Abstract
Ricin is a potent ribotoxin that is considered a bioterror threat due to its ease of isolation and possibility of aerosolization. In yeast, mutation of arginine residues away from the active site results in a ricin toxin A chain (RTA) variant that is unable to bind the ribosome and exhibits reduced cytotoxicity. The goal of the present work was to determine if these residues contribute to ribosome binding and cytotoxicity of RTA in mammalian cells. The RTA mutant R193A/R235A did not interact with mammalian ribosomes, while a G212E variant with a point mutation near its active site bound ribosomes similarly to wild-type (WT) RTA. R193A/R235A retained full catalytic activity on naked RNA but had reduced activity on mammalian ribosomes. To determine the effect of this mutant in intact cells, pre R193A/R235A containing a signal sequence directing it to the endoplasmic reticulum and mature R193A/R235A that directly targeted cytosolic ribosomes were each expressed. Depurination and protein synthesis inhibition were reduced by both pre- and mature R193A/R235A relative to WT. Protein synthesis inhibition was reduced to a greater extent by R193A/R235A than by G212E. Pre R193A/R235A caused a greater reduction in caspase activation and loss of mitochondrial membrane potential than G212E relative to WT RTA. These findings indicate that an RTA variant with reduced ribosome binding is less toxic than a variant with less catalytic activity but normal ribosome binding activity. The toxin-ribosome interaction represents a novel target for the development of therapeutics to prevent or treat ricin intoxication.
Keywords: ricin A chain, ribosome binding, mammalian cells, cytotoxicity, depurination, therapeutic target
1. Introduction
Ricin is a type-2 ribosome inactivating protein (RIP) produced by the castor bean plant (Ricinus communis) that is highly toxic to mammalian cells. It consists of two subunits; a catalytic ricin toxin A chain (RTA) and a galactose-binding B chain (RTB) covalently linked by a single disulfide bond (Robertus and Monzingo, 2004). Following endocytosis, a small amount of ricin holotoxin enters the endoplasmic reticulum (ER) by retrograde translocation from the trans-Golgi network. Cleavage of the disulfide bond between RTA and RTB is required for RTA to enter the cytosol where it depurinates a universally conserved adenine residue within the sarcin/ricin loop (SRL) of the 28S rRNA to inhibit protein synthesis (Spooner and Lord, 2012). Due to its high toxicity, ricin has been classified as a category B bioterrorism agent (Audi et al., 2005). While two RTA-based vaccines have been tested in Phase I clinical trials (Smallshaw and Vitetta, 2012; Vitetta et al., 2012; Pittman et al., 2015), no post-exposure therapeutics for ricin intoxication are currently available.
During protein synthesis, elongation factors and other translational (t)GTPases interact with ribosomes at the GTPase-associated center, which includes the SRL and a lateral protuberance referred to as the ribosomal stalk (Uchiumi and Kominami, 1992; Wahl and Moller, 2002; Clementi and Polacek, 2010; Voorhees et al., 2010; Shi et al., 2012). The latter consists of ribosomal proteins that are phosphorylated and hence referred to as ribosomal P-proteins (Tchorzewski, 2002). The eukaryotic stalk is composed of two heterodimers of P1 and P2 proteins which bind to the base of the stalk (P0), forming a pentameric structure (Ballesta and Remacha, 1996; Guarinos et al., 2003; Krokowski et al., 2006; Grela et al., 2010). Early studies showed that the rate of depurination of native ribosomes by ricin was dramatically greater than that of naked rRNA (Endo and Tsurugi, 1988). Subsequent work indicated that RTA (Chiou et al., 2008; McCluskey et al., 2008; May et al., 2012), the catalytic subunit of the type 2 RIP Shiga toxin 1 (Stx1) (McCluskey et al., 2008; McCluskey et al., 2012), and the type-I RIP trichosanthin (TCS) (Chan et al., 2001; Chan et al., 2007; Too et al., 2009) bind to ribosomal P-proteins. Deletion of P1 or P2 protein decreases enzymatic activity and toxicity of RTA, Stx1 or Shiga toxin 2 (Stx2) in yeast (Chiou et al., 2008; Chiou et al., 2011) and decreases enzymatic activity and toxicity of RTA in mammalian cells (May et al., 2012). Collectively, these studies indicate that interaction with ribosomal P-proteins is required for full enzymatic activity of these RIPs and suggest a role for the ribosomal stalk in localizing the toxins near the SRL on the ribosome.
We have previously examined the ability of different RTA mutants to affect depurination activity, protein synthesis inhibition and apoptosis in mammalian cells (Jetzt et al., 2012). These studies showed that a relatively low level of depurination by RTA is sufficient to inhibit protein synthesis and activate stress-induced cell signaling and apoptosis such that a substantial reduction in the depurination activity of RTA is required to prevent its inhibitory effect on protein synthesis. All of the mutated residues in our study were on the side of RTA where the active site is located (Jetzt et al., 2012). However, several studies have shown that arginine residues in RTA located away from the active site cleft affect its enzymatic activity in vitro as well as its ability to bind the ribosome (Watanabe and Funatsu, 1986; Watanabe et al., 1994; Kitaoka, 1998; Li et al., 2013). In yeast, mutation of arginine residues 193 and 235 to alanine (R193A/R235A) results in a variant that folds similarly to wild-type (WT) but is unable to bind the ribosome or the stalk pentamer (Li et al., 2013). This mutant exhibits reduced cytotoxicity even though it depurinates naked RNA at a similar catalytic rate as WT RTA (Li et al., 2013). Whether these specific arginine residues contribute to binding of RTA to mammalian ribosomes and affect the toxicity of RTA in mammalian cells in vivo has not been determined. Here we examine the ribosome binding mutant R193A/R235A, as well as the G212E mutant, which contains a point mutation near the active site, to determine whether ribosome binding is important for ribosome depurination, translation inhibition and apoptosis in mammalian cells. Since the 35-residue signal sequence would target precursor (pre) RTA to the ER and thus mimic retrograde trafficking while the mature form of RTA would represent catalytic activity in the absence of retrograde trafficking, both the pre- and mature forms of RTA were expressed.
2. Materials and methods
2.1 Reagents
Dulbecco’s modified Eagle’s medium (DMEM) containing 4.5 g/L D-glucose and penicillin/streptomycin were purchased from Life Technologies (Carlsbad, CA). Phenol red-free DMEM with low glucose, gentamicin, insulin, D-(+)-glucose, fetal bovine serum (FBS), GTP, puromycin, heparin, PMSF, aprotinin, leupeptin, and trypsin inhibitor were obtained from Sigma-Aldrich (St. Louis, MO).
2.2 Recombinant RTA protein purification
Wild type (WT) RTA and RTA mutants were cloned into the E. coli expression vector, pET28, with an N-terminal His10 followed by the mature RTA (Li et al., 2013). WT RTA and His10-tagged EGFP were purified using nickel-nitrilotriacetic acid (NTA)-agarose from Qiagen (Li et al., 2013). RTA mutants were purified by the Northeast Biodefense Center protein expression core facility (Wadsworth Center, Albany, NY).
2.3 Isolation of bovine ribosomes
MAC-T cells were lysed in buffer A [20 mM HEPES-KOH pH 7.6, 5 mM Mg (CH3COO)2, 50 mM KCl, 10% glycerol, 1 mg/mL heparin, 1 mM PMSF, 1 mM DTT and 10 µg/mL leupeptin, trypsin inhibitor and aprotinin]. Cells were then homogenized by 10 passages through a 23 gauge needle. After addition of Triton X-100 to a final concentration of 1%, lysates were mixed gently and centrifuged at 30,000 x g for 20 min at 4°C. The resulting supernatant was applied to 1 mL buffer C cushion [50mM HEPES-KOH pH 7.6, 5 mM Mg (CH3COO)2, 50 mM NH4Cl, 0.1 mM PMSF, 0.1 mM DTT and 25% glycerol] and centrifuged at 303,800 x g for 2 hr at 4°C. The pellet was washed with buffer C and resuspended in buffer B [20 mM HEPES-KOH pH 7.6, 20 mM Mg (CH3COO)2, 0.5 M KCl, 10% glycerol, 1 mg/mL heparin, 1 mM PMSF, 1 mM DTT and 10 µg/mL leupeptin, trypsin inhibitor and aprotinin] by shaking on a vortex at 4°C for 1.5 h. Puromycin and GTP, both at 1 mM final concentration, were added, and the sample was incubated at 30°C for 30 min. After centrifugation at 14,000 rpm for 10 min at 4°C, the supernatant was applied to 1 mL buffer B cushion [20 mM HEPES-KOH pH 7.6, 20 mM Mg (CH3COO)2, 0.5 M KCl, 25% glycerol, 1 mg/mL heparin, 1 mM PMSF, 1 mM DTT and 10 µg/mL leupeptin, trypsin inhibitor and aprotinin] and centrifuged at 303,800 x g for 2 hr at 4°C. The ribosome pellet was washed and resuspended in buffer C.
2.4 Isolation of rat ribosomes
Animal care was performed in accordance with guidelines from the Rutgers Institutional Animal Care and Use Committee (IACUC) and complied with NIH policy. Rat livers were collected following decapitation, flash frozen in liquid nitrogen and stored at −80°C. Approximately 3–4 grams of tissue were cut into smaller pieces using scissors, and homogenized in Buffer A using a glass homogenizer. After addition of Triton X-100 to a final concentration of 2%, lysates were mixed gently and centrifuged at 30,000 x g for 20 min at 4°C. The resulting supernatant was applied to 4 mL buffer C cushion and centrifuged at 199,600 x g for 2 h at 4°C. The pellet was washed with buffer C and resuspended in buffer B by mixing gently. Subsequent puromycin treatment and salt washing of ribosomes was as described above for bovine ribosomes.
2.5 Interaction of wild type and mutant RTA with bovine and rat ribosomes
A Biacore T200 (GE Healthcare) was used to measure RTA-ribosome interactions as previously described (Basu et al., 2016). Briefly, an NTA chip was used to capture His10-tagged RTA and its mutants. R193A R235A was captured on flow cell (Fc) 2 between 937 and 1284 resonance units (RU), WT RTA and G212E were captured either on Fc3 or Fc4 between 879 and 970 RU. His10-tagged EGFP was captured on the reference channel (Fc1) between 1113 and 1183 RU as the control. The running buffer contained 10 mM Hepes pH 7.4, 150 mM NaCl, 5 mM MgCl2, 50 µM EDTA and 0.003% Surfactant P20. Bovine or rat ribosomes at 0.5, 1.0, 2.0, 4.0 and 8.0 nM were passed over the surface at 50 µL/min using the single kinetic injection method. Association was 2 min and dissociation was 5 min. The surface was regenerated with a 1 min injection of 0.35 M EDTA plus a 1 min injection of 0.3% SDS. The RTA was freshly captured at each cycle. The experiments were repeated 3 times. The apparent affinity of interaction was fitted by Biacore T200 Evaluation Software.
2.6 In vitro depurination assay
For RNA depurination, purified MAC-T RNA (1 µg) was diluted in 18 µL of reaction buffer containing 20 mM citrate, pH 5.0. The reaction was started by adding 2 µL RTA and then incubating at 37°C for 30 min. RNA was precipitated with ethanol and the extent of depurination was determined by qRT-PCR as described below using 375 ng total RNA in the reverse transcription reaction. For ribosome depurination, the reaction mixture contained 30 nM bovine or rat ribosomes, 1X reaction buffer [10 mM Tris-HCl, pH 7.4, 60 mM KCl, and 10 mM MgCl2], and different concentrations of RTA ranging from 0 to 205 nM in a total reaction of 100 µL. The reaction was started by adding RTA, and then incubated at 30°C for 3 min. The reaction was stopped by adding 100 µL of 2X RNA extraction buffer [50 mM Tris-HCl, pH 8.8, 240 mM NaCl, 20 mM EDTA, and 2% SDS]. The RNA was extracted once with phenol, and again with phenol/chloroform, and then precipitated twice with ethanol. The extent of depurination was determined by qRT-PCR as described below using 375 ng rRNA in the reverse transcription reaction.
2.7 Mutant RTA plasmid construction
Site-directed mutagenesis was performed using the coding sequence of RTA from Ricinus communis using the QuikChange Lightning Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA). The RTA sequence was previously optimized for production in a bovine system (Jetzt et al., 2012). Mutant RTA in both the pre (containing the 35-residue leader sequence) and mature (lacking the leader sequence) forms were constructed. Mutagenesis was confirmed by sequencing (Genewiz, South Plainfield, NJ). Plasmid DNA constructs for transfection were prepared using an EndoFree Plasmid Maxi kit (Qiagen, Valencia, CA).
2.8 Transfection
The bovine mammary epithelial cell line MAC-T (Huynh et al., 1991) was routinely maintained as described previously (Fleming et al., 2005). For experiments, cells were plated in phenol red-free DMEM containing 4.5 g/L D-glucose, 10% FBS, 20 U/mL penicillin, 20 µg/mL streptomycin, and 50 µg/mL gentamicin. Cells were cultured in a humidified environment at 37°C with 5% CO2. For transfection with RTA constructs or vector alone, cells were plated at 3.5 × 104 cells per cm2 and transfected the following day with endotoxin-free plasmid DNA and Superfect (Qiagen) as previously described (Jetzt et al., 2012). Cells were assayed 19 h after the start of transfection for all analyses except protein synthesis inhibition, which was performed 21 h after the start of transfection.
2.9 Western immunoblotting
Cell lysates were collected as previously described (Jetzt et al., 2012). Protein concentrations were determined using the Bradford method (Bio-Rad, Hercules, CA). Proteins were separated by SDS-PAGE and transferred to 0.2 µm nitrocellulose (Bio-Rad). Antibodies against RTA (polyclonal anti-ricin toxin A chain, NR-863; NIH Biodefense and Emerging Infections Research Resources Repository, NIAID, NIH) and HSP60 (Abcam, Cambridge, MA) were used to detect the corresponding proteins. Membranes were incubated with horseradish peroxidase-conjugated donkey anti-rabbit IgG secondary antibody (GE Healthcare, Piscataway, NJ) and peroxidase activity was detected using ECL Prime (GE Healthcare).
2.10 qRT-PCR depurination assay
Cell lysates were collected in lysis buffer provided with the NucleoSpin RNA kit (Macherey-Nagel, Bethlehem PA) and total RNA was purified according to manufacturer’s instructions. For bovine samples, rRNA depurination was analyzed by qRT-PCR as described previously (Jetzt et al., 2012). A similar procedure was used for rat samples with the following primers: 28S-F, 5’- GATGTCGGCTCTTCCTATCATTGT −3’; 28S-R, 5’-CCAGCTCACG TTCCCTATTAGTG −3’; dep-F, 5’-GCCATGGTAATCCTGCTCAGTA −3’; dep-R, 5’-TCTGAACCTGCGGTTCCACA −3’. Each primer set was validated by constructing standard curves using serial dilutions (1:50 to 1:500,000) of an RT reaction derived from RNA purified from rat liver ribosomes (40 nM) treated with WT RTA (0.2 nM). The calculated amplification efficiencies were 95.6 ± 0.7% for the 28S primer pair and 95.7 ± 1.1% for the depurination primer pair (mean ± SD for two independent curves).
2.11 Caspase 3/7 assay and protein synthesis assay
Caspase 3/7 activation was measured using the SensoLyte Homogeneous AMC Caspase 3/7 Assay Kit (AnaSpec, San Jose, CA) as described by the manufacturer. To measure protein synthesis, cells were plated in black 96-well plates as described above and transfected the following day with equal amounts of RTA mutant and pEGFP-N1 plasmid. Twenty-one hours after the start of transfection the EGFP fluorescence signal was measured in a plate reader (BioTek) with excitation filter 485/20 and emission filter 530/25. Fluorescence measured in cells co-transfected with GFP and empty vector was considered 100% (Jetzt et al., 2012). To ensure that the inhibition of protein synthesis was not affected by non-specific effects of cell death the assay was also performed in the presence of a pan caspase inhibitor, Z-VAD-FMK (BD-Biosciences, San Jose CA). The inhibitor (10 µM final concentration) or DMSO vehicle control was added to cells 3 hours after the start of transfection when the transfection complex was removed and replaced with fresh media.
2.12 Mitotracker Red assay
Spent media were collected to retain floating cells, then cells were washed twice with EDTA-PBS, trypsinized and combined with the media. Samples were centrifuged at 600 rpm for 4 min. The pellets were washed one time with PBS prior to resuspending in 0.3 ml of 100 nM Mitotracker Red (Thermo Fisher) diluted in PRF-DMEM-H containing 10% FBS, and incubated at 37°C for 30 min. Cells were then centrifuged at 500 rpm for 4 min, resuspended in 0.3 ml PRF-DMEM-H containing 10% FBS, placed on ice, and analyzed using an Accuri C6 flow cytometer.
2.13 Statistical analysis
Data were analyzed by two-way ANOVA with appropriate posttest comparisons. Differences were considered significant with P<0.05. Statistical analyses were performed using GraphPad Prism (7.00).
3. Results
3.1 R193A/R235A does not interact with the mammalian ribosome
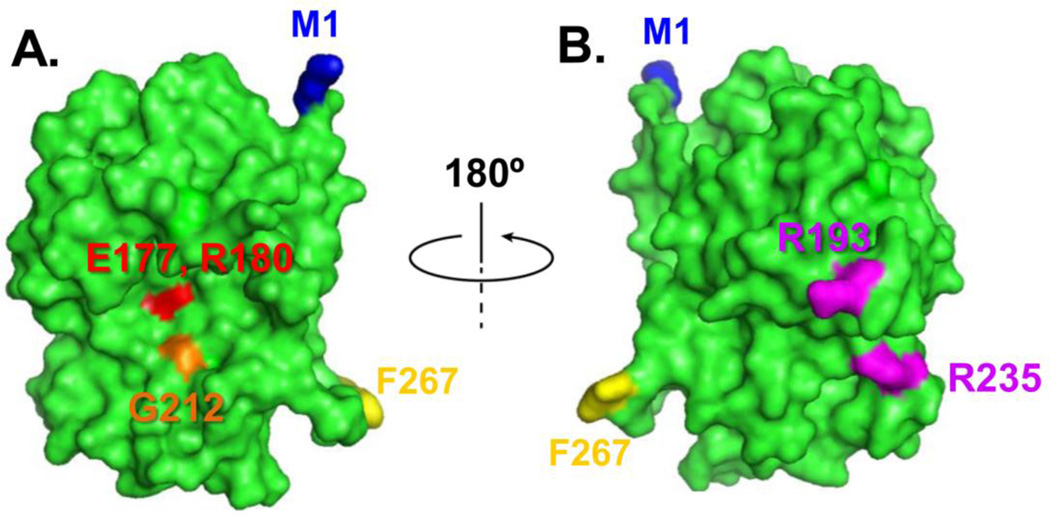
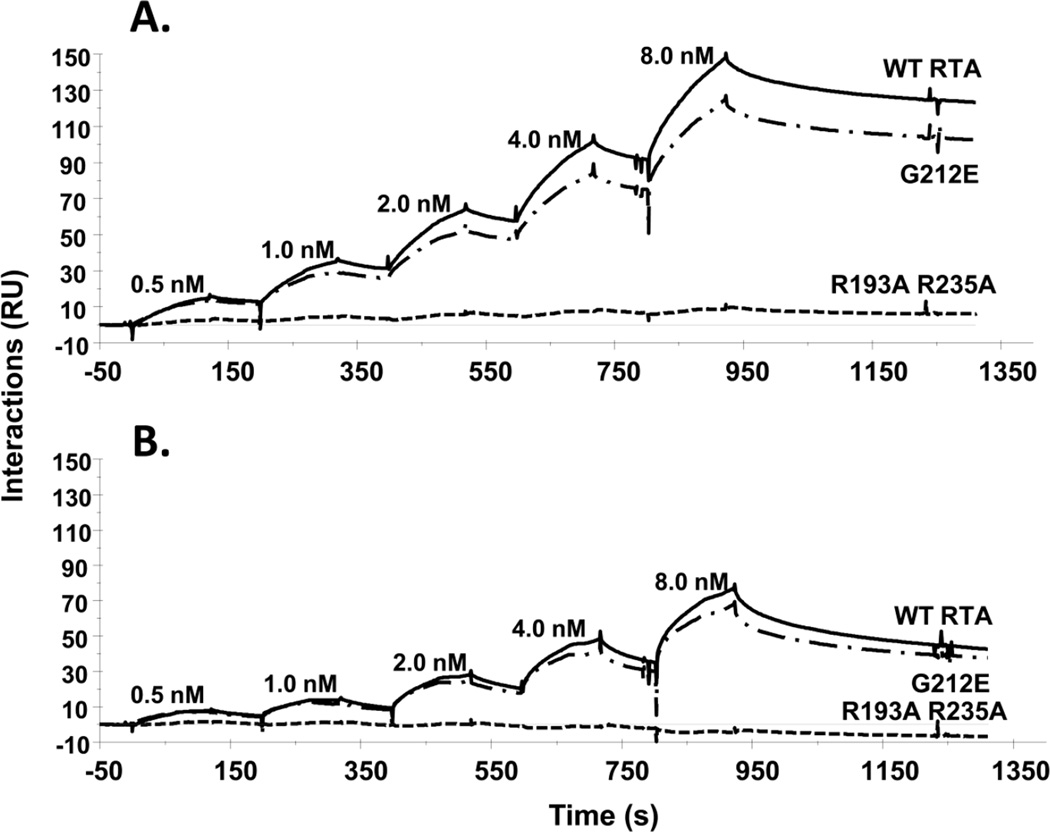
The present work examines the roles of arginine (R) residues 193 and 235 and glycine (G) 212 of RTA on ribosome binding and cytotoxicity in mammalian cells. As shown in Figure 1, G212 resides near the active site of RTA while R193 and R235 reside on the opposite side of the molecule. In order to determine if mutation of arginine residues affects ribosome binding in mammalian cells, Biacore analysis was performed using the single cycle kinetic injection method (Basu et al., 2016). Results showed that WT RTA and the active site mutant G212E bound ribosomes isolated from the MAC-T cell line similarly (Figure 2A). To ensure this was not a species- or cell line-specific effect, the analysis was also conducted with ribosomes isolated from rat liver. Ribosomes isolated from this source also showed similar binding between WT RTA and G212E (Figure 2B), indicating that the glycine residue at position 212 is not critical for the interaction of RTA with mammalian ribosomes. The interaction of immobilized WT RTA or mutant G212E with ribosomes did not follow a single step binding model. Both the association and dissociation phases of the interaction had an initial faster component that was followed by a slower component. Because the interactions did not reach steady state and did not follow a one-to-one interaction, the KD values are referred to as apparent affinities. As shown in Table 1, the apparent KD values were not different between WT RTA and G212E using ribosomes from either source. In contrast, no ribosome binding was detected for R193A/R235A (Fig. 2A and B), indicating that these residues are critical for interaction of RTA with the mammalian ribosome.
Figure 1.
Crystallographic structure of RTA showing the active site and the ribosome binding site. The RTA structure (green) is modeled from the Protein Data Bank ID: 1RTC using PyMol. Panel A, RTA is oriented to show Glu177 and Arg180 (red) at the active site. Gly212 is shown in orange. The first amino acid Met1 is shown in blue and the last amino acid Phe267 is shown in yellow. Panel B, RTA structure from panel A is rotated 180 degrees to show Arg193 and Arg235 at the ribosome binding side.
Figure 2.
Interaction of wild-type, G212E, and R193A/R235A RTA with mammalian ribosomes by Biacore analysis. RTA and RTA mutants were immobilized on an NTA chip. MAC-T ribosomes (A) and rat liver ribosomes (B) (0.5–8 nM) were passed over the surface at 50 µl/min using the single kinetic injection method. Association was 2 min and dissociation was 5 min. Data from representative experiments are shown in A and B. The experiments were repeated 3 times. In (A) WT, G212E, and R193A/R235A RTA were immobilized on the chip at 947 resonance units (RU), 934 RU and 1134 RU, respectively. In (B) WT, G212E, and R193A/R235A RTA were immobilized on the chip at 970 RU, 954 RU and 1031 RU, respectively.
Table 1.
The apparent dissociation constants (KD) of wild type and G212E RTA.
| KD (M)a | ||
|---|---|---|
| RTA | MAC-T Ribosomes | Rat Liver Ribosomes |
| wild type | 5.98 ± 0.63 × 10−9 | 8.81 ± 1.43 × 10−9 |
| G212E | 5.58 ± 0.68 × 10−9 | 7.34 ± 2.14 × 10−9 |
Values were determined using Biacore analysis with wild type or mutant RTA as ligand and MAC-T or rat liver ribosomes as analyte. Data are presented as mean ± standard error of three experiments.
3.2 Double arginine mutant retains full catalytic activity on RNA but has reduced activity on mammalian ribosomes
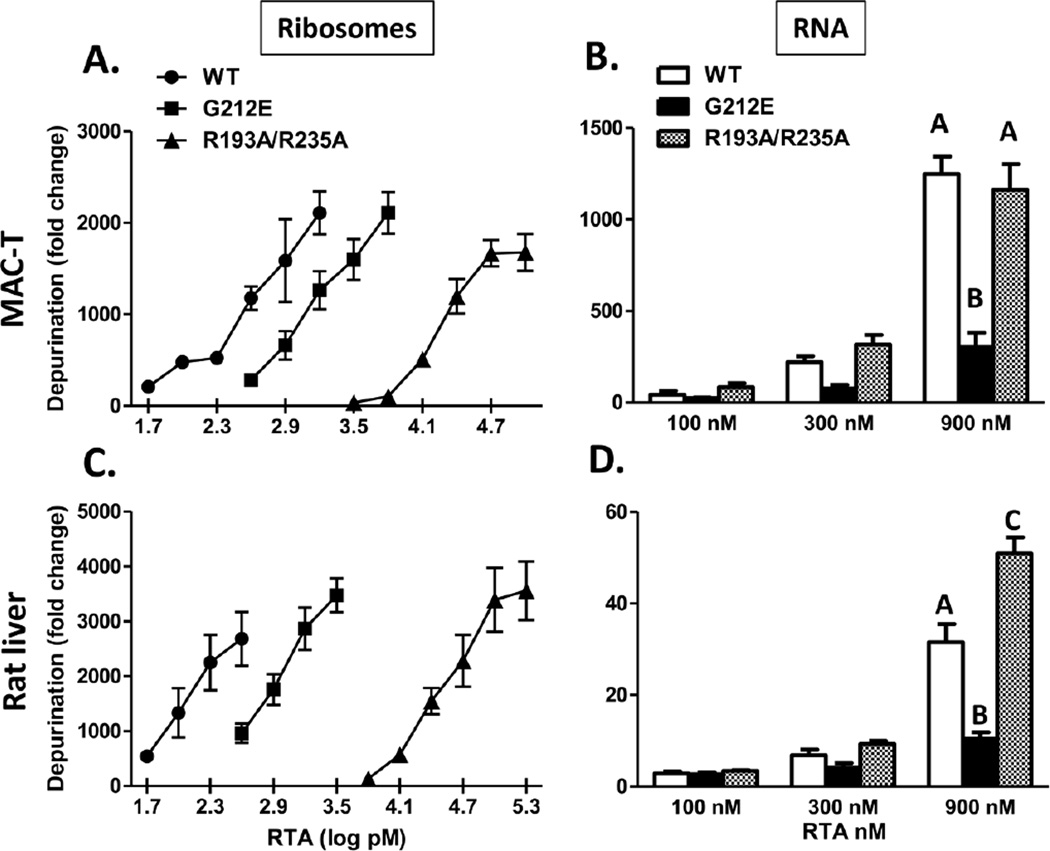
To determine if the alteration in ribosome binding observed for the double R mutant could adversely affect its ability to depurinate the ribosome, in vitro depurination assays were conducted. As shown in Figure 3A, the depurination activity of R193A/R235A was approximately 60-fold less than that of WT RTA when MAC-T ribosomes were used as a substrate, indicating that mutation of the two arginine residues caused a dramatic reduction in depurination activity. In contrast, the activity of G212E was only reduced 4-fold relative to WT RTA. Since a reduction in depurination activity could result from either a decrease in ribosome binding or a decrease in catalytic activity, assays were also conducted with naked RNA as a substrate, where ribosome binding would not be a factor. As shown in Fig. 3B, the depurination activity of R193A/R235A was similar to that of WT RTA when RNA isolated from MAC-T cells served as the substrate, indicating that this mutant retained full enzymatic activity under these conditions. In contrast, the ability of G212E to depurinate MAC-T RNA was significantly reduced relative to WT, indicating that the Gly212 is important for full catalytic activity. Since catalytic activity of R193A/R235A was not affected when RNA served as the substrate (Figure 3B), the reduction in depurination observed with ribosomes was likely due to the inability of R193A/R235A to bind the ribosome. In vitro depurination assays conducted using ribosomes isolated from rat liver produced similar results to those obtained with ribosomes isolated from MAC-T cells i.e. the depurination activity of R193A/R235A was 300-fold less than that of WT RTA while the activity of G212E was reduced by only 7-fold (Fig. 3C). Interestingly, R193A/R235A was significantly more active than WT RTA when rat liver RNA served as the substrate while the activity of G212E was again reduced (Figure 3D). Collectively these data indicate that the reduced depurination activity of R193A/R235A on ribosomes is caused by its decreased ability to bind the ribosome and not by a reduction in catalytic activity.
Figure 3.
In vitro depurination of RNA and intact ribosomes by RTA and RTA mutants. Ribosomes or RNA were isolated from MAC-T cells (A and B, respectively) or rat liver (C and D, respectively) and incubated with RTA and RTA mutants. The extent of depurination was quantified by qRT-PCR. For A and C, bars represent mean ± standard error of 3–5 experiments. For B and D, error bars represent standard error of 3 experiments. Data were analyzed by two-way ANOVA with Bonferroni posttests. Different letters indicate significant difference within each concentration, P<0.001.
3.3 Double arginine mutant has reduced depurination activity and toxicity in mammalian cells
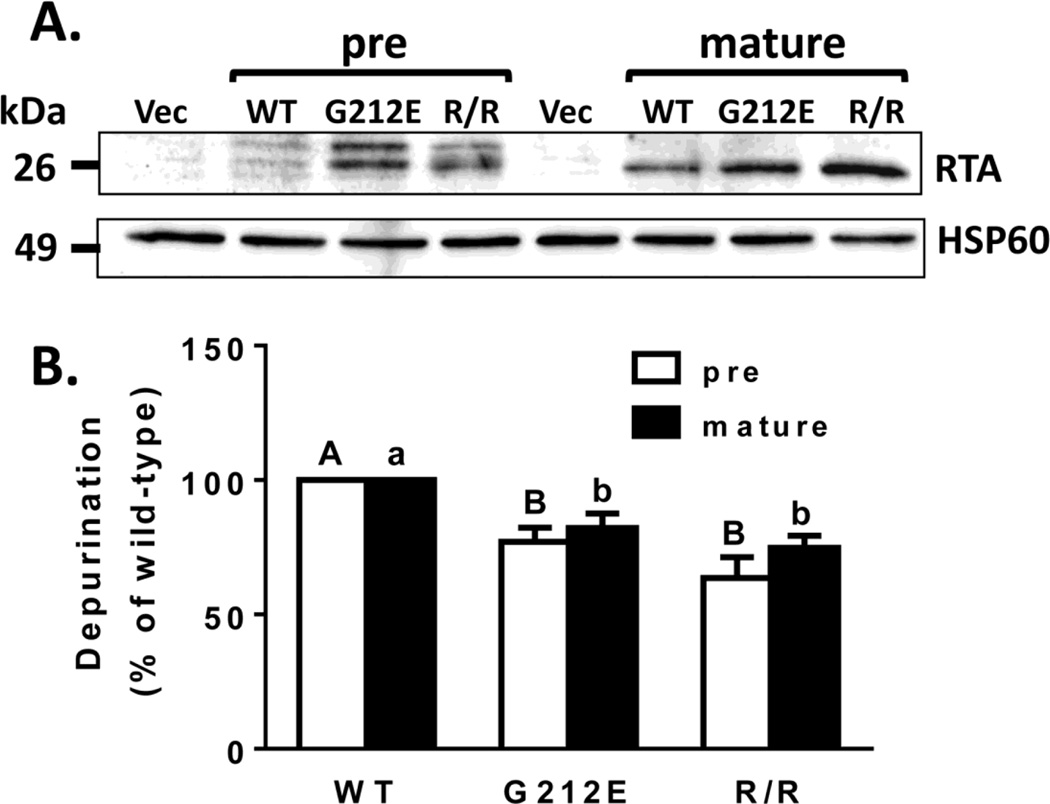
To study the depurination activity and toxicity of the double arginine mutant in vivo, we utilized a previously described mammalian expression system (Jetzt et al., 2012). In this system the Ricinus communis gene encoding RTA is modified to utilize preferred codons for the bovine in order to increase expression in the MAC-T cell line. Site-directed mutagenesis was used to produce the double arginine mutant (R193A/R235A). This mutant was expressed in both the pre-RTA form (containing the 35-residue N-terminal leader peptide) and the mature RTA form (lacking the leader peptide). We have previously shown that transfection of pre-RTA results in glycosylated RTA as expected since the leader peptide directs the precursor form to the ER (Jetzt et al., 2012). MAC-T cells were transiently transfected with WT and mutant forms of RTA. As shown in Figure 4A, expression of both pre- and mature WT RTA as well as both forms of the two mutants was detectable by western blot. WT RTA was expressed at a lower level possibly due to its higher toxicity as previously observed in yeast (Li et al. 2013).
Figure 4.
Expression of RTA and RTA mutants in transfected MAC-T cells and their effects on ribosome depurination. Total cell lysates or RNA were collected 19 h after transfection. (A) Total cell lysates (50 µg) were separated by SDS-PAGE and immunoblotted with RTA antibody. HSP60 served as a loading control. Vec, vector; WT, wild type; R/R, R193A/R235A. (B) Ribosome depurination was determined by qRT-PCR. Bars represent mean ± standard error of at least 4 experiments. Data were analyzed by a two-way ANOVA with Tukey’s multiple comparisons tests. Different letters denote significance (P<0.05) between pre RTAs (capital letters) or mature RTAs (small letters). There was no significant difference between the pre- and mature forms of WT RTA or either mutant.
To determine the depurination activity of G212E and R193A/R235A in vivo, MAC-T cells were transiently transfected with pre- and mature WT RTA or mutants. Total RNA was collected 19 h after transfection and quantitative RT-PCR (qRT-PCR) was used to measure the extent of depurination relative to the vector control. The pre-and mature forms of G212E reduced depurination 23 and 18%, respectively, while the pre and mature forms of R193A/R235A reduced depurination 36% and 25%, respectively, relative to WT RTA (Figure 4B).
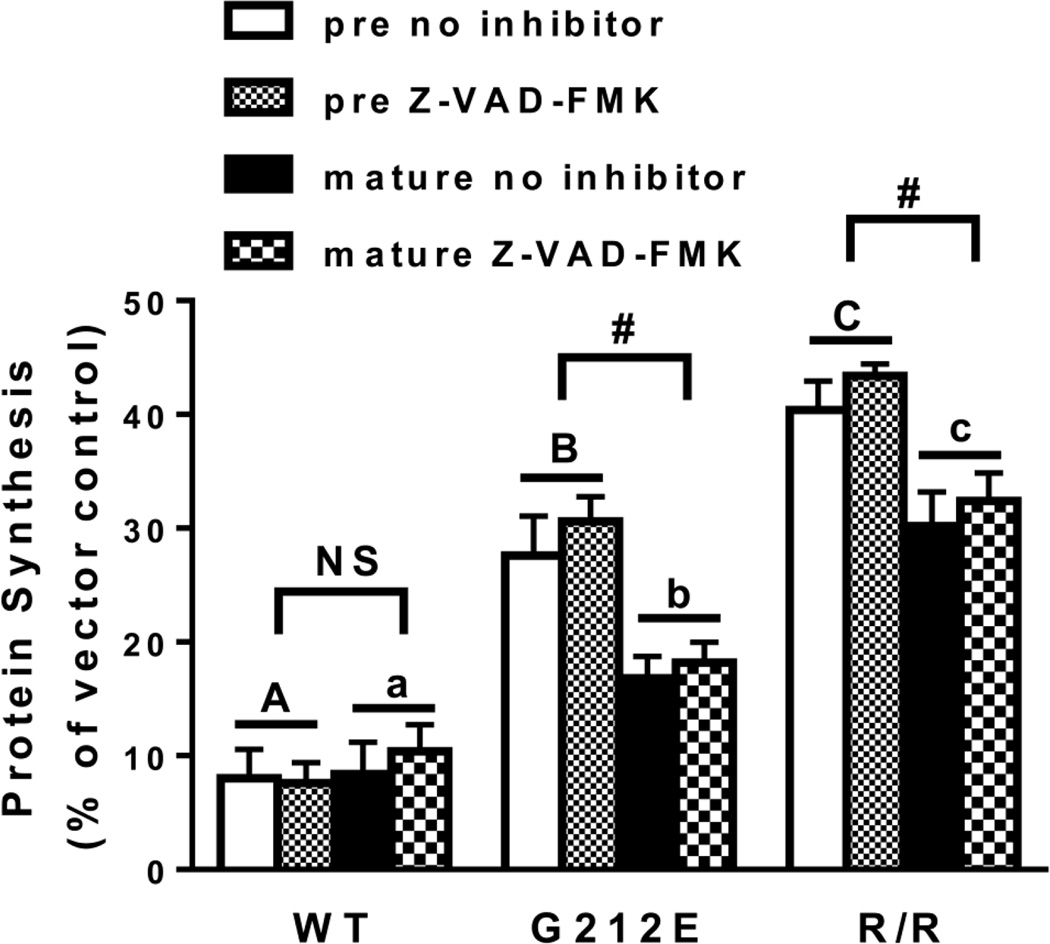
To measure the extent of protein synthesis inhibition by the two mutants, an EGFP (enhanced green fluorescent protein) transfection assay was used (Redmann et al., 2011; Jetzt et al., 2012). In this assay the GFP protein must be translated for fluorescence to be detected. If protein synthesis is inhibited, fluorescence will decrease. This assay as well as a similar assay that detects luminescence (Zhao and Haslam, 2005) were developed for transection assays because the traditional protein synthesis assay that depends on radiolabeled amino acid incorporation is not sensitive enough to detect toxin effects on smaller numbers of cells. To ensure the decrease in fluorescence was not influenced by apoptotic cell death, the assay was also performed in the presence of the caspase inhibitor Z-VAD-FMK. As shown in Figure 5, protein synthesis was inhibited to the same extent in the presence or absence of the inhibitor for pre- and mature forms of WT or mutant RTA, therefore data were combined. Protein synthesis was almost completely inhibited by WT pre- and mature RTA (92 and 91% inhibition, respectively). Both mutants inhibited protein synthesis significantly less than their WT counterparts i.e. pre- and mature G212E inhibited protein synthesis 71 and 83%, respectively while R193A/R235A inhibited protein synthesis 58 and 69%, respectively. In addition, pre-R193A/R235A inhibited protein synthesis less than pre-G212E while mature R193A/R235A inhibited protein synthesis less than mature G212E (P<0.05), indicating that the ribosome binding mutant was less toxic than the G212E mutant. Additionally, the pre forms of both mutants were less toxic than their mature counterparts (P<0.05).
Figure 5.
Protein synthesis in MAC-T cells transfected with RTA and RTA mutants. Cells were co-transfected with pEGFP-N1 and wild-type RTA, RTA mutants or vector alone. Three hours after transfection media was replenished ± 10 µM Z-VAD-FMK. Fluorescence was measured 21 h after transfection. Signal in cells co-transfected with pEGFP-N1 and vector was considered 100%. Bars represent mean ± standard error of 5 experiments. Analysis of the data by three-way ANOVA indicated no effect of the caspase inhibitor. Therefore data were consolidated and analyzed by two-way ANOVA. Different letters denote significance (P<0.05; Tukey’s multiple comparisons test) between pre RTAs (capital letters) or mature RTAs (small letters). In addition, pre- and mature RTA differed (P<0.05; Sidak’s multiple comparisons test) for each mutant.
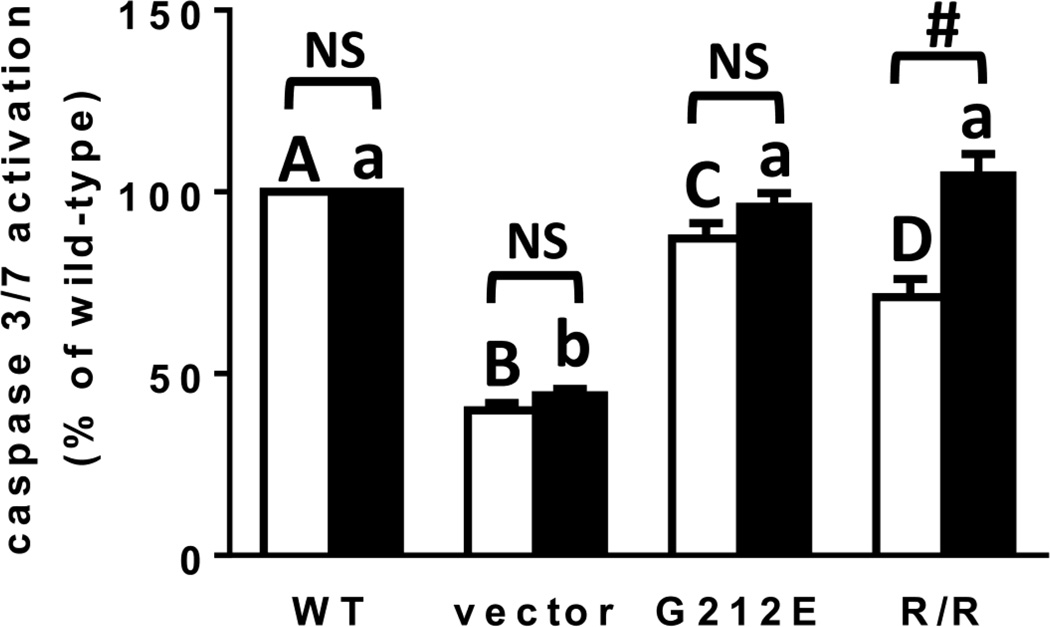
To determine if apoptosis was reduced by either mutant relative to WT RTA, activation of caspase 3/7 was measured. As shown in Figure 6, caspase activation was reduced by pre-R193A/R235A and pre-G212E relative to pre-WT RTA (29 and 13%, respectively), with pre-R193A/R235A reducing caspase activation significantly more than G212E. Interestingly, the mature forms of both mutants activated caspases to the same degree as mature WT RTA. For R193A/R235A, the ability of the pre- and mature forms of the protein to activate caspases was significantly different.
Figure 6.
Caspase 3/7 activation in MAC-T cells transfected with RTA and RTA mutants. Caspase 3/7 activity was measured 19 h after transfection using the SensoLyte Homogeneous AMC Caspase 3/7 assay kit (Anaspec). Data were analyzed by a two-way ANOVA with Bonferroni posttests. Different letters denote significance (P<0.05) between pre RTAs (capital letters) or mature RTAs (small letters). # indicates significance (P<0.001) between pre- and mature RTA.
To determine if similar effects were observed with a different indicator of apoptosis, cells were transfected and incubated with the lipophilic cationic fluorescent dye Mitotracker Red, which accumulates in mitochondria of actively respiring cells (Galluzzi et al., 2007). The percentage of dead cells was analyzed by flow cytometry. Similar to the results obtained with caspase 3/7 activation, pre R193A/R235A was the least toxic relative to pre WT RTA (Figure 7A). Using this measure of cell death, the pre G212E exhibited intermediate toxicity, while the mature form of R193A/R235A was also less toxic than mature RTA (Figure 7B).
Figure 7.
Analysis of mitochondrial membrane potential in transfected MAC-T cells. Nineteen hours after transfection, cells were incubated with Mitotracker Red and analyzed by flow cytometry. Panel (A) shows a histogram of a representative experiment. Panel (B) shows a bar graph depicting the mean ± standard error of 5 experiments. Data were analyzed by a two-way ANOVA with Tukey’s multiple comparisons test. Different letters denote significance (P<0.05) between pre RTAs (capital letters) or mature RTAs (small letters). There was no significant difference between the pre and mature forms of WT or either mutant.
4. Discussion
4.1 Ribosome binding is critical for depurination of mammalian ribosomes in vitro
A considerable body of work with different RIPs indicates that binding to the P proteins of the ribosomal stalk plays a role in their cytotoxicity by affecting their ability to access the SRL. The eukaryotic P proteins P0, P1 and P2 are characterized by a conserved sequence in their C-terminal domains (CTD) which interacts with translation factors during protein synthesis (Remacha et al., 1995; Nomura et al., 2012). A peptide corresponding to the last 11 amino acids of P-proteins (P11) has been shown to interact with TCS (Chan et al., 2007), RTA and Stx1A (McCluskey et al., 2008; McCluskey et al., 2012). The crystal structure of P11 complexed to TCS shows that the N-terminus of the peptide interacts with positively charged residues Lys173, Arg174 and Lys177 of TCS while the hydrophobic residues of the CTD insert into a hydrophobic pocket (Too et al., 2009). However, the amino acids in RTA that are responsible for binding the mammalian ribosome have not been identified. Early studies suggested that arginine residues located on the opposite side of the active site cleft may be involved in ribosome binding as modification of arginines at positions 193, 196, 213 and 234/235 decreased depurination rate and the affinity for the ribosome without causing a detectable change in the conformation of the catalytic site (Watanabe and Funatsu, 1986). A recent study showed that Arg193 and Arg235 are both critical for RTA to bind to the ribosome and to the ribosomal stalk in yeast, but not for the catalytic activity of RTA (Li et al., 2013). Based on this work we predicted that the double R mutant, R193A/R235A, would have reduced affinity for mammalian ribosomes. In the present work the mutant G212E was used for comparison because Gly212 resides near the active site and has reduced catalytic activity on an RNA mimic of the SRL and on naked yeast RNA, but binds to yeast ribosomes similarly to WT RTA (Li et al., 2013). These two mutants have been shown to have structures similar to WT RTA (Li et al., 2013). Biacore analysis using ribosomes isolated from a bovine cell line or from rat liver confirmed that G212E bound ribosomes similarly to WT RTA and the apparent KD values between the two proteins did not differ. In contrast, R193A/R235A failed to exhibit any interaction with ribosomes. This corresponded with a drastic reduction in its ability to depurinate ribosomes in vitro. Since R193A/R235A exhibited full enzymatic activity with naked RNA we conclude that the reduced depurination activity on ribosomes is likely due to its reduced ribosome binding, similar to results observed in yeast (Li et al., 2013). By comparison, since G212E had greatly reduced catalytic activity when naked RNA was the substrate, we expected this mutant to also exhibit dramatically reduced depurination activity on ribosomes, as was observed with yeast (Li et al., 2013). However, when ribosomes served as substrate, catalytic activity was only reduced 4-fold by G212E compared to 64-fold by R193A/R235A. It is possible that G212E may cause a higher level of depurination on ribosomes than R193A/R235A in vitro because it is able to bind the ribosomal stalk, which may stimulate catalytic activity by orienting its active site towards the SRL. The finding that R193A/R235A elicited a greater reduction in depurination of mammalian ribosomes in the present study compared to its effect on yeast ribosomes (Li et al., 2013) suggests that ribosome binding may be more critical for depurination of mammalian ribosomes by RTA than for depurination of yeast ribosomes.
4.2 Ribosome binding is critical for ribosome depurination, translation inhibition and toxicity of RTA in mammalian cells in vivo
While in vitro experiments allow for testing specific interactions between RTA mutants and their substrates, these experiments are performed in the absence of an intact cellular environment. To study the effect of the RTA mutants in an intact cellular environment, we expressed R193A/R235A in MAC-T cells as either a pre form containing the signal peptide that directs transport through the ER to reach the ribosome in the cytosol or the mature form which lacks this signal and thus remains in the cytosol. Quantitation of ribosome depurination by qRT-PCR showed that both G212E and R193A/R235A reduced depurination similarly (23 to 36% for pre and 18 to 25% for mature, relative to WT forms). This is interesting in view of our in vitro data showing that depurination by R193A/R235A was dramatically reduced relative to G212E. A possible explanation is that in vivo, R193A/R235A may have better access to the ribosome because chaperone proteins help fold RTA (Spooner and Lord, 2015), while in vitro the interaction of RTA with the ribosome would be solely dependent on electrostatic interactions. In yeast, the expression of RTA is controlled by the galactose inducible GAL1 promoter and it is therefore possible to see early differences in depurination (at zero or 1 h after induction). However, in the present work RTA is transiently expressed and the depurination level is measured at 19 h following transfection, precluding determination of greater differences in depurination at earlier time points which could be occurring.
We also tested whether G212E and R193A/R235A inhibit protein synthesis and induce apoptosis to different degrees in vivo. Results of protein synthesis inhibition supported a greater role for ribosome binding relative to catalytic activity as R193A/R235A reduced this parameter significantly more than G212E. By two different measures of apoptosis, caspase 3/7 activation and using a mitochondrial membrane potential sensitive dye, we showed that R193A/R235A was less toxic than WT RTA, with G212E exhibiting intermediate toxicity. Interestingly, both forms of R193A/R235A were less toxic relative to WT RTA using the Mitotracker Red assay while only the pre form was less toxic when the caspase 3/7 assay was used. The pre form may be more biologically relevant as it has to travel through the ER to get to the ribosomes in the cytosol, similar to the situation that occurs in ricin toxicity. Therefore disruption of ribosome interaction reduces RTA-induced apoptosis in mammalian cells.
4.3 Relevance of R193A/R235A to development of new therapies for ricin intoxication
Due to its wide availability and ease of production, ricin has been exploited as an agent of bioterrorism and biological warfare (Christopher et al., 1997; Audi et al., 2005). This has led to intensive investigation into the development of preventative and therapeutic agents to treat ricin toxicity. While two vaccines have been developed, the wide-spread adoption of a vaccination program outside of the U.S. military is unlikely, therefore it is important to continue the search for pre- or post-exposure therapeutics (Reisler and Smith, 2012). A large effort has been aimed at designing small molecular inhibitors that block ricin toxicity (Wahome et al., 2012). The large size and polarity of the RTA active site make it a difficult target for drug development and the effectiveness of inhibitors that bind in or near this site has also been limited by issues of solubility and cell toxicity (Bai et al., 2010). Structure-based approaches have been used to identify inhibitors that block substrate binding to the active site (Bai et al., 2009; Pruet et al., 2011) or prevent an active-site residue from adopting an active confirmation (Pang et al., 2011). Other inhibitors have been identified using computer modeling approaches coupled with cell-based high throughput screens (Bai et al., 2010; Wahome et al., 2010). However, to date, none of the small molecules that have been identified exhibit potent (nanomolar) protection. A second approach has been to develop ricin-specific neutralizing monoclonal antibodies. While a number of these have been shown to partially rescue mice from ricin intoxication (Hu et al., 2012; O’Hara et al., 2012; Sully et al., 2014), none have been tested in Phase I clinical trials to date. Several neutralizing antibodies have been shown to act by targeting ricin uptake and trafficking (Herrera et al., 2016; Yermakova et al., 2016). Interestingly, the ricin-neutralizing antibody 6C2 recognizes an epitope distinct from the catalytic active site of RTA and is more potent in its neutralizing ability relative to antibodies that bind the active site (Dai et al., 2011). These researchers further proposed that this antibody may inhibit binding of RTA to the ribosome (Zhu et al., 2013). We show here for the first time that targeting specific arginine residues of RTA can reduce its toxicity in mammalian cells by inhibiting its interactions with the ribosome while leaving the active site intact. Furthermore, inhibiting ribosome binding may provide protection against ribotoxic stress which activates cellular signaling that leads to apoptosis and inflammation at the local level (Thorpe et al., 1999; Thorpe et al., 2001; Lindauer et al., 2009) which is not provided by therapeutics that solely block the enzymatic active site. Therefore the inhibition of ribosome binding represents a new target that can be used in the design of small molecule inhibitors or neutralizing antibodies to treat ricin toxicity. In addition, since the effectiveness of vaccines against RTA is limited by incomplete protection against local tissue damage (Olson et al., 2004; Smallshaw et al., 2007; McLain et al., 2012; Roy et al., 2015; Brey et al., 2016), a small molecule inhibitor that targets ribosome binding could potentially be used for added protection in vaccinated individuals exposed to ricin.
Highlights.
Arginines 193 and 235 of RTA are critical for binding to the mammalian ribosome.
R193A/R235A has full catalytic activity on RNA but not on mammalian ribosomes.
R193A/R235A is less toxic than a mutant that targets the active site.
The toxin-ribosome interaction is a therapeutic target for ricin intoxication.
Acknowledgments
The authors thank Dr. Michael Pierce and the School of Environmental and Biological Sciences Core Facility for assistance with flow cytometry. This work was supported by National Institutes of Health Grant AI072425 (to N.E.T. and W.S.C.).
Abbreviations
- CTD
C-terminal domain
- EGFP
enhanced green fluorescent protein
- ER
endoplasmic reticulum
- qRT-PCR
quantitative reverse transcription PCR
- RIP
ribosome-inactivating protein
- RTA
ricin toxin A chain
- RTB
ricin B-chain
- RU
resonance units
- SRL
sarcin-ricin loop
- Stx1
Shiga toxin 1
- Stx2
Shiga toxin 2
- TCS
trichosanthin
- WT
wild-type
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
All authors certify that they have no conflict of interest relevant to this work.
References
- Audi J, Belson M, Patel M, Schier J, Osterloh J. Ricin poisoning: a comprehensive review. JAMA. 2005;294:2342–2351. doi: 10.1001/jama.294.18.2342. [DOI] [PubMed] [Google Scholar]
- Bai Y, Monzingo AF, Robertus JD. The X-ray structure of ricin A chain with a novel inhibitor. Arch Biochem Biophys. 2009;483:23–28. doi: 10.1016/j.abb.2008.12.013. [DOI] [PubMed] [Google Scholar]
- Bai Y, Watt B, Wahome PG, Mantis NJ, Robertus JD. Identification of new classes of ricin toxin inhibitors by virtual screening. Toxicon. 2010;56:526–534. doi: 10.1016/j.toxicon.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballesta JP, Remacha M. The large ribosomal subunit stalk as a regulatory element of the eukaryotic translational machinery. Prog Nucleic Acid Res Mol Biol. 1996;55:157–193. doi: 10.1016/s0079-6603(08)60193-2. [DOI] [PubMed] [Google Scholar]
- Basu D, Li XP, Kahn JN, May KL, Kahn PC, Tumer NE. The A1 Subunit of Shiga Toxin 2 Has Higher Affinity for Ribosomes and Higher Catalytic Activity than the A1 Subunit of Shiga Toxin 1. Infect Immun. 2016;84:149–161. doi: 10.1128/IAI.00994-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brey RN, 3rd, Mantis NJ, Pincus SH, Vitetta ES, Smith LA, Roy CJ. Recent Advances in the Development of Vaccines against Ricin. Hum Vaccin Immunother. 2016;12:1196–1201. doi: 10.1080/21645515.2015.1124202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan DS, Chu LO, Lee KM, Too PH, Ma KW, Sze KH, Zhu G, Shaw PC, Wong KB. Interaction between trichosanthin, a ribosome-inactivating protein, and the ribosomal stalk protein P2 by chemical shift perturbation and mutagenesis analyses. Nucleic Acids Res. 2007;35:1660–1672. doi: 10.1093/nar/gkm065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SH, Hung FS, Chan DS, Shaw PC. Trichosanthin interacts with acidic ribosomal proteins P0 and P1 and mitotic checkpoint protein MAD2B. Eur J Biochem. 2001;268:2107–2112. doi: 10.1046/j.1432-1327.2001.02091.x. [DOI] [PubMed] [Google Scholar]
- Chiou JC, Li XP, Remacha M, Ballesta JP, Tumer NE. The ribosomal stalk is required for ribosome binding, depurination of the rRNA and cytotoxicity of ricin A chain in Saccharomyces cerevisiae. Mol Microbiol. 2008;70:1441–1452. doi: 10.1111/j.1365-2958.2008.06492.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou JC, Li XP, Remacha M, Ballesta JP, Tumer NE. Shiga toxin 1 is more dependent on the P proteins of the ribosomal stalk for depurination activity than Shiga toxin 2. Int J Biochem Cell Biol. 2011;43:1792–1801. doi: 10.1016/j.biocel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christopher GW, Cieslak TJ, Pavlin JA, Eitzen EM., Jr Biological warfare. A historical perspective. Jama. 1997;278:412–417. [PubMed] [Google Scholar]
- Clementi N, Polacek N. Ribosome-associated GTPases: the role of RNA for GTPase activation. RNA Biol. 2010;7:521–527. doi: 10.4161/rna.7.5.12467. [DOI] [PubMed] [Google Scholar]
- Dai J, Zhao L, Yang H, Guo H, Fan K, Wang H, Qian W, Zhang D, Li B, Wang H, Guo Y. Identification of a novel functional domain of ricin responsible for its potent toxicity. J Biol Chem. 2011;286:12166–12171. doi: 10.1074/jbc.M110.196584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo Y, Tsurugi K. The RNA N-glycosidase activity of ricin A-chain. The characteristics of the enzymatic activity of ricin A-chain with ribosomes and with rRNA. J Biol Chem. 1988;263:8735–8739. [PubMed] [Google Scholar]
- Fleming J, Leibowitz BJ, Kerr DE, Cohick WS. IGF-I differentially regulates IGF binding protein expression in primary mammary fibroblasts and epithelial cells. J Endocrinol. 2005;186:165–178. doi: 10.1677/joe.1.06164. [DOI] [PubMed] [Google Scholar]
- Galluzzi L, Zamzami N, de La Motte Rouge T, Lemaire C, Brenner C, Kroemer G. Methods for the assessment of mitochondrial membrane permeabilization in apoptosis. Apoptosis. 2007;12:803–813. doi: 10.1007/s10495-007-0720-1. [DOI] [PubMed] [Google Scholar]
- Grela P, Krokowski D, Gordiyenko Y, Krowarsch D, Robinson CV, Otlewski J, Grankowski N, Tchorzewski M. Biophysical properties of the eukaryotic ribosomal stalk. Biochemistry. 2010;49:924–933. doi: 10.1021/bi901811s. [DOI] [PubMed] [Google Scholar]
- Guarinos E, Santos C, Sanchez A, Qiu DY, Remacha M, Ballesta JP. Tag-mediated fractionation of yeast ribosome populations proves the monomeric organization of the eukaryotic ribosomal stalk structure. Mol Microbiol. 2003;50:703–712. doi: 10.1046/j.1365-2958.2003.03733.x. [DOI] [PubMed] [Google Scholar]
- Herrera C, Klokk TI, Cole R, Sandvig K, Mantis NJ. A Bispecific Antibody Promotes Aggregation of Ricin Toxin on Cell Surfaces and Alters Dynamics of Toxin Internalization and Trafficking. Plos One. 2016;11:e0156893. doi: 10.1371/journal.pone.0156893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu WG, Yin J, Chau D, Negrych LM, Cherwonogrodzky JW. Humanization and characterization of an anti-ricin neutralization monoclonal antibody. PLoS One. 2012;7:e45595. doi: 10.1371/journal.pone.0045595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh HT, Robitaille G, Turner JD. Establishment of bovine mammary epithelial cells (MAC-T): an in vitro model for bovine lactation. Exp. Cell Res. 1991;197:191–199. doi: 10.1016/0014-4827(91)90422-q. [DOI] [PubMed] [Google Scholar]
- Jetzt AE, Cheng JS, Li XP, Tumer NE, Cohick WS. A relatively low level of ribosome depurination by mutant forms of ricin toxin A chain can trigger protein synthesis inhibition, cell signaling and apoptosis in mammalian cells. Int J Biochem Cell Biol. 2012;44:2204–2211. doi: 10.1016/j.biocel.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitaoka Y. Involvement of the amino acids outside the active-site cleft in the catalysis of ricin A chain. Eur J Biochem. 1998;257:255–262. doi: 10.1046/j.1432-1327.1998.2570255.x. [DOI] [PubMed] [Google Scholar]
- Krokowski D, Boguszewska A, Abramczyk D, Liljas A, Tchorzewski M, Grankowski N. Yeast ribosomal P0 protein has two separate binding sites for P1/P2 proteins. Mol Microbiol. 2006;60:386–400. doi: 10.1111/j.1365-2958.2006.05117.x. [DOI] [PubMed] [Google Scholar]
- Li XP, Kahn PC, Kahn JN, Grela P, Tumer NE. Arginine residues on the opposite side of the active site stimulate the catalysis of ribosome depurination by ricin A chain by interacting with the P-protein stalk. J Biol Chem. 2013;288:30270–30284. doi: 10.1074/jbc.M113.510966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindauer ML, Wong J, Iwakura Y, Magun BE. Pulmonary inflammation triggered by ricin toxin requires macrophages and IL-1 signaling. J Immunol. 2009;183:1419–1426. doi: 10.4049/jimmunol.0901119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May KL, Li XP, Martinez-Azorin F, Ballesta JP, Grela P, Tchorzewski M, Tumer NE. The P1/P2 proteins of the human ribosomal stalk are required for ribosome binding and depurination by ricin in human cells. Febs J. 2012;279:3925–3936. doi: 10.1111/j.1742-4658.2012.08752.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCluskey AJ, Bolewska-Pedyczak E, Jarvik N, Chen G, Sidhu SS, Gariepy J. Charged and hydrophobic surfaces on the a chain of shiga-like toxin 1 recognize the C-terminal domain of ribosomal stalk proteins. PLoS One. 2012;7:e31191. doi: 10.1371/journal.pone.0031191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCluskey AJ, Poon GM, Bolewska-Pedyczak E, Srikumar T, Jeram SM, Raught B, Gariepy J. The catalytic subunit of shiga-like toxin 1 interacts with ribosomal stalk proteins and is inhibited by their conserved C-terminal domain. J Mol Biol. 2008;378:375–386. doi: 10.1016/j.jmb.2008.02.014. [DOI] [PubMed] [Google Scholar]
- McLain DE, Lewis BS, Chapman JL, Wannemacher RW, Lindsey CY, Smith LA. Protective effect of two recombinant ricin subunit vaccines in the New Zealand white rabbit subjected to a lethal aerosolized ricin challenge: survival, immunological response, and histopathological findings. Toxicol Sci. 2012;126:72–83. doi: 10.1093/toxsci/kfr274. [DOI] [PubMed] [Google Scholar]
- Nomura N, Honda T, Baba K, Naganuma T, Tanzawa T, Arisaka F, Noda M, Uchiyama S, Tanaka I, Yao M, Uchiumi T. Archaeal ribosomal stalk protein interacts with translation factors in a nucleotide-independent manner via its conserved C terminus. Proc Natl Acad Sci U S A. 2012;109:3748–3753. doi: 10.1073/pnas.1112934109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Hara JM, Whaley K, Pauly M, Zeitlin L, Mantis NJ. Plant-based expression of a partially humanized neutralizing monoclonal IgG directed against an immunodominant epitope on the ricin toxin A subunit. Vaccine. 2012;30:1239–1243. doi: 10.1016/j.vaccine.2011.12.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson MA, Carra JH, Roxas-Duncan V, Wannemacher RW, Smith LA, Millard CB. Finding a new vaccine in the ricin protein fold. Protein Eng Des Sel. 2004;17:391–397. doi: 10.1093/protein/gzh043. [DOI] [PubMed] [Google Scholar]
- Pang YP, Park JG, Wang S, Vummenthala A, Mishra RK, McLaughlin JE, Di R, Kahn JN, Tumer NE, Janosi L, Davis J, Millard CB. Small-molecule inhibitor leads of ribosome-inactivating proteins developed using the doorstop approach. PLoS One. 2011;6:e17883. doi: 10.1371/journal.pone.0017883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittman PR, Reisler RB, Lindsey CY, Guerena F, Rivard R, Clizbe DP, Chambers M, Norris S, Smith LA. Safety and immunogenicity of ricin vaccine, RVEc, in a Phase 1 clinical trial. Vaccine. 2015;33:7299–7306. doi: 10.1016/j.vaccine.2015.10.094. [DOI] [PubMed] [Google Scholar]
- Pruet JM, Jasheway KR, Manzano LA, Bai Y, Anslyn EV, Robertus JD. 7-Substituted pterins provide a new direction for ricin A chain inhibitors. Eur J Med Chem. 2011;46:3608–3615. doi: 10.1016/j.ejmech.2011.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redmann V, Oresic K, Tortorella LL, Cook JP, Lord M, Tortorella D. Dislocation of ricin toxin A chains in human cells utilizes selective cellular factors. J Biol Chem. 2011;286:21231–21238. doi: 10.1074/jbc.M111.234708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisler RB, Smith LA. The need for continued development of ricin countermeasures. Adv Prev Med. 2012;2012:149737. doi: 10.1155/2012/149737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remacha M, Jimenez-Diaz A, Santos C, Briones E, Zambrano R, Rodriguez Gabriel MA, Guarinos E, Ballesta JP. Proteins P1, P2, and P0, components of the eukaryotic ribosome stalk. New structural and functional aspects. Biochem Cell Biol. 1995;73:959–968. doi: 10.1139/o95-103. [DOI] [PubMed] [Google Scholar]
- Robertus JD, Monzingo AF. The structure of ribosome inactivating proteins. Mini Rev Med Chem. 2004;4:477–486. doi: 10.2174/1389557043403837. [DOI] [PubMed] [Google Scholar]
- Roy CJ, Brey RN, Mantis NJ, Mapes K, Pop IV, Pop LM, Ruback S, Killeen SZ, Doyle-Meyers L, Vinet-Oliphant HS, Didier PJ, Vitetta ES. Thermostable ricin vaccine protects rhesus macaques against aerosolized ricin: Epitope-specific neutralizing antibodies correlate with protection. Proc Natl Acad Sci U S A. 2015;112:3782–3787. doi: 10.1073/pnas.1502585112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Khade PK, Sanbonmatsu KY, Joseph S. Functional role of the sarcin-ricin loop of the 23S rRNA in the elongation cycle of protein synthesis. J Mol Biol. 2012;419:125–138. doi: 10.1016/j.jmb.2012.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smallshaw JE, Richardson JA, Vitetta ES. RiVax, a recombinant ricin subunit vaccine, protects mice against ricin delivered by gavage or aerosol. Vaccine. 2007;25:7459–7469. doi: 10.1016/j.vaccine.2007.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smallshaw JE, Vitetta ES. Ricin vaccine development. Curr Top Microbiol Immunol. 2012;357:259–272. doi: 10.1007/82_2011_156. [DOI] [PubMed] [Google Scholar]
- Spooner RA, Lord JM. How ricin and Shiga toxin reach the cytosol of target cells: retrotranslocation from the endoplasmic reticulum. Curr Top Microbiol Immunol. 2012;357:19–40. doi: 10.1007/82_2011_154. [DOI] [PubMed] [Google Scholar]
- Spooner RA, Lord JM. Ricin trafficking in cells. Toxins (Basel) 2015;7:49–65. doi: 10.3390/toxins7010049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sully EK, Whaley KJ, Bohorova N, Bohorov O, Goodman C, Kim do H, Pauly MH, Velasco J, Hiatt E, Morton J, Swope K, Roy CJ, Zeitlin L, Mantis NJ. Chimeric plantibody passively protects mice against aerosolized ricin challenge. Clin Vaccine Immunol. 2014;21:777–782. doi: 10.1128/CVI.00003-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchorzewski M. The acidic ribosomal P proteins. Int J Biochem Cell Biol. 2002;34:911–915. doi: 10.1016/s1357-2725(02)00012-2. [DOI] [PubMed] [Google Scholar]
- Thorpe CM, Hurley BP, Lincicome LL, Jacewicz MS, Keusch GT, Acheson DW. Shiga toxins stimulate secretion of interleukin-8 from intestinal epithelial cells. Infect Immun. 1999;67:5985–5993. doi: 10.1128/iai.67.11.5985-5993.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorpe CM, Smith WE, Hurley BP, Acheson DW. Shiga toxins induce, superinduce, and stabilize a variety of C-X-C chemokine mRNAs in intestinal epithelial cells, resulting in increased chemokine expression. Infect Immun. 2001;69:6140–6147. doi: 10.1128/IAI.69.10.6140-6147.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Too PH, Ma MK, Mak AN, Wong YT, Tung CK, Zhu G, Au SW, Wong KB, Shaw PC. The C-terminal fragment of the ribosomal P protein complexed to trichosanthin reveals the interaction between the ribosome-inactivating protein and the ribosome. Nucleic Acids Res. 2009;37:602–610. doi: 10.1093/nar/gkn922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiumi T, Kominami R. Direct evidence for interaction of the conserved GTPase domain within 28 S RNA with mammalian ribosomal acidic phosphoproteins and L12. J Biol Chem. 1992;267:19179–19185. [PubMed] [Google Scholar]
- Vitetta ES, Smallshaw JE, Schindler J. Pilot phase IB clinical trial of an alhydrogel-adsorbed recombinant ricin vaccine. Clin Vaccine Immunol. 2012;19:1697–1699. doi: 10.1128/CVI.00381-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorhees RM, Schmeing TM, Kelley AC, Ramakrishnan V. The mechanism for activation of GTP hydrolysis on the ribosome. Science. 2010;330:835–838. doi: 10.1126/science.1194460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl MC, Moller W. Structure and function of the acidic ribosomal stalk proteins. Curr Protein Pept Sci. 2002;3:93–106. doi: 10.2174/1389203023380756. [DOI] [PubMed] [Google Scholar]
- Wahome PG, Bai Y, Neal LM, Robertus JD, Mantis NJ. Identification of small-molecule inhibitors of ricin and shiga toxin using a cell-based high-throughput screen. Toxicon. 2010;56:313–323. doi: 10.1016/j.toxicon.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahome PG, Robertus JD, Mantis NJ. Small-molecule inhibitors of ricin and Shiga toxins. Curr Top Microbiol Immunol. 2012;357:179–207. doi: 10.1007/82_2011_177. [DOI] [PubMed] [Google Scholar]
- Watanabe K, Dansako H, Asada N, Sakai M, Funatsu G. Effects of chemical modification of arginine residues outside the active site cleft of ricin A-chain on its RNA N-glycosidase activity for ribosomes. Biosci Biotechnol Biochem. 1994;58:716–721. doi: 10.1271/bbb.58.716. [DOI] [PubMed] [Google Scholar]
- Watanabe K, Funatsu G. Involvement of arginine residues in inhibition of protein synthesis by ricin A-chain. FEBS Lett. 1986;204:219–222. doi: 10.1016/0014-5793(86)80815-8. [DOI] [PubMed] [Google Scholar]
- Yermakova A, Klokk TI, O’Hara JM, Cole R, Sandvig K, Mantis NJ. Neutralizing Monoclonal Antibodies against Disparate Epitopes on Ricin Toxin’s Enzymatic Subunit Interfere with Intracellular Toxin Transport. Sci Rep. 2016;6:22721. doi: 10.1038/srep22721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Haslam DB. A quantitative and highly sensitive luciferase-based assay for bacterial toxins that inhibit protein synthesis. J Med Microbiol. 2005;54:1023–1030. doi: 10.1099/jmm.0.46143-0. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Dai J, Zhang T, Li X, Fang P, Wang H, Jiang Y, Yu X, Xia T, Niu L, Guo Y, Teng M. Structural insights into the neutralization mechanism of monoclonal antibody 6C2 against ricin. J Biol Chem. 2013;288:25165–25172. doi: 10.1074/jbc.M113.480830. [DOI] [PMC free article] [PubMed] [Google Scholar]