Abstract
Human mesenchymal stem cells (hMSCs) receive differentiation cues from a number of stimuli, including extracellular matrix (ECM) stiffness. The pathways used to sense stiffness and other physical cues are just now being understood and include proteins within focal adhesions. To rapidly advance the pace of discovery for novel mechanosensitive proteins, we employed a combination of in silico and high throughput in vitro methods to analyze 47 different focal adhesion proteins for cryptic kinase binding sites. High content imaging of hMSCs treated with small interfering RNAs for the top 6 candidate proteins showed novel effects on both osteogenic and myogenic differentiation; Vinculin and SORBS1 were necessary for stiffness-mediated myogenic and osteogenic differentiation, respectively. Both of these proteins bound to MAPK1 (also known as ERK2), suggesting that it plays a context-specific role in mechanosensing for each lineage; validation for these sites was performed. This high throughput system, while specifically built to analyze stiffness-mediated stem cell differentiation, can be expanded to other physical cues to more broadly assess mechanical signaling and increase the pace of sensor discovery.
Keywords: SORBS1, ponsin, stiffness, hydrogel
Graphical Abstract
Introduction
Although physical properties of the niche have become widely recognized for their influence on a host of cell behaviors 1–3, significant attention has been paid to the influence of extracellular matrix (ECM) stiffness on stem cells 4–6. While initially reported to be myosin contractility sensitive 7, their upstream mechanisms have remained unclear. Recently, however, mechanisms have been proposed involving the nucleus 8, translocation of factors to the nucleus 9, Rho GTPases 10, stretch activated channels 11, and focal adhesions, i.e. “molecular strain gauges” 12. While numerous mechanisms may overlap, it is clear from these examples that many sensors within each category are still undetermined.
High throughput systems 13 to assess mechano-signaling have yet to play as significant a role as they have in other biomedical and engineering contexts, e.g. biomaterial microarrays to explore niche conditions 14,15 and microcontact printing to explore the influence of cell shape 16; this may be due to fabrication limitations with small volume hydrogels, imaging limitations with thick hydrogels at high magnification, and biological limitations with high throughput molecular screening in stem cells. For example, hydrogels are often fabricated in larger 6- and 24-well formats 7,17,18 and have been used to investigate how a variety of niche properties influence cells 19. Creating physiologically relevant substrates in small volumes to elicit appropriate cell behaviors is challenging but not unprecedented 20; ensuring that the imaging plane is flat in such small wells, however, has proven difficult and has limited high resolution imaging required for many stem cell applications. Several groups have pursued high throughput imaging of cells on soft surfaces 21,22, although these efforts were performed in open culture systems where media interacting with cells on one surface condition was free to diffuse to cells on other surface conditions. Despite these challenges, it is clear that discovery of novel proteins that convert mechanical forces into biochemical signals, e.g. phosphorylation, will require screening due to the sheer number of proteins that could be involved in each mechanism type 8–12.
To create a high throughput screen of potential mechanosensing proteins and determine their effects on stem cells, high content screening analysis of multiple cell parameters for phenotyping 23,24 is required in addition to high throughput screening systems 25. While this combination has been used in pre-fabricated small interfering RNA (siRNA) 26 or polymer arrays 15 to examine stem cell pluripotency, their combination in a high throughput array to study mechanically sensitive stem cell differentiation has been technically challenging. Attempts to leverage high throughput hydrogel systems with high content imaging has been limited by an inability to perform high magnification single cell imaging or investigate the immunofluorescence expression of individual transcription factors 20,27.
Here, we have overcome the imaging challenges associated with the 96 well hydrogel array format 20 and combined it with a focal adhesion siRNA screen to determine novel proteins that convert mechanical forces into biochemical responses, whether acting as direct or indirect transducers of force. We report the identification of several protein hits that may regulate lineage-specific, substrate stiffness dependent differentiation.
Experimental
Cell Culture and Reagents
Human mesenchymal stem cells (Lonza) were maintained in growth medium (DMEM, 10% FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin) which was changed every four days (except in 96 well plates). Only low passage hMSCs were used for experimental studies, i.e. less than passage 9. For MAPK1 inhibition, the MAPK1 inhibitor pyrazolylpyrrole, dissolved in DMSO, was used at a final concentration of 2 nM and added to cells immediately post-plating. At 2 nM, pyrazolylpyrrole is extremely selective and has only been shown to inhibit MAPK1, limiting potential off-target effects 28. Non-differentiation based experiments, including western blots and durotaxis assays, were performed after 24 hours while siRNA-induced protein knockdown was at a maximum. Conversely, differentiation experiments took place over the course of four days, since differentiation occurs as the integration of cues over time. Cells were plated at a density of 500 cells/cm2, a sparse density that reduces the likelihood of density-dependent cell signaling over the course of the experiment.
Polyacrylamide Hydrogel Fabrication in 6- and 96-Well Formats
For MAPK1 inhibitor experiments performed in six well plates, acrylamide was polymerized on aminosilanized coverslips. A solution containing the crosslinker N, N′ methylene-bis-acrylamide, the monomer acrylamide, 1/100 volume 10% Ammonium Persulfate and 1/1000 volume of N, N, N′, N′-Tetramethylethylenediamine was mixed. Two different combinations of acrylamide and bis-acrylamide were used to make hydrogels of 11 and 34 kilopascal (kPa; a unit of stiffness). Approximately 50 μL of the mixed solution was placed between 25 mm diameter aminosilanized coverslips and a chlorosilanized glass slide for 6-well plates. 100 μg/mL collagen I was chemically crosslinked to the substrates using the photoactivatable crosslinker Sulfo-SANPAH (Pierce). Custom 96 well plates containing collagen type I-conjugated polyacrylamide hydrogels crosslinked to glass bottom surfaces (Matrigen) were fabricated containing equal numbers of 15 kPa wells and 42 kPa hydrogels to induce myogenesis and osteogenesis, respectively (Figure S1A). Stiffness values were verified using an MFP3D-Bio atomic force microscope (Asylum Research, Santa Barbara, CA) using previously established methods (Figure S1B) 29,30. Polyacrylamide gel thickness was also verified using a BD CARV II confocal microscope (Figure S1C,D) and found to be approximately 250 μm, which is thick enough that the cells are unable to feel the glass substrate below the gel 31.
siRNA Transfection
siRNA oligonucleotides against human Vinculin, p130Cas, SORBS1 (Ponsin), SORBS3 (Vinexin), Palladin, Paxillin, and Filamin (ON-TARGETplus SMARTpool; Thermo Fisher Scientific, Waltham, MA) and a pool of four non-targeting siRNAs control oligonucleotides (ON-TARGETplus siControl; Dharmacon), diluted in DEPC water (OmniPure, EMD) and 5X siRNA buffer (Thermo Fisher Scientific, Waltham, MA), were transiently transfected into human hMSCs using Dharmafect 1 (Thermo Fisher Scientific, Waltham, MA) at an optimized concentration of 50 nM in low serum antibiotic free growth media, according to the manufacturers’ protocols. Specific siRNA sequences can be found in Supplemental Table 1. Protein knockdown was characterized by western blot and immunofluorescence. After 24 hours of transfection in antibiotic-free media (2% FBS), media was replaced with standard hMSC growth media and cells replated onto appropriate substrates.
Plasmid Transfection
pEGFP-C1 sub-cloned with complete Vinculin cDNA, which had been originally excised from p1005 with EcoRI and inserted in EcoRI digested pEGFP-C1 (labeled as FL), was obtained from Dr. Susan Craig 32. L765I mutant Vinculin plasmids were obtained via site-directed mutagenesis on FL Vinculin plasmids. All plasmids were purified using QIAGEN Plasmid Midi Kit (Qiagen). hMSCs were transfected in antibiotic-free medium with 1 mg of plasmid precomplexed with 2 μl of Lipofectamine 2000 (Life Technologies) in 100 μl of DMEM. After 24 hours of transfection in antibiotic-free media with 2% FBS, media was replaced with standard hMSC growth media.
Immunofluorescence
hMSCs were fixed with 3.7% formaldehyde for 30 minutes at 4°C and permeabilized with 1% Triton-X for 5 minutes at 37°C. The cells were then stained with primary antibodies against human MyoD (sc-32758, Santa Cruz), Myf5 (sc-302, Santa Cruz, Dallas, TX), Osterix (ab22552, Abcam), CBFA1 (RUNX2) (sc-101145, Santa Cruz), Vinculin (ab129002, Abcam), p130Cas (ab108320, Abcam), SORBS1 (ab4551, Abcam), SORBS3 (GTX-115362, Genetex), Filamin (ab51217, Abcam), or Paxillin (ab32084, Abcam). Corresponding secondary antibodies were conjugated to Alexa Fluor 488 (FITC) or Alexa Fluor 647 (Cy5) (Invitrogen). Nuclei were counterstained with Hoechst dye (Sigma), and the actin cytoskeleton was stained with rhodamine-conjugated phalloidin (Invitrogen). Cells not plated in 96 well plates were imaged with a Nikon Eclipse Ti-S inverted fluorescence microscope equipped with a BD Carv II camera.
High Content Imaging and Analysis
96 well plates were imaged on a CV1000 Cell Voyager (Yokogawa). Briefly, images were acquired through 5 z-positions with 10 μm step sizes at 25 different points in each well with three different filter sets (FITC, TXRD, and DAPI). Maximum Intensity Projections (MIPs) were constructed from the resulting stitched z-stacks to account for uneven, slanted, or differentially swollen hydrogel surfaces and analyzed using a semiautomated image analysis pipeline in CellProfiler 33. Nuclear outlines were obtained as primary objects with automatic Otsu Global thresholding (Figure S2A) and cell outlines were obtained using the TXRD channel as secondary objects using a Watershed Gradient algorithm (Figure S2B). The pipeline calculated morphological attributes (such as cell area, aspect ratio, and eccentricity) for each cell, as well as the mean and integrated density of the FITC channel signal in nuclei, cell outlines, and cytoplasm outlines. From these data, one could distinguish cells with nuclear expression only, cytoplasm expression only, uniform positive expression, and uniform negative expression, as shown with example cells in Figure S2C. Data analysis was performed with Microsoft Excel, GraphPad Prism, and CellAnalyst 34.
Western Blots
Cell lysates were collected by rinsing samples with cold PBS, followed by a five minute lysis in mRIPA buffer (50 mM HEPES pH 7.5, 150 mM NaCl, 1.5 mM MgCl2, 1% Triton, 1% Na-DOC, 0.1% SDS) with 1 mM EGTA, 1 mM Na3VO4, 10 mM Na4P2O7, and 1 mM PMSF (protease inhibitors). Cell lysates were separated via SDS-PAGE, transferred to PVDF membranes (Bio-Rad), and washed in Buffer A (25 mM Tris-HCl, 150 mM NaCl, 0.1% Tween-20) + 4% SeaBlock (Thermo Fisher Scientific, Waltham, MA) overnight at 4°C. Membranes were incubated with anti-Vinculin (ab18058, Abcam), GAPDH (ab8245, Abcam), ERK2 (ab7948, Abcam), p130Cas (ab108320, Abcam), SORBS1 (ab4551, Abcam), SORBS3 (GTX-115362, Genetex), Filamin (ab51217, Abcam), or Paxillin (ab32084, Abcam) antibodies for 1 hour, washed with Buffer A containing SeaBlock, and incubated in streptavidin horseradish-peroxidase-conjugated secondary antibodies (Bio-Rad) for 30 minutes at room temperature. Immunoblots were visualized using ECL reagent (Pierce). All western blot antibodies were obtained from Abcam (Cambridge, England).
Quantitative PCR
mRNA was isolated from hMSCs grown after 4 days with Trizol, and subsequently treated with chloroform and precipitated with isopropanol. The cell lysate was centrifuged and the pellet washed in ethanol twice, after which the pellet was allowed to dry before resuspension in DEPC water. cDNA was assembled through reverse transcriptase polymerase chain reaction (RT-PCR) for one hour at 37°C, followed by a 5 minute inactivation step at 99°C. 1 μL of the resulting cDNA mixture was added to 12.5 μL SYBR Green Real Time PCR Master Mix (Thermo Fisher Scientific, Waltham, MA) containing 0.25 nM forward and reverse primers (Supplemental Table 2) and enough DEPC water to bring the total reaction volume per well to 25 μL.
Immunoprecipitation
Cell lysates were collected with a non-denaturing lysis buffer (20 mM Tris-HCl pH 8, 127 mM NaCl, 1% Nonidet P-40, 2 mM EDTA). Anti-ERK2 antibody (Abcam ab124362) was bound to protein G-conjugated Dynabeads (Life Technologies, Carlsbad, CA, USA) for 1 hour at 4°C with gentle agitation. Beads were magnetically captured, the supernatant removed, and the pellet incubated overnight at 4°C before Western Blot analysis.
Statistics
All experiments were performed in triplicate with the indicated number of cells analyzed per condition. Error bars shown are the standard error of the mean (SEM). Significance was assessed with Bonferroni’s Multiple Comparison Test at a significance threshold of p<0.05 or lower as indicated. Values less than 0.1 were noted. For instances where data is not significantly different, N.S. is stated.
Results
Bioinformatic Assessment of Focal Adhesion-based Mechanosensing Reveals that MAPK1 Binding is Frequent and Cryptic
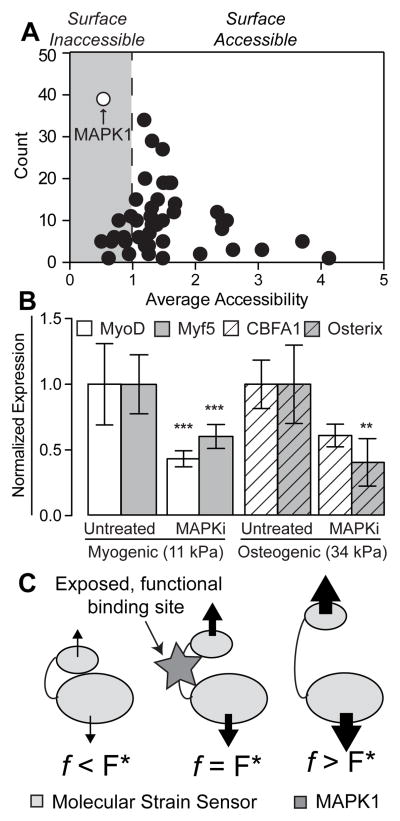
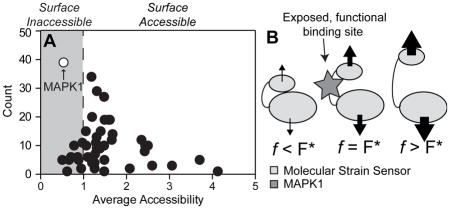
We selected 47 focal adhesion proteins 35,36 (Supplemental Table 3) based on their ability to bind multiple proteins at their N- and C-terminal ends such that they could potentially be unfolded when one end of the protein is displaced relative to the other, i.e. a “molecular strain sensor” 12. These candidates were analyzed with ScanSite 37, a tool designed to identify short protein sequence binding motifs and predict whether the motif is surface accessible. After analyzing all 47 proteins, a scatter plot showing the number of times a predicted binding site was found versus the average accessibility of the identified sites was constructed (Figure 1A, Supplemental Table 3). Interestingly, predicted MAPK1 binding sites were found most frequently and with the second-lowest average accessibility, implying that MAPK1 is the most likely candidate to affect stem cell differentiation across a wide variety of cellular pathways in a manner that requires a change in surface accessibility of the MAPK1 binding site.
Figure 1. ScanSite Results for 47 Different Focal Adhesion Proteins.
(A) Each data point represents a predicted binding partner. The y-axis displays the number of times this binding partner was identified during the analysis of the 47 focal adhesion proteins, while the x-axis shows the average accessibility of the binding site. Predicted surface inaccessible binding sites have accessibility values below 1 (gray region). (B) MAPK1 inhibitor pyrazolylpyrrole (MAPKi) was applied to cells at the beginning of the 4-day time course on both (A) 11 kPA and (B) 34 kPa substrates and stained for (A) MyoD (white) or Myf5 (gray) and (B) CBFA1 (white barred) or Osterix (gray barred) as indicated on day 4. Mean nuclear fluorescence is plotted normalized to untreated cells. **p<0.01 and ***p<0.001 relative to untreated cells stained for the same transcription factor. (C) Schematic of force-induced conformational changes by a “molecular strain sensor” where proteins bound to the sensor stretch the it by transmitting a force across the protein. The resulting conformational change exposes the once cryptic binding site at an optimal force, F* (middle schematic). Above or below that value results in excessive deformation of the binding site to prevent binding or not enough stretch causing the site to remain cryptic, respectively.
MAPK1 Inhibition Prevents Mechanosensitive Stem Cell Myogenesis and Osteogenesis
To analyze the effect of MAPK1 inhibition on substrate stiffness-directed hMSC differentiation, MAPK1 was inhibited with pyrazolylpyrrole, an extremely potent and selective MAPK1 inhibitor 38, immediately post plating in order to limit any early mechanosensing events, which can occur on the time scale of minutes 39. Consistent with previous results 17, we found that hMSCs exhibited a 50% reduction in nuclear-localized myogenic transcription factors MyoD and Myf5 after 4 days in culture (Figure 1B). On 34 kPa substrates, pyrazolylpyrrole-treated hMSCs also exhibited reduced osteogenic transcription factor expression and localization (Figure 1B). However, since pyrazolylpyrrole is a global MAPK1 inhibitor, it may inadvertently reduce lineage commitment through non-stiffness mediated mechanisms.
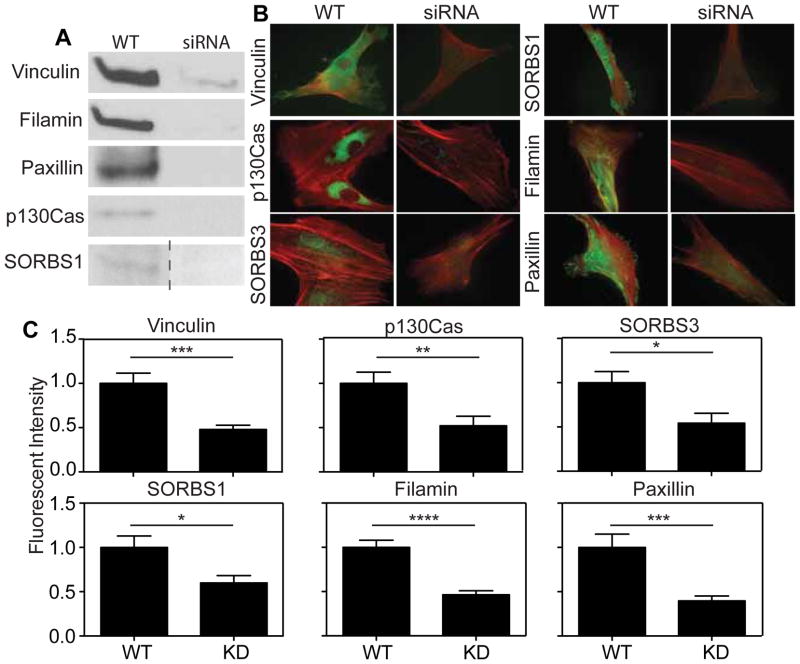
To address this, the potentially upstream focal adhesion proteins identified by ScanSite were investigated further. Of the proteins analyzed, five–Vinculin, p130Cas, Filamin, SORBS1 (Ponsin), SORBS3 (Vinexin)–had a predicted cryptic MAPK1 binding site and terminal multiple binding sites to other proteins, which would allow the protein to be strained and change configuration under an appropriate amount of force, F* (Figure 1C). This conformation change could then expose the MAPK1 binding site predicted to be cryptic, but only under the appropriate amount of force. Paxillin was selected as a control protein because it did not have a cryptic MAPK1 binding site (Figure S3). siRNAs were used to transiently knock down candidate proteins, which was verified by western blot (Figure 2A) and immunofluorescence (Figure 2B–C). siRNA-induced knockdown of these proteins did not affect endogenous expression of MAPK1 (Figure S5).
Figure 2. Confirmation of siRNA-induced Knockdown.
(A) Western blots of lysates collected 2 days post siRNA treatment. (B) Immunofluorescence images of proteins being knocked down. (C) Quantification of mean immunofluorescence intensity from knockdown cells. For Vinculin, p130Cas, SORBS3, SORBS1, Filamin, and Paxillin in (C), n > 10 cells in triplicate.
To analyze whether siRNA-induced knockdown of the five candidate proteins could alter mechanically-sensitive myogenic and osteogenic differentiation, hMSCs were cultured in 96 well plates containing polyacrylamide hydrogels of roughly 250 μm thickness and stiffness of either 15 kPa (myogenic) or 42 kPa (osteogenic) for four days (Figure S1). These stiffness values are within the characteristic ranges of myogenic- and osteogenic-inducing 2D substrates 7,40–42. To analyze osteogenesis, cells in the 42 kPa wells were fixed and stained for the osteogenic transcription factors Osterix and CBFA1, while for myogenesis, cells in the 15 kPa wells were fixed and stained for the myogenic transcription factors MyoD and Myf5. Expression levels for the transcription factors were compared with those in untreated cells at day 0 (negative control) and at day 4 (positive control) on the corresponding stiffness hydrogels. Transcription factors were specifically chosen as outputs for identifying mechanosensitivity because both the expression and nuclear localization could be used as criteria for lineage commitment (Figure S2). To further reduce the false discovery rate, we only classified a protein as a mechanosensor if their knockdown impaired stiffness-induced differentiation as assessed by both transcription factors. As transcription factor expression is often sequential, this reduces the likelihood that the assay simply missed the time when the transcription factor was active.
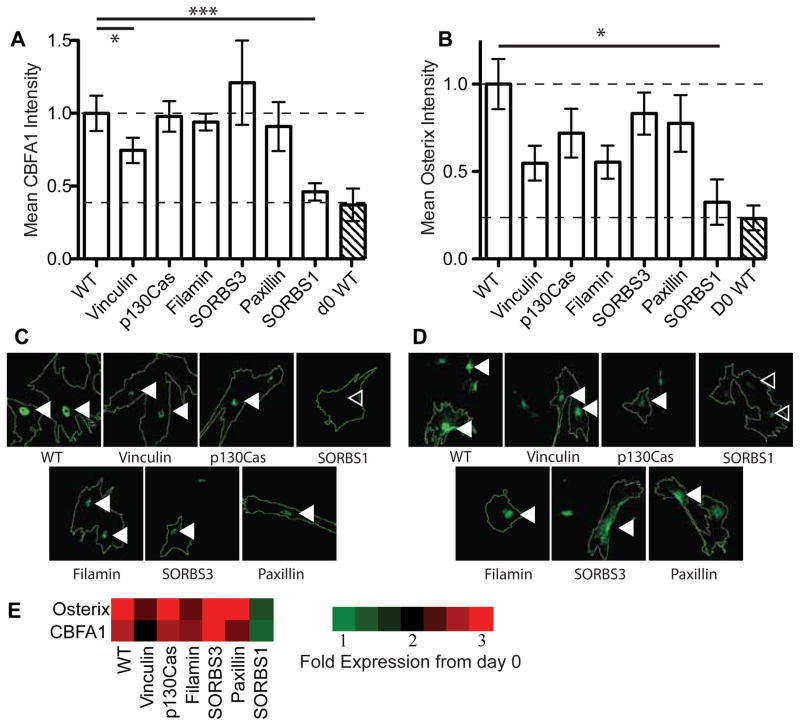
In the osteogenesis assay, we found that p130Cas, Filamin, Paxillin, and SORBS3 (Vinexin) knockdown did not affect osteogenic differentiation signals after 4 days relative to day 0 expression and localization. Conversely, the knockdown of SORBS1 (Ponsin), which interacts with Vinculin 43 and plays a role in insulin signaling 44, reduced both CBFA1 and Osterix nuclear expression by over 50%. Vinculin knockdown, which was previously shown to not affect CBFA1 expression 17, slightly reduced CBFA1 but not Osterix expression (Figure 3); no myogenic expression was found in these cells (data not shown). Thus, we concluded that SORBS1 could act as a unique stiffness-mediated sensor for osteogenic differentiation.
Figure 3. Osteogenic Differentiation and Focal Adhesion Protein Knockdown.
Normalized mean intensity levels of (A) CBFA1 and (B) Osterix immunofluorescence staining after four days of culture of osteogenically favorable 42 kPa substrates. Representative images show cell outlines along with (C) CBFA1 and (D) Osterix expression. Filled arrowheads indicate nuclei that maintained transcription factor expression whereas open arrowheads indicate nuclei that lost expression. (E) Heat map indicating fold-change in expression of the indicated osteogenic markers from day 0 wild type cells. For WT, Vinculin, p130Cas, Filamin, SORBS3, Paxillin, SORBS1, and d0 WT in (A) and (B), n=298, 35, 28, 44, 29, 28, 20, and 40, respectively.
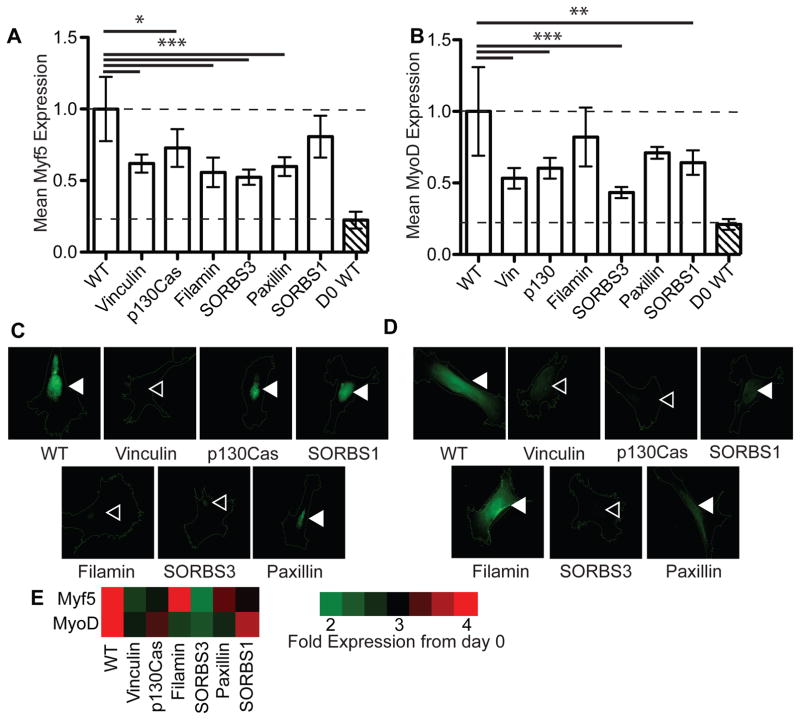
In the myogenesis assay, siRNA knockdown of Vinculin, p130Cas, or SORBS3 resulted in a loss of stiffness-induced expression of both MyoD and Myf5 at day 4. This is in agreement with recent reports of Vinculin-mediated SORBS3 mechanosensing 45. However, Filamin, SORBS1, and Paxillin only reduced expression of one of the two myogenic markers (Figure 4). Paxillin does not contain a cryptic MAPK1 binding site, so Myf5 reduction may be due to other predicted cryptic binding domains that it contains, e.g. MAPK3; no osteogenic expression was found in these cells (data not shown). Thus, we concluded that Vinculin could act as a unique stiffness-mediated sensor for myogenic differentiation, consistent with prior reports 17.
Figure 4. Myogenic Differentiation and Focal Adhesion Protein Knockdown.
Normalized mean intensity levels of (A) Myf5 and (B) MyoD immunofluorescence staining after four days of culture of myogenically favorable 15 kPa substrates. Representative images show cell outlines along with (C) Myf5 and (D) MyoD expression. Filled arrowheads indicate nuclei that maintained transcription factor expression whereas open arrowheads indicate nuclei that lost expression. (E) Heat map indicating fold-change in expression of the indicated myogenic markers from day 0 wild type cells. For WT, Vinculin, p130Cas, Filamin, SORBS3, Paxillin, SORBS1, and d0 WT in (A) and (B), n=39, 31, 43, 24, 30, 35, 29, and 9, respectively.
If knockdown of the candidate focal adhesion proteins disrupts not just mechanosensitive signaling but also other normal cell behaviors, stiffness-mediated differentiation differences may not solely be related to signaling. High content image analysis was performed with CellProfiler to measure cell area and morphology, i.e. eccentricity, of cells from all conditions. Neither area nor morphology was altered by any of the siRNA treatments (Figure S4A–B). Cell migration speed was also unaffected by siRNA knockdown, although SORBS3 knockdown appeared to increase migration persistence (Figure S4C). Perhaps most importantly, focal adhesion assembly in terms of size and distribution appeared unaffected in single knockdown experiments; outside of the expected loss of expression of the proteins being knocked down, no changes were observed in these focal adhesion characteristics (Figure S4D). Differentiation changes could also be due to off-target effects of the siRNA on MAPK1 expression, thus depleting the endogenous pool of the sensor’s binding partner and inadvertently preventing differentiation. However, MAPK1 western blots indicated that knockdown did not impact endogenous expression (Figure S5), reinforcing the concept that individual mechanosensing proteins regulated transcription factor expression.
Validation of Candidate Mechanosensor Hits for MAPK1 Interaction
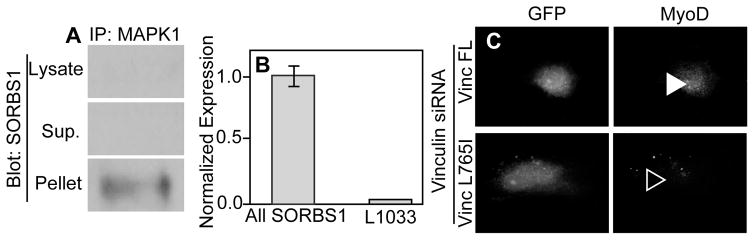
To verify hits directly using more targeted molecular methods, SORBS1 was immunoprecipated via MAPK1. For hMSCs cultured for 24 hours on 34 kPa PA gels, SORBS1 was detected in the pellet but not the unconcentrated whole cell lysate, suggesting that, although expressed at low levels, SORBS1 and MAPK1 interact in cells cultured on physiological-stiffness gels (Figure 5A). SORBS1 contains two predicted binding sites for MAPK1 at L500 and L1033 (Figure S2B), but among the twelve SORBS1 isoforms, only two contain the predicted L1033 binding site 46–48. qPCR indicated that undifferentiated cells cultured on 34 kPa substrates for 24 hours did not significantly express SORBS1 isoforms containing L1033 (Figure 5B). Lacking other kinase binding domains predicted with high confidence to be inaccessible (i.e. Scansite accessibility prediction less than 0.5), the MAPK1 binding site found on SORBS1 at L500 is the most likely candidate to act as a stretch sensitive mechanosensor. For Vinculin, which pulls MAPK1 down with immunoprecipitation on 11 kPa substrates 17, MAPK1 binding was predicted at L765 (Figure S2B). To confirm that L765 is specifically required for myogenic differentiation on 11 kPa substrates, a plasmid containing L765I-mutated Vinculin and Green Fluorescent Protein (GFP) was added back to cells that had been treated with Vinculin siRNA. While Vinculin knockdown was sufficient to reduce myogenic transcription factor expression in hMSCs, addback of full-length Vinculin rescued expression whereas addback of L765I-mutated Vinculin was insufficient to fully rescue expression (Figure 5C, filled vs. open arrowhead, respectively).
Figure 5. Molecular validation of Mechanosensitive Protein Interactions.
(A) SORBS1 blots of lysates without (top) or with immunoprecipitation (middle and bottom) via a MAPK1 antibody. Supernatant and pellet fractions of the immunoprecipitation are shown (middle and bottom, respectively). Prior to lysis, cells were cultured on 34 kPa substrates. (B) qPCR of SORBS1 using primers that target a conserved portion of the gene (labeled All SORBS1) versus a region only found in the two full length isoforms (labeled L1033). Data is normalized to the GAPDH and then the All SORBS1 condition. Input RNA was collected from hMSCs on 34 kPa substrates for 24 hours. (C) Add back of Full-Length (FL) or mutated Vinculin plasmid (L765I) to Vinculin siRNA-treated cells showing GFP and MyoD expression after 4 days on 11 kPa substrates. Filled and open arrowheads indicate where nuclear localized MyoD expression is or should be.
Discussion
While these data specifically focus on screening 47 focal adhesion proteins with a “molecular strain sensor”-like structure as predicted by ScanSite, some of which have never been identified as mechanically sensitive, the list of proteins comprising focal adhesions is much larger and dynamic. Current estimates implicate as many as 232 different components, of which 148 are intrinsic and 84 are transient 49, as a common signature of adhesions. Recent analyses of focal adhesions have even identified more than 1300 distinct proteins within isolated adhesion complexes 50, suggesting exceedingly complex adhesion-based mechanisms for cells that must actively sense their surroundings. Focal adhesion composition and structure have also recently been shown to be relatively stable to external perturbation, including siRNA knockdown or chemical inhibition of components, suggesting that signaling transduction occurs independently of structural integrity 51. That said, our data also focused on proteins with relatively little functional data, e.g. SORBS1, to establish proof-of-principle that we can use a high content imaging based platform to identify candidate sensors via their influence on stem cell differentiation.
Prior to this work, SORBS1, also known as Ponsin, Sorbin, CAP, or c-Cbl associated protein, has not been implicated in mechanosensitive differentiation, although it has been shown to affect actin cytoskeleton organization via Dynamin GTPases 52, bind to vinculin43, and be overexpressed and phosphorylated in response to endogenous PYK2 expression, a focal adhesion complex-localized kinase capable of suppressing osteogenesis 53.
Even with a fairly well studied focal adhesion protein like Vinculin, questions about its force-sensitive behavior remain. Vinculin undergoes a conformational change from its autoinhibited state to an ‘activated’ state in which it can bind F-actin, allowing it to transmit force from the cytoskeleton 54. Studies have shown that vinculin is under mechanical tension within focal adhesions, although the activating conformational change is separable from the application of force across the protein 55. Recent work has revealed that this tension is independent of substrate stiffness56, suggesting that vinculin’s upstream binding partner talin may bear the brunt of force sensing. Intriguingly, talin’s unfolding under force is sufficient to expose differential amounts of cryptic vinculin binding sites 57, meaning that differential amounts of (potentially force-sensitive) vinculin activation can initiate different differentiation pathways. Thus, it is possible that the exposure of the cryptic MAPK1 domain in vinculin occurs after activation, and after talin and actin binding, in a force dependent manner. While adding a Talin knockdown to our screen would serve as an effective positive control, attempts at siRNA-induced talin knockdown have led to a loss in normal cell morphology (data not shown), likely because of the key structural role it plays in linking the cytoskeleton to focal adhesions.
Beyond stem cell differentiation assays, several alternative high throughput techniques have been adapted for mechanobiology sensor identification 58 though they do not utilized biomimetic substrates. For example, mass spectroscopy “cysteine shotgun” assays use cysteine-binding dyes to assess differential protein labeling under stress 59 but this approach focuses on the conformational change itself and may overlook downstream signaling changes. Even when applied directly to differential unfolding in response to mechanical signals 60, one could miss transient protein unfolding during signal transduction, especially if cryptic binding domains do not contain cysteine residues. While this RNAi screening approach is more targeted, it can be adapted to fit any instance in which immunofluorescence is used to measure an output, e.g. a response to change in substrate stiffness, and can be specific for nuclear or cytoplasmic expression (Figure S2).
Conclusions
A computational approach was used to select candidate proteins that could potentially play a role in MAPK1-based mechanosensitive differentiation based on an analysis of their binding partners and presence of cryptic signaling sites, i.e. the “molecular strain gauge” structure 12. A high throughput, high content analysis based system capable of finding hits much more quickly and efficiently was then constructed to test these candidates, with which we identified SORBS1 and Vinculin as potential mechanosensors in hMSCs. While this method was applied specifically to the mechanical influence of stiffness on stem cells differentiation, it can be applied to a number of applications in cell biology in which an immunofluorescently-labeled marker is differentially up- or down-regulated in response to a physical stimulus, e.g. stiffness, etc.
Supplementary Material
It has become widely recognized in the field of mechanobiology that cell behavior is regulated by physical parameters of the niche, including its stiffness. While critical observations have been made regarding the molecular details of this regulation, e.g. translocation of YAP/TAZ, the proteins or complexes that actually convert biophysical to biochemical signals that the cell can interpret remain uncertain. Here we have developed an assay that enables one to predict which proteins could be mechanically sensitive by determining their effect on stem cell differentiation (although other metrics could be substituted). We then identify several focal adhesion mechanosensors and validate them using conventional molecular biology methods.
Acknowledgments
The authors would like to acknowledge Dr. Jamie Kasuboski for assistance with high content imaging and Dr. Brenton Hoffman for helpful comments on the manuscript. This work was supported by grants from the National Institutes of Health (DP2OD006460 to A.J.E.) and National Science Foundation (1463689 to A.J.E) as well as a National Science Foundation predoctoral fellowship (to L.G.V.)
References
- 1.Curtis A, Wilkinson C. Biomaterials. 1997;18:1573–1583. doi: 10.1016/s0142-9612(97)00144-0. [DOI] [PubMed] [Google Scholar]
- 2.Sun Y, Chen CS, Fu J. Annu Rev Biophys. 2012;41:519–542. doi: 10.1146/annurev-biophys-042910-155306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Discher DE, Janmey P, Wang YL. Science. 2005;310:1139–1143. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- 4.Murphy WL, McDevitt TC, Engler AJ. Nat Mater. 2014;13:547–557. doi: 10.1038/nmat3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gattazzo F, Urciuolo A, Bonaldo P. Biochimica et Biophysica Acta (BBA) - General Subjects. 2014;1840:2506–2519. doi: 10.1016/j.bbagen.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Discher DE, Mooney DJ, Zandstra PW. Science. 2009;324:1673–1677. doi: 10.1126/science.1171643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Engler AJ, Sen S, Sweeney HL, Discher DE. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 8.Swift J, Ivanovska IL, Buxboim A, Harada T, Dingal PCDP, Pinter J, Pajerowski JD, Spinler KR, Shin JW, Tewari M, Rehfeldt F, Speicher DW, Discher DE. Science. 2013;341:1240104–1240104. doi: 10.1126/science.1240104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M, Bicciato S, Elvassore N, Piccolo S. Nature. 2011;474:179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- 10.Cameron AR, Frith JE, Gomez GA, Yap AS, Cooper-White JJ. Biomaterials. 2014;35:1857–1868. doi: 10.1016/j.biomaterials.2013.11.023. [DOI] [PubMed] [Google Scholar]
- 11.Pathak MM, Nourse JL, Tran T, Hwe J, Arulmoli J, Le DTT, Bernardis E, Flanagan LA, Tombola F. Proc Natl Acad Sci USA. 2014;111:16148–16153. doi: 10.1073/pnas.1409802111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holle AW, Engler AJ. Current Opinion in Biotechnology. 2011;22:648–654. doi: 10.1016/j.copbio.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang JH, Chung TDY, Oldenburg KR. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 14.Ranga A, Gobaa S, Okawa Y, Mosiewicz K, Negro A, Lutolf MP. Nature Communications. 2014;5 doi: 10.1038/ncomms5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mei Y, Saha K, Bogatyrev SR, Yang J, Hook AL, Kalcioglu ZI, Cho SW, Mitalipova M, Pyzocha N, Rojas F, Van Vliet KJ, Davies MC, Alexander MR, Langer R, Jaenisch R, Anderson DG. Nat Mater. 2010;9:768–778. doi: 10.1038/nmat2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee J, Abdeen AA, Zhang D, Kilian KA. Biomaterials. 2013;34:8140–8148. doi: 10.1016/j.biomaterials.2013.07.074. [DOI] [PubMed] [Google Scholar]
- 17.Holle AW, Tang X, Vijayraghavan D, Vincent LG, Fuhrmann A, Choi YS, Álamo JC, Engler AJ. Stem Cells. 2013;31:2467–2477. doi: 10.1002/stem.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lakins JN, Chin AR, Weaver VM. In: Cell Migration- Textbook. Wells CM, Parsons M, editors. Vol. 916. Humana Press; Totowa, NJ: 2012. pp. 317–350. [Google Scholar]
- 19.Ayala R, Zhang C, Yang D, Hwang Y, Aung A, Shroff SS, Arce FT, Lal R, Arya G, Varghese S. Biomaterials. 2011;32:3700–3711. doi: 10.1016/j.biomaterials.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 20.Mih JD, Sharif AS, Liu F, Marinkovic A, Symer MM, Tschumperlin DJ. PLoS ONE. 2011;6:e19929. doi: 10.1371/journal.pone.0019929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gobaa S, Hoehnel S, Roccio M, Negro A, Kobel S, Lutolf MP. Nature Methods. 2011;8:949–955. doi: 10.1038/nmeth.1732. [DOI] [PubMed] [Google Scholar]
- 22.Unadkat HV, Hulsman M, Cornelissen K, Papenburg BJ, Truckenmüller RK, Carpenter AE, Wessling M, Post GF, Uetz M, Reinders MJT, Stamatialis D, van Blitterswijk CA, de Boer J. Proc Natl Acad Sci USA. 2011;108:16565–16570. doi: 10.1073/pnas.1109861108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gasparri F. Expert Opin Drug Discov. 2009;4:643–657. doi: 10.1517/17460440902992870. [DOI] [PubMed] [Google Scholar]
- 24.Conrad C, Erfle H, Warnat P, Daigle N, Lörch T, Ellenberg J, Pepperkok R, Eils R. Genome Res. 2004;14:1130–1136. doi: 10.1101/gr.2383804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mayr LM, Bojanic D. Current Opinion in Pharmacology. 2009;9:580–588. doi: 10.1016/j.coph.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 26.Erfle H, Neumann B, Liebel U, Rogers P, Held M, Walter T, Ellenberg J, Pepperkok R. Nature Protocols. 2007;2:392–399. doi: 10.1038/nprot.2006.483. [DOI] [PubMed] [Google Scholar]
- 27.Yang YH, Hsieh T-L, Ji AT-Q, Hsu W-T, Liu CY, Lee OKS, Ho JHC. Stem Cells. 2016 doi: 10.1002/stem.2405. [DOI] [PubMed] [Google Scholar]
- 28.Aronov Alex M, Baker Christopher, Bemis Guy W, Cao Jingrong, Chen Guanjing, Ford Pamella J, Germann Ursula A, Green Jeremy, Hale Michael R, Jacobs Marc, Janetka James W, Maltais Francois, Martinez-Botella Gabriel, Namchuk Mark N, Straub Judy, Tang A Qing, Xie X. J Med Chem. 2007;50:1280–1287. doi: 10.1021/jm061381f. [DOI] [PubMed] [Google Scholar]
- 29.Kaushik G, Fuhrmann A, Cammarato A, Engler AJ. Biophys J. 2011;101:2629–2637. doi: 10.1016/j.bpj.2011.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Radmacher M. Cell Mechanics. Vol. 83. Elsevier; 2007. pp. 347–372. [DOI] [PubMed] [Google Scholar]
- 31.Buxboim A, Rajagopal K, Brown AEX, Discher DE. Journal of physics Condensed matter: an Institute of Physics journal. 2010;22:194116. doi: 10.1088/0953-8984/22/19/194116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cohen DM, Chen H, Johnson RP, Choudhury B, Craig SW. The Journal of biological chemistry. 2005;280:17109–17117. doi: 10.1074/jbc.M414704200. [DOI] [PubMed] [Google Scholar]
- 33.Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang I, Friman O, Guertin DA, Chang J, Lindquist RA, Moffat J, Golland P, Sabatini DM. Genome Biology. 2006;7:10. doi: 10.1186/gb-2006-7-10-r100. 2006, 7, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jones TR, Kang IH, Wheeler DB, Lindquist RA, Papallo A, Sabatini DM, Golland P, Carpenter AE. BMC Bioinformatics. 2008;9:1. doi: 10.1186/1471-2105-9-482. 2008, 9, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zamir E, Geiger B. J Cell Sci. 2001;114:3583–3590. doi: 10.1242/jcs.114.20.3583. [DOI] [PubMed] [Google Scholar]
- 36.von Wichert G, Jiang G, Kostic A, De Vos K, Sap J, Sheetz MP. J Cell Biol. 2003;161:143–153. doi: 10.1083/jcb.200211061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Obenauer JC, Cantley LC, Yaffe MB. Nucl Acids Res. 2003;31:3635–3641. doi: 10.1093/nar/gkg584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Junttila MR, Li SP, Westermarck J. FASEB J. 2007;22:954–965. doi: 10.1096/fj.06-7859rev. [DOI] [PubMed] [Google Scholar]
- 39.Li J, Han D, Zhao Y-P. Sci Rep. 2014;4 doi: 10.1038/srep03910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lanniel M, Huq E, Allen S, Buttery L, Williams PM, Alexander MR. Soft Matter. 2011;7:6501–6514. [Google Scholar]
- 41.Wen JH, Vincent LG, Fuhrmann A, Choi YS, Hribar KC, Taylor-Weiner H, Chen S, Engler AJ. Nat Mater. 2014;13:979–987. doi: 10.1038/nmat4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huebsch N, Arany PR, Mao AS, Shvartsman D, Ali OA, Bencherif SA, Rivera-Feliciano J, Mooney DJ. Nat Mater. 2010;9:518–526. doi: 10.1038/nmat2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mandai K, Nakanishi H, Satoh A, Takahashi K, Satoh K, Nishioka H, Mizoguchi A, Takai Y. J Cell Biol. 1999;144:1001–1017. doi: 10.1083/jcb.144.5.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin WH, Huang CJ, Liu MW, Chang HM, Chen YJ, Tai TY, Chuang LM. Genomics. 2001;74:12–20. doi: 10.1006/geno.2001.6541. [DOI] [PubMed] [Google Scholar]
- 45.Yamashita H, Ichikawa T, Matsuyama D, Kimura Y, Ueda K, Craig SW, Harada I, Kioka N. J Cell Sci. 2014;127:1875–1886. doi: 10.1242/jcs.133645. [DOI] [PubMed] [Google Scholar]
- 46.Vandenbroere I, Paternotte N, Dumont JE, Erneux C, Pirson I. Biochemical and Biophysical Research Communications. 2003;300:494–500. doi: 10.1016/s0006-291x(02)02894-2. [DOI] [PubMed] [Google Scholar]
- 47.Consortium TU. Nucl Acids Res. 2014;43:gku989-D212. [Google Scholar]
- 48.Lebre AS, Jamot L, Takahashi J, Spassky N, Leprince C, Ravisé N, Zander C, Fujigasaki H, Kussel-Andermann P, Duyckaerts C, Camonis JH, Brice A. Hum Mol Genet. 2001;10:1201–1213. doi: 10.1093/hmg/10.11.1201. [DOI] [PubMed] [Google Scholar]
- 49.Winograd-Katz SE, Fässler R, Geiger B, Legate KR. Nature Reviews Molecular Cell Biology. 2014;15:273–288. doi: 10.1038/nrm3769. [DOI] [PubMed] [Google Scholar]
- 50.Ajeian JN, Horton ER, Astudillo P, Byron A, Askari JA, Millon-Frémillon A, Knight D, Kimber SJ, Humphries MJ, Humphries JD. Prot Clin Appl. 2015;10:51–57. doi: 10.1002/prca.201500033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Horton ER, Humphries JD, Stutchbury B, Jacquemet G, Ballestrem C, Barry ST, Humphries MJ. J Cell Biol. 2016;212:349–364. doi: 10.1083/jcb.201508080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tosoni D, Cestra G. FEBS Letters. 2009;583:293–300. doi: 10.1016/j.febslet.2008.12.047. [DOI] [PubMed] [Google Scholar]
- 53.Bonnette PC, Robinson BS, Silva JC, Stokes MP, Brosius AD, Baumann A, Buckbinder L. Journal of Proteomics. 2010;73:1306–1320. doi: 10.1016/j.jprot.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 54.Chen H, Cohen DM, Choudhury DM, Kioka N, Craig SW. J Cell Biol. 2005;169:459–470. doi: 10.1083/jcb.200410100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grashoff C, Hoffman BD, Brenner MD, Zhou R, Parsons M, Yang MT, McLean MA, Sligar SG, Chen CS, Ha T, Schwartz MA. Nature. 2010;466:263–266. doi: 10.1038/nature09198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kumar A, Ouyang M, Van den Dries K, McGhee EJ, Tanaka K, Anderson MD, Groisman A, Goult BT, Anderson KI, Schwartz MA. J Cell Biol. 2016;213:371–383. doi: 10.1083/jcb.201510012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.del Rio A, Perez-Jimenez R, Liu R, Roca-Cusachs P, Fernandez JM, Sheetz MP. Science. 2009;323:638–641. doi: 10.1126/science.1162912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Otto O, Rosendahl P, Mietke A, Golfier S, Herold C, Klaue D, Girardo S, Pagliara S, Ekpenyong A, Jacobi A, Wobus M, Töpfner N, Keyser UF, Mansfeld J, Fischer-Friedrich E, Guck J. Nature Methods. 2015;12:199–202. doi: 10.1038/nmeth.3281. [DOI] [PubMed] [Google Scholar]
- 59.Johnson CP, Tang HY, Carag C, Speicher DW, Discher DE. Science (New York, NY) 2007;317:663–666. doi: 10.1126/science.1139857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Krieger CC, An X, Tang HY, Mohandas N, Speicher DW, Discher DE. PNAS. 2011;108:8269–8274. doi: 10.1073/pnas.1018887108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.