Abstract
Aims
Ischemic cardiomyopathy (ICM) resulting from myocardial infarction is a major cause of heart failure (HF). Recently, thousands of long non-coding RNAs (lncRNAs) have been discovered and implicated in a variety of biological processes. However, the role of most lncRNAs in HF remains largely unknown. The aim of this study is to test the hypothesis that the expression and function of lncRNAs are differentially regulated in diseased hearts.
Methods and results
In this study, we performed RNA deep sequencing of protein-coding and non-coding RNAs from cardiac samples of patients with ICM (n = 15) and controls (n = 15). Genome-wide transcriptome analysis confirmed that many protein-coding genes previously known to be involved in HF were altered in ICM hearts. Among the 145 differentially expressed lncRNAs identified in ICM hearts, we found a set of 35 lncRNAs that display strong positive expression correlation. Expression correlation coefficient analyses of differentially expressed lncRNAs and protein-coding genes revealed a strong association between lncRNAs and extracellular matrix (ECM) protein-coding genes. We overexpressed or knocked down selected lncRNAs in cardiac fibroblasts and our results suggest that lncRNAs are important regulators of fibrosis and the expression of ECM synthesis genes. Moreover, we show that lncRNAs participate in the TGF-β pathway to modulate the expression of ECM genes and myofibroblast differentiation.
Conclusion
Our studies demonstrate that the expression of many lncRNAs is dynamically regulated in ICM. lncRNAs regulate the expression and function of ECM and cardiac fibrosis during the development of ICM. Our results further indicate that lncRNAs may represent novel regulators of heart function and cardiac disorders, including ICM.
Keywords: Ischemic cardiomyopathy , lncRNA , Heart disease , Non-coding RNAs , Extracellular matrix
1. Introduction
Ischemic cardiomyopathy (ICM), often resulting from the narrowing of coronary arteries, is a leading cause of heart failure (HF).1,2 More than 1 million people suffer heart attacks each year in the USA alone. Multiple genetic and environmental factors have been linked to ICM.3 However, the molecular events that lead to this disorder remain elusive.
One surprise of the human genome project was that only about 20 000 to 25 000 protein-coding genes exist in our species. Astonishingly, less than 2% of the sequences in the human genome are used for coding proteins. It is now recognized that the majority of the genome is actively transcribed to produce thousands of non-coding transcripts, including long non-coding RNAs (lncRNAs), which are RNAs >200 nt in length that do not encode proteins. The function of lncRNAs in diverse biological processes and diseases has begun to be investigated.4,5 Genome-scale profiling has identified numerous lncRNAs in normal and diseased conditions.5,6
Recent studies have linked several lncRNAs to cardiovascular development and disease. Braveheart, a novel lncRNA, has been defined to be a critical regulator of cardiovascular commitment from embryonic stem cells (ESCs).7,8 Mechanistically, braveheart mediates the epigenetic regulation of cardiac commitment by interacting with SUZ12, a component of the polycomb repressive complex 2 (PRC2).7,9 Fendrr, another novel cardiac lncRNA, has been shown to be an essential regulator of heart and body wall development.8,10 Most recently, several studies have documented that expression and function of lncRNAs are associated with a variety of cardiovascular conditions. For example, it was reported that angiotensin II induced the expression of lncRNAs in vascular smooth muscle cells.11 Another study documented dysregulated lncRNAs in infarcted mouse hearts.12 Interestingly, Yang et al.,13 performed RNA deep sequencing and they identified HF associated lncRNAs in human patients. Together, these studies suggest that lncRNAs participate in cardiac disease pathogenesis. However, these pioneering studies have likely revealed the tip of the iceberg, and it remains largely unknown how lncRNAs participate in human heart disease, including ischemia-induced cardiomyopathy and HF.
In order to gain insight into cardiomyopathy-related lncRNAs, in this study we investigated the expression of lncRNAs in heart samples from human patients with ICM. Compared to controls, we found that the expression of lncRNAs is dysregulated in hearts of ICM patients. We performed experiments to verify the regulation of lncRNAs in diseased hearts and, most importantly, linked the function of dysregulated lncRNAs to the TGF-β pathway and the differentiation of cardiac fibroblasts (CF). Our studies therefore identify novel mechanisms by which lncRNAs regulate cardiac function and disease.
2. Methods
2.1 Cardiac tissues, RNA isolation, RNA-seq, and data analyses
Left ventricular myocardial samples of 15 patients with ICM and 15 non-ICM controls were the same as those used in our prior study.14 Details regarding the signs and symptoms of the ICM patients were reported.14 RNA was extracted with Trizol (Invitrogen) and depleted of rRNA using the Ribominus kit (Life Technologies). Illumina RNA-seq libraries were prepared using the standard Illumina protocol. Each of the 30 heart samples was used to generate a separate RNA-seq library that was analyzed on one lane of an Illumina GAIIx (36 nt paired-end reads). Data analyses were performed as described.15–17 Additional myocardial samples were obtained from HF patients and controls for RNA extraction and qRT-PCR analysis.18 For overexpression and knockdown of lncRNAs, CF were isolated from adult or neonatal mouse hearts using a previously described procedure.19 Details of materials and methods are provided in the Supplementary material online.
2.2 Study approval and ethical aspects of the study
Experiments of isolation of CF from adult and neonatal mice were approved by the IACUC of Boston Children’s Hospital, Boston, MA, USA. All animal experiments were performed conform the NIH guidelines-Guide for the Care and Use of Laboratory Animals. CO2 was used to sacrifice the adult mice for the isolation of CF. The flow of CO2 is slow enough not to make a noise or frighten the animals. Slower flow also allows the animals a more gradual induction of unconsciousness and is thus less stressful. Animals were left in the chamber for a sufficient time (4–5 min) so that complete asphyxia has been attained. For neonatal mice, decapitation was used to sacrifice the animals for the isolation of CF.
Acquisition and investigation of de-identified myocardial samples was performed under protocols approved by Institutional Review Boards of The University of Sydney, University of Utah, Comenius University, and Boston Children’s Hospital. All patients provided written informed consent. The human study was performed conform the declaration of Helsinki.
3. Results
3.1 Systematic determination of the expression of protein-coding genes in ICM
We obtained left ventricular myocardial samples from 30 human patients, 15 with ICM and 15 sex- and age-matched controls without HF as previously described.14 Total RNA purified from each myocardial sample was depleted of ribosomal RNA, fragmented, and converted into libraries for high throughput sequencing (see Methods). The 30 libraries were sequenced on an Illumina Genome Analyzer IIx, yielding 36 nucleotide reads from each end of cDNA fragments (Figure 1A and see Supplementary material online, Figure S1). Each sample yielded at least 20 million reads (range: 20.77–38.18 million). About 90% of the reads were useable reads that aligned uniquely to the human genome (see Supplementary material online, Table S1).
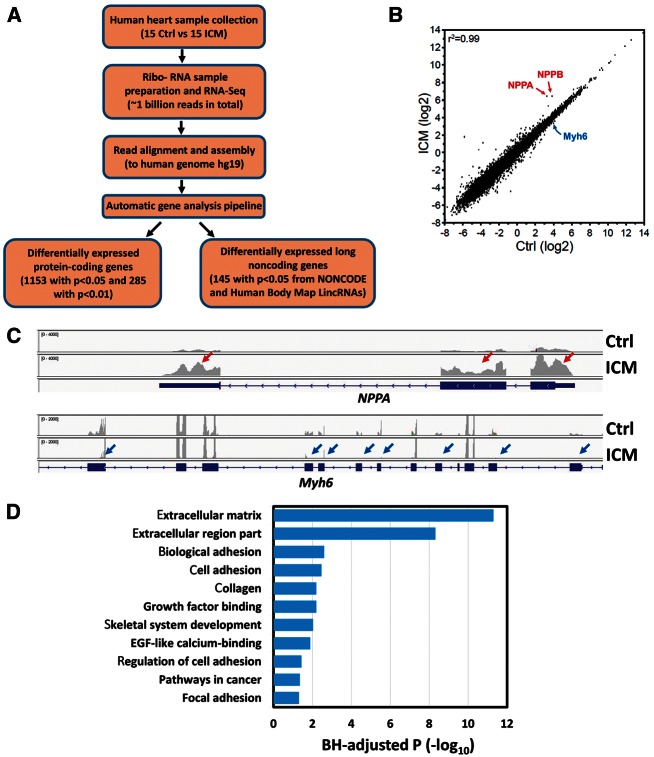
Figure 1.
RNA-Seq analyses of gene expression in control and ICM human heart samples. (A) Workflow of RNA-Seq and data analysis; (B) Scatter plot of the expression of protein-coding genes between control and ICM samples indicates that the overall transcriptome for protein-coding genes is similar between control and ICM hearts (R2 =0.99); (C) Comparison of total reads from control and ICM hearts for NPPA and Myh6 loci. The RNA-Seq reads were plotted and visualized with IGV browser. Red and blue arrows indicate up- or down-regulated expression of exons, respectively, in ICM. (D) Gene ontology analysis of 1051 up-regulated protein-coding genes (P<0.05) in ICM samples. The GO terms are ranked by the Benjamini–Hochberg (BH)-adjusted P-value.
We first measured the expression level of protein-coding genes by transcript read density, quantified as reads per kilobase per million mapped reads (RPKM). Overall intragroup correlation coefficients (Pearson) for all pairwise comparisons of mean expression values >1 RPKM were 0.97 ± 0.02 (mean ± SD) in control and 0.97 ± 0.03 in ICM. Intergroup correlations between ICM and control likewise showed a high correlation of 0.99 (Figure 1B), suggesting that overall the transcriptome is similar between ICM and control hearts. Hierarchical clustering analyses clearly documented differentially expressed genes between ICM and control samples. A total of 1153 genes are differentially expressed in ICM, with 102 down- and 1051 up-regulated genes when compared to controls (P < 0.05) (see Supplementary material online, Figure S2, Table S2). We then applied a more stringent statistical cutoff (P < 0.01) to the profiling in order to avoid potential false positives. As a result, a total of 285 genes were differentially expressed in ICM (15 down- and 270 up-regulated; see Supplementary material online, Figure S3). The results of our analysis are consistent with a recently reported similar study.13
We inspected the expression of signature marker genes that are associated with HF in the two groups. Indeed, many marker genes were found to be differentially expressed. NPPA and NPPB, well-known HF markers, were up-regulated in ICM samples (Figure 1B, C). Consistent with previous reports, MYH6, a transcript encoding a key myosin isoform that mediates heart contraction, was down-regulated in ICM (Figure 1B, C). In addition, genes associated with other functions, such as apoptosis (PHLDA1), cell growth (FRZB, SFRP4, SPOCK, CTGF), and cell cycle control (G0S2) were differentially expressed (see Supplementary material online, Figure S2, Table S2), consistent with a previous report on human dilated cardiomyopathy.20 We performed Gene Ontology (GO) term analysis and found that the differentially regulated genes were enriched for functional annotations related to extracellular matrix (ECM), cell adhesion, collagen, and growth factor binding, as demonstrated by Benjamini–Hochberg (BH)-adjusted P-values (Figure 1D). Together, these data show the genome-wide dynamics of protein-coding gene expression in human ICM.
3.2 The expression of lncRNAs is altered in ICM
We examined the expression of lncRNAs in ICM and control hearts. We used two lncRNA data sources for our analysis: the NONCODE 3.0 non-coding RNA database21 and the Human Body Map lincRNAs Catalog.22 Together, a collective lncRNA gene list containing over 30 000 annotated human lncRNA genes was used in this study.21,22 All single-exon transcripts as well as any transcripts overlapping RefSeq genes and known protein-coding genes were removed to exclude potential protein-coding genes (Figure 2A). To define a stringent set of candidate lncRNAs, transcripts were selected from the catalogue of expressed transcripts if they fulfilled both of the following criteria: (i) >200 bp long and (ii) expression of >0.5 RPKM in the sample (Methods; Figure 2A). Out of these transcripts, 145 lncRNAs were differentially expressed between ICM and control (multiple testing corrected P-value <0.05; Figure 2B and see Supplementary material online, Figure S4, Table S3). Of these lncRNAs, 37 and 108 were down- and up-regulated, respectively, in ICM. We selected the significance threshold of P < 0.05 for this analysis because we reasoned that a more stringent threshold (e.g. P < 0.01) might lead to loss of some true positive, novel lncRNAs. Comparing these 145 differentially expressed lncRNAs to 679 lncRNAs differentially expressed in ICM patients in an earlier study,13 we found that 19 were dysregulated in both studies. Genome browser views of representative differentially expressed lncRNAs are shown in Figure 2C.
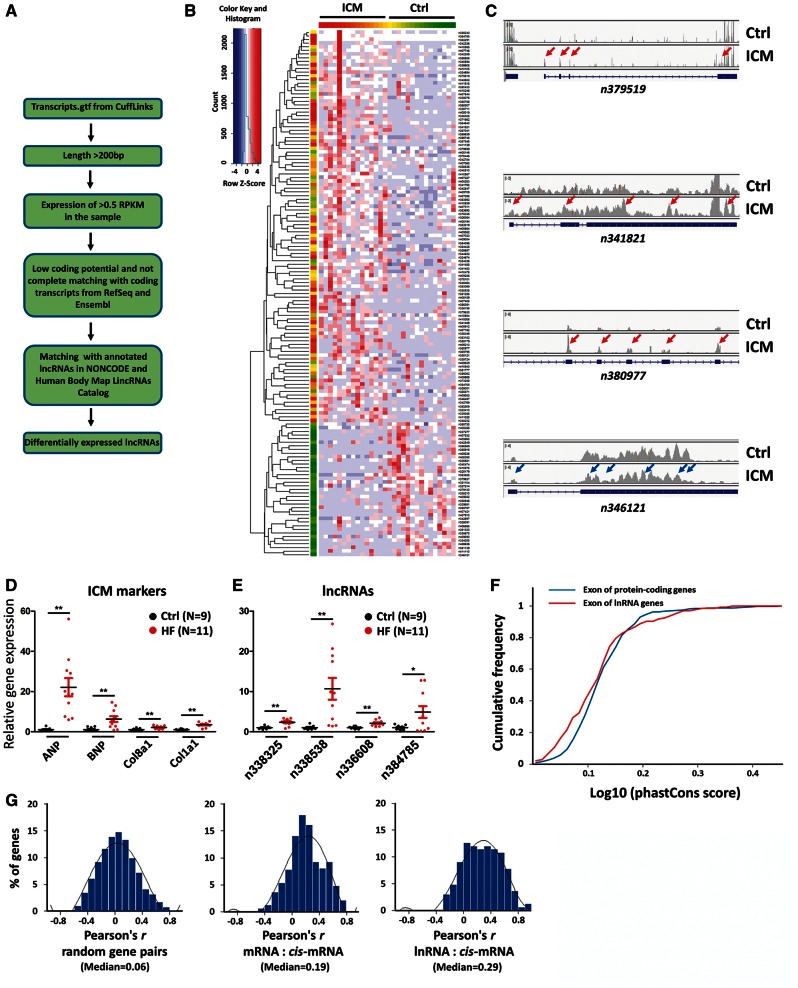
Figure 2.
Analyses of differentially expressed lncRNAs in control and ICM human heart samples. (A) Workflow of identification of differentially expressed lncRNAs; (B) Hierarchical clustering of 145 differentially expressed lncRNA genes between control and ICM hearts (P<0.05); (C) Comparison of total reads from control and ICM hearts for n379519, n341821, n380977 and n346121 loci. The RNA-Seq reads were plotted and visualized with IGV browser. Red and blue arrows indicate up- or down-regulated expression of exons, respectively, in ICM. (D, E) qRT-PCR analysis of the expression of (D) cardiomyopathy marker genes, and (E) dysregulated lncRNAs in control and ICM hearts. N number for each group is indicated.*P<0.05; **P<0.01. (F) Evolutionary conservation of lncRNAs and mRNAs. Cumulative distribution of sequence conservation across 11 species for differentially expressed lncRNAs (red) and protein-coding genes (blue). (G) The Pearson correlation coefficient analyses of random gene pairs, mRNA:cis-mRNA pairs, and pairs of lncRNAs and neighbour protein-coding genes (lncRNA:cis-mRNAs).
To validate the RNA-seq results, we next measured the expression of dysregulated lncRNAs by quantitative RT-PCR (qRT-PCR). Total RNA from control (Ctrl, n=9) and heart failure (HF, n=11) patient heart samples were used. We first verified an increased expression of cardiomyopathy markers, including ANP, BNP, and fibrosis markers, including Col8a1 and Col1a1, in the hearts of HF patients (Figure 2D). qRT-PCR confirmed altered expression of candidate lncRNAs tested (Figure 2E).
Next, we analysed the sequence conservation of lncRNAs and compared it with that of protein-coding genes. We computed phastCons scores from the exons of lncRNAs and protein-coding genes that were significantly dysregulated in patient hearts (P < 0.05 for lncRNAs and P < 0.01 for protein-coding genes), using 11 primate species plus mouse. As shown in Figure 2F, both lncRNAs and mRNAs are conserved. However, the distribution of protein-coding mRNA phastCons scores was shifted towards higher values compared to lncRNAs (Figure 2F). Specifically, phylopCons scoring shows that 80.5% of 145 differentially expressed lncRNAs are conserved, while 92.2% of 285 differentially expressed protein-coding genes are conserved. Our results support the view that, in general, lncRNAs are less conserved than protein-coding genes at the primary sequence level, which is consistent with previous reports,15,16 although some lncRNAs do display high primary sequence conservation.
lncRNAs frequently regulate their neighbouring genes. To test the hypothesis that levels of differentially expressed lncRNAs are coupled to their neighboiring genes, we analysed the correlation of a gene and its neighbouring coding genes. Comparing differentially expressed lncRNA-coding gene pairs to differentially expressed coding-coding or random coding-coding pairs, we found that lncRNAs were more correlated to their neighbouring coding (Figure 2G and see Supplementary material online, Table S4). Using a Pearson correlation coefficient of greater than 0.5, 24% of lncRNA-coding gene pairs were correlated, compared to 16% for coding-coding gene pairs (P < 0.001, Fisher’s exact test).
3.3 Functional correlation between lncRNA expression and ICM
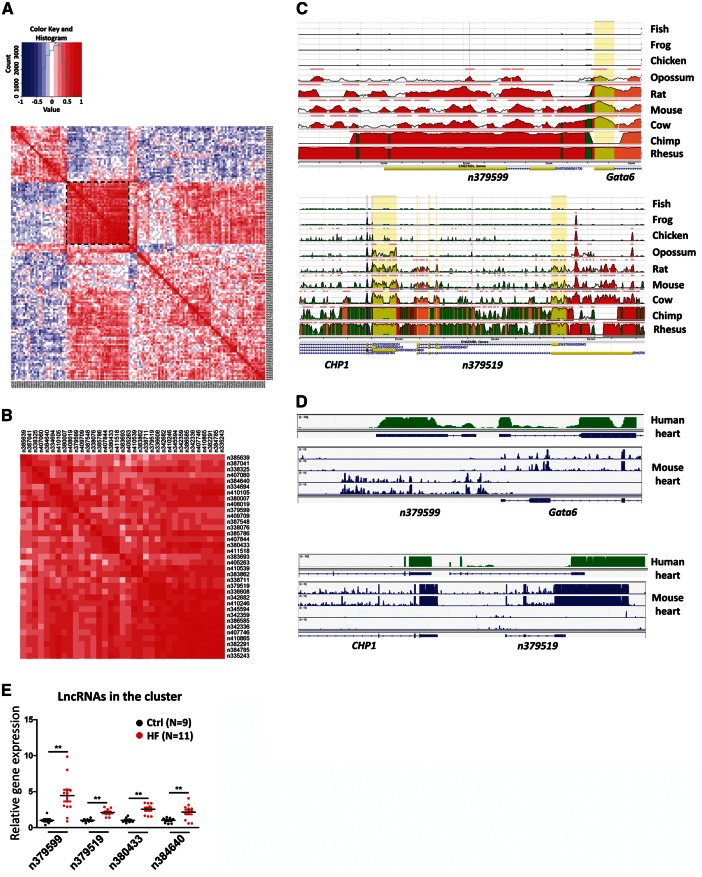
Having identified lncRNAs that are differentially expressed between ICM and control groups, we next attempted to infer the function of these lncRNAs. We first created an lncRNA–lncRNA expression correlation coefficient matrix for the 145 lncRNAs that were differentially expressed in ICM hearts (Figure 3A). An lncRNA cluster containing 35 up-regulated lncRNAs in the ICM group showed strong positive expression correlation (Figure 3A, boxed; Figure 3B and see Supplementary material online, Figure S5), suggesting that they work together and regulate a common biological process or signalling pathway. Importantly, some of the lncRNAs in these clusters are highly conserved across species. We found that the genomic sequences for lncRNAs n379599 and n379519 are highly conserved among mammalian species (Figure 3C), consistent with the result of prior analysis (Figure 2F). lncRNAs frequently regulate adjacent genes,23 and our Pearson correlation coefficient analysis indicates that lncRNAs and their cis-mRNAs display strong correlations (Figure 2G). Intriguingly, several of these differentially expressed lncRNA genes are located in the genomic loci adjacent to some known important regulators of cardiac disease. For example, n379599 is located less than 1 kb away from the transcriptional start site of the GATA6 gene, a member of the GATA family of transcription factors, which plays a key role in the regulation of cardiogenesis and cardiac hypertrophy24; n379519 is located less than 3 kb away from the transcriptional start site of the CHP1 gene, which encodes a calcineurin homologous protein25 (Figure 3D). It will be interesting to experimentally determine whether the expression and function of these neighbouring lncRNAs and protein coding genes are correlated to participate in the regulation of cardiac gene expression and heart function. Using qRT-PCR assays, we verified that the expression of lncRNAs from the cluster was elevated in the hearts of patients with HF (Figure 3E). Together, these analyses indicate that subsets of lncRNAs are associated with common regulatory pathways involving ICM.
Figure 3.
Expression correlation of differentially expressed lncRNAs in ICM. (A) Matrix for expression correlation of 145 differentially expressed lncRNAs. Positive (red) and negative (blue) expression correlations are shown. A cluster sub-matrix is boxed and shown in panel (B). (C) DNA sequence conservation of lncRNAs n379599 and n379519 and their neighbouring protein-coding genes in human and other species was analysed and visualized by GCR (Evolutionary Conservation of Genomes) browser. (D) The homologous loci for n379599 and n379519 are transcribed in both human and mouse hearts. RNA-Seq data from human and mouse hearts are visualized by IGV browser. The RNA Seq data of mouse hearts (8-week-old hearts of C57BL/6J mice) come from ENCODE/Cold Spring Harbor Lab (GSM900199). (E) qRT-PCR analysis of the expression of lncRNAs from the cluster in control and ICM hearts. N number for each group is indicated.*P<0.05; **P<0.01.
3.4 The expression of lncRNAs is linked to ECM gene expression and cardiac fibrosis
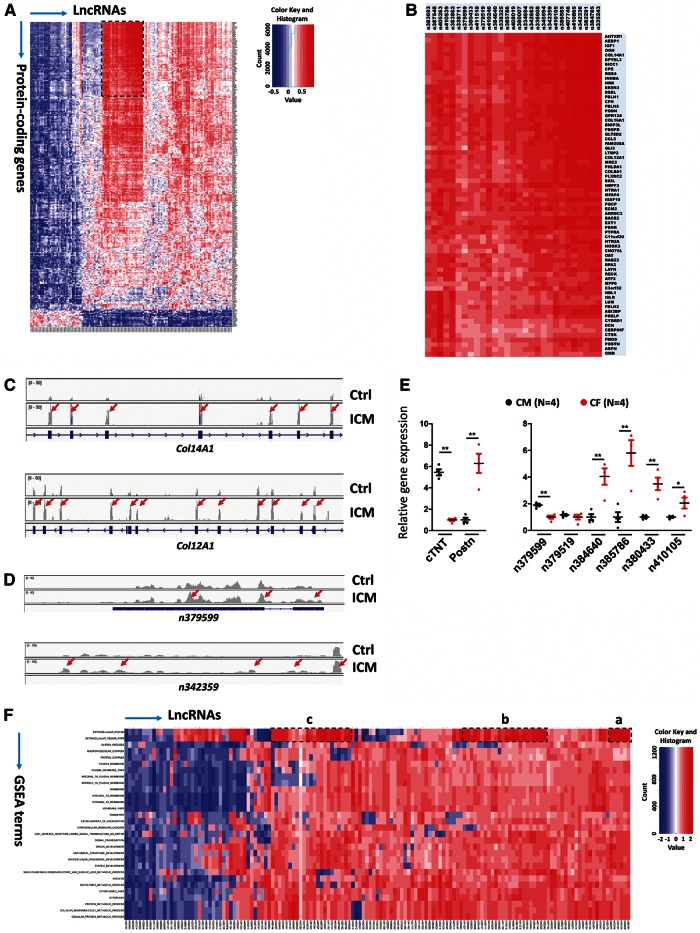
Little is known about the biological function of lncRNAs. We hypothesized that the expression patterns and functions of lncRNAs and protein-coding genes are correlated. To establish the potential function of lncRNAs that are differentially expressed in ICM, we created a lncRNA–mRNA expression correlation coefficient matrix between 145 differentially expressed (P < 0.05) lncRNAs and 285 differentially expressed (P < 0.01) mRNAs, using the Pearson correlation method as previously reported15 (Figure 4A). Interestingly, 29 out of the 35 lncRNAs (83%) in the cluster defined above showed a strong positive expression correlation with a cluster of 69 protein-coding genes (Figure 4B and see Supplementary material online, Figure S6). Further analysis of functional annotation clustering using Database for Annotation, Visualization and Integrated Discovery (DAVID) indicates that this cluster of 69 protein-coding genes is highly enriched for functional annotations related to signalling of ECM composition and turnover (Enrichment Score: 15.3). Multiple collagen genes, such as COL14A1, COL16A1, COL12A1, COL8A1, and the gene encoding the ECM protein, periostin, were present in this cluster (Figure 4C and see Supplementary material online, Figure S7). Consistent with this observation, we found that the expression of the lncRNAs n379599 and n342359 was increased in ICM hearts (Figure 4D), further supporting the view that these lncRNAs may function in the regulation of ECM components and fibrosis. Similarly, we compared the expression of additional lncRNAs of this cluster in a genome browser and confirmed that they are differentially expressed in ICM samples (see Supplementary material online, Figure S8).
Figure 4.
The expression of lncRNAs is associated with ECM gene expression in ICM. (A) Association matrix of 145 differentially expressed lncRNAs and 285 differentially expressed protein-coding genes. Positive (red) and negative (blue) expression correlations are shown. Clustered lncRNAs and 69 protein-coding genes are boxed and shown in (B). (C, D) Comparison of total reads from control and ICM hearts for protein-coding gene loci Col14A1 and Col12A1 and lncRNA gene loci n379599 and n342359, respectively. RNA-Seq reads were plotted and visualized with IGV browser. Red arrows indicate up-regulated exons in ICM samples. (E) qRT-PCR analysis of the expression of mouse lncRNAs in adult mouse CM and CF. Cardiac troponin T (cTNT) or periostin (Postn) marks CM and CF, respectively (left panel). N number for each group is indicated.*P <0.05; **P <0.01. (F) Association matrix of 145 differential expressed lncRNAs and 30 functional gene sets based on GSEA. Positive (red) and negative (blue) expression correlations are shown. Three clusters with positive expression correlation are boxed and labelled as a–c.
It is well known that the expression of these ECM genes are dramatically increased and closely related to cardiac fibrosis during cardiac remodelling and dilation.26,27 Given that the expression of dysregulated lncRNAs is closely correlated with that of ECM genes, we tested the expression of lncRNAs in cardiomyocytes (CM) and CF. We isolated CM and CF from adult mouse hearts. Molecular marker gene expression confirmed that cardiac troponin (cTNT) is enriched in CM, whereas periostin (Postn) expression marks CF (Figure 4E, left panel). We examined the expression of mouse homologs of human lncRNAs that are highly conserved between the two species, and we found that several lncRNAs, including n384640, n385786, n380433, and n410105, are enriched in CF (Figure 4E, right panel). These data suggest that lncRNAs participate in the regulation of cardiac fibrosis in ICM.
In order to assign potential functions to these dysregulated lncRNAs, we adapted the ‘guilt-by-association’ analyses, which have been introduced for functional predictions of mammalian lncRNAs.15,16,22,28 We examined the expression correlation coefficient between individual lncRNAs and a given functional gene set. The 285 differentially expressed protein-coding genes were classified into 30 functional gene sets, using Gene Set Enrichment Analysis (GSEA). These include Extracellular Region, Organ Development, Multicellular Organismal Development, Cellular Protein Metabolic Process, etc. We then generated a heat map of the expression correlation coefficient between the 145 differentially expressed lncRNAs and the above 30 functional gene sets (GSEA terms) (Figure 4F). Remarkably, 32 out of 53 lncRNAs (32/53, 60%), which were previously grouped in the cluster, showed very strong positive expression correlation with the GSEA functional terms Extracellular Region and Extracellular Region Part and were present in three sub-sets (Figure 4F, boxes a–c, and see Supplementary material online, Figure S9). These results further support the view that this cluster of lncRNAs is likely involved in ECM and fibrosis. Collectively, these analyses suggest that the expression and function of differentially expressed lncRNAs in ICM hearts may contribute to the pathophysiology of fibrosis associated HF.
3.5 lncRNAs regulate fibrosis and the expression of ECM genes
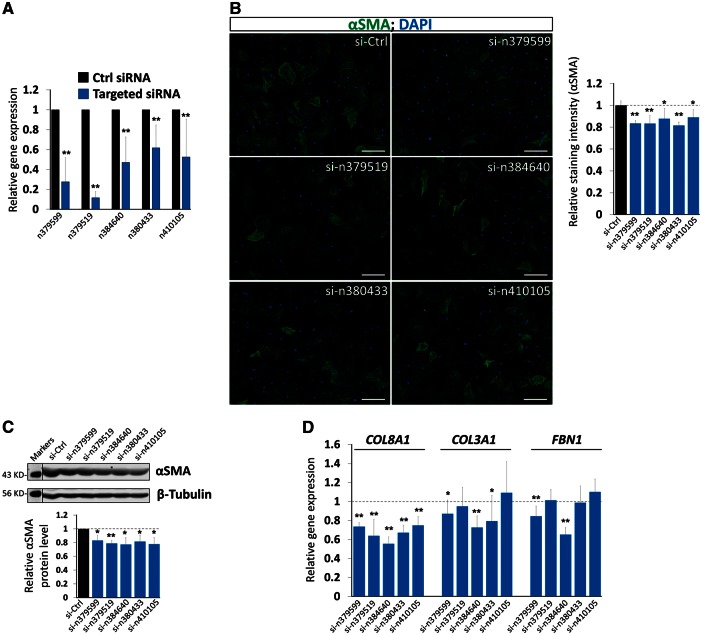
To test the hypothesis that lncRNAs are directly involved in the regulation of the expression of genes associated with fibrosis, we took both gain- and loss-of function approaches in isolated CF. We chose from above five highly conserved and CF-enriched lncRNAs (n379599, n379519, n384648, n380433, and n410105) and designed two independent siRNAs for each lncRNA and confirmed that these lncRNAs were knocked down efficiently in mouse CF (Figure 5A). We performed immunostaining for α-smooth muscle actin (αSMA), which is expressed in activated CF and a hallmark of fibrotic cardiac diseases,29 comparing lncRNA knockdown to control (Figure 5B). Indeed, inhibition of endogenous lncRNAs resulted in a modest yet statistically significant reduction of αSMA expression (Figure 5B). Decreased αSMA expression was further confirmed by western blotting analysis and quantification (Figure 5C). Furthermore, quantitative real-time PCR demonstrated the lncRNA knockdown reduced expression of fibrosis genes, including collagen 3A1 (COL3A1), collagen 8A1 (COL8A1), and fibronectin (FBN1) (Figure 5D). We have also examined the expression of additional genes related to ECM and fibrosis and found that their expression not altered when lncRNAs were knocked down (see Supplementary material online, Figure S10)
Figure 5.
Knockdown of lncRNAs reduced the expression of ECM genes. (A) qRT-PCR detecting the knockdown of lncRNA expression in isolated mouse CF. The result of each lncRNA knockdown was the combination of the results from two independent siRNAs. Data were obtained from four independent experiments. (B) Immunochemistry detecting the expression of αSMA in lncRNA knockdown CF. DAPI marks the nucleus. Bar=10 µm. αSMA immunostaining signal was quantified and presented as the Relative Staining Intensity (right panel). Data were obtained from three independent experiments. (C) Western blotting of αSMA in lncRNA knockdown CF. β-tubulin serves as a loading control. Molecular weight marker was provided. Relative αSMA protein levels were quantified (lower panel). Data were obtained from three independent experiments. (D) qRT-PCR detecting the expression of marker genes for ECM synthesis, FBN1, and for collagen synthesis, Col3A1, Col8A1 in lncRNA knockdown CF. Each CF isolation was treated as one independent experiment. Data were obtained from four independent experiments. *P <0.05; **P <0.01.
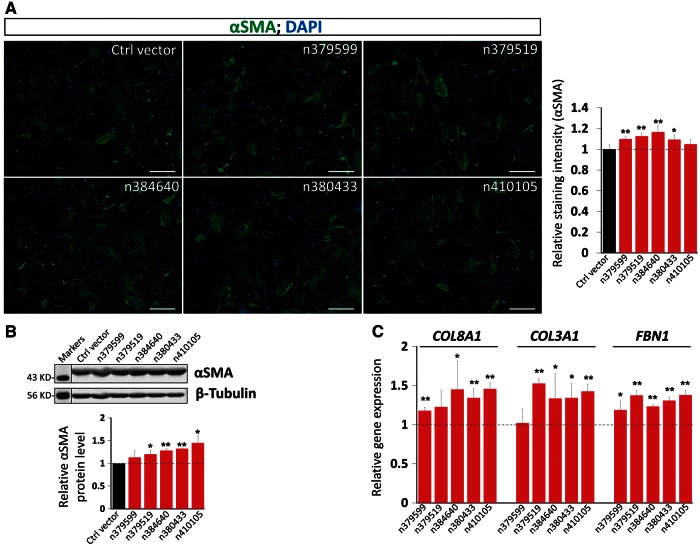
Conversely, we overexpressed these lncRNAs in CF (see Supplementary material online, Figure S11). Immunostaining for αSMA and quantification of staining intensity demonstrated that overexpression of lncRNAs modestly yet reproducibly induced the expression of αSMA (Figure 6A). Western blotting analysis confirmed the above observation (Figure 6B). Finally, we performed qRT-PCR analysis of the expression of ECM and fibrosis genes and found that overexpression of lncRNAs enhanced the expression of Col8A1, Col3A1, and FBN1 (Figure 6C), but not that of collagen 1A1 (Col1A1), periostin (POSTN) or elastin (ELN) (see Supplementary material online, Figure S12). Together, our gain- and loss-of function investigation indicated that lncRNAs are involved in the regulation of fibrosis related genes.
Figure 6.
Overexpression of lncRNAs induced the expression of ECM genes. (A) Immunochemistry detecting the expression of αSMA in CF overexpressing the indicated lncRNA. DAPI marks the nucleus. Bar=10 µm. αSMA immunostaining signal was quantified and presented as the Relative Staining Intensity (right panel). Data were obtained from 3 independent experiments. (B) Western blotting of αSMA in lncRNA overexpressed CF. β-tubulin serves as a loading control. Molecular weight marker was provided. Relative αSMA protein levels were quantified (lower panel). Data were obtained from 3 independent experiments. (C) qRT-PCR detecting the expression of marker genes for ECM synthesis, FBN1, and for collagen synthesis, Col3A1, Col8A1, after overexpression of indicated lncRNAs in CF. Each CF isolation was treated as one independent experiment. Data were obtained from 4 independent experiments. *P <0.05; **P <0.01.
3.6 lncRNAs participate in the TGF-beta signalling pathway
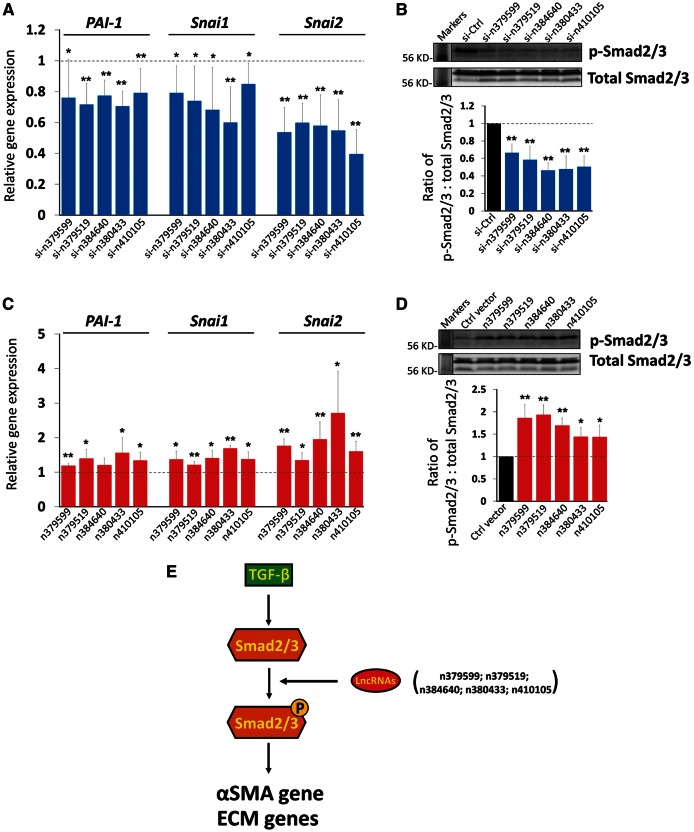
We sought to further understand the molecular mechanism by which these lncRNAs regulate cardiac fibrosis and the expression of ECM genes. The TGF-β signalling pathway has been shown essential for fibrosis and related gene expression.30 We examined the expression levels of several TGF-β downstream targets that are involved in fibrosis and cell migration. We found that the expression of PAI-1, Snai1, and Snai2 was reduced when lncRNAs were inhibited (Figure 7A). Next, we examined the expression of Smad2/3, major mediators of the TGF-β signalling pathway. The expression levels of Smad2/3 protein were not altered; however, the levels of phosphorylated Smad2/3 (p-Smad2/3) were decreased when lncRNAs were knockdown (Figure 7B and see Supplementary material online, Figure S13). Conversely, we examined the expression of TGF-β targets PAI-1, Snai1, and Snai2 in CF overexpressing lncRNAs and confirmed that lncRNAs induced the expression of these target genes (Figure 7C). Consistent with this result, we showed that levels of phosphorylated Smad2/3, but not that of total Smad2/3 protein, were induced by lncRNAs (Figure 7D and see Supplementary material online, Figure S14). It is worthy to note that the changes of the phosphorylated Smad2/3 levels were not enormous yet statistically significant. Together, these data suggest a molecular mechanism by which lncRNAs modulate the expression of ECM genes, at least in part, through regulating the expression and function of the TGF-β signalling pathway and its downstream targets involved in cardiac fibrosis.
Figure 7.
lncRNAs participate in the TGF-β pathway in CF. (A) qRT-PCR detecting the expression of PAI-1, Snai1 and Snai2 in lncRNA knockdown CF. Data were obtained from four independent experiments. (B) Western blotting of total and phosphorylated Smad2/3 (p-Smad2/3) in lncRNA knockdown CF. The ratio of total Smad2/3 and p-Smad2/3 was quantified using both bands (lower panel). Molecular weight marker was provided. Data were obtained from three independent experiments. (C) qRT-PCR detecting the expression of PAI-1, Snai1 and Snai2 in CF overexpressing lncRNA. Data were obtained from four independent experiments. (D) Western blotting of total and phosphorylated Smad2/3 (p-Smad2/3) in lncRNA overexpressed CF. The ratio of total Smad2/3 and p-Smad2/3 was quantified using both bands (lower panel). Molecular weight marker was provided. Data were obtained from three independent experiments. (E) A working modelling postulating the role of lncRNAs in the TGF-β pathway in CF. Each CF isolation was treated as one independent experiment. *P <0.05; **P <0.01.
4. Discussion
In this study, we discovered that the expression of lncRNAs is altered in the hearts of ICM patients, and that the expression of subsets of lncRNAs positively or negatively correlates with differential expression of ECM genes that participate in cardiac fibrosis and HF. Gain- and loss-of function investigations further suggest a direct role of lncRNAs in ECM gene expression.
lncRNAs have emerged as a novel class of regulators in diverse biological processes.4,5 Thousands of lncRNAs have been reported; many of them are expressed in a tissue-specific manner.5 Our study links the expression of a subset of lncRNAs to heart disease. While this study was in preparation, a similar study was published. In that study, the investigators profiled lncRNA expression from paired non-ischemic and ischemic human failing left ventricles before and after left ventricular assist device (LVAD).13,31 The authors suggested that lncRNAs could be used to discriminate failing hearts of different pathologies. Interestingly, another recent study identified a novel circulating lncRNA, LIPCAR, as a molecular marker for HF.32 Our study, together with these prior reports, indicates that lncRNAs are a novel class of regulators and biomarkers for cardiovascular biology and diseases.
RNA-sequencing results of lncRNAs demonstrate that the expression levels of most lncRNAs are substantially lower than those of protein-coding genes in the heart. This observation is consistent with most previous reports.5 Nevertheless, our overexpression and knockdown experiments in cultured CF indicate that lncRNAs, though expressed at low levels, play an important role in regulating the expression of ECM related genes. We recognized that the overall changes of lncRNA-mediated ECM gene expression appeared to be modest, suggesting that lncRNAs may not directly participate in turning on or off gene expression programs like that of transcription factors. The observation that lncRNA expression is associated with fibrosis could be indirect, resulted from an increased fibrosis in diseased hearts. Our studies further suggest that lncRNAs may impact important signalling pathways, such as the TGF-β pathway, in the heart (Figure 7E). Although our results showed that lncRNAs could modulate the levels of phosphorylated Smad2/3, at present it remains to be answered about how exactly lncRNAs achieved it and how the specificity is determined. Clearly, future studies will be needed to provide molecular insights into how lncRNAs function in vitro and in vivo. Nonetheless, our studies highlight the potential functional significance of lncRNAs in fundamental biological processes.
Recent studies have linked several lncRNAs to cardiovascular disease. ANRIL was identified as a risk factor for coronary disease.33 Though it is still not fully understood how ANRIL functions, evidence suggests that this lncRNA may participate in the regulation of histone methylation.34 Another lncRNA, MIAT (myocardial infarction-associated transcript) (or Gomafu/RNCR2) was identified as a risk factor associated with patients with myocardial infarction.35 However, how MIAT controls the status of myocardial infarction remains largely unknown. In another case, the genetic loci that encode MYH6 and MYH7, the main myosin heavy chain genes in cardiac muscle, appear to produce a non-coding anti-sense transcript (Myh7-as). Myh7-as transcription may regulate the ratio of Myh6 and Myh7, altering the function of muscle contraction.36 Intriguingly, a recent report described a cluster of lncRNA transcripts from Myh7 loci, Myheart (Mhrt) in the regulation of pathological hypertrophy and cardiac remodeling.37,38 Our current study reveals that the expression of a subset of lncRNAs is associated with fibrosis in the hearts of ICM patients. Future investigations will focus on the in vivo function of these lncRNAs in cardiac function and cardiac disease.
Supplementary material
Supplementary material is available at Cardiovascular Research online.
Acknowledgements
We thank members of the Wang laboratory for advice and support.
Conflict of interest: Yan Ding is a cofounder of Genomic Future, which provides genome-based data analyses services. None declared for other authors.
Funding
This work was supported by the March of Dimes Foundation, Muscular Dystrophy Association and the NIH (HL085635, HL116919 to D.Z.W., HL095712, HL094683 to W.T.P.), Innovative Grant from Chinese Central Government of Human Resources (Y.D.), and Merit Grant for Extraordinary Oversea Talent from Dalian Government, China (Y.D.). Z.P.H. is supported by the NIH training grant T32 HL007572. D.Z.W. and W.T.P. are Established Investigators of the American Heart Association. Grants from National Natural Science Foundation of China (No. 81470382 to J.C.).
References
- 1.Sanganalmath SK, Bolli R. Cell therapy for heart failure: a comprehensive overview of experimental and clinical studies, current challenges, and future directions. Circ Res 2013;113:810–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vander Heide RS, Steenbergen C. Cardioprotection and myocardial reperfusion: pitfalls to clinical application. Circ Res 2013;113:464–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morita H, Seidman J, Seidman CE. Genetic causes of human heart failure. J Clin Invest 2005;115:518–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell 2009;136:629–641. [DOI] [PubMed] [Google Scholar]
- 5.Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell 2013;152:1298–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol 2011;21:354–361. [DOI] [PubMed] [Google Scholar]
- 7.Klattenhoff CA, Scheuermann JC, Surface LE, Bradley RK, Fields PA, Steinhauser ML, Ding H, Butty VL, Torrey L, Haas S, Abo R, Tabebordbar M, Lee RT, Burge CB, Boyer LA. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell 2013;152:570–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kataoka M, Huang ZP, Wang DZ. Build a braveheart: the missing linc (RNA). Circ Res 2013;112:1532–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sauvageau M, Goff LA, Lodato S, Bonev B, Groff AF, Gerhardinger C, Sanchez-Gomez DB, Hacisuleyman E, Li E, Spence M, Liapis SC, Mallard W, Morse M, Swerdel MR, D'Ecclessis MF, Moore JC, Lai V, Gong G, Yancopoulos GD, Frendewey D, Kellis M, Hart RP, Valenzuela DM, Arlotta P, Rinn JL. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. eLife 2013;2:e01749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grote P, Wittler L, Hendrix D, Koch F, Wahrisch S, Beisaw A, Macura K, Blass G, Kellis M, Werber M, Herrmann BG. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev Cell 2013;24:206–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leung A, Trac C, Jin W, Lanting L, Akbany A, Saetrom P, Schones DE, Natarajan R. Novel long noncoding RNAs are regulated by angiotensin II in vascular smooth muscle cells. Circ Res 2013;113:266–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ounzain S, Micheletti R, Beckmann T, Schroen B, Alexanian M, Pezzuto I, Crippa S, Nemir M, Sarre A, Johnson R, Dauvillier J, Burdet F, Ibberson M, Guigo R, Xenarios I, Heymans S, Pedrazzini T. Genome-wide profiling of the cardiac transcriptome after myocardial infarction identifies novel heart-specific long non-coding RNAs. Eur Heart J 2015;36:353–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang KC, Yamada KA, Patel AY, Topkara VK, George I, Cheema FH, Ewald GA, Mann DL, Nerbonne JM. Deep RNA sequencing reveals dynamic regulation of myocardial noncoding RNAs in failing human heart and remodeling with mechanical circulatory support. Circulation 2014;129:1009–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kong SW, Hu YW, Ho JW, Ikeda S, Polster S, John R, Hall JL, Bisping E, Pieske B, dos Remedios CG, Pu WT. Heart failure-associated changes in RNA splicing of sarcomere genes. Circ Cardiovasc Genet 2010;3:138–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature 2012;482:339–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, Cabili MN, Jaenisch R, Mikkelsen TS, Jacks T, Hacohen N, Bernstein BE, Kellis M, Regev A, Rinn JL, Lander ES. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 2009;458:223–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, Regev A, Lander ES, Rinn JL. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA 2009;106:11667–11672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang ZP, Kataoka M, Chen J, Wu G, Ding J, Nie M, Lin Z, Liu J, Hu X, Ma L, Zhou B, Wakimoto H, Zeng C, Kyselovic J, Deng ZL, Seidman CE, Seidman JG, Pu WT, Wang DZ. Cardiomyocyte-enriched protein CIP protects against pathophysiological stresses and regulates cardiac homeostasis. J Clin Invest 2015;125:4122–4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O'Connell TD, Swigart PM, Rodrigo MC, Ishizaka S, Joho S, Turnbull L, Tecott LH, Baker AJ, Foster E, Grossman W, Simpson PC. Alpha1-adrenergic receptors prevent a maladaptive cardiac response to pressure overload. J Clin Invest 2006;116:1005–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barth AS, Kuner R, Buness A, Ruschhaupt M, Merk S, Zwermann L, Kaab S, Kreuzer E, Steinbeck G, Mansmann U, Poustka A, Nabauer M, Sultmann H. Identification of a common gene expression signature in dilated cardiomyopathy across independent microarray studies. J Am Coll Cardiol 2006;48:1610–1617. [DOI] [PubMed] [Google Scholar]
- 21.Bu D, Yu K, Sun S, Xie C, Skogerbo G, Miao R, Xiao H, Liao Q, Luo H, Zhao G, Zhao H, Liu Z, Liu C, Chen R, Zhao Y. NONCODE v3.0: integrative annotation of long noncoding RNAs. Nucleic Acids Res. 2012;40:D210–D215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev 2011;25:1915–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell 2013;154:26–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brewer A, Pizzey J. GATA factors in vertebrate heart development and disease. Expert Rev Mol Med 2006;8:1–20. [DOI] [PubMed] [Google Scholar]
- 25.Di Sole F, Vadnagara K, Moe OW, Babich V. Calcineurin homologous protein: a multifunctional Ca2+-binding protein family. Am J Physiol Renal Physiol 2012;303:F165–F179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pfeffer MA, Braunwald E. Ventricular remodeling after myocardial infarction. Experimental observations and clinical implications. Circulation 1990;81:1161–1172. [DOI] [PubMed] [Google Scholar]
- 27.Dorn GW., 2nd Novel pharmacotherapies to abrogate postinfarction ventricular remodeling. Nat Rev Cardiol 2009;6:283–291. [DOI] [PubMed] [Google Scholar]
- 28.Dinger ME, Amaral PP, Mercer TR, Pang KC, Bruce SJ, Gardiner BB, Askarian-Amiri ME, Ru K, Solda G, Simons C, Sunkin SM, Crowe ML, Grimmond SM, Perkins AC, Mattick JS. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res 2008;18:1433–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leask A. Getting to the heart of the matter: new insights into cardiac fibrosis. Circ Res. 2015;116:1269–1276. [DOI] [PubMed] [Google Scholar]
- 30.Piersma B, Bank RA, Boersema M. Signaling in fibrosis: TGF-beta, WNT, and YAP/TAZ converge. Front Med (Lausanne) 2015;2:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiao XG, Touma M, Wang Y. Decoding the noncoding transcripts in human heart failure. Circulation 2014;129:958–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kumarswamy R, Bauters C, Volkmann I, Maury F, Fetisch J, Holzmann A, Lemesle G, de Groote P, Pinet F, Thum T. Circulating long noncoding RNA, LIPCAR, predicts survival in patients with heart failure. Circ Res 2014;114:1569–1575. [DOI] [PubMed] [Google Scholar]
- 33.Broadbent HM, Peden JF, Lorkowski S, Goel A, Ongen H, Green F, Clarke R, Collins R, Franzosi MG, Tognoni G, Seedorf U, Rust S, Eriksson P, Hamsten A, Farrall M, Watkins H, consortium Susceptibility to coronary artery disease and diabetes is encoded by distinct, tightly linked SNPs in the ANRIL locus on chromosome 9p. Hum Mol Genet 2008;17:806–814. [DOI] [PubMed] [Google Scholar]
- 34.Yap KL, Li S, Munoz-Cabello AM, Raguz S, Zeng L, Mujtaba S, Gil J, Walsh MJ, Zhou MM. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell 2010;38:662–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ishii N, Ozaki K, Sato H, Mizuno H, Saito S, Takahashi A, Miyamoto Y, Ikegawa S, Kamatani N, Hori M, Saito S, Nakamura Y, Tanaka T. Identification of a novel non-coding RNA, MIAT, that confers risk of myocardial infarction. J Hum Genet 2006;51:1087–1099. [DOI] [PubMed] [Google Scholar]
- 36.Pandya K, Smithies O. Beta-MyHC and cardiac hypertrophy: size does matter. Circ Res 2011;109:609–610. [DOI] [PubMed] [Google Scholar]
- 37.Han P, Li W, Lin CH, Yang J, Shang C, Nurnberg ST, Jin KK, Xu W, Lin CY, Lin CJ, Xiong Y, Chien HC, Zhou B, Ashley E, Bernstein D, Chen PS, Chen HS, Quertermous T, Chang CP. A long noncoding RNA protects the heart from pathological hypertrophy. Nature 2014;514:102–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu J, Wang DZ. An epigenetic “LINK(RNA)” to pathological cardiac hypertrophy. Cell Metab 2014;20:555–557. [DOI] [PMC free article] [PubMed] [Google Scholar]