of 22 of these families have known tertiary structures designated by CATH or the Structural Classification of Proteins (SCOP) database,27 such as three‐layer α/β/α sandwiches (Rossmann folds), α/β‐hydrolase structures, patatin‐like folds, α/β‐TIM barrels, seven‐bladed β‐propellers, three‐solenoid folds, three‐layer β/β/α sandwiches, β‐barrels, two‐layer sandwich folds, and β‐sandwiches (Table 1). A further six families have tertiary structures that are not classified by either CATH or SCOP.
Of the most studied CEHs, large numbers of named carboxylesterases or EC 3.1.1.1 designations are found in families 12 (clan A), 26, 28, 29 (clan C), 62, and 85, of those not in any clan (Table 1). Families 4 (clan A), 21, 24 (clan B), 39, 77, 79, 81, and 90 (no clan) have triacylglycerol lipases (EC 3.1.1.3). Phospholipase A2s (EC 3.1.1.4) exist in families 23 (clan B), 34–36 (clan E), 58, 65, 67, and 68 (no clan). Lysophospholipases (EC 3.1.1.5) are in families 27 (clan C), 42, and 74 (no clan). Acetylcholinesterases (EC 3.1.1.7) and butyrylcholinesterases (often simply called cholinesterases) (EC 3.1.1.8) are found only in family 12 (clan A), as are cocaine esterases (EC 3.1.1.84). Cutinases (EC 3.1.1.74) are in family 17 (clan B). Enzyme name and EC number assignments tend to be somewhat elastic and arbitrary, causing some spurious variation, and some families now not assigned to clans will eventually find their way into them once they have known tertiary structures. Therefore, these lists are subject to future revision. However, it is evident by the number of sequences in each family that carboxylesterases are mainly in families 26, 28, and 29 (clan C), triacylglycerol lipases exist largely in families 21, 24 (clan B), and 39 (no clan), and phospholipase A2s are found predominantly throughout clan E and in family 58 (no clan).
CEH Mechanisms
Table 2 presents catalytic residues from representative families from each of the 64 families that have known tertiary structures, gathered from the articles corresponding to the PDB structures listed there. Clans A, B, and C all have catalytic residues characteristic of serine protease mechanisms, with catalytic serine residues followed by catalytic aspartate or glutamate residues and then catalytic histidine residues. In only four families in these three clans is this pattern not followed. There is not enough information to identify characteristic catalytic residues in families 10 and 14, a full set of three catalytic residues is not available for family 21, and a catalytic cysteine residue replaces a serine residue in family 25.
Table 2.
Catalytic Residues of Carboxylic Ester Hydrolase Families with Known Tertiary Structures
| Clan | Family | PDB designation | Producing species and enzyme | Catalytic residues |
|---|---|---|---|---|
| A | 1 | 1QIT | Moorea producens (Lyngbya majuscula) decarboxylating thioesterase | S100/E124/H266 |
| A | 2 | 4CCY | Bacillus subtilus carboxylesterase CesB | S130/E245/H274 |
| A | 3 | 4KRX | Escherichia coli acetyl esterase Aes | S165/D262/H292 |
| A | 4 | 4CG1 | Thermobifida fusca polyethylene terephthalate degrading hydrolase | S130/D176/H208 |
| A | 5 | 2XLB | Bacillus pumilus (Bacillus mesentericus) acetyl xylan esterase | S181/D269/H298 |
| A | 6 | 3C5V | Human PP2A‐specific methylesterase | S156/D181/H349 |
| A | 7 | 3D59 | Human plasma platelet‐activating factor acetylhydrolase | S273/D296/H351 |
| A | 8 | 4G4G | Sporotrichum thermophile (Myceliophthora thermophila) glucuronyl esterase | S213/E236/H346 |
| A | 9 | 1K8Q | Dog gastric lipase | S153/D324/H353 |
| A | 10 | – | – | – |
| A | 11 | 4UYU | Human WNT deacylase notum | S232/D340/H389 |
| A | 12 | 2JGJ | Mouse acetylcholinesterase | S203/E334/H447 |
| A | 13 | 1JJF | Clostridium thermocellum feruloyl esterase | S172/D230/H260 |
| B | 14 | – | – | – |
| B | 15 | 1U8U | Escherichia coli thioesterase/protease/lysophospholipase L1 | S10/D154/H157 |
| B | 16 | 1I6W | Bacillus subtilis lipase | S77/D133/H156 |
| B | 17 | 4PSD | Trichoderma reesei cutinase | S164/D216/H229 |
| B | 18 | 1VYH | Mouse PAF‐AH holoenzyme | S48/D/193/H196 |
| B | 19 | 1DEO | Aspergillus aculeatus rhamnogalacturonan acetylesterase | S9/D192/H195 |
| B | 20 | 1EB8 | Manihot esculenta hydroxynitrile lyase | S80/D208/H236 |
| B | 21 | 4HYQ | Streptomyces albidoflavus phospholipase A1 | S11/H218 |
| B | 22 | 1CVL | Chromobacterium viscosum lipase | S87/D263/H285 |
| B | 23 | 4X92 | Human lysosomal phospholipase A2 | S165/D327/H359 |
| B | 24 | 4FDM | Bacillus L2 lipase | S113/D317/H358 |
| C | 25 | 1DIN | Pseudomonas knackmussi dienelactone hydrolase | C123/D171/H202 |
| C | 26 | 4ETW | Shigella flexneri enzyme/ACP substrate gatekeeper | S82/D207/H235 |
| C | 27 | 3HJU | Human monoglyceride lipase | S121/D239/H269 |
| C | 28 | 3CN9 | Pseudomonas aeruginosa carboxylesterase | S113/D166/H197 |
| C | 29 | 1TQH | Geobacillus stearothermophilus carboxylesterase Est30 | S94/D193/H223 |
| C | 30 | 3BF7 | Escherichia coli esterase | S89/D113/S206/H234 |
| C | 31 | 1LBS | Moesziomyces antarticus triacylglycerol hydrolase | S105/D187/H224 |
| D | 32 | 2DG0 | Staphylococcus aureus lactonase | D138/D236 |
| D | 33 | 1V04 | Human/rabbit/mouse/rat serum paraoxonase | H115/H134 |
| E | 34 | 1PPA | Agkistrodon piscivorus lysine 49 phospholipase A2 | H48/Y73/D99 |
| E | 35 | 1POC | Apis mellifera venom phospholipase A2 | H34/D35 |
| E | 36 | 2WG7 | Rice class X1b phospholipase A2 | H61/D62 |
| – | 37 | 1RZW | Archaeglobus fulgidis peptidyl‐tRNA hydrolase | H20/D93/H113 |
| – | 38 | 3EB9 | Trypanosoma brucei 6‐phosphogluconolactonase | D163/H165 |
| – | 39 | 1RP1 | Dog pancreatic lipase related protein | S152/D176/H263 |
| – | 40 | 1TGL | Rhizomucor michei triacylglycerol lipase | S144/D203/H257 |
| – | 41 | – | – | – |
| – | 42 | 1CJY | Human cytosolic phospholipase A2 | S228/D549 |
| – | 43 | 3KVN | Pseudomonas aeruginosa autotransporter EstA | S14/D286/H289 |
| – | 44 | 1UWC | Aspergillus niger feruloyl esterase | S133/D194/H247 |
| – | 45 | 4HOY | Acinetobacter baumanni peptidyl‐tRNA hydrolase | H22/N70/D95/N116 |
| – | 46 | 1TIA | Penicillium camemberti lipase | S145/D199/H259 |
| – | 47 | 1CHD | Salmonella enterica chemotaxis receptor methylesterase | S164/H190/D286 |
| – | 48 | 4PKB | Solanum cardiophyllum patatin‐17 | S77/D215 |
| – | 49 | 3CL6 | Pseudomonas fluorescens allantoinase | E36/H259 |
| – | 50 | 4DI9 | Sphingomonas paucimobilisi 2‐pyrone‐4,6‐dicarboxylic acid carboxylase | R124/D248 |
| – | 51 | – | – | – |
| – | 52 | 2NSP | Dickeya dadantii (Erwinia chrysanthami) pectin methylesterase | Q177/D178/D199 |
| – | 53 | 2WYM | Escherichia coli l‐ascorbate‐6‐phospholactonase | D121/H122/D226/H281 |
| – | 54 | 3KO9 | Plasmodium falciparum d‐amino acid deacylase | T90 |
| – | 55 | 2ERV | Pseudomonas aeruginosa 3‐O‐deacylase | H126/S128/E140 |
| – | 56 | 1ILZ | Escherichia coli phospholipase A | H142/S144/N156 |
| – | 57 | – | – | – |
| – | 58 | 1CJY | Human cytosolic phospholipase A2 | S228/D549 |
| – | 59 | 2JY9 | Thermus thermophilus YaeJ bound to ribosome | Gln28 |
| – | 60 | 1FLC | Influenza C virus hemagglutinin‐esterase fusion glycoprotein | S57/D352/H355 |
| – | 61 | 2Z8X | Pseudomonas sp. family I.3 lipase | S207/D255/H313 |
| – | 62 | 4NFU | Arabidopsis thaliana signaling node | S123/D187/H317 |
| – | 63 | 3WMT | Aspergillus oryzae feruloyl esterase B | D17/S203/H457 |
| – | 64 | – | – | – |
Clans D and E present different catalytic residue patterns than Clans A, B, and C. Clan D, with six‐bladed β‐propeller folds, has two characteristic patterns, two aspartate residues in family 32 and two histidine residues in family 33. Clan E enzymes, with three‐α‐helix up‐down bundle tertiary structures, have histidine and aspartate catalytic residues.
Identities and locations of catalytic residues are more variable in families 37–64. Families 37–47, less families 41, 45, and 47, which have various α/β‐hydrolase structures, have all or some of the catalytic residues characteristic of serine proteases. Families 60–62, which at present do not have settled tertiary structures, have Ser/Asp/His catalytic residue patterns, suggesting that they may be composed of serine proteases when more is known about them.
Database Construction
The information covered earlier has been gathered into a freely accessible database labeled CASTLE (CArboxylic eSTer hydroLasEs, http://www.castle.cbe.iastate.edu). This database at present is divided into the 91 CEH families defined earlier, each of them containing a listing of enzyme names or other identifiers, EC numbers, producing organism genus and species names, and GenBank, RefSeq, UniProt, and PDB accession codes in columns (Fig. 2). Most entries are of proteins gathered from genomic studies that have not undergone experimental verification, and therefore, only a minority have EC numbers. Producing organisms are separated into archaea, bacteria, and eukaryota. Some families have entries in all three kingdoms, while others have entries in one or two. Entries in each kingdom are alphabetically ordered by the genus of the producing organism. In general there are 500 entries on each page.
Figure 2.
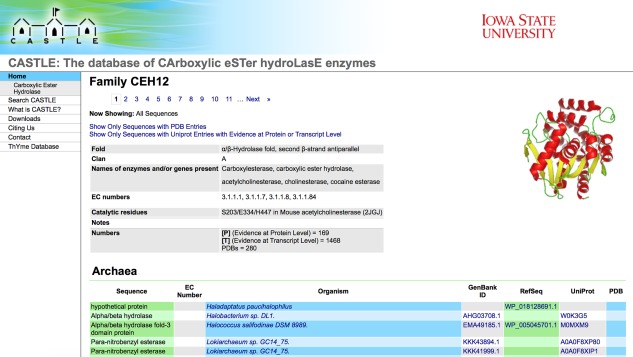
Screen shot of the family 12 (clan A) introductory page.
The top of each page (Fig. 2) shows an addressable list of pages, prompts to see only sequences with PDB entries or only sequences with UniProt entries having evidence at protein level (labeled by [P]) or transcript level (labeled by [T]). Other information is the characteristic fold of the family (if known), dominant names of enzymes and/or genes present, dominant EC numbers, catalytic residues (if known), and numbers of sequences with evidence at protein and transcript levels and with PDB entries. All enzyme names and EC numbers are taken from either the UniProt database or the NCBI protein database; we do not assign enzyme names or EC numbers.
Entries in all columns except that of enzyme names or identifiers are linked to other electronic resources: those of EC numbers to the ExPASy SwissProt enzyme nomenclature database (http://enzyme.expasy.org/EC/ x), those of producing organisms to the NCBI taxonomy database (http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id = y), those of GenBank and RefSeq identifiers to the NCBI protein database (http://www.ncbi.nlm.nih.gov/protein/ y), those of UniProt identifiers to the UniProt database (http://www.uniprot.org/uniprot/ y/), and those of PDB identifiers to the PDB database (http://www.rcsb.org/pdb/explore/explore.do?structureId=y), where x is the EC number and y is the NCBI taxonomy, GenBank, RefSeq, UniProt, or PDB accession codes.
Two features ease the task of navigating and retrieving information in CASTLE. A search tool allows keywords, EC numbers and GenBank, RefSeq, UniProt, or PDB accession codes to be searched. Furthermore, each family can be downloaded into a comma‐separated value file, which can be viewed in a spreadsheet. These files can be obtained by contacting the authors.
Conclusions
Classifying CEH enzymes into families and clans provides valuable insights about them. Several observations may be made about their structural classifications:
Often CEHs with the same enzyme function appear in multiple families and clans. For instance, phospholipase A2s are found in eight families. Four of these families are in two clans, with a fifth family having a known tertiary structure but not being in a clan. Three further families have no known tertiary structures. Carboxylesterases occur in six families, four of them in two clans. A fifth has a known but not firmly described structure, while a sixth has no known tertiary structure.
On the other hand, some major enzyme groups are found in single families. Acetyl‐ and butylrylcholinesterases and cocaine esterases all occupy family 12 in clan A.
All clans except clan E include diverse enzymes with various functions, as do families not in clans. The clan with the largest number of families (clan A) includes enzymes with eight different EC numbers, plus the generic EC 3.1.1.–, and many more than eight enzyme functions. Clan B contains enzymes with seven EC numbers plus EC 3.1.1.–.
It is common for some families to contain enzymes with not only EC numbers denoting CEHs, but also with other EC numbers.
Some families show little experimental work on their enzymes. Twenty‐seven families (those from 65 to 91) at present have no known tertiary structures. Eight of them, plus two others, have only one sequence with evidence at protein level for each family. These families need more attention from researchers, because they may have novel substrate specificities that will be useful or important to industrial or medical applications.
To emphasize the sharp difference between classifying enzymes by function and classifying them by structure, only enzymes from 33 EC numbers out of the total 91 EC numbers starting with EC 3.1.1 (1–5, 7, 8, 11, 13, 14, 17, 20, 22, 24, 29, 31, 32, 45, 47, 53, 57, 58, 61, 72–77, 81, 84, 89, and 96) are found in Table 1. This indicates that the other 68 supposed CEHs either do not actually exist, have been combined with other CEHs, or have been researched very rarely.
To extend the utility of this work, it has been gathered into a freely accessible database, entitled CASTLE (CArboxylic eSTer hydroLasEs, http://www.castle.cbe.iastate.edu).
Supporting information
Supporting Information 1
Supplementary Table SI. Tertiary structure similarity within families. Families (1–91) vs. multiple sequence alignment count for total conservation of identical and chemically similar but not identical amino acid residues, along with RMSD and P avg values for each family.
Supplementary Table SII. Tertiary structural similarity within clans. Clans (A–E) vs. number of families, RMSD and P avg values, and characteristic folds for each clan.
Supplementary Figure S1. Multiple sequence alignments of each family.
Supplementary Figure S2. Secondary structures (wire diagrams) of those families that have known tertiary structures.
Acknowledgments
The authors thank Google for the use of its cloud platform to allow the intensive computation conducted in this project. They declare no conflict of interest.
References
- 1. Nomenclature Committee of the International Union of Biochemistry and Molecular Biology (NC–IUBMB) (1992) Enzyme Nomenclature. San Diego, CA: Academic Press. [Google Scholar]
- 2. Sillitoe I, Lewis TE, Cuff A, Das S, Ashford P, Dawson NL, Furnham N, Laskowski RA, Lee D, Lees JG, Lehtinen S, Studer RA, Thornton J, Orengo CA (2015) CATH: comprehensive structural and functional annotations for genome sequences. Nucleic Acids Res 43:D376–D381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ollis DL, Cheah E, Cygler M, Dijkstra B, Frolow F, Franken SM, Harel M, Remington SJ, Silman I, Schrag J, Sussman JL, Verschueren KHG, Goldman A (1992) The α/β hydrolase fold. Protein Eng 5:197–211. [DOI] [PubMed] [Google Scholar]
- 4. Carr PD, Ollis DL (2009) Alpha/beta hydrolase fold: an update. Protein Pept Lett 16:1137–1148. [DOI] [PubMed] [Google Scholar]
- 5. Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B (2014) The carbohydrate‐active enzymes database (CAZy) in 2013. Nucleic Acids Res 42:D490–D495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cantu DC, Chen Y, Lemons ML, Reilly PJ (2011) ThYme: a database for thioester‐active enzymes. Nucleic Acids Res 39:D342–D346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cousin X, Hotelier T, Giles K, Lievin P, Toutant J‐P, Chatonnet A (1997) The α/β fold family of proteins database and the cholinesterase gene server ESTHER. Nucleic Acids Res 25:143–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lenfant N, Hotelier T, Velluet E, Bourne Y, Marchot P, Chatonnet A (2013) ESTHER, the database of the α/β‐hydrolase fold superfamily of proteins: tools to explore diversity of functions. Nucleic Acids Res 41:D423–D429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Widmann M, Juhl PB, Pleiss J (2010) Structural classification by the lipase engineering database: a case study of Candida antarctica lipase A. BMC Genomics 11:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI‐BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kang H‐Y, Kim JF, Kim MH, Park SH, Oh T‐K, Hur C‐G (2006) MELDB: a database for microbial esterases and lipases. FEBS Lett 580:2736–2740. [DOI] [PubMed] [Google Scholar]
- 12. Cantu DC, Chen Y, Reilly PJ (2010) Thioesterases: a new perspective based on their primary and tertiary structures. Protein Sci 19:1281–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen Y, Kelly EE, Masluk RP, Nelson CL, Cantu DC, Reilly PJ (2011) Structural classification and properties of ketoacyl synthases. Protein Sci 20:1659–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. The UniProt Consortium (2015) UniProt: a hub for protein information. Nucleic Acids Res 43:D204–D212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador‐Vegas A, Salazar GA, Tate J, Bateman A (2016) The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res 44:D279–D285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pruitt KD, Brown GR, Hiatt SM, Thibaud‐Nissen F, Astashyn A, Ermolaeva O, Farrell CM, Hart J, Landrum MJ, McGarvey KM, Murphy MR, O'Leary NA, Pujar S, Rajput B, Rangwala SH, Riddick LD, Shkeda A, Sun H, Tamez P, Tully RE, Wallin C, Webb D, Weber J, Wu W, DiCuccio M, Kitts P, Maglott DR, Murphy TD, Ostell JM (2014) RefSeq: an update on mammalian reference sequences. Nucleic Acids Res 42:D756–D763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Benson DA, Cavenaugh M, Clark K, Karsch‐Mizrachi I, Lipman DJ, Ostell J, Sayers EW (2013) GenBank. Nucleic Acids Res 41:D36–D42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) The Protein Data Bank. Nucleic Acids Res 28:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wu CH, Yeh L‐SL, Huang H, Arminski L, Castro‐Alvear J, Chen Y, Hu Z, Kourtesis P, Ledley RS, Suzek BE, Vinayaka CR, Zhang J, Barker WC (2003) The protein information resource. Nucleic Acids Res 31:D345–D347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.The Protein Research Foundation. Available at: http://www.prf.or.jp.
- 21. Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. [DOI] [PubMed] [Google Scholar]
- 22. Shatsky M, Nussinov R, Wolfson HJ (2004) A method for simultaneous alignment of multiple protein structures. Proteins 56:143–156. [DOI] [PubMed] [Google Scholar]
- 23. Kabsch W, Sander C (1983) Dictionary of protein secondary structure: pattern recognition of hydrogen‐bonded and geometrical features. Biopolymers 22:2577–2637. [DOI] [PubMed] [Google Scholar]
- 24. Touw G, Baakman C, Black J, te Beek TAH, Krieger E, Joosten RP, Vriend G (2015) A series of PDB‐related databases for everyday needs. Nucleic Acids Res 43:D364–D368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. DeLano WL (2002) The PyMOL molecular graphics system, version 1.8. Schrödinger LLC. Available at: http://www.pymol.org/.
- 26. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Murzin AG, Brenner SE, Hubbard T, Chothia C (1995) SCOP: a structural classification of proteins database for the investigation of sequences and structures. J Mol Biol 247:536–540. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information 1
Supplementary Table SI. Tertiary structure similarity within families. Families (1–91) vs. multiple sequence alignment count for total conservation of identical and chemically similar but not identical amino acid residues, along with RMSD and P avg values for each family.
Supplementary Table SII. Tertiary structural similarity within clans. Clans (A–E) vs. number of families, RMSD and P avg values, and characteristic folds for each clan.
Supplementary Figure S1. Multiple sequence alignments of each family.
Supplementary Figure S2. Secondary structures (wire diagrams) of those families that have known tertiary structures.


