Abstract
Aims
Vascular smooth muscle cells (SMCs) are major precursors contributing to osteochondrogenesis and calcification in atherosclerosis. Runt-related transcription factor-2 (Runx2) has been found essential for SMC differentiation to an osteochondrogenic phenotype and subsequent calcification in vitro. A recent study using a conditional targeting allele that produced a truncated Runx2 protein in SMCs of ApoE−/− mice showed reduced vascular calcification, likely occurring via reduction of receptor activator of nuclear factor-κB ligand (RANKL), macrophage infiltration, and atherosclerotic lesion formation. Using an improved conditional Runx2 knockout mouse model, we have elucidated new roles for SMC-specific Runx2 in arterial intimal calcification (AIC) without effects on atherosclerotic lesion size.
Methods and results
We used an improved targeting construct to generate LDLr−/− mice with floxed-Runx2 alleles (LDLr−/−:Runx2f/f) such that Cre-mediated recombination (LDLr−/−:Runx2ΔSM) does not produce functional truncated Runx2 protein, thereby avoiding off-target effects. We found that both LDLr−/−:Runx2f/f and LDLr−/−:Runx2ΔSM mice fed with a high fat diet developed atherosclerosis. SMC-specific Runx2 deletion did not significantly reduce atherosclerotic lesion size, macrophage number, or expression of RANKL, MCP-1, and CCR2. However, it significantly reduced AIC by 50%. Mechanistically, Sox9 and type II collagen were unaltered in vessels of LDLr−/−:Runx2ΔSM mice compared to LDLr−/−:Runx2f/f counterparts, while type X collagen, MMP13 and the osteoblastic marker osteocalcin were significantly reduced.
Conclusions
SMC autonomous Runx2 is required for SMC differentiation towards osteoblast-like cells, SMC-derived chondrocyte maturation and AIC in atherosclerotic mice. These effects were independent of systemic lipid metabolism, RANKL expression, macrophage infiltration, and atheromatous lesion progression.
Keywords: Arterial intimal calcification • Atherosclerosis • Chondrocyte maturation • Runx2 • Smooth muscle cells
1. Introduction
Arterial intimal calcification (AIC) is prevalent and progressive in patients with atherosclerosis.1,2 AIC is highly correlated with plaque burden3 and predicts coronary events, such as plaque instability and risk of myocardial infarction,2,4–6 as well as risk of stroke.7 During early stages of the disease, microcalcification may occur within the fibrous caps of atherosclerotic plaques, resulting in compliance mismatch that increases local stress and chances of plaque rupture.8 Although initially considered a passive, uncontrollable process, recent evidence supports AIC as an actively regulated process with similarities to bone development, including the appearance of calcified cartilage- and bone-like structures within lesions.9–11 Lineage tracing studies in mouse models have shown that vascular smooth muscle cells (SMCs) can transdifferentiate into osteochondrogenic cells,12,13 implicating these cells as important mediators of AIC. However, the mechanisms by which SMCs transdifferentiate and contribute to plaque mineralization are not well understood.
Runt-related transcription factor 2 (Runx2), a master regulator of bone development, has been detected in calcified atherosclerotic lesions in people,14–17 and is upregulated in SMCs undergoing transdifferentiation to osteochondrogenic cells in mouse models of AIC.12,13 In humans, mutations in the Runx2 gene are associated with cleidocranial dysplasia, a disorder characterized by delayed ossification of midline structures, especially the collarbone and cranium.18 In mice, the global deletion of Runx2 is perinatal lethal due to the lack of skeletal mineralization and respiratory failure.19,20 With regard to its role in atherosclerosis, a recent study used a conditional targeting construct to generate a truncated, Runt-homology domain containing Runx2 protein in SMCs in ApoE−/− mice and found dramatically decreased RANKL expression and macrophage infiltration in the arteries. However, the role of Runx2 on SMC transdifferentiation and mineralization could not be adequately addressed with this model, as atherosclerotic lesion formation was largely inhibited in these mice.21
In the present study, we used an improved targeting construct to generate LDLr−/− mice with floxed-Runx2 alleles (Runx2f/f ) in which Cre-mediated recombination does not produce functional truncated Runx2 proteins (Runx2ΔSM), thereby minimizing potential off target effects. This model allowed us to genetically separate atherosis and calcific sclerosis. Indeed, SMC-specific deletion of Runx2 in LDLr−/−:Runx2ΔSM mice did not reduce RANKL expression, macrophage infiltration, or atherosclerotic lesion size compared to LDLr−/−; Runx2f/f control mice. Instead, we observed a decrease in lesion mineralization in addition to substantially decreased expression of osteoblast markers, osteocalcin (OCN), osterix (OSX), alkaline phosphatase (ALP), and chondrocyte maturation markers, type X collagen (Col X) and matrix metallopeptidase 13 (MMP13). These data suggest that SMC autonomous Runx2 is required for osteoblastic differentiation, chondrocyte maturation, and mineralization in atherosclerotic lesions but is required not for atherogenesis or atheromatous lesion progression.
2. Methods
2.1 Animal models
Mice carrying conditional alleles that target the runt homology domain of the Runx2 gene (Runx2f/f) were generated as previously described.22 Mice were backcrossed with C57BL/6J mice three times prior to breeding with congenic C57BL/6J SM22-Cre mice and LDLr−/− mice to generate LDLr−/−:Runx2f/f:SM22-Cre+/o (LDLr−/−:Runx2ΔSM) and LDLr−/−: Runx2f/f:SM22-Creo/o (LDLr−/−:Runx2f/f) experimental mice. Both male and female littermates were used for study. These mice were fed a high fat diet (HFD; Research Diets Inc., 1.25% cholesterol, 39.9% kcal fat, 40% kcal carbohydrate) at 10 weeks of age to induce atherosclerosis, vascular cartilaginous metaplasia, and calcification as previously described.12 Normal chow (NC) was used as a dietary control for each genotype. Body weight was recorded and fasting serum collected before the diet challenge and at termination. After 18–24 weeks of HFD feeding, a total of 87 mice were euthanized via intraperitoneal injection of pentobarbital (150 mg/kg) followed by exsanguination through cardiac puncture. All protocols were in compliance with the NIH Guideline for the Care and Use of Laboratory Animals and have been approved by the Institutional Animal Care and Use Committee, University of Washington.
2.2 Quantification of aortic arch calcium content
Approximately 6 mm of the aortic arch from each LDLr−/− mouse was collected and lyophilized to a constant weight. Calcium was extracted from the lyophilized tissue with 0.6 N HCl and determined colorimetrically using the o-Cresolphthalein complexone kit (Teco Diagnostics, C503-480) as previously described.13 Values were normalized to the dry weight of the arches.
2.3 Histochemical and immunohistochemical staining
Specimens were fixed with modified Clark’s fixative, processed, and embedded in paraffin. Serial sections were made in 4 μm thickness and subject to various histochemical and immunohistochemical staining. Alizarin red S (0.5%, pH 9.0) was used to visualize calcium deposition. Movat pentachrome staining was used to visualize atherosclerotic lesion and cartilaginous metaplasia. Antibodies recognizing Runx2 (MAB2006, R&D System), Sox9 (SC-20095, Santa Cruz), type II collagen (Col II; AB761, Millipore), Col X (Col X; LSL-LB-0092, Cosmo Bio Co., LTD.), OCN (OCN; SC-30045, Santa Cruz), and MOMA2 (MCA519G, AbD Serotec) were used to immunohistochemically detect osteochondrogenic precursors, prechondroblasts, chondrocytes, osteoblasts, and macrophages. Signals were either visualized with fluorescent secondary antibodies (Jackson ImmunoRes), or biotinylated secondary antibodies, Vectastain ABC Elite kit (PK-6100, Vector Lab), and 3, 3’-diaminobenzidine peroxidase substrate (D-0426, Sigma). All the immunohistochemical staining was validated with non-specific IgG controls as well as negative and positive control tissues.
2.4 Morphometric analysis of atherosclerosis and cartilaginous metaplasia
To measure atherosclerotic lesion size and cartilaginous metaplasia of the lesions, eight aortic arch longitudinal sections, 40 µm apart over 320 μm in depth, starting at the appearance of the three major branches on the greater curvature of the vessels, and 8 brachiocephalic artery cross sections, 100 µm apart over 800 μm in length, starting at the bifurcation of the aortic arch, were collected. Sections were coded and stained using the Movat pentachrome method. Intimal lesion areas were measured blindly and normalized to the length of internal elastic lamina (for longitudinal sections of aortic arches) or area of arterial media [for cross sections of brachiocephalic arteries (BCA)] using Metamorph software (Molecular Device).23 Cartilaginous metaplasia was identified by yellow (collagen rich) and blue (proteoglycan rich) stain consisting of chondrocyte-like cells,13 and expressed as percent intimal area. Macrophages were identified by MOMA2 antibody, quantified with the same sampling scheme, and expressed as percent intimal area. To assess osteoblastic and chondrocytic differentiation in aortic arches, Sox9. Runx2, OCN, collagen type II, and X protein distributions were determined immunohistochemically using the same sampling scheme. Sox9, Runx2, and OCN levels were determined as percentage of intimal lesion area. Collagen type II and X positive matrices were scored as percentage of cartilaginous metaplasia (0, 10, 25, 50, and 75%) blindly and independently by two investigators, and expressed as Col X to Col II ratio using adjacent sections.
2.5 Complete blood count (CBC) and serum analyses
Whole blood was collected from unfasted mice for CBC by Department of Laboratory Medicine, University of Washington. For serum analysis, mice were fasted for 4 h prior to blood collection into serum separator tubes. Sera were analysed for cholesterol, triglyceride, and phosphate levels via bioanalyzer. Calcium levels were determined colorimetrically using the o-Cresolphthalein complexone kit (Teco Diagnostics, C503-480). Blood urea nitrogen (BUN) was measured using QuantiChrom Urea Assay Kit (BioAssay System, DIUR-500).24
2.6 Quantitative reverse transcription-polymerase chain reaction (RT-PCR)
Total RNA was extracted from aortic arches using RNeasy Mini kit. The contaminating genomic DNA was digested by RNase-free DNase I (Qiagen). Total RNA was used to synthesize first-strand cDNA using Omniscript (Qiagen) at 37 °C for 1 h, and the cDNA produced was used to determine expression of the osteochondrogenic transcription factor Runx2, the chondrogenic transcription factor Sox9, the chondrocyte marker Pro-Col II, the hypertrophic chondrocyte markers, Pro-Col X and matrix metalloproteinase 13 (MMP 13), and the osteoblastic marker genes, osterix (OSX), alkaline phosphatase (ALP) and OCN using Taqman quantitative RT-PCR. We also determined expression levels of genes associated with macrophage recruitment, including monocyte chemotactic protein 1 (MCP1), C–C chemokine receptor type 2 (CCR2), and receptor activator of nuclear factor kappa-B ligand (RANKL) quantitatively. Primer and probe sequences are: Runx2 forward primer: 5’ CACCG ACAGTCCCAACTTCCT 3’, Runx2 reverse primer: 5’ACGGTAAC CA CAGTCCCATCTG 3’, and Runx2 probe: 5’ FAM-CCTTCAAGGT TGTAGCCCT-MGB 3’, Sox9 forward primer: 5’ GCGGAGCTC AGCA AGACTCT 3’, Sox9 reverse primer: 5’ GGTGGTCTTT CTTGTGCT GCA 3’, and Sox9 probe: FAM-TCTGGAGGCTGCTGAA-MGB, Pro-Col II forward primer: 5’ GGCAACAGCAGGTTCACATACA 3’, Pro-Col II reverse primer: 5’ CTTCTGTGATCGGTACTCGATGAC 3’, and Pro-Col II probe: 5’ FAM-TGGCTGCACGAAACA-MGB 3’, OSX forward: 5’ GGTTCTCTCCATCTGCCTGACT 3’, OSX reverse: 5’ CAGGGGACTGGAGCCATAGT 3’, OSX probe: 5’ FAM-CTGC TTG A GGAAGAAG-MGB 3’, ALP forward: 5’ CAAGGACATCGCATAT CAGCTAA 3’, ALP reverse: 5’CAGTTCTGTTCTTCGGGTACATGT 3’, and ALP probe: 5’ FAM-AGGATATCGACGTGATCAT-MGB 3’, OCN forward: 5’ CTGGCTGCGCTCTGTCTCT 3’, OCN reverse: 5’ GACATGAAGGCTTTGTCAGACTCA 3’, and OCN probe: 5’ FAM-TGACCTCACAGATGCCAA-MGB 3’. Primers/probes of Pro-Col X (Mm00487041_m1), MMP 13 (Mm00439491_m1), MCP1 (Mm00441242_m1), CCR2 (Mm00438270_m1), and RANKL (Mm00441906_m1) were purchased from Life Technologies and the assay IDs are listed in parentheses. All the probe sequences span exon-exon junctions to avoid amplification of residual genomic DNA. 18s ribosomal RNA expression was determined using Taqman Ribosomal RNA Control Reagents (ABI) to control sample loading. Gene expression levels were determined in triplicates, normalized to 18s ribosomal RNA levels, and expressed as a fold change over control samples. The Runx2 primers and probe were designed to amplify exon 4, the exon excised through Cre recombination, to determine the deletion efficiency in the Runx2ΔSM alleles.22
2.7 Statistics
Normality of distribution was assessed with D’Agostino-Pearson normality test. Normally distributed data are shown as means ± standard error of the mean (S.E.M.), and were analysed with Student’s t-test for comparison of two groups and two-way analysis of variance (ANOVA) and Bonferroni’s post hoc test for comparison of multiple groups. Non-normally distributed data were analysed with Mann-Whitney test. Data are considered statistically significant at a P value < 0.05.
3. Results
In the present study, we examined mice in which the Runx2 gene was specifically deleted in vascular SMCs (Runx2ΔSM) without producing truncated Runx2 proteins containing the Runt homology domain.22 These mice were bred onto an LDLr−/− background and fed with either NC or a HFD to determine whether Runx2 expression in SMCs was required for atherosclerotic lesion formation, SMC osteochondrogenic differentiation and/or AIC.
3.1 Runx2 deletion in SMCs does not alter blood cell counts, serum lipid or mineral profiles in NC and HFD fed LDLr−/− mice
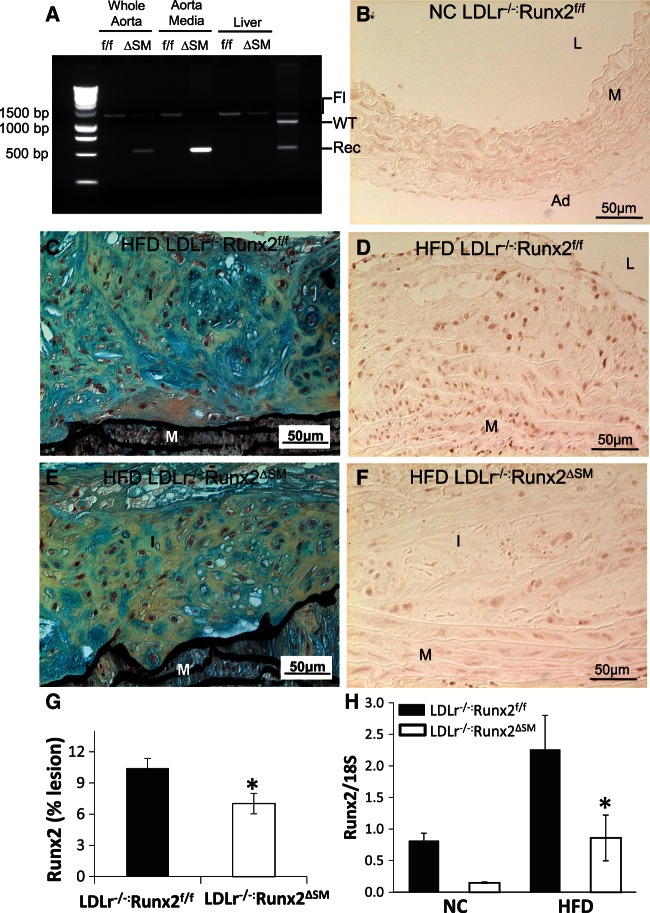
LDLr−/−:Runx2ΔSM and LDLr−/−:Runx2f/f mice were viable, normal in size, and fertile (data not shown). Similar to Runx2ΔSM mice,22 SM22Cre-directed removal of Runx2 in blood vessels of LDLr−/−:Runx2ΔSM mice was efficient. As shown in Figure 1A, Runx2 removal was efficient in aortas of the LDLr−/−:Runx2ΔSM mice and was completely deleted in the aortic media compared to the LDLr−/−:Runx2f/f counterparts. In contrast, no SM22α–Cre mediated DNA rearrangement of the Runx2 gene was observed in the liver of the LDLr−/−:Runx2ΔSM mice. Removal of Runx2 in SMCs did not affect blood counts, including white blood cells, red blood cells, hemoglobin, hematocrit, mean corpuscular volume and haemoglobin, and platelets (data now shown). Feeding with HFD induced significant hypertriglyceridemia and hypercholesterolemia in both genotypes compared to the NC fed mice (Table 1). No significant differences in serum calcium and phosphate levels were observed between HFD and NC groups for either genotype. BUN levels were slightly higher in LDLr−/−:Runx2ΔSM mice in comparison to LDLr−/−:Runx2f/f mice, but were well within the reported normal range.12,25 These data suggest that removal of Runx2 in SMCs does not affect systematic lipid metabolism or mineral homeostasis in LDLr−/− mice.
Figure 1.
Removal of Runx2 in vascular SMCs of LDLr−/− mice. (A) Tissue-specific deletion of Runx2 was determined by PCR using primers that amplify the loxP-flanked targeting sequence. Genomic DNA isolated from aorta, aortic media, and liver of LDLr−/−:Runx2f/f (f/f) and LDLr−/−:Runx2ΔSM (ΔSM) mice was used as a template. (B–G) LDLr−/−:Runx2f/f (B–D) and LDLr−/−:Runx2ΔSM mice (E and F) were fed with NC (B) or HFD (C–G) for 18 weeks. Aortic arches were collected for Movat pentachrome staining (C and E; yellow = collagen rich, blue = proteoglycan rich) and Runx2 immunohistochemical staining (B, D, F; brown nuclear stain). L = lumen, I = intima, M = media, Ad = adventitia. (G) Quantitation of Runx2 immunostaining in HFD LDLr−/−:Runx2f/f and LDLr−/−:Runx2ΔSM vessels, mean ± S.E.M., n = 4, student t-test, *P < 0.05. (H) Total RNA was extracted from pooled 18-week aortic arches (3–4 arches were pooled for each RNA sample, n = 3 RNA samples/group) and used to determine expression of Runx2 by Taqman quantitative RT-PCR, mean ± S.E.M., n = 3, two-way ANOVA with Bonferroni’s test, *P < 0.05 (HFD LDLr−/−:Runx2f/f vs. HFD LDLr−/−:Runx2ΔSM).
Table 1.
Blood chemistry of LDLr−/−:Runx2f/f and LDLr−/−:Runx2ΔSM mice fed with HFD or NC for 24 weeks
| Normal chow |
High fat diet |
|||
|---|---|---|---|---|
| LDL−/−:Runx2f/f | LDLr−/−:Runx2ΔSM | LDL−/−:Runx2f/f | LDLr−/−:Runx2ΔSM | |
| Triglyceride (mmol/L) | 0.65 ± 0.06 | 0.79 ± 0.05 | 7.08 ± 1.40† | 4.96 ± 0.79† |
| Cholesterol (mmol/L) | 6.26 ± 0.42 | 5.95 ± 0.36 | 42.74 ± 2.83† | 36.50 ± 1.90† |
| Phosphate (mmol/L) | 2.79 ± 0.15 | 3.27 ± 0.14 | 2.67 ± 0.23 | 3.07 ± 0.18 |
| Calcium (mmol/L) | 2.62 ± 0.14 | 2.57 ± 0.02 | 2.57 ± 0.06 | 2.58 ± 0.04 |
| BUN (mmol/L) | 13.67 ± 0.45 | 15.62 ± 0.74* | 11.57 ± 0.78 | 14.16 ± 0.51* |
Data represented as mean ± S.E.M., N = 8–12, and analysed by two-way ANOVA.
*P < 0.05 (vs. LDL−/−:Runx2f/f),
†P < 0.0001 (vs. NC).
3.2 Runx2 deletion in SMCs does not block atherosclerotic lesion formation in LDLr−/− mice fed a HFD
We have previously confirmed vascular smooth muscle-specific removal of Runx2 in the newly created Runx2ΔSM mouse model.22 These mice, when bred onto the atherogenic LDLr−/− background and fed with a NC diet, showed normal arterial morphology with no Runx2 positive cells detected in either LDLr−/−:Runx2f/f or LDLr−/−:Runx2ΔSM vessels (Figure 1B, and data not shown). HFD feeding induced atherosclerotic lesion formation (Figure 1C) in the arteries of LDLr−/−:Runx2f/f mice, with abundant Runx2 positive cells found throughout the lesions and in medial cells underlying the lesions (Figure 1D). Consistent with previous genetic fate mapping studies,12 the majority of Runx2 positive cells no longer express their original lineage marker proteins, as determined by dual fluorescent immunohistochemistry for Runx2 and SM22α (see Supplementary material online, Figure 1A–C). While comparable intimal lesions also formed in the arteries of HFD fed LDLr−/−:Runx2ΔSM mice (Figure 1E), removal of Runx2 in vascular SMCs reduced the number of Runx2 positive cells (Figure 1F and see Supplementary material online, Figure 1E), notably in deep intima and the inner layers of media underneath atherosclerotic lesions, areas where osteochondrogenic differentiation of vascular SMCs initiates.12 Quantitation of percent Runx2 immunoactivity (Figure 1G) and Runx2 mRNA levels (Figure 1H) in aortic arches of these mice verified statistically lower levels of Runx2 in LDLr−/−:Runx2ΔSM vessels in comparison to LDLr−/−:Runx2f/f vessels. HFD feeding induced Runx2 mRNA levels in aortic arch by ∼2.8 fold compared to NC feeding. Knockout of Runx2 in SMCs led to ∼62% reduction of Runx2 mRNA levels and ∼33% reduction of Runx2 cells in HFD vessels. These data also suggest that other cell types, in addition to SM22 positive vascular SMCs, expressed Runx2 in the vessels of HFD fed mice. Indeed, the Runx2 positive cells observed in LDLr−/−:Runx2ΔSM vessels were primarily located on the luminal side of the atherosclerotic lesions, consistent with the localization of bone marrow-derived osteochondrogenic cells,12 and some of them were positive for macrophage marker proteins, e.g. CD68, (see Supplementary material online, Figure 2D–E).
To determine whether deletion of Runx2 in SMCs affected lesion formation, histomorphometric analyses were performed on aortic arches and BCA of LDLr−/−:Runx2f/f and LDLr−/−:Runx2ΔSM mice fed a HFD. As shown in Figure 2, arteries from LDLr−/−:Runx2f/f and LDLr−/−:Runx2ΔSM mice had typical atherosclerotic lesions, containing foam cells, cartilaginous metaplasia, necrotic core, and fibrous caps. Quantitation of lesion size in either arterial site showed no significant difference between genotypes, indicating that Runx2 expression in SMCs was not required for atherosclerotic lesion formation in HFD fed LDLr−/− mice.
Figure 2.
SMC-specific Runx2 removal does not alter atherosclerotic lesion progression. Serial sections were taken from aortic arches (A–C, eight longitudinal sections cover 320 μm) and BCA (D–F, eight cross-sections cover 800 µm) of 24 weeks HFD-fed LDLr−/−:Runx2f/f and LDLr−/−:Runx2ΔSM mice and processed with Movat pentachrome staining to evaluate atherosclerotic lesion progression. Intima area was measured morphometrically as described in method. Data are normally distributed (D’Agostino-Pearson test) and shown as mean ± S.E.M. n = 8 and 9. Differences between genotypes were analysed by student t-test, N.S.= not significant. L = lumen, I = intima, M = media, Ad = adventitia.
3.3 Runx2 in SMCs is required for AIC in HFD fed LDLr−/− mice
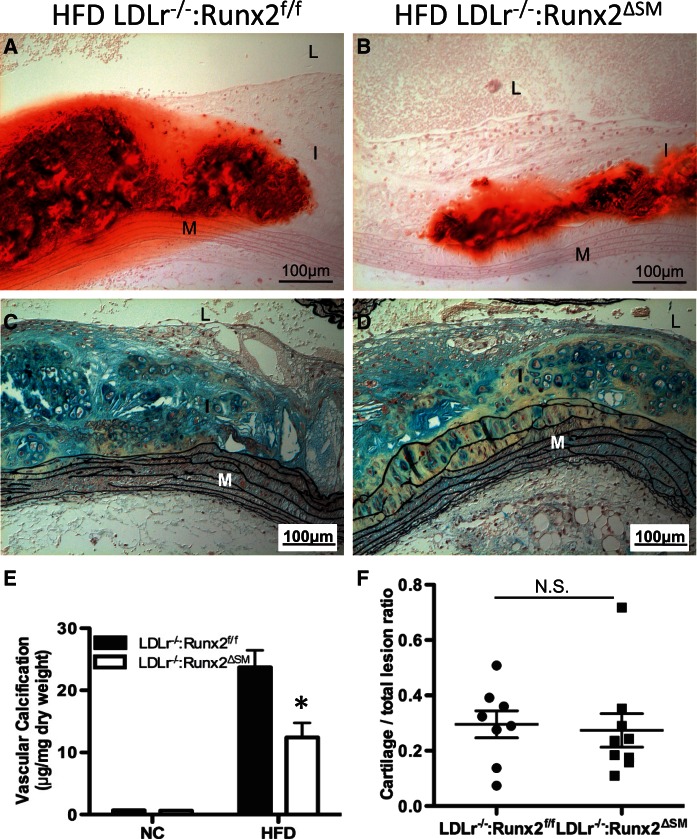
Atherosclerotic lesions in both LDLr−/−:Runx2f/f and LDLr−/−:Runx2ΔSM vessels contained AIC as measured by alizarin red S staining (Figure 3A and B). Mineral deposits were predominantly found in regions containing cartilaginous metaplasia, as shown in Figure 3C and D. AIC was quantitated using aortic arches collected from LDLr−/−:Runx2f/f and LDLr−/−:Runx2ΔSM mice following 24 weeks of HFD diet challenge. As shown in Figure 3E, removal of Runx2 in vascular SMCs resulted in a 50% reduction of calcium deposition in atherosclerotic intima of LDLr−/−:Runx2ΔSM vessels.
Figure 3.
Removal of Runx2 in SMCs reduces AIC in LDLr−/− mice fed with HFD diet. LDLr−/−:Runx2f/f and LDLr−/−:Runx2ΔSM mice were fed with NC or HFD for 24 weeks. Aortic arches were collected for Alizarin red S staining of mineral (A and B; red), Movat pentachrome staining (C and D; yellow = collagen rich, blue = proteoglycan rich), and biochemical quantification of the arch calcium amounts (E, n = 10–12). Movat pentachrome-stained sections of HFD arches were also used to quantify intimal cartilaginous metaplasia morphometrically (F, n = 8 and 9). Data are normally distributed (D’Agostino-Pearson test) and shown as mean ± S.E.M. Differences between genotypes were analysed by two-way ANOVA with Bonferroni’s test, *P < 0.05 (LDLr−/−: Runx2f/f vs. LDLr−/−:Runx2ΔSM) for (E) and by student t-test, N.S.=not significant for (F). L = lumen, I = intima, M = media, Ad = adventitia.
3.4 Runx2 deletion in SMCs does not block transdifferentiation of SMCs to early chondrocytes, but substantially inhibits chondrocyte maturation
Since AIC was predominantly associated with areas of cartilaginous metaplasia, we postulated that a reduction in SMC transdifferentiation to chondrocytes might explain the decreased calcification observed in LDLr−/−:Runx2ΔSM mice compared to LDLr−/−:Runx2f/f mice. However, quantitation of Movat pentachrome-stained specimens indicated that removal of Runx2 in SMCs did not alter the size of cartilaginous metaplasia in atherosclerotic Runx2ΔSM vessels in comparison to the Runx2f/f counterparts (Figure 3F).
To determine whether SMC-specific removal of Runx2 affects specific stages of chondrocyte differentiation, thereby preventing AIC development, we determined expression levels of major marker genes along the chondrocytic differentiation pathway: Sox9, a master transcription factor critical for prechondrogenic fate decision, Col II, a marker for proliferating chondrocytes at early differentiation state, and Col X and MMP 13, markers for mature, hypertrophic chondrocytes.26–28 NC vessels showed no Sox9, Col II or Col X expression (data not shown), while HFD feeding promoted Sox9, Col II, Col X, and MMP13 (Figure 4A, D, F, and I). Levels of Sox9-positive cells and Col II-positive matrices were similarly elevated in LDLr−/−:Runx2f/f and LDLr−/−:Runx2ΔSM vessels, and not altered by SMC-specific Runx2 deletion (Figure 4A–E), suggesting that the initial commitment of SMC to chondrocyte differentiation in calcifying blood vessels was largely unaffected by Runx2 deficiency. In contrast, the hypertrophic chondrocyte marker protein, Col X, was greatly reduced in HFD LDLr−/−:Runx2ΔSM vessels compared to HFD LDLr−/−:Runx2f/f vessels (Figure 4F and G). Further morphometric analysis of Col II and Col X in cartilaginous matrices in adjacent sections confirmed a significantly reduced Col X/Col II ratio (Figure 4H). Similar findings at the mRNA level were observed by qRT-PCR of aortic arch samples (see Supplementary material online, Figure 3A–C). Finally, a significant reduction in MMP13 mRNA, another hypertrophic chondrocyte marker, was found in LDLr−/−:Runx2ΔSM vessels using qRT-PCR (Figure 4I). Together, these data suggest a critical role for Runx2 in maturation of SMC-derived chondrocytes to hypertrophic chondrocytes. Indeed, Col X and MMP13 are typically seen at the terminal stage of chondrocyte differentiation and strongly associated with endochondral ossification, representing a plausible mechanism for the AIC protection seen in Runx2ΔSM mice, even though cartilaginous lesion size was largely unaffected in these vessels (Figure 3C, D and F).
Figure 4.
Role of Runx2 in transdifferentiation of vascular SMCs into chondrocytes. LDLr−/−:Runx2f/f and LDLr−/−:Runx2ΔSM mice were fed with HFD for 24 weeks (A–G) and with NC or HFD for 18 weeks (I). 24-week HFD aortic arches were collected for immunohistochemical staining (brown) of Sox9 (A–C), Col II (D, E, H) and Col X (F, G, H), and morphometric analysis (C and H). Data are normally distributed (D’Agostino-Pearson test) and shown as mean ± S.E.M., *P < 0.05 (Student t-test). (I) MMP13 expression in 18-week aortic arches was measure with pooled triplicate RNA extractions as described in Figure 1H, n = 3, two-way ANOVA with Bonferroni’s test, **P < 0.01 (LDLr−/−:Runx2f/f vs. LDLr−/−:Runx2ΔSM). L = lumen, I = intima, M = media, Ad = adventitia.
3.5 Runx2 deletion in SMCs blocks osteoblastic differentiation in atherosclerotic blood vessels
In addition to its role in chondrocytic differentiation and maturation, Runx2 is also a well-known regulator for osteoblastic differentiation in skeleton.19,20 To determine whether Runx2 was required for SMC differentiation into osteoblast-like cells in atherosclerotic blood vessels, we determined levels of the osteoblast marker gene, OCN, in aortic arches of these animals. Immunohistochemical staining confirmed a significant, ∼83% reduction in OCN protein in HFD LDLr−/−:Runx2ΔSM vessels compared to HFD LDLr−/−:Runx2f/f vessels; (Figure 5A–C). Likewise, removal of Runx2 in vascular SMCs led to a reduction in OCN mRNA expression by ∼75% (HFD fed LDLr−/−:Runx2ΔSM mice vs. HFD LDLr−/−: Runx2f/f mice), though this result did not attain statistical significance (P = 0.0659). A similar trend was found in the expression of OSX and ALP mRNAs (see supplementary material online, Figure 3D and E). These results identified an essential role of Runx2 in SMC transdifferentiation to osteogenic progenitors and osteoblasts during AIC.
Figure 5.
SMC-specific Runx2 removal prevents osteogenic differentiation in atherosclerotic vasculature of LDLr−/− mice. Specimens collected as in Figure 4 were used to determine osteogenic differentiation of SMCs immunohistochemically (brown; A–C) and via quantitative RT-PCR for OCN (D) Data shown are mean ± S.E.M., n = 4, student t-test, **P < 0.01 for (C), and mean ± S.E.M., n = 3, two-way ANOVA with Bonferroni’s test for (D). L = lumen, I = intima, M = media, Ad = adventitia.
3.6 Runx2 deletion in SMCs does not inhibit RANKL expression or macrophage content of atherosclerotic lesions in HFD fed LDLr−/− mice
Macrophage infiltration and foam cell formation are major processes required for atherosclerotic lesion formation in mouse models, including ApoE−/− and LDLr−/− mice.29,30 Previous studies showed that Runx2 deficiency in SMCs led to a dramatic decrease in RANKL expression and macrophage infiltration in ApoE−/− arteries.21 In contrast to those studies, we observed abundant monocyte/macrophage accumulation in aortic arches and BCA of HFD fed LDLr−/−:Runx2f/f mice (Figure 6A and D), and comparable levels in LDLr−/−:Runx2ΔSM mice that do not produce functional, truncated Runx2 protein22 (Figure 6B, C E and F). Furthermore, expression of RANKL, MCP1, and CCR2 was not significantly altered in LDLr−/−:Runx2f/f mice compared to LDLr−/−:Runx2ΔSM mice (Figure 6G–I).
Figure 6.
SMC-specific Runx2 removal does not alter monocyte/macrophage infiltration and RANKL expression. Monocyte/macrophage infiltration was visualized using MOMA2 antibody in 18-week aortic arches (A, B) and BCA (D, E) of HFD LDLr−/−:Runx2f/f (A and D) and LDLr−/−:Runx2ΔSM (B and E) mice. L = lumen, I = intima, M = media, Ad = adventitia. Quantification of MOMA2 positive area for both 18 week aortic arches as well as 18 and 24 week BCA were shown in (C) and (F) respectively (n = 4–8). Data shown are mean ± S.E.M., differences between genotypes were determined by Mann–Whitney test. (G–I) Total RNA was extracted from pooled aortic arches (3–4 arches were pooled for each RNA sample, n = 3 RNA samples/group). Expression of RANKL, MCP1, and CCR2, was determined by Taqman quantitative RT-PCR. Data shown are mean ± S.E.M, n = 3. Differences between genotypes were analysed by two-way ANOVA with Bonferroni’s test.
4. Discussion
In this study, we utilized an improved conditional Runx2 knockout mouse model to investigate the role of SMC-specific Runx2 in atherosclerotic vascular calcification. We developed a mouse model that targets exon 4 of the Runx2 gene and hence no functional, truncated Runx2 protein could be produced upon Cre-mediated gene deletion.22 After introduction of this transgene into atherosclerotic LDLr−/− mice, we found that SMC-specific removal of Runx2 inhibited not only osteoblastic differentiation of vascular SMCs but also SMC-derived chondrocyte maturation, leading to a 50% reduction of AIC. Furthermore, we found that removal of Runx2 in vascular SMCs did not affect systematic lipid metabolism, RANKL expression, monocyte/macrophage recruitment, or atherosclerotic lesion size. These findings demonstrate a cell autonomous and pathway specific nature of Runx2 in AIC and are the first to present a genetic separation of the calcific sclerotic process from the lipid laden, atherogenic process, suggesting a likelihood of different mechanisms in regulating formation and progression of calcification and atheroma, two major pathological features often seen in advanced atherosclerotic lesions.
Mechanistically, a decrease in osteoblastic gene expression in calcifying atherosclerotic vessels was observed following SMC-specific Runx2 deletion. These findings are consistent with Runx2’s role in skeletal development, where it acts as a critical transcriptional factor in the development and maturation of osteoblasts leading to intramembranous ossification and bone formation.19,20 In cardiovasculature, Runx2 and its downstream genes have long been found in cells surrounding mineral nodules of human and animal arterial medial calcification (AMC).15,24,31,32 Through a genetic fate mapping strategy, these cells were identified as transdifferentiated SMCs on their way to an osteoblastic fate prior to matrix calcification.33 Deletion of Runx2 in vascular SMCs, therefore, prohibits osteoblastic differentiation and AMC,22 consistent with the current findings of osteoblastic differentiation in LDLr−/− mice.
Given previous studies suggesting a role for Runx2 in osteochondrogenic differentiation of SMCs in vitro34,35 and in vivo,12,21,33 we were surprised that deletion of Runx2 in SMCs did not change the overall content of chondrocyte-like cells in the intima. Our studies show that while SMC expression of Runx2 was not required for initial chondrogenesis, it was necessary for maturation of chondrocytes to a pro-mineralizing, hypertrophic state, leading to endochondral ossification. Runx2 expression has been observed in chondrocyte-like cells within atherosclerotic lesions and its role in chondrocytic differentiation during skeletal development has been well-documented.12,27,36,37 Initial chondrocyte differentiation through Sox9, a master chondrogenic transcriptional factor, is absolutely critical and has been shown to act upstream of Runx2, suggesting possible Runx2-independent mechanisms during early-stage chondrogenesis.38,39 In a Runx2 global knockout mouse model, isolated chondrocytes proliferated normally but were unable to maintain chondrocyte phenotype in culture.36 Similarly, in our study, we found that Runx2 was not required for SMC-specific chondrocyte differentiation, further supporting a Runx2-independent mechanism for initial chondrocyte commitment. Other factors, including Sox9, may play a role in Runx2-independent chondrocyte differentiation, although additional work is necessary to clearly identify these factors.40 Thus, our data support the hypothesis that SMC-specific removal of Runx2 prevents both endochondral and intramembranous ossification in atherosclerotic lesions.
In contrast to our studies, Sun et al.21 reported a role for SMC autonomous Runx2 in macrophage accumulation and atheromatous lesion formation using a very different Runx2 targeting construct.41 Their construct targeted exon 8 of the Runx2 gene and produced a functional, truncated Runx2 protein upon Cre-mediated recombination of the floxed-Runx2 allele.21,41 Importantly, this truncated protein contained an intact Runt DNA binding domain and domains interacting with Runx2 partner proteins, such as CBF and Smad3/4. 21,42 Using SM22αCre to remove Runx2 in vascular SMCs of ApoE−/− mice with this model, the authors found significantly reduced SMC elaboration of RANKL, a tumour necrosis factor family member important for monocyte infiltration and differentiation of mineral-resorbing osteoclasts,43,44 along with a near complete blockage of monocyte/macrophage recruitment and atherosclerosis.21 AIC was also completely blocked likely due to the inhibition of oxidative stress-dependent osteogenic differentiation and osteoblast-osteoclast crosstalk.21 These findings are likely explained by the truncated Runx2 protein acting via a dominant negative effect42,45 on the Runt-related family members in addition to Runx2, especially Runx1 and Runx3 that share the same highly conserved Runt DNA binding domain and are involved in regulating monocyte/macrophage biology.46–48
Along these lines, recent studies have shown transdifferentiation of SMCs into a lipid-loaded, foam cell-like state during atherosclerosis.49–51 These studies suggest that SMCs may not only contribute to fibrous cap formation and calcification but also directly participate in the process of atheroma formation through macrophage foam cell formation, accounting for ∼18–40% Mac2/CD68-positive cells observed in human and mouse atherosclerotic lesions.49,50 Since Runx1 and Runx3 are known to control myeloid differentiation necessary for monocyte/macrophage and osteoclast formation,46–48 their transcriptional activities may also be critical in controlling the SMC to macrophage/foam cell-like differentiation. Therefore, the dominant negative effect of truncated Runx2 in both bone marrow-derived cells and SMCs could act synergistically to prevent atheromatous plaque formation as seen in the Sun et al.21 study, though a detailed mechanism would require further investigation. Finally, macrophages have been found to release matrix vesicles, the initial sites of microcalcification observed in early atherosclerotic lesion formation.29 Therefore, the decreased monocyte/macrophage infiltration and differentiation observed in the Sun et al.21 model may also contribute to the reduced vascular calcification. While we also employed SM22αCre for tissue specific deletion, our Runx2 targeting construct did not produce detectable truncated proteins.22 In addition, removal of exon 4 ensures the loss of functionality of the Runt homology domain should a truncated protein be made, thereby minimizing off target effects from Runx1 and Runx3 in myeloid differentiation.
In conclusion, we have demonstrated that SMC-specific Runx2 plays critical roles in both osteoblastic differentiation and chondrocytic maturation during atherosclerosis-induced vascular calcification. This protective effect is independent of systemic lipid metabolism, mineral balance, RANKL expression, monocyte/macrophage infiltration, and atherosclerotic progression. Given its role in the stepwise progression of lesion formation, targeting the Runx2 signalling pathway in the vasculature may provide a novel therapeutic method for prevention of AIC in this disease.
Supplementary material
Supplementary material is available at Cardiovascular Research online.
Conflict of interest: none declared.
Funding
This work was supported by the National Institutes of Health grants R01 HL081785 (CMG), R01 HL62329 (CMG), and K01 DK075665 (MYS). T.C. was supported by the National Science Foundation Graduate Research Fellowship, C.S. was supported by the Fulbright Visiting Scholarship, and S.Y. was supported by the JSPS Postdoctoral Fellowship for Research Abroad.
References
- 1.Rumberger JA, Simons DB, Fitzpatrick LA, Sheedy PF, Schwartz RS. Coronary artery calcium area by electron-beam computed tomography and coronary atherosclerotic plaque area. A histopathologic correlative study. Circulation 1995;92:2157–2162. [DOI] [PubMed] [Google Scholar]
- 2.Ehara S, Kobayashi Y, Yoshiyama M, Shimada K, Shimada Y, Fukuda D, Nakamura Y, Yamashita H, Yamagishi H, Takeuchi K, Naruko T, Haze K, Becker AE, Yoshikawa J, Ueda M. Spotty calcification typifies the culprit plaque in patients with acute myocardial infarction: an intravascular ultrasound study. Circulation 2004;110:3424–3429. [DOI] [PubMed] [Google Scholar]
- 3.Sangiorgi G, Rumberger JA, Severson A, Edwards WD, Gregoire J, Fitzpatrick LA, Schwartz RS. Arterial calcification and not lumen stenosis is highly correlated with atherosclerotic plaque burden in humans: a histologic study of 723 coronary artery segments using nondecalcifying methodology. J Am Coll Cardiol 1998;31:126–133. [DOI] [PubMed] [Google Scholar]
- 4.Taylor AJ, Burke AP, O'Malley PG, Farb A, Malcom GT, Smialek J, Virmani R. A comparison of the Framingham risk index, coronary artery calcification, and culprit plaque morphology in sudden cardiac death. Circulation 2000;101:1243–1248. [DOI] [PubMed] [Google Scholar]
- 5.Beadenkopf WG, Daoud AS, Love BM. Calcification in the coronary arteries and its relationship to arteriosclerosis and myocardial infarction. Am J Roentgenel 1964;92:865–871. [PubMed] [Google Scholar]
- 6.Locker TH, Schwartz RS, Cotta CW, Hickman JR. Fluoroscopic coronary artery calcification and associated coronary disease in asymptomatic young men. J Am Coll Cardiol 1992;19:1167–1192. [DOI] [PubMed] [Google Scholar]
- 7.Vliegenthart R, Hollander M, Breteler MM, van der Kuip DA, Hofman A, Oudkerk M, Witteman JC. Stroke is associated with coronary calcification as detected by electron-beam CT: the Rotterdam Coronary Calcification Study. Stroke 2002;33:462–465. [DOI] [PubMed] [Google Scholar]
- 8.Hutcheson JD, Maldonado N, Aikawa E. Small entities with large impact: microcalcifications and atherosclerotic plaque vulnerability. Curr Opin Lipidol 2014;25:327–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hunt JL, Fairman R, Mitchell ME, Carpenter JP, Golden M, Khalapyan T, Wolfe M, Neschis D, Milner R, Scoll B, Cusack A, Mohler ER., III Bone formation in carotid plaques: a clinicopathological study. Stroke 2002;33:1214–1219. [DOI] [PubMed] [Google Scholar]
- 10.Qiao JH, Mertens RB, Fishbein MC, Geller SA. Cartilaginous metaplasia in calcified diabetic peripheral vascular disease: morphologic evidence of enchondral ossification. Hum Pathol 2003;34:402–407. [DOI] [PubMed] [Google Scholar]
- 11.Sage AP, Tintut Y, Demer LL. Regulatory mechanisms in vascular calcification. Nat Rev Cardiol 2010;7:528–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Naik V, Leaf EM, Hu JH, Yang HY, Nguyen NB, Giachelli CM, Speer MY. Sources of cells that contribute to atherosclerotic intimal calcification: an in vivo genetic fate mapping study. Cardiovasc Res 2012;94:545–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nguyen NB, Naik V, Speer MY. Diabetes mellitus accelerates cartilaginous metaplasia and calcification in atherosclerotic vessels of LDLr mutant mice. Cardiovasc Pathol 2013;22:167–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moe SM, Duan D, Doehle BP, O'Neill KD, Chen NX. Uremia induces the osteoblast differentiation factor Cbfa1 in human blood vessels. Kidney Int 2003;63:1003–1011. [DOI] [PubMed] [Google Scholar]
- 15.Tyson KL, Reynolds JL, McNair R, Zhang Q, Weissberg PL, Shanahan CM. Osteo/chondrocytic transcription factors and their target genes exhibit distinct patterns of expression in human arterial calcification. Arterioscler Thromb Vasc Biol 2003;23:489–494. [DOI] [PubMed] [Google Scholar]
- 16.Aikawa E, Nahrendorf M, Sosnovik D, Lok VM, Jaffer FA, Aikawa M, Weissleder R. Multimodality molecular imaging identifies proteolytic and osteogenic activities in early aortic valve disease. Circulation 2007;115:377–386. [DOI] [PubMed] [Google Scholar]
- 17.Bobryshev YV. Transdifferentiation of smooth muscle cells into chondrocytes in atherosclerotic arteries in situ: implications for diffuse intimal calcification. J Pathol 2005;205:641–650. [DOI] [PubMed] [Google Scholar]
- 18.Mundlos S, Otto F, Mundlos C, Mulliken JB, Aylsworth AS, Albright S, Lindhout D, Cole WG, Henn W, Knoll JH, Owen MJ, Mertelsmann R, Zabel BU, Olsen BR. Mutations involving the transcription factor CBFA1 cause cleidocranial dysplasia. Cell 1997;89:773–779. [DOI] [PubMed] [Google Scholar]
- 19.Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson RT, Gao YH, Inada M, Sato M, Okamoto R, Kitamura Y, Yoshiki S, Kishimoto T. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell 1997;89:755–764. [DOI] [PubMed] [Google Scholar]
- 20.Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR, Stamp GW, Beddington RS, Mundlos S, Olsen BR, Selby PB, Owen MJ. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell 1997;89:765–771. [DOI] [PubMed] [Google Scholar]
- 21.Sun Y, Byon CH, Yuan K, Chen J, Mao X, Heath JM, Javed A, Zhang K, Anderson PG, Chen Y. Smooth muscle cell-specific runx2 deficiency inhibits vascular calcification. Circ Res 2012;111:543–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin M, Chen T, Leaf EM, Speer MY, Giachelli CM. Runx2 expression in smooth muscle cells is required for arterial medial calcification in mice. Am J Pathol 2015;185:1958–1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Renard CB, Kramer F, Johansson F, Lamharzi N, Tannock LR, von Herrath MG, Chait A, Bornfeldt KE. Diabetes and diabetes-associated lipid abnormalities have distinct effects on initiation and progression of atherosclerotic lesions. J Clin Invest 2004;114:659–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pai A, Leaf EM, El Abbadi M, Giachelli CM. Elastin degradation and vascular smooth muscle cell phenotype change precede cell loss and arterial medial calcification in a uremic mouse model of chronic kidney disease. Am J Pathol 2011;178:764–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stender RN, Engler WJ, Braun TM, Hankenson FC. Establishment of blood analyte intervals for laboratory mice and rats by use of a portable clinical analyzer. J Am Assoc Lab Anim Sci 2007;46:47–52. [PubMed] [Google Scholar]
- 26.Akiyama H, Chaboissier MC, Martin JF, Schedl A, de Crombrugghe B. The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes Dev 2002;16:2813–2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fujita T, Azuma Y, Fukuyama R, Hattori Y, Yoshida C, Koida M, Ogita K, Komori T. Runx2 induces osteoblast and chondrocyte differentiation and enhances their migration by coupling with PI3K-Akt signaling. J Cell Biol 2004;166:85–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dy P, Wang W, Bhattaram P, Wang Q, Wang L, Ballock RT, Lefebvre V. Sox9 directs hypertrophic maturation and blocks osteoblast differentiation of growth plate chondrocytes. Dev Cell 2012;22:597–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.New SE, Goettsch C, Aikawa M, Marchini JF, Shibasaki M, Yabusaki K, Libby P, Shanahan CM, Croce K, Aikawa E. Macrophage-derived matrix vesicles: an alternative novel mechanism for microcalcification in atherosclerotic plaques. Circ Res 2013;113:72–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Averill MM, Barnhart S, Becker L, Li X, Heinecke JW, LeBoeuf RC, Hamerman JA, Sorg C, Kerkhoff C, Bornfeldt KE. S100A9 differentially modifies phenotypic states of neutrophils, macrophages, and dendritic cells: implications for atherosclerosis and adipose tissue inflammation. Circulation 2011;123:1216–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shanahan CM, Cary NR, Salisbury JR, Proudfoot D, Weissberg PL, Edmonds ME. Medial localization of mineralization-regulating proteins in association with Monckeberg's sclerosis: evidence for smooth muscle cell-mediated vascular calcification. Circulation 1999;100:2168–2176. [DOI] [PubMed] [Google Scholar]
- 32.Steitz SA, Speer MY, Curinga G, Yang HY, Haynes P, Aebersold R, Schinke T, Karsenty G, Giachelli CM. Smooth muscle cell phenotypic transition associated with calcification—upregulation of Cbfa1 and downregulation of smooth muscle lineage markers. Circ Res 2001;89:1147–1154. [DOI] [PubMed] [Google Scholar]
- 33.Speer MY, Yang HY, Brabb T, Leaf E, Look A, Lin W-L, Frutkin AD, Dichek DA, Giachelli CM. Smooth muscle cells give rise to osteochondrogenic precursors and chondrocytes in calcifying arteries. Circ Res 2009;104:733–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Speer MY, Li X, Hiremath PG, Giachelli CM. Runx2/Cbfa1, but not loss of myocardin, is required for smooth muscle cell lineage reprogramming toward osteochondrogenesis. J Cell Biochem 2010;110:935–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Byon CH, Javed A, Dai Q, Kappes JC, Clemens TL, Darley-Usmar VM, McDonald JM, Chen Y. Oxidative stress induces vascular calcification through modulation of the osteogenic transcription factor Runx2 by AKT signaling. J Biol Chem 2008;283:15319–15327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Enomoto H Furuichi T Zanma A Yamana K Yoshida C Sumitani S Yamamoto H Enomoto-Iwamoto M Iwamoto M Komori T. Runx2 deficiency in chondrocytes causes adipogenic changes in vitro. J Cell Sci 2004;117:417–425. [DOI] [PubMed] [Google Scholar]
- 37.Hinoi E, Bialek P, Chen YT, Rached MT, Groner Y, Behringer RR, Ornitz DM, Karsenty G. Runx2 inhibits chondrocyte proliferation and hypertrophy through its expression in the perichondrium. Genes Dev 2006;20:2937–2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamashita S, Andoh M, Ueno-Kudoh H, Sato T, Miyaki S, Asahara H. Sox9 directly promotes Bapx1 gene expression to repress Runx2 in chondrocytes. Exp Cell Res 2009;315:2231–2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hartmann C. Transcriptional networks controlling skeletal development. Curr Opin Genet Dev 2009;19:437–443. [DOI] [PubMed] [Google Scholar]
- 40.Liu T, Gao Y, Sakamoto K, Minamizato T, Furukawa K, Tsukazaki T, Shibata Y, Bessho K, Komori T, Yamaguchi A. BMP-2 promotes differentiation of osteoblasts and chondroblasts in Runx2-deficient cell lines. J Cell Physiol 2007;211:728–735. [DOI] [PubMed] [Google Scholar]
- 41.Chen H, Ghori-Javed FY, Rashid H, Adhami MD, Serra R, Gutierrez SE, Javed A. Runx2 regulates endochondral ossification through control of chondrocyte proliferation and differentiation. J Bone Miner Res 2014;29:2653–2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adhami MD, Rashid H, Chen H, Clarke JC, Yang Y, Javed A. Loss of Runx2 in committed osteoblasts impairs postnatal skeletogenesis. J Bone Miner Res 2015;30:71–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boyce BF, Xing L. Functions of RANKL/RANK/OPG in bone modeling and remodeling. Arch Biochem Biophys 2008;473:139–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Breuil V, Schmid-Antomarchi H, Schmid-Alliana A, Rezzonico R, Euller-Ziegler L, Rossi B. The receptor activator of nuclear factor (NF)-kappaB ligand (RANKL) is a new chemotactic factor for human monocytes. FASEB J 2003;17:1751–1753. [DOI] [PubMed] [Google Scholar]
- 45.Ducy P, Starbuck M, Priemel M, Shen J, Pinero G, Geoffroy V, Amling M, Karsenty G. A Cbfa1-dependent genetic pathway controls bone formation beyond embryonic development. Genes Dev 1999;13:1025–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rosenbauer F, Tenen DG. Transcription factors in myeloid development: balancing differentiation with transformation. Nat Rev Immunol 2007;7:105–117. [DOI] [PubMed] [Google Scholar]
- 47.Estecha A, Aguilera-Montilla N, Sanchez-Mateos P, Puig-Kroger A. RUNX3 regulates intercellular adhesion molecule 3 (ICAM-3) expression during macrophage differentiation and monocyte extravasation. PLoS One 2012;7:e33313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Woolf E, Xiao C, Fainaru O, Lotem J, Rosen D, Negreanu V, Bernstein Y, Goldenberg D, Brenner O, Berke G, Levanon D, Groner Y. Runx3 and Runx1 are required for CD8 T cell development during thymopoiesis. Proc Natl Acad Sci U S A 2003;100:7731–7736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shankman LS, Gomez D, Cherepanova OA, Salmon M, Alencar GF, Haskins RM, Swiatlowska P, Newman AA, Greene ES, Straub AC, Isakson B, Randolph GJ, Owens GK. KLF4-dependent phenotypic modulation of smooth muscle cells has a key role in atherosclerotic plaque pathogenesis. Nat Med 2015;21:628–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Allahverdian S, Chehroudi AC, McManus BM, Abraham T, Francis GA. Contribution of intimal smooth muscle cells to cholesterol accumulation and macrophage-like cells in human atherosclerosis. Circulation 2014;129:1551–1559. [DOI] [PubMed] [Google Scholar]
- 51.Vengrenyuk Y, Nishi H, Long X, Ouimet M, Savji N, Martinez FO, Cassella CP, Moore KJ, Ramsey SA, Miano JM, Fisher EA. Cholesterol loading reprograms the microRNA-143/145-myocardin axis to convert aortic smooth muscle cells to a dysfunctional macrophage-like phenotype. Arterioscler Thromb Vasc Biol 2015;35:535–546. [DOI] [PMC free article] [PubMed] [Google Scholar]